- 1Department of Genetics and Plant Production, Estación Experimental de Aula Dei, Consejo Superior de Investigaciones Científicas, Zaragoza, Spain
- 2Fundación ARAID, Zaragoza, Spain
Flowering time in plants is a tightly regulated process. In barley (Hordeum vulgare L.), HvFT1, ortholog of FLOWERING LOCUS T, is the main integrator of the photoperiod and vernalization signals leading to the transition from vegetative to reproductive state of the plant. This gene presents sequence polymorphisms affecting flowering time in the first intron and in the promoter. Recently, copy number variation (CNV) has been described for this gene. An allele with more than one copy was linked to higher gene expression, earlier flowering, and an overriding effect of the vernalization mechanism. This study aims at (1) surveying the distribution of HvFT1 polymorphisms across barley germplasm and (2) assessing gene expression and phenotypic effects of HvFT1 alleles. We analyzed HvFT1 CNV in 109 winter, spring, and facultative barley lines. There was more than one copy of the gene (2–5) only in spring or facultative barleys without a functional vernalization VrnH2 allele. CNV was investigated in several regions inside and around HvFT1. Two models of the gene were found: one with the same number of promoters and transcribed regions, and another with one promoter and variable number of transcribed regions. This last model was found in Nordic barleys only. Analysis of HvFT1 expression showed that association between known polymorphisms at the HvFT1 locus and the expression of the gene was highly dependent on the genetic background. Under long day conditions the earliest flowering lines carried a sensitive PpdH1 allele. Among spring cultivars with different number of copies, no clear relation was found between CNV, gene expression and flowering time. This was confirmed in a set of doubled haploid lines of a population segregating for HvFT1 CNV. Earlier flowering in the presence of several copies of HvFT1 was only seen in cultivar Tammi, which carries one promoter, suggesting a relation of gene structure with its regulation. HvCEN also affected to a large extent flowering time.
Introduction
In temperate cereals, like barley (Hordeum vulgare L.) and wheat (Triticum aestivum L.), flowering is regulated by the integration of two seasonal signals (Laurie, 2009): photoperiod (day length) and vernalization (prolonged exposures to low temperatures). Flowering time is also closely linked with agronomic performance. Plants must flower at the appropriate time of the year, when conditions are most favorable for pollination, seed development and high grain yield.
The responses to day length and temperature serve to classify barley varieties according to their adaptation pattern. Based on the response to day length, varieties are divided into photoperiod-sensitive (long days accelerate flowering) or -insensitive (plants flower almost independently of the day length). Based on the response to vernalization, barley varieties are classified as winter (vernalization is required for timely flowering) or spring (flowering irrespective of vernalization), although the presence of an allelic series at VrnH1 produces intermediate genotypes (Hemming et al., 2009). Usually, winter varieties are sown in autumn, spring varieties in winter and spring, and there is a third category known as facultative varieties, that can be sown anytime. Several major genes are the main responsible for the responses to photoperiod and vernalization, which are described next.
Allelic differences in the photoperiod genes PpdH1 and PpdH2 are associated with natural variation in the response to day length. PpdH1 (a member of the Pseudo Response Regulator family) is part of the circadian clock of the plant and promotes flowering under long days (Turner et al., 2005). Recessive mutations in the PpdH1 gene result in delayed flowering under long days (Turner et al., 2005; Hemming et al., 2008). The PpdH1 gene acts in parallel to HvCO1 (Campoli et al., 2012), which is one of the barley homologs of the Arabidopsis (Arabidopsis thaliana) CONSTANS (CO) gene. Overexpression of HvCO1 results in the up-regulation of HvFT1 (the ortholog in barley of the Arabidopsis FLOWERING LOCUS T, or FT) and the acceleration of flowering (Campoli et al., 2012). PpdH2 has been long acknowledged as the responsible for acceleration of flowering in response to short photoperiod, although its role is being re-defined (Casao et al., 2011). It is a paralog of HvFT1, (alternatively named HvFT3 by Faure et al., 2007 and Kikuchi et al., 2009). Its effect on flowering is not as strong as HvFT1 and it seems to be restricted to winter genotypes under short days or long days without vernalization (Casao et al., 2011). Another paralog of HvFT1 with a large effect on time to flowering, particularly at Mediterranean latitudes (Boyd et al., 2003; Cuesta-Marcos et al., 2008), is HvCEN, and its two main haplotypes are differentially distributed over spring and winter barley varieties (Comadran et al., 2012).
Natural variation in barley vernalization requirement is mainly found in the vernalization loci VrnH1 (Trevaskis et al., 2003; Yan et al., 2003), VrnH2 (Yan et al., 2004), and VrnH3 (Yan et al., 2006). The VrnH1 gene is closely related to the Arabidopsis gene APETALA1 (AP1), responsible for the transition from the vegetative to the reproductive stage (Trevaskis et al., 2007). Different alleles of VrnH1 have been identified, with insertions or deletions in the first intron of the gene (von Zitzewitz et al., 2005; Cockram et al., 2007; Hemming et al., 2009), affecting the length of the optimum vernalization period. Alleles lacking large sections of the ~11 kb intron are more active and are associated with earlier flowering without vernalization, whereas alleles lacking small segments are associated with only a moderate increase in VrnH1 activity and weaker promotion of flowering (Szűcs et al., 2007; Hemming et al., 2009). VrnH2 includes three closely related genes designated as HvZCCTa-c, which are characterized by a putative zinc finger and a CCT-domain. VrnH2 is considered to play the role of repressor of flowering and it has been shown that deletions of all the three HvZCCT genes result in spring growth habit (Karsai et al., 2005; Trevaskis et al., 2006). Finally, VrnH3 was shown to correspond to HvFT1, the ortholog of the Arabidopsis FT gene (Yan et al., 2006; Faure et al., 2007).
FT is considered as the main flowering integrator of the photoperiod and vernalization pathways in both monocot and dicot species (Turck et al., 2008). In barley, the most accepted hypothesis for the regulation of HvFT1 establishes that, during the fall, when temperate cereals germinate, HvFT1 is repressed by VrnH2 (Yan et al., 2006; Hemming et al., 2008; Distelfeld et al., 2009). During winter, vernalization up-regulates VrnH1 (Trevaskis et al., 2006; Oliver et al., 2009), which results in the repression of VrnH2 in the leaves and, consequently, the activation of HvFT1 transcription in the spring (Loukoianov et al., 2005; Trevaskis et al., 2006; Hemming et al., 2008; Chen and Dubcovsky, 2012). The precise molecular mechanisms of action of these genes are still the object of numerous studies in barley and other cereals. FT transcription is induced in the leaves and it has been demonstrated in different species that the encoded protein travels through the phloem to the stem apical meristem, where it plays a central role in triggering flowering (Corbesier et al., 2007; Tamaki et al., 2007). In Arabidopsis, FT interacts with the bZIP transcription factor FD and up-regulates the expression of the meristem identity gene AP1 at the shoot apex (Abe et al., 2005; Wigge et al., 2005). A similar interaction has been reported in wheat, where the orthologous FT protein interacts with an FD-like protein and has the ability to bind in vitro the promoter of VrnH1, the wheat homolog of AP1 (Li and Dubcovsky, 2008).
Ample natural variation in the HvFT1 gene has been found, with polymorphisms reported in the promoter and in the first intron. This variation has been linked to differences in flowering phenotypes in a number of studies (Yan et al., 2006; Hemming et al., 2008; Casas et al., 2011). It seems clear now that, in Arabidopsis, the FT promoter and first intron contain cis-regulatory sites that are important for its transcriptional regulation (Tiwari et al., 2010). However, the FT1 regulatory regions of barley and wheat are not as well characterized. Yan et al. (2006) found an association between growth habit and mutations in the first intron, but the sequencing of the HvFT1 alleles from populations previously used to map QTL for flowering time (Hemming et al., 2008) failed to reveal any significant association between the two linked SNPs in intron 1 and flowering time. Further results reported in other surveys of HvFT1 allelic variation (Cuesta-Marcos et al., 2010; Casas et al., 2011) were also in disagreement with Yan et al. (2006) regarding the direction of the effect assigned to the functional polymorphism in the first intron. Yan et al. (2006) also identified two promoter haplotypes, characterized by seven linked SNPs and two indels in the first 550 bp upstream of the start codon. Using primers specific to differentiate those indels, Casas et al. (2011) analyzed natural variation for the promoter haplotypes (135–146 vs. 139–142 bp) and the intron 1 haplotypes (AG/TC) in a collection of barley landraces. In that study the intron AG haplotype was clearly associated with later flowering than the TC haplotype. The results for the promoter haplotypes hinted at a role of these polymorphisms on flowering time, but of lesser magnitude than intron polymorphism. The combination of the 135–146 promoter with the TC intron was associated with earliest flowering (Casas et al., 2011; Ponce-Molina et al., 2012). Further evidence from other populations (Nitcher et al., 2013) confirmed the description of promoter haplotypes as “early” (135–146) and “late” (139–142). Another SNP in the promoter of the HvFT1 gene, upstream of the studied region was also suggested to have an additional role on flowering time (Cuesta-Marcos et al., 2010; Casas et al., 2011).
