- 1Wageningen UR Plant Breeding, Wageningen University and Research Centre, Wageningen, Netherlands
- 2Biosystematics Group, Wageningen University, Wageningen, Netherlands
Next Generation Sequencing (NGS) may enable estimating relationships among genotypes using allelic variation of multiple nuclear genes simultaneously. We explored the potential and caveats of this strategy in four genetically distant Lilium cultivars to estimate their genetic divergence from transcriptome sequences using three approaches: POFAD (Phylogeny of Organisms from Allelic Data, uses allelic information of sequence data), RAxML (Randomized Accelerated Maximum Likelihood, tree building based on concatenated consensus sequences) and Consensus Network (constructing a network summarizing among gene tree conflicts). Twenty six gene contigs were chosen based on the presence of orthologous sequences in all cultivars, seven of which also had an orthologous sequence in Tulipa, used as out-group. The three approaches generated the same topology. Although the resolution offered by these approaches is high, in this case there was no extra benefit in using allelic information. We conclude that these 26 genes can be widely applied to construct a species tree for the genus Lilium.
Introduction
The preponderance of data used in plant molecular phylogenetics over the last decade comes from chloroplast DNA and nuclear rDNA (notably rDNA ITS) (APG, 2003, 2009; Chase and Reveal, 2009). Chloroplast DNA has the advantage of straightforward genetics: haploid, non-recombinant and highly conserved with respect to gene content and arrangement, notably among closely related species (Olmstead and Palmer, 1992). However, cpDNA reveals only half of the phylogenetic origin of a plant-lineage since it is uni-parentally inherited and its substitution rates are generally low compared with bi-parentally inherited nuclear DNA (Small et al., 2004). As a special case rDNA has been used extensively in Angiosperm (and fungal) phylogenetic reconstruction, especially using the Internal Transcribed Spacer regions (White et al., 1990; Baldwin, 1992). However, when not all rDNA copies are fully homogenized as was observed for instance in tulip and peonies (Sang et al., 1995; Booy et al., 2000; Lim et al., 2001; Alvarez and Wendel, 2003), the risk of using paralogs in phylogenetic reconstruction becomes large (Kim and Jansen, 1994; Sang et al., 1995; Alvarez and Wendel, 2003) and hence rDNA has been disregarded as phylogenetic marker in species-level Angiosperm phylogenetics [8]. Multi-locus, low copy nuclear DNA sequences have been used in plant phylogenetic studies since the late nineties (De La Torre et al., 2006; Hughes et al., 2006; Sanderson and McMahon, 2007; Griffin et al., 2011) and, because of their bi-parental inheritance and wealth of long and independently-inherited genes (Small et al., 2004), became the focus of plant phylogenetic reconstruction in general. Also, the ability to identify heterozygosity within individuals and hybrids (allelic variation) is considered a distinct advantage of using nuclear DNA over that from organelles. Using two alleles instead of one can give, in principle, better estimations of phylogenetic relationships between closely related taxa (Joly and Bruneau, 2006; Liu et al., 2008), or in case of species hybrids, enable establishing correct gene trees, in which both alleles are placed within the germplasm that they are derived from Zhang et al. (2013).
The availability of Next Generation Sequencing (NGS) data in plants opens the door to phylogenetic studies using a wide set of loci, representing truly genome-wide coverage. Commonly-used techniques for estimating phylogenetic trees from multiple-loci data are: concatenation or “super matrix” methods (Nylander et al., 2004), super tree construction (Beninda-Emonds, 2004) and gene tree parsimony (Page, 1998). On the other hand, Bayesian Estimation of Species Trees (Liu et al., 2008) and Bayesian Evolutionary Analysis by Sampling Trees (Drummond and Rambaut, 2007; Heled and Drummond, 2010) estimate species trees from separate gene trees and deal with the multi-allelic nature of genes by enabling incorporation of several genes separately in estimating effective population size and tree topology. This is implemented by using a Monte Carlo Markov Chain to find a posterior distribution of species trees. In this way concatenation is no longer necessary and differences in mutation rate between genes can be included in the analyses, or accommodated using appropriate priors. However, SNPs between the alleles are treated as ambiguous (IUPAC) bases in consensus sequences in this approach, obviously discarding part of the available data. Use of NGS data for phylogenetic reconstruction requires choices between trade-offs, in particular so when dealing with data derived from cultivated plants, which often have a complex genetic background that may or may not be well-documented.
Here we explore NGS data originally generated for genetic resource and SNP marker retrieval in Lilium (Shahin et al., 2012) in a phylogenetic context. Lilium L. was ranked among the top seven of the most popular flower bulb genera (Benschop et al., 2010). Lilium is classified into seven sections based on 13 morphological and two germination characteristics (Comber, 1949), and into four hybrid groups: Asiatic (A, Sinomartagon section), Oriental (O, Archelirion section), Longiflorum (L, Leucolirion subsection b), and Trumpet hybrid groups (T, Leucolirion subsection a). Phylogenetic relationships within Lilium were reconstructed using molecular markers (Dubouzet and Shinoda, 1999; Nishikawa et al., 1999, 2001; Arzate-Fernandez et al., 2005; Muratović et al., 2010). Most of the species clustered into clades correlating with their morphological classification of Comber (1949), but a few behaved differently. Species of section Leucolirion (subsection a and b) that were supposed to cluster closely according to Comber (Comber, 1949), grouped separately. Species of Leucolirion (subsection b) were closer to section Sinomartagon, and species of Leucolirion (subsection a) were closer to section Archelirion in both studies (Nishikawa et al., 1999; Arzate-Fernandez et al., 2005). Lily breeding dates back about 200 years (Shimizu, 1987), significant breakthroughs are only 50 years old however, starting with the breeding of Asiatic hybrids (McRae, 1998). It has only been since the 1970's that the lily has become, after tulip, the most important flower bulb and cut flower (Lim and Van Tuyl, 2006).
