- 1The Australian Research Council Centre of Excellence in Plant Energy Biology, The University of Western Australia, Perth, WA, Australia
- 2Centre of Excellence in Computational Systems Biology, The University of Western Australia, Perth, WA, Australia
- 3Centre for Comparative Analysis on Biomolecular Networks, The University of Western Australia, Perth, WA, Australia
Sub-functionalization during the expansion of gene families in eukaryotes has occurred in part through specific subcellular localization of different family members. To better understand this process in plants, compiled records of large-scale proteomic and fluorescent protein localization datasets can be explored and bioinformatic predictions for protein localization can be used to predict the gaps in experimental data. This process can be followed by targeted experiments to test predictions. The SUBA3 database is a free web-service at http://suba.plantenergy.uwa.edu.au that helps users to explore reported experimental data and predictions concerning proteins encoded by gene families and to define the experiments required to locate these homologous sets of proteins. Here we show how SUBA3 can be used to explore the subcellular location of the Deg protease family of ATP-independent serine endopeptidases (Deg1–Deg16). Combined data integration and new experiments refined location information for Deg1 and Deg9, confirmed Deg2, Deg5, and Deg8 in plastids and Deg 15 in peroxisomes and provide substantial experimental evidence for mitochondrial localized Deg proteases. Two of these, Deg3 and Deg10, additionally localized to the plastid, revealing novel dual-targeted Deg proteases in the plastid and the mitochondrion. SUBA3 is continually updated to ensure that researchers can use the latest published data when planning the experimental steps remaining to localize gene family functions.
Introduction
The expansion of gene families in eukaryotes has divided function between members in a variety of ways (Massingham et al., 2001; Rutter et al., 2012; Wang et al., 2012). One such division has been the diversification of localization of protein family members to different parts of the cell (e.g., for isoprenoid metabolism; Beck et al., 2013). Interconnected metabolic and regulatory pathways operate in distinct subcellular compartments and the proteins that perform these processes are restricted by compartment boundaries. An important step toward defining the biochemical role of a given protein family member is therefore to identify the intracellular location in which it accumulates and functions.
Researchers can determine the subcellular location of proteins by a number of approaches. These include in silico prediction methods and experimental approaches. Computational prediction programs are often based on machine-learning algorithms that search for sequence features in a primary amino acid sequence to predict the likelihood that a protein is found in a specific subcellular location. These computer programs have become critical tools for annotating newly sequenced genomes on a large scale. Experimental approaches that are available for confirming subcellular location include in vitro protein import studies into isolated organelles, in vivo protein tagging by fluorescent markers, enzyme activity measurements, immunolocalization, or cell fractionation followed by protein detection using mass spectrometry (Millar et al., 2009). It is important to note that localization data sets obtained from such experiments form the basis of both the determination of subcellular localization and the set up of training sets that are used to create prediction programs.
Proteomic studies employ mass spectrometry to identify proteins in enriched subcellular compartments and lead to large, information-rich datasets. Purification techniques have improved rapidly over the last decade and have allowed better identification of more specific subcellular locations. For example, the combination of density gradient centrifugation with free-flow electrophoresis was employed to improve the separation of tonoplast from plasma membranes (Bardy et al., 1998), mitochondria from peroxisomes and plastids (Eubel et al., 2007), and the isolation of Golgi membranes (Parsons et al., 2012). In addition, novel analysis strategies have been developed, such as intelligent data-dependent acquisition (IDDA), that can increase the number of peptide ions analyzed in the mass spectrometer and consequently improve the identification of peptides and proteins relative to previous methods (Eubel et al., 2008; Hoopmann et al., 2009).
Another experimental approach that is widely used to localize proteins in the cell is the expression and visualization of fluorescent proteins (FPs) that are attached to the proteins of interest. Notably, in vivo FP tagging is the only subcellular location method that provides data for intact, living cells. However, the positioning of the FP in a chimeric construct is important as it can mask the targeting ability of a protein signal peptide and this can greatly affect the accuracy of the localization results. For example, an N-terminally tagged mitochondrial protein is likely to be mistargeted and so is a C-terminally tagged peroxisomal protein. Often this method is referred to as green FP (GFP) tagging because GFP is one of the most frequently used FPs (Chiu et al., 1996). Thousands of Arabidopsis proteins have been visualized using this direct approach (including some high-throughput GFP screens) and these form an important resource for determining subcellular location (Tian et al., 2004; Koroleva et al., 2005; Li et al., 2006; Carrie et al., 2009; Van Aken et al., 2009; Boruc et al., 2010; Lee et al., 2011; Narsai et al., 2011; Inze et al., 2012).
Predicted and experimental localisation data are scattered in the literature and researchers can spend large amounts of time and effort to ensure all published localization information for a given protein has been collated. In fact, despite best efforts, published data can easily be overlooked as large number of protein localizations can be reported in an article but not listed in the title, abstract or text. In addition, curated subcellular proteomes and catalogs of GFP targeting information are not readily available as defined data sets for specific cellular locations.
The SUBcellular localization database for Arabidopsis proteins (SUBA; Heazlewood et al., 2005, 2007; Tanz et al., 2013) aggregates these datasets to combine prediction of protein localization for Arabidopsis proteins with experimental data and annotations. SUBA3 also includes a naive Bayesian classifier (SUBAcon) to provide a likely consensus location of a protein within the cell (Tanz et al., 2013). SUBA has previously been used for assessing targeting prediction programs (Heazlewood et al., 2004; Ryngajllo et al., 2011), for building metabolic network models (de Oliveira Dal’Molin et al., 2010; Mintz-Oron et al., 2012), and for analyzing co-expression and protein–protein interaction (PPI) data (Cui et al., 2011; Ryngajllo et al., 2011). Here we highlight features of SUBA3 that can be used to explore protein families by using the Deg protease family in Arabidopsis as an example. The Deg protease family was chosen because experimental localization data for some members of this family were complex, including conflicting data and the absence of any experimental data for a range of family members. This analysis is used for prioritizing and performing experiments highlighted by SUBA3, which were required to complete the localization of this protein family.
Material and Methods
SUBA3 Database and Data Sources
SUBA3 can easily be queried through a web-browser based graphical user interface (GUI) that is freely available at http://suba.plantenergy.uwa.edu.au. The interface works best via the Mozilla Firefox, Google Chrome, or Safari web browsers but will work on Microsoft Explorer (6 and above). Currently, 24,142 entries are based on subcellular proteomic studies (7891 distinct proteins), 4110 entries are based on FP tagging studies (2647 distinct proteins), and 13,164 entries are based on PPI studies (4999 distinct proteins). SUBA3 also contains bioinformatic predictions for protein localization from the output of 22 prediction programs. Details of the database structure and sources have been described previously (Tanz et al., 2013). To best estimate a protein’s location in the cell, SUBA3 also contains a consensus location (SUBAcon) based on Bayesian probabilities calculated from all the experimental localization data and predictions available for each protein.
