- 1School of Agricultural and Wine Sciences, Faculty of Science, Charles Sturt University, Wagga Wagga, NSW, Australia
- 2Graham Centre for Agricultural Innovation, Charles Sturt University, Wagga Wagga, NSW, Australia
- 3Faculty of Science, Charles Sturt University, Wagga Wagga, NSW, Australia
- 4New South Wales Department of Primary Industries, Wagga Wagga, NSW, Australia
Allelopathy is one crop attribute that could be incorporated in an integrated weed management system as a supplement to synthetic herbicides. However, the underlying principles of crop allelopathy and secondary metabolite production are still poorly understood including in canola. In this study, an allelopathic bioassay and a metabolomic analysis were conducted to compare three non-allelopathic and three allelopathic canola genotypes. Results from the laboratory bioassay showed that there were significant differences among canola genotypes in their ability to inhibit root and shoot growth of the receiver annual ryegrass; impacts ranged from 14% (cv. Atr-409) to 76% (cv. Pak85388-502) and 0% (cv. Atr-409) to 45% (cv. Pak85388-502) inhibition respectively. The root length of canola also differed significantly between genotypes, there being a non-significant negative interaction (r = -0.71; y = 0.303x + 21.33) between the root length of donor canola and of receiver annual ryegrass. Variation in chemical composition was detected between organs (root extracts, shoot extracts) and root exudates and also between canola genotypes. Root extracts contained more secondary metabolites than shoot extracts while fewer compounds were recorded in the root exudates. Individual compound assessments identified a total of 14 secondary metabolites which were identified from the six tested genotypes. However, only Pak85388-502 and Av-opal exuded sinapyl alcohol, p-hydroxybenzoic acid and 3,5,6,7,8-pentahydroxy flavones in agar growth medium, suggesting that the synergistic effect of these compounds playing a role for canola allelopathy against annual ryegrass in vitro.
Introduction
Weed control options for canola in Australia have been improved considerably with the development of a wide range of herbicide–tolerant cultivars with resistance to triazine, imidazolinone or glyphosate herbicides. The implementation of glyphosate-tolerant canola has changed the pattern of herbicide use, decreasing the use of other herbicides, and has given growers an efficient and simple solution for weed control worldwide (Harker et al., 2000; Beckie et al., 2011). Unfortunately, the use of herbicides in herbicide-tolerant canola cultivars has encouraged weeds to evolve herbicide-resistance (Powles et al., 1998; Heap, 2002). The ubiquitious weed annual ryegrass (Lolium rigidum L.) has already shown resistance to glyphosate in Australia (Pratley et al., 1999). Thus, herbicide resistance of weeds is a major threat to sustainable crop production. Consequently, alternatives to conventional synthetic herbicide application have become a focus of much research in Australia and worldwide. The potential use of crop allelopathy as part of a weed control program is one option gaining attention of the researchers (Kathiresan, 2005).
Rice (1984) defined allelopathy as the direct or indirect (harmful or beneficial) effect of a plant, and microbes, on another plant through the release of compounds into the environment. Allelochemicals have usually been considered to be secondary metabolites or waste products of the main metabolic pathways in plants (Swain, 1977) and released via several mechanisms (Seigler, 1996; Singh et al., 2003; Weston and Duke, 2003) including leaching (by dew and rain), residue decomposition (Putnam and DeFrank, 1983; Purvis et al., 1985) and exudation from living plants (Rice, 1984; Blum, 2011; Thorpe et al., 2011). Furthermore, the production and the release of biologically active compounds differ between species and between cultivars (Jeffery et al., 2003; Bennett et al., 2006; Keurentjes et al., 2006; Abdel-Farid et al., 2007), although relatively few have strong allelopathic properties (Bhowmik and Inderjit, 2003; Khanh et al., 2005; Xuan et al., 2005). The potential role of crop allelopathy in weed control has been the focus of much research and has been extensively reviewed (e.g., Einhellig and Leather, 1988; Purvis, 1990; Wu et al., 1999). Results from allelopathic assessment of canola cultivars against weeds in vitro and under field condition showed that canola allelopathy is genetically controlled (Asaduzzaman et al., 2014a,b). Canola allelopathy also seems to be independent from the competitive traits in the above ground morphology growth and phenology of the crop (Asaduzzaman et al., 2014c,d). However, there are no reports that holistically analyze the canola allelochemicals complex.
Plant secondary metabolites are generally present in plant tissue but few are exuded into the environment (Weston and Duke, 2003; Badri and Vivanco, 2009). To establish the involvement of any root exudates in crop plant allelopathy, it is important to demonstrate their phytotoxic effect by direct release to the growth medium (Inderjit, 1996). The exudation of allelochemicals by plant roots is an active metabolic process (Overland, 1966) and seems to be universal in the plant kingdom (Martin, 1957; Fay and Duke, 1977; Abdul-Rahman and Habib, 1989; Einhellig and Souza, 1992). Brassicaceae plants possess several groups of secondary metabolites including phenylpropanoids (hydroxycinnamates), flavonoids, as well as Brassicaceae-specific metabolites such as glucosinolates. The characterisation of these phytochemicals between strong and weak allelopathic cultivars is very important, as it will help to understand the chemical basis of canola allelopathy. Appropriate advanced tools, such as metabolomics, can be used for identifying and characterizing the potential metabolites responsible for the allelopathic defenses recently demonstrated in canola (Asaduzzaman et al., 2014a,b).
Metabolomics is an approach that allows a biochemical analysis of the total metabolite complement of a given plant tissue (Rinu et al., 2005; Kim et al., 2011). It is being used as an important procedure for identifying compounds involved in allelopathic interactions (D’Abrosca et al., 2013). Through mass spectral (MS) analysis of metabolomes in plant organs and principal component analysis (PCA), relative variability between organs can be explored. In addition, due to complex interactions, the field assessment of crop allelopathy is challenging (Inderjit and del Moral, 1997; Olofsdotter et al., 1999; Inderjit and Weston, 2000; Bertin et al., 2003; Bais et al., 2006) and difficult to separate from competition (Olofsdotter et al., 1999). Hence, laboratory screening of crop cultivars, coupled with advanced multivariate statistical analysis of metabolomes, offers new insights into the subterranean biology of plant allelopathy (Rinu et al., 2005).
The present research aimed to determine the metabolite composition of different organs (namely shoot, root) and root exudates of canola by using time-of-flight (TOF–MS) analysis technique and to establish a platform for understanding canola allelopathy.
Materials and Methods
Plant materials
Six canola (Brassica napus, rapeseed, oilseed rape) genotypes were selected for this study namely: Av-opal, Pak85388-502, Av-garnet, Barossa, Cb-argyle and Atr-409. Previous field and in vitro screening results showed that Av-opal and Pak85388-502 were strongly allelopathic against annual ryegrass in vitro, and against the background weed populations (over 2 years: 2012 and 2013) under field conditions, whereas, Atr-409 and Barossa were weakly allelopathic genotypes (Asaduzzaman et al., 2014a,b). Two other genotypes were chosen based on a previous canola competitiveness field study conducted by Lemerle et al. (2014): Av-garnet was reported to be strongly competitive and Cb-argyle weakly competitive on weed species and associated total weed biomass. Seeds of these canola genotypes were obtained from the National Brassica Germplasm Improvement Program, located at NSW Department of Primary Industries, Wagga Wagga, NSW, Australia. Agar (technical grade) was purchased from Sigma Aldrich (St. Louis, MO, USA).
Sterilization and germination
Canola seeds were surface-sterilized by soaking in 2% sodium hypochlorite (NaOCl) for 5 min, then rinsed six times in sterilized distilled water. The seeds were transferred to a petri dish with one sheet of Whatman No. 1 filter paper, moistened with 5 ml sterilized distilled water, and sealed with parafilm. The surface-sterilized seeds of Brassica and ryegrass were kept in a 12-h light/12-h dark, 20/15°C controlled environment for 36 h and 48 h respectively.
