- 1Division of Biological Science, Graduate School of Science, Nagoya University, Nagoya, Japan
- 2Graduate School of Bioscience and Biotechnology, Chubu University, Kasugai, Japan
Crown gall tumors are formed mainly by actions of a group of genes in the T-DNA that is transferred from Agrobacterium tumefaciens and integrated into the nuclear DNA of host plants. These genes encode enzymes for biosynthesis of auxin and cytokinin in plant cells. Gene 6b in the T-DNA affects tumor morphology and this gene alone is able to induce small tumors on certain plant species. In addition, unorganized calli are induced from leaf disks of tobacco that are incubated on phytohormone-free media; shooty teratomas, and morphologically abnormal plants, which might be due to enhanced competence of cell division and meristematic states, are regenerated from the calli. Thus, the 6b gene appears to stimulate a reprogramming process in plants. To uncover mechanisms behind this process, various approaches including the yeast-two-hybrid system have been exploited and histone H3 was identified as one of the proteins that interact with 6b. It has been also demonstrated that 6b acts as a histone H3 chaperon in vitro and affects the expression of various genes related to cell division competence and the maintenance of meristematic states. We discuss current views on a role of 6b protein in tumorigenesis and reprogramming in plants.
Introduction
Agrobacterium tumefaciens strains that harbor tumor inducible (Ti) plasmids cause crown gall tumors upon infection of dicot plants. Ti plasmids of Agrobacterium transfer their T-DNAs to the chromosomal DNAs of plants. These T-DNAs contain three gene loci involved in this tumorigenesis, tmr, tms (tms1 and tms2), and tml. The tmr and tms loci contain genes involved in the biosynthesis of cytokinins and auxins, respectively. The tml locus was originally identified as the mutated gene that causes tumors larger than those induced by the wild-type T-DNA (Garfinkel et al., 1981). It contains two genes 6a and 6b: the former encodes an unknown protein whereas the molecular characteristics of the latter had also remained undefined until Terakura et al. (2007) identified them recently. Gene 6b in the tml loci is conserved in the T-DNA of all strains of Agrobacterium and is actively transcribed in the tumor cells (Willmitzer et al., 1983; Otten and De Ruffray, 1994). In addition, some plant species that were infected with Agrobacterium stains having T-DNAs containing only the 6b genes showed the formation of small but significantly sized tumors at the infection sites (Hooykaas et al., 1988; Tinland et al., 1989). When 6b was introduced into leaf disks, which were then incubated on hormone-free medium, unorganized calli were formed (Wabiko and Minemura, 1996). Transgenic plants regenerated from these calli exhibited morphological defects in various organs (Tinland et al., 1992; Wabiko and Minemura, 1996; Gális et al., 1999, 2002; Helfer et al., 2003; Terakura et al., 2006). The defects appear to be due to increased meristematic states and cell division potentials. These observations suggest that the 6b proteins play roles in the maintenance of tumorigenic states as well as the induction of dedifferentiation and differentiation (reprogramming process) of plant organs.
In the present mini review, we will summarize the results of recent studies, including our own, to present a novel understanding of the molecular characteristics of 6b protein and discuss how 6b might act as a reprogramming factor. We focus here on the subcellular localization of 6b protein; relationships between such localization and the ability of 6b to induce tumorigenic states; plant genes that are affected by 6b; plants proteins that interact with 6b; and, finally, a molecular function of 6b.
Structural Features of 6b Protein
The 6b gene of A. tumefaciens AKE10 strain (Wabiko and Minemura, 1996) encodes a protein with 208 amino acid residues that contains an acidic region (Figure 1A: residues 164–184) near the carboxyl terminus, suggestive of interactions with other proteins to generate complexes. The acidic region is essential for the generation of abnormal phenotypes by 6b (Terakura et al., 2006). It is worth pointing out that when the GAL4 DNA binding domain is fused to 6b protein, the fusion protein has a transactivation activity, which requires the acidic region (Kitakura et al., 2002).