Subcellular Localization by GFP Tagging
The full-length coding sequences where ever possible, or the first 222–300 bp of the coding sequences of Deg1–Deg16 were amplified according to the manufacturer’s instructions using the Expand High Fidelity PCR system (Roche Diagnostics) with primers listed in Supplemental Table 1 containing the attB sites for Gateway® cloning. The PCR products of Deg1–Deg15 were cloned into the Gateway® vector pDONR207 (Invitrogen) and sequenced. The entry clone and a Gateway® cloning cassette (pDest/pgem/CGFP; Carrie et al., 2009) were recombined to clone the full-length or the first 222–300 bp in frame with the coding region of the GFP at their N- or C-terminus. For co-localization studies, the small subunit (SSU) of Arabidopsis ribulose-1,5-bisphosphate carboxylase oxygenase fused to the N-terminus of the red FP (SSU-RFP; Carrie et al., 2009) was used as a plastid control, the mitochondrial targeting sequence of yeast ScCox4 fused to the N-terminus of mCherry in pBIN20 (mt-rk CD3-991; Nelson et al., 2007) was used as a mitochondrial control, and the peroxisomal targeting signal 1 (PTS1, Ser-Lys-Leu) fused to the C-terminus of mCherry (px-rk CD3-983; Nelson et al., 2007) was used as peroxisome control. The fusion constructs were biolistically transformed into cultured Arabidopsis cells. The GFP and RFP/mCherry plasmids (5 μg of each) were co-precipitated onto 1-μm gold particles and transformed using the biolistic PDS-1000/He system (Bio-Rad). Particles were bombarded onto 2 mL of cultured Arabidopsis cells resting on filter paper on osmoticum plates (2.17 g/L Murashige and Skoog Modified Basal Salt Mixture, 30 g/L sucrose, 0.5 mg/L naphthalene acetic acid, 0.05 mg/L kinetin, 36.44 g/L mannitol). After bombardment, the cells were placed in the dark at 22°C. Fluorescence images were obtained 24 h after transformation using an Olympus BX61 epifluorescence microscope with excitation wavelengths of 460/480 nm (GFP) and 535/555 nm (RFP and mCherry), and emission wavelengths of 495–540 nm (GFP) and 570–625 nm (RFP and mCherry). Subsequent images were captured using Cell® imaging software.
Results
Querying SUBA3 to Locate Data on Members of Protein Families
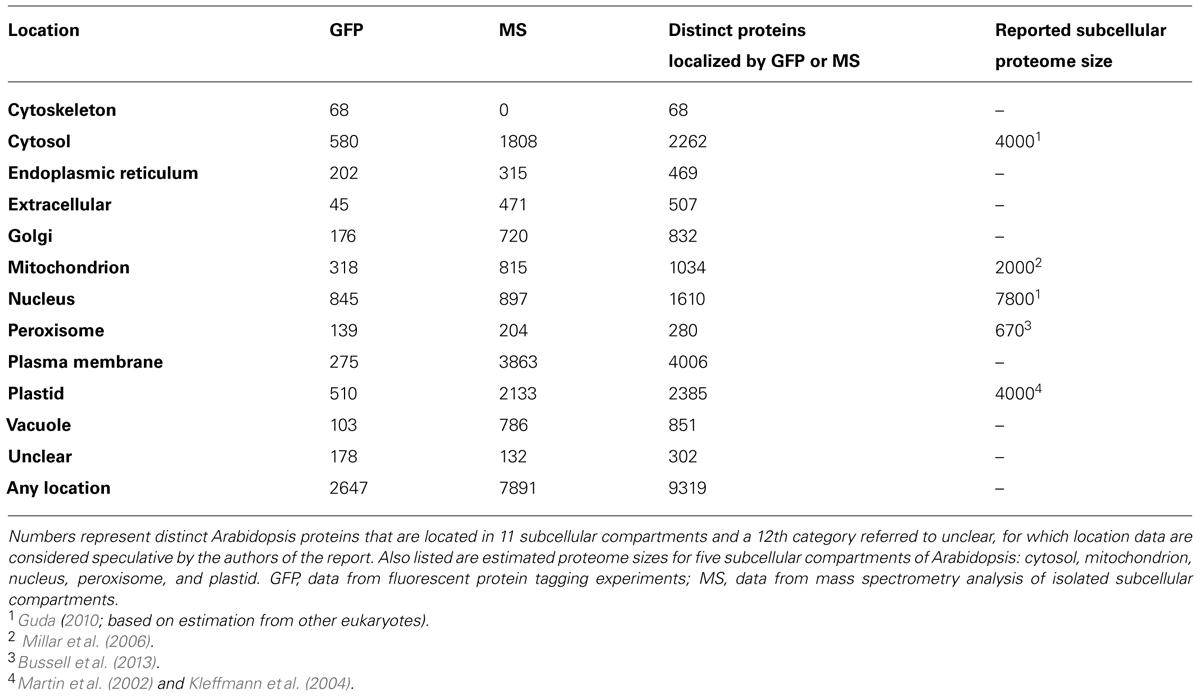
Users can operate the SUBA3 interface1 to ask simple questions about one protein at a time or to construct moderately complex SQL queries using drop down menus and buttons to perform powerful Boolean queries (AND, OR, NOT) across all entries for a protein family. The primary “Search” tab allows the development of the query (Figure 1A). Once a query has been submitted, the “Results” page shows a table, which contains the Arabidopsis genome initiative (AGI) identifier, a short description, summary localization information from SUBAcon, predictions, annotations, GFP, mass spectrometry, and PPI data (Figure 1B). Nearly all retrieved data are linked to a reference in PubMed2. A variety of information and helpful links for each AGI in the result page is hyperlinked by access to the “SUBA flatfile” (Figure 1C). These data include detailed information on the subcellular localization predictions, a cell cartoon displaying the probability values of the consensus location (SUBAcon), access to GFP images for localizations if available, and links to other resources for this AGI (Figure 1C).
FIGURE 1. The SUBA3 interface. Screenshots showing the SUBA3 search page (A) where complex queries can be build using pull down menus in combination with AND, OR, and bracketing functions. The SUBA3 Results page (B) of a query is shown with seven default columns and a “Download All Results” button in the left top corner. An example of a SUBA3 flatfile (C) showing detailed localization information of predictions, annotations, and experimental data (GFP, mass spectrometry, protein–protein interaction data), the SUBAcon call displayed as pictographic heat map of a plant cell, GFP images, and other details such as the description, amino acid sequence, hydropathy plot and links to other useful websites at the bottom of the page.
To query a particular gene family, the full list of AGI identifiers can be entered in the “AGI” input box and directly queried in SUBA3. Alternatively, SUBA3 can be searched by entering a descriptor in the “keyword” input box, such as the name of a particular gene family in the TAIR10 genome annotation. For many gene families, this type of search can be used to rapidly assess the predicted location and, if applicable, collate the experimental location information for each member. The importance of multiple pieces of independent evidence for high confidence assessments of subcellular location has been highlighted (Millar et al., 2009). SUBA3 rapidly provides clarity on the available evidence and the methods used, allowing researchers to make decisions to augment what is currently known with independent experimental approaches.
Using the search term “DegP protease” as a keyword in a SUBA3 search (as indicated in Figure 1A) yields 18 proteins (Figure 2). Note, the nomenclature of “DegP proteases” has recently changed and we will now refer to them collectively as “Deg proteases” (Huesgen et al., 2005). This family of ATP-independent serine endopeptidases (originally named for “degradation of periplasmic proteins”) functions in various proteolytic events in the cell. Sixteen different Deg protease gene loci are known in Arabidopsis (Deg1–Deg16). Two of the 18 result outputs from SUBA3, At2g47940.2 and At5g39830.2, are alternative splice variants of Deg2 and Deg8, respectively (Figure 2). Examining the SUBAcon results for the remaining 16 Deg proteases shows that they are predicted to eight of the 11 different physical locations in SUBA3 (namely cytosol, endoplasmic reticulum, Golgi, mitochondrion, nucleus, peroxisome, plasma membrane, and plastid). Eight Deg proteases have been identified in subcellular proteomic studies, and two of those eight have additionally been localized by GFP tagging (Figure 2). Studying these data shows there are agreements and disagreements between predictors and/or between experimental datasets for the location of Deg proteases. This raises a series of issues that can be addressed in turn by further analysis in SUBA. Firstly, when experimental data are absent but prediction is clear, is the experimental coverage of a given location extensive enough to expect the protein to have been found? Secondly, when experimental data disagree could specific experiments be erroneous? Thirdly, when experimental data disagree, could the protein be multi-targeted?
