- Functional and Applied Genomics Laboratory, National Institute of Plant Genome Research, New Delhi, India
Growth hormone auxin regulates various cellular processes by altering the expression of diverse genes in plants. Among various auxin-responsive genes, GH3 genes maintain endogenous auxin homeostasis by conjugating excess of auxin with amino acids. GH3 genes have been characterized in many plant species, but not in legumes. In the present work, we identified members of GH3 gene family and analyzed their chromosomal distribution, gene structure, gene duplication and phylogenetic analysis in different legumes, including chickpea, soybean, Medicago, and Lotus. A comprehensive expression analysis in different vegetative and reproductive tissues/stages revealed that many of GH3 genes were expressed in a tissue-specific manner. Notably, chickpea CaGH3-3, soybean GmGH3-8 and -25, and Lotus LjGH3-4, -5, -9 and -18 genes were up-regulated in root, indicating their putative role in root development. In addition, chickpea CaGH3-1 and -7, and Medicago MtGH3-7, -8, and -9 were found to be highly induced under drought and/or salt stresses, suggesting their role in abiotic stress responses. We also observed the examples of differential expression pattern of duplicated GH3 genes in soybean, indicating their functional diversification. Furthermore, analyses of three-dimensional structures, active site residues and ligand preferences provided molecular insights into function of GH3 genes in legumes. The analysis presented here would help in investigation of precise function of GH3 genes in legumes during development and stress conditions.
Introduction
Auxin is an important phytohormone which regulates various aspects of plant growth and development. Most of these processes are regulated by auxin-responsive genes, namely auxin/indole-3-acetic acid (Aux/IAA), auxin-response factor (ARF), small auxin-up RNAs (SAUR) and Gretchen Hagen3 (GH3; Hagen and Guilfoyle, 2002). Auxin-responsiveness to these genes is conferred by auxin-responsive elements (AuxREs, TGTCTC) present in their promoters (Hagen et al., 1991; Li et al., 1994; Ulmasov et al., 1995; Hagen and Guilfoyle, 2002). To understand molecular mechanism of auxin action, several auxin-responsive genes have been isolated and characterized from many plant species, such as pea, soybean, tobacco, and cucumber (Hagen and Guilfoyle, 2002).
Gretchen Hagen3 family of proteins maintain auxin level by catalyzing conjugation of amino acids with indole-3-acetic acid, salicylic acid (SA), and jasmonic acid (JA; Staswick et al., 2002, 2005). The first GH3 gene was identified by Hagen et al. (1984) as an early auxin-responsive gene in soybean. Since then, a large number of GH3 homologs have been identified in numerous plant species ranging from mosses to angiosperms (Jain et al., 2006; Terol et al., 2006; Ludwig-Müller et al., 2008; Kumar et al., 2012; Yuan et al., 2013). The studies on GH3 proteins have revealed their regulatory function in plant growth, organ development, light signaling, abiotic stress tolerance, and plant defense responses (Woodward and Bartel, 2005; Park et al., 2007; Jain and Khurana, 2009; Ludwig-Muller, 2011; Du et al., 2012; Kumar et al., 2012; Yuan et al., 2013). In Arabidopsis, GH3 gene family has been classified into three groups (I–III) based on sequence similarity and substrate specificities (Staswick et al., 2002). Group I GH3 proteins of Arabidopsis are JA-amido synthetases (Staswick et al., 2002, 2005). AtGH3-11, a group I GH3 protein, was characterized based on analysis of jar1 mutant, which was insensitive to JA and was required for the formation of bioactive jasmonate JA-isoleucine (Staswick et al., 2002). A different allele of this gene (FIN219) was identified as a phytochrome A signaling component, having crucial role in photomorphogenesis (Hsieh et al., 2000). Group II GH3 proteins of Arabidopsis are involved in conjugation of IAA to various amino acids (Staswick et al., 2002, 2005). AtGH3-2 gain-of-function mutant, Ydk1-D, was shown to be responsible for short primary root, reduced lateral root number, and apical dominance (Takase et al., 2004). In another report AtGH3-6 mutant, dfl1, was shown to regulate shoot elongation and lateral root formation negatively, but positively regulate the light responses to hypocotyl length (Nakazawa et al., 2001). Some Group II GH3 proteins of rice (TLD1/OsGH3-13, OsGH3-2, and OsGH3-8) have also been characterized that conjugate IAA with aspartate or alanine (Chen et al., 2009, 2010; Zhang et al., 2009). A gain-of-function mutant of rice OsGH3-13 gene, tld1-D, resulted in increased tillers, enlarged leaf angles, dwarfism and improved drought tolerance (Zhang et al., 2009). AtGH3-12/PBS3 is the only characterized member of group III, which catalyzes the conjugation of glutamic acid (Glu) to 4-aminobenzoate and 4-hydroxybenzoate and is involved in SA signaling (Jagadeeswaran et al., 2007; Nobuta et al., 2007; Okrent et al., 2009). Recently, the crystal structure and mechanism of catalytic action of AtGH3-12 and JAR1/AtGH3-11 (Westfall et al., 2012) in Arabidopsis, and VvGH3-1 in grapevine (Peat et al., 2012) have also been reported.
Legumes are nutritionally important crop plants, which serve as a rich source of proteins and fibers. Although the first auxin-responsive gene was identified from soybean (Hagen et al., 1984), genome-wide analysis of GH3 genes in legumes is still lacking. This may be attributed to scarcity of genomic resources for legumes until recently. However, in recent years several genomic resources have been generated for various legumes. The genome and transcriptome sequences of desi and kabuli chickpea (Cicer arietinum), soybean (Glycine max), Medicago (Medicago truncatula), and Lotus (Lotus japonicus) have been published (Sato et al., 2008; Schmutz et al., 2010; Garg et al., 2011; Young et al., 2011; Jain et al., 2013; Varshney et al., 2013). The availability of genome annotation provides an opportunity for characterization of GH3 gene family in legumes, which can help in better understanding of their function in various cellular processes. The availability of crystal structures of GH3 proteins (Peat et al., 2012; Westfall et al., 2012) provides a resource to identify substrate specificity determining motifs/residues in the GH3 proteins, which can help in understanding auxin-mediated regulation of cellular processes in legumes.
Here, we performed genome-wide identification and analysis of GH3 gene family in four legume species, including chickpea, soybean, Medicago, and Lotus. We report their genomic organization, chromosomal distribution, sequence homology, and phylogenetic relationship in/among different legumes. Comprehensive gene expression analyses in various tissues/stages and abiotic stress conditions have also been performed to gain insight into their putative function. Putative promoter sequences of the GH3 genes were also analyzed for identification of cis-regulatory elements, which may be involved in various development processes and stress responses. In addition, their ligand preferences were predicted based on the protein structure and sequence analysis. These data provide a framework for further in-depth functional analyses of GH3 genes in legumes.
Material and Methods
Identification of GH3 Genes
Chickpea genome annotation was downloaded from Chickpea Genome Analysis Project (CGAP v1.0; Jain et al., 2013), soybean and Medicago genome annotations were downloaded from Phytozome (v9.01), and Lotus genome annotation was taken from miyakazusa.jp database (v2.52). A total of 19 protein sequences of GH3 family members of Arabidopsis and 13 protein sequences of rice GH3 family members were downloaded from TAIR3 and RGAP database4, respectively. The rice and Arabidopsis GH3 proteins were searched in chickpea, soybean, Medicago and Lotus proteomes individually, using BLASTP with an e-value cutoff of 1e-05. Further, the HMM profile of GH3 domain was downloaded from pfam database5 and HMMER was used to search proteomes of chickpea, soybean, Medicago, and Lotus for GH3 domain. All the tentative gene lists obtained from these two searches were combined to make a non-redundant gene list for each legume, and their protein sequences were searched in pfam database to confirm the presence of conserved GH3 domain. Using the similar strategies, we investigated the chickpea transcriptome sequence (Garg et al., 2010) as well for identification of any additional GH3 gene family member that may not be represented in chickpea genome annotation.
Sequence Analysis and Phylogenetic Tree Construction
Multiple sequence alignment of all the GH3 protein sequences of chickpea, soybean, Medicago and Lotus with Arabidopsis GH3 protein sequences was carried out using MAFFT and phylogenetic tree was constructed by UPGMA method using CLC Genomics Workbench (v4.7.2). Bootstrap analysis was performed by taking 1,000 replicates and the generated tree was viewed using FigTree (v1.3.1).
Gene Duplication Analysis
Synteny analysis was performed using Plant Genome Duplication Database6. Syntenic blocks were evaluated using Circos tool. Information about the chromosome locations was obtained from Phytozome database. Genes were regarded as segmentally duplicated if they found to be coparalogs located on duplicated blocks, as proposed by Wei et al. (2007). Tandem duplication was characterized as multiple genes of one family located within the same or neighboring intergenic region (Du et al., 2013a).
Promoter Sequence Analysis
Genomic co-ordinates of coding sequences were determined using GFF files obtained from chickpea and soybean genome annotation. The regions of 2,000 bp upstream from start codon were extracted from genomic DNA sequences. Cis-regulatory elements on both strands of promoter sequences were scanned using PLACE web server7 .
Homology Modeling
The 3-D protein structures of AtGH3-11 (Protein Data Bank code 4EPL; Westfall et al., 2012) and Vv-GH3-1 (Protein Data Bank code 4B2G; Peat et al., 2012) were downloaded from Protein Data Bank8. Phyre2 (Protein Homology/AnalogY Recognition Engine9) was used for predicting the protein structure by homology modeling under ‘intensive’ mode (Kelley and Sternberg, 2009). The protein structures modeled with >90% confidence were selected. The core of predicted protein structure or allowed area in the plot showing the preferred region for psi/phi angles pair for residues was determined through Ramachandran plot using RAMPAGE server10 and models were viewed using Chimera (V1.9). Only those structures representing >95% of residues in favored region were considered for further analysis. For substrate binding site prediction, templates and model were superimposed using MatchMaker of Chimera (V1.9) and ligands were transferred on model from templates.