General bioassay and growing conditions
The equal-compartment-agar-method (ECAM), described previously by Wu et al. (2000a) was chosen for bioassay. The method was developed based on the plant box method and relay seedling technique and separates competition and allelopathy phenomena between two simultaneously growing species. In this method, each species was placed into separate regions in the same container, where each species received equal space for its root system development. Briefly, glass beakers (600 mL, 12 cm depth, 8 cm diameter) containing 30 mL of 0.3% agar-medium (no nutrients, 1.3 cm depth) were autoclaved. The previous bioassay of 70 canola genotypes showed that 30 seedlings/beaker allelopathically gave greatest inhibition of the root length of annual ryegrass (Asaduzzaman et al., 2014a). Hence for each genotype, 30 uniform seedlings per beaker were chosen and aseptically transplanted from the germination dish onto one half of the agar surface, with the embryo up. The beaker tops were sealed with parafilm to prevent contamination and evaporation from the agar surface, and the beakers were placed in a controlled growth incubator with a daily 12-h light/12-h dark, 20/15°C cycle. Canola plants were grown for 6 days, 15 pre-germinated uniform seeds of annual ryegrass were aseptically sown on the other half of the agar surface at a distance of 4 cm from the canola seedlings. A piece of pre-autoclaved white paperboard was inserted across the center and down the middle of the beaker with the lower edge of the paperboard kept 1 cm above the agar surface. The beaker was divided into two equal compartments to minimize competition for space and light between the canola and ryegrass seedlings. The roots of canola freely entered the ryegrass compartment so that any allelochemicals produced and released by the canola seedlings can diffuse throughout the entire agar medium to influence ryegrass root growth. After ryegrass sowing, the beakers were again wrapped with parafilm and placed back in the growth chamber for 7 days. The receiver species, annual ryegrass, was also grown alone as a control. After 7 days, each annual ryegrass and canola seedling was carefully removed from the agar to avoid root breakage, and the root and the shoot lengths of 10 randomly selected plants within each beaker of both species were measured.
Experimental design and statistical analysis
A randomized complete block design was used for the experiment described. For each genotype 30 replicates were used in three different experimental units (beakers). The inhibited root and shoot length of annual ryegrass (mm) was converted as percentage of control. To determine the percentage change a percent ration was calculated between the mean root/shoot length of all (n = 30) ryegrass seedlings and the root/shoot length of every singly seedling. Further, to evaluate the equivalence of shoot and root inhibition of ryegrass with root length of canola, Pearson correlation co-efficient values were calculated. A linear regression analysis (y = mx + c) was also performed between root length (mm) of canola (independent) and of annual ryegrass (dependent) to know their mutual relationship. All data were subjected to analysis of variance using Genstat v13 (VSN International, Hemel Hempstead, UK) and the treatment means compared using the least significance difference (LSD) at a 5% level of probability. Plots of residual versus fitted values were examined for all traits to ensure the normality and homogenecity.
Biochemical Analysis by Metabolomics Approach
Preparation of shoot and root extracts
Canola seedlings of each genotype were grown alone at a density of 30 seedlings/beaker for 13 days, as described in the above laboratory bioassay (see General bioassay and growing conditions). The roots and the shoots were cut from the canola seedlings and were immediately stored at -80°C in a sealed container. The frozen tissue was then freeze-dried for 24 h (Alpha 2–4 LD plus; John Morris). To extract metabolites, the freeze-dried tissue was then crushed to a fine powder using liquid nitrogen-chilled mortar and pestle. Sixty mg of the root and the shoot tissue of each canola genotype were placed separately into a 2 mL tube chilled in liquid nitrogen. The tube was filled with 400 μL 100% methanol solution containing internal standards 13C6-sorbitol (0.5 mg/mL); 13C515N-valine (0.5 mg/mL); penta-fluorobenzoic acid (0.25 mg/mL) and 2-aminoanthracene (0.25 mg/mL; Roessner and Dias, 2013). The tubes were vortexed for 30 s and centrifuged for 15 min at 13000 rpm at 4°C. The supernatant was transferred to a new pre-labeled 2 mL tube. An amount of 400 μL MQ water was added to the remaining pellet and vortexed, centrifuged and the supernatant was combined with the previous methanol containing supernatant. Three aliquots of each tissue containing 650 μL were prepared and stored at -80°C until analysis.
Collection of root exudates
Canola seedlings were carefully uprooted from their nutrient-free agar medium and the roots were rinsed twice with 5 mL portions of distilled water to remove any adhering agar and root exudates. The washings were pooled with the agar medium (30 mL). The agar medium was stirred carefully and extracted three times using 5 mL of 80% methanol. The extracted samples were vortexed and centrifuged and filtered through a 0.22 μm syringe filter into 2mL labeled tubes. Three aliquots of 650 μL of each genotype were prepared and stored at -80°C before analysis.
Metabolites profiling by LC-QTOF-MS
To assess the metabolite composition differences among the organs and root exudates of canola genotypes, non-targeted and targeted metabolite profiling of extracted material was conducted. The compounds of canola shoots, roots extracts, and root exudates were separated on an Agilent 6520 LC-QTOF-MS system (Santa Clara, CA, USA, Agilent Mass Hunter Qualitative Analysis Build 6.0), with a dual sprayer ESI source, and attached to an Agilent 1200 series HPLC system (Santa Clara, CA, USA) consisting of a vacuum degasser, binary pump, with a thermo stated auto-sampler, column compartment, and diode array detector. The MS was operated in the negative mode using the following conditions: nebuliser pressure 45 psi, gas flow-rate 10 L/min, gas temperature 300°C, capillary voltage 3500V, fragmentor 150 and skimmer 65V. The instrument was operated in the extended dynamic range mode with data collected in mass-to-charge ratio (m/z), range 70–1700 amu.
Chromatography
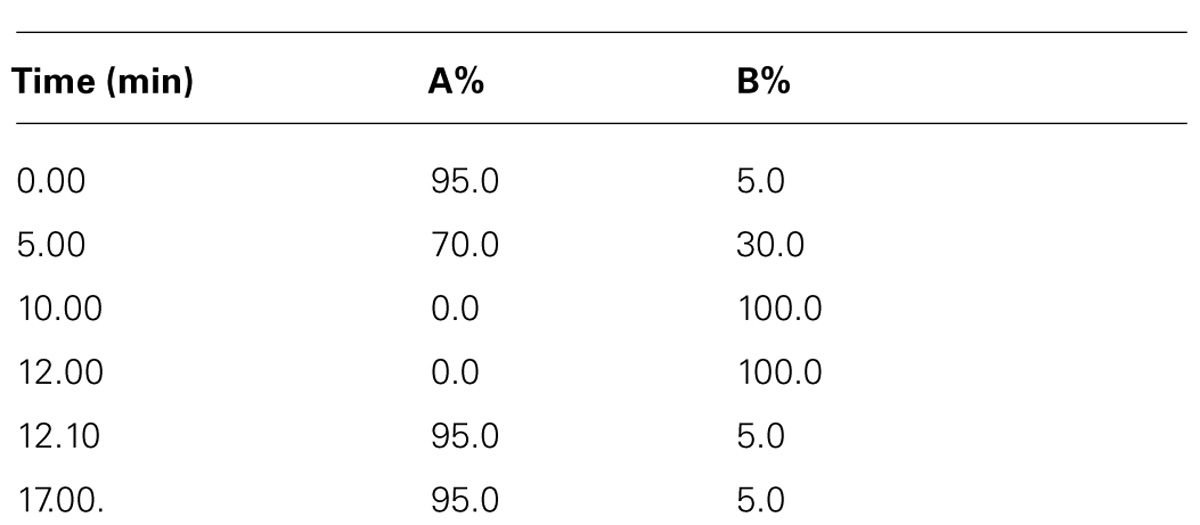
An Agilent Zorbax Eclipse XDB-C18, 2.1 × 100 mm, 1.8 μm (Agilent) column was used with a flow rate of 400 μL/min maintained at ambient temperature (35 ± 1°C), resulting in operating pressures below 600 bar with a 12 min run time. A gradient LC-QTOF-MS method (Table 1) was used with mobile phases comprised of (A) 0.1% formic acid in de-ionized water and (B) 0.1% formic acid in acetonitrile. The sample run was conducted first for the 5 min by using linear gradient from 5% solvent (B) to 30% solvent (B), followed by a 5 min linear gradient to 30% solvent (B) to 100% solvent (B), then a 2 min hold at 100% solvent (B) and a 5 min re-equilibration at 5% solvent (B). Total time = 17 min. Three replications were run for each category of samples of each genotype.