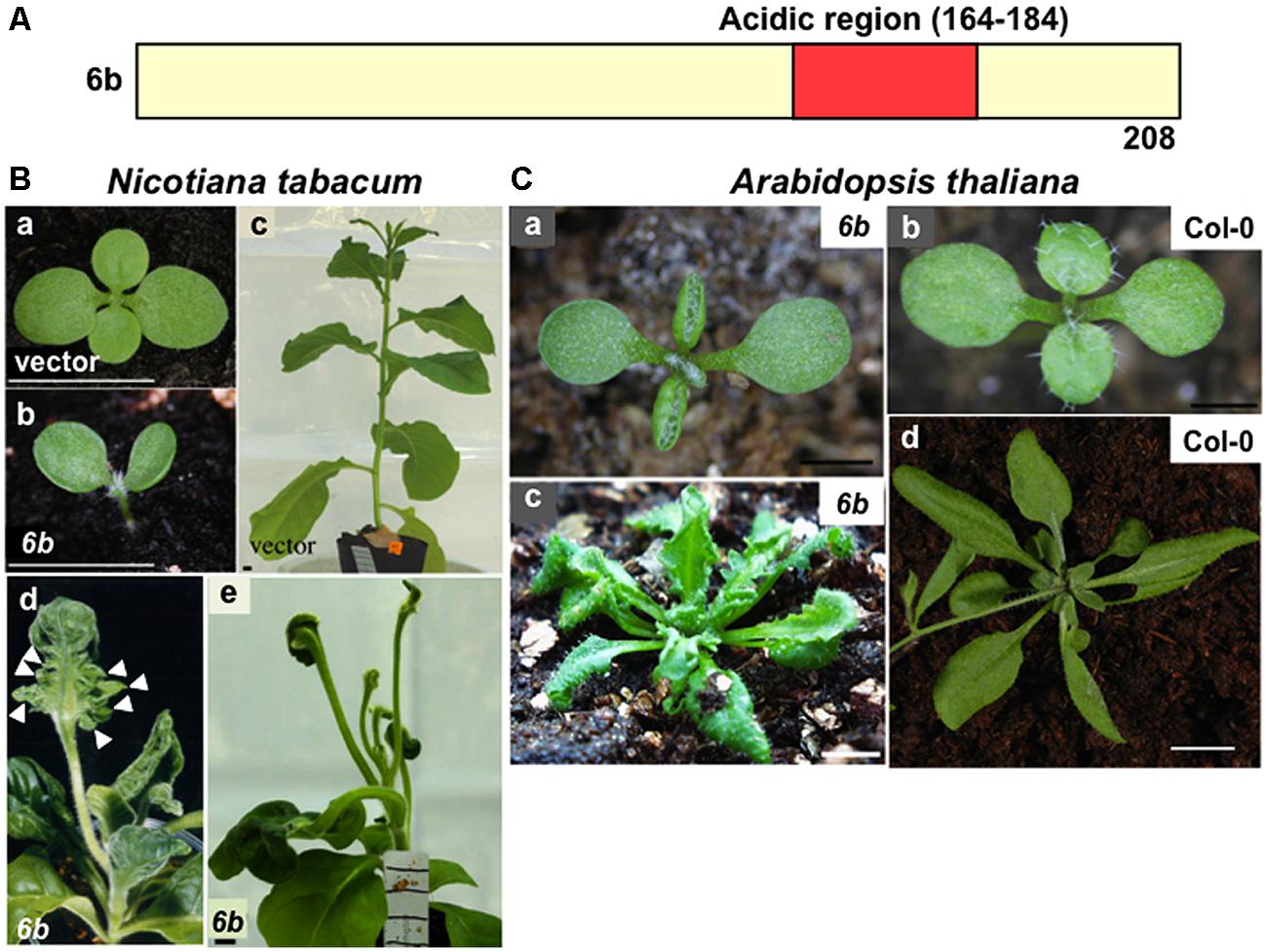
FIGURE 1. Morphological defects in the transgenic plants that express 6b protein. (A) Schematic domain organization of 6b protein. The acidic region (residues 164–184) is present in 6b. (B) Photographs showing clearly the reported phenotypes of transgenic tobacco plants. Transgenic plants transformed with the empty vector (a,c) or 6b gene (b,d,e). The 6b-transgenic seedling (b) and young plant (d) exhibiting mild defects and the 6b-plant (e) exhibiting severe defects. Arrowheads indicate protrusions on the abaxial side of leaves. Scale bars: 10 mm. (C) Photographs showing clearly the reported phenotypes of non-transgenic and transgenic Arabidopsis plants. The 6b-transgenic Arabidopsis plants (a,c); non-transgenic Col-0 plants (b,d). Scale bars: 1 mm (a,b), 10 mm (c,d). Some of pictures were published previously ( Terakura et al., 2006).
The remaining amino acid sequence of 6b exhibits weak similarity to those sequences of several proteins encoded by other genes in the Ti and Ri plasmids of A. rhizogenes T-DNAs, rolB, rolC, ORF13, and ORF14 that belong to the plast family (Spena et al., 1987; Levesque et al., 1988; Stieger et al., 2004). Despite of their weak similarities, they also induce commonly abnormal growth and morphology of both roots and shoots, and aberrant accumulation of sugars and metabolites (Cardarelli et al., 1987; Spena et al., 1987; Lemcke and Schmülling, 1998; Helfer et al., 2002; Stieger et al., 2004; Mohajjel-Shoja et al., 2011). The amino acid sequence of the ORF13 protein contains a retinoblastoma (Rb)-binding motif (LXCXE), and it binds to the maize Rb protein (Stieger et al., 2004); however, 6b does not have this motif.
Although 6b protein contains no motif similar to the nuclear localization signal (NLS), 6b protein localizes to the nucleus (Kitakura et al., 2002). Only the RolB protein in the plast family has also been shown to localize to plant nuclei (Moriuchi et al., 2004). Tobacco plants that express a glucocorticoid-receptor-fused 6b protein show the abnormal phenotypes described above only after being treated with dexamethasone, which stimulates nuclear import of the protein (Terakura et al., 2006). This result suggests that the nuclear localization of 6b is required for the appearance of morphological defects of plants and abnormal cell proliferation. Mechanisms of nuclear localization of the 6b protein, however, have remained unknown. There are two possible explanations for such a localization of 6b. (1) The relative molecular mass of 6b protein is sufficiently small enough (approximately 20,000) to be transported into the nucleus by passive diffusion and it might be trapped there by a certain nuclear protein. (2) As mentioned below, 6b interacts with the NtSIP1 and NtSIP2 proteins, which are tobacco transcription factor-like nuclear proteins (Kitakura et al., 2002, 2008). Therefore, 6b proteins might be transported into the nucleus through an interaction with these endogenous nuclear proteins. Regardless of the molecular mechanisms involved, however, 6b appears to play a role in the expression of some genes that might be involved in cell proliferation and differentiation.