Plant Material and Stress Treatments
Chickpea (C. arietinum L. genotype ICC4958) seeds were grown in culture room and field for collection of various tissue samples. Mature leaf, young leaf, young pod, flower buds (FB), flower bud opened (FBO), unopened flowers (UOF), and mature flower (MF) were harvested from field grown plants. Root and shoot tissues were collected from 15-day-old chickpea seedlings grown in autoclaved mixture (1:1) of agropeat and vermiculite in plastic pots in the culture room maintained at 22 ± 1°C with a photoperiod of 14 h, as described (Garg et al., 2010). Germinating seedlings (GS) were collected after 5 days of seed germination on wet Whatman paper sheet in Petri dishes as described (Singh et al., 2013). Two stages of flower bud development (FB 4 mm and FBO 8–10 mm) were collected on the basis of size and morphological differences (Singh et al., 2013). Two stages of flower development, including young flower with closed petals (UOF) and MF with opened petals were also collected. For stress treatments, 10-day-old chickpea seedlings were kept in water for control, 150 mM solution of NaCl for salt stress, at 4°C for cold stress and between folds of tissue paper for desiccation stress. Root and shoot tissues were harvested separately after 5 h of treatments as described (Garg et al., 2010). All samples were quickly frozen into liquid nitrogen after harvesting and stored at -80°C till RNA isolation.
RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was extracted using TRI reagent (Sigma Life Science, St. Louis, MO, USA) following the manufacturer’s instructions. RNA quality and quantity was determined using Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). RNA samples with 260/280 ratio between 1.8 and 2.1 and 260/230 ratio between 2.0 and 2.5 were used for cDNA synthesis. Primers were designed for all genes using Primer Express (v3.0) software (Applied Biosystems, Foster City, CA, USA). Specificity of each pair of primers was determined via BLAST search. All the primer sequences used have been listed in Supplemental Table S1. For each tissue, at least two independent biological replicates and three technical replicates of each biological replicate were taken for the analysis. Real time PCR analysis was performed using the 7500 Detection System (Applied Biosystems) as described (Garg et al., 2010). The expression of elongation factor-1 alpha gene was used as internal control for normalization of sample input variance (Garg et al., 2010).
RNA-seq and Microarray Data Analysis
The expression patterns of chickpea and soybean GH3 genes were analyzed using RNA-seq data from various tissue/stages of development. For chickpea, we mapped our RNA-seq data (Singh et al., 2013) on the genome using TopHat (v2.0.6), assembled with Cufflinks (v2.1.1), and merged with Cuffmerge to estimate read count in FPKM. For soybean, normalized gene expression data (RPKM) was downloaded from SoySeq11. Medicago and Lotus GH3 gene expression data were downloaded from MtGEA12 and LjGEA13, respectively. Probsets corresponding to MtGH3 and LjGH3 genes were identified using BLASTN search with best hits.
Results and Discussion
GH3 Gene Family in Legumes
The availability of genome sequences provides an opportunity to identify and analyze GH3 gene family in legumes. We investigated members of GH3 gene family in four legumes, including chickpea, soybean, Medicago, and Lotus, using two strategies, BLASTP and HMM profile search. For chickpea, we selected genome sequence of desi genotype (ICC4958), because of the availability of comprehensive expression (RNA-seq) data from various tissues/developmental stages (Jain et al., 2013) and abiotic stress conditions (Garg et al., 2014), which can provide better insights into the functions of GH3 genes (as described in latter sections). The GH3 gene family members identified via these two searches were combined and a unique gene list was obtained for each legume species. In total, 11, 28, 10, and 18 GH3 gene members were identified in chickpea, soybean, Medicago and Lotus, respectively, after analyzing their protein sequences in pfam database for the presence of conserved GH3 domain. To identify additional members of GH3 gene family in chickpea, which may not be represented in the genome annotation, the published chickpea transcriptome (Garg et al., 2011) was also analyzed using similar strategies. This resulted in the identification of one additional GH3 gene family member for a total of 12 in chickpea. A list of GH3 genes and their identifiers in different legumes along with their genomic co-ordinates is given in Supplemental Table S2.
The number of GH3 proteins identified in chickpea, Medicago and Lotus were comparable to Arabidopsis (10; excluding group III members, which are exclusively present in Arabidopsis), rice (13), tomato (15), and sorghum (16; Jain et al., 2006; Wang et al., 2010; Kumar et al., 2012). Whereas, the number of GmGH3 proteins are found to be approximately double as compared to other legume plants. The soybean genome has undergone two rounds of whole genome duplication, including an ancient duplication prior to the divergence of papilionoids (58–60 Mya) and a soybean-specific duplication that is estimated to have occurred ∼13 Mya (Schmutz et al., 2010), which might have resulted into duplication of members of this gene family.
Genomic Organization and Chromosomal Distribution
All GH3 proteins identified in legumes showed the presence of characteristic GH3 domain, and sequence conservation in the core region in multiple sequence alignment of proteins. The gene structure (exon–intron organization) analysis of CaGH3 and GmGH3 genes revealed that number of introns varied from one to four except for CaGH3-11 and GmGH3-24, which do not have any intron (Figure 1). Most of the CaGH3 and GmGH3 had similar intron-phasing distribution (Figure 1) and followed the pattern reported earlier for rice GH3 genes (Jain et al., 2006). Next, we analyzed the distribution of GH3 genes on the chromosomes in different legumes. Only two CaGH3 genes could be located on the linkage groups, whereas others were located on scaffolds (Supplemental Table S2). This may be due to availability of incomplete draft genome sequence and unanchored scaffold as of now (Jain et al., 2013). For soybean, all the 28 GmGH3 genes were distributed on 14 of 20 chromosomes, with six GH3 genes located on chromosome 12, four and three being present on chromosome 6 and 13, respectively, two each on chromosome 2, 3, 15, and 17, and one each on chromosome 1, 5, 7, 10, 11, 16, and 19 (Supplemental Table S2). In soybean, many GH3 genes were clustered, such as adjacent genes on chromosome 6 (GmGH3-7, -8, -9, and -10), chromosome 12 (GmGH3-14 and -15, and GmGH3-17, -18 and -19), chromosome 13 (GmGH3-20, -21, and -22), and chromosome 17 (GmGH3-26 and -27). The amino acid sequences of these genes [GmGH3-7 to -10 (7–8% identity), GmGH3-14, and -15 (32% identity), GmGH3-17, -18, -19 (28–57% identity), GmGH3-20 to -22 (30–68%), and GmGH3-26 and -27 (70% identity)] showed very low (7%) to high (70%) similarity (Supplemental Table S3B), indicating that these GmGH3 genes probably resulted from tandem duplication and some of them diverged during course of evolution. In Medicago, 7 of 10 GH3 genes were distributed on 5 of 8 chromosomes and three MtGH3 genes were located on scaffolds (Supplemental Table S2). Chromosome 5 and 8 of Medicago harbored two MtGH3 genes each and one each resided on chromosome 2, 3, and 7. In Lotus, out of 18 LjGH3 genes, only eight were located on 4 of 6 chromosomes; three located on chromosome 3, two each on chromosomes 2 and 4, and one on chromosome 1 (Supplemental Table S2). Altogether, it appears that tandem gene duplication resulted in the amplification of GH3 gene family members in legumes and low homology between them suggested their divergence during course of evolution.
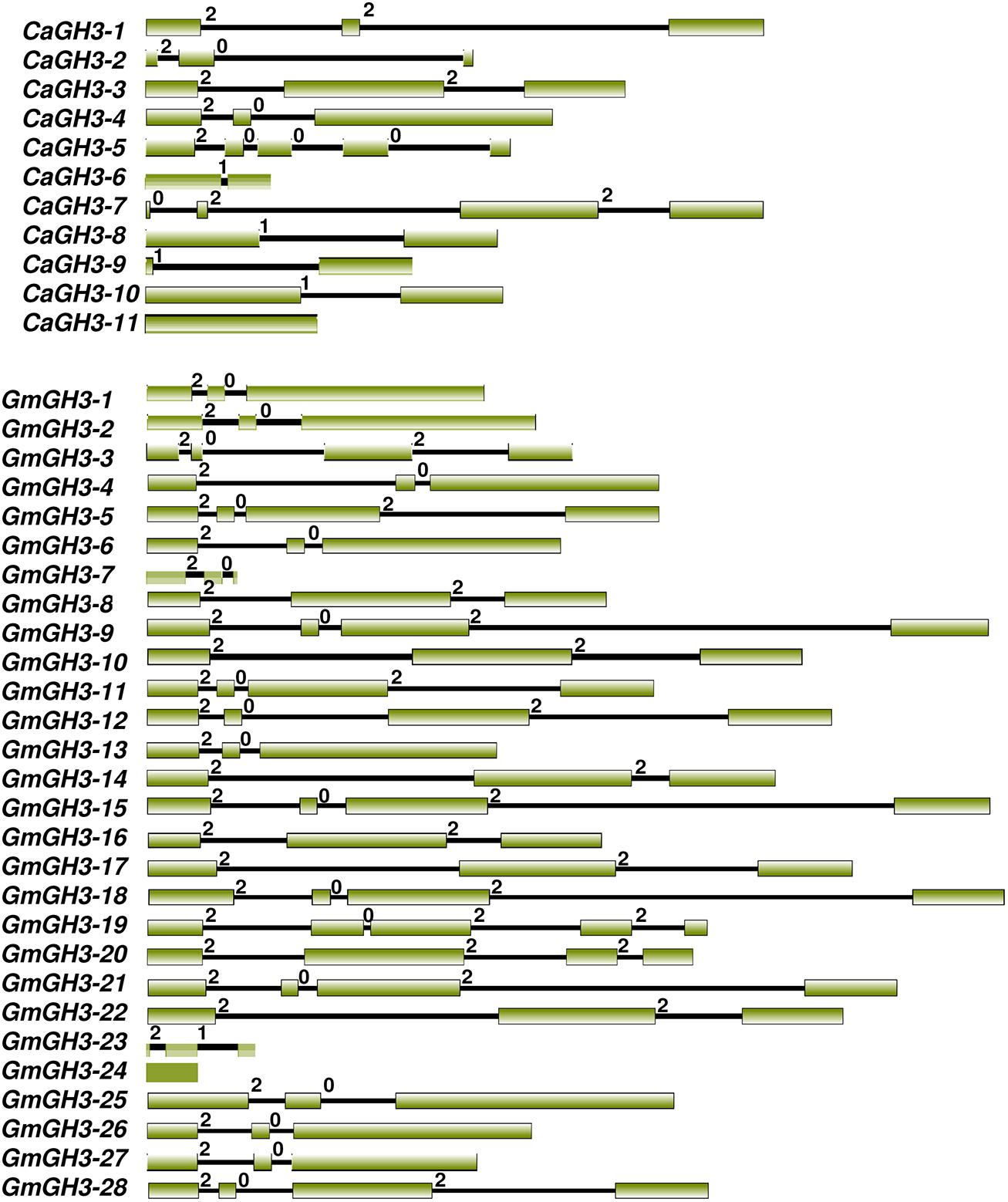
FIGURE 1. Exon-intron organization of chickpea and soybean GH3 genes. Boxes and lines represent exons and introns, respectively. The numbers 0, 1, and 2 represent phase 0, 1, and 2 introns, respectively.