Mass spectrum data processing
Relative qualitative analyses of the metabolites in the six canola genotypes were performed using Mass Hunter data analysis software (Agilent Technologies, USA). The extracted molecular features of each detected compound were matched with two different data bases (METLIN-AM-PCDL and HMDB-KEGG), plus the mass of the reference compounds from commercial standards. The individual compounds were also determined through assessing the outcomes of score (>70), hit count (total number of hits in the database) and mass differences (<5.0).
Chemoassays Using Reference Compounds
Preparation of the different concentrations
Stock solutions (10000 μM) of sinapyl alcohol, p-hydroxybenzoic acid and 3,5,6,7, 8-pentahydroxy flavones were prepared separately. A mixture of these three compounds (10000 μM) was also made by using 1:1:1 ratio. The stock solutions of individual compounds and of their mixture were diluted to concentrations of 5000, 100, and 50 μM in HPLC-grade methanol.
Annual ryegrass bioassay with reference compounds
The modified chemical bioassay described by Seal et al. (2004b) was used to evaluate the phytotoxic effects of three reference compounds on annual ryegrass. One milliliter of each of the above concentrations (50, 100, 5000 and 10000 μM) was added to 600 ml beakers lined with Whatman #1 filter paper (Micro science, grade: MS 2 85 mm, size: 85 mm, Quality: 100) at the base. For the control, 1 ml of pure methanol was added. After the methanol had completely evaporated using the method described by Seal et al. (2004b), 5 ml of sterile double distilled water was added. Ten annual ryegrass seeds were sown directly into the water and the beaker was covered with parafilm. Three replicates of each treatment were arranged in a randomized complete block design in a growth chamber described in “General bioassay and growing conditions.” 7 days later the annual ryegrass seedlings were removed from the system and both their root and shoot lengths were measured to the nearest 0.5 mm.
Statistical analysis
All dose-response curves were subjected to two-way ANOVA using Genstat v13 (VSN International, Hemel Hempstead, UK). Annual ryegrass root length (mm) was converted as percentage of control as described in “Experimental design and statistical analysis.” The treatment means were compared using the LSD at a 5% level of probability. Plots of residual versus fitted values were examined for all traits to ensure that the assumptions of analysis of variance were met.
Results
Laboratory Bioassay
Genotypes differed significantly (P < 0.001) in their ability to suppress the root and the shoot growth of annual ryegrass (Figure 1). Genotypes Atr-409, Cb-argyl and Barossa showed less inhibitory effects on annual ryegrass while Av-opal, Pak85388-502 and Av-garnet were more inhibitive. In all collections, root growth (14–76%) of annual ryegrass was inhibited more than shoot growth (0–15%). The most suppressive genotype Pak85388-502 resulted in 76% root growth control of annual ryegrass followed by genotype Av-opal (74%) and Av-garnet (46%). The weakest genotype cv. Atr-409 inhibited the root length of annual ryegrass by only about 14%.
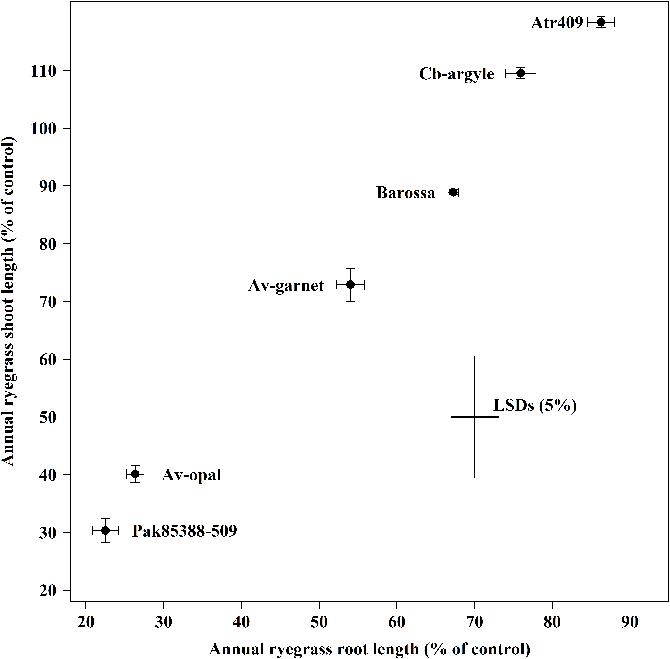
FIGURE 1. Laboratory bioassays (ECAM) of canola (Brassica napus) seedling allelopathy against annual ryegrass (Lolium rigidum) seedlings. Data shown are the means (n = 30; ±SE) of the root and the shoot length (% of control) of annual ryegrass seedlings. Data were pooled of three experimental units (beakers). The cross indicates significant difference between the genotypes within two variables (root and shoot; P < 0.05, ANOVA with post hoc Fisher-LSD-test). The Pearson correlation coefficient (r) is 0.99 (P < 0.001).
The average root length of canola seedlings differed significantly (P < 0.001) between genotypes (Figure 2). Genotypes Av-opal and Pak85388-502 produced the longest root; in contrast Cb-argyle and Atr-409 produced the shortest roots. The regression analysis (r = -0.71; y = 0.303x + 21.33) showed that annual ryegrass root growth (mm) was not decreased (P > 0.05) with increased canola root growth (mm).
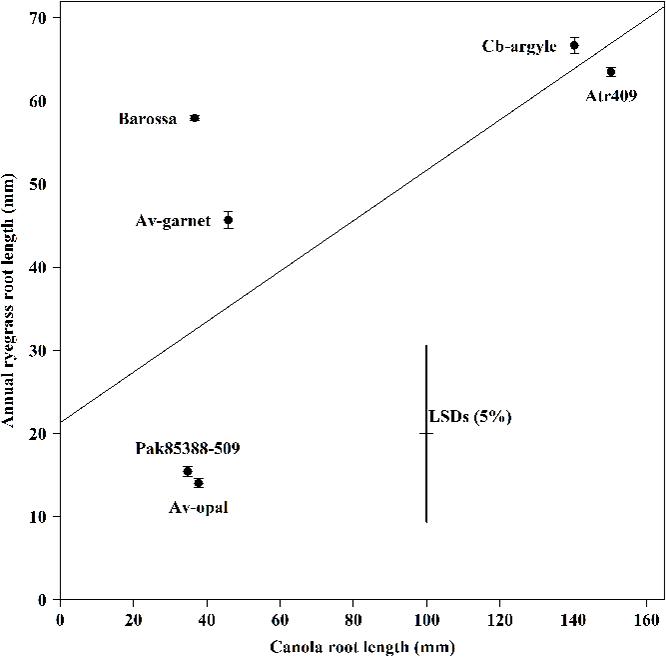
FIGURE 2. The root length of canola (Brassica napus) and annual ryegrass (Lolium rigidum) seedlings when grown together in the ECAM bioassay. Data shown are means (n = 30;±SE) of the root length (mm) of annual ryegrass seedlings and of the root length (mm) of canola seedlings. Data were pooled of three experimental units (beakers). The cross indicates significant differences between the genotypes within one variable (P < 0.05, ANOVA with post hoc Fisher-LSD-test). The Pearson correlation coefficient (r) is -071 (P > 0.05) and were determined between the two variables across all genotypes and the regression equation is y = 0.303x + 21.33, with R2 = 0.50, P > 0.05).
