- Laboratory of Plant Molecular Biology, Instituto Potosino de Investigación Científica y Tecnológica, San Luis Potosí, Mexico
mRNA accumulation is tightly regulated by diverse molecular pathways. The identification and characterization of enzymes and regulatory proteins involved in controlling the fate of mRNA offers the possibility to broaden our understanding of posttranscriptional gene regulation. Processing bodies (P bodies, PB) are cytoplasmic protein complexes involved in degradation and translational arrest of mRNA. Composition and dynamics of these subcellular structures have been studied in animal systems, yeasts and in the model plant Arabidopsis. Their assembly implies the aggregation of specific factors related to decapping, deadenylation, and exoribonucleases that operate synchronously to regulate certain mRNA targets during development and adaptation to stress. Although the general function of PB along with the flow of genetic information is understood, several questions still remain open. This review summarizes data on the composition, potential molecular roles, and biological significance of PB and potentially related proteins in Arabidopsis.
Introduction
The diversity of eukaryotic mRNA decay pathways illustrates the importance of nullifying transcripts as a mechanism to ensure proper course of gene expression and prevent the accumulation of aberrant mRNA (Houseley and Tollervey, 2009; Christie et al., 2011). These molecular mechanisms that abrogate accumulation of transcripts and proteins depend on the formation of RNA-protein complexes (RNPs). Under specific conditions or developmental cues, eukaryotic cells sort proteins, and mRNA in microscopically detectable complexes, commonly called cytoplasmic foci, categorized depending on the protein composition. Immunodetection of foci of the mice 5′-exoribonuclease XRN1 indicated that some pathways of RNA degradation might occur in protein speckles (Bashkirov et al., 1997). The 5′-exoribonuclease activity depends on the removal of 5′ cap structure 7-methyl-Guanosine-diphosphate (m7GDP). Interestingly, subsequent localization studies in yeast demonstrated that decapping-associated proteins are also localized in cytoplasmic foci, which were called Processing bodies, P bodies (PB; Sheth and Parker, 2003) or GW bodies in mammals (Eystathioy et al., 2003). Further microscopical and biochemical studies ended up proposing the composition of PB, which is conserved in eukaryotic organisms (Parker and Sheth, 2007). Similar structures have been identified in the protists Trypanosoma cruzi and Entamoeba histolytica (Holetz et al., 2007; López-Rosas et al., 2012). Electronic microscopy observations in human cells revealed that PB are clustered strands of 10–15 nm of diameter (Souquere et al., 2009).
Formation of PB depends on the availability of free cytoplasmic mRNA or mRNA not associated with polysomes (Teixeira et al., 2005). This correlation has been reported in Arabidopsis by trapping mRNA into polysomes by short treatments with cycloheximide (CHX; Weber et al., 2008). However, endogenous signaling events influence the formation of PB in Saccharomyces cerevisiae (Kilchert et al., 2010; Ramachandran et al., 2011).
The mRNAs aggregated into PB are translationally repressed and potentially degraded, even their re-incorporation in ribosomes is plausible. Decapping, deadenylation and 5′-exonucleolytic decay are biochemically related enzymatic modifications catalyzed in PB. Besides their involvement in RNA decay, these proteins may secure the translational arrest. Accumulation of related decay factors in PB implicates spatially ordered mechanisms of post-transcriptional regulation. As the activity of such factors in modifying mRNA is most likely irreversible, other mechanisms, possibly occurring outside PB and before the transcript is loaded into them, are necessary to accomplish a selective and controlled delivery, degradation, or translational arrest. It is crucial for the living cell because the knockout of some of those components causes lethality or severely abnormal phenotypes in Arabidopsis.
The intrinsic dynamics of PB in controlling the mRNA fate could lead a functional link between PB and other mRNA-containing cytoplasmic foci, for example, stress granules (sg) and heat shock granules (HSG). In human cells, SG are composed by proteins related to initiation of translation such as eukaryotic initiation factors (eIF3, eIF4E, eIFG), ribosomal subunits, and poly(a)-binding protein1 (PABP-1; Anderson and Kedersha, 2008). Assembly of SG is triggered by environmental stress and requires mRNA associated with 48S complexes, thus, assembly can be disrupted by CHX. The HSG described in Arabidopsis and tomato are formed upon prolonged heat treatment, they contain heat shock proteins and polyadenylated mRNA (Nover et al., 1989; Weber et al., 2008). The physical proximity between SG/HSG and PB has been evidenced in human cells and Arabidopsis (Kedersha et al., 2005; Weber et al., 2008). Such proximity might be necessary for the proper sorting of mRNAs after release form polysomes or to facilitate the exchange of mRNAs and proteins in a confined subcellular space. As SG formation is triggered by stress, they might protect transcripts during the lapse of stress and re-initiate translation once conditions get more favorable. If the translation of some transcripts aggregated into SG is not required, the proximity between foci might facilitate transport and decay into PB. Although some proteins related to RNA metabolism can associate with SG (Anderson and Kedersha, 2008), modification of mRNA targets into SG should be demonstrated. In contrast, the battery of proteins involved in RNA decay contained in PB favors a scenario where target mRNAs are translationally repressed and degraded, either they are released from polysomes, SG, or directly delivered after nuclear export.
Composition of P Bodies
Subunits of decapping complex, the 5′-exoribonuclease, and deadenylases are prominent markers of PB in eukaryotic organisms. Such core decay factors are distinguishable from other accessory proteins because they have the potential to modify the structure of mRNA. RNPs accessing into PB may be decorated with proteins that can interact with decay factors to mediate their activity within PB. Those proteins are distributed in different subcellular locations, including PB. Their frequency of association with PB might be synchronized with endogenous or exogenous signals that control the fate of specific mRNAs as a mechanism to fulfill requirements of the cell. The microRNA (miRNA)-dependent endonuclease argonaute1 (AGO1), components of nonsense-mediated mRNA decay (NMD) and RNA-binding proteins such as tandem zinc finger proteins occasionally localize in PB, as so far, their functional connection to PB is hypothetical in Arabidopsis. The Figure 1 illustrates reported and putative PB components in Arabidopsis.