The scope of HvFT1 polymorphism has been recently widened even further by including copy number variation (CNV), first described by Nitcher et al. (2013). Recently, this type of polymorphism has been proposed as a key contributor to intra-species genetic variation, along with SNPs and indel polymorphisms. Some data suggest that CNV mainly affects the members of large families of functionally redundant genes and that the effects of individual CNV events on phenotype are usually modest (Zmieńko et al., 2014). Nevertheless, there are many cases in which CNVs for specific genes have been linked to important traits such as flowering time and plant height and resistance (Zmieńko et al., 2014). Regarding HvFT1, it has been reported recently that a genotype with high gene copy number (BGS213, derived from cultivar Tammi) was responsible for early flowering and an epistatic override of winter growth habit, caused by the combination of vrnH1 and VrnH2 alleles (Nitcher et al., 2013). This study pursues to carry out a comprehensive survey of HvFT1 polymorphisms, including CNV, in barley accessions of different origins and germplasm groups. Also, we aim to provide further new information on gene expression and phenotypic effects of contrasting genotypes at HvFT1.
Materials and Methods
Plant Materials
HvFT1 polymorphism
A set of 109 genotypes was used to survey the polymorphisms present at HvFT1, 89 cultivars, mainly European, and 20 inbred lines derived from Spanish landraces (Igartua et al., 1998). They were classified into 60 winter and 49 spring types, according to their genetic constitution at vernalization and photoperiod genes (Table S1). These genotypes constitute a representative sample of barley germplasm available to European breeders.
HvFT1 phenotypic effect
Several biparental populations used in past studies were reassessed to account for possible phenotypic effects of the polymorphisms at HvFT1 (Table 1). In some cases, these effects were already described in the references cited. In others, further genotyping allowed a better resolution of the QTLs or the discovery of previously unknown polymorphism. QTL x QTL interaction analyses were done using the unbalanced analysis of variance option implemented in Genstat 14 (Payne et al., 2009), following a factorial model with the markers closest to the QTL peaks and “environment” as factors. The field experiments of the population Beka x Mogador are explained in the publication by Cuesta-Marcos et al. (2008).
Gene expression analysis
Seven spring cultivars and two landrace-derived inbred lines, selected to represent HvFT1 CNV types, were used for gene expression analysis. Also, eight doubled haploid (DH) lines of the population Beka x Mogador (Table 2) were used for this purpose. Several major flowering time genes were segregating in this population (Cuesta-Marcos et al., 2008). To focus only on variation at HvFT1, DH lines were selected with spring (Beka) alleles at VrnH1, VrnH2, and PpdH2. Variation in HvCEN, the most important factor determining flowering time in this population, was also considered. We aimed at having two plants per HvCEN-HvFT1 haplotype, but found only one for the Mogador-Mogador class (an extra plant was allocated to the Beka-Mogador class). PpdH1 was not segregating in this cross. Both parents carry the recessive, long-day insensitive allele.
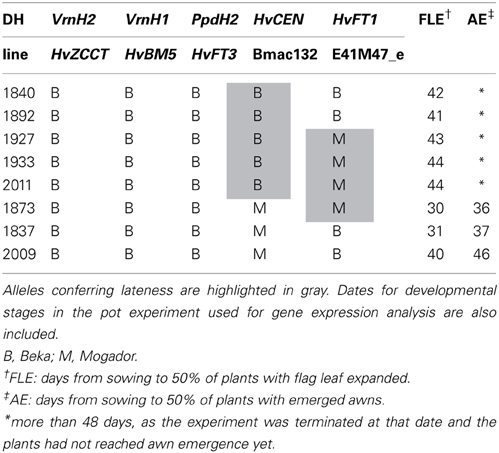
Table 2. Haplotypes for major flowering time genes and markers closest to QTLs (see Cuesta-Marcos et al., 2008) for selected doubled haploid (DH) lines of the population Beka × Mogador.
Sequence Polymorphisms at Major Genes
DNA sequence polymorphisms for the 109 accessions were screened with allele-specific primers of candidate genes. VrnH1 was scored based on the size of the first intron of its candidate HvBM5A (Yan et al., 2003; von Zitzewitz et al., 2005). Alleles were classified according to Hemming et al. (2009); VrnH2 was evaluated as presence of HvZCCT-Ha and HvZCCT-Hb (Karsai et al., 2005). PpdH1 was genotyped using SNP22 in the CCT (Constans, Constans-like, TOC1) domain of its candidate gene HvPRR7, after digestion with BstU I (Turner et al., 2005). PpdH2 was scored as presence of the HvFT3 gene as reported by Casao et al. (2011). Regarding VrnH3, two indels in the promoter and two SNPs in the first intron of the HvFT1 gene were assessed (Casas et al., 2011). HvCEN, candidate gene for Mat-c or Eam6 was partially sequenced in 24 genotypes. The haplotypes are identified as reported by Comadran et al. (2012).
It was not possible to assess HvFT1 polymorphism directly at the Beka x Mogador DH population (Cuesta-Marcos et al., 2008) because the sequences of the parental alleles were conserved and they differed only in copy number (2 Beka, 1 Mogador). The population was reanalyzed based on the new information found (parents polymorphic at HvFT1 for CNV). Two microsatellite markers, in the HvFT1 region, were mapped (EBmac0603 and AF022725A), to provide better resolution of the flowering time QTL found in this region.
HvFT1 CNV Analysis by qPCR
Genomic DNA was isolated from frozen barley leaves using the NucleoSpin Plant II kit (Macherey-Nagel, Germany) and used as template for CNV analysis by qPCR in an ABI 7500, essentially as described by Nitcher et al. (2013) with some modifications. Briefly, 100 ng genomic DNA were mixed with 2 μM of each primer and 10 μl of 2X Power SYBR Green Supermix (Thermo Fisher Scientific, Waltham, MA). The PCR program comprised 10 min at 95°C, 40 cycles of 10 s 95°C and 50 s 60°C, and a melting curve stage. Number of copies of the first exon of HvFT1 was tested in all 109 genotypes. Morex was selected as the calibrator genotype and SNF2 as the housekeeping gene (Yan et al., 2002). Then, in a subset of lines, two other HvFT1 regions (promoter and exon 3) and three other genes close to HvFT1 (UCW118, UCW123, and UCW120) were tested as reported by Nitcher et al. (2013). Efficiency for each primer pair was obtained by serial dilutions of barley genomic DNA and it was taken into account for CNV calculation (Weaver et al., 2010). Efficiencies for SNF2, HvFT1-promoter, HvFT1-exon1, HvFT1-exon3, UCW118, UCW120, and UCW123 were 0.95, 0.96, 0.92, 0.95, 0.96, 1.03, and 0.85, respectively.
Growth Conditions for Expression Studies
Barley plants used for expression analysis were grown in pots of 11.5 (diameter) × 16.0 (height) cm with a mix of peat, sand, and perlite. Six seeds of one genotype were sown per pot. After emergence, they were thinned to three seedlings per pot. The plants were grown in a growth chamber, under long-day conditions (16 h light, 250 μE m−2 s−1, 20°C, 60% relative humidity/8 h dark, 16°C, 65% relative humidity) for 7 weeks. There were three pots per genotype, which were used as replicates.
Two experiments were carried out with samplings at two different times. In the first experiment, leaf tissue was harvested in the middle of the light period, after 8 h light, as reported by Kikuchi et al. (2009). In the second experiment, to maximize circadian expression of HvFT1 (Turner et al., 2005) harvesting took place 2 h before dark, after 14 h light. In both cases, leaf tissue (last expanded or flag leaf) was harvested and frozen immediately in liquid nitrogen before tissue homogenization (Mixer Mill model MM 400, Retsch, Germany). At each sampling time, three samples were analyzed per treatment and genotype. Each sample came from a different plant and pot.
mRNA Extraction, cDNA Synthesis, and Gene Expression Analysis
For qPCR analysis of HvFT1 expression levels, 1 μg of total RNA (purified using the NucleoSpin RNA Plant kit, Macherey-Nagel) was transcribed to cDNA by using the SuperScript III reverse transcriptase and 2.5 μM poly(dT)20 primer according to the manufacturer's instructions (Invitrogen). The reaction mixture for qPCR and the PCR program have been previously described. cDNA was quantified using a Nanodrop system (Thermo Fisher Scientific) and equal amounts were used for all samples. Actin was selected as the housekeeping gene (Trevaskis et al., 2006), and expression of HvFT1 and HvCEN was analyzed using the same primers as in Yan et al. (2006) and Comadran et al. (2012), respectively. Efficiencies for Actin, HvFT1, and HvCEN, were 0.97, 0.86, and 0.96, respectively.
Results
Survey of HvFT1 CNV
Winter and spring genotypes were classified as follows: winter genotypes carry a functional VrnH2 allele and a winter or intermediate allele in VrnH1 (wild type vrnH1, VrnH1-6, or VrnH1-4). Spring or facultative lines have been classified as those with a spring allele in VrnH1 or lines in which VrnH2 is absent. With regard to HvFT1, there were accessions representing all possible combinations of intron and promoter polymorphisms, both within the winter and spring groups (Table 3). The combination of the “late” promoter with the “early” intron, however, was the most frequent. HvFT1 exon 1 copy number ranged from 0.35 ± 0.13 in the winter cultivar Igri to 5.12 ± 0.28 in the spring cultivar Zaida (Figure 1). The results for several genotypes were intermediate between two classes, and were assigned to classes based on pedigree information when possible. Genotypes with several copies of HvFT1 presented allele combinations typical of both winter and spring cultivars at all the Vrn and Ppd genes, with one exception, VrnH2. No genotype with the VrnH2 gene present had more than one HvFT1 copy.