The aim of this study was to use transcriptome data for estimating both genetic divergence and relationships among four Lilium cultivars, and for comparing, for those orthologous sequences available, the data to a set of cultivars in Tulipa, the closest related cultivar group with transcriptome sequence data available (Shahin et al., 2012). We use three approaches that differ with respect to optimality criteria and type of data used and compare their results: (i) separate allelic data using distance analysis, as implemented in POFAD (Joly and Bruneau, 2006), (ii) concatenated analysis of consensed sequences, i.e., between the alleles, using maximum likelihood (RAxML, Stamatakis), and (iii) Consensus Networks (Holland et al., 2005) of separate parsimony gene trees derived from consensed sequences. Whereas RAxML is a tree building method, both the POFAD and Consensus Networks approach construct and visualize comparative data in networks. Consensus Networks are reconstructed by converting trees into splits to summarize possible among-tree conflict in a reticulate structure, where edge lengths are proportional to the occurrence of splits. In contrast, the POFAD algorithm calculates a pairwise distance matrix of all (separate) haplotypes, followed by conversion of this matrix into an organism-level distance matrix by taking the average of distances between the haploids. This matrix is then visualized in a Neighbor Network (Bryant and Moulton, 2004) allowing “non-treelike” patterns in the data. By using this algorithm we can combine the distance matrices of different loci without the need to concatenate the loci or to construct a (artificial) consensus allele per locus.
Materials and Methods
Plant Material
Transcriptome sequence data of four Lilium and five Tulipa cultivars (all diploid) used for this study were from Shahin et al. (2012). The four Lilium cultivars, representing the four main hybrid groups of the genus Lilium, are: “Star Gazer” (Oriental, Archelirion section), breeding line “Trumpet 061099” (Trumpet, Leucolirion subsection a), “White Fox” (Longiflorum, Leucolirion subsection b), and “Connecticut King” (Asiatic, Sinomartagon section) (Figure 1). The five Tulipa cultivars are: “Cantata” and “Princeps,” which belong to T. fosteriana (Eichleres section), and “Bellona,” “Kees Nelis” and “Ile de France,” which belong to T. gesneriana (Tulipa section).
Methodology
For RNA isolation, library processing and 454 sequencing protocols used see Shahin et al. (2012). The sequence data of the four Lilium cultivars were assembled using the CLC assembler (Shahin et al., 2012). As a result of the assembly step, an Ace file was generated that contains contigs (i.e., the consensus of all assembled ESTs that belong to one locus) which were used as starting point in this analysis. Contigs with high coverage (>100 reads per contig and at least 4 reads for each individual cultivar) were picked for further analysis. All the individual haplotype consensus sequences (e.g., Trumpet_A, Trumpet_B) for each gene were aligned in SeqMan and trimmed to the same size for all cultivars. If a contig showed more than two haplotypes/alleles per cultivar which indicates either assembled paralogs or sequencing errors, such contig was discarded. BlastX was used for annotation of contig consensus sequences. The number of polymorphic sites for each contig (27 contigs) were calculated using TOPALi v. 2 (Milne et al., 2009).
The 27 Lilium contigs were blasted to the Tulip-ALL assembly (BLASTn, 1E-20) (Shahin et al., 2012), in order to select tulip as out-group for subsequent tree building (see below). Seven of the 27 contigs showed to have orthologous sequence in the five Tulipa cultivars that have the same criteria (high coverage >100 reads per contig and at least 4 reads for each individual cultivar, and only 2 haplotypes per cultivar). These seven orthologous genes were analyzed using the same steps explained above. The number of polymorphic sites for each contig were calculated using TOPALi v. 2 (Milne et al., 2009).
Recombination Test
In order to use these gene contigs for phylogenetic or distance tree construction, recombination tests should be applied to avoid using any sequence that is putatively recombined, (e.g., Vriesendorp and Bakker, 2005). This was done on the different haplotypes within the cultivars using PDM (Probabilistic Divergence Measure) and DSS (Difference of Sum of Squares), both implemented in TOPALi v. 2 (Milne et al., 2009). The test operates by sliding a fixed-size window (e.g., 500 bp wide) along the alignment, comparing the left-hand part with the right-hand part in terms of phylogenetic topologies based on either part. In PDM the marginal posterior distributions of topologies are compared, whereas SSD fits pairwise genetic distances of each part to a phylogenetic tree based on the other part. Upon moving into a recombinant site, marginal distributions or SSD resp. should change. We used the default options of the program except for the nucleotide substitution model, where we replaced the (default) Jukes-Cantor model by Felsenstein84. Parametric bootstrapping was applied to estimate the significance of the predictions (100 reps). Observed values of DSS and PDM methods beyond the 95% point of this distribution may well correspond to a recombination event. Contigs with a putative recombination site were discarded for further study.
Tree Building and Network Analysis
POFAD
The edited and trimmed haplotypes of every locus were imported in MEGA 5 (Tamura et al., 2011) and an uncorrected genetic distance matrix (p-distance) was generated for each contig. Reweighting the individual matrices, which is essential to insure their equal contribution in the estimation of the genetic distance, was done by the algorithm implemented in POFAD (Joly and Bruneau, 2006). The genotypes' reweighted matrices for each gene contig individually was transferred to SplitsTree v.4 (Huson and Bryant, 2006) to construct Neighbor Networks. Similarly, the matrices of the 7 orthologous Lilium and Tulipa sequences were also transferred to SplitsTree v.4 for constructing Neighbor Networks.