Sequence Analysis and Phylogenetic Relationship
Pairwise analysis of the full-length protein sequences of chickpea and soybean GH3 proteins showed very high homology, 76.9% between paralogous pair, CaGH3-7 and CaGH3-8 (Figure 2; Supplemental Table S3A), and 97.4% between paralogous pair, GmGH3-8 and GmGH3-16 (Figure 2; Supplemental Table S3B). Such high homologies suggest that they may perform similar functions (Jain et al., 2006).
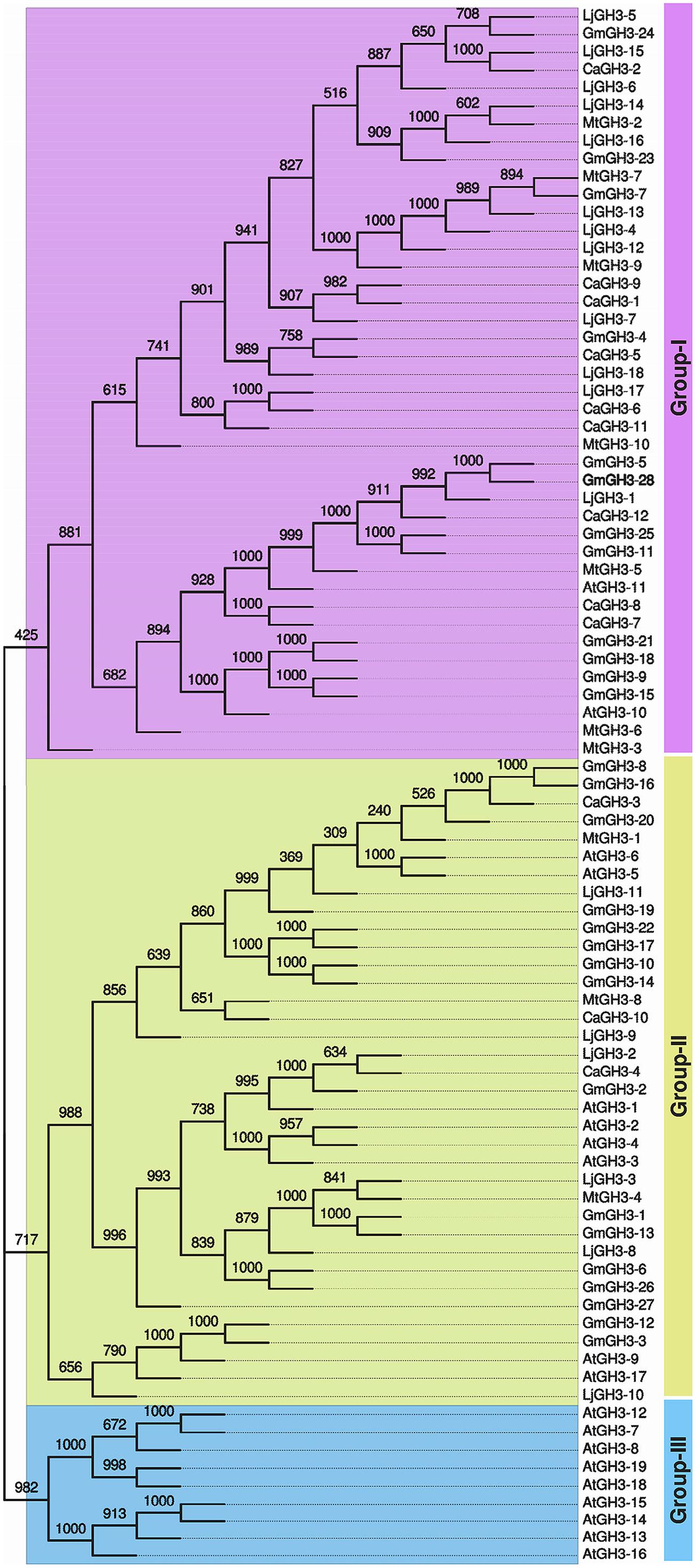
FIGURE 2. Phylogenetic relationship among chickpea, soybean, Medicago, Lotus, and Arabidopsis GH3 proteins. Multiple sequence alignment of all GH3 proteins from chickpea (CaGH3), soybean (GmGH3), Medicago (MtGH3), Lotus (LjGH3) and Arabidopsis (AtGH3) was performed and tree was generated by UPGMA method. FigTree was used for visualization of the tree. The value at the nodes represents bootstrap values from 1000 replicates. Different groups of GH3 proteins are labeled.
In Arabidopsis, GH3 proteins have been classified into three groups on the basis of sequence similarity and specificity to adenylate plant hormones (Staswick et al., 2002). We also analyzed the phylogenetic relationship among GH3 proteins identified in legumes and classified them into different groups. Phylogenetic analysis of legume GH3 proteins showed clustering into only two groups, I and II. Group III GH3 proteins were found absent in all the legumes (Figure 2). This observation is consistent to previous reports (Jain et al., 2006; Kumar et al., 2012; Yuan et al., 2013) and suggested that group III GH3 proteins might have been lost in legumes during the course of evolution. The group I consisted of nine members of CaGH3 proteins, 12 GmGH3 proteins, seven MtGH3 proteins, and 12 LjGH3 proteins (Figure 2). Group II included three CaGH3 proteins, 16 GmGH3 proteins, three MtGH3 proteins, and six LjGH3 proteins (Figure 2).
Phylogenetic tree comprising CaGH3, GmGH3, MtGH3, LjGH3, and AtGH3 proteins showed a total of 26 sister pairs. Group I comprised of 12 sister pairs, four of GmGH3-GmGH3 proteins, two each of CaGH3-CaGH3 and CaGH3-LjGH3 proteins, and one each of CaGH3-GmGH3, GmGH3-MtGH3, GmGH3-LjGH3, MtGH3-LjGH3 proteins. Group II consisted of eleven sister pairs, including six GmGH3-GmGH3 proteins, two each of AtGH3-AtGH3 proteins, and one each of CaGH3-MtGH3, CaGH3-LjGH3, and MtGH3-LjGH3 proteins. In Group III, three sister pairs of only Arabidopsis GH3 proteins were present. To gain further insight into structural diversity of GH3 genes, we compared exon/intron organization of individual GH3 gene in chickpea and soybean. Most of the sister pairs shared similar exon/intron structures, intron numbers and intron phasing (Figure 2; Supplemental Tables S3A,B). However, all closely located GmGH3 genes, such as GmGH3-7, -8, -9, -10 on chromosome 6, GmGH3-17, -18, and -19 on chromosome 12, GmGH3-20, -21, and -22 on chromosome 13, and GmGH3-26 and -27 on chromosome 17, were not paired together (Figure 2). This suggested that these genes might have diverged substantially during evolution. Most of the AtGH3/GmGH3 proteins showed 1:4 orthologous relationship, such as AtGH3-10/GmGH3-9, -15, -21, and -18 (Figure 2). Presence of such orthologous relationship between AtGH3/GmGH3 pairs is also in agreement with the fact that soybean whole genome duplication happened twice in the past (Schmutz et al., 2010). Some Arabidopsis and legume GH3 protein pairs (AtGH3-5 and -6/CaGH3-3 and -10, AtGH3-9, and -17/GmGH3-3, and -12, AtGH3-11/MtGH3-5, LjGH3-1) exhibited n:n orthologous relationship, which suggest that members of this family have diversified both in Arabidopsis and legumes independently (Wang et al., 2007; Wu et al., 2011; Liu and Hu, 2013; Yuan et al., 2013).
Differential Expression Patterns of GH3 Genes During Development
Phytohormone auxin is required for plant morphogenesis, including tropistic growth, root patterning, vascular tissue differentiation, axillary bud formation, and floral organ development (Zhao, 2010). Expression analysis of GH3 genes in various tissue-types during different developmental stages in different plant species have suggested their diverse roles in plants (Gee et al., 1991; Nakazawa et al., 2001; Takase et al., 2004; Khan and Stone, 2007; Jain and Khurana, 2009; Zhang et al., 2009; Böttcher et al., 2010; Kuang et al., 2011; Kumar et al., 2012). Therefore, we performed expression analysis of GH3 genes in various tissue/stages of development in legumes to know their putative functions. Availability of gene expression atlas covering various tissues/organs and stages of development (Benedito et al., 2008; Libault et al., 2010; Severin et al., 2010; Singh et al., 2013; Verdier et al., 2013), serves as resource to profile expression of candidate genes in legumes. We analyzed the expression of chickpea GH3 genes using our RNA-seq data (Singh et al., 2013) and validated the results via qRT-PCR analysis (Figure 3). This analysis revealed that CaGH3 genes were differentially expressed in various tissues/stages of development. CaGH3-3 and CaGH3-5 genes exhibited higher expression in root, which was also confirmed via qRT-PCR, suggesting their role in chickpea root development (Figure 3). CaGH3-3 orthologs in Arabidopsis, AtGH3-2, and AtGH3-6, were found to have role in root development (Nakazawa et al., 2001; Takase et al., 2004). In addition, CaGH3-1 and CaGH3-11 exhibited preferential expression in unopened flower, indicating that these genes might be involved in auxin homeostasis during a specific developmental stage of flower (Figure 3). In rice, OsGH3-1, -4, -5, -8, and -11 genes displayed highest expression level in flower (Jain et al., 2006) and OsGH3-8 has been reported as the downstream target of rice MADS-box transcription factor (OsMADS1), which is involved in patterning of inner whorl floral organ (Prasad et al., 2005). Expression of CaGH3-10 was also distinctly higher in unopened flower, suggesting its role in flower development. CaGH3-10 was found to be in same phylogenetic clade with AtGH3-5 and -6, whose orthologs in rice OsGH3-1 and -4 have higher expression in flower (Jain et al., 2006; Jain and Khurana, 2009), validating our observation. Paralogous gene pair, CaGH3-7 and -8 exhibited significantly higher expression in open flower bud, indicating their possible role in auxin homeostasis in early stages of flower development and support the notion that paralogs might have similar expression patterns and function. Transcript level of CaGH3-2 could not be detected via qRT-PCR, suggesting it might be expressed in a specific tissue/stages of development. These findings highlight the role of CaGH3 genes in overall plant development including various stages of reproductive development.