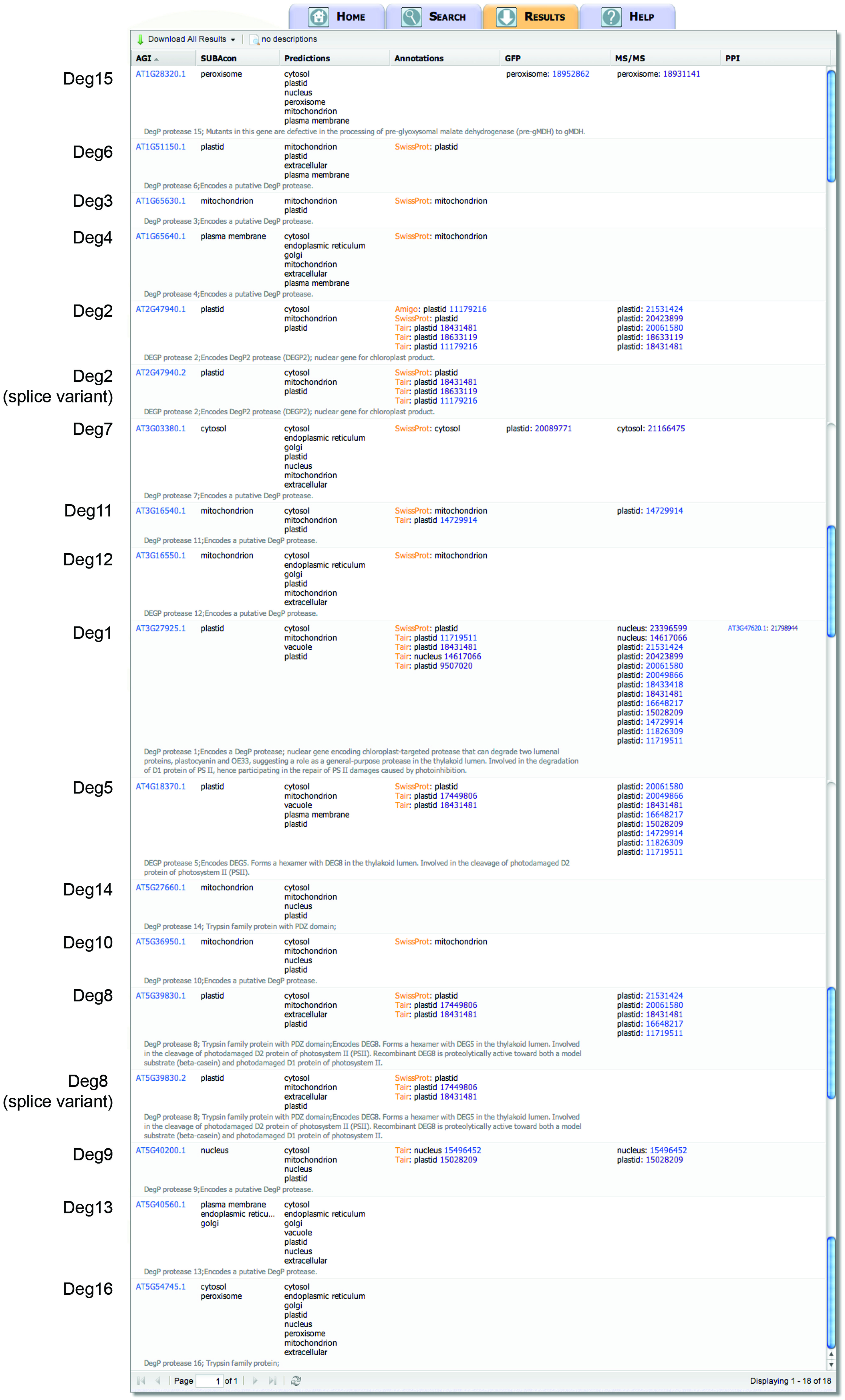
FIGURE 2. SUBA3 result output for Deg proteases in Arabidopsis. Querying the SUBA3 database by entering “DegP protease” as descriptor in the “keyword” input box returns results for 18 gene products with location evidence. Displayed are the location calls by SUBAcon, the predicted locations determined by the 22 predictors in SUBA3 (column “Predictions”), the locations from the annotators TAIR, AmiGO, and UniProt/SwissProt (column “Annotations”), the experimentally determined locations by GFP tagging and mass spectrometry, as well as protein–protein interaction (PPI) data. Short TAIR descriptions are displayed in gray and the name of each Deg family member is indicated on the left.
Defining the Size of Experimental vs Predicted Datasets for a Specific Subcellular Location
To determine in which subcellular location a protein without experimental localization data can be expected to be found, it is often very beneficial to generate a list of the subcellular locations of proteins by different experimental methods (Table 1). An additional step supported by SUBA is combining experimental sets like mass spectrometry and GFP tagging (“MS/MS assay” OR “GFP assay”). Using this approach, plastid and mitochondrial sets can be defined that contain 2385 and 1034 proteins respectively. These sets can be expanded by adding other proteins that are predicted to be in the relevant organelle to construct the total predicted proteome. For example, by including proteins predicted by the three predictors TargetP, Predotar and YLoc [“MS/MS assay” OR “GFP assay” OR (“TargetP” AND “Predotar” AND “YLoc”)], the plastid proteome set is 3026 proteins and the mitochondrial set is 1651. The sizes of a variety of organelle proteomes in plants and other eukaryotes have been previously reported in the literature (Table 1).
Table 1 shows that over 2000 proteins have been located to the plastid by MS analysis, which represents almost a third of all proteins located by this method. Similarly, proteins located to the plastid by GFP tagging represent almost a fifth of all the proteins located by GFP tagging. Taken together, these datasets give a 60% experimental coverage of the estimated plastid proteome size in Arabidopsis (Table 1). Thus, it can be expected that experimental data are likely to exist for Deg proteases located in plastids. In fact, seven of the eight Deg proteases, for which experimental data exist, have been localized to the plastid by the MS or GFP approach (Figure 2). In comparison, only one Deg protease has been localized to the peroxisome. Significantly fewer proteins have been localized to the peroxisome experimentally and the coverage of the estimated peroxisome proteome is only 42% (Table 1) leaving more room for unidentified Deg proteases to be found in this location.
Conflicts between Published Localization Datasets
As new reports of subcellular locations of proteins accumulate in the literature discrepancies with previous observations inevitably accumulate as well. SUBA3 gives an overview of these data sets and by using the “Literature referenced location is/is not described in …” option on the “Search” page, SUBA3 gives access to data sets from each individual paper. To directly compare these individual data sets and to determine whether claimed locations have also been reported by other groups, the AND, OR, and bracketing connectors in the “Search” window can be used. For example, Reumann et al. (2007) and Eubel et al. (2008) began to define the peroxisomal proteome by mass spectrometry and GFP analysis, listing 79 and 115 proteins, respectively. The common set between the two studies is 53 (Reumann et al., 2007 AND Eubel et al., 2008). In 2009, Reumann et al. published a larger protein set of 151 peroxisomal proteins and the common set between the two groups rose to 73 proteins [(Reumann et al., 2007 OR Reumann et al., 2009) AND Eubel et al., 2008]. Similarly, Ferro et al. (2010) and Olinares et al. (2010) published plastid proteome sets of 1321 and 586, respectively. The common set of 473 proteins shows that 81% of the proteins listed in Olinares et al. (2010) were also found by Ferro et al. (2010).