Metabolite Profiling
The different metabolite patterns were observed by simple visual inspection of the MS traces of the three different organs. A total of 2806 mass signals were recorded in three different sample types. The number of metabolites in the root and the shoot extracts varied between genotypes. Metabolites were highly enriched in root extracts followed by shoot extracts and root exudates (Table 2). Over 1807 compounds were found in roots, with Av-opal, Pak85388-502, Barossa and Atr-409 assigned 1586, 1532, 1471 and 1525 compounds respectively.
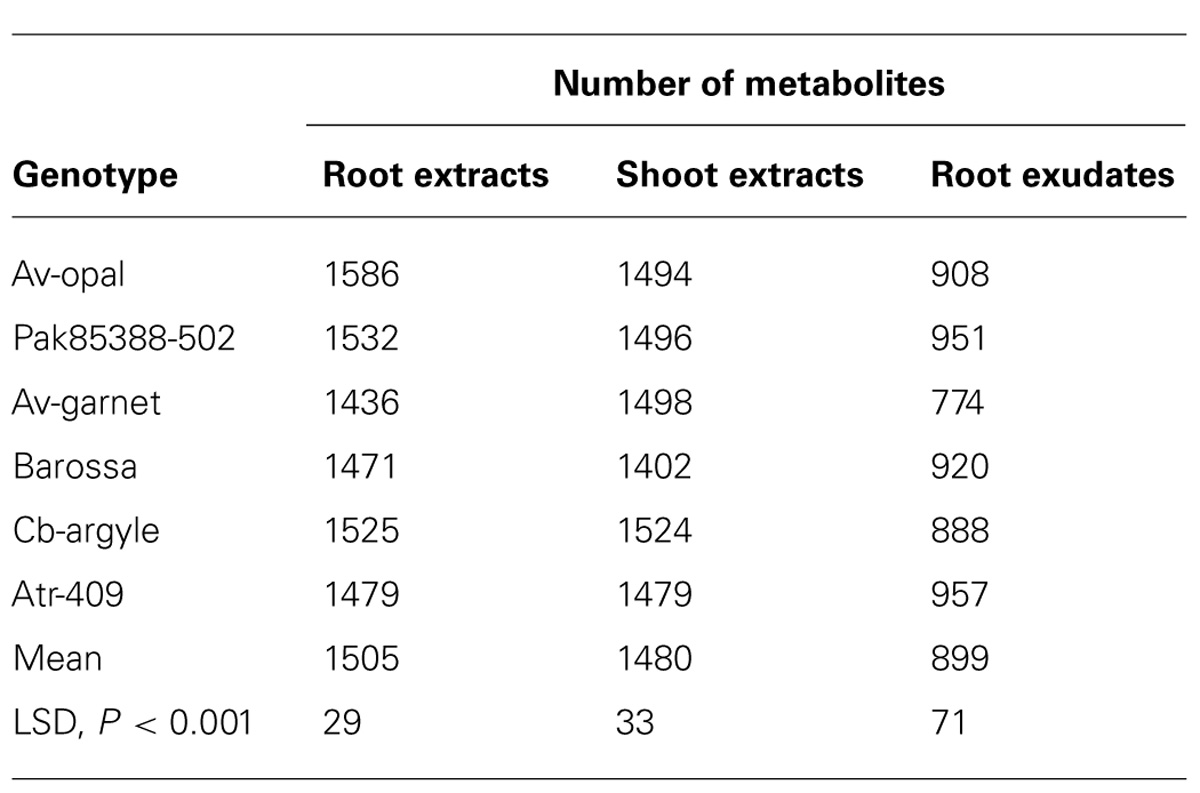
TABLE 2. Total numbers of metabolites identified in root and shoot extracts and root exudates of six canola genotypes.
Identification of Phytochemicals in Canola Genotypes
Fourteen secondary metabolites, including two internal signaling molecules, namely jasmonic acid and methyl-jasmonate, were detected across the samples of the six canola genotypes (Table 3). Only eight metabolites were identified in the root exudates.
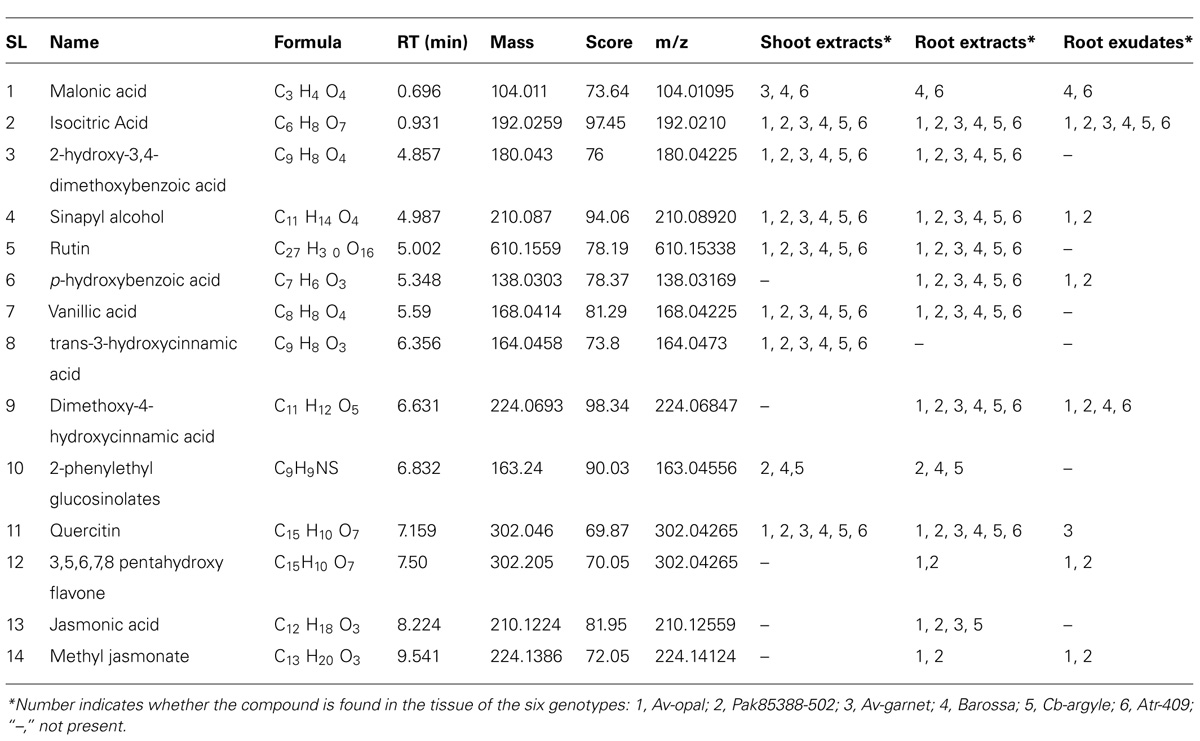
TABLE 3. Phytochemicals identified in shoot and root extracts and root exudates of six canola genotypes using LC-QTOF-MS in negative mode and matched with data from two data bases.
The three interested metabolites were only found in the root exudates of highly allelopathic genotypes (Av-opal, Pak85388-502, and possibly Av-garnet). Five metabolites (or some mixture of these) were the most likely candidates for an allelopathic effect; sinapyl alcohol, p-hydroxybenzoic acid, quercitin, 3,5,6,7,8- pentahydroxy flavones, and methyl-jasmonate. Of these five, quercitin was formed only in the exudates of Av-garnet, and sinapyl alcohol was found only in the exudates of Av-opal and Pak85388-502.