RAxML
To compare the average distance-based POFAD output with that from a character-based tree-building analysis we first merged allelic/haplotype sequences for each individual cultivar by calculating their consensus (including IUPAC bases) using Bio Edit version 7 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and then aligning them with other cultivars and concatenate the alignments of all contigs using Mesquite version 2.75 (Maddison and Maddison, 2011). The resulting super-matrix, see Supplementary Materials, was then analyzed in RAxML (Stamatakis et al., 2008) at the XSEDE Teragrid of the CIPRES science Gateway (Miller et al., 2010), including 100 replicates of fast-bootstrapping using the GTR-CAT substitution model (Stamatakis, 2006). Similarly, and to determine optimal rooting of our lily cultivars relationships, a super-matrix was generated for the seven genes orthologous between Lilium and Tulipa and then analyzed in RAxML (Stamatakis et al., 2008) using the same parameters.
Consensus network
The alignments of all gene contigs that were built using Mesquite version 2.75, were used to construct separate gene trees by standard heuristic search in PAUP* (Swofford, 2003). All resulting parsimony trees, including multiple equally parsimonious trees from the same alignment, were pooled and input into SplitsTree v. 44 (Huson and Bryant, 2006), where they were decomposed into splits and assembled using the Consensus Network option. We applied various (split) conflict thresholds in order to assess among tree conflict.
Results
From NGS sequences generated from leaf transcriptomes of the lily cultivars (Shahin et al., 2012), 27 contigs, with the highest overall sequence depth, were selected for this study. The length of these contigs ranged between 372 and 1230 bp (Table 1), and the number of polymorphic sites (on average) varied from one in contig_22926 to 71 in contig_36700 (Table 1). There were very few BlastX hits to known genes (Table 1, only the highest hit is shown). The total length of Lilium sequence data used for this study was 18,275 bp, containing 623 polymorphic sites, i.e., an average of one substitution event every 29 bp.
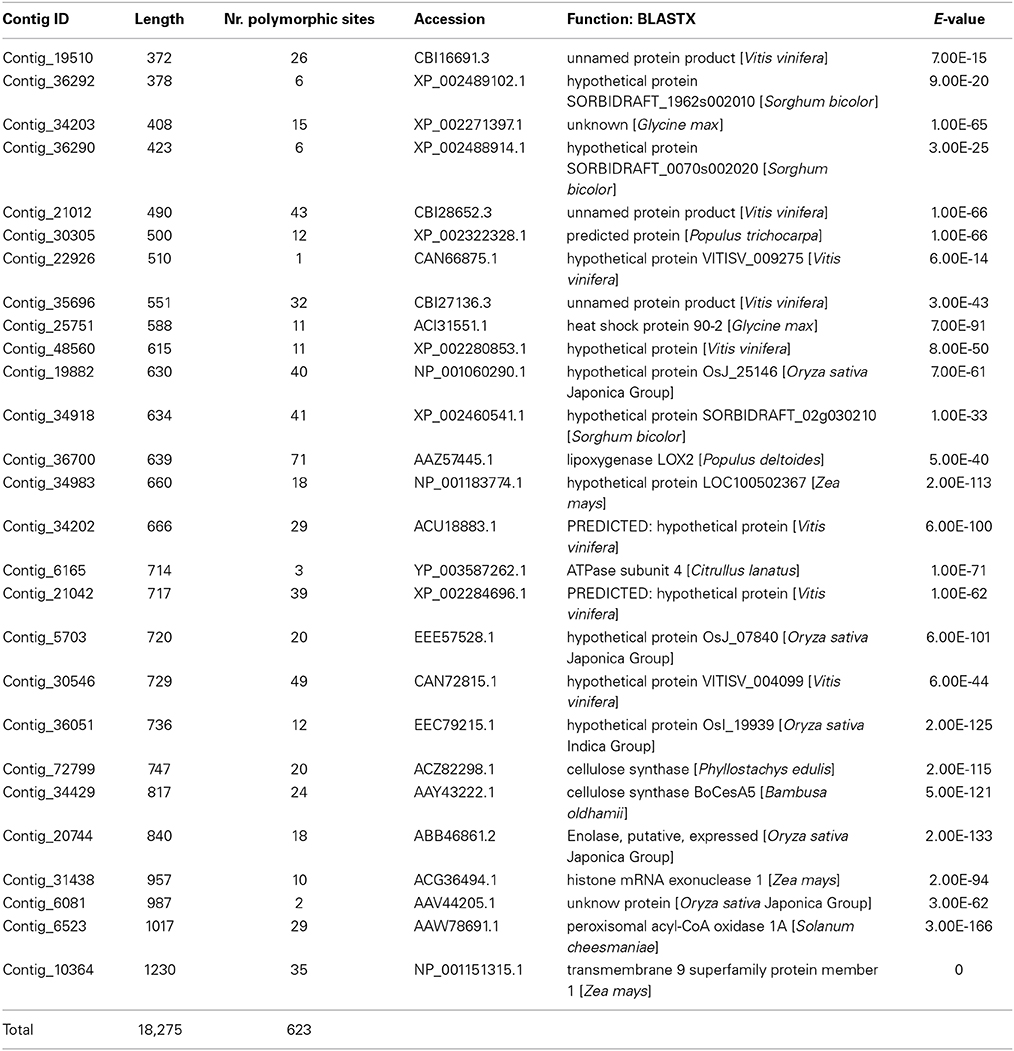
Table 1. Description of the 27 Lilium contigs used in this study: length, informative sites (calculated using TOPALi), the top hit result of blasting them to gene bank is presented.