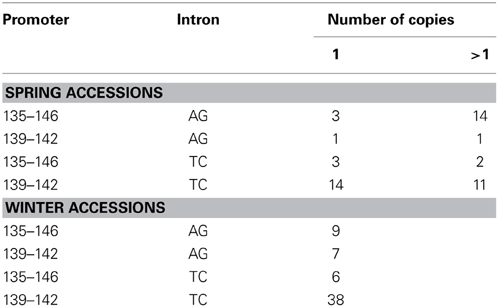
Table 3. Number of spring and winter accessions classified according to HvFT1 haplotypes defined by polymorphisms at the promoter (indel 1- indel 2), first intron and number of copies of exon 1.
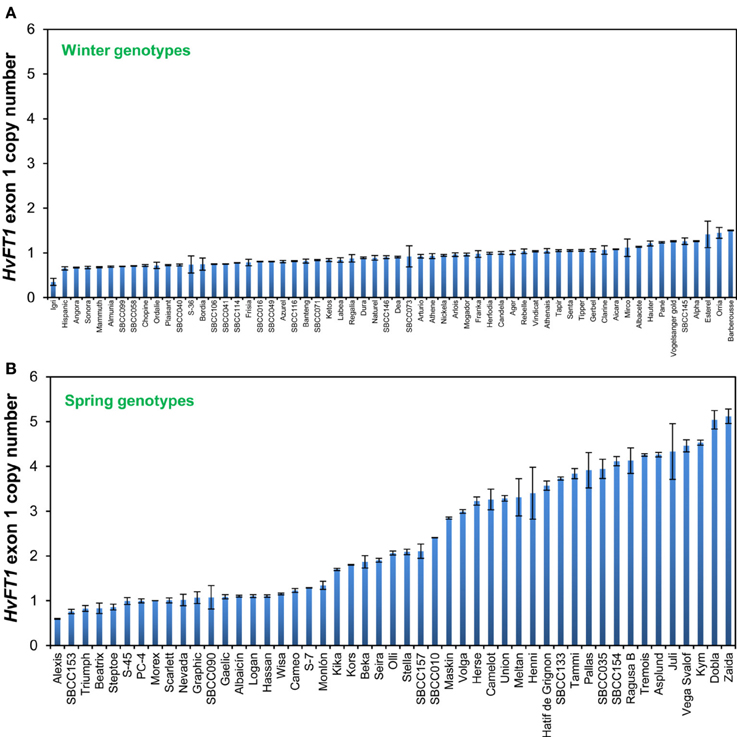
Figure 1. Determination of copy number variation for the first exon of HvFT1 in winter (A) and spring or facultative (B) barleys. Bars represent means ± s.e.m. Morex was used as the calibrator genotype (copy number = 1). SBCC, Spanish Barley Core Collection.
It is remarkable that all of the winter barley cultivars, i.e., with a dominant VrnH2 allele, had only one copy of the first exon. Barberousse, the last winter genotype in Figure 1, had 1.50 ± 0.01 copies, which we considered as a single copy. On the other hand, 28 out of the 49 spring cultivars analyzed contained more than 1.70 copies of exon 1 of HvFT1, which we have considered as multiple copies.
The study was extended to analyze CNV in other areas within or near the HvFT1 gene, as in Nitcher et al. (2013). Thus, as well as qPCR primers for HvFT1 exon 1 (amplifying the region of +200 to +293 bp downstream from the ATG start codon), we used primers for UCW118 (nearest known gene in the flanking Morex BAC 455J22 upstream from HvFT1), HvFT1 promoter (−727 to −656 bp upstream from the ATG), HvFT1 exon 3 (+778 to +902 bp downstream from the ATG), UCW123 (+6.6 kbp downstream from HvFT1), and UCW120 (nearest known gene in the flanking Morex BAC 761F04 downstream from UCW123). This analysis was carried out in 10 barley varieties from Northern Europe (Figure 2A), 18 Spanish landraces (Figure 2B), and in another nine varieties from diverse origins (Figure 2C). Although all genotypes were not analyzed for all the genes, several results merit further attention. Two main patterns of HvFT1 promoter/exon 1/exon 3 copy number were found in Northern European barleys: Asplund, Olli, Herse, Stella, Tammi, and Maskin contained one single copy of the promoter and multiple copies of exon 1 and exon 3, whereas Henni, Meltan, Pallas, and Juli contained multiple copies of promoter, exon 1, and exon 3. These different gene structures may affect gene functionality, as we will discuss later. CNV in other genes around HvFT1 was only analyzed in 11 accessions. UCW118 (next to the promoter), and UCW123 and UCW120 (next to the exon 3) were similar to the corresponding CNV in the adjacent regions, with the exception of Tammi, which contained multiple copies of UCW118 but only one promoter. We do not provide results for UCW120 in Meltan since the primers gave no amplification. Regarding the Spanish lines, only 4 out of 18 genotypes analyzed (SBCC157, SBCC154, SBCC133, and SBCC035) contained multiple copies of the promoter, exon 1 and exon 3. For the other genotypes analyzed, only Beka and Dobla presented several gene copies in the HvFT1 promoter, exon 1, and exon 3.
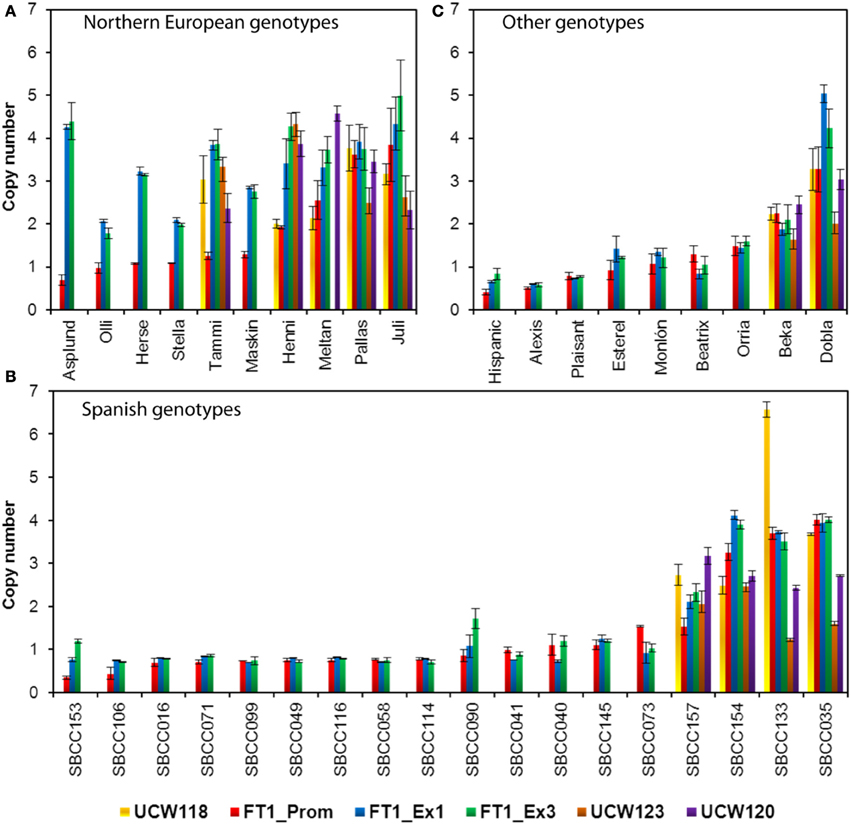
Figure 2. Analysis of copy number variation for HvFT1(promoter, exon 1, and exon 3), and for regions upstream (UCW118) or downstream (UCW123 and UCW120) of HvFT1 on barley genotypes of Northern European (A), Spanish (B), and other origins (C). Bars represent means ± s.e.m. Morex was used as the calibrator genotype (copy number = 1). SBCC, Spanish Barley Core Collection.
Effect of HvFT1 Polymorphism in Populations
We have reassessed several populations to illustrate the effect of sequence and CNV polymorphism at HvFT1.
The population Beka x Mogador (Cuesta-Marcos et al., 2008) is a spring x winter population with very large flowering time variation. It has been reassessed because we have found that, although the sequence of HvFT1 is conserved in both genotypes (1 distal mismatch in 2547 bp sequenced), it still presents CNV polymorphism: Beka has two copies of the gene, whereas Mogador has one. A small flowering time QTL in the vicinity of HvFT1 was already reported, although the region was not well covered with markers in the original study. Close markers were now identified and genotyped in the population to increase coverage. The QTL already detected on 7HS in this cross was made more conspicuous with the markers introduced for this study (Figure 3), with the peak hinting at the HvFT1 position, and a high significance [-log10(P) above 11]. In this case, Beka contributed the early allele, about 2.2 days earlier than the Mogador allele. If this QTL is truly due to the effect of HvFT1, it must be caused by differential effect of the number of copies. The possible effect of HvFT1 CNV on the vernalization mechanism, as described by Nitcher et al. (2013), should have been evident in this population as an interaction of the QTL found at the VrnH1, VrnH2 regions with the QTL at the HvFT1 region, in trials without enough vernalization. This could have occurred in the three late sowings, in which the number of cooling degree days (a measure of vernalizing potential) was much lower than at the fall sowings (Table S1 in Cuesta-Marcos et al., 2008). The six field trials were reanalyzed for interactions between the five major QTL (VrnH1, VrnH2, HvCEN, PpdH2, HvFT1), using the closest marker to each peak, and dividing the six trials into fall sowings (November) and winter-spring sowings (late February to late March). The effect of HvFT1 was clear across all trials, but did not interact much with other genes. There was just one possible interaction of HvFT1 with VrnH1 but in the fall-sown trials and not in the late sown trials. It was caused by a significant difference for the HvFT1 alleles (with Beka, the one with two copies, inducing earliness) only in the presence of the winter (Mogador) allele at VrnH1 (Table 4). The expected interaction between vernalization and HvFT1 at the late-sown trials, however, was not detected at all. Therefore, we can conclude from this result that the possible promoting effect of the double HvFT1 gene of parent Beka was not strong enough to affect the vernalization requirement of the winter lines of this population.