Characteristics of Transgenic Plants that Express 6b
Phenotypic Characteristics of Tobacco Plants that Express 6b
The shoots and leaves of transgenic tobacco plants derived from calli, which are induced by the expression of 6b and cultured on hormone-free-medium, show various morphological defects (Figure 1B; Tinland et al., 1992; Wabiko and Minemura, 1996; Helfer et al., 2003; Terakura et al., 2006). In the tobacco transgenic plants that show mild defects, cotyledons and leaves are curled upwardly at an early growth stage (Figure 1Bb,d) and generate a number of outgrowths from the abaxial surface (Figure 1Bd). Transgenic plants that show the severe phenotype produce leaves with long petioles and an unexpanded lamina associated with rod-shaped protrusions (Figure 1Be). Expression of 6b in transgenic tobacco plants also induces abnormal venation patterns of cotyledons, which sometimes include veins with inverted adaxial–abaxial polarity (Kakiuchi et al., 2007). In hypocotyles of these transgenic tobacco seedlings, basipetal auxin transport was reduced, which might be related to morphological abnormalities as described above (Kakiuchi et al., 2006). Takahashi et al. (2013) suggested that 6b protein is involved in modulating auxin and cytokinin localization and affects proliferation of tumorigenic cells expressing 6b.
To understand the molecular basis for the upward curling of leaves and the protrusion at the leaf abaxial side of tobacco transgenic plants with the 6b gene, Terakura et al. (2006) measured levels of Cyclin B1 and NACK1 transcripts (as markers for the cell cycle) in mature leaves of 6b-transgenic plants and carried out in situ hybridization with sections of mature leaves and cDNAs of NACK1. Transcript levels of these genes were increased and these genes were ectopically transcribed on the abaxial side of mature leaves, suggesting increased cell division competence in the abaxial domain of 6b-transgenic leaves.
In addition to aberrant morphology and patterns of cell division, expression of 6b in tobacco plants stimulates sugar uptake and retention, which might cause expansion of leaf and root tissues of transgenic tobacco (Clément et al., 2006, 2007). Expressions of rolC and tobacco endogenous rolC homolog also induce similar effects on sucrose uptake, suggesting a functional relationship between rolC and 6b (Mohajjel-Shoja et al., 2011).
In several Arabidopsis mutants that have defects in the establishment of leaf adaxial–abaxial polarity, similar leaf morphological defects are observed (McConnell and Barton, 1998; Eshed et al., 2001; Ueno et al., 2007; Kojima et al., 2011; Ishibashi et al., 2012). A model has been proposed whereby the appropriate cell differentiation of leaf adaxial and abaxial domains is required for the development of flat leaf lamina, and thus defects in the establishment of leaf adaxial–abaxial polarity cause inhibition of the leaf laminar expansion (Waites and Hudson, 1995; Emery et al., 2003). In the transgenic tobacco plants that express 6b, the adaxial–abaxial polarity of leaves might be affected as a consequence of aberrant expression of tobacco genes that are involved in the establishment of such a polarity.
Phenotypes of Arabidopsis Plants that Express 6b
In the transgenic Arabidopsis plants that express the 6b gene, morphological defects were also observed, such as the upwardly curled leaves, protrusion at the leaf abaxial side, and apparent serrations in the leaf lamina (Figure 1C; Terakura et al., 2006). In addition, the Arabidopsis transgenic plants that express 6b genes from the Agrobacterium vitis AB4 strain or A. tumefaciens Tm4 strain generate rod-like shaped leaves with short lamina along the longitudinal axis and a long petiole (Helfer et al., 2003). These phenotypes are somewhat similar to those of the Arabidopsis plants in which the ASYMMETRIC LEAVES2 (AS2) gene is ectopically overexpressed (Iwakawa et al., 2002).
To understand the molecular bases for the above mentioned phenotypes of Arabidopsis transgenic plants, Terakura et al. (2006) measured the transcript levels of a number of genes related to leaf morphology and cell division. The results showed increases in levels of transcripts of the Cyclin B1 and AtNACK1 genes (cell cycle markers); class 1 KNOX genes (markers for meristem maintenance: STM, BP, KNAT2, and KNAT6); and CUC genes (markers for meristem maintenance and separation of organs from the meristem: CUC1, CUC2, and CUC3). These data suggest that the meristematic state and cell division competence are increased in 6b-transgenic Arabidopsis plants. Transcript levels of HD-ZIP III and ASYMMETRIC LEAVES1 (markers for adaxial development) or of KAN1 and FIL (markers for abaxial development), however, were not significantly affected by 6b expression (Terakura et al., 2006). The results of microarray analyses do not allow us to simply explain the molecular basis of the abnormal leaf morphology of the 6b-transgenic Arabidopsis.
The 6b Gene Enhances Developmentally Indeterminate States and Cell Proliferation Potential in Plants
The tobacco leaf disks that are transcribed under control of the Cauliflower mosaic virus 35S promoter generate white or green abnormal cell proliferation. During the callus formation by 6b expression, the amounts of endogenous phytohormones, cytokinin and auxin, are not changed (Wabiko and Minemura, 1996). It seems likely that the 6b gene might directly or indirectly induce the expression of genes required for formation of calli. Gene 6b also suppresses both the shoot formation induced by cytokinins in vitro (Spanier et al., 1989; Wabiko and Minemura, 1996) and the root formation induced by auxin and rolABC (Tinland et al., 1990), suggesting that 6b protein not only induces the cell proliferation under hormone-free conditions but also suppresses the cell differentiation induced by cytokinins and auxins.
Transcripts levels of genes for meristem (stem cell) maintenance, class 1 KNOX genes, are increased in Arabidopsis and that the expression of class 1 KNOX genes of tobacco (NTH15, NTH1, NTH20, and NTH22) is also increased in the leaves of 6b-transgenic tobacco plants (Terakura et al., 2006). These results suggest an increase in the undifferentiated states of cells in these 6b-transgenic plants.