Chemoassays Using Reference Compounds
The root growth of annual ryegrass seedlings differed significantly (P < 0.001) between compounds and their concentrations (Figures 3 and 4). Among the compounds 3,5,6,7,8- pentahydroxy flavones showed greater toxicity, while sinapyl alcohol was less toxic in all tested concentrations. When all tested compounds were considered together in mixture, the root growth of ryegrass was inhibited more compared to the individual effect of each compound, even in medium concentrations. Under the mixture of three compounds at, medium-to-high concentrations (100μM–10000μM) the germination ability of most of the ryegrass seeds was restricted.
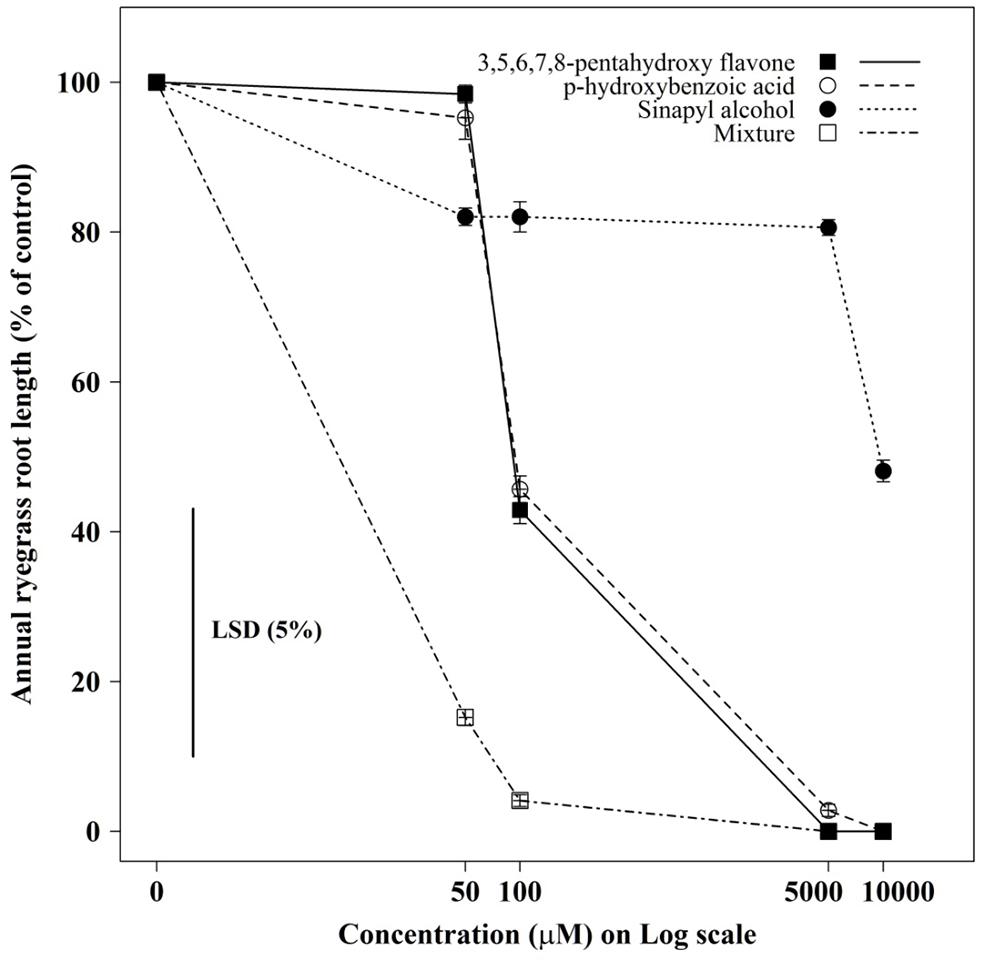
FIGURE 3. The individual, and combined, effect of sinapyl alcohol, p-hydroxybenzoic acid, and 3,5,6,7,8-pentahydroxy flavone on the root growth of annual ryegrass seedlings. Data shown are the means (n = 30; ±SE) of the root length of annual ryegrass seedlings. Data were pooled of three experimental units (beakers). The cross indicates significant difference between the genotypes × concentration interaction within one variables (P < 0.05, ANOVA with post hoc Fisher-LSD-test).
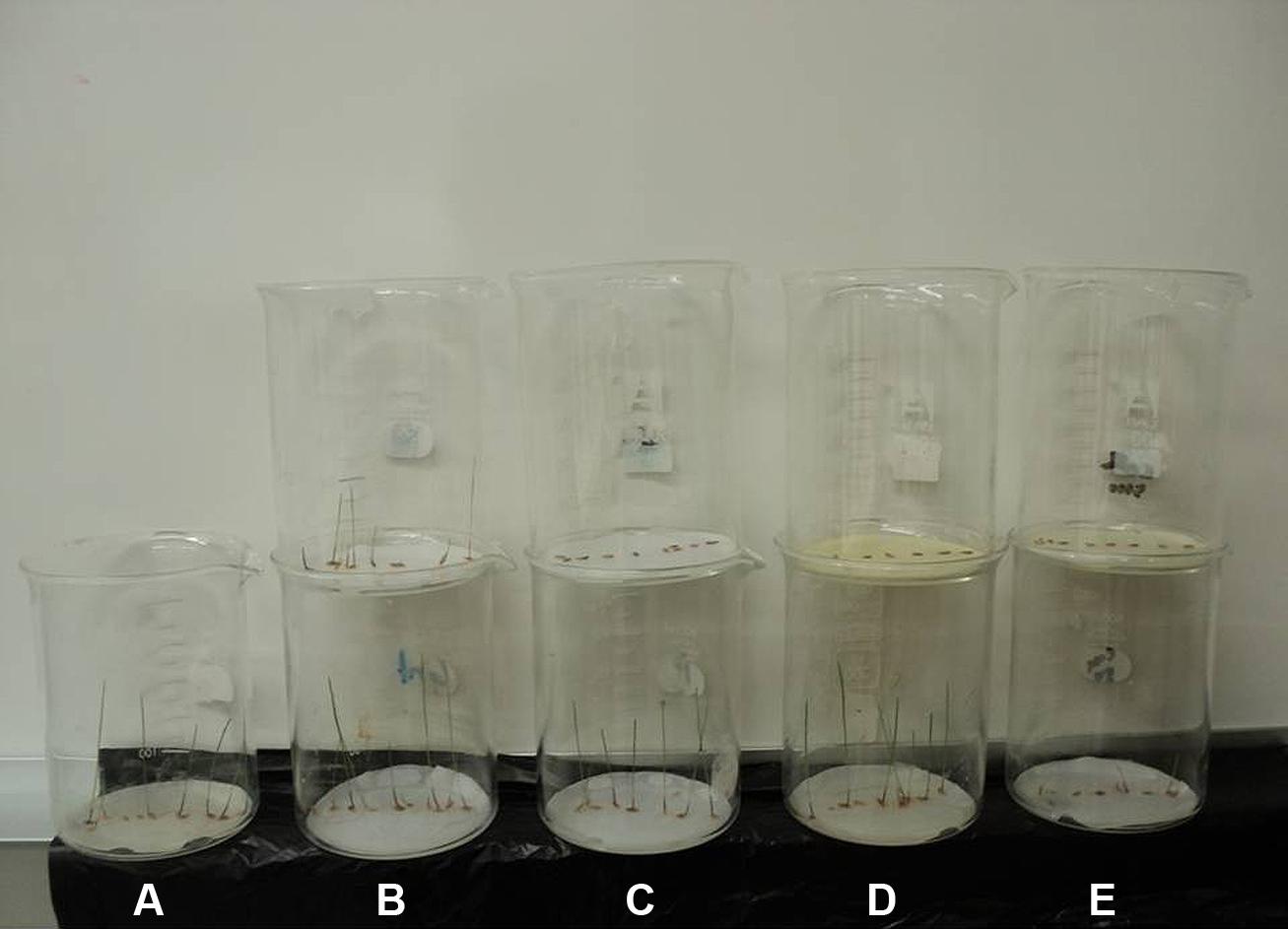
FIGURE 4. Comparison of the seedlings growth of annual ryegrass affected by (A) control, (B) sinapyl alcohol, (C) p-hydroxybenzoic acid, (D) 3,5,6,7,8-pentahydroxy flavone and (E) their mixture. The beakers in the bottom were treated with low concentration (50 μM), while beakers on the top were treated with high concentration (10000 μM).