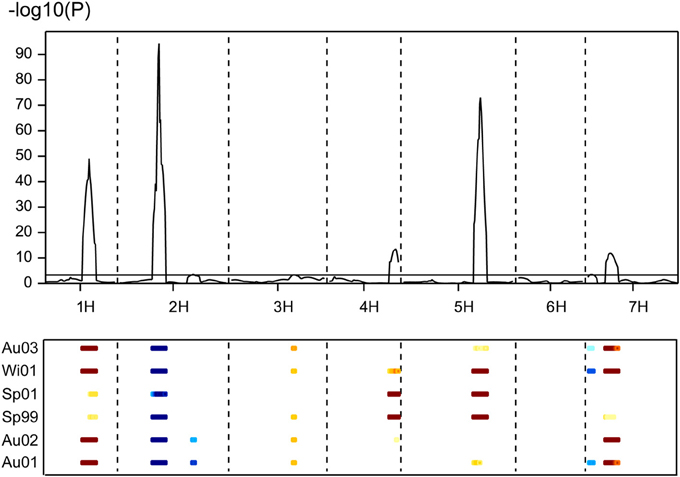
Figure 3. Multi-environment QTL analysis for days to flowering from January 1st for six field experiments carried out with 120 doubled haploid lines and the parents of the population Beka x Mogador. The peaks above the threshold (dashed line) indicate presence of QTL significantly affecting the trait. Data taken from Cuesta-Marcos et al. (2008), enriched with new markers on the 7HS chromosome. In the lower part of the figure, field trials are coded with Au (autumn sowing), Wi (winter sowing), or Sp (spring sowing) and two digits for the year; the colored dashes indicate the extent of the QTL and its direction: blue means that the early allele came from Mogador, yellow-brown from Beka, with intensity proportional to the size of the effect.
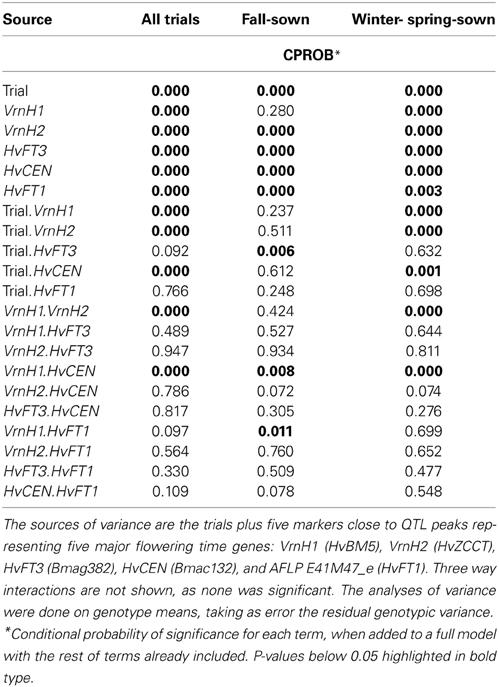
Table 4. Probabilities of significance from three analyses of variance of flowering date recorded at six field trials, three fall-sown and three winter- or spring-sown in the Beka x Mogador population.
SBCC154 x Beatrix is a cross of two spring genotypes. The population was described by Hofmann et al. (2013), but the flowering time data have not been reported yet. SBCC154 has four copies of HvFT1, whereas Beatrix has only one. Both have the putatively late (139–142) promoter (Nitcher et al., 2013), whereas Beatrix has the early intron (TC) and SBCC154 the late one (AG). The HvFT1 marker detected a significant QTL with an effect of 2.5 days, with Beatrix as the early allele (Figure 4).
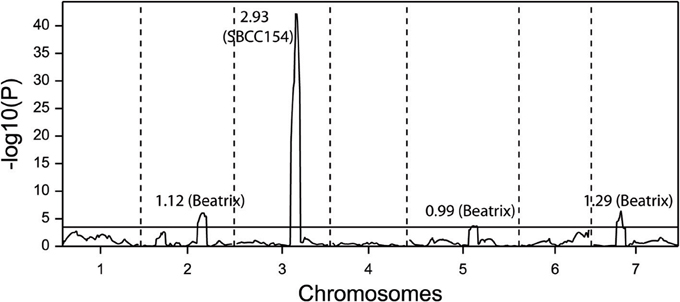
Figure 4. QTL scan for flowering time at a field trial for a population of 168 doubled haploid lines from the cross SBCC154 x Beatrix. The figures besides the QTL peaks indicate the size of the effect, with the early allele indicated in parentheses.
The population Henni x Meltan (Borràs-Gelonch et al., 2010), a cross of two spring cultivars, was an example of polymorphism just at the promoter. Each genotype has four copies of HvFT1 and, although they may have just two copies of the promoter; the parents present the same number of copies across the whole gene. The only polymorphism found was at the promoter, with Meltan carrying the early promoter (135–146) and Henni the late one (139–142). Confirming this expectation, the early allele of the QTL in Borràs-Gelonch et al. (2010) was contributed by Meltan, the difference being 23°C d (around 1–2 days).
Expression Analysis of HvFT1 in Selected Spring Barleys
After finding CNV variation for HvFT1, the next step was to evaluate the effect of CNV variation on gene expression. If the effect of several copies of the gene was always as large as reported by Nitcher et al. (2013), then it should be detectable as a large increase of gene expression and a very early phenotype, overriding the effect of any other polymorphisms at HvFT1. This hypothesis was tested by evaluating gene expression on a set of spring and facultative cultivars representative of different HvFT1 copy number alleles: Morex (reference genotype for one copy of HvFT1) and Beatrix as single HvFT1 copy number, SBCC154, SBCC157, Dobla, Beka, Pallas, and Juli as examples of multiple promoter and exon 1 copies, and Tammi as representative of the genotypes with one promoter but multiple exon 1 copies (Table S1 and Figure 5A). HvFT1 expression was found, in general, to increase according the developmental stage of the plants, although the expression levels differed widely among genotypes. At the second sampling date all genotypes, except Dobla, had not reached awn emergence yet. At this point, SBCC154, SBCC157, and Dobla, the earliest genotypes, displayed the highest HvFT1 mRNA levels (awn emergence dates are included in Figure 5A). Apart from this observation, we did not observe much correlation between HvFT1 expression and time to awn appearance. Interestingly, these three varieties had the dominant allele for PpdH1, whereas the rest had the recessive allele at this gene. Also, we could not find a clear correlation between number of HvFT1 copies or sequence polymorphism and HvFT1 mRNA expression. For example, Juli and Tammi contained multiple copies of HvFT1 (four promoters and four genes in Juli, one promoter and four genes in Tammi), but both of them showed lower HvFT1 expression than SBCC154, SBCC157, or Dobla, at the same sampling date. The influence of the difference in HvFT1 promoter copies on HvFT1 expression observed in Tammi with respect to Juli and Pallas will be discussed later. Thus, we can conclude that CNV has not a prevailing effect on HvFT1 expression, over other types of polymorphisms at the same gene. It is not the main factor controlling HvFT1 expression and flowering time (measured as awn appearance), and it depends largely on the genetic background, and polymorphisms at other genes, especially PpdH1, as has been reported previously in the literature in studies done before taking into account CNV (Turner et al., 2005; Hemming et al., 2008).
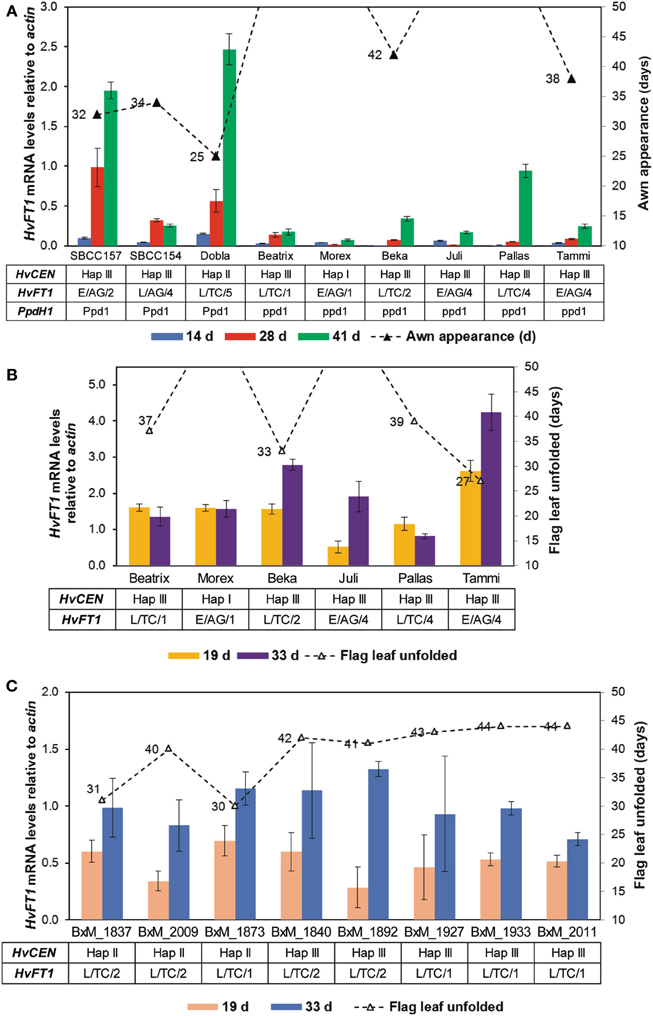
Figure 5. HvFT1 expression analysis by qPCR. In a first experiment (A), leaf tissue was harvested in the middle of the light period (after 8 h light) 14, 28, or 41 days after sowing. Days until awn appearance (triangles) are shown for the whole duration of the experiment (50 days). HvCEN, HvFT1, and PpdH1 haplotypes for each plant are also indicated. In the case of HvFT1, polymorphisms for promoter (E, “early”; L, “late”), intron 1 (AG or TC) and copy number variation (for exon 1) are shown. In the second experiment (B,C), harvesting took place 2 h before dark (after 14 h light), 19 or 33 days after sowing. Days until full unfolding of the flag leaf (triangles) are shown for the whole duration of the experiment (50 days). As previously, HvCEN and HvFT1 haplotypes for each plant are included. (C) Expression in selected doubled haploid lines of the Beka x Mogador population (BxM, see Table 2). Bars represent means ± s.e.m.