Molecules Interacting with 6b Protein
To unveil the molecular functions of 6b protein, Kitakura et al. (2002, 2008) and Terakura et al. (2007) have sought for 6b-interacting proteins of the tobacco cultured cell line BY-2. They screened tobacco cDNA libraries by yeast two-hybrid systems and eventually identified three positive cDNAs of interest in terms of tumorigenicity, cell division competency, and the formation of morphologically malformed leaves. Kitakura et al. (2002, 2008) identified two of these genes designated Nicotiana Six-b Interacting Protein 1 and 2 (NtSIP1 and NtSIP2); and Terakura et al. (2007) identified a third gene, NtSIP3.
NtSIP1 is Localized to Nuclei and Enhances the Nuclear Localization of the 6b Protein in Tobacco Cells
NtSIP1 encodes a protein that consists of 318 amino acid residues with a molecular mass of 34.8 kD (Kitakura et al., 2002). The region from residue 72 to residue 131 of NtSIP1 is predicted to form three-helices with short intervening loops. This region is similar to the triple helix motif of rice transcription factor GT-2, which controls the transcription level of the PHYA gene (Dehesh et al., 1992). There are two basic regions that resemble a NLS. In the Arabidopsis genome, several homologs of the NtSIP1 gene exist. The amino acid residues of NtSIP1 are 43% identical to those of the predicted protein of F14P22.220, 36% identical to those of MOP10_9, 34% identical to those of F9F8.9, and 27% identical to those of F11B9.6. Phylogenetic tree analysis indicates that NtSIP1 is closely related to F14P22.220 of Arabidopsis. Amino acid sequences corresponding to a triple helix motif and NLSs are conserved in all of these deduced sequences. In BY-2 cells, NtSIP1 localizes in the nuclei. Accumulated NtSIP1 transcripts are detected in roots, stems, mature leaves, and shoot apices that contain the shoot apical meristem, and those transcript levels are significantly higher in shoot apices than in other organs (Kitakura et al., 2002). NtSIP1 enhances the nuclear localization of 6b protein in BY-2 cells, and the acidic region of 6b is necessary for the interaction with NtSIP1 and the enhancement of the nuclear localization of 6b (Kitakura et al., 2002).
NtSIP2 has Homology to TNP1 Encoded by the Transposable Element Tam1 of Antirrhinum majus
The NtSIP2 gene is predicted to encode a protein with 424 amino acid residues, and the predicted protein exhibits homology to TNP1 encoded by the transposable element Tam1 of A. majus (Kitakura et al., 2008). The predicted amino acid sequence of NtSIP2 is 30% identical and 50% similar to that of the TNP1 protein (Nacken et al., 1991). NtSIP2 has three short stretches of basic amino acid residues that are similar to the NLS sequence. The NtSIP2 transcripts are detected in a variety of plant tissues without significant quantitative difference. Almost all NtSIP2 protein is localized to the nucleolus. Since significant amounts of the 6b protein are also detected in the nucleolus, NtSIP2 in the nucleolus might be interacted with the 6b protein. NtSIP2 might play a role in transporting 6b into the nucleolus by binding activity and the 6b-NtSIP2 complex might act as a transcriptional repressor within the nucleolus.
NtSIP3
NtSIP3 encodes histone H3
By using the yeast two-hybrid system with a 6b sequence lacking the DNA region corresponding to the acidic region (Figure 1A), Terakura et al. (2007) identified two tobacco cDNAs for members of the tobacco histone H3 protein family. The predicted amino acid sequences of the two tobacco clones are identical to those of histone H3.1 (DNA replication-dependent H3) and H3.2 (replication-independent H3) of Arabidopsis (Johnson et al., 2004), respectively, which are collectively designated as NtSIP3. The tobacco histone H3.2 clone was used for in vitro binding assay and in vivo binding by BiFC (bimolecular fluorescence complementation) analysis. The results of these experiments have shown that the 6b protein directly interacts with tobacco histone H3.2 in vitro and in vivo. Protein 6b also binds to Arabidopsis histone H3.1 (At3g27360.1) and H3.2 (At4g40030.1) in vitro. In the tobacco BY-2 cells, 6b protein interacts with chromatin. The amino acid sequence of 6b without the acidic region binds to the histone fold region of histone H3.2 (60–135 residues) and the C-terminal region of 6b (185–208 residues) is required for the binding (Terakura et al., 2007).
It was examined whether 6b can bind other core histones (H2A, H2B, and H4). The results show that it binds to histone H3 specifically (Terakura et al., 2007). Since the histone fold consists of three α helices that are involved in the formation of nucleosomes, 6b protein might have some effect on the formation of the nucleosome in vivo (Figure 2).