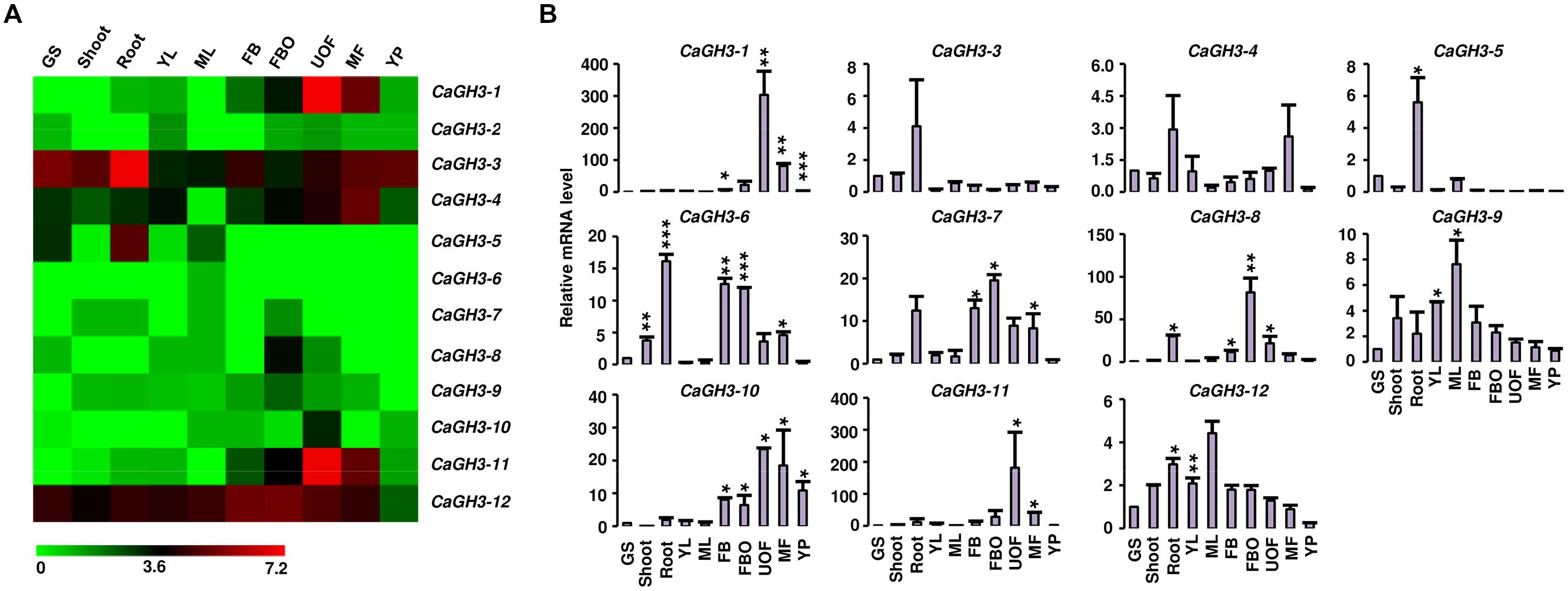
FIGURE 3. Expression profiles of CaGH3 genes during development. (A) Heatmap showing expression profiles of CaGH3 genes based on RNA-seq data in various tissues/development stages. Heatmap was generated based on log2 FPKM. (B) Real-time PCR analysis of CaGH3 genes in various tissue/stages of development. Expression of germinating seedling (GS) was taken as a reference to determine relative mRNA level in other tissues for each gene. Error bars indicate SE of mean. YL, young leaf; ML, mature leaf; FB, flower bud; UOF, unopened flower; FBO, flower bud open; MF, mature flower; YP, young pod. Data points marked with asterisk (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001) indicate statistically significant difference between control (GS) and other tissues.
Furthermore, we analyzed the expression profiles of GmGH3, MtGH3, and LjGH3 genes in different vegetative and reproductive tissues, utilizing expression data from published RNA-seq atlas of soybean (Severin et al., 2010), Medicago (Benedito et al., 2008), and Lotus (Verdier et al., 2013), respectively. Expression analysis of GmGH3 genes revealed their dynamic regulation in various tissues and stages of development (Figure 4A). GmGH3-8 and GmGH3-25 showed distinctly higher expression in root, GmGH3-4 and GmGH3-13 were up-regulated in nodule, GmGH3-14 and GmGH3-18 exhibited flower-specific expression, GmGH3-9 showed specific expression in young leaf and GmGH3-20 expression was higher in stages of seed development (Figure 4A). Previously, it has been reported that GH3 genes in soybean exhibit transient expression during floral development and higher expression in ovule and ovary at later stages of floral development (Gee et al., 1991). Reports also suggested role of GH3 genes during seed development, for example, GH3 gene (YDK1) was found to be specifically up-regulated at heart stage during embryogenesis of Solanum chacoense (Tebbji et al., 2010). In rice, involvement of GH3 genes in seed development has also been reported. For instance, OsGH3-13 overexpressing rice exhibited smaller seeds (Zhang et al., 2009) and OsGH3-4 have higher expression during various stages of seed development (Jain and Khurana, 2009). These findings indicated that GmGH3 genes could play an important role in seed development.
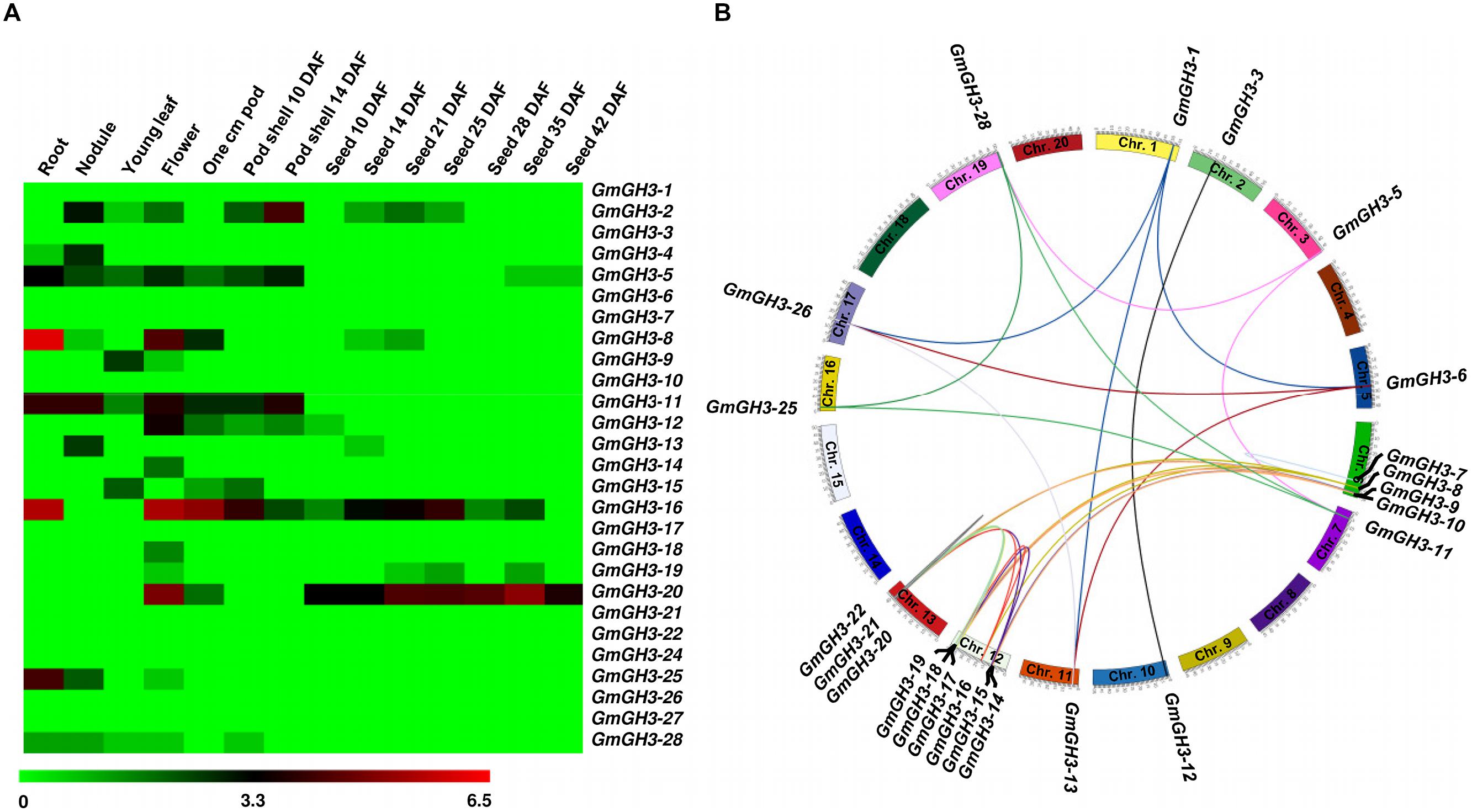
FIGURE 4. Expression profiles and gene duplication of GmGH3 genes. (A) Heatmap showing expression profiles of soybean GH3 genes at various stages of development. Heatmap was generated based on log2 RPKM. (B) Mapping of GmGH3 genes and duplication between them are shown on the soybean chromosomes. Duplication was determined using Plant Genome Duplication Database. Genes and their duplications were mapped on chromosomes using Circos tool. Soybean chromosomes have been arranged in circle and duplications are represented by lines.
The paralogous GmGH3 genes, GmGH3-6 and GmGH3-26, GmGH3-8 and GmGH3-16, GmGH3-11 and GmGH3-25, GmGH3-17 and GmGH3-22, and GmGH3-23 and GmGH3-24 localized on duplicated chromosomal segments, exhibited similar expression patterns in various tissues/stages of development (Figures 4A,B), suggesting their similar function. However, duplicated genes are also known to have a great degree of expression and functional divergence due to selection pressure and need for diversification (Prince and Pickett, 2002). Many duplicated GmGH3 genes exhibited expression divergence as well, such as GmGH3-1 and GmGH3-13, GmGH3-3 and GmGH3-12, GmGH3-5 and GmGH3-28, GmGH3-9 and GmGH3-15, and GmGH3-11 and GmGH3-25 (Figures 4A,B). These results suggested that chromosomal duplication events not only facilitated the amplification of the GmGH3 gene family members, but also resulted into expression divergence between duplicated genes, which might have contributed in the establishment of gene functional diversity during evolution.