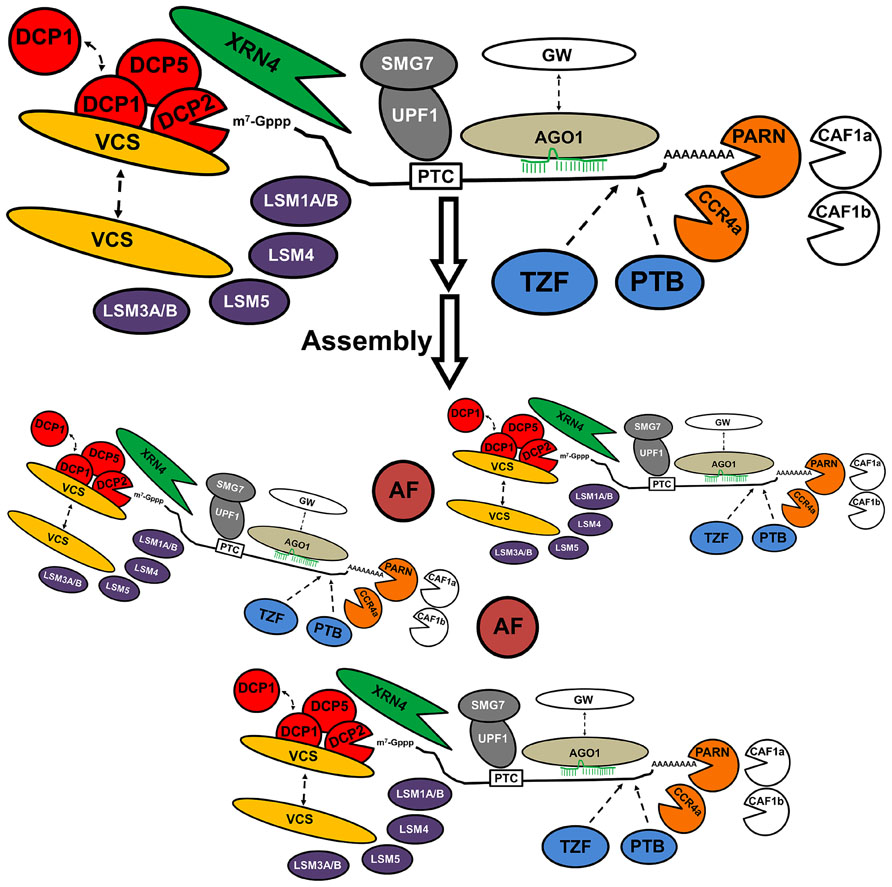
FIGURE 1. Composition of P body in Arabidopsis thaliana. Proteins involved in decapping (DCP1, DCP2, DCP5, VCS, LSM); deadenylases (PARN, CCR4a); the 5′-exoribonulease 4 (XRN4), microRNA-dependent endonuclease (AGO1); proteins of non-sense mediated RNA decay (UPF1, SMG7), and RNA-binding proteins such as tandem zinc fingers proteins (TZF) and polypyrimidine-binding proteins (PTB) have been detected in PB. Localization of the Deadenylases CAF1a, CAF1b, and GW proteins has to be experimentally confirmed. Unknown “Aggregation Factors” (AF), such as Q-rich proteins, might mediate the assembly of the structure, possibly by joining individual components or fusion of independent PB. The mRNA trapped into the PB is translationally arrested, and is eventually degraded by the decay factors, or return to polysomes to be translated.
Decapping Complex
The cleavage of the 5′ cap compromises mRNA to 5′ to 3′ exonucleolytic decay, apparently in an irreversible way, hence, decapping activity is tightly regulated (Li and Kiledjian, 2010). In yeast and animals, decapping complex consists on several subunits such as DCP1, DHH1p, EDC3, SCD6, PAT1, and LSM1-7 which mediate the activity of the catalytic subunit DCP2 (Kulkarni et al., 2010; Zheng et al., 2011). The decapping complex in Arabidopsis is formed by the catalytic subunit DCP2, and the subunits DCP1, DCP5, and varicose (VCS; Xu et al., 2006), although other proteins can potentially affect decapping activity.
The Arabidopsis DCP2 harbors a conserved nudix hydrolase domain essential for the decapping activity, and physically interacts with DCP5, and VCS (Xu et al., 2006). In the same report. Xu et al. (2006) demonstrated that DCP1 stimulates decapping activity in vivo. DCP1 contains an EVH1-like domain potentially involved in protein-protein interactions (She et al., 2004). DCP1 interacts with itself, and experimental evidences in Nicotiana benthamiana demonstrated DCP1 does not interact with DCP2 (Iwasaki et al., 2007), indicating that additional components are required to link the function of DCP1 to the catalysis of DCP2. DCP1 and DCP2 may be physically close each other due to their interaction with VCS. Interaction between VCS, DCP1, and DCP2 was demonstrated in vivo while yeast two hybrid experiments revealed interaction between VCS and DCP2 and VCS with itself (Xu et al., 2006; Goeres et al., 2007). VCS contains two protein–protein interaction WD40 motifs, although they are not necessary to the interaction with DCP1 and DCP2. Deletion of WD40 repeats in Ge-1, the human homolog of VCS, did not compromised the localization into PB, then, WD40 repeats probably function in the recruitment of another mediators of decapping (Yu et al., 2005).
The Arabidopsis DCP5 is required for decapping and repression of translation in vivo (Xu and Chua, 2009). PB visualized by expressing DCP1-CFP are smaller in knockdown dcp5-1 compared to Col-0. This comparison indicates DCP5-mediated assembly of PB is necessary for proper decapping. Impaired translational repression or decapping in dcp5-1 might cause the release of some mRNA form PB, thus, smaller size of PB in dcp5-1 might be a consequence of their decreased capacity to retain mRNA. Interaction between DCP5 and the RNA chaperone cold shock domain protein 3 AtCSP3 was reported (Kim et al., 2013) and possibly indicates that unwinding secondary structures of RNA might be necessary to facilitate decapping or inhibition of translation. The yeast DEAD box helicase DHH1p and homolog proteins in animals promote decapping and inhibit translation (Kulkarni et al., 2010). Arabidopsis lacks a homolog of DHH1p, but other proteins might replace its function through analogous processes.
Pat1 (protein-associated with topoisomerases) is conserved in different organisms, involved in activation of decapping and inhibition of translation (Coller and Parker, 2005; Scheller et al., 2007). In concordance with the formation of PB and inhibition of translation, nutrient deprivation causes nuclear accumulation of tRNAs in yeast. The yeast dhh1Δ and pat1Δ mutants failed to accumulate tRNAs into nucleus during starvation, but xrn1Δ showed normal tRNA nuclear accumulation (Hurto and Hopper, 2011). These evidences suggest that the formation of PB is accompanied by the nuclear accumulation of tRNAs in a DHH1p-Pat1-dependent manner as an mechanism to ensure inhibition of translation. The Arabidopsis genes At1g79090, At3g22270, and At4g14990 are annotated as Pat1 (http://www.arabidopsis.org), experimental evidences are required to confirm their localization and molecular function.
Sm-like proteins (LSM) interact each other to form complexes to activate decapping and modulate splicing in yeast (Tharun et al., 2000). The Arabidopsis LSM1-7 proteins form a cytoplasmic heptameric complex. LSM1A-GFP, LSM1B-GFP, LSM3A-GFP, LSM3B-GFP, and LSM4-GFP localize to cytoplasmic foci in root tips of stably transformed plants (Perea-Resa et al., 2012). Formation of these foci is sensitive to CHX, and the co-localization with RFP-DCP1 confirmed their physical association with PB. Heat stress facilitates the detection of LSM1-4-GFP containing cytoplasmic foci, it tempts to consider that LSM proteins are simultaneously incorporated in distinct cytoplasmic foci. Given that cytoplasmic foci localization of GFP-DCP2 and GFP-VCS is depleted in lsm1alsm1b, both proteins might participate in the assembly of PB or, at least influence the localization of VCS and DCP2.
Decapping activity is prevented by the translation initiation factor eIF4E, a 5′ cap-binding protein (von der Haar et al., 2004). Its localization in PB and SG is documented in humans and yeast (Andrei et al., 2005; Brengues and Parker, 2007), but the homolog of Arabidopsis was only detected in SG (Weber et al., 2008).