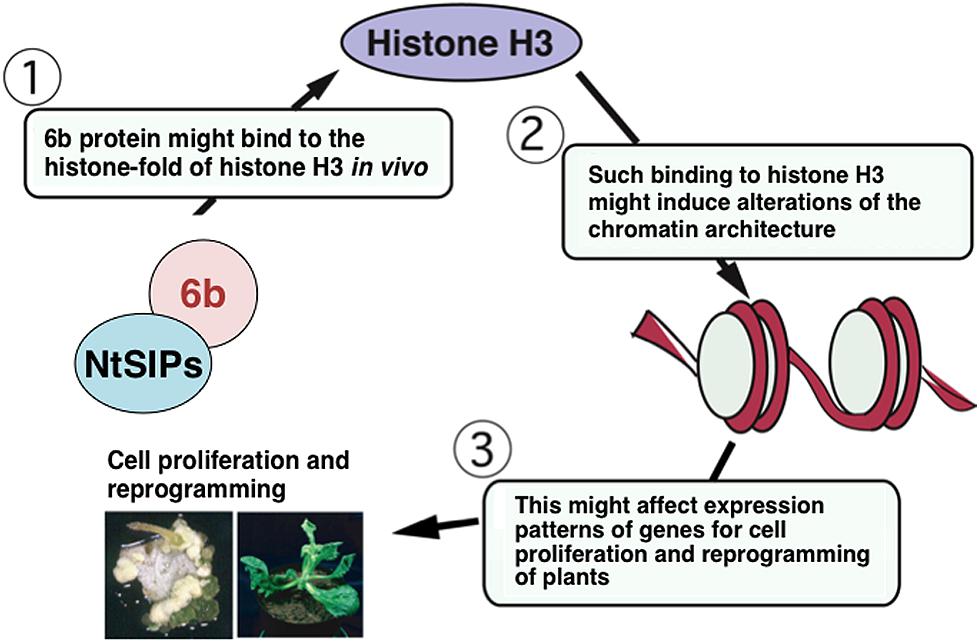
FIGURE 2. Working hypothesis of 6b action. See details in text. NtSIPs represent NtSIP1 or NtSIP2. They might independently bind to 6b and/or both might simultaneously bind to 6b.
6b protein has histone chaperone activity
Histone chaperones are known to be factors that enhance the formation of nucleosomes in vitro (Loyola and Almouzni, 2004). Histone chaperones, such as HIRA (histone regulator A) and ASF1 (antisilencing factor 1), bind to the histone fold domains of core histones (Munakata et al., 2000; Ray-Gallet et al., 2002). Furthermore, histone chaperones, such as NAP-1 (nucleosome assembly protein-1; Ishimi and Kikuchi, 1991), nucleophosmin/B23 (Okuwaki et al., 2001), yeast FK506bp (Kuzuhara and Horikoshi, 2004), and nucleolin (Angelov et al., 2006), contain acidic regions of amino acid sequence. The β sheet structure of ASF1 is required for the interaction with histone, and other histone chaperones, such as NAP-1, RbAp46/48, p60 (a component of Chromatin Assemby Factor-1 complex) and HIRA, are also predicted to contain similar β sheet structures (English et al., 2006). Intriguingly, the C-terminal amino acids that are required for the interaction with histone H3 are predicted to form the β-sheet structure. The 6b protein is similar to known histone chaperones in terms of the following three points: (1) the physical association of 6b with the histone fold of histone H3; (2) the presence of the β-sheets in the C-terminus of 6b (Wang et al., 2011); and (3) the presence of the acidic region in 6b.
By using a supercoiling assay, 6b protein was demonstrated to have histone chaperone-like activity in vitro (Terakura et al., 2007). The C-terminal region is required for the cell division-stimulating activity, interaction with histone H3, and histone chaperon-like activity of 6b, suggesting that such activity is related to the ability of 6b to induce cell proliferation. AB6b from A. vitis AB4 strain also binds to histone H3 and has histone chaperone-like activity. Similarly as for AK6b, AB6b also induces severe morphological defects in transfected plants (Helfer et al., 2003). Histone chaperones affect the histone dynamics, such as histone conservation (Dutta et al., 2001), histone transport (Ito et al., 1996; Mosammaparast et al., 2002), and chromatin structure conversion (Smith and Stillman, 1989; Belotserkovskaya et al., 2003; Adkins et al., 2004; Boeger et al., 2004; Schermer et al., 2005) to control the transcription and DNA replication in the nucleus (Loyola and Almouzni, 2004; Polo and Almouzni, 2006). The 6b protein might act as a histone chaperone in vivo and regulate the chromatin structure to affect gene transcription for cell proliferation and plant differentiation (Figure 2).
Future Prospects
Through the analyses of its interacting molecules, Agrobacterium 6b protein functions in the nucleus and the nucleolus of plant cells, in which it might interact with NtSIP1, NtSIP2, and histone H3 and act as a histone chaperone to alter expression patterns of genes related to tumorigenicity, cell proliferation, and reprogramming of plant cells (Figure 2). It is generally accepted that histone chaperons affect epigenetic status of chromatin mainly by replacing canonical histones with replacement histones (differing in several amino acid residues from canonical ones), which can alter patterns of gene expression (Skene and Henikoff, 2013). Formation of crown gall tumors largely depends on expression of genes for enzymes involved in cytokinin and auxin biosynthesis. The histone chaperone activity of 6b might affect chromatin structures to regulate the expression levels of the genes related to auxin and cytokinin biosynthesis. Plants have various silencing mechanisms to repress expression of transgenes, such as those from pathogens, by chromatin remodeling, DNA methylation, histone modification, and/or the RNAi system (Zilberman and Henikoff, 2005). Expression of the phytohormone biosynthesis genes in the T-DNA region might be down-regulated by the silencing mechanisms of host plants. Histone chaperone activity of 6b might compromise actions of the plant silencing systems. Suppression of the gene silencing systems by 6b might promote expression of genes for auxin and cytokinin biosynthesis, which might affect the host range of Agrobacterium strains to induce crown gall tumors (Hooykaas et al., 1988). Genetic manipulation of the 6b gene might contribute to improving Agrobacterium-mediated gene transfer technologies.
Apart from the histone chaperone activity of 6b, the crystal structure of this protein exhibits a certain extent of the structural similarity to an ADP ribosylation factor of Arabidopsis, and the 6b protein bears a biochemical role in ADP ribosylation under conditions used by Wang et al. (2011). Relationships between this activity, phenotypes and alteration of patterns of gene expression generated by 6b remain to be elucidated.