When a conflict between experimental data exists for a gene family (e.g., as seen in Figure 2 for Deg1, Deg7, and Deg9), SUBA3 users can directly compare the individual research publications that reported the data sets and determine whether similar conflicts/contaminations have also been reported by other groups. Deg9, for example, has been localized by MS to two different locations by two independent research groups. Pendle et al. (2005) found Deg9 in the nucleus, whereas Kleffmann et al. (2004) claim a plastid location for this protein. Using the “Literature reference location” search with the AND connector (Pendle et al., 2005 AND Kleffmann et al., 2004), results in a shared set of 61 proteins that are claimed to be located in both compartments, the nucleus and plastid. Examining this set of 61 proteins further, showed that 20 of the 26 proteins for which independent GFP tagging data exist were confirmed to be located in the nucleus, whereas a plastid location could only be verified for two proteins. This suggests that a nuclear location for Deg9 is more likely than a plastid location.
Potentially Multi-Targeted Proteins
Multi-targeting of proteins was originally expected to be a rare event, given the specialized function of the different subcellular compartments in a cell. However, the number of proteins that have been shown to be multi-targeted has greatly increased. Currently, more than 100 proteins are known to be dual-targeted to mitochondria and plastids in Arabidopsis (Carrie and Small, 2013). This is due to the ease by which dual-targeting can be confirmed by GFP tagging and because of an increased interest in processes expected to be common to these two organelles. These processes include DNA replication and repair, transcription, translation, and proteolysis. Many proteins have been found to be dual-targeted between other organelles in plants, including mitochondria, plastids and cytosol (Small et al., 1998), mitochondrion, plastid and endoplasmic reticulum (Lee et al., 2011), nucleus and cytosol (Inze et al., 2012), plastids and nucleus (Schwacke et al., 2007), mitochondria and nucleus (Krause and Krupinska, 2009), plastids and peroxisomes (Reumann et al., 2007; Sapir-Mir et al., 2008), mitochondria and peroxisomes (Carrie et al., 2008, 2009), plastid and cytosol (Kiessling et al., 2004; Thatcher et al., 2007), mitochondria and cytosol (Duchene et al., 2001), Golgi-like vesicles and cytosol (Rautengarten et al., 2011), plastids and endoplasmic reticulum (Levitan et al., 2005), and mitochondrion and endoplasmic reticulum (Lee et al., 2011). In addition, some proteins associate with the exterior of an organelle but do not penetrate the hydrophobic membrane and thus, whilst actually cytosolic, appear localized to a specific organelle (Rautengarten et al., 2011).
SUBA3 can assist by systematically searching for putatively dual-targeted proteins. Using the information from the prediction programs and the experimental data, lists of candidates for dual-targeted proteins can be generated. To compile a list of candidate proteins targeted to both plastid and mitochondrion, for example, SUBA3 can be queried for proteins that have been observed in these two locations by GFP tagging or mass spectrometry [(by “GFP” to be in “mitochondrion” OR by “MS/MS” to be in “mitochondrion”) AND (by “GFP” to be in “plastid” OR by “MS/MS” to be in “plastid”)]. This results in a list of 315 proteins that have been observed in both locations. Using SQL or manually analyzing the SUBA3 result file shows that in many of these cases (93 proteins) both locations are reported within the same publication. Therefore, these are known dual-targeted proteins. However, for over 200 proteins, the two locations were reported in different publications, and thus these are potentially unrecognized dual-targeted proteins. Repeating this process for plastids and peroxisomes gives a list of 86 proteins and for mitochondria and peroxisomes a list of 41 proteins. These potential examples of dual-targeting can be tested experimentally by GFP tagging, import studies or other approaches.
Figure 2 shows that for three Deg proteases experimental localization data disagree. Both Deg1 and Deg9 have been localized to the plastid and nucleus using mass spectrometry (Figure 2). Similarly, Deg7 has been localized to the plastid by GFP tagging and to the cytosol by mass spectrometry (Figure 2). Searching GFP and MS data sets in SUBA using the procedure described above results in a list of 398 proteins localized to the plastid and nucleus and a list of 254 proteins for plastid and cytosol. Analyzing these two lists further shows that only four proteins for plastid/nucleus and twelve proteins for plastid/cytosol are reported by the same research group. Thus, for the majority of proteins the plastid/nucleus or plastid/cytosol dual locations were described in different publications, including Deg1, Deg7, and Deg9, and thus, these Deg proteases are potentially unrecognized dual-targeted proteins.
Expanding Deg Protease Localization Data from SUBA3 with Novel GFP Tagging Data
When considering the Deg protease dataset as a whole, it became clear that the reasons for discrepancies could not be resolved without new data. The key experiments missing in order to determine if the proteomics data could be independently confirmed, and to resolve conflicts or multi-targeting, were to systematically fuse the 16 Deg proteases to GFP and observe localization by fluorescence microscopy. To do this, the full-length coding sequences were amplified from cDNA and used for the fusion with GFP. Due to the low expression of some of the Deg proteases or the possibility of being pseudogenes (Schuhmann and Adamska, 2012), their full-length coding sequence could not be amplified from cDNA. Where possible the first 300 bp were amplified from genomic DNA or, if the first exon was shorter than 300 bp, the longest possible region within the first exon was amplified (see schematic representations in Figure 3). These Deg sequences were C-terminally tagged with GFP as most of the Deg proteases were expected to be localized to the mitochondrion or plastid, based on prediction, and N-terminal fusions would have masked the targeting ability of the plastid and mitochondrial signal peptides. On the contrary, Deg15 contains a C-terminal peroxisomal targeting signal 1 (PTS1, Ser-Lys-Leu) and a C-terminally tagged Deg15 would be mistargeted. Thus, the GFP was fused to the N-terminus of its particular coding sequence. The data obtained are presented in Figure 3 with additional co-localizations available in Supplemental Figure 1.