Seven out of 27 contigs have orthologous sequences with five Tulipa cultivars, and were included in our study as a separate analysis (Table 2). The contig length ranged between 423 and 1230 bp. The number of polymorphic sites among the nine cultivars (on average) was very low in some contigs (8 sites in contig_6081), but much higher in others (200 sites in contig_10364) (Table 2). A total of 5790 bp with 587 polymorphic sites were available for this part of the study, of which 395 sites were polymorphic only between Lilium and Tulipa, 124 sites were also polymorphic within Lilium, and 68 were also polymorphic within Tulipa. This is equivalent to a substitution rate of 0.021 substitutions per site in Lilium, 0.012 in Tulipa, and 0.1 between Lilium and Tulipa.
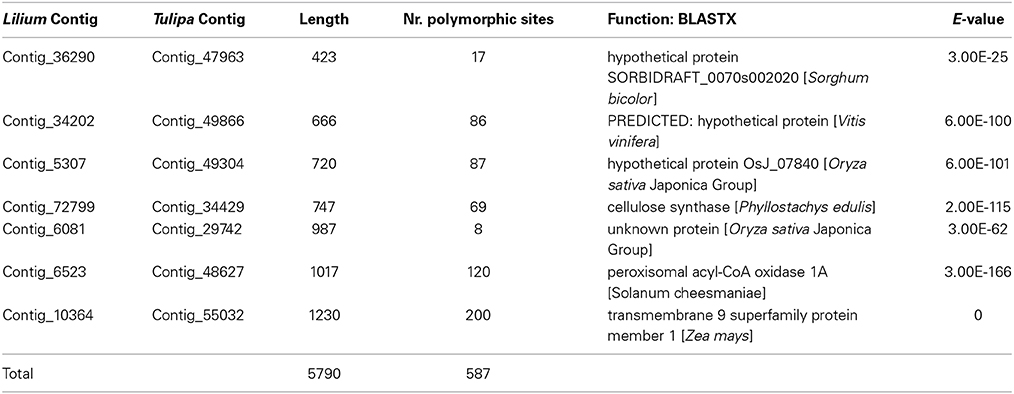
Table 2. As Table 1 but for the seven orthologous contigs between Lilium and Tulipa used in this study.
Recombination Test
In case a recombination event is detected in a contig this would indicate that more than one evolutionary history is present in this sequence. Therefore, any recombinant sequences, as detected by our TOPALi analysis, were discarded from further phylogenetic analysis. This turned out to be only one of the 27 Lilium contigs (contig_30546), which showed a possible recombination event between positions 157 and 220 bp.
Tree Building and Network Analysis
POFAD
Gene trees were constructed for each contig separately using POFAD. In 23 of the 26 gene trees, “Connecticut King” and “White Fox” grouped together, as well as “Star Gazer” and “Trumpet” (the exceptions being Contig_25751, contig_6165, and contig_34202). The same clustering resulted from constructing the Neighbor Network of the combined weighted genetic distance matrices of the 26 gene contigs (Figure 2A). As expected, introducing Tulipa as an out-group to the analysis did not introduce changes in the clustering among the Lilium cultivars (Figure 3A). The four cultivars are connected to multiple edges in the Network (Figure 3A), possibly indicating “non tree-like” behavior of the sequences involved. As for Tulipa, “Cantata” and “Princeps” that belong to T. fosteriana grouped together and “Ile de France,” “Kees Nelis” and “Bellona” that belong to T. gesneriana clustered together as well with multiple edges (Figure 3A).
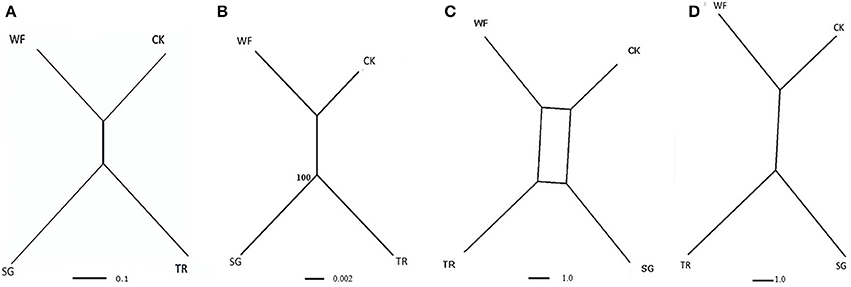
Figure 2. Representation of the relationships of the four Lilium cultivars: “Star Gazer” referred to as “SG,” “Trumpet 061099” referred to as “TR,” “White Fox” referred to as “WF,” and “Connecticut King” referred to as “CK” obtained from the combined analysis of all 26 non-recombinant contigs. (A) Neighbor Network based on 26 Lilium contigs, using the POFAD approach. (B) RAxML tree (with 100 rep. bootstrap support values) of the 26 Lilium concatenated consensus, (C) Consensus Network based on parsimony trees (see text) using a threshold of 0.33 split conflict, and (D) Consensus Network for the same trees using a threshold of 10%. Branch length is proportional to the genetic divergence among genotypes (A,B, with the scale bar indicating numbers of substitutions per site), and proportional to the occurrence of splits in the consensus network analysis (D,C).