It is notable that transcript levels of cell division- and meristem-related genes such as class 1 KNOX and CYCLIN genes are increased. These data support the hypothesis that the cell proliferation potential and meristematic states might be enhanced by 6b. Accumulation levels of transcripts of a number of auxin-inducible genes, such as those of the GH3, IAA, and ACS family genes, are decreased in 6b-expressing transgenic Arabidopsis plants (Terakura et al., 2007). These results might imply that various physiological and cellular statuses that are controlled by such auxin-inducible genes might be altered by 6b expression. It should be critical to identify direct target genes of 6b to uncover molecular frameworks of cell reprogramming and intriguing to examine whether plant endogenous histone chaperons might have reprogramming activities.
Author Contributions
Nanako Ishibashi and Yasunori Machida designed the outline of this article. Nanako Ishibashi wrote Abstract, Introduction, Section “Characteristics of Transgenic Plants that Express 6b,” and References. Saeko Kitakura wrote the Section “NtSIP1 is Localized to Nuclei and Enhances the Nuclear Localization of the 6b Protein in Tobacco Cells.” Chiyoko Machida wrote the Sections “Structural Feature of 6b Protein” and “NtSIP2 has Homology to TNP1 Encoded by the Transposable Element Tam1 of Antirrhinum majus,” and contributed to revise the original title by the addition of “Reprogramming” to the present form. Chiyoko Machida wrote the Section “Future Prospects” to revise the original version. Shinji Terakura and Yasunori Machida wrote the Section “NtSIP3.” Yasunori Machida wrote the original part of the Section “Future prospects,” and review and edit the entire manuscript. All authors are accountable for all aspects of the description of this article in ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences (Japan) and by Grants-in-Aid for Scientific Research on Priority Areas (nos. 10182102, 14036216, 15028208, and 19060003 to Yasunori Machida) from the Ministry of Education, Science, Culture and Sport (Japan).
References
Adkins, M. W., Howar, S. R., and Tyler, J. K. (2004). Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14, 657–666. doi: 10.1016/j.molcel.2004.05.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Angelov, D., Bondarenko, V. A., Almagro, S., Menoni, H., Mongélard, F., Hans, F.,et al. (2006). Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25, 1669–1679. doi: 10.1038/sj.emboj.7601046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M., and Reinberg, D. (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093. doi: 10.1126/science.1085703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boeger, H., Griesenbeck, J., Strattan, J. S., and Kornberg, R. D. (2004). Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14, 667–673. doi: 10.1016/j.molcel.2004.05.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cardarelli, M., Mariotti, D., Pomponi, M., Spanò, L., Capone, I., and Costantino, P. (1987). Agrobacterium rhizogenes T-DNA genes capable of inducing hairy root phenotype. Mol. Gen. Genet. 209, 475–480. doi: 10.1007/BF00331152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clément, B., Perot, J., Geoffroy, P., Legrand, M., Zon, J., and Otten, L. (2007). Abnormal accumulation of sugars and phenolics in tobacco roots expressing the Agrobacterium T-6b oncogene and the role of these compounds in 6b-inducible growth. Mol. Plant Microbe Interact. 20, 53–62. doi: 10.1094/MPMI-20-0053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clément, B., Pollmann, S., Weiler, E., Urbanczyk-Wochniak, E., and Otten, L. (2006). The Agrobacterium vitis T-6b oncoprotein induces auxin-independent cell expansion in tobacco. Plant J. 45, 1017–1027. doi: 10.1111/j.1365-313X.2006.02663.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dehesh, K., Hung, H., Tepperman, J. M., and Quail, P. H. (1992). GT-2: a transcription factor with twin autonomous DNA-binding domains of closely related but different target sequence specificity. EMBO J. 11, 4131–4144.
Dutta, S., Akey, I. V., Dingwall, C., Hartman, K. L., Laue, T., Nolte, R. T.,et al. (2001). The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8, 841–853. doi: 10.1016/S1097-2765(01)00354-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A.,et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E., and Tyler, J. K. (2006). Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508. doi: 10.1016/j.cell.2006.08.047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eshed, Y., Baum, S. F., Perea, J. V., and Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. doi: 10.1016/S0960-9822(01)00392-X
Gális, I., Simek, P., Van Onckelen, H. A., Kakiuchi, Y., and Wabiko, H. (2002). Resistance of transgenic tobacco seedlings expressing the Agrobacterium tumefaciens C58-6b gene, to growth-inhibitory levels of cytokinin is associated with elevated IAA levels and activation of phenylpropanoid metabolism. Plant Cell Physiol. 43, 939–950. doi: 10.1093/pcp/pcf112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gális, I. I., Simek, P., Macas, J., Zahradnickova, H., Vlasak, J., Wabiko, H.,et al. (1999). The Agrobacterium tumefaciens C58-6b gene confers resistance to N(6)-benzyladenine without modifying cytokinin metabolism in tobacco seedlings. Planta 209, 453–461. doi: 10.1007/s004250050748
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garfinkel, D. J., Simpson, R. B., Ream, L. W., White, F. F., Gordon, M. P., and Nester, E. W. (1981). Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27, 143–153. doi: 10.1016/0092-8674(81)90368-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Helfer, A., Clement, B., Michler, P., and Otten, L. (2003). The Agrobacterium oncogene AB-6b causes a graft-transmissible enation syndrome in tobacco. Plant Mol. Biol. 52, 483–493. doi: 10.1023/A:1023962121894
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Helfer, A., Pien, S., and Otten, L. (2002). Functional diversity and mutational analysis of Agrobacterium 6B oncoproteins. Mol. Gen. Gen. 267, 577–586. doi: 10.1007/s00438-002-0707-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hooykaas, P. J. J., der Dulk-Ras, H., and Schilperoort, R. A. (1988). The Agrobacterium tumefaciens T-DNA gene 6b is an onc gene. Plant Mol. Biol. 11, 791–794. doi: 10.1007/BF00019519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ishibashi, N., Kanamaru, K., Ueno, Y., Kojima, S., Kobayashi, T., Machida, C.,et al. (2012). ASYMMETRIC-LEAVES2 and an Ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 1, 197–207. doi: 10.1242/bio.2012406
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ishimi, Y., and Kikuchi, A. (1991). Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266, 7025–7029.