5′-Exoribonuclease
Decapped mRNA is prone to 5′ to 3′ decay. The Arabidopsis XRN4 complements the yeast xrn1Δ mutant (Olmedo et al., 2006). XRN4 co-localizes to DCP1 in Arabidopsis, furthermore, PB visualized with DCP1 antiserum were quite observable in the knockout xrn4 line compared to wild type plants (Weber et al., 2008). This interesting comparison suggests the loss of 5′-exoribonuclease activity in xrn4 enhances the assembly of PB by over accumulation of potentially uncapped mRNA. XRN4 might prevent accumulation of uncapped intermediates potentially involved in short interfering (siRNA) biogenesis (Gregory et al., 2008), and also participates in the degradation of selected 3′ end products from miRNA-mediated decay (Rymarquis et al., 2011).
Deadenylation
Deadenylases and their regulatory proteins play an essential role both in 5′ to 3′ RNA decay and formation of PB (Zheng et al., 2008). Polyadenylated mRNA is aggregated in SG and HSG of tomato cells, thus, deadenylation might be necessary to sort mRNAs into different foci. Shortening of poly-A might exclude poly-A binding proteins and proteins associated with translation to decrease the efficiency of translation and produce mRNAs accessible to PB components. The deadenylation is catalyzed by the poly-A-specific ribonuclease AtPARN and the carbon catabolite repressor 4-CCR4 associated factor 1-CAF1 complex (CCR4–CAF1). Both AtPARN and CCR4a co-localize to DCP1 in Agro-infiltrated tobacco leaves (Moreno et al., 2013). The activity of CCR4–CAF1 complex is regulated by the NOT1-5 proteins in yeast and humans, nevertheless their function has not been confirmed in Arabidopsis (Zheng et al., 2011; Abbasi et al., 2013).
Argonaute1
ARGONAUTE (AGO) proteins have miRNA-dependent RNA slicer activity and are part of the RNA interference effector complex (RISC). Interaction between AGO proteins and the PB-associated GW182 (TNRC6A) is critical to gene silencing in humans (Lazzaretti et al., 2009). Early immunofluorescence studies of GW182 (TNRC6A) in mammalian cells revealed co-localization with proteins of decapping in foci, were initially known as GW-bodies (Eystathioy et al., 2003). Human AGO1 and AGO2 co-localize to DCP1 (Liu et al., 2005). GW182 and its isoforms contain a GW motif and a Q-rich region which contributes to the localization into PB, but is not required for silencing (Eulalio et al., 2007). Q or N-rich regions in other PB-related proteins might provoke a general aggregation of other components (Reijns et al., 2008). GW-containing proteins have been identified Arabidopsis by a computational approach. However, experimental evidence is required to confirm the potential association with AGO proteins (Karlowski et al., 2010; Gu et al., 2012). Arabidopsis AGO1 is associated with microRNAs, endogenous, and transgene derived siRNAs (Baumberger and Baulcombe, 2005), its potential localization in PB is documented (Pomeranz et al., 2010a). The accumulation of 3′-end products of miRNA-mediated decay in the Arabidopsis xrn4 mutant suggests that AGO slicer activity acts upstream XRN4, possibly associated with PB (Rymarquis et al., 2011). Moreover, the increase of potential targets of miRNA in dcp1, dcp2, and vcs suggests a functional link between decapping and AGO1 activity (Motomura et al., 2012).
Localization of AGO1 varies under different conditions. GFP-AGO1 driven by the native AGO1 promoter is detectable in cytosol, nuclear envelope and the proximity to Golgi apparatus (Derrien et al., 2012). The viral suppressor P0 triggers the localization of GFP1-AGO1 into vesicles as a necessary early step in its degradation by autophagy.
The translational repression activity of AGO1 has been uncovered for specific miRNA targets. For example, the superoxide dismutase CSD2 mRNA is targeted by the miR398 to repress translation under “low Cu++” conditions, this mechanism requires VCS (Brodersen et al., 2008). Both miRNA and AGO1 are detectable in polysomes to repress translation of the targets independent of the AGO1 slicer activity (Lanet et al., 2009). These results proposed that AGO1 and VCS sequester mRNA into PB to prevent their translation. Nevertheless, the translational repression activity of AGO1 rather requires the integral Endoplasmic reticulum (ER) protein altered meristem program1 (ATM1) and possibly its paralog LAMP1 (Li et al., 2013). Increased amounts of miRNA target transcripts such as AGO1, PHB, and CSD2 in membrane bound polysomes of the double mutant atm1lamp1 suggests that ATM1 and LAMP1 displace miRNA target transcripts from translation in membrane bound polysomes. The specificity in the inhibition might depend in the miRNA-dependent ability of AGO1 to recognize unique mRNAs. Although VCS participates in translational arrest, the evidences illustrate that the AGO1-dependent translational inhibition is related to ER and not to PB.
Nonsense-Mediated mRNA Decay
The NMD refers a surveillance system that detects and eliminates mRNAs containing premature termination codons (PTC) and also controls expression of wild type genes with long 3′-UTR, mRNAs harboring 3′-UTR-located introns, and upstream open reading frames (uORF)-containing genes, defined as small ORFs encoded in the 5′-UTR of major ORFs (mORF; Hayden and Jorgensen, 2007; Kerényi et al., 2008). The up-frameshift proteins UPF1, UPF2, and UPF3 are essential factors of NMD. In yeast, UPF1-3 are involved in recognition and targeting PTC-containing mRNA to PB (Sheth and Parker, 2006; Nyikó et al., 2009). UPF1-3 are cytoplasmic, but they co-localize to DCP2 in the dcp1Δ strain, theoretically by accumulation of non-degraded aberrant mRNAs or other substrates of NMD.
Arabidopsis UPF1 is also cytoplasmic, and its incorporation in PB depends on 14-3-3-like protein SMG7 (Mérai et al., 2012). A RNP formed by the NMD target, the presumably phosphorylated UPF1, UPF2, and UPF3 could be transported to PB by SMG7 to promote the degradation of the target. As several components of NMD -such as SMG5 and SMG6- are absent in Arabidopsis, the localization of UPF1 and SMG7 in PB would indicate that 5′ to 3′ decay could have a predominant role in NMD. XRN4-silenced tobacco leaves accumulated PTC-containing mRNA at equal levels than the control leaves (Mérai et al., 2012). This results contradicts the model in yeast, where XRN1 is involved in NMD. So far, the role of decapping complex in plant NMD is not demonstrated, and evidences of 3′ to 5′ decay in PB are lacking. If decapping is dispensable for plant NMD, XRN4 would be dispensable as well. Plant XRN4-independent events of NMD and the role of decapping in this pathway have to be clarified in the future.