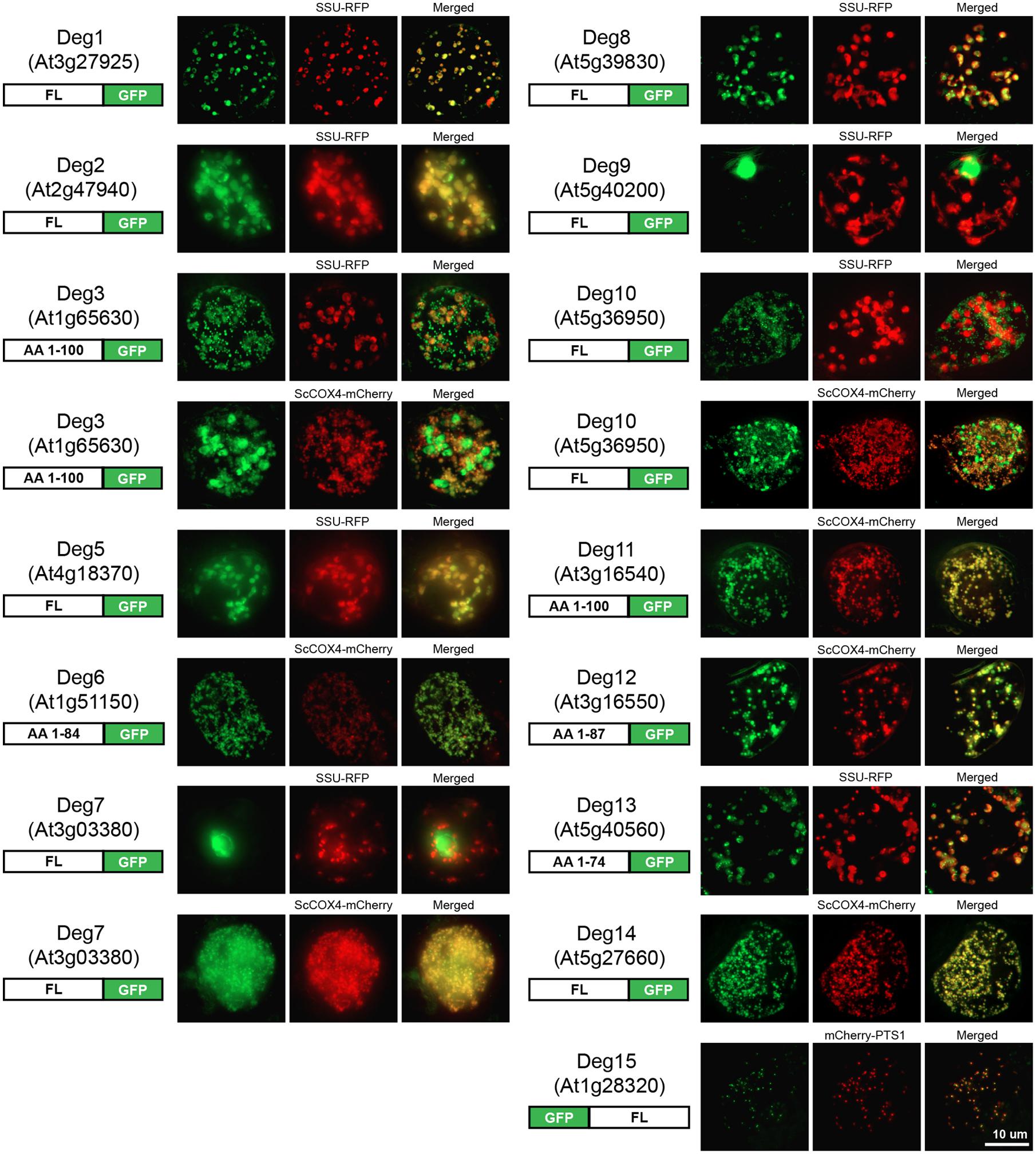
FIGURE 3. Fluorescence images of the subcellular localization of Deg proteases by GFP tagging. N- and/or C-terminal GFP fusion proteins were constructed. Either the full-length (FL) or part of the protein with the number of amino acids (AA) indicated in the schematic representation of each construct (left) were fused to GFP. Targeting ability was tested in Arabidopsis suspension cells using SSU-RFP as a marker for plastid targeting, ScCOX4-mCherry as a marker for mitochondrial targeting, and mCherry-PTS1 as a marker for peroxisome targeting. Multiple co-localizations are presented for Deg3, Deg7, and Deg10 as these Deg proteases localized to more than one subcellular compartment in this study suggesting dual-targeting or conflicting localization results. Scale as indicated.
Plastid and mitochondrial Deg proteases
The plastid location of Deg1, Deg2, Deg5, and Deg8 reported by multiple proteomic studies was confirmed by GFP tagging (Figure 3). These proteases have been shown to function either in the chloroplast stroma (Deg2) or in the thylakoid lumen (Deg1, Deg5, Deg8) and it has been suggested that they are involved in the biogenesis of photosystem II, maintaining protein homeostasis in the thylakoid lumen, and degrading and repairing damaged proteins in the thylakoid lumen and the stroma (reviewed in Schuhmann and Adamska, 2012). The nuclear location of Deg1 found by MS studies could not be confirmed by GFP tagging, despite using a full-length fusion that should contain any nuclear localization signal of Deg1. The PPI data show that Deg1 interacts with TCP14 (At3g47620), a transcription factor localized to the nucleus that regulates seed germination and shows elevated expression level just prior to germination. The PPI study was a large high-throughput study based on the yeast-two hybrid system (Arabidopsis Interactome Mapping Consortium, 2011) and awaits independent confirmation. As we cannot exclude a location of Deg1 to the nucleus, perhaps during germination, this PPI data may be informative. However, both proteomics studies that found Deg1 localized to the nucleus were performed on adult plants so do not directly support the germination hypothesis.
The plastid location of Deg11 reported by a single proteomic study (Friso et al., 2004) could not be confirmed by GFP tagging. Instead Deg11 was found to be located in the mitochondrion (Figure 3). A mitochondrial location of this protease is also predicted by 14 of the 22 predictors in SUBA3, including TargetP (Emanuelsson et al., 2000), Predotar (Small et al., 2004), and YLoc (Briesemeister et al., 2010).
No experimental data on the location of Deg proteases Deg3, Deg6, Deg10, Deg12, Deg13, and Deg14 (Figure 2 and Table 2) have been published, and nothing is known about their physiological roles. At least eight predictors in SUBA3 predict a mitochondrial location for Deg3, Deg6, Deg10, Deg12, and Deg14 and indeed we found these five proteases are targeted to the mitochondrion by our GFP tagging approach (Figure 3). Surprisingly, a fluorescence signal was also detected in the plastid when Deg3-GFP and Deg10-GFP fusion constructs were used to transform Arabidopsis suspension cells (Figure 3), suggesting that Deg3 and Deg10 are dual-targeted to the mitochondrion and to the plastid. Interestingly, Deg3 and Deg10 were also predicted to both the plastid and the mitochondrion when querying SUBA3 using Predotar, TargetP and WoLF PSORT [(“is” predicted by “Predotar” to be in “plastid” OR “is” predicted by “TargetP” to be in “plastid” OR “is” predicted by “WoLF PSORT” to be in “plastid”) AND (“is” predicted by “Predotar” to be in “mitochondrion” OR “is” predicted by “TargetP” to be in “mitochondrion” OR “is” predicted by “WoLF PSORT” to be in “mitochondrion”)]. Very recently, a semi-quantitative proteomic approach has detected Deg3 in low abundance in the stroma-lamellae of the thylakoid (Tomizioli et al., 2014) supporting a plastid location for this Deg protease without excluding a mitochondrial location. In addition, Deg13 was localized to the plastid in our study (Figure 3). Given these results we should bear in mind that the full-length coding sequence could only be amplified from cDNAs of Deg10 and Deg14. For Deg3, Deg6, Deg12, and Deg13 the first 300 bp or less were amplified from genomic DNA and fused to GFP. With proteomics data existing for Deg3 and Deg12 (Baerenfaller et al., 2008; Tomizioli et al., 2014), but transcript and protein data being absence from publicly accessible databases for Deg6 and Deg13, could indicate that these two Deg proteases are pseudogenes (Schuhmann and Adamska, 2012).
Peroxisome Deg protease
The location of Deg15 in peroxisomes was previously demonstrated by both subcellular proteomic and GFP tagging studies (Eubel et al., 2008; Schuhmann et al., 2008). As shown in Figure 3, the peroxisomal location of Deg15 was confirmed again by this study. In vivo and in vitro analysis has shown that Deg15 is responsible for processing PTS2-containing proteins and plants lacking Deg15 display a phenotype potentially linked to reduced fatty acid β-oxidation due to lack of enzyme processing (Schuhmann et al., 2008).