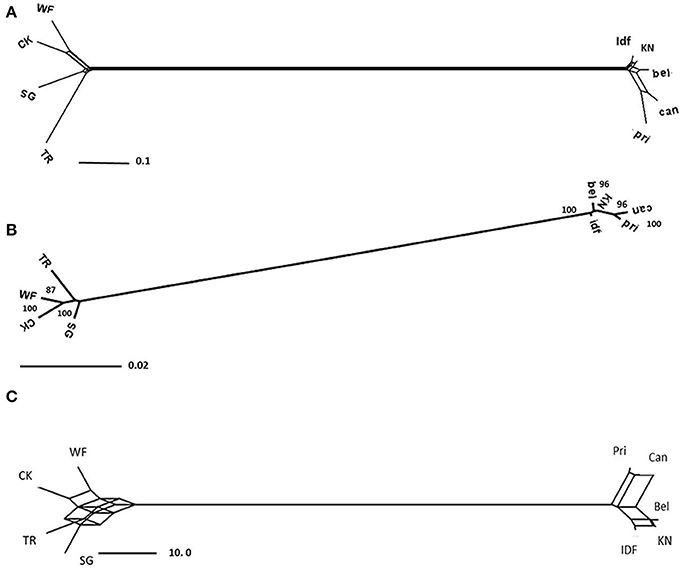
Figure 3. As Figure 2 but with five Tulipa cultivars included (see text).
RAxML
RAxML analysis of the consensus sequences (alleles had been merged into consensus sequences, see M&M) of the concatenated 26 contigs (best tree) resulted in grouping “Connecticut King” and “White Fox” together without bootstrap support, and in grouping “Star Gazer” and “Trumpet” together with bootstrap value 100 (Figure 2B), yielding the same tree topology as POFAD Network. RAxML tree of the seven concatenated gene alignments of Lilium and Tulipa showed also a comparable topology and branch lengths as found using POFAD for both Lilium and Tulipa (Figure 3B) but with relatively high bootstrap values (Figure 3B).
Consensus network
After parsimony analyses of the separate 26 alignments (excluding the potentially recombinant contig_30546), all resulting 68 equally parsimonious reconstructions were combined in a Consensus Network that resulted in the same topology as the POFAD and RAxML tree (Figure 2C). Using a (default) split-conflict threshold of 0.33, the Consensus Network was tree-shaped (Figure 2D), whereas lowering this threshold to 5% resulted in a box structure separating the 4 cultivars at equidistance. For the seven ortholog analysis, for both all 4 Lilium and 5 Tulipa cultivars, we obtained a Consensus Network (Figure 3C) that was congruent with the POFAD Median Network showing comparable resolution.
Discussion
Multi-locus genomic data obtained by NGS technology are rapidly becoming the main sources for inferring evolutionary relationships (Haussler et al., 2009; Emerson et al., 2010; Griffin et al., 2011) providing factors such as taxon sampling (Graybeal, 1998), polymorphic sites (Lopez et al., 2002) and hybridization (Naumov et al., 2000) have been properly accommodated. Nuclear DNA is more dynamic and evolves faster than plastid DNA, thus it provides a rich source of polymorphisms compared with plastid DNA; however, depending on what taxonomic level is targeted, we expect more nuclear genes to be required for constructing phylogenetic relationships due to the bi-parentally inherited and recombinant nature of nuclear DNA. In yeasts, a minimum of 8–20 genes were found to be sufficient to generate a stable species tree topology with bootstrap values of at least 70–95% (Rokas et al., 2003) which is largely influenced by number of informative sites present in these genes (Rokas et al., 2003) as well as the species tree estimation method used (Edwards et al., 2007). In our study, using 7 or 26 contigs genes resulted in the same tree topology. We did not further explore possible among gene tree incongruences as our Consensus Network analysis yielded patterns congruent or identical to those obtained by POFAD and RAxML. However, the use of TOPALi could in principle be extended from detecting candidate recombinant sites among haplotypes to the concatenated contigs, in order to detect among gene tree incongruence, when relevant. This could then provide valuable additional insight into the relationships between the cultivars.
Genetic variation among the four Lilium cultivars (0.034 substitutions per site) was higher than that detected for Tulipa (0.021 substitutions per site, data not shown). The same result was obtained using the seven orthologous genes between Lilium and Tulipa, which are expected to be conserved genes (0.021 subst/site in Lilium and 0.012 subst/site in Tulipa). As the cultivars used are thought to represent overall diversity and classification in these genera (see Introduction) we feel these rate differences are not affected by (taxonomic) sampling artifacts and may actually reflect genetic divergence within the respective genera.
Both Lilium and Tulipa are outcrossing species, and vegetatively reproduced. Thus, the apparent difference in evolutionary rate can probably be explained by generation time and breeding history. Generation time (2.5x faster in lily compared with tulip) is considered to be negatively correlated with substitution rate, while breeding and selection probably influences the fixation of substitutions over generations (Buschiazzo et al., 2012). In addition, sequence divergence rates are considered to be governed by life span, i.e., short-lived species are capable of changing more quickly than those that have a longer life span and reproduce less often, and indeed, higher evolution rates have been observed in annuals compared with perennials (Yue et al., 2010). Another possible explanation for the different evolutionary rates between Tulipa and Lilium is their breeding history, though documentation is limited due to the fact that breeding historically was widely done by amateurs and private companies before professional institutes took over (Benschop et al., 2010). However, it is known that the breeding history of lily is more complex than tulip since more species were involved, which might explain the difference in evolution rate between both cultivar groups, as it could reflect actual difference in Ne.