Tandem Zinc Finger Proteins and Other RNA-Binding Proteins
RNPs shuttle between cytoplasm-polysomes-PB and other cytoplasmic foci as a dynamic mechanism to regulate the fate of mRNAs. Microscopical observations and analysis of polysomal profiles revealed movement of mRNA to PB triggered under glucose deprivation in yeast; and selected mRNAs are newly detectable in polysomal fractions after readdition of glucose (Brengues et al., 2005). The mRNAs harbor diverse cis-elements potentially recognized by RNA-binding proteins, siRNAs or microRNAs that regulate their maturation, stability, translation rate, as well as localization elements or Zipcodes, involved in the delivery of transcripts to specific subcellular compartments (Ahmed et al., 2011). Specificity in the reactions catalyzed in PB might be conferred by RNA-binding proteins involved in the recognition of targets prior to their delivery in PB.
The human tandem CCCH-type zinc finger proteins such as TTP, are detectable in PB. TTP exists in complex with the proteins DCP1, DCP2, and XRN1 (Lykke-Andersen and Wagner, 2005; Franks and Lykke-Andersen, 2007). The RNA-binding activity of TTP and similar proteins is conferred by the CCCH-type zinc finger motifs. TTP binds to the class II AU-rich sequence 5′-UUAUUUAUU-3′ (class II ARE) located in the 3′-UTR of specific mRNAs. TTP enhances decapping of ARE-containing RNA in vitro, which suggests that targets of TTP are prone to decapping after their delivery into PB (Fenger-Grøn et al., 2005). Beyond its role in ARE-containing mRNA decay, TTP has been postulated as a nucleation factor of PB since microscopic detection of GFP-HsDCP1 increases by overexpression (OE) of TTP (Franks and Lykke-Andersen, 2007).
The Arabidopsis CCCH-containing Tandem Zinc Finger protein family consists of 11 members. Transient expression in protoplasts revealed all of them localize to cytoplasmic foci, and the co-localization with PB and SG markers has been demonstrated for specific members (Pomeranz et al., 2010a,b; Bogamuwa and Jang, 2013; Maldonado-Bonilla et al., 2014).
Localization of TZF proteins is highly dependent on the experimental conditions that influence the physiology and requirements of the cell. Nuclear, cytoplasmic, and nucleocytoplasmic distribution was reported to SZF1/TZF11, SZF2/TZF10, OXS2/TZF7, TZF2, and TZF3, by using either transient expression in onion epidermal cells or stable expression in transgenic plants (Sun et al., 2007; Blanvillain et al., 2011; Lee et al., 2012). Those reports ruled out any localization in cytoplasmic foci or PB.
In planta detection of TZF1 in putative PB depends on Methyl Jasmonate (MeJA) treatment or wounding (Pomeranz et al., 2010a) unlike TZF4, TZF5, and TZF6, whose localization in putative PB is detectable in intact plants (Bogamuwa and Jang, 2013). TZF1 accumulates into the nucleus by inhibiting XPO1-mediated nuclear export with Leptomycin B (LMB; Pomeranz et al., 2010a), evidencing a nucleocytoplasic shuttling. Once in cytoplasm, TZF1 has potential to be incorporated in PB or SG, though the signals, factors or functional domains of TZF1 that promote assembly in PB are still unknown. The localization of TZF9 is also affected by LMB in Arabidopsis protoplasts, however, further experiments has to be performed in order to confirm the nucleocytoplasmic shuttling (Maldonado-Bonilla et al., 2014). The nucleocytoplasmic shuttling of TZF proteins might indicate they have different functions both in nucleus and cytoplasm, or perhaps, that the proteins recognize the potential mRNA target during or after transcription. A hypothetical RNP formed in the nucleus might determine the fate of the mRNA once exported in the cytoplasm.
In vitro evidences indicate that TZF proteins bind to RNA, DNA, or even possess ribonuclease activity (Pomeranz et al., 2010a; Blanvillain et al., 2011; Lee et al., 2012; Maldonado-Bonilla et al., 2014). Directs mRNA targets of Arabidopsis TZF proteins have not been reported so far, and critical amino acid substitutions in the zinc finger motifs might entail their inability to recognize the class II ARE sequence. However, the arginine-rich region preceding CCCH zinc fingers also contributes in the RNA-binding activity (Qu et al., 2014).
Arabidopsis polypyrimidine tract-binding proteins PTB1, PTB2, and PTB3 are involved in alternative splicing -a nucleus-localized process-, however they are also detectable in PB (Stauffer et al., 2010). As alternative splicing can produce PTC-containing mRNAs (Wang and Brendel, 2006), these proteins might traffic to PB as part of the mechanism of degradation of substrates of NMD.
The human poly-C binding protein 2 (PCBP2) shuttles between nucleus and cytoplasm, and is detectable in PB and SG. Different RNA-binding KH domains are critical to confer its localization in both cytoplasmic foci (Fujimura et al., 2008). This changing localization may be necessary to join different protein complexes to regulate translation or transcript stability. Arabidopsis possesses 26 KH-containing proteins, the arrangement and architecture of KH domains of PEPPER (At4g26000) and At1g14170 are reminiscent to PCBP2 (Lorkoviæ and Barta, 2002; Ripoll et al., 2006). Subcellular localization studies on these proteins might complement their characterization.
The subcellular dynamics of RNA-binding proteins mentioned above might be related to direct or indirect interaction with cytoskeletal proteins. Myosin Va co-localizes to PB markers, co-precipitates with the 5′cap-binding protein eIF4E, and is required to the assembly of PB in HeLa cells (Lindsay and McCaffrey, 2011). Although eIF4E is not accumulated in PB in plants, direct interaction between myosins and DCP1 has been recently reported in Arabidopsis (Steffens et al., 2014). This evidence might motivate to further investigate movement of PB and RNP along actin filaments or microtubules.
Biological Relevance of PB Components in Development and Response to Stress
Analysis of the phenotype of mutant lines has uncovered the importance of PB machinery for plant development and responses to stress. The Table 1 summarizes Arabidopsis PB-related genes and the phenotypes of the corresponding knockout and OE lines.