Nuclear Deg protease
The SUBA3 query reported that Deg9 has been located by different proteomic studies to the plastid (Kleffmann et al., 2004) and to the nucleus (Pendle et al., 2005). The nuclear location of Deg9 could be confirmed here by GFP tagging (Figure 3). However, the plastid location could not be confirmed in this study. From the analysis of conflicts between published localization datasets (see Conflicts Between Published Localization Datasets) a nuclear location for Deg9 is also suggested to be more likely than a plastid location. As the physiological role of Deg9 remains to be elucidated, the possible plastid location of Deg9 will need to be proven by further independent analysis.
Deg7 protease
Deg7, the last of the proteases for which proteomic data are available in SUBA3, was reported to be located in the cytosol (Ito et al., 2011). Another study has found this protease in the chloroplast stroma by GFP tagging and immunoblotting (Sun et al., 2010). Neither of these two locations could be verified by GFP tagging in this study, instead Deg7 was localized to the nucleus and the mitochondrion (Figure 3). A nuclear location is plausible because Deg7 is putatively orthologous to the only fungal Deg protease, which is located in the nucleus (Schuhmann et al., 2011). The nuclear location of Deg7 could have been missed by Sun et al. (2010) as the authors fused only the N-terminal 243 amino acids of Deg7 to GFP, whereas in this study the full-length coding sequence of Deg7 was fused to the N-terminus of GFP. Interestingly, NLS Mapper (Kosugi et al., 2009) predicts a bipartite nuclear localization signal from 845 to 873 amino acids. The analysis of deg7 mutant plants revealed that the mutant is more sensitive to high light stress than wild type as demonstrated by an inhibited growth phenotype of deg7 mutants when exposed to high light. Additionally, it was shown that Deg7 interacts directly with PSII (Sun et al., 2010). Thus, a plastid location of this protease is likely and may have been missed in this study for a reason associated with the GFP fusion or the intensity of fluorescence in the plastid. Ito et al. (2011) localized Deg7 to the cytosol using a proteomic approach. Although purification techniques have improved immensely and the authors rigorously selected for cytosolic proteins, it is very difficult to avoid contamination of the cytosol by soluble proteins from organelles. Thus, the cytosolic location of Deg7 could be due to contamination during the cytosolic sample preparation. In fact, other soluble chloroplast proteins have been reported as cytosolic in this study, such as a chloroplast form of ATP sulfurylase (AT1G19920), a plastidic triose phosphate isomerase (AT2G21170), a chloroplast beta-amylase (AT3G23920), a plastidic uracil phosphoribosyltransferase (AT3G53900), and an f-type thioredoxin localized in the chloroplast stroma (AT5G16400). The localization of Deg7 represents an example of how contradictory location data can arise even when the same localization method was used.
Unknown location Deg proteases
Using GFP tagging we were unable to define a subcellular location for Deg4 and Deg16 (Table 2). Both proteases have been suggested to be potential pseudogenes, because no transcript or protein data could be found in publicly available databases (Schuhmann and Adamska, 2012). Indeed, we were only able to amplify the first 300 bp from genomic DNA of Deg4 and Arabidopsis suspension cells transformed with the Deg4-GFP fusion construct showed a GFP signal but the subcellular compartment could not be determined (Supplemental Figure 1).
Discussion
SUBA3 integrates protein localization information for Arabidopsis proteins from various sources, including data from bioinformatics prediction programs, from annotators and from experimental data sources. SUBAcon was developed to estimate a protein’s consensus location based on Bayesian probabilities calculated from all the experimental data and predictions available for each protein. By collating localizations from 22 different predictors and including experimental data as location evidence, SUBA3 overcomes the limitations of each of the individual predictors. In addition, SUBA3 allows the building of complex queries to investigate many different aspects of protein location and can be used effectively to select candidate lists of proteins for further experimental analysis as exemplified with the example of the Deg proteases presented here.
Searching SUBA3, no previous experimental evidence existed for eight of the 16 Deg proteases. This study systematically analyzed the subcellular localization of the Deg protease family and provided the first experimental evidence for six Deg proteases. Furthermore, it resulted in substantial experimental evidence for mitochondrial localized Deg proteases. Indeed, six Deg proteases were targeted to the mitochondrion by our GFP tagging approach, namely Deg3, Deg6, Deg10, Deg11, Deg12, and Deg14. Two of these, Deg3 and Deg10, additionally localized to the plastid, exposing novel dual-targeted Deg proteases in the plastid and the mitochondrion. In addition, Deg13 was localized to the plastid and the subcellular locations of Deg1 and Deg9 were further refined. Previously localized to both the plastid and nucleus, a sole plastid location was verified for Deg1, whereas a sole nuclear location was confirmed for Deg9. This study also confirmed previous results obtained for Deg2, Deg5, and Deg8 to be located in the plastid and a peroxisome location for Deg15, whereas contradictory results were obtained for Deg7.
The number of proteins for which localization information is available has increased dramatically over the last few years as well as the number of organelles for which at least part of the proteome is known. For example, the plastid proteome shows a 60% experimental coverage of the estimated plastid proteome size in Arabidopsis and seven Deg proteases were already experimentally located in plastids prior to this study (see Defining the Size of Experimental vs Predicted Datasets for a Specific Subcellular Location). However, this still leaves many more proteins to be discovered in this subcellular location as shown by the targeted localizations of Deg3, Deg10, and Deg13 to the plastid. Similarly, the experimental coverage of other subcellular locations is still limited (as indicated in Table 1) and there is room for many proteins to be experimentally identified in these locations even using available methodologies.
With the increase of localization information, the amount of contradictory information has also increased. For example, Deg9 has been localized by proteomic studies to the plastid and the nucleus by two independent research groups. Further analysis in SUBA3 suggests a nuclear location to be more likely than a plastid location (see Conflicts Between Published Localization Datasets). Accordingly, the nuclear location could be verified in this study, whereas the plastid location of Deg9 could not be confirmed.
Not only do different experimental approaches result in such discrepancies, even using the same approach by different researchers gives rise to different results as shown by the GFP localization of the Deg7 protease. Some of these discrepancies are yet to be recognized dual-targeted proteins. Thus, the identification of more multi-localized proteins (such as Deg3 and Deg10) is required to resolve these discrepancies. In addition, we need to find better ways of describing the net or final location of some proteins that may move around in the cell and are present for a time at various locations as in the case of proteins that enter the secretory system. However, there are real discrepancies and these will not be resolved by adding more and more of the same type of information. By adding useful additional experimental datasets, such as PPI data, and using predictions available for each protein, SUBAcon estimates a protein’s consensus location and also assists in finding missing organellar proteins. There is also published data from quantitative analysis of MS and GFP data that could be used as evidence for proteins not being in certain locations and this could be used to counteract more qualitative data from earlier published reports.
As seen from the example of the Deg protease family presented here, using SUBA to determine the subcellular location of members of a protein family can assist in defining the function of a particular protein family member. This gives insight into processes such as sub-functionalization, where gene families have expanded and then divided their functions between protein family members located in different parts of the cell. For example, bacterial genomes usually encode three Deg proteases, yeasts own one (occasionally duplicated), and four to five genes coding for Deg proteases are present in mammalian genomes (Kim and Kim, 2005; Rawlings et al., 2008; Huesgen et al., 2011). In plants, however, this family has expanded and plant genomes contain many more genes encoding Deg proteases. Arabidopsis thaliana possesses 16 Deg protease genes (Huesgen et al., 2005), Oryza sativa 15 (Tripathi and Sowdhamini, 2006), and Populus trichocarpa 20 (Garcia-Lorenzo et al., 2006). This relatively high number of Deg proteases in plants is mainly due to gene duplications (Schuhmann et al., 2012) and some additional Deg genes may result from a gene transfer from the plastid, which is of prokaryotic origin, to the nucleus. Combining subcellular location data in the future with sequence similarity information, evidence of more recent duplication events in genomes and synteny across related species will be needed to provide this insight systematically across gene families.