Likewise, the expression of MtGH3 and LtGH3 genes was also found to be variable in various tissue/stages of development. For instance, MtGH3-4 exhibited significantly higher expression in seed at 36 day after pollination (DAP), MtGH3-8 showed greater expression in root and various stages of seed development (Supplemental Figure S2). LjGH3-2 was found to be up-regulated in root, whereas LjGH3-1, -6, and -12 showed distinctly higher expression in leaf, and LjGH3-3, -4, -5, and -18 were seen to be up-regulated in root and nodule (Supplemental Figure S4). Expression of other GH3 genes of legumes was also found to be variable in various tissue/stages of development elucidating their involvement in various growth and development processes (Wang et al., 2010; Kuang et al., 2011).
Furthermore, we analyzed expression patterns of paralogous/orthologous GH3 genes to investigate their functional conservation across legumes. Although the available expression data represented diverse tissues/developmental stages in different legumes, we made an effort to define correlation in expression profiles of GH3 genes in different legumes. Some of paralogous/orthologous GH3 genes exhibited similar expression patterns in different legumes, such as CaGH3-3, GmGH3-8, -16, -20, MtGH3-1 and LjGH3-11; CaGH3-12, GmGH3-5, -11, -25, -28 and LjGH3-1; CaGH3-5 and GmGH3-4; CaGH3-4 and LjGH3-2; GmGH3-13 and LjGH3-2; MtGH3-2 and LjGH3-14; and MtGH3-3 and -6, suggesting their conserved function across legumes (Figures 3–5; Supplemental Figures S2 and S3). Some of these paralogous/orthologous genes harbor similar cis-regulatory elements in their promoter regions (Supplemental Table S4). For instance, CaGH3-3, GmGH3-8, -16, and -20 contain cis-regulatory elements, S000037, S000270, S000273, S000390, S000414, S000453, and S000461, conserved in their promoter sequences (Supplemental Table S4). An earlier study revealed similarity of gene expression profiles in various organs for a significant number of paralogous/orthologous gene pairs in Medicago and Arabidopsis (Benedito et al., 2008). Moreover, comparison of soybean transcriptome with Medicago and Lotus demonstrated similar tissue-specificity for 45% of the genes analyzed (Libault et al., 2010). Overall, these findings provide insights into the putative roles of GH3 genes in legumes in various aspects of plant growth and development.
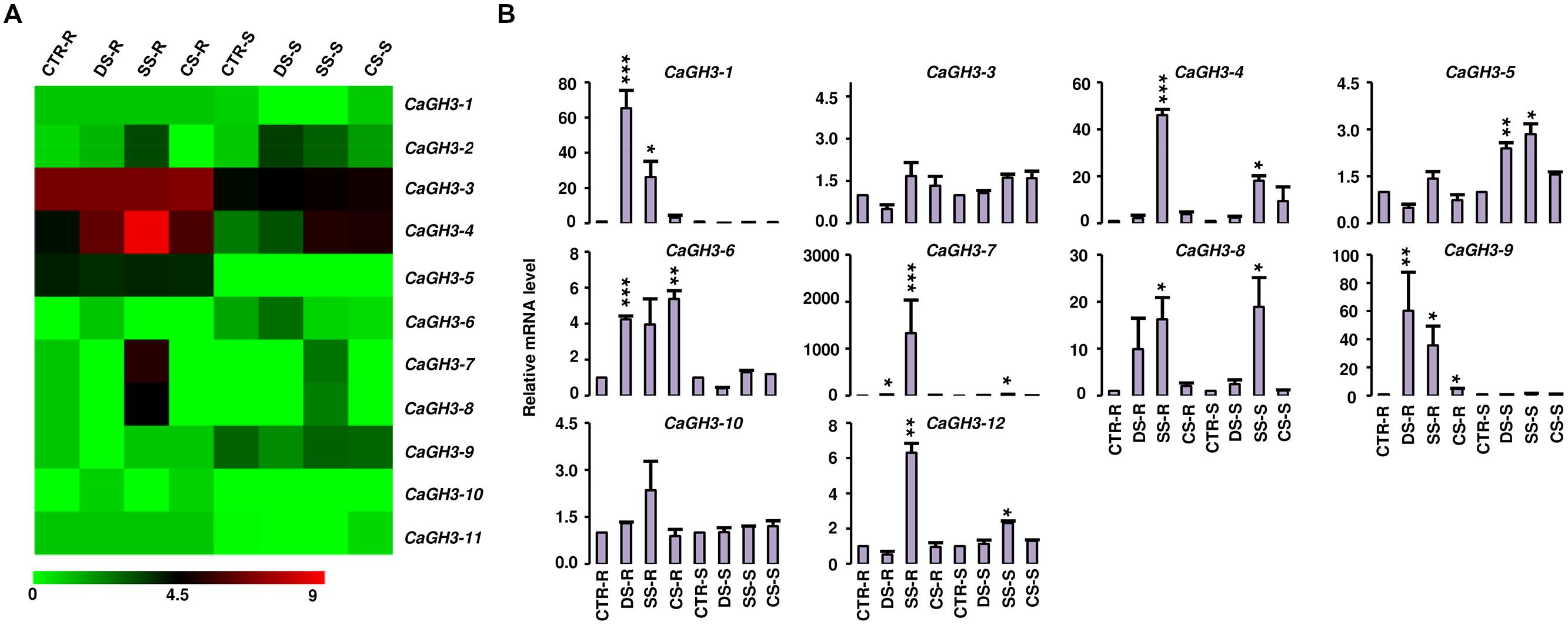
FIGURE 5. Expression profiles of CaGH3 genes under abiotic stress conditions. (A) Heatmap showing expression of CaGH3 genes based on RNA-seq data. Heatmap was generated based on log2 FPKM. (B) Real-time PCR analysis of CaGH3 genes under various stress treatments. Root control (CTR-R) and shoot control (CTR-S) was taken as a reference to determine relative mRNA level under stress conditions. Error bars indicate standard error of mean. DS-R: desiccation stressed root, SS-R, salt stressed root; CS-R, cold stressed root; DS-S, desiccation stressed shoot; SS-S, salt stressed shoot; CS-S, cold stressed shoot. Data points marked with asterisk (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001) indicate statistically significant difference between control and stress treatments.
Differential Expression Patterns of GH3 Genes Under Abiotic Stresses
Plants are constantly exposed to various abiotic stresses in their life cycle. Several recent studies have implicated auxin in abiotic stress responses (Jain and Khurana, 2009; Wang et al., 2010; Du et al., 2012; Kumar et al., 2012; Yuan et al., 2013). Some studies have revealed that GH3 genes are regulated by abiotic stresses, like drought, salt, and cold stresses (Park et al., 2007; Jain and Khurana, 2009). The transcript level of AtGH3-5 (WES1) has been shown to be induced by various abiotic stress conditions, like drought, high salt, and cold (Park et al., 2007). In rice, the transcript abundance of OsGH3-1, OsGH3-2, OsGH3-8, and OsGH3-13 were markedly higher in seedlings subjected to salt, drought and cold stresses (Jain and Khurana, 2009; Zhang et al., 2009; Du et al., 2012). In Sorghum, at least six GH3 genes were found to be induced upon salt and drought treatments in leaf (Wang et al., 2010).
To investigate the role of legume GH3 genes in abiotic stress responses, we performed scanning of cis-acting regulatory DNA elements within promoter regions (2 kb upstream from the start codon) using PLACE database. This analysis predicted several elements responsive to auxin (IAA), abscisic acid (ABA), SA, JA, drought, salinity, and disease (Supplemental Table S4), suggesting that the function of these genes may be associated with various phytohormone signals and/or environmental stresses. Considering regulatory role of cis-elements, we analyzed expression of GH3 genes under abiotic stress conditions to know their function during abiotic stresses. For chickpea, we analyzed RNA-seq data from root and shoot tissues subjected to desiccation, salinity and cold conditions (Garg et al., 2014), and performed real-time PCR analysis for validation. In our analysis, paralogous gene pair, CaGH3-1 and -9, showed induction under both desiccation and salinity stresses in root (Figures 5A,B), and also their promoter sequences harbor desiccation (S000414) and salinity (S000453) responsive cis-regulatory elements (Supplemental Table S4), indicating their role in desiccation and salinity stress. Recently, rice group-I gene, OsGH3-12, has also been found to be markedly induced by drought stress (Du et al., 2013b). Similarly, promoter of CaGH3-4 harbor salinity responsive cis-element (S000453) and showed higher expression in root under salt stress (Figures 5A,B). Its ortholog, AtGH3-1, has also been found to be up-regulated under salt stress (Sani et al., 2013), corroborating our result. Group-I paralogous genes, CaGH3-7 and -8, were found to be induced in root under salinity stress (Figures 5A,B), implying their involvement in homeostasis of auxin under salinity stress in root. CaGH3-5 and -6 showed enhanced expression under desiccation, salt and cold stresses in shoot and root (Figures 5A,B), respectively, suggesting their role during multiple abiotic stress responses.
In Medicago, MtGH3-8 and -9 genes were induced under salt stress in root, and MtGH3-7 was induced under drought stress in root (Supplemental Figure S2). Previous reports suggest that IAA, SA, JA, ethylene, and ABA regulate the protective responses of plants against both biotic and abiotic stress responses via signaling crosstalk (Bostock, 2005; Lorenzo and Solano, 2005; Mauch-Mani and Mauch, 2005; Ding et al., 2008; Domingo et al., 2009; Fu et al., 2011). In addition, orthologous genes, CaGH3-10 and MtGH3-8, showed induced expression under salt stress in root (Figure 5; Supplemental Figure S2); suggesting their conserved function in both legumes. Taken together, these findings indicated that members of GH3 gene family might be involved in stress adaptation in legumes.
Homology Modeling and Substrate Preferences
The availability of crystal structures of two Arabidopsis GH3 proteins: AtGH3-12, which conjugate benzoate substrate and JA-specific AtGH3-11/JAR1 (Westfall et al., 2012); and grapevine IAA-amido synthetase GH3-1 (VvGH3-1) gave us an exciting opportunity to determine three-dimensional structure of GH3 members in legumes by homology modeling.