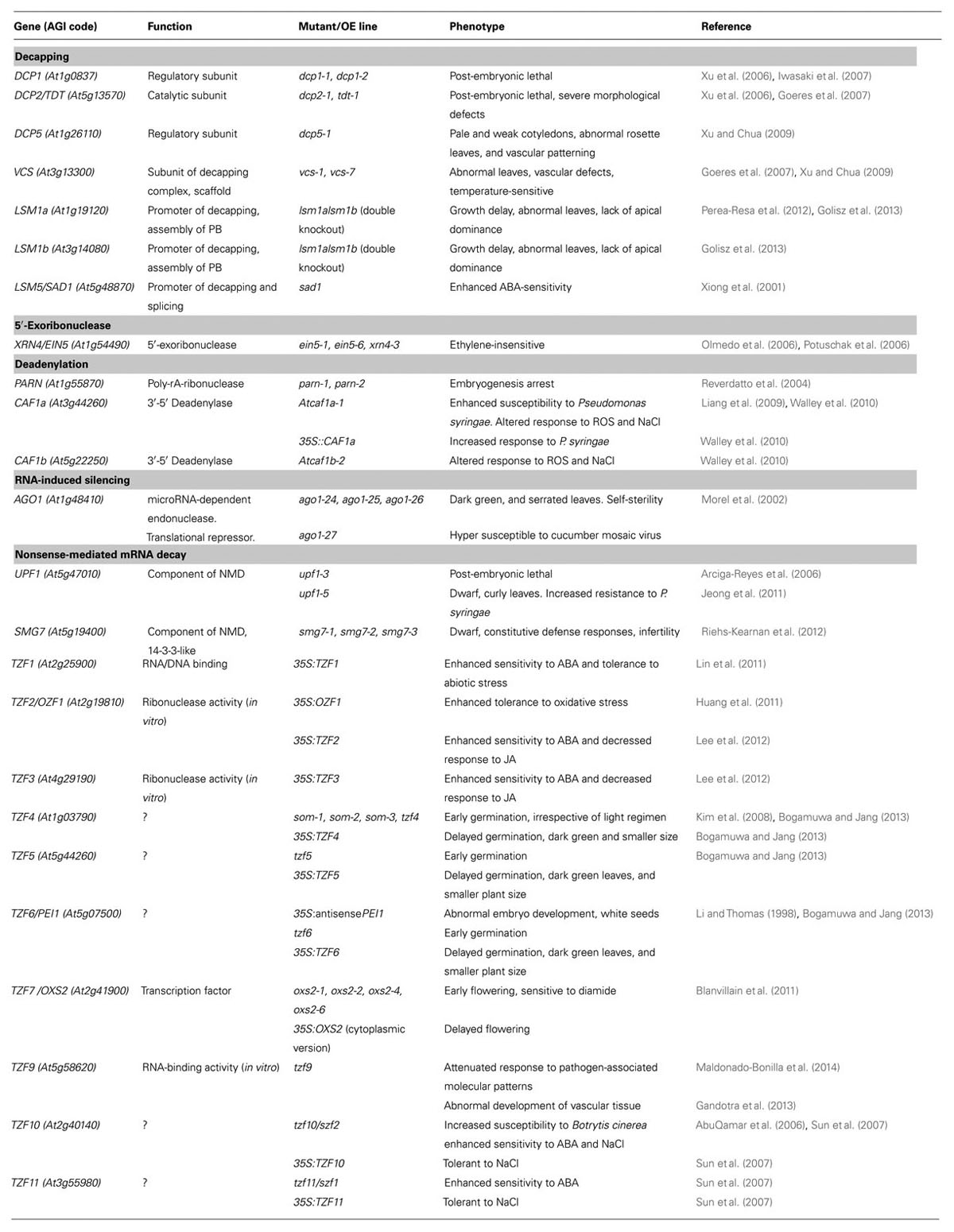
TABLE 1. Knock-out and over expression (OE) of P body-related genes alters development and adaptation to stress (view text for details).
Post-embryonic lethality of Arabidopsis dcp1 and dcp2 mutants, and severe phenotypic defects in vcs and dcp5 mutants such as abnormal patterning of vascular tissue, defects in the shoot apical meristem, chlorosis, slow germination, and temperature sensitivity illustrate functional decapping components are essential for optimal plant development (Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007; Xu and Chua, 2009). Enhanced decapping activity is also necessary to adaptation to abiotic stress. Arabidopsis sad1(lsm5) is hypersensitive to ABA and drought (Xiong et al., 2001). Arabidopsis DCP1 is phosphorylated by the stress-related MAP kinase MPK6 in response to dehydration. Comparison of capped to uncapped transcripts ratios, and global transcript profiling in different mutants and transgenic lines unveiled that MPK6-mediated phosphorylation of DCP1 increases decapping activity in concert with DCP5 (Xu and Chua, 2012). As MAP kinase activity is triggered by other signals, it is worth to investigate whether decapping activity is modulated via phosphorylation of DCP1 in response to other adverse conditions.
The function of LSM proteins is crucial for plant growth (Perea-Resa et al., 2012). In Arabidopsis, some transcripts with prolonged half-life in xrn4 mutant are also stabilized in lsm1alsm1b and lsm5 mutants. Since capping is increased in some of the stabilized transcripts, LSM1A, LSM1B, and LSM5 might stimulate decapping (Golisz et al., 2013). The stabilization of mRNAs such as WRKY40, WRKY33, and ACS6 in lsm1alsm1b and lsm5 could be considered as evidence that modulation of decapping activity takes part during execution of plant defense to pathogens or response to stress (Golisz et al., 2013).
Disruption of XRN4/EIN5 in Arabidopsis affects the response to ethylene. Reduction in the accumulation of the transcription factor EIN3 in ein5 mutant is consequence of the increased levels of F-box proteins EBF1 and EBF2 which target EIN3 to degradation in an Ubiquitin-dependent manner (Olmedo et al., 2006; Potuschak et al., 2006).
Knockout of the deadenylase AtPARN, caused embryogenesis arrest, indicating this enzyme might be indispensable to deadenylate bulk mRNA during embryo development (Reverdatto et al., 2004). Selective deadenylation mediated by CCR4-CAF1 proteins is necessary to regulate responses to stress. Knockout mutant lines atcaf1a and atcaf1b display enhanced susceptibility to Pseduomonas syringae and altered response to abiotic stress (Liang et al., 2009; Walley et al., 2010). AtCAF1A and AtCAF1B are necessary to respond to P. syringae, possibly by deadenylating jasmonic acid-responsive mRNAs or negative regulators of plant defense. The higher sensitivity to reactive oxygen species (ROS) of atcaf1a and atcaf1b could be helpful to investigate the potential link between AtCAF1A/B, the response to ROS and the defense against P. syringae.
Severe vegetative and reproductive defects in ago1 mutants confirms the importance of AGO1 for plant development. Hypomorphic ago1 plants permitted higher accumulation of Cucumber Mosaic Virus (CMV) and displayed enhanced symptoms, which evidences the role of AGO1 in posttranscriptional gene silencing, virus resistance and degradation of exogenous RNAs (Morel et al., 2002).
Potential Functional Link Between NMD Pathway and PB
NMD pathway is essential for seed development. Homozygous upf1-2 and smg7 are lethal (Yoine et al., 2006; Riehs-Kearnan et al., 2012). Weak mutant allele of UPF1 (upf1-5) and upf3 showed anomalies in vegetative growth and flowering (Arciga-Reyes et al., 2006). Elevated defense-related gene expression and salicylic acid (SA) accumulation of upf3 may reflect the reduction in the growth of P. syringae (Jeong et al., 2011). A selected group of WRKYs is up-regulated in upf1-5 and upf3, suggesting UPF1/3 are involved in the decay of WRKY transcripts. Coincidently, CHX promotes accumulation of those WRKY mRNAs in wild type plants. The effect of CHX might imply the arrest of mRNAs in polysomes that impedes the recognition of UPF1 as an early step in NMD. XRN4 is dispensable for decay of PTC-containing transcripts (Mérai et al., 2012). Hence, targets of NMD could be transiently delivered within PB by UPF1 and SMG7 as a mechanism to inhibit translation, and, in some point, the target might be released from PB and degraded by other nucleolytic pathways. The link between UPF1 and the transport of mRNA targets to PB should be investigated in the future.