Author Contributions
Sandra K. Tanz collected the subcellular localization data from the literature, carried out the localization studies by GFP tagging, analyzed the data and drafted the manuscript. Ian Castleden developed the SUBA3 database and helped analyzing the data. Cornelia M. Hooper participated in collecting the subcellular localization data from the literature. Ian Small participated in the design of the study. A. Harvey Millar participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank James Whelan for kindly providing the Gateway® cloning cassette (pDest/pgem/CGFP) and the small subunit of Arabidopsis ribulose-1,5-bisphosphate carboxylase oxygenase fused to RFP (SSU-RFP) construct that were used in the GFP tagging studies. This work was supported by the Australian Research Council [CE140100008 to A. Harvey Millar and Ian Small, FT110100242 to A. Harvey Millar, DE120100307 to Sandra K. Tanz]; and the Government of Western Australia through funding for the WA Centre of Excellence for Computational Systems Biology [DIR WA CoE].
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00396/abstract
Footnotes
References
Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. doi: 10.1126/science.1203877
Baerenfaller, K., Grossmann, J., Grobei, M. A., Hull, R., Hirsch-Hoffmann, M., Yalovsky, S.,et al. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941. doi: 10.1126/science.1157956
Bardy, N., Carrasco, A., Galaud, J. P., Pont-Lezica, R., and Canut, H. (1998). Free-flow electrophoresis for fractionation of Arabidopsis thaliana membranes. Electrophoresis 19, 1145–1153. doi: 10.1002/elps.1150190715
Beck, G., Coman, D., Herren, E., Ruiz-Sola, M. A., Rodriguez-Concepcion, M., Gruissem, W.,et al. (2013). Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 82, 393–416. doi: 10.1007/s11103-013-0070-z
Boruc, J., Mylle, E., Duda, M., De Clercq, R., Rombauts, S., Geelen, D.,et al. (2010). Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 152, 553–565. doi: 10.1104/pp.109.148643
Briesemeister, S., Rahnenfuhrer, J., and Kohlbacher, O. (2010). YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38, W497–W502. doi: 10.1093/nar/gkq477
Bussell, J. D., Behrens, C., Ecke, W., and Eubel, H. (2013). Arabidopsis peroxisome proteomics. Front. Plant Sci. 4:101. doi: 10.3389/fpls.2013.00101
Carrie, C., Kuhn, K., Murcha, M. W., Duncan, O., Small, I. D., O’Toole, N.,et al. (2009). Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 57, 1128–1139. doi: 10.1111/j.1365-313X.2008.03745.x
Carrie, C., Murcha, M. W., Kuehn, K., Duncan, O., Barthet, M., Smith, P. M.,et al. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079. doi: 10.1016/j.febslet.2008.07.061
Carrie, C., and Small, I. (2013). A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta 1833, 253–259. doi: 10.1016/j.bbamcr.2012.05.029
Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. doi: 10.1016/S0960-9822(02)00483-9
Cui, J., Liu, J., Li, Y., and Shi, T. (2011). Integrative identification of Arabidopsis mitochondrial proteome and its function exploitation through protein interaction network. PLoS ONE 6:e16022. doi: 10.1371/journal.pone.0016022
de Oliveira Dal’Molin, C. G., Quek, L. E., Palfreyman, R. W., Brumbley, S. M., and Nielsen, L. K. (2010). AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 152, 579–589. doi: 10.1104/pp.109.148817
Duchene, A. M., Peeters, N., Dietrich, A., Cosset, A., Small, I. D., and Wintz, H. (2001). Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J. Biol. Chem. 276, 15275–15283. doi: 10.1074/jbc.M011525200
Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Eubel, H., Lee, C. P., Kuo, J., Meyer, E. H., Taylor, N. L., and Millar, A. H. (2007). Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J. 52, 583–594. doi: 10.1111/j.1365-313X.2007.03253.x
Eubel, H., Meyer, E. H., Taylor, N. L., Bussell, J. D., O’Toole, N., Heazlewood, J. L.,et al. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148, 1809–1829. doi: 10.1104/pp.108.129999
Ferro, M., Brugiere, S., Salvi, D., Seigneurin-Berny, D., Court, M., Moyet, L.,et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084. doi: 10.1074/mcp.M900325-MCP200
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q.,et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Garcia-Lorenzo, M., Sjodin, A., Jansson, S., and Funk, C. (2006). Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 6:30. doi: 10.1186/1471-2229-6-30
Guda, C. (2010). “Towards cataloguing the subcellular proteomes of eukaryotic organisms,” in Sequence and Genome Analysis: Methods and Applications, ed. Z. Zhao (Hong Kong: iConcept Press Ltd), 259–269.
Heazlewood, J. L., Tonti-Filippini, J., Verboom, R. E., and Millar, A. H. (2005). Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 139, 598–609. doi: 10.1104/pp.105.065532
Heazlewood, J. L., Tonti-Filippini, J. S., Gout, A. M., Day, D. A., Whelan, J., and Millar, A. H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256. doi: 10.1105/tpc.016055
Heazlewood, J. L., Verboom, R. E., Tonti-Filippini, J., Small, I., and Millar, A. H. (2007). SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 35, D213–D218. doi: 10.1093/nar/gkl863
Hoopmann, M. R., Merrihew, G. E., Von Haller, P. D., and Maccoss, M. J. (2009). Post analysis data acquisition for the iterative MS/MS sampling of proteomics mixtures. J. Proteome Res. 8, 1870–1875. doi: 10.1021/pr800828p
Huesgen, P., Schuhmann, H., and Adamska, I. (2005). The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 123, 413–420. doi: 10.1111/j.1399-3054.2005.00458.x
Huesgen, P. F., Miranda, H., Lam, X., Perthold, M., Schuhmann, H., Adamska, I.,et al. (2011). Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics and mechanism. Biochem. J. 435, 733–742. doi: 10.1042/BJ20102131
Inze, A., Vanderauwera, S., Hoeberichts, F. A., Vandorpe, M., Van Gaever, T., and Van Breusegem, F. (2012). A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 35, 308–320. doi: 10.1111/j.1365-3040.2011.02323.x
Ito, J., Batth, T. S., Petzold, C. J., Redding-Johanson, A. M., Mukhopadhyay, A., Verboom, R.,et al. (2011). Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 10, 1571–1582. doi: 10.1021/pr1009433
Kiessling, J., Martin, A., Gremillon, L., Rensing, S. A., Nick, P., Sarnighausen, E.,et al. (2004). Dual targeting of plastid division protein FtsZ to chloroplasts and the cytoplasm. EMBO Rep. 5, 889–894. doi: 10.1038/sj.embor.7400238
Kim, D. Y., and Kim, K. K. (2005). Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38, 266–274. doi: 10.5483/BMBRep.2005.38.3.266
Kleffmann, T., Russenberger, D., Von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W.,et al. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362. doi: 10.1016/j.cub.2004.02.039
Koroleva, O. A., Tomlinson, M. L., Leader, D., Shaw, P., and Doonan, J. H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41, 162–174. doi: 10.1111/j.1365-313X.2004.02281.x
Kosugi, S., Hasebe, M., Tomita, M., and Yanagawa, H. (2009). Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176. doi: 10.1073/pnas.0900604106