Ito, T., Bulger, M., Kobayashi, R., and Kadonaga, J. T. (1996). Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16, 3112–3124.
Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H.,et al. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. doi: 10.1093/pcp/pcf077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, L., Mollah, S., Garcia, B. A., Muratore, T. L., Shabanowitz, J., Hunt, D. F.,et al. (2004). Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 32, 6511–6518. doi: 10.1093/nar/gkh992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kakiuchi, Y., Gàlis, I., Tamogami, S., and Wabiko, H. (2006). Reduction of polar auxin transport in tobacco by the tumorigenic Agrobacterium tumefaciens AK-6b gene. Planta 223, 237–247. doi: 10.1007/s00425-005-0080-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kakiuchi, Y., Takahashi, S., and Wabiko, H. (2007). Modulation of the venation pattern of cotyledons of transgenic tobacco for the tumorigenic 6b gene of Agrobacterium tumefaciens AKE10. J. Plant Res. 120, 259–268. doi: 10.1007/s10265-006-0049-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kitakura, S., Fujita, T., Ueno, Y., Terakura, S., Wabiko, H., and Machida, Y. (2002). The protein encoded by oncogene 6b from Agrobacterium tumefaciens interacts with a nuclear protein of tobacco. Plant Cell 14, 451–463. doi: 10.1105/tpc.010360
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kitakura, S., Terakura, S., Yoshioka, Y., and Machida, Y. (2008). Interaction between Agrobacterium tumefaciens oncoprotein 6b and a tobacco nucleolar protein that is homologous to TNP1 encoded by a transposable element of Antirrhinum majus. J. Plant Res. 121, 425–433. doi: 10.1007/s10265-008-0160-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kojima, S., Iwasaki, M., Takahashi, H., Imai, T., Matsumura, Y., Fleury, D.,et al. (2011). Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52, 1259–1273. doi: 10.1093/pcp/pcr083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuzuhara, T., and Horikoshi, M. (2004). A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 11, 275–283. doi: 10.1038/nsmb733
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lemcke, K., and Schmülling, T. (1998). Gain of function assays identity non-rol genes from Agrobacterium rhizogenes TL-DNA that alter plant morphogenesis or hormone sensitivity. Plant J. 15, 423–433. doi: 10.1046/j.1365-313X.1998.00223.x
Levesque, H., Delepelaire, P., Rouze, P., Slightom, J., and Tepfer, D. (1988). Common evolutionaly origin of the central portions of the Ri TL-DNA of Agrobacterium rhizogenes and the Ti T-DNAs of Agrobacterium tumefaciens. Plant Mol. Biol. 11, 731–744. doi: 10.1007/BF00019514
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loyola, A., and Almouzni, G. (2004). Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677, 3–11. doi: 10.1016/j.bbaexp.2003.09.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McConnell, J. R., and Barton, M. K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942.
Mohajjel-Shoja, H., Clément, B., Perot, J., Aliua, M., and Otten, L. (2011). Biological activity of the Agrobacterium rhizogenes-derived trolC gene of Nicotiana tabacum and its functional relation to other plast genes. Mol. Plant Microbe Interact. 24, 44–53. doi: 10.1094/MPMI-06-10-0139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moriuchi, H., Okamoto, C., Nishihama, R., Yamashita, I., Machida, Y., and Tanaka, N. (2004). Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J. 38, 260–275. doi: 10.1111/j.1365-313X.2004.02041.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mosammaparast, N., Ewart, C. S., and Pemberton, L. F. (2002). A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538. doi: 10.1093/emboj/cdf647
Munakata, T., Adachi, N., Yokoyama, N., Kuzuhara, T., and Horikoshi, M. (2000). A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5, 221–233. doi: 10.1046/j.1365-2443.2000.00319.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nacken, W. K., Piotrowiak, R., Saedler, H., and Sommer, H. (1991). The transposable element Tam1 from Antirrhium majus shows structural homology to the maize transposon En/Spm and has no sequences specificity of insertion. Mol. Gen. Gent. 228, 201–208. doi: 10.1007/BF00282466
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okuwaki, M., Matsumoto, K., Tsujimoto, M., and Nagata, K. (2001). Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506, 272–276. doi: 10.1016/S0014-5793(01)02939-8
Otten, L., and De Ruffray, P. (1994). Agrobacterium vitis nopaline Ti plasmid pTiAB4: relationship to other Ti plasmids and T-DNA structure. Mol. Gen. Genet. 245, 493–505. doi: 10.1007/BF00302262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Polo, S. E., and Almouzni, G. (2006). Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 16, 104–111. doi: 10.1016/j.gde.2006.02.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ray-Gallet, D., Quivy, J. P., Scamps, C., Martini, E. M., Lipinski, M., and Almouzni, G. (2002). HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100. doi: 10.1016/S1097-2765(02)00526-9
Schermer, U. J., Korber, P., and Horz, W. (2005). Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19, 279–285. doi: 10.1016/j.molcel.2005.05.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Skene, P. J., and Henikoff, S. (2013). Histone variants in pluripotency and disease. Development 140, 2513–2524. doi: 10.1242/dev.091439
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, S., and Stillman, B. (1989). Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25. doi: 10.1016/0092-8674(89)90398-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spanier, K., Schell, J., and Schreier, P. H. (1989). A functional analysis of T-DNA gene 6b: the fine tuning of cytokinin effects on shoot development. Mol. Gen. Genet. 219, 209–216. doi: 10.1007/BF00261179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spena, A., Schumülling, T., Koncz, C., and Schell, J. S. (1987). Independent and synergistic activity of rol A, B and C loci in stimulating abnormal growth in plants. EMBO J. 6, 3891–3899.