Conserved uORF homology groups have been identified in Arabidopsis and rice by following a in silico approach (Hayden and Jorgensen, 2007). Some genes encoding transcription factors, kinases, or enzymes of polyamine metabolism harbor uORF that potentially induces ribosome stalling or reduces stability of the mORF. If the translation or accumulation of uORF perturbates the translation of the mORF, the latter one gets prone to be recognized by components of the NMD pathway. Depletion of GFP expression driven by uORF in Agro-infiltrated Nicotiana benthamiana strengthen this potential mechanism, although not all the uORF-containing transcripts would be substrates of NMD (Nyikó et al., 2009). It is worth to investigate whether the decay of mORF depends on decapping, deadenylation, and 5′-exoribonuclease activity.
Is the Function TZF Proteins Associated with PB?
Knockout and OE of TZF genes has revealed their involvement in ABA signaling and response to stress (AbuQamar et al., 2006; Sun et al., 2007; Blanvillain et al., 2011; Huang et al., 2011; Lin et al., 2011; Lee et al., 2012). The changing localization and in vitro RNA-binding and ribonuclease activity of TZF proteins suggests that their occasional association with PB might be biologically relevant. However, there are not evidences that indicate the phenotype of such mutants and OE lines is a consequence of mis-regulation of transcripts aggregated in PB.
The human CCCH protein TTP promotes mRNA decay by interaction with DCP2. Simultaneous presence of the target and TTP in PB has been demonstrated (Fenger-Grøn et al., 2005; Franks and Lykke-Andersen, 2007). The rice TZF1–GFP is detectable in PB upon salt stress, the recombinant protein binds to the 3′-UTR of selected stress related genes which are down-regulated in the OE line (Jan et al., 2013), indicating that OsTZF1 might lead the decay of these mRNA targets. Additional experiments have to be designed to investigate whether OsTZF1 destabilizes targets by stimulation of decapping. If this hypothesis is validated, similar experiments could be performed in Arabidopsis as starting point to demonstrate the functional link between TZF proteins and the decapping activity localized in PB.
Concluding Remarks and Perspectives
The aggregation of related decay factor in PB enables an organized set of mechanisms to prevent translation and catalyze the decay of specific transcripts. The core of plant PB is consensually known and different shuttling proteins might be involved in delivering mRNA targets and promoting degradation or storage via their interactions with core components, which is critical in regulating plant development and response to stress.
Current availability of genomes and transcriptome data encourages us to use the well-known information of Arabidopsis to in silico identify genes encoding components of PB as a first approach to study these foci in other plant models. The in silico analysis also allows the identification of cis-elements in potential mRNA targets.
Several methodologies to investigate RNA-protein interactions, isolation of polysomal mRNA and high resolution cell imaging used in yeast and animal systems have to be set up in model plants. Such technical package is necessary to further molecular characterization of proteins associated to PB, as well as the identification mRNA targets and RNA sequences recognized by RNA binding proteins. By overcoming technical challenges, we will broaden our understanding about the dynamics, plasticity, and mechanisms related to the PB function.
Finally, as knockout or overexpression of some PB-related proteins alters the response to environmental stresses, we can use those genes to design novel strategies of post-transcriptional gene regulation to improve crop protection and production.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Council of Science, Mexico (Postdoctoral Fellowship No.118915). Special thanks to Alok K. Sinha and Itzel López-Rosas by critically reading the manuscript.
References
Abbasi, N., Park, Y.-I., and Choi, S.-B. (2013). RNA deadenylation and decay in plants. J. Plant Biol. 56, 198–207. doi: 10.1007/s12374-013-0201-8
AbuQamar, S., Chen, X., Dhawan, R., Bluhm, B., Salmeron, J., Lam, S., et al. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48, 28–44. doi: 10.1111/j.1365-313X.2006.02849.x
Ahmed, F., Benedito, V. A., and Zhao, P. X. (2011). Mining functional elements in messenger RNAs: overview, challenges, and perspectives. Front. Plant Sci. 2:84. doi: 10.3389/fpls.2011.00084
Anderson, P., and Kedersha, N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150. doi: 10.1016/j.tibs.2007.12.003
Andrei, M. A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Lührmann, R. (2005). A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11, 717–727. doi: 10.1261/rna.2340405
Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47, 480–489. doi: 10.1111/j.1365-313X.2006.02802.x
Bashkirov, V. I., Scherthan, H., Solinger, J. A., Buerstedde, J. M., and Heyer, W. D. (1997). A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136, 761–773. doi: 10.1083/jcb.136.4.761
Baumberger, N., and Baulcombe, D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 11928–11933. doi: 10.1073/pnas.0505461102
Blanvillain, R., Wei, S., Wei, P., Kim, J. H., and Ow, D. W. (2011). Stress tolerance to stress escape in plants: role of the OXS2 zinc-finger transcription factor family. EMBO J. 30, 3812–3822. doi: 10.1038/emboj.2011.270
Bogamuwa, S., and Jang, J.-C. (2013). The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant. Cell Environ. 36, 1507–1519. doi: 10.1111/pce.12084
Brengues, M., and Parker, R. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4F and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2592–2602. doi: 10.1091/mbc.E06
Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489. doi: 10.1126/science.1115791
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
Christie, M., Brosnan, C. A., Rothnagel, J. A., and Carroll, B. J. (2011). RNA decay and RNA silencing in plants: competition or collaboration? Front. Plant Sci. 2:99. doi: 10.3389/fpls.2011.00099
Coller, J., and Parker, R. (2005). General translational repression by activators of mRNA decapping. Cell 122, 875–886. doi: 10.1016/j.cell.2005.07.012
Derrien, B., Baumberger, N., Schepetilnikov, M., Viotti, C., De Cillia, J., Ziegler-Graff, V., et al. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 109, 15942–15946. doi: 10.1073/pnas.1209487109
Eulalio, A., Behm-Ansmant, I., Schweizer, D., and Izaurralde, E. (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970–3981. doi: 10.1128/MCB.00128-07
Eystathioy, T., Jakymiw, A., and Chan, E. K. L. (2003). The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9, 1171–1173. doi: 10.1261/rna.5810203
Fenger-Grøn, M., Fillman, C., Norrild, B., and Lykke-Andersen, J. (2005). Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20, 905–915. doi: 10.1016/j.molcel.2005.10.031