Discussion
Different inhibition activities against ryegrass seedlings were observed among the tested canola genotypes. This is in accordance with previous observations in rice (Seal et al., 2004a), wheat (Wu et al., 2000b), and rapeseed (Uremis et al., 2009), leading to the general conclusion that allelopathy is genetically controlled. The most allelopathic genotypes in this study were Av-opal and Pak85388-502, then competitive genotype Av-garnet. This suggests that root exudation from Av-opal and Pak85388-502 might also have played a significant role for its allelopathic activity in the bioassay. These two genotypes were previously characterized as highly allelopathic in vitro testing (Asaduzzaman et al., 2014a) and were also highly weed suppressive in the field (Asaduzzaman et al., 2014b).
The negative relationship between the root length of canola and annual ryegrass suggests that long roots of canola seedlings might produce more allelochemicals than short roots. Hence, despite vigorous shoot growth, Barossa and Av-garnet showed less root-exuded allelopathic activity, whereas the short vegetative growth but longer root growth of Av-opal still inhibited the root growth of annual ryegrass to a greater extent. Such findings also infer that the inhibition effects on the receiver plant were due to chemical interactions between the roots and that such chemicals were exuded into the agar by the canola roots. It seems possible that the allelopathy potential of any particular genotype depends upon firstly, the chemical composition of the root exudates, and secondly, the amount of chemical exuded which may be a function of root system length or surface area particular at later growth satge.
The biochemical analysis of canola organs and root exudates showed differences between genotypes in the production of their total metabolomes. It is to be expected that different canola genotypes will produce varying types and amounts of phytotoxic compounds since this has been shown to occur in various other crop species (Guenzi and McCalla, 1966; Fay and Duke, 1977; Wu et al., 2001; Jeffery et al., 2003; Fang et al., 2012; Farag et al., 2012). Gardiner et al. (1999) reported that the roots of rapeseed (B. napus L) contained more compounds than did the shoot. The root also contributed more to the total chemical pool for allelopathic activity (Gardiner et al., 1999). Similarly, in this study, the number of metabolites was generally higher in the root than in the shoot and in root exudates. Allelopathic research findings have also revealed that the allelochemical concentrations were higher in the roots than in the shoots of wheat (Wu et al., 2001). It is not clear whether the higher amounts of these allelochemicals in the roots result from their direct synthesis in situ, from their translocation from the shoots to the roots, or both. The presence of chemicals in the root exudates does not infer that they play any role in the observed phytotoxicity. However, it suggests that roots and shoots contain many compounds but only some are released as root exudates, depending upon particular conditions in the rhizosphere (Badri and Vivanco, 2009).
In previous Brassica allelopathy research, glucosinolates and their derivatives were proposed as potential allelochemicals of the crop’s residue (Gardiner et al., 1999). These compounds were detected only in the root and the shoot extracts of three genotypes in this study. Possibilities for their non-detection in root exudates include: they remained locked inside the vacuole of fresh tissue of living plant; or they could not be detected due to their complex volatile nature. Glucosinolates were not detected in the root exudates from living tissue of any genotypes showing high allelopathy in our study. Therefore it seems unlikely that they are responsible for allelopathy. This conclusion is most striking when comparing the consistent results from the three replications of the tested genotypes, including Av-opal and Pak85388-502. Both are highly allelopathic but Av-opal is low in glucosinolates in the seed while Pak85388-502 is high in glucosinolates in the seed (Asaduzzaman et al., 2014b). Glucosinolates and their breakdown products are significant in the phytotoxic effects observed for canola stubble and stubble leachates after harvest (Boydston and Hang, 1995; Brown and Morra, 1996; Al-Khatib et al., 1997). It may be that senescence (aging) and fallen leaves may make a contribution to weed suppression during the life cycle of the crop but this has not been specially recorded. The cut and green manure rapeseed suppressed weeds (Boydston and Hang, 1995) but this may be due to physical smothering rather than chemical effects.
Several potential allelopathic compounds were found in the root and the shoot tissue in this study but were not detected in root exudates. This suggests that the expression of the allelopathic effect not only depends on particular compounds being synthesized but also on the ability of the genotypes to actively exude these into their rhizosphere. For instance, Barossa and Atr-409, the two weakly allelopathic genotypes, contained potential phytotoxic metabolites in the roots and the shoots but their inhibitory effect on annual ryegrass was weak. Dicarboxylic malonic acid was found only in the root exudates of these two weakly allelopathic genotypes and this compound may act as a buffering agent to reduce the threshold levels of other potential allelochemicals in the rhizosphere. Similar results have been also reported in rice (Seal et al., 2004b), where the amounts of dicarboxylic acids was high in root exudates of non-allelopathic rice cultivars.
Sinapyl alcohol, p-hydroxybenzoic acid and 3,5,6,7,8-pentahydroxy flavone were isolated from root exudates of the two strongly allelopathic canola genotypes, suggesting that they were at least partly responsible for the observed allelopathic activity. The detection of two signal molecules (jasmonic acid and methyl-jasmonate) in the allelopathic genotypes also supports the proposition that they are also involved in canola allelopathy. Jasmonic acid and methyl-jasmonate act as secondary messengers in signal transduction events in the cell and have inhibitory effects on many plant physiological processes (Sembdner and Parthier, 1993). Abdel-Farid et al. (2007) reported that the accumulation of these signal molecules is connected with demand or synthesis of the secondary metabolites sinapyl alcohol and p-hydroxy benzoic acid in Brassica rapa. Furthermore, 3,5,6,7,8- pentahydroxyflavone was also detected previously in root exudates of another member of the Brassicaceae, Brassica alba (Ponce et al., 2004). p-hydroxybenzoic acid has been reported as a potential allelochemical in other crops including, Glycine max (Barkosky and Einhellig, 1993), Camelina alyssum (Grummer and Beyer, 1960), and several members of the genus Althaea (Gude and Bieganowski, 1990). Some of the reduction in root and coleoptile growth of wheat seedlings caused by wild oat (Avena fatua) root exudates is attributed to this compound (Perez and Ormeno-Nunez, 1991).
It has been postulated that allelopathic effects are most likely due to the combination and interaction of a complex mixture of compounds (Rizvi and Rizvi, 1992; An et al., 2001). The chemobioassay results of the present study revealed that, the allelopathic activity of canola cultivars resulted from the synergistic effects of sinapyl alcohol, p-hydroxybenzoic acid and 3,5,6,7,8-pentahydroxy flavones. It is possible that multiple compounds present at low concentrations can have pronounced allelopathic effects through their joint action, though evidence for this elusive. Joint allelopathic interactions between compounds have also been reported in several tested species including rice (Chou et al., 1991; Seal et al., 2004b) and vulpia (An et al., 2001).
The phytotoxicity observed among the tested canola genotypes indicates that allelopathy plays a role in inhibiting the annual ryegrass weed species. Field experiments (Asaduzzaman et al., 2014b) support this conclusion. The comprehensive chemical analysis reported here revealed that sinapyl alcohol, p-hydroxybenzoic acid and 3,5,6,7,8-pentahydroxy flavones in most suppressive genotypes (cv. Av-opal and Pak85388-502) are likely allelopathic agents via root exudates in canola.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The senior author is grateful to Charles Sturt University for an International Postgraduate Research Scholarship, an Australian Postgraduate Award, and Writing Up award. We are also grateful to Metabolomics Australia, University of Melbourne for their technical supports. We thank to Nusrat Subhan for providing information of chemical standards.