Expression Analysis of HvFT1 in Selected Lines of the Beka x Mogador Population
In a second gene expression experiment, eight DH lines of the Beka x Mogador population were analyzed, together with some genotypes in common with the first experiment (Figures 5B,C). This population was found to contain a QTL for flowering time in HvFT1 gene, as shown above. Individuals were selected according to CNV in HvFT1: Beka contained two copies while Mogador contained only one. qPCR analysis were performed as before, but this time the material was harvested 2 h before dark. Sampling time actually had a major effect on the detection of HvFT1 expression: under these conditions, HvFT1 mRNA levels were, in general, clearly higher than at the first experiment (Figure 5A), in which samples were harvested 8 h after the commencement of the light period, instead of 14 h. The varieties used in the first experiment, which had a recessive ppdH1 allele, were also included for comparison. Apart from the clear induction of expression from a dominant PpdH1 allele, we were not able to establish a clear relationship between HvFT1 copy number, mRNA levels and flowering time for these plants (Figure 5B). HvFT1 expression in the DHs was similar and stable during the experiment after 19 and 33 days. No significant differences were observed in HvFT1 mRNA levels between the two alleles as main effects, although there was a significant interaction (P = 0.022) between the alleles at HvCEN and HvFT1. At the second sampling date, the plants with the Beka allele in HvFT1 showed significantly higher expression than plants with Mogador allele, only if the allele at HvCEN also came from Beka. The Mogador allele at HvCEN had a major effect on earliness, and these lines (1837, 1873, and 2009) flowered markedly earlier than the others (flag leaf unfolding dates are included in Figure 5C).
Another interesting result of the second experiment was that Tammi showed the largest HvFT1 expression after 33 days, and also flowered the earliest. It seems that Tammi has a more effective HvFT1 gene to promote flowering than the others. This could be related to the fact that Tammi, as indicated above, presented four copies of the HvFT1 gene but only one copy of the promoter, whereas Juli and Pallas, for example, have four copies of both promoter and exon 1.
Expression Analysis of HvCEN in the Beka x Mogador Population and in Selected Spring Barley Genotypes
As we have shown previously, CNV and different haplotypes for HvFT1 are not sufficient to establish a clear relationship between them, HvFT1 mRNA levels and awn appearance. For example, the presence of a dominant PpdH1 allele was found to enhance HvFT1 expression in different spring genotypes (Figure 5A). Additionally, CNV in HvFT1 did not have a clear role to determine flag leaf unfolding in the Beka x Mogador DHs (Figure 5C). For these reasons, we decided to analyze the mRNA levels of another gene involved in the establishment of flowering time, HvCEN, a paralog of FT and TFL1 in Arabidopsis (Kobayashi et al., 1999). We used the same cDNAs as for the previous expression studies (Figure 6). We included haplotype information for some genotypes about HvCEN in Table S1 for comparison. As observed for HvFT1, our results suggest that HvCEN mRNA levels are not the main responsible to explain awn appearance (Figure 6). HvCEN expression dramatically increased when samples were harvested 14 h after dawn instead of 8 h (Figures 6A–C), as we observed for HvFT1. Another remarkable observation is that HvCEN expression also changed during the development, in disagreement with previously observed results for HvCEN expression (Comadran et al., 2012). For example, Morex contained significantly higher HvCEN mRNA levels than Tammi, but Morex did not flower after 50 days and awns in Tammi appeared after 33 days.
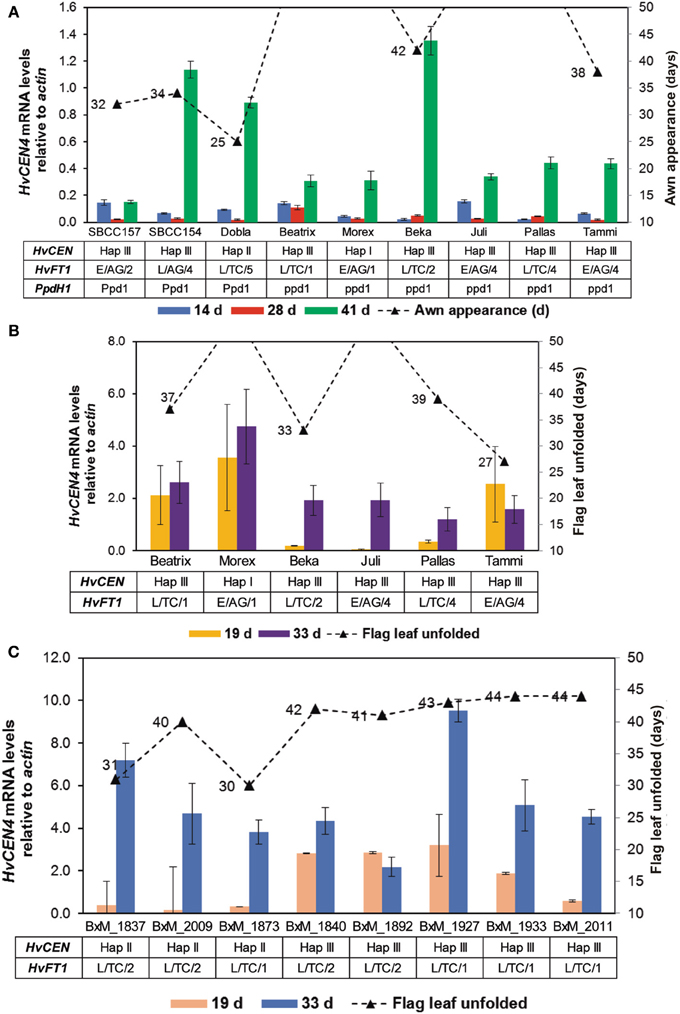
Figure 6. HvCEN expression analysis by qPCR. HvCEN mRNA levels were quantified using the same conditions as for Figure 5. (A) leaf tissue was harvested in the middle of the light period (after 8 h light) 14, 28, or 41 days after sowing. (B,C), harvesting took place after 14 h light, 19 or 33 days after sowing. (C) Expression in selected doubled haploid lines of the Beka x Mogador population (BxM, see Table 2). Bars represent means ± s.e.m.
Discussion
Distribution and Phenotypic Effect of HvFT1 Polymorphisms Over Barley Germplasm
In this paper, we analyzed the extent of HvFT1 CNV and its effect on HvFT1 expression in more than 100 different spring and winter barley genotypes, mainly European, including some landraces from the Spanish Barley Core Collection (SBCC) (Igartua et al., 1998). Recently, Nitcher et al. (2013) demonstrated that the FT1 allele present in the barley genetic stock BGS213, which carried several copies of HvFT1, showed earlier transcriptional up-regulation of FT1, and was associated with a dominant spring growth habit. Cultivar Tammi, from Finland, was the donor of the mutation present in that genetic stock (Franckowiak and Konishi, 1997). This allele is not frequent and was reportedly found only in spring cultivars originating from regions of extremely high latitude or high altitude (Takahashi and Yasuda, 1971). We confirmed the finding by Nitcher et al. (2013) that the Tammi/BGS213 allele was characterized by having a single copy of the promoter and several copies of the transcribed region. This kind of allele was present in a group of 6 six row Northern European cultivars, some of them related by pedigree (Baumer and Cais, 2000; Chiapparino et al., 2006). The origin of this allele could be traced back to primitive cultivar Asplund, which is in the parentage of Tammi and Herse, both showing alleles with multiple exon copies and one promoter (Figure 2). This allele seems to be particularly beneficial at high latitudes because both parents of Tammi (Asplund and Olli) come from the Northern limits of the barley cultivation range (Manninen and Nissila, 1997).