Krause, K., and Krupinska, K. (2009). Nuclear regulators with a second home in organelles. Trends Plant Sci. 14, 194–199. doi: 10.1016/j.tplants.2009.01.005
Lee, J., Lee, H., Kim, J., Lee, S., Kim, D. H., Kim, S.,et al. (2011). Both the hydrophobicity and a positively charged region flanking the C-terminal region of the transmembrane domain of signal-anchored proteins play critical roles in determining their targeting specificity to the endoplasmic reticulum or endosymbiotic organelles in Arabidopsis cells. Plant Cell 23, 1588–1607. doi: 10.1105/tpc.110.082230
Levitan, A., Trebitsh, T., Kiss, V., Pereg, Y., Dangoor, I., and Danon, A. (2005). Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 102, 6225–6230. doi: 10.1073/pnas.0500676102
Li, S., Ehrhardt, D. W., and Rhee, S. Y. (2006). Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 141, 527–539. doi: 10.1104/pp.106.078881
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T.,et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251. doi: 10.1073/pnas.182432999
Massingham, T., Davies, L. J., and Lio, P. (2001). Analysing gene function after duplication. Bioessays 23, 873–876. doi: 10.1002/bies.1128
Millar, A. H., Carrie, C., Pogson, B., and Whelan, J. (2009). Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21, 1625–1631. doi: 10.1105/tpc.109.066019
Millar, A. H., Whelan, J., and Small, I. (2006). Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 9, 610–615. doi: 10.1016/j.pbi.2006.09.002
Mintz-Oron, S., Meir, S., Malitsky, S., Ruppin, E., Aharoni, A., and Shlomi, T. (2012). Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 339–344. doi: 10.1073/pnas.1100358109
Narsai, R., Law, S. R., Carrie, C., Xu, L., and Whelan, J. (2011). In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 157, 1342–1362. doi: 10.1104/pp.111.183129
Nelson, B. K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Olinares, P. D., Ponnala, L., and Van Wijk, K. J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9, 1594–1615. doi: 10.1074/mcp.M000038-MCP201
Parsons, H. T., Christiansen, K., Knierim, B., Carroll, A., Ito, J., Batth, T. S.,et al. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159, 12–26. doi: 10.1104/pp.111.193151
Pendle, A. F., Clark, G. P., Boon, R., Lewandowska, D., Lam, Y. W., Andersen, J.,et al. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16, 260–269. doi: 10.1091/mbc.E04-09-0791
Rautengarten, C., Ebert, B., Herter, T., Petzold, C. J., Ishii, T., Mukhopadhyay, A.,et al. (2011). The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23, 1373–1390. doi: 10.1105/tpc.111.083931
Rawlings, N. D., Morton, F. R., Kok, C. Y., Kong, J., and Barrett, A. J. (2008). MEROPS: the peptidase database. Nucleic Acids Res. 36, D320-325. doi: 10.1093/nar/gkm954
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E.,et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. doi: 10.1105/tpc.107.050989
Reumann, S., Quan, S., Aung, K., Yang, P., Manandhar-Shrestha, K., Holbrook, D.,et al. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150, 125–143. doi: 10.1104/pp.109.137703
Rutter, M. T., Cross, K. V., and Van Woert, P. A. (2012). Birth, death and subfunctionalization in the Arabidopsis genome. Trends Plant Sci. 17, 204–212. doi: 10.1016/j.tplants.2012.01.006
Ryngajllo, M., Childs, L., Lohse, M., Giorgi, F. M., Lude, A., Selbig, J.,et al. (2011). SLocX: predicting subcellular localization of Arabidopsis proteins leveraging gene expression data. Front. Plant Sci. 2:43. doi: 10.3389/fpls.2011.00043
Sapir-Mir, M., Mett, A., Belausov, E., Tal-Meshulam, S., Frydman, A., Gidoni, D.,et al. (2008). Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 148, 1219–1228. doi: 10.1104/pp.108.127951
Schuhmann, H., and Adamska, I. (2012). Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol. Plant 145, 224–234. doi: 10.1111/j.1399-3054.2011.01533.x
Schuhmann, H., Huesgen, P. F., and Adamska, I. (2012). The family of Deg/HtrA proteases in plants. BMC Plant Biol. 12:52. doi: 10.1186/1471-2229-12-52
Schuhmann, H., Huesgen, P. F., Gietl, C., and Adamska, I. (2008). The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 148, 1847–1856. doi: 10.1104/pp.108.125377
Schuhmann, H., Mogg, U., and Adamska, I. (2011). A new principle of oligomerization of plant DEG7 protease based on interactions of degenerated protease domains. Biochem. J. 435, 167–174. doi: 10.1042/BJ20101613
Schwacke, R., Fischer, K., Ketelsen, B., Krupinska, K., and Krause, K. (2007). Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics 277, 631–646. doi: 10.1007/s00438-007-0214-4
Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. doi: 10.1002/pmic.200300776
Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. doi: 10.1023/A:1006081903354
Sun, X., Fu, T., Chen, N., Guo, J., Ma, J., Zou, M.,et al. (2010). The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 152, 1263–1273. doi: 10.1104/pp.109.150722
Tanz, S. K., Castleden, I., Hooper, C. M., Vacher, M., Small, I., and Millar, H. A. (2013). SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41, D1185–D1191. doi: 10.1093/nar/gks1151
Thatcher, L. F., Carrie, C., Andersson, C. R., Sivasithamparam, K., Whelan, J., and Singh, K. B. (2007). Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. J. Biol. Chem. 282, 28915–28928. doi: 10.1074/jbc.M702207200
Tian, G. W., Mohanty, A., Chary, S. N., Li, S., Paap, B., Drakakaki, G.,et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135, 25–38. doi: 10.1104/pp.104.040139
Tomizioli, M., Lazar, C., Brugiere, S., Burger, T., Salvi, D., Gatto, L.,et al. (2014). Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol. Cell. Proteomics doi: 10.1074/mcp.M114.040923 [Epub ahead of print].
Tripathi, L. P., and Sowdhamini, R. (2006). Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7:200. doi: 10.1186/1471-2164-7-200
Van Aken, O., Zhang, B., Carrie, C., Uggalla, V., Paynter, E., Giraud, E.,et al. (2009). Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2, 1310–1324. doi: 10.1093/mp/ssp053
Keywords: subcellular localization, database, Arabidopsis, protein, Deg protease
Citation: Tanz SK, Castleden I, Hooper CM, Small I and Millar AH (2014) Using the SUBcellular database for Arabidopsis proteins to localize the Deg protease family. Front. Plant Sci. 5:396. doi: 10.3389/fpls.2014.00396
Received: 26 May 2014; Accepted: 24 July 2014;
Published online: 12 August 2014.
Edited by:
Katja Baerenfaller, Swiss Federal Institute of Technology Zurich, SwitzerlandReviewed by:
Harriet T. Parsons, University of Copenhagen, DenmarkBerit Ebert, Lawrence Berkeley Laboratory Joint BioEnergy Institute, USA
Myriam FERRO, Commissariat à l’Energie Atomique et aux Energies Alternatives, France
Copyright © 2014 Tanz, Castleden, Hooper, Small and Millar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra K. Tanz, The Australian Research Council Centre of Excellence in Plant Energy Biology, The University of Western Australia, M316, 35 Stirling Highway, Crawley, Perth, WA 6009, Australia e-mail:c2FuZHJhLnRhbnpAdXdhLmVkdS5hdQ==
 Sandra K. Tanz
Sandra K. Tanz Ian Castleden2
Ian Castleden2 Ian Small
Ian Small A. Harvey Millar
A. Harvey Millar