References
Abdel-Farid, I. B., Kim, H. K., Choi, Y. H., and Verpoorte, R. (2007). Metabolomic characterization of Brassica rapa leaves by NMR spectroscopy. J. Agric. Food Chem. 55, 7936–7943. doi: 10.1021/jf071294b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Abdul-Rahman, A. A., and Habib, S. A. (1989). Allelopathic effects of alfalfa (Medicago sativa) on blade grass (Imperata cylindrica). J. Chem. Ecol. 15, 2289–2300. doi: 10.1007/BF01012082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Al-Khatib, K., Libbey, C., and Boydston, R. A. (1997). Weed suppression with Brassica green manure crops in green pea. Weed Sci. 45, 439–445. doi: 10.1007/BF01012082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
An, M., Pratley, J. E., and Haig, T. (2001). Phytotoxicity of Vulpia residues: III. Biological activity of identified allelochemicals from Vulpia myuros. J. Chem. Ecol. 27, 383–394. doi: 10.1023/A:1005640708047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Asaduzzaman, M., An, M., Pratley, J. E., Luckett, D. J., and Lemerle, D. (2014a). Canola (Brassica napus) germplasm shows variable allelopathic effects against annual ryegrass (Lolium rigidum). Plant Soil 380, 47–56. doi: 10.1007/s11104-014-2054-4
Asaduzzaman, M., Luckett, D. J., Cowley, R. B., Min, A., Pratley, J. E., and Lemerle, D. (2014b). Canola cultivar performance in weed-infested field plots confirms allelopathy ranking from in vitro testing. Biocontrol Sci.Technol. 24, 1394–1411. doi: 10.1080/09583157.2014.942596
Asaduzzaman, M., Luckett, D. J., Min, A., Pratley, J. E., and Lemerle, D. (2014c). Management of Paterson’s curse (Echium plantagineum) through canola interference. Paper Presented at the 19th Australasian Weeds Conference, Sep. 2014,Hobart, TAS.
Asaduzzaman, M., Pratley, J. E., An, M., Luckett, D. J., and Lemerle, D. (2014d). Canola interference for weed control. Springer Sci. Rev. 2, 63–74. doi: 10.1007/s40362-014-0022–22
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2008.01926.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bais, H. P., Weir, T. L., Perry, L. G., Gilory, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barkosky, R. R., and Einhellig, F. A. (1993). Effects of salicylic acid on plant-water relationships. J. Chem. Ecol. 19, 237–247. doi: 10.1007/BF00993692
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beckie, H. J., Harker, K. N., Legere, A., Morrison, M. J., Swartz, G. S., and Falk, K. C. (2011). GM canola: the Canadian experiance. Farm Pol. J. 8, 43–49.
Bennett, R. N., Rosa, E. A. S., Mellon, F. A., and Kroon, P. A. (2006). Ontogenic profiling of glucosinolates, flavonoids and other secondary metabolites in Eruca sative, Diplotaxis erucoides, Diplotaxis tenuifolia and Bunias orientalis. J. Agric. Food Chem. 54, 4005–4015. doi: 10.1021/jf052756t
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bertin, C., Yang, X., and Weston, L. (2003). The role of root exudates and allelochemicals in rhizosphere. Plant Soil 256, 67–83. doi: 10.1023/A:1026290508166
Bhowmik, P. C., and Inderjit. (2003). Challenges and opportunities in implementing allelopathy for natural weed management. Crop Prot. 22, 661–671. doi: 10.1016/S0261-2194(02)00242-9
Blum, U. (2011). “Phenolic acids, cover crops and weed emergence,” in Plant–Plant Allelopathic Interactions, ed. U. Blum (New York, NY: Springer), 1–7.
Boydston, R. A., and Hang, A. (1995). Rapeseed (Brassica napus) green manure crop suppresses weeds in potato (Solanum tuberosum). Weed Technol. 669–675.
Brown, P. D., and Morra, M. J. (1996). Hydrolysis products of glucosinolates in Brassica naupus tissue as inhibitors of seed germination. Plant Soil 181, 307–316. doi: 10.1007/BF00012065
Chou, C. H., Change, F. J., and Oka, H. I. (1991). Allelopathic potential of a wild rice Oryza perennis. Taiwania 36, 201–210.
D’Abrosca, B., Scognamiglio, M., Fiumano, V., Esposito, A., Choi, Y. H., Verpoorte, R.,et al. (2013). Plant bioassay to assess the effects of allelochemicals on the metabolome of the target species Aegilops geniculata by an NMR-based approach. Phytochemistry 93, 27–40. doi: 10.1016/j.phytochem.2013.03.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Einhellig, F. A., and Leather, G. A. (1988). Potential for exploiting allelopathy to enhance crop production. J. Chem. Ecol. 14, 1829–1844. doi: 10.1007/BF01013480
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Einhellig, F. A., and Souza, I. F. (1992). Phytotoxicity of sorgoleone found in grain Sorghum root exudates. J. Chem. Ecol. 18, 1–11. doi: 10.1007/BF00997160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fang, T., Reichelt, M., Hidalgo, W., Agnolet, S., and Chneider, B. (2012). Tissue-specific distribution of secondary metabolites in rapeseed (Brassica napus L.). PLoS ONE 7:e48006. doi: 10.1371/journal.pone.0048006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farag, M. A., Eldin, M. G. S., Kassem, H., and Fetouch, M. A. (2012). Metabolomics classification of Brassica napus L. tissue via UPLC-QTOF-PDA-MS and their anti-oxidant potential. Phytochem. Anal 24, 277–287. doi: 10.1002/pca.2408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fay, A. K., and Duke, W. B. (1977). An assessment of allelopathic potential in Avena germplasm. Weed Res. 25, 224–228.
Gardiner, J. B., Morra, M. J., Eberlein, C. V., Brown, P. D., and Borek, V. (1999). Allelochemicals released in soil following incoporation of rapeseed (Brassica napus) green manures. J. Agric. Food Chem. 47, 3387–3842. doi: 10.1021/jf9812679
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grummer, G., and Beyer, H. (1960). The influence exerted by species of Camelina on flax by means of toxic substances. Symp. Brit. Ecol. Soc. 26, 456–458.
Gude, J., and Bieganowski, M. L. (1990). Chromatographic investigations of phenolic acids and coumarins in the leaves and flowers of some species of the genus Althaea. J. Liq. Chromatogr. 13, 4081–4092. doi: 10.1080/01483919008049590
Guenzi, W. D., and McCalla, T. M. (1966). Phenolic acids in oats, wheat, sorghum and corn residues. Agron. J. 58, 303–304. doi: 10.2134/agronj1966.00021962005800030017x
Harker, K. N., Blackshaw, R. E., Kirkland, K. J., Derksen, D. A., and Wall, D. (2000). Herbicide-tolerant canola: weed control and yield comparisons in western Canada. Can. J. Plant Sci. 80, 647–654. doi: 10.4141/P99-149
Heap, I. (2002). Herbicide resistance– Australia versus the rest of the world. Paper Presented at 13th Australian Weeds Conference, Perth, WA.
Inderjit, and del Moral, R. (1997). Is separating resource competition form allelopathy realistic. Bot. Rev. 63, 221–230. doi: 10.1007/BF02857949
Inderjit, and Weston, L. A. (2000). Are laboratory bioassays for allelopathy suitable for prediction of field responses? J. Chem. Ecol. 26, 2111–2118. doi: 10.1023/A:1005516431969
Jeffery, E. H., Brown, A. F., Kurilich, A. C., Keck, A. S., Matusheski, N., Klein, B. P.,et al. (2003). Variation in content of bioactive components in broccoli. J. Food Compost. Anal. 16, 323–330. doi: 10.1016/S0889-1575(03)00045-0
Kathiresan, R. M. (2005). Evaluation of allelopathic plant materials for aquatic weed control. Paper Presented at 4th World Congress on Allelopathy, Wagga Wagga, NSW.