Stieger, P. A., Meyer, A. D., Kathmann, P., Frundt, C., Niederhauser, I., Barone, M.,et al. (2004). The orf13 T-DNA gene of Agrobacterium rhizogenes confers meristematic competence to differentiated cells. Plant Physiol. 135, 1798–1808. doi: 10.1104/pp.104.040899
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takahashi, S., Sato, R., Takahashi, M., Hashiba, N., Ogawa, A., Toyofuku, K.,et al. (2013). Ectopic localization of auxin and cytokinin in tobacco seedlings by the plant-oncogenic AK-6b gene of Agrobacterium tumefaciens AKE10. Planta 238, 753–770. doi: 10.1007/s00425-013-1930-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terakura, S., Kitakura, S., Ishikawa, M., Ueno, Y., Fujita, T., Machida, C.,et al. (2006). Oncogene 6b from Agrobacterium tumefaciens induces abaxial cell division at late stages of leaf development and modifies vascular development in petioles. Plant Cell Physiol. 47, 664–672. doi: 10.1093/pcp/pcj036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terakura, S., Ueno, Y., Tagami, H., Kitakura, S., Machida, C., Wabiko, H.,et al. (2007). An oncoprotein from the plant pathogen Agrobacterium has histone chaperone-like activity. Plant Cell 19, 2855–2865. doi: 10.1105/tpc.106.049551
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tinland, B., Fournier, P., Heckel, T., and Otten, L. (1992). Expression of a chimaeric heat-shock-inducible Agrobacterium 6b oncogene in Nicotiana rustica. Plant Mol. Biol. 18, 921–930. doi: 10.1007/BF00019206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tinland, B., Huss, B., Paulus, F., Bonnard, G., and Otten, L. (1989). Agrobacterium tumefaciens 6b genes are strain specific and affect the activity of auxin as well as cytokinin genes. Mol. Gen. Genet. 219, 217–224. doi: 10.1007/BF00261180
Tinland, B., Rohfritsch, O., Michler, P., and Otten, L. (1990). Agrobacterium tumefaciens T-DNA gene 6b stimulates rol-induced root formation, permits growth at high auxin concentrations and increases root size. Mol. Gen. Genet. 223, 1–10. doi: 10.1007/BF00315790
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ueno, Y., Ishikawa, T., Watanabe, K., Terakura, S., Iwakawa, H., Okada, K.,et al. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19, 445–457. doi: 10.1105/tpc.106.042325
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wabiko, H., and Minemura, M. (1996). Exogenous phytohormone-independent growth and regeneration of tobacco plants transgenic for the 6b gene of Agrobacterium tumefaciens AKE10. Plant Physiol. 112, 939–951. doi: 10.1104/pp.112.3.939
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waites, R., and Hudson, A. (1995). Phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154.
Wang, M., Soyano, T., Machida, S., Yang, J. Y., Jung, C., Chua, N. H.,et al. (2011). Molecular insights into plant cell proliferation disturbance by Agrobacterium protein 6b. Gene Dev. 25, 64–76. doi: 10.1101/gad.1985511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Willmitzer, L., Dhaese, P., Schreier, P. H., Schmalenbach, W., Van Montagu, M., and Schell, J. (1983). Size, location and polarity of T-DNA-encoded transcripts in nopaline crown gall tumors; common transcripts in octopine and nopaline tumors. Cell 32, 1045–1056. doi: 10.1016/0092-8674(83)90289-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zilberman, D., and Henikoff, S. (2005). Epigenetic inheritance in Arabidopsis: selective silence. Curr. Opin. Genet. Dev. 15, 557–562. doi: 10.1016/j.gde.2005.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: tumorigenesis, oncogene 6b, meristematic states, cell division potential, reprogramming of plant cells
Citation: Ishibashi N, Kitakura S, Terakura S, Machida C and Machida Y (2014) Protein encoded by oncogene 6b from Agrobacterium tumefaciens has a reprogramming potential and histone chaperone-like activity. Front. Plant Sci. 5:572. doi: 10.3389/fpls.2014.00572
Received: 29 August 2014; Accepted: 05 October 2014;
Published online: 28 October 2014.
Edited by:
Toshihiko Komari, Japan Tobacco Inc., JapanReviewed by:
Gong-yin Ye, Zhejiang University, ChinaToshihiko Komari, Japan Tobacco Inc., Japan
Nobukazu Tanaka, Hiroshima University, Japan
Copyright © 2014 Ishibashi, Kitakura, Terakura, Machida and Machida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasunori Machida, Division of Biological Science, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya , Aichi 464-8602, Japan e-mail:eWFzQGJpby5uYWdveWEtdS5hYy5qcA==
 Nanako Ishibashi
Nanako Ishibashi Saeko Kitakura
Saeko Kitakura Shinji Terakura1
Shinji Terakura1 Yasunori Machida
Yasunori Machida