In our analyses Lilium cultivars “Connecticut King” and “White Fox,” belonging to sections Sinomartagon and Leucolirion (subsection b) respectively, always grouped together, while “Star Gazer” and “Trumpet” (sections Archelirion and Leucolirion subsection a) clustered together as well (Figures 2A–D). Similar results were reported in other phylogenetic studies (Nishikawa et al., 1999; Arzate-Fernandez et al., 2005), based on cpDNA sequence comparisons. This is not in agreement with Comber's (Comber, 1949) classification, based on morphological and germination characteristics, in which “White Fox” and “Trumpet” belong to the same section (Leucolirion). On the other hand, crosses of Longiflorum hybrids (L, Leucolirion subsection b) with Trumpet hybrids (T, Leucolirion subsection a) are less successful compared with crosses of Trumpet hybrids with Oriental hybrids (O, Archelirion) and compared to crosses of Longiflorum hybrids with Asiatic hybrids (Sinomartagon) (Alex van Silfhout, Wageningen UR Plant Breeding, personal observations). In the latter there are even combinations in which hybrids are fertile on the diploid level and can be used for analytic breeding (Khan et al., 2009). Thus, patterns derived from crossability and molecular markers appear to support each other in Lilium.
Comparison of Methods
Given the ongoing increase of generating comparative transcriptome data, at and below the plant species-level, comparing analytical approaches in terms of performance and accuracy is more important than ever. In this paper we demonstrate the relative performance of commonly-used tree and network building methods.
The POFAD algorithm implements allelic information for inferring genetic distances in cultivars. Using POFAD helped to include the variation between haplotypes in estimating their relationships by taking their average (i.e., un-observed) distances. However, the standard POFAD pipeline does not allow inferring node-support, for instance by bootstrap values. This could be overcome by bootstrapping the sequence alignments, then following the POFAD procedure for each bootstrapped (pseudo) alignment and summarizing the occurrence of groups (bootstrap frequencies), similar to Neighbor Joining bootstrapping.
Three lily gene contigs presented deviating Neighbor Joining topologies in our analyses. These reflect either artifacts due to the low number of samples used (long branches and short internode) (Wiens, 2005), the NJ algorithm itself, or biological deviation which can be explained by assuming that each genomic region underwent an unique array of evolutionary events such as recombination, selection, mutation and/or gene flow (Buerkle et al., 2011). If such fragments are highly informative for their own phylogenetic history, it might in principle be possible to track every genomic segment to its origin and thus visualize species hybridization events (Zhang et al., 2013).
The three approaches generated the same topology, be it at different resolutions. Neighbor Network and the Consensus Network approaches suggested some non-tree like evolution in our gene contig sequences, possibly reflecting reticulate breeding histories within Lilium and Tulipa (Figures 3A,C). On the other hand, the concatenated approach (RAxML) generated one tree that may actually simplify evolutionary history (Figure 3B).
Obviously, the limited “taxon” sampling of the cultivars used in our study could limit the generality of our findings. For instance, using bi-allelic data did not appear to add significantly to our estimation of cultivar relationships. It will be interesting to extend a comparative study using bi-allelic data of nDNA in order to assess evolutionary relationships between other, hybrid species, using these approaches. Limited “taxon” sampling combined with increased character-sampling can easily result in long-branch attraction artifacts (Wiens, 2005). However, our results in terms of topologies obtained by the three approaches was in agreement with Nishikawa et al. (1999), who used 55 Lilium species. This may be related to the availability of a large sequence data set rich in polymorphic sites (26 gene contigs sequences: more than 18 kb yielded around 600 polymorphic sites in Lilium) in the present study. These 26 contigs could therefore be an excellent set of genes to study the phylogeny of Lilium in depth by comparing to many other species and construct gene trees and species trees. Similarly, the seven orthologous sequences among the nine Lilium and Tulipa sequence provide promising material to build generic-level trees.
Conclusions
Our study demonstrates the applicability of sequence data generated by next generation technology for estimating genetic divergence using the most commonly-used tree and network building methods. However, the benefit of the allelic nature of nuclear DNA in estimating the phylogeny of hybrids is still to be further established. The high number of polymorphic sites identified showed to be an effective tool for measuring genetic divergence, and the possible wide usage of these genes for phylogeny study for Lilium and Tulipa genus. The strategy to determine genetic distances based on a random set of genes for which orthologous sequences are retrieved from transcriptome sequencing, can be broadly applied. As the number of transcriptome datasets keeps increasing exponentially this will enable studies of the genetic relationships in many species complexes.
Author Contributions
Arwa Shahin participated in designing the work, generation, analysis and interpretation of the data. Freek T. Bakker contributed to the conception of the work, analysis and interpretation of the data. Jaap M. van Tuyl participated in the conception of the work. Marinus J. M Smulders participated in designing the work and interpretation of data. Paul Arens participated in the conception of the work and interpretation of data. The MS is written by Arwa Shahin, and revised critically by all co-authors. The co-authors approved the final version of the MS, and they agree to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Technological Top Institute Green Genetics (TTI GG) is thanked for their financial support in sequence data production.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00567/abstract
References
Alvarez, I., and Wendel, J. F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/S1055-7903(03)00208-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
APG. (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linnean Soc. 141, 399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x
APG. (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linnean Soc. 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
Arzate-Fernandez, A. M., Mejía-González, C. O., Nakazaki, T., Okumoto, Y., and Tanisaka, T. (2005). Isozyme electrophoretic characterization of 29 related cultivars of lily (Lilium spp.). Plant Breed. 124, 71–78. doi: 10.1111/j.1439-0523.2004.01046.x
Baldwin, B. (1992). Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol. Phylogenet. Evol. 1, 3–16. doi: 10.1016/1055-7903(92)90030-K
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beninda-Emonds, O. R. P. (2004). Phylogenetic Super Trees: Combining Information to Reveal the Tree of Life. New York, NY: Springer Verlag. doi: 10.1007/978-1-4020-2330-9
Benschop, M., Kamenetsky, R., Le Nard, M., Okubo, H., and De Hertogh, A. (2010). “The global flower bulb industry: production, utilization, research,” in Horticultural Reviews, ed J. Janick (Hoboken, NJ: John Wiley & Sons, Inc.), 1–115.