Franks, T. M., and Lykke-Andersen, J. (2007). TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21, 719–735. doi: 10.1101/gad.1494707
Fujimura, K., Kano, F., Murata, M., and Fujimura, K. E. N. (2008). Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14, 425–431. doi: 10.1261/rna.780708
Gandotra, N., Coughlan, S. J., and Nelson, T. (2013). The Arabidopsis leaf provascular cell transcriptome is enriched in genes with roles in vein patterning. Plant J. 74, 48–58. doi: 10.1111/tpj.12100
Goeres, D. C., Van Norman, J. M., Zhang, W., Fauver, N. A., Spencer, M., and Sieburth, L. E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19, 1549–1564. doi: 10.1105/tpc.106.047621
Golisz, A., Sikorski, P. J., Kruszka, K., and Kufel, J. (2013). Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 41, 6232–6249. doi: 10.1093/nar/gkt296
Gregory, B. D., O’Malley, R. C., Lister, R., Urich, M. A., Tonti-Filippini, J., Chen, H., et al. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14, 854–866. doi: 10.1016/j.devcel.2008.04.005
Gu, X.-L., Wang, H., Huang, H., and Cui, X.-F. (2012). SPT6L encoding a putative WG/GW-repeat protein regulates apical-basal polarity of embryo in Arabidopsis. Mol. Plant 5, 249–259. doi: 10.1093/mp/ssr073
von der Haar, T., Gross, J. D., Wagner, G., and McCarthy, J. E. G. (2004). The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 503–511. doi: 10.1038/nsmb779
Hayden, C. A., and Jorgensen, R. A. (2007). Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 5:32. doi: 10.1186/1741-7007-5-32
Holetz, F. B., Correa, A., Avila, A. R., Nakamura, C. V., Krieger, M. A., and Goldenberg, S. (2007). Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356, 1062–1067. doi: 10.1016/j.bbrc.2007.03.104
Houseley, J., and Tollervey, D. (2009). The many pathways of RNA degradation. Cell 136, 763–776. doi: 10.1016/j.cell.2009.01.019
Huang, P., Chung, M.-S., Ju, H.-W., Na, H.-S., Lee, D. J., Cheong, H.-S., et al. (2011). Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 124, 699–705. doi: 10.1007/s10265-010-0397-3
Hurto, R. L., and Hopper, A. K. (2011). P-body components, Dhh1 and Pat1, are involved in tRNA nuclear-cytoplasmic dynamics. RNA 17, 912–924. doi: 10.1261/rna.2558511
Iwasaki, S., Takeda, A., Motose, H., and Watanabe, Y. (2007). Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 581, 2455–2459. doi: 10.1016/j.febslet.2007.04.051
Jan, A., Maruyama, K., Todaka, D., Kidokoro, S., Abo, M., Yoshimura, E., et al. (2013). OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216. doi: 10.1104/pp.112.205385
Jeong, H.-J., Kim, Y. J., Kim, S. H., Kim, Y.-H., Lee, I.-J., Kim, Y. K., et al. (2011). Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 52, 2147–2156. doi: 10.1093/pcp/pcr144
Karlowski, W. M., Zielezinski, A., Carrère, J., Pontier, D., Lagrange, T., and Cooke, R. (2010). Genome-wide computational identification of WG/GW Argonaute-binding proteins in Arabidopsis. Nucleic Acids Res. 38, 4231–4245. doi: 10.1093/nar/gkq162
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. doi: 10.1083/jcb.200502088
Kerényi, Z., Mérai, Z., Hiripi, L., Benkovics, A., Gyula, P., Lacomme, C., et al. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27, 1585–1595. doi: 10.1038/emboj.2008.88
Kilchert, C., Weidner, J., Prescianotto-baschong, C., and Spang, A. (2010). Defects in the Secretory Pathway and high Ca 2+ induce multiple P-bodies. Mol. Biol. Cell 21, 2624–2638. doi: 10.1091/mbc.E10
Kim, D. H., Yamaguchi, S., Lim, S., Oh, E., Park, J., Hanada, A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20, 1260–1277. doi: 10.1105/tpc.108.058859
Kim, M.-H., Sonoda, Y., Sasaki, K., Kaminaka, H., and Imai, R. (2013). Interactome analysis reveals versatile functions of Arabidopsis COLD SHOCK DOMAIN PROTEIN 3 in RNA processing within the nucleus and cytoplasm. Cell Stress Chaperones 18, 517–525. doi: 10.1007/s12192-012-0398-3
Kulkarni, M., Ozgur, S., and Stoecklin, G. (2010). On track with P-bodies. Biochem. Soc. Trans. 38, 242–251. doi: 10.1042/BST0380242
Lanet, E., Delannoy, E., Sormani, R., Floris, M., Brodersen, P., Crété, P., et al. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21, 1762–1768. doi: 10.1105/tpc.108.063412
Lazzaretti, D., Tournier, I., and Izaurralde, E. (2009). The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of argonaute proteins. RNA 15, 1059–1066. doi: 10.1261/rna.1606309
Lee, S., Jung, H. J., Kang, H., and Kim, S. Y. (2012). Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 53, 673–686. doi: 10.1093/pcp/pcs023
Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., et al. (2013). microRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574. doi: 10.1016/j.cell.2013.04.005.microRNAs
Li, Y., and Kiledjian, M. (2010). Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA 1, 253–265. doi: 10.1002/wrna.15
Li, Z., and Thomas, T. L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embr yo formation in Arabidopsis. Plant Cell 10, 383–398. doi: 10.1105/tpc.10.3.383
Liang, W., Li, C., Liu, F., Jiang, H., Li, S., Sun, J., et al. (2009). The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 19, 307–316. doi: 10.1038/cr.2008.317
Lin, P.-C., Pomeranz, M. C., Jikumaru, Y., Kang, S. G., Hah, C., Fujioka, S., et al. (2011). The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 65, 253–268. doi: 10.1111/j.1365-313X.2010.04419.x
Lindsay, A. J., and McCaffrey, M. W. (2011). Myosin Va is required for P body but not stress granule formation. J. Biol. Chem. 286, 11519–11528. doi: 10.1074/jbc.M110.182808
Liu, J., Valencia-Sanchez, M. A., Hannon, G. J., and Parker, R. (2005). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719–723. doi: 10.1038/ncb1274
López-Rosas, I., Orozco, E., Marchat, L. A., García-Rivera, G., Guillen, N., Weber, C., et al. (2012). mRNA decay proteins are targeted to poly(A)+ RNA and dsRNA-containing cytoplasmic foci that resemble P-bodies in Entamoeba histolytica. PLoS ONE 7:e45966. doi: 10.1371/journal.pone.0045966
Lorkoviæ, Z. J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. doi: 10.1093/nar/30.3.623
Lykke-Andersen, J., and Wagner, E. (2005). Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19, 351–361. doi: 10.1101/gad.1282305
Maldonado-Bonilla, L. D., Eschen-Lippold, L., Gago-Zachert, S., Tabassum, N., Bauer, N., Scheel, D., et al. (2014). The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol. 55, 412–425. doi: 10.1093/pcp/pct175
Mérai, Z., Benkovics, A. H., Nyikó, T., Debreczeny, M., Hiripi, L., Kerényi, Z., et al. (2012). The late steps of plant nonsense-mediated mRNA decay. Plant J. 73, 50–62. doi: 10.1111/tpj.12015
Morel, J., Godon, C., Mourrain, P., Béclin, C., Boutet, S., Feuerbach, F., et al. (2002). Fertile Hypomorphic ARGONAUTE (ago1) Mutants Impaired in Post-Transcriptional Gene Silencing and Virus Resistance. Plant Cell 14, 629–639. doi: 10.1105/tpc.010358
Moreno, A. B., Martínez de Alba, A. E., Bardou, F., Crespi, M. D., Vaucheret, H., Maizel, A., et al. (2013). Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 41, 4699–4708. doi: 10.1093/nar/gkt152