Keurentjes, J. J. B., Fu, J. Y., De Vos, R. C. H., Lommen, A., Hall, R. D., Bino, R. J.,et al. (2006). The genetics of plant metabolisms. Nat. Genet. 38, 842–849. doi: 10.1038/ng1815
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khanh, T. D., Chung, M. I., Xuan, T. D., and Tawata, S. (2005). The exploitation of crop allelopathy in sustainable agricultural production. J. Agron. Crop Sci. 191, 172–184. doi: 10.1111/j.1439-037X.2005.00172.x
Kim, H. K., Choi, Y. H., and Verpoorte, R. (2011). NMR-based metabolomics: where do we stand, where do we go? Trends Biotechnol. 29, 267–275. doi: 10.1016/j.tibtech.2011.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lemerle, D., Luckett, D. J., Lockley, P., Koetz, E., and Wu, H. (2014). Competitive ability of Australian canola (Brassica napus) genotypes for weed management. Crop Pasture Sci. 65, 1300–1310. doi: 10.1071/CP14125
Martin, P. (1957). Die abgabe von organischen verbindungen insbesondere von scopletin ans den keimwureln des hafers. Z. Bot. 45, 475–506.
Olofsdotter, M., Navarez, D., Rebulanan, M., and Streibig, J. C. (1999). Weed-suppressing rice cultivars-does allelopathy play a role? Weed Res. 39, 441–454. doi: 10.1046/j.1365-3180.1999.00159.x
Overland, P. (1966). The role of allelopathic substances in the smother crop barley. Am. J. Bot. 53, 423–442. doi: 10.2307/2440341
Perez, F. J., and Ormeno-Nunez, J. (1991). Root exudates of wild oats: allelopathic effect on spring wheat. Phytochemistry 30, 2199–2202. doi: 10.1016/0031-9422(91)83614-Q
Ponce, M. A., Scervino, J. M., Eva-Balsells, R., Ocampo, J. A., and Godeas, A. M. (2004). Flavonoids from shoots, roots and roots exudates of Brassica alba. Phytochemistry 65, 3131–3134. doi: 10.1016/j.phytochem.2004.08.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Powles, S. B., Lorraine-Colwill, D. F., Dellow, J. J., and Preston, J. (1998). Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci. 46, 604–607.
Pratley, J., Urwin, N., Stanton, R., Baines, P., Broster, J., Cullis, K.,et al. (1999). Resistance to glyphosate in Lolium rigidum. I. Bioevaluation. Weed Sci. 47, 405–411.
Purvis, C., Jessop, R., and Lovett, J. (1985). Selective regulation of germination and growth of annual weeds by crop residues. Weed Res. 25, 415–421. doi: 10.1111/j.1365-3180.1985.tb00664.x
Putnam, A. R., and DeFrank, J. (1983). Use of phytotoxic plant residues for selective weed control. Crop Prot. 2, 173–181. doi: 10.1016/0261-2194(83)90042-X
Rinu, G., Luis, A. J., and Jones, G. (2005). Advancing our understanding of allelopathy through metabolomics. Paper Presented at 4th World Congress on Allellopathy, Wagga Wagga, NSW.
Rizvi, S. J. H., and Rizvi, V. (1992). Allelopathy: Basic and Applied Aspect. London: Chapman and Hall.
Roessner, U., and Dias, D. A. (2013). “Plant tissue extraction for metabolomics,” in Metabolomics Tools for Natural Product Discovery Methods in Molecular Biology, eds U. Roessner and D. A. Dias (New York, NY: Humanna Press),21–28.
Seal, A. N., Pratley, J. E., and Haig, T. (2004a). Screening rice cultivars for allelopathic potential against arrowhead (Sagittaria montevidensis), an aquatic weed infesting Australian Riverina rice crops. Aust. J. Agric. Res. 59, 183–188. doi: 10.1071/AR03238
Seal, A. N., Haig, T., and Pratley, J. E. (2004b). Evaluation of putative allelochemicals in rice root exudates for their role in the suppression of arrowhead root growth. J. Chem. Ecol. 30, 1663–1678. doi: 10.1023/B:JOEC.0000042075.96379.71
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seigler, D. D. (1996). Chemistry and mechanisms of allelopathic interactions. Agron. J. 88, 876–885. doi: 10.2134/agronj1996.00021962003600060006x
Sembdner, G., and Parthier, B. (1993). The biochemistry and the physiological and molecular actions of jasmonates. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 569–589. doi: 10.1146/annurev.pp.44.060193.003033
Singh, H. P., Batish, D. R., and Kohli, R. K. (2003). Allelopathic interactions and allelochemicals: New possibilities for sustainable weed management. CRC Crit. Rev. Plant Sci. 22, 239–311. doi: 10.1080/713610858
Swain, T. (1977). Secondary compounds as protective agents. Annu. Rev. Plant Physiol. 28, 479–482. doi: 10.1146/annurev.pp.28.060177.002403
Thorpe, A. S., Aschehoug, E. T., Atwater, D. Z., and Callaway, R. M. (2011). Interactions among plants and evolution. J. Ecol. 99, 729–740. doi: 10.1111/j.1365-2745.2011.01802.x
Uremis, I., Arslan, M., Sangun, M. K., Uygur, V., and Isler, N. (2009). Allelopathic potential of rapeseed cultivars on germination and seedling growth of weeds. Asian J. Chem. 21, 2170–2184.
Weston, L. A., and Duke, S. O. (2003). Weed and crop allelopathy. CRC Crit. Rev. Plant Sci. 22, 367–389. doi: 10.1080/713610861
Wu, H., Pratley, J. E., Lemerle, D., and Haig, T. (1999). Crop cultivars with allelopathic capability. Weed Res. 39, 171–181. doi: 10.1046/j.1365-3180.1999.00136.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, H., Pratley, J. E., Lemerle, D., and Haig, T. (2000a). Laboratory screening for allelopathic potential of wheat (Triticum aestivum) accessions against annual ryegrass (Lolium rigidum). Aust. J. Agric. Res. 51, 259–266. doi: 10.1071/AR98183
Wu, H., Pratley, J. E., Lemerle, D., and Haig, T. (2000b). Evaluation of seedling allelopathy in 453 wheat (Triticum aestivum) accessions agasint annual ryegrass (Lolium rigidum) by the equal-compartment-agar method. Crop Pasture Sci. 51, 937–944. doi: 10.1071/AR00017
Wu, H., Pratley, J. E., Lemerle, D., Haig, T., and An, M. (2001). Distribution and exudation of allelochemicals in wheat Triticum aestivum. J. Chem. Ecol. 26, 2141–2154. doi: 10.1023/A:1005520500110
Keywords: Brassica napus, rapeseed, weed, root exudates, LC-QTOF-MS and metabolomics
Citation: Asaduzzaman M, Pratley JE, An M, Luckett DJ and Lemerle D (2015) Metabolomics differentiation of canola genotypes: toward an understanding of canola allelochemicals. Front. Plant Sci. 5:765. doi: 10.3389/fpls.2014.00765
Received: 12 August 2014; Accepted: 10 December 2014;
Published online: 09 January 2015.
Edited by:
Judy Simon, University of Konstanz, GermanyReviewed by:
Judy Simon, University of Konstanz, GermanyDoerte Randewig, University of Konstanz, Germany
Bartosz Adamczyk, Finnish Forest Research Institute (Metla), Finland
Copyright © 2015 Asaduzzaman, Pratley, An, Luckett and Lemerle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Asaduzzaman, School of Agricultural and Wine Sciences, Faculty of Science, Charles Sturt University, Boorooma Street, Wagga Wagga, NSW 2650, Australia e-mail:bWFzYWR1enphbWFuQGNzdS5lZHUuYXU=
 M. Asaduzzaman
M. Asaduzzaman James E. Pratley
James E. Pratley Min An2,3
Min An2,3