Booy, G., Schoot, J., and Vosman, B. (2000). Heterogeneity of the internal transcribed spacer 1 (ITS1) in Tulipa (Liliaceae). Plant Syst. Evol. 225, 29–41. doi: 10.1007/BF00985457
Bryant, D., and Moulton, V. (2004). Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265. doi: 10.1093/molbev/msh018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buerkle, C. A., Gompert, Z., and Parchman, T. L. (2011). The n = 1 constraint in population genomics. Mol. Ecol. 20, 1575–1581. doi: 10.1111/j.1365-294X.2011.05046.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buschiazzo, E., Ritland, C., Bohlmann, J., and And Ritland, K. (2012). Slow but not low: genomic comparisons reveal slower evolutionary rate and higher dN/dS in conifers compared to angiosperms. BMC Evol. Biol. 12:8. doi: 10.1186/1471-2148-12-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chase, M. W., and Reveal, J. L. (2009). A phylogenetic classification of the land plants to accompany APG III. Bot. J. Linnean Soc. 161, 122–127. doi: 10.1111/j.1095-8339.2009.01002.x
Comber, H. F. (1949). A new classification of the genus Lilium. Lily Yearb. R. Hort. Soc. 13, 86–105.
De La Torre, J., Egan, M., Katari, M., Brenner, E., Stevenson, D., Coruzzi, G., et al. (2006). ESTimating plant phylogeny: lessons from partitioning. BMC Evol. Biol. 6:48. doi: 10.1186/1471-2148-6-48
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Drummond, A., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dubouzet, J. G., and Shinoda, K. (1999). Phylogenetic analysis of the internal transcribed spacer region of Japanese Lilium species. Theor. Appl. Genet. 98, 954–960. doi: 10.1007/s001220051155
Edwards, S. V., Liu, L., and Pearl, D. K. (2007). High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. U.S.A. 104, 5936–5941. doi: 10.1073/pnas.0607004104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Emerson, K. J., Merz, C. R., Catchen, J. M., Hohenlohe, P. A., Cresko, W. A., Bradshaw, W. E., et al. (2010). Resolving postglacial phylogeography using high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 107, 16196–16200. doi: 10.1073/pnas.1006538107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Graybeal, A. (1998). Is it better to add taxa or characters to a difficult phylogenetic problem? Syst. Biol. 47, 9–17. doi: 10.1080/106351598260996
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Griffin, P., Robin, C., and Hoffmann, A. (2011). A next-generation sequencing method for overcoming the multiple gene copy problem in polyploid phylogenetics, applied to Poa grasses. BMC Biol. 9:19. doi: 10.1186/1741-7007-9-19
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haussler, D., O'brien, S., Ryder, O., Barker, F., Clamp, M., Crawford, A., et al. (2009). Genome 10K: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J. Hered. 100, 659–674. doi: 10.1093/jhered/esp086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heled, J., and Drummond, A. J. (2010). Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. doi: 10.1093/molbev/msp274
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Holland, B. R., Delsuc, F., Moulton, V., and Baker, A. (2005). Visualizing conflicting evolutionary hypotheses in large collections of trees: using consensus networks to study the origins of placentals and hexapods. Syst. Biol. 54, 66–76. doi: 10.1080/10635150590906055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hughes, C. E., Eastwood, R. J., and Bailey, C. D. (2006). From famine to feast? Selecting nuclear DNA sequence loci for plant species-level phylogeny reconstruction. Philos. Trans. Biol. Sci. 361, 211–225. doi: 10.1098/rstb.2005.1735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joly, S., and Bruneau, A. (2006). Incorporating allelic variation for reconstructing the evolutionary history of organisms from multiple genes: an example from rosa in North America. Syst. Biol. 55, 623–636. doi: 10.1080/10635150600863109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khan, N., Ramanna, M. S., Visser, R. G. F., and Van Tuyl, J. M. (2009). Potential for analytic breeding in allopolyploids: an illustration from Longiflorum x Asiatic hybrid lilies (Lilium). Euphytica 166, 399–409. doi: 10.1007/s10681-008-9824-0
Kim, K. J., and Jansen, R. K. (1994). Comparisons of phylogenetic hypotheses among different data sets in dwarf dandelions (Krigia, Asteraceae): additional information from internal transcribed spacer sequences of nuclear ribosomal DNA. Plant Syst. Evol. 190, 157–185. doi: 10.1007/BF00986191
Lim, K. B., and Van Tuyl, J. M. (2006). “Lilium hybrids,” in Flower Breeding and Genetics: Issues, Challenges and Opportunities for the 21st Century, ed N. O. Anderson (Dordrecht: Springer), 517–537. doi: 10.1007/978-1-4020-4428-1_20
Lim, K.-B., Wennekes, J., Jong, J. H. D., Jacobsen, E., and Van Tuyl, J. M. (2001). Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridisation. Genome 44, 911–918. doi: 10.1139/gen-44-5-911
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, L., Pearl, D., Brumfield, R., and Edwards, S. (2008). Estimating species trees using multiple-allele DNA sequence data. Evolution 62, 2080–2091. doi: 10.1111/j.1558-5646.2008.00414.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopez, P., Casane, D., and Philippe, H. (2002). Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19, 1–7. doi: 10.1093/oxfordjournals.molbev.a003973
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maddison, W. P., and Maddison, D. R. (2011). “Mesquite: A Modular System for Evolutionary Analysis. Version 2.75. Availabe online at: http://mesquiteproject.org.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Gateway Computing Environments Workshop (GCE) (New Orleans, LA), 1–8.