We found several copies of HvFT1 in accessions from apparently disconnected germplasm groups, like Scandinavian and Spanish landraces. Although the sample examined is not enough to derive definitive conclusions, the distribution of CNV alleles over type of growth habit haplotypes is intriguing. Multiple copies at HvFT1 occurs only in genotypes that do not have winter growth habit. This suggests a possible disadvantageous agronomic effect of the presence of several copies of the gene in interaction with VrnH2. A possible mechanism that provides a plausible explanation for this is that the increased copy number of HvFT1 is epistatic to winter alleles for VrnH1 and VrnH2 (as reported by Nitcher et al., 2013). Muñoz-Amatriaín et al. (2013) have recently shown that high levels of CNVs are found in the barley genome, around 9.5% in coding genes, at increasing frequencies as distance to the centromere increases. This is a widespread phenomenon, contributing to phenotypic variation in barley. HvFT1 CNV may be neutral in spring genotypes, but breeders could have selected against this duplication when breeding winter barley to avoid early transition to reproductive growth and exposure of reproductive tissues to damaging low temperatures. However, we have shown that this epistatic mechanism is not universal, as it does not occur in the Beka x Mogador population.
The reassessment of three biparental populations representing polymorphisms of different kind identified QTL in the region of HvFT1. Previous studies also shed some light on the possible phenotypic effect of HvFT1. The QTL found in the cross SBCC145 x Beatrix (Ponce-Molina et al., 2012), whose peak coincides with the gene, must be due to polymorphisms at the promoter region, as both parents have one copy of the gene and same first intron sequence. The early allele was contributed by SBCC145, a Spanish landrace from the Canary Islands, which carries the “early” promoter. It must be pointed out, however, that SBCC145 has a distinct promoter, with additional polymorphisms compared to others, similar to cultivars Dairokkaku (Casas et al., 2011) and Meltan. The cross SBCC154 x Beatrix offered the opportunity to contrast two different polymorphisms (promoter for SBCC145 x Beatrix, first intron, and CNV for SBCC154 x Beatrix) against a common parent (Beatrix). This population was tested at a single trial sown in autumn, in the same field as the population SBCC145 x Beatrix, and the result was the presence of a QTL, exactly at the marker used to genotype HvFT1, with SBCC154 as the late allele. In this case, high copy number (SBCC154) was late compared to one copy, but this could be due to an effect of SBCC154 carrying the “late” intron. Therefore, any acceleration of flowering that might be produced as a consequence of high copy number was secondary to the lateness associated to the sequence polymorphism. It must be noted that PpdH1 was segregating in this population. However, flowering occurred too early to allow for a significant effect of PpdH1, which was not detected as QTL.
In the cross SBCC016 x Esterel (Casas et al., 2011), each genotype had one copy of HvFT1, Esterel has the “early” intron (TC) and SBCC016 the “late” one (AG). Accordingly, the early allele of the QTL, which also peaked at the gene itself, was contributed by Esterel. Hayes et al. (1993) and Borràs-Gelonch et al. (2012) reported a QTL in this region for the population Steptoe x Morex. Although the size of the effect was not large, in both studies the Morex allele was significantly later. Both parents have just one copy of the gene, and a mixture of late and early polymorphisms at the promoter and the intron. It seems that the intron effect is stronger, as Steptoe carries the early (TC) allele at this position.
A QTL in the region of HvFT1 was also detected in the classical studies carried out in the Igri x Triumph population (Laurie et al., 1995), with the Triumph allele conferring earlier flowering, although the nucleotide sequences were identical (Yan et al., 2006). CNV has been recently identified in this population, with two copies of the HvFT1 gene in Triumph (R. Nitcher, personal communication). This result would match our findings in the Beka x Mogador population. Nevertheless, we could not reproduce this result, since the Triumph seed we analyzed had only one copy of the gene. We cannot discard that the samples analyzed are different, because this cultivar is known under two different names, Trumpf in Germany and Triumph in the UK, where it was reselected from somewhat heterogeneous seed (van Harten, 1998).
In summary, these findings reported in the literature, combined with the results presented in this study, reveal that detection of flowering time QTL in the region of HvFT1 in biparental populations representing all kinds of polymorphisms at HvFT1 (promoter, first intron, and CNV) is common. There is no functional proof that this gene is responsible for all the QTL, but it is a good candidate. An alternative explanation could be the presence, at least in some cases, of an additional flowering time gene closely linked to HvFT1. In any case, we have to wait until there are either functional proofs or increasing evidence from other populations to declare that HvFT1 is the responsible for the variation observed. We expect that the catalog of polymorphisms presented in this study will help other researchers to contribute information to clarify the issue.
The survey of HvFT1 polymorphisms allows us to conclude that HvFT1 (VrnH3) is far from being effectively fixed in cultivated barley, as stated up to now (Stracke et al., 2009; Comadran et al., 2012). This statement is probably true if one only considers the allele responsible for the huge phenotypic effect observed by Nitcher et al. (2013) on a winter barley, which confirmed observations of the seminal work by Takahashi and Yasuda (1971). The effects that we have found in different populations (this study), association studies (Casas et al., 2011) and also reported in the literature (previous paragraphs) point at smaller phenotypic effects that are linked to all types of polymorphisms in this gene (promoter, first intron, CNV). These effects, however, cannot be easily extrapolated to different genetic backgrounds.
CNV and Gene Expression
The commencement of the reproductive stage in barley and the duration of the time period until flowering are controlled by a variety of factors that act interactively. In temperate cereals like wheat and barley, flowering is promoted by long days. In barley, the up-regulation of VRN3/FT1 appears to be the main trigger for the initiation of flowering (Faure et al., 2007), although an alternative view is that its role may be to accelerate inflorescence development and reduce the time taken from double-ridge to head emergence (Sasani et al., 2009). Its expression is tightly regulated, repressed by VRN2 under long days (Hemming et al., 2008), which actually competes with CO to bind to protein complexes that activate FT (Li et al., 2011). Also in response to long days, FT1 expression is promoted by CO-like proteins (Li et al., 2011; Campoli et al., 2012) and the pseudo response regulator PpdH1 (Turner et al., 2005; Faure et al., 2007). This promotion may occur by interaction of CO with the promoter of FT, as happens in Arabidopsis (Tiwari et al., 2010; Li et al., 2011; Andrés and Coupland, 2012).
The findings of this study add to mounting evidence pointing at a complex control of the timing of head emergence that cannot be easily reduced to a simple scheme. On one hand, HvFT1 displays a variety of polymorphisms in regions that are compatible with a regulatory role. On the other hand, its expression pattern is compounded by its nodal position, downstream of the vernalization and long day pathways, whose genes also have large influence on duration of plant development.
The first gene expression experiment results confirmed the induction of HvFT1 expression by the long day pathway, irrespective of the polymorphisms present at HvFT1. The genotypes that reached first awn appearance in that experiment were all those having a sensitive/dominant PpdH1 allele, particularly at the second sampling date. This date is probably the most meaningful because all PpdH1 genotypes reached awn appearance just a few days later. This result agrees with other reports where HvFT1 expression in ppdH1 background was markedly lower (Turner et al., 2005; Hemming et al., 2008; Kikuchi et al., 2009). Therefore, we can conclude that gene duplication of HvFT1 does not always ensure higher expression and, in any case, the scale of its effect is minor compared to the induction by PpdH1. The case of high HvFT1 expression in presence of ppdH1 described in Nitcher et al. (2013) seems an exception, probably due to a unique genetic background.
Our results confirm that the triggering of events at the meristem leading to flowering is not determined just by HvFT1 expression. There must be additional factors that probably need the presence of HvFT1 product to interact with. We cannot rule out, however, that copy number is related to a dosage effect, precisely by the different genetic background in each genotype, which may lead to differences in the induction of HvFT1. A beneficial increase in dosage is actually one of the evolutionary forces explaining the conservation of gene duplications, particularly for genes that mediate the interaction between the organism and the environment, as reported for Ppd-B1 and Vrn-A1 in wheat by Díaz et al. (2012), or for genes with dosage-sensitive functions, owing to protein-protein interaction (Innan and Kondrashov, 2010). HvFT1 corresponds to the first class, and may as well be part of the second. CNV is widespread in plants (Zmieńko et al., 2014), and certainly in barley (Muñoz-Amatriaín et al., 2013), but its effects vary in each case. For instance, in barley, the powdery mildew resistance allele mlo-11 acts by disrupting its expression through accumulation of non-functional copies of the gene upstream of the wild-type copy (Piffanelli et al., 2004). On the other hand, the effect of the freezing tolerance locus FrH2 in barley depends on the number of CBF genes transcripts produced, that are proportional to the number of copies present in the gene cluster identified as responsible for this QTL (Pasquariello et al., 2014). The plant immunity locus GER4, also in barley, is also a cluster of tandemly duplicated genes. In this case, the enhanced transcript dosage was proposed as the evolutionary driving force for the local expansion and functional redundancy of the locus (Himmelbach et al., 2010).
The results of Nitcher et al. (2013) and the earliness induced by the double copy “Beka” allele in the Beka x Mogador population point in the direction of a dosage effect. However, the direct comparison of HvFT1 expression figures for different alleles is hampered by the large influence of the alleles at PpdH1 on HvFT1 and at HvCEN on flowering time. The analysis of the eight sister lines of the Beka x Mogador population indicates a slightly earlier flowering and higher HvFT1 expression of lines with the Beka allele only in presence of the Beka allele at HvCEN (spring, Hap III), but the evidence is too small to declare that we have found a clear dosage effect.