Group-I protein, CaGH3-3 and GmGH3-8 of chickpea and soybean, respectively, were modeled using structure of AtGH3-11 (Protein Data Bank code 4EPL; Westfall et al., 2012) and Group-II proteins, CaGH3-12 and GmGH3-25 were modeled using grapevine, Vv-GH3-1 (Protein Data Bank code 4B2G; Peat et al., 2012). The homology modeling revealed high degree of conservation in the protein structure of these proteins. To predict active sites, we transferred ligands from template to model by superimposing structures. Ligands for group-I proteins are JA-Ile and AMP (amino acid mono phosphate), and group-II proteins are adenosine-5′-[2-(1H-indole-3-yl)ethyl]phosphate (AIEP), which mimics the adenylated intermediate of the IAA conjugation reaction (Figures 6 and 7; Böttcher et al., 2012; Westfall et al., 2013). By comparing sequences of model and template, we also identified the residues forming acyl acid/hormone-binding site and nucleotide binding site (Figures 7 and 8). Most of these residues were found to be conserved between the model and template. For example, hormone-binding residues of CaGH3-12 and GmGH3-25 with AtGH3-11 (JA-conjugating), Ca-Leu137, Gm-Leu115 to At-Leu117; Ca-Thr141, Gm-Thr119 to At-Thr121; Ca-Thr185, Gm-Thr163 to At-Thr166; Ca-Val188, Gm-Val168 to At-Val169; Ca-Ile323, Gm-Ile301 to At-Ile304; and Ca-Trp355, Gm-Trp333 to At-Trp336 are conserved (Figures 7 and 8). Similarly, conservation was found between hormone-binding residues of CaGH3-3 and GmGH3-8 with VvGH3-1(IAA-conjugating), Ca-Val167, Gm-Val167 to Vv-Val172; Ca-Leu168, Gm-Leu168 to Vv-Leu173; Ca-Ala332, Gm-Ala332 to Vv-Ala337; and Ca-Tyr337, Gm-Tyr337 to Vv-Tyr342 (Figures 7 and 8). In addition, we also found nucleotide-binding residues, Ser, Thr, Phe, and Tyr conserved in all the structures (Figures 7 and 8), which is in agreement with earlier studies that nucleotide binding residues conserved in not only GH3 proteins but also the ANL superfamily (Gulick, 2009; Peat et al., 2012; Westfall et al., 2012, 2013).
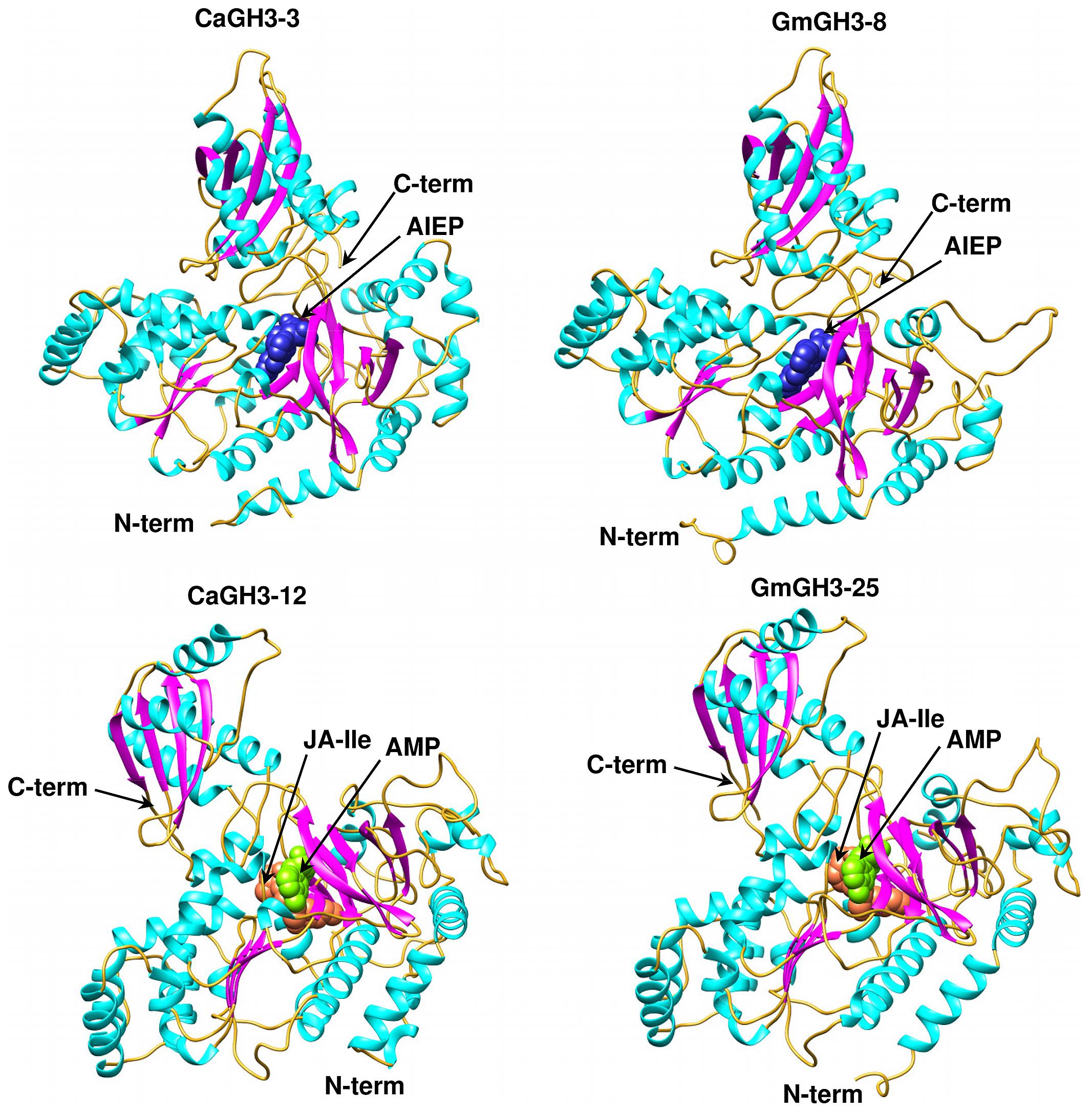
FIGURE 6. Predicted structures of GH3 proteins. Ribbon diagram showing the N- and C-terminal domains of chickpea (CaGH3-3 and CaGH3-8) and soybean (GmGH3-8 and GmGH3-25) GH3 protein with α-helices, β-strands and loops colored cyan, magenta, and gold, respectively. Ligands AIEP, JA-Ile, AMP are shown as space-filling model in blue, coral, and green colors, respectively.
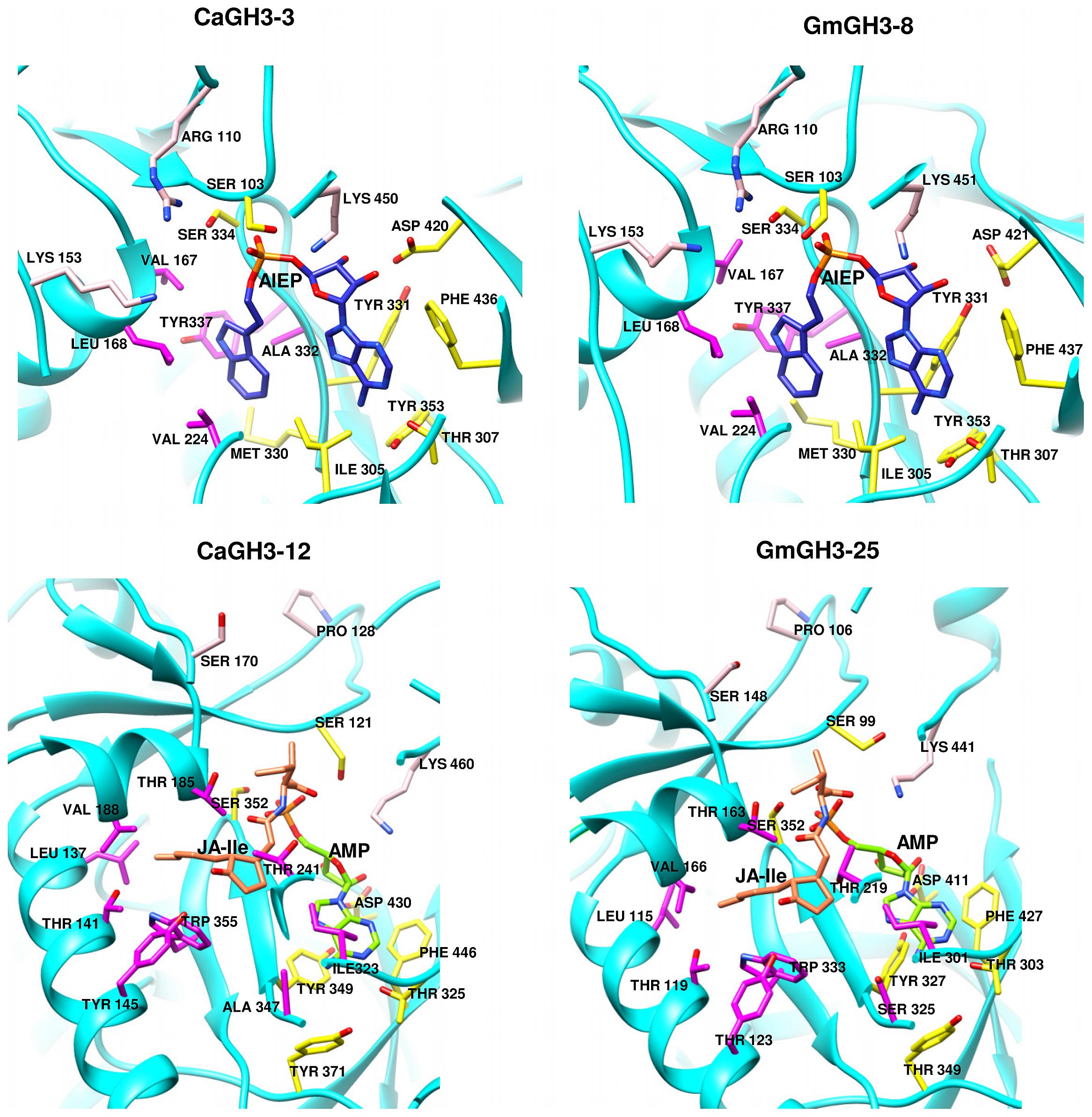
FIGURE 7. Hormone and nucleotide binding residues in GH3 proteins. Ribbon diagram showing hormone binding residues in magenta, nucleotide (ATP/AMP) binding residues in yellow, and residues in pink determine amino-acid preferences.