Milne, I., Lindner, D., Bayer, M., Husmeier, D., McGuire, G., Marshall, D. F., et al. (2009). TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25, 126–127. doi: 10.1093/bioinformatics/btn575
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muratović, E., Hidalgo, O., Garnatje, T., and Siljak-Yakovlev, S. (2010). Molecular phylogeny and genome size in European lilies (Genus Lilium, Liliaceae). Adv. Sci. Lett. 3, 180–189. doi: 10.1166/asl.2010.1116
Naumov, G. I., James, S. A., Naumova, E. S., Louis, E. J., and Roberts, I. N. (2000). Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50, 1931–1942.
Nishikawa, T., Okazaki, K., Arakawa, K., and Nagamine, T. (2001). Phylogenetic analysis of section sinomartagon in genus lilium using sequences of the internal transcribed spacer region in nuclear ribosomal DNA. Breed. Sci. 51, 39–46. doi: 10.1270/jsbbs.51.39
Nishikawa, T., Okazaki, K., Uchino, T., Arakawa, K., and Nagamine, T. (1999). A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J. Mol. Evol. 49, 238–249. doi: 10.1007/PL00006546
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nylander, J. A., Ronquist, F., Huelsenbeck, J. P., and Nieves-Aldrey, J. (2004). Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67. doi: 10.1080/10635150490264699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olmstead, R., and Palmer, J. (1992). A chloroplast DNA phylogeny of the Solanaceae: subfamilial relationships and character evolution. Ann. Mo. Bot. Gard. 79, 346–360. doi: 10.2307/2399773
Page, R. D. (1998). GeneTree: comparing gene and species phylogenies using reconciled trees. Bioinformatics 14, 819–820. doi: 10.1093/bioinformatics/14.9.819
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rokas, A., Williams, B., King, N., and Carroll, S. (2003). Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425, 798–804. doi: 10.1038/nature02053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanderson, M., and McMahon, M. (2007). Inferring angiosperm phylogeny from EST data with widespread gene duplication. BMC Evol. Biol. 7:S3. doi: 10.1186/1471-2148-7-S1-S3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sang, T., Crawford, D. J., and Stuessy, T. F. (1995). Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 6813–6817. doi: 10.1073/pnas.92.15.6813
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahin, A., Van Kaauwen, M., Esselink, D., Bargsten, J., Van Tuyl, J., Visser, R., et al. (2012). Generation and analysis of expressed sequence tags in the extreme large genomes Lilium and Tulipa. BMC Genomics 13:640. doi: 10.1186/1471-2164-13-640
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Small, R. L., Cronn, R. C., and Wendel, J. F. (2004). Use of nuclear genes for phylogeny reconstruction in plants. Aust. Syst. Bot. 17, 145–170. doi: 10.1071/SB03015
Stamatakis, A. (2006). “Phylogenetic models of rate heterogeneity: a high performance computing perspective,” in 20th International Parallel and Distributed Processing Symposium, 2006. IPDPS 2006 (Los Alamitos, CA: IEEE Computer Society Press).
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Swofford, D. L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vriesendorp, B., and Bakker, F. T. (2005). Reconstructing patterns of reticulate evolution in angiosperms: what can we do? Taxon 54, 593–604. doi: 10.2307/25065417
White, T., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. Innis, D. Gelfand, J. Shinsky, and T. White (New York, NY: Academic Press), 315–322.
Wiens, J. J. (2005). Can incomplete taxa rescue phylogenetic analyses from long-branch attraction? Syst. Biol. 54, 731–742. doi: 10.1080/10635150500234583
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yue, J.-X., Li, J., Wang, D., Araki, H., Tian, D., and Yang, S. (2010). Genome-wide investigation reveals high evolutionary rates in annual model plants. BMC Plant Biol. 10:242. doi: 10.1186/1471-2229-10-242
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Lilium, allelic variation, POFAD, RAxML, Consensus Network, genetic divergence, Tulipa
Citation: Shahin A, Smulders MJM, van Tuyl JM, Arens P and Bakker FT (2014) Using multi-locus allelic sequence data to estimate genetic divergence among four Lilium (Liliaceae) cultivars. Front. Plant Sci. 5:567. doi: 10.3389/fpls.2014.00567
Received: 11 June 2014; Accepted: 01 October 2014;
Published online: 20 October 2014.
Edited by:
Jaime Prohens, Universitat Politècnica de València, SpainReviewed by:
Sergio Lanteri, University of Turin, ItalyJose Blanca, Universidad Politecnica de Valencia, Spain
Copyright © 2014 Shahin, Smulders, van Tuyl, Arens and Bakker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Freek T. Bakker, Biosystematics Group, Wageningen University, Droevendaalsesteeg 1, 6700 AP Wageningen, Netherlands e-mail:ZnJlZWsuYmFra2VyQHd1ci5ubA==
 Arwa Shahin
Arwa Shahin Marinus J. M. Smulders
Marinus J. M. Smulders Jaap M. van Tuyl
Jaap M. van Tuyl Paul Arens
Paul Arens Freek T. Bakker
Freek T. Bakker