These results led us to speculate that the most efficient version of the barley gene could be one with a single conserved promoter, and variable number of transcribed regions, as seen in cultivar Tammi, or one promoter and one copy of the gene. Other versions of the gene, with several promoters and transcribed regions may take longer to be induced. Indeed, we do not know whether the multiple copies of HvFT1 are functional or not. They seem to be expressed in Tammi/BGS213, but there is no evidence for other alleles. If they are, the presence of several full and functional copies of the gene may affect differently the expression of the gene, depending on the concentration of the promoting signal. If it is low, its dilution over several copies may delay transcription, whereas an abundant promoting signal would enhance transcription proportionally in alleles with several copies. The special case of Tammi/BGS213 indicates either that the transcription of several copies is triggered by a single promoter, or that the additional copies of the gene are placed under the control of other promoters and thus escape the regulatory control of the wild-type gene. For other multiple copy alleles, with equal number of promoters and transcribed regions, we can speculate that the copies are full and perfect replicates of the original gene, and that the expression of them all is affected by the same processes.
The difference in overall gene expression between the two experiments agrees with the reports on pattern of diurnal oscillation of FT1 expression during the day (Turner et al., 2005; Kikuchi et al., 2009). Actually, the same observation can be made for HvCEN (Figure 6). This information, combined with the sequence analogy between the two genes, suggests also that HvCEN could be subject to circadian rhythm.
Interactions of HvFT1
The DH lines (Figures 5C, 6C) presented very different dates to reach a developmental stage (flag leaf unfolding in this case) in the presence of rather similar levels of HvFT1 expression. As pointed out before, in this case the cause for differences in development seems to lie on the allele present at HvCEN. It is clear that accelerated development in the presence of the Mogador HvCEN allele (Hap II) did not depend on higher expression of HvFT1. Therefore, early development in these lines must be caused by some mechanism acting in parallel to HvFT1, either through protein-protein interaction with HvFT1 or by a combined effect with HvFT1 by focusing on the same targets.
Actually, the interaction between HvFT1 and HvCEN is by no means unexpected. HvCEN and HvFT1 are probably paralogs as suggested by their high sequence identity (59%), and indeed have been annotated as members of closely related protein families before (Higgins et al., 2010; Andrés and Coupland, 2012; Comadran et al., 2012). Therefore, it is likely that their products display similar interactive patterns with other proteins. There are many evidences in the literature pointing at the interaction of FT proteins with other proteins in the flowering promotion pathway. Ahn et al. (2006) reported that the family of small proteins coded by FT and homologous genes act as “either scaffolds or regulators of signaling complexes” in Arabidopsis. The bZIP transcription factor FD also seems to play a central role in flowering (Wigge et al., 2005). More recently, Jaeger et al. (2013) put forward a model to explain flowering control in Arabidopsis in which TFL1 (the product of another paralog gene) competed with FT to form the complex with the FD gene product, needed to trigger floral meristem specification. Some evidence for this kind of process in cereals was found by Li and Dubcovsky (2008), who detected interaction between the proteins of TaFT (the FT homolog of bread wheat) and FD-like2, and also in rice (Taoka et al., 2011), although in this case the interaction needed the intermediation of a third protein. In barley, however, FD orthologs can only be predicted in terms of sequence similarity.
A search in the Protein Data Bank revealed that, besides the FT-FD interaction, Arabidopsis homodimers of FT and also TFL1 have been reported. Thus, it is also conceivable that proteins coming from genes as similar as HvFT1 and HvCEN may form heterodimers as well, making the speculation about the interaction between the products of these two genes more plausible. The HvCEN polymorphism, which translates to a non-synonymous mutation (Pro135Ala), is located in a solvent-exposed protein loop (Comadran et al., 2012). However, with the structural evidence at hand, this loop does not directly participate in any dimeric interface and therefore nothing can be concluded about its molecular role in the interface.
The role of HvFT1 may actually go beyond the duration of developmental phases. There is recent evidence about dramatic agronomic effects of the ortholog of FT in tomato, SFT in interaction with SP (SELF PRUNING, itself an ortholog of HvCEN and HvTFL1). SFT is in a “dose-dependent epistatic interaction” with SP which results in a modification of plant architecture that can be optimized to produce higher yields (Jiang et al., 2013). This dose-dependent action of SFT also opens the ground for speculation about possible dose-dependent action of multiple copies of HvFT1 and its agronomic outcome.
Conclusions and Further Work
The main conclusions of this study are:
• A wide survey of barley germplasm revealed that HvFT1 duplication was only observed in spring and facultative barleys that do not possess a functional VrnH2 allele.
• Two models of HvFT1 duplication were observed, one that includes the promoter and the gene, the other only the transcribed region. Higher gene expression seems associated only to the second one.
• There are flowering time QTL on the region of HvFT1 in different populations, representing all types of polymorphism at HvFT1, promoter, first intron and CNV.
• Analysis of HvFT1 expression and phenotypic effects showed that they depend on gene polymorphisms but also on genetic background.
Plant breeders must be able to fully harness the development of cereal plants to be able to respond to the challenges of climate change. In this study, we present the case to state that HvFT1 has been a hot spot to fine tune barley adaptation to environmental conditions, and will have to be given due consideration by breeders to create future cultivars.
The role of other genes in triggering flowering initiation, possibly in interaction with HvFT1, seems a very rich area which deserves more research. In particular, further avenues to attempt to resolve the precise sequence or structural variation in HvFT1 which causes early or late flowering,- particularly for the Tammi allele will require crossing different HvFT1 alleles into a common genetic background, maintaining fixed PpdH1 and HvCEN. This could be combined with BAC sequencing for the entire HvFT1 region, for key alleles.
Author Contributions
Ernesto Igartua, Ana M. Casas and M. P. Gracia conceived the experiments. Jorge Loscos performed qPCR analysis. Ana M. Casas and Jorge Loscos characterized the flowering time genes. Ernesto Igartua and Jorge Loscos performed computational analysis. Bruno Contreras-Moreira performed the bioinformatics searches and helped with the interpretation of the data. All authors wrote and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the Spanish Ministry of Science and Innovation (MICINN), within the project AGL2010-21929; multiplication of plant materials was funded by project RFP-00015-00-00; all authors were supported by Aragon government project A06. We appreciate helpful comments by Jorge Dubcovsky and Rebecca Nitcher during the design of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00251/abstract
Table S1. Barley accessions surveyed in this study classified according to the genetic constitution at vernalization and photoperiod genes. Countries of origin and row number are also indicated.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Baumer, M., and Cais, R. (2000). Abstammung der Gerstensorten. Freising: Bayerische Landesanstalt für Bodenkultur und Pflanzenbau.
Borràs-Gelonch, G., Denti, M., Thomas, W. T. B., and Romagosa, I. (2012). Genetic control of pre-heading phases in the Steptoe x Morex barley population under different conditions of photoperiod and temperature. Euphytica 183, 303–321. doi: 10.1007/s10681-011-0526-7
Borràs-Gelonch, G., Slafer, G. A., Casas, A. M., van Eeuwijk, F., and Romagosa, I. (2010). Genetic control of pre-heading phases and other traits related to development in a double-haploid barley (Hordeum vulgare L.) population. Field Crops Res. 119, 36–47. doi: 10.1016/j.fcr.2010.06.013
Boyd, W. J. R., Li, C. D., Grime, C. R., Cakir, M., Potipibool, S., Kaveeta, L., et al. (2003). Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L) genotypes grown over a mild winter growing season. Aust. J. Agric. Res. 54, 1277–1301. doi: 10.1071/AR03014
Campoli, C., Drosse, B., Searle, I., Coupland, G., and von Korff, M. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69, 868–880. doi: 10.1111/j.1365-313X.2011.04839.x
Casao, M. C., Igartua, E., Karsai, I., Lasa, J. M., Gracia, M. P., and Casas, A. M. (2011). Expression analysis of vernalization and day-length response genes in barley (Hordeum vulgare L.) indicates that VRNH2 is a repressor of PPDH2 (HvFT3) under long days. J. Exp. Bot. 62, 1939–1949. doi: 10.1093/jxb/erq382
Casas, A. M., Djemel, A., Ciudad, F. J., Yahiaoui, S., Ponce, L. J., Contreras-Moreira, B., et al. (2011). HvFT1 (VrnH3) drives latitudinal adaptation in Spanish barleys. Theor. Appl. Genet. 122, 1293–1304. doi: 10.1007/s00122-011-1531-x
Chen, A., and Dubcovsky, J. (2012). Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 8:e1003134. doi: 10.1371/journal.pgen.1003134
Chiapparino, E., Donini, P., Reeves, J., Tuberosa, R., and O'Sullivan, D. M. (2006). Distribution of β-amylase I haplotypes among European cultivated barleys. Mol. Breeding 18, 341–354. doi: 10.1007/s11032-006-9035-0
Cockram, J., Chiapparino, E., Taylor, S., Stamati, K., Donini, P., Laurie, D., et al. (2007). Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor. Appl. Genet. 115, 993–1001. doi: 10.1007/s00122-007-0626-x
Comadran, J., Kilian, B., Russell, J., Ramsay, L., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1392. doi: 10.1038/ng.2447
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cuesta-Marcos, A., Igartua, E., Ciudad, F. J., Codesal, P., Russell, J. R., Molina-Cano, J. L., et al. (2008). Heading date QTL in a spring x winter barley cross evaluated in Mediterranean environments. Mol. Breeding 21, 455–471. doi: 10.1007/s11032-007-9145-3
Cuesta-Marcos, A., Szucs, P., Close, T. J., Filichkin, T., Muehlbauer, G. J., Smith, K. P., et al. (2010). Genome-wide SNPs and re-sequencing of growth habit and inflorescence genes in barley: implications for association mapping in germplasm arrays varying in size and structure. BMC Genomics 11:707. doi: 10.1186/1471-2164-11-707
Díaz, A., Zikhali, M., Turner, A. S., Isaac, P., and Laurie, D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 7:e33234. doi: 10.1371/journal.pone.0033234
Distelfeld, A., Li, C., and Dubcovsky, J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12, 178–184. doi: 10.1016/j.pbi.2008.12.010
Faure, S., Higgins, J., Turner, A., and Laurie, D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
Franckowiak, J. D., and Konishi, T. (1997). Stock number BGS213: spring growth habit 3. Barley Genet. Newsl. 26, 212.