Motomura, K., Le, Q. T. N., Kumakura, N., Fukaya, T., Takeda, A., and Watanabe, Y. (2012). The role of decapping proteins in the miRNA accumulation in Arabidopsis thaliana. RNA Biol. 9, 644–652. doi: 10.4161/rna.19877
Nover, L., Scharf, K., and Neumann, D. (1989). Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9, 1298–1308. doi: 10.1128/MCB.9.3.1298
Nyikó, T., Sonkoly, B., Mérai, Z., Benkovics, A. H., and Silhavy, D. (2009). Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71, 367–378. doi: 10.1007/s11103-009-9528-4
Olmedo, G., Guo, H., Gregory, B. D., Nourizadeh, S. D., Aguilar-Henonin, L., Li, H., et al. (2006). Ethylene-insensitive5 encodes a 5′ -> 3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. U.S.A. 103, 13286–13293. doi: 10.1073/pnas.0605528103
Parker, R., and Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646. doi: 10.1016/j.molcel.2007.02.011
Perea-Resa, C., Hernández-Verdeja, T., López-Cobollo, R., del Mar Castellano, M., and Salinas, J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24, 4930–4947. doi: 10.1105/tpc.112.103697
Pomeranz, M. C., Hah, C., Lin, P.-C., Kang, S. G., Finer, J. J., Blackshear, P. J., et al. (2010a). The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 152, 151–165. doi: 10.1104/pp.109.145656
Pomeranz, M., Lin, P., Finer, J., and Jang, J. (2010b). AtTZF gene family localizes to cytoplasmic foci. Plant Signal. Behav. 5, 190–192. doi: 10.1104/pp.109.145656.n
Potuschak, T., Vansiri, A., Binder, B. M., Lechner, E., Vierstra, R. D., and Genschik, P. (2006). The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18, 3047–3057. doi: 10.1105/tpc.106.046508
Qu, J., Kang, S. G., Wang, W., Musier-Forsyth, K., and Jang, J. C. (2014). Arabidopsis thaliana tandem zinc finger 1 (AtTZF1) protein in RNA binding and decay. Plant J. 78, 452–467. doi: 10.1111/tpj.12485
Ramachandran, V., Shah, K. H., and Herman, P. K. (2011). The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43, 973–981. doi: 10.1016/j.molcel.2011.06.032
Reijns, M. A., Alexander, R. D., Spiller, M. P., and Beggs, J. D. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472. doi: 10.1242/jcs.024976
Reverdatto, S. V., Dutko, J. A., Chekanova, J. A., and Hamilton, D. A. (2004). mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10, 1200–1214. doi: 10.1261/rna.7540204.mRNAs
Riehs-Kearnan, N., Gloggnitzer, J., Dekrout, B., Jonak, C., and Riha, K. (2012). Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 40, 5615–5624. doi: 10.1093/nar/gks195
Ripoll, J. J., Ferrándiz, C., Martínez-Laborda, A., and Vera, A. (2006). PEPPER, a novel K-homology domain gene, regulates vegetative and gynoecium development in Arabidopsis. Dev. Biol. 289, 346–359. doi: 10.1016/j.ydbio.2005.10.037
Rymarquis, L. A., Souret, F. F., and Green, P. J. (2011). Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17, 501–511. doi: 10.1261/rna.2467911
Scheller, N., Resa-Infante, P., de la Luna, S., Galao, R. P., Albrecht, M., Kaestner, L., et al. (2007). Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta 1773, 1786–1792. doi: 10.1016/j.bbamcr.2007.08.009
She, M., Decker, C. J., Sundramurthy, K., Liu, Y., Chen, N., Parker, R., et al. (2004). Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 11, 249–256. doi: 10.1038/nsmb730
Sheth, U., and Parker, R. (2003). Deccaping and decay of RNAs occur in cytoplasmic processing bodies. Science 300, 805–808. doi: 10.1126/science.1082320
Sheth, U., and Parker, R. (2006). Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109. doi: 10.1016/j.cell.2006.04.037
Souquere, S., Mollet, S., Kress, M., Dautry, F., Pierron, G., and Weil, D. (2009). Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122, 3619–3626. doi: 10.1242/jcs.054437
Stauffer, E., Westermann, A., Wagner, G., and Wachter, A. (2010). Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 64, 243–255. doi: 10.1111/j.1365-313X.2010.04321.x
Steffens, A., Jaegle, B., Tresch, A., Hülskamp, M., and Jakoby, M. (2014). Processing body movement in Arabidopsis thaliana depends on an interaction between myosins and DCP1. Plant Physiol. 164, 1879–1892. doi: 10.1104/pp.113.233031
Sun, J., Jiang, H., Xu, Y., Li, H., Wu, X., Xie, Q., et al. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48, 1148–1158. doi: 10.1093/pcp/pcm088
Teixeira, D., Sheth, U., Valencia-Sanchez, M. A., Brengues, M., and Parker, R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382. doi: 10.1261/rna.7258505
Tharun, S., He, W., Mayes, A. E., Lennertz, P., Beggs, J. D., and Parker, R. (2000). Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404, 515–518. doi: 10.1038/35006676
Walley, J. W., Kelley, D. R., Nestorova, G., Hirschberg, D. L., and Dehesh, K. (2010). Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 152, 866–875. doi: 10.1104/pp.109.149005
Wang, B.-B., and Brendel, V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 103, 7175–7180. doi: 10.1073/pnas.0602039103
Weber, C., Nover, L., and Fauth, M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56, 517–530. doi: 10.1111/j.1365-313X.2008.03623.x
Xiong, L., Gong, Z., Rock, C. D., Subramanian, S., Guo, Y., Xu, W., et al. (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1, 771–781. doi: 10.1016/S1534-5807(01)00087-9
Xu, J., and Chua, N.-H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21, 3270–3279. doi: 10.1105/tpc.109.070078
Xu, J., and Chua, N.-H. (2012). Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 31, 1975–1984. doi: 10.1038/emboj.2012.56
Xu, J., Yang, J.-Y., Niu, Q.-W., and Chua, N.-H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18, 3386–3398. doi: 10.1105/tpc.106.047605
Yoine, M., Nishii, T., and Nakamura, K. (2006). Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 47, 572–580. doi: 10.1093/pcp/pcj035
Yu, J. H., Yang, W., Gulick, T. O. D., and Bloch, K. D. (2005). Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11, 1795–1802. doi: 10.1261/rna.2142405.cleavage
Zheng, D., Chen, C.-Y. A., and Shyu, A.-B. (2011). Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA 17, 1619–1634. doi: 10.1261/rna.2789611
Keywords: P bodies, decapping, deadenylation, mRNA decay, post-transcriptional gene regulation, RNA-protein complex, stress
Citation: Maldonado-Bonilla LD (2014) Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 5:201. doi: 10.3389/fpls.2014.00201
Received: 08 February 2014; Accepted: 24 April 2014;
Published online: 14 May 2014.
Edited by:
Marisa Otegui, University of Wisconsin at Madison, USAReviewed by:
Douglas Muench, University of Calgary, CanadaAlexis Maizel, Heidelberg University, Germany
Copyright © 2014 Maldonado-Bonilla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis D. Maldonado-Bonilla, Laboratory of Plant Molecular Biology, Instituto Potosino de Investigación Científica y Tecnológica, Camino a la Presa San José 2055, San Luis Potosí 78216, Mexico e-mail:bHVpcy5tYWxkb25hZG9AaXBpY3l0LmVkdS5teA==
 Luis D. Maldonado-Bonilla
Luis D. Maldonado-Bonilla