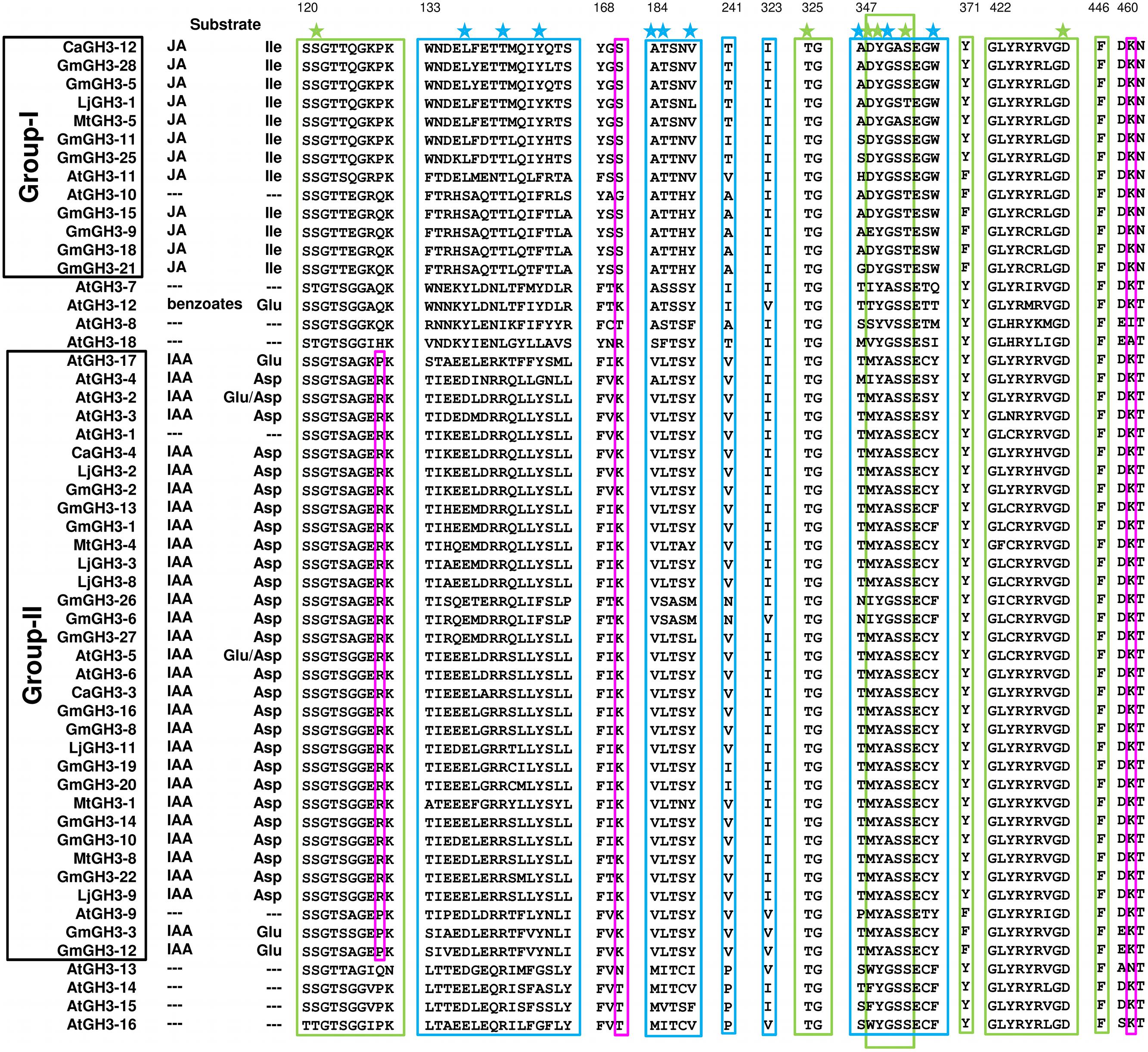
FIGURE 8. Proposed substrates of GH3 proteins based on conserved amino acid residues. Protein sequences of all the identified GH3 genes were aligned using MAFFT. Green and blue boxes represent nucleotide (ATP/AMP) and hormone-binding motifs/residues, respectively. Magenta boxes represent residues determining amino-acid preferences. Only sequences with complete C- and N-terminal domains were included. Star across the top of the alignment indicates conserved residues in pocket forming active site. Numbering at the top corresponds to CaGH3-12.
Further, to determine amino-acid specificities, we identified residues involved in discrimination of apolar (i.e., Ile) and acidic (i.e., Asp/Glu) substrates in the transferase reaction by comparing structures. Within the active site, a lysine residue (Lys450 in CaGH3-3, Lys460 in CaGH3-12, Lys451 in GmGH3-8 and Lys441 in GmGH3-25) was conserved at same position (Figures 7 and 8). This residue was also found to be highly conserved in GH3 proteins with known amino-acid preference (Westfall et al., 2012). Also, it has been found that Lys428 (AtGH3-12) is conserved in GH3-proteins that accept acidic-amino acid, whereas Ser151 (AtGH3-11) at the same position is conserved in enzymes specific to isoleucine conjugation (Westfall et al., 2012). We also found Lys153 in CaGH3-3 and GmGH3-8, suggesting CaGH3-3 and GmGH3-8 may accept acidic-amino acid (i.e., Asp/Glu) and at the same position Ser170 in CaGH3-12 and Ser148 in GmGH3-25, indicating their preferences for isoleucine (Figures 7 and 8). In addition, conservation of another residue Arg110 in IAA-specific CaGH3-3 and GmGH3-8 (Figures 7 and 8), further specified their Asp-conjugating nature. This was also corroborated by another study in grapevine, where residue Arg115 in VvGH3-1 (IAA-specific) was conserved in all the four Asp-conjugating GH3s, whereas Glu-conjugating GH3s had a Proline at that position (Peat et al., 2012). Also, the same pattern was found in IAA-conjugating GH3 enzymes with known amino-acid substrate preferences from Arabidopsis (Staswick et al., 2005) and rice (Zhang et al., 2009; Chen et al., 2010). Next, we also found similar conservation in residues determining amino-acid preferences for other members of GH3 proteins, which led us to propose substrates for them (Figure 8). Group-I proteins with conserved Ser and Lys at similar position as that of CaGH3-12 (Ser170 and Lys461; magenta boxes; Figure 8) are proposed to have Ile as substrate (Figure 8). For group-II proteins, Asp will be the substrate when Arg at 128 and Lys at positions 170 and 461 (magenta boxes; Figure 8); and Glu, will be the substrate when Arg is replaced by Pro at the same position (Figure 8). Altogether, the structures presented here showed conservation of residues at hormone-binding site, nucleotide-binding site, and amino-acid preferences determining residues, indicating similar function.
Several previous studies have reported the differential expression of GH3 genes in various tissues/developmental stages and in response to various stimuli, including auxin, jasmonic acid, salicyclic acid, and abiotic/biotic stresses in different plants (Park et al., 2007; Zhang et al., 2007; Jain and Khurana, 2009; Kumar et al., 2012; Yuan et al., 2013; Wu et al., 2014). Our results also revealed preferential/tissue-specific and stress-responsive expression of many GH3 genes in different legumes. The knowledge of motifs/residues of GH3 proteins that determine substrate preferences and conjugation to auxin may help modulate their binding efficiency and substrate preferences for engineering plants with desired agronomic traits.
Conclusions
We performed a genome-wide analysis of GH3 gene family in legumes to reveal gene structure, phylogenetic relationship, and expression profiles during various developmental stages and abiotic stress conditions. Some GH3 genes exhibited preferential/specific expression in a particular tissue and/or under abiotic stress condition(s). Our analysis revealed that GH3 genes seem to be involved in biology of various tissues or organs and actively participate in stress responses in legumes. The analysis of protein structures of few members identified key features of substrate recognition, which might help in investigation of their molecular functions in legumes. The data generated in this study will serve as a foundation for functional characterization of GH3 gene family members in legumes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the Department of Biotechnology, Government of India, under the Next Generation Challenge Programme on Chickpea Genomics (grant number BT/PR12919/AGR/02/676/2009 from 2009 to 2014) to Mukesh Jain. Rohini Garg acknowledges INSPIRE faculty award from the Department of Science and Technology, Government of India. Vikash K. Singh acknowledges the award of research fellowship from the Department of Biotechnology, Government of India.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00789/abstract
Footnotes
- ^http://www.phytozome.net
- ^http://www.kazusa.or.jp/lotus/index.html
- ^http://www.arabidopsis.org/
- ^http://rice.plantbiology.msu.edu/
- ^http://pfam.sanger.ac.uk/
- ^http://chibba.agtec.uga.edu/duplication/
- ^http://www.dna.affrc.go.jp/PLACE/
- ^http://www.rcsb.org/pdb/home/home.do
- ^http://www.sbg.bio.ic.ac.uk/phyre2
- ^http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php
- ^http://soybase.org/soyseq/
- ^http://mtgea.noble.org/v3/
- ^http://ljgea.noble.org/v2/
References
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K.,et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bostock, R. M. (2005). Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu. Rev. Phytopathol. 43, 545–580. doi: 10.1146/annurev.phyto.41.052002.095505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Böttcher, C., Dennis, E. G., Booker, G. W., Polyak, S. W., Boss, P. K., and Davies, C. (2012). A novel tool for studying auxin-metabolism: the inhibition of grapevine indole-3-acetic acid-amido synthetases by a reaction intermediate analogue. PLoS ONE 7:e37632. doi: 10.1371/journal.pone.0037632
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Böttcher, C., Keyzers, R. A., Boss, P. K., and Davies, C. (2010). Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3–1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 61, 3615–3625. doi: 10.1093/jxb/erq174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Q., Westfall, C. S., Hicks, L. M., Wang, S., and Jez, J. M. (2010). Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285, 29780–29786. doi: 10.1074/jbc.M110.146431
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Q., Zhang, B., Hicks, L. M., Wang, S., and Jez, J. M. (2009). A liquid chromatography-tandem mass spectrometrybased assay for indole-3-acetic acid-amido synthetase. Anal. Biochem. 390, 149–154. doi: 10.1016/j.ab.2009.04.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, X., Cao, Y., Huang, L., Zhao, J., Xu, C., Li, X.,et al. (2008). Activation of the indole-3- acetic acid-amidosynthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate- independent basal immunity in rice. Plant Cell 20, 228–240. doi: 10.1105/tpc.107.055657
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Domingo, C., Andrés, F., Tharreau, D., Iglesias, D. J., and Talón, M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 22, 201–210. doi: 10.1094/MPMI-22-2-0201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, D., Hao, R., Cheng, T., Pan, H., Yang, W., Wang, J.,et al. (2013a). Genome-wide analysis of the AP2/ERF gene family in Prunus mume. Plant Mol. Biol. Rep. 31, 741–750. doi: 10.1007/s11105-012-0531-6
Du, H., Liu, H., and Xiong, L. (2013b). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4:397. doi: 10.3389/fpls.2013.00397
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, H., Wu, N., Fu, J., Wang, S., Li, X., Xiao, J.,et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63, 6467–6480. doi: 10.1093/jxb/ers300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fu, J., Liu, H., Li, Y., Yu, H., Li, X., Xiao, J.,et al. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155, 589–602. doi: 10.1104/pp.110.163774
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garg, R., Bhattacharjee, A., and Jain, M. (2014). Genome-scale transcriptomic insights into molecular aspects of abiotic stress responses in chickpea. Plant Mol. Biol. Rep. doi: 10.1007/s11105-014-0753-x
Garg, R., Patel, R. K., Jhanwar, S., Priya, P., Bhattacharjee, A., Yadav, G.,et al. (2011). Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 156, 1661–1678. doi: 10.1104/pp.111.178616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garg, R., Sahoo, A., Tyagi, A. K., and Jain, M. (2010). Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem. Biophys. Res. Commun. 396, 283–288. doi: 10.1016/j.bbrc.2010.04.079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gee, M. A., Hagen, G., and Guilfoyle, T. J. (1991). Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3, 419–430. doi: 10.1105/tpc.3.4.419
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gulick, A. M. (2009). Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827. doi: 10.1021/cb900156h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hagen, G., Kleinschmidt, A., and Guilfoyle, T. (1984). Auxin-regulated gene expression in intact soybean hypocotyls and excised hypocotyls sections. Planta 162, 147–153. doi: 10.1007/BF00410211
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hagen, G., Martin, G., Li, Y., and Guilfoyle, T. J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. doi: 10.1007/BF00040658
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, H. L., Okamoto, H., Wang, M., Ang, L. H., Matsui, M., Goodman, H.,et al. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970.
