- 1Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Beijing Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops, Department of Vegetable Science, China Agricultural University, Beijing, China
Methylthioalkylmalate synthases (MAMs) encoded by MAM genes are central to the diversification of the glucosinolates, which are important secondary metabolites in Brassicaceae species. However, the evolutionary pathway of MAM genes is poorly understood. We analyzed the phylogenetic and synteny relationships of MAM genes from 13 sequenced Brassicaceae species. Based on these analyses, we propose that the syntenic loci of MAM genes, which underwent frequent tandem duplications, divided into two independent lineage-specific evolution routes and were driven by positive selection after the divergence from Aethionema arabicum. In the lineage I species Capsella rubella, Camelina sativa, Arabidopsis lyrata, and A. thaliana, the MAM loci evolved three tandem genes encoding enzymes responsible for the biosynthesis of aliphatic glucosinolates with different carbon chain-lengths. In lineage II species, the MAM loci encode enzymes responsible for the biosynthesis of short-chain aliphatic glucosinolates. Our proposed model of the evolutionary pathway of MAM genes will be useful for understanding the specific function of these genes in Brassicaceae species.
Introduction
Plants synthesize an immense number of defensive compounds to attack other organisms. An important model system for studying the role of chemical defenses in plants is the glucosinolates, a group of sulfur-rich secondary metabolites largely found in the plant family Brassicaceae (or Cruciferae). Glucosinolates and their degradation products play an important roles against microbial pathogens and herbivorous insects (Kroymann et al., 2003; Clay et al., 2009), and are responsible for the special flavors of Brassica vegetables such as turnip (Brassica rapa ssp. rapa), broccoli (Brassica oleracea var. italica), and caulifiower (B. oleracea var. botrytis) (Schonhof et al., 2004; Padilla et al., 2007). Furthermore, glucosinolates are of particular interests because of their cancer-preventing properties to human beings. They can inhibit carcinogen activation (Hecht, 2000; Nakajima et al., 2001) and carcinogenesis by triggering cell cycle arrest and stimulating apoptosis (Wittstock et al., 2003; Hayes et al., 2008).
Glucosinolates are derived from amino acids, and can be classified as aliphatic, aromatic, or indole glucosinolates according to their precursor amino acids (Halkier and Gershenzon, 2006; Sonderby et al., 2010). There are three independent processes for glucosinolate biosynthesis: (1) chain elongation of the precursor amino acid; (2) formation of the core structure; and (3) side chain modification. Differences in the degree of elongation and modification of the side chains lead to diverse glucosinolate structures. To date, more than 30 different glucosinolates have been identified in