Hayes, P. M., Liu, B. H., Knapp, S. J., Chen, F., Jones, B., Blake, T., et al. (1993). Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor. Appl. Genet. 87, 392–401. doi: 10.1007/BF01184929
Hemming, M. N., Fieg, S., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomics 282, 107–117. doi: 10.1007/s00438-009-0449-3
Hemming, M. N., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2008). Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
Higgins, J. A., Bailey, P. C., and Laurie, D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5:e10065. doi: 10.1371/journal.pone.0010065
Himmelbach, A., Liu, L., Zierold, U., Altschmied, L., Maucher, H., Beier, F., et al. (2010). Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell 22, 937–952. doi: 10.1105/tpc.109.067934
Hofmann, K., Silvar, C., Casas, A. M., Herz, M., Büttner, B., Gracia, M. P., et al. (2013). Fine mapping of the Rrs1 resistance locus against scald in two large populations derived from Spanish barley landraces. Theor. Appl. Genet. 126, 3091–3102. doi: 10.1007/s00122-013-2196-4
Igartua, E., Gracia, M. P., Lasa, J. M., Medina, B., Molina-Cano, J. L., Montoya, J. L., et al. (1998). The spanish barley core collection. Genet. Resour. Crop Evol. 45, 475–481. doi: 10.1023/A:1008662515059
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
Jaeger, K. E., Pullen, N., Lamzin, S., Morris, R. J., and Wigge, P. A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25, 820–833. doi: 10.1105/tpc.113.109355
Jiang, K., Liberatore, K. L., Park, S. J., Alvarez, J. P., and Lippman, Z. B. (2013). Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet. 9:e1004043. doi: 10.1371/journal.pgen.1004043
Karsai, I., Szucs, P., Meszaros, K., Filichkina, T., Hayes, P. M., Skinner, J. S., et al. (2005). The Vrn-H2 locus is a major determinant of flowering time in a facultative x winter growth habit barley (Hordeum vulgare L.) mapping population. Theor. Appl. Genet. 110, 1458–1466. doi: 10.1007/s00122-005-1979-7
Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T., and Handa, H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353. doi: 10.1104/pp.108.132134
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Laurie, D. A. (2009). “Developmental and reproductive traits in the Triticeae,” in Genetics and Genomics of the Triticeae. Plant Genetics and Genomics: Crops and Models 7, eds C. Feuillet and G. J. Muehlbauer (Netherlands: Springer Science Business Media), 591–609. doi: 10.1007/978-0-387-77489-3_20
Laurie, D. A., Pratchett, N., Bezant, J. H., and Snape, J. W. (1995). RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome 38, 575–585. doi: 10.1139/g95-074
Li, C., Distelfeld, A., Comis, A., and Dubcovsky, J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67, 763–773. doi: 10.1111/j.1365-313X.2011.04630.x
Li, C., and Dubcovsky, J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55, 543–554. doi: 10.1111/j.1365-313X.2008.03526.x
Loukoianov, A., Yan, L., Blechl, A., Sanchez, A., and Dubcovsky, J. (2005). Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 138, 2364–2373. doi: 10.1104/pp.105.064287
Manninen, O., and Nissila, E. (1997). Genetic diversity among Finnish six-rowed barley cultivars based on pedigree information and DNA markers. Hereditas 126, 87–93. doi: 10.1111/j.1601-5223.1997.00087.x
Muñoz-Amatriaín, M., Eichten, S. R., Wicker, T., Richmond, T. A., Mascher, M., Steuernagel, B., et al. (2013). Distribution, functional impact, and origin mechanisms of copy number variation in the barley genome. Genome Biol. 14, R58. doi: 10.1186/gb-2013-14-6-r58
Nitcher, R., Distelfeld, A., Tan, C., Yan, L., and Dubcovsky, J. (2013). Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Genet. Genomics 288, 261–275. doi: 10.1007/s00438-013-0746-8
Oliver, S. N., Finnegan, E. J., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U.S.A. 106, 8386–8391. doi: 10.1073/pnas.0903566106
Pasquariello, M., Barabaschi, D., Himmelbach, A., Steuernagel, B., Ariyadasa, R., Stein, N., et al. (2014). The barley Frost resistance-H2 locus. Funct. Integr. Genomics 14, 85–100. doi: 10.1007/s10142-014-0360-9
Payne, R. W., Murray, D. A., Harding, S. A., Baird, D. B., and Soutar, D. M. (2009). GenStat for Windows, 12th Edn., Introduction. Hemel Hempstead: VSN International.
Piffanelli, P., Ramsay, L., Waugh, R., Benabdelmouna, A., D'Hont, A., Hollricher, K., et al. (2004). A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430, 887–891. doi: 10.1038/nature02781
Ponce-Molina, L. J., Casas, A. M., Gracia, M. P., Silvar, C., Mansour, E., Thomas, W. B. T., et al. (2012). Quantitative trait loci and candidate loci for heading date in a large population of a wide barley cross. Crop Sci. 52, 2469–2480. doi: 10.2135/cropsci2012.01.0029
Sasani, S., Hemming, M. N., Oliver, S. N., Greenup, A., Tavakkol-Afshari, R., Mahfoozi, S., et al. (2009). The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J. Exp. Bot. 60, 2169–2178. doi: 10.1093/jxb/erp098
Stracke, S., Haseneyer, G., Veyrieras, J. B., Geiger, H. H., Sauer, S., Graner, A., et al. (2009). Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor. Appl. Genet. 118, 259–273. doi: 10.1007/s00122-008-0896-y
Szűcs, P., Skinner, J. S., Karsai, I., Cuesta-Marcos, A., Haggard, K. G., Corey, A. E., et al. (2007). Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol. Genet. Genomics 277, 249–261. doi: 10.1007/s00438-006-0195-8
Takahashi, R., and Yasuda, S. (1971). “Genetics of earliness and growth habit in barley,” in Barley Genetics II. Proceeding 2nd International Barley Genetics Symposium, ed R. A. Nilan (Pullman: Washington State University Press), 388–408.
Tamaki, S., Matsuo, S., Wong, H., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Taoka, K. I., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Tiwari, S. B., Shen, Y., Chang, H.-C., Hou, Y., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U.S.A. 100, 13099–13104. doi: 10.1073/pnas.1635053100
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Trevaskis, B., Hemming, M. N., Peacock, W. J., and Dennis, E. S. (2006). HvVRN2 responds to day length, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140, 1397–1405. doi: 10.1104/pp.105.073486
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
van Harten, A. M. (1998). “Mutation breeding,” in Theory And Practical Applications, ed A. M. van Harten (Cambridge: Cambridge University Press), 353.
von Zitzewitz, J., Szűcs, P., Dubcovsky, J., Yan, L., Francia, E., Pecchioni, N., et al. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59, 449–467. doi: 10.1007/s11103-005-0351-2
Weaver, S., Dube, S., Mir, A., Qin, J., Sun, G., Ramakrishnan, R., et al. (2010). Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 50, 271–276. doi: 10.1016/j.ymeth.2010.01.003
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Yan, L., Echenique, V., Busso, C., SanMiguel, P., Ramakrishna, W., Bennetzen, J. L., et al. (2002). Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol. Genet. Genomics 268, 488–499. doi: 10.1007/s00438-002-0765-3
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19851–19856. doi: 10.1073/pnas.0607142103
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Keywords: barley, flowering time, copy number variation, HvFT1, HvCEN
Citation: Loscos J, Igartua E, Contreras-Moreira B, Gracia MP and Casas AM (2014) HvFT1 polymorphism and effect—survey of barley germplasm and expression analysis. Front. Plant Sci. 5:251. doi: 10.3389/fpls.2014.00251
Received: 19 March 2014; Accepted: 16 May 2014;
Published online: 06 June 2014.
Edited by:
Maria Von Korff Schmising, Max Planck Society, GermanyReviewed by:
Ben Trevaskis, Commonwealth Scientific and Industrial Research Organisation, AustraliaMuhammad Aman Mulki, Max Planck Institute for Plant Breeding Research, Germany
Copyright © 2014 Loscos, Igartua, Contreras-Moreira, Gracia and Casas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana M. Casas, Department of Genetics and Plant Production, Estación Experimental de Aula Dei, Consejo Superior de Investigaciones Científicas, Avda. Montañana 1005, 50059 Zaragoza, Spain e-mail:YWNhc2FzQGVlYWQuY3NpYy5lcw==
 Jorge Loscos
Jorge Loscos Ernesto Igartua
Ernesto Igartua Bruno Contreras-Moreira
Bruno Contreras-Moreira M. Pilar Gracia
M. Pilar Gracia Ana M. Casas
Ana M. Casas