Jagadeeswaran, G., Raina, S., Acharya, B. R., Maqbool, S. B., Mosher, S. L., Appel, H. M.,et al. (2007). Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 51, 234–246. doi: 10.1111/j.1365-313X.2007.03130.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jain, M., Kaur, N., Tyagi, A. K., and Khurana, J. P. (2006). The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 36–46. doi: 10.1007/s10142-005-0142-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jain, M., and Khurana, J. P. (2009). An expression compendium of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 276, 3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jain, M., Misra, G., Patel, R. K., Priya, P., Jhanwar, S., Khan, A. W.,et al. (2013). A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 74, 715–729. doi: 10.1111/tpj.12173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kelley, L. A., and Sternberg, M. J. E. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. doi: 10.1038/nprot.2009.2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khan, S., and Stone, J. M. (2007). Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226, 21–34. doi: 10.1007/s00425-006-0462-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuang, J. F., Zhang, Y., Chen, J. Y., Chen, Q. J., Jiang, Y. M., Lin, H. T.,et al. (2011). Two GH3 genes from longan are differentially regulated during fruit growth and development. Gene 485, 1–6. doi: 10.1016/j.gene.2011.05.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, R., Agarwal, P., Tyagi, A. K., and Sharma, A. K. (2012). Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genomics 287, 221–235. doi: 10.1007/s00438-011-0672-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Liu, Z. B., Shi, X., Hagen, G., and Guilfoyle, T. J. (1994). An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 106, 645–657. doi: 10.1104/pp.106.1.37
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D.,et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99.
Liu, S. Q., and Hu, H. F. (2013). Genome-wide analysis of the auxin response factor gene family in cucumber. Genet. Mol. Res. 12, 4317–4331. doi: 10.4238/2013.April.2.1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signaling network. Curr. Opin. Plant Biol. 8, 532–554. doi: 10.1016/j.pbi.2005.07.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ludwig-Muller, J. (2011). Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773. doi: 10.1093/jxb/erq412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ludwig-Müller, J., Jülke, S., Bierfreund, N. M., Decker, E. L., and Reski, R. (2008). Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 181, 323–338. doi: 10.1111/j.1469-8137.2008.02677.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mauch-Mani, B., and Mauch, F. (2005). The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant. Biol. 8, 409–414. doi: 10.1016/j.pbi.2005.05.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y. Y., Yoshizumi, T., Hasunuma, K.,et al. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. doi: 10.1046/j.1365-313x.2001.00957.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nobuta, K., Okrent, R. A., Stoutemyer, M., Rodibaugh, N., Kempema, L., and Wildermuth, M. C.,et al. (2007). The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156. doi: 10.1104/pp.107.097691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okrent, R. A., Brooks, M. D., and Wildermuth, M. C. (2009). Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754. doi: 10.1074/jbc.M806662200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, J. E., Park, J. Y., Kim, Y. S., Staswick, P. E., Jeon, J., Yun, J.,et al. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282, 10036–10046. doi: 10.1074/jbc.M610524200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peat, T. S., Bottcher, C., Newman, J., Lucent, D., Cowieson, N., and Davies, C. (2012). Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 24, 4525–4538. doi: 10.1105/tpc.112.102921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prasad, K., Sriram, P., and Usha, V. (2005). OsMADS1, a rice MADS box factor, controls differentiation of specific cell type in lemma and plea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928. doi: 10.1111/j.1365-313X.2005.02504.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prince, V. E., and Pickett, F. B. (2002). Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. doi: 10.1038/nrg928
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sani, E., Herzyk, P., Perrella, G., Colot, V., and Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14:R59. doi: 10.1186/gb-2013-14-6-r59
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Kato, T., Nakamura, Y.,et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15, 227–239. doi: 10.1093/dnares/dsn008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W.,et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Severin, A. J., Woody, J. L., Bolon, Y. T., Joseph, B., Diers, B. W., Farmer, A. D.,et al. (2010). RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. doi: 10.1186/1471-2229-10-160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, V. K., Garg, R., and Jain, M. (2013). A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol. J. 11, 691–701. doi: 10.1111/pbi.12059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C.,et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Staswick, P. E., Tiryaki, I., and Rowe M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. doi: 10.1105/tpc.000885
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N.,et al. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483. doi: 10.1046/j.1365-313X.2003.01973.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tebbji, F., Nantel, A., and Matton, D. P. (2010). Transcription profiling of fertilization and early seed development events in a solanaceous species using a 7.7 K cDNA microarray from Solanum chacoense ovules. BMC Plant Biol. 10:174. doi: 10.1186/1471-2229-10-174
Terol, J., Domingo, C., and Talon, M. (2006). The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371, 279–290. doi: 10.1016/j.gene.2005.12.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ulmasov, T., Liu, Z. B., Hagen, G., and Guilfoyle, T. J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. doi: 10.1105/tpc.7.10.1611
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G.,et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. doi: 10.1038/nbt.2491
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Verdier, J., Torres-Jerez, I., Wang, M., Andriankaja, A., Allen, S. N., He, J.,et al. (2013). Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74, 351–362. doi: 10.1111/tpj.12119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H.,et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, S. K., Bai, Y. H., Shen, C. J., Wu, Y. R., Zhang, S. N., Jiang, D. A.,et al. (2010). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics 10, 533–546. doi: 10.1007/s10142-010-0174-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wei, F., Coe, E., Nelson, W., Bharti, A. K., Engler, F., Butler E.,et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3:e123. doi: 10.1371/journal.pgen.0030123
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Westfall, C. S., Muehler, A. M., and Jez, J. M. (2013). Enzyme action in the regulation of plant hormone responses. J. Biol. Chem. 288, 19304–19311. doi: 10.1074/jbc.R113.475160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Westfall, C. S., Zubieta, C., Herrmann, J., Kapp, U., Nanao, M. H., and Jez, J. M. (2012). Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711. doi: 10.1126/science.1221863
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, J., Liu, S., Guan, X., Chen, L., He, Y., Wang, J.,et al. (2014). Genome-wide identification and transcriptional profiling analysis of auxin response-related gene families in cucumber. BMC Res. Notes 7:218. doi: 10.1186/1756-0500-7-218
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, J., Wang, F., Cheng, L., Kong, F., Peng, Z., Liu, S.,et al. (2011). Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 30, 2059–2073. doi: 10.1007/s00299-011-1113-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Young, N. D., Debellé, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K.,et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuan, H. Z., Zhao, K., Lei, H. J., Shen, X. J., Liu, Y., Liao, X.,et al. (2013). Genome-wide analysis of the GH3 family in apple (Malus×domestica). BMC Genomics 14:297. doi: 10.1186/1471-2164-14-297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, S. W., Li, C. H., Cao, J., Zhang, Y. C., Zhang, S. Q., Xia, Y. F.,et al. (2009). Altered architecture and enhanced drought tolerance in rice via the downregulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901. doi: 10.1104/pp.109.146803
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z. Q., Li, Q., Li, Z. M., Staswick, P. E., Wang, M. Y., Zhu, Y.,et al. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Phsiol. 145, 450–464. doi: 10.1104/pp.107.106021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: gene family, GH3 genes, legumes, RNA-seq, homology modeling, substrate specificities
Citation: Singh VK, Jain M and Garg R (2015) Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 5:789. doi: 10.3389/fpls.2014.00789
Received: 21 October 2014; Paper pending published: 10 December 2014;
Accepted: 18 December 2014; Published online: 14 January 2015.
Edited by:
Jun Yu, Beijing Institute of Genomics, ChinaReviewed by:
Swarup Kumar Parida, National Institute of Plant Genome Research, IndiaM. Awais Khan, International Potato Center, Peru
Copyright © 2015 Singh, Jain and Garg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohini Garg, Functional and Applied Genomics Laboratory, National Institute of Plant Genome Research, New Delhi 110067, India email:cm9oaW5pQG5pcGdyLmFjLmlu
 Vikash K. Singh
Vikash K. Singh Mukesh Jain
Mukesh Jain Rohini Garg
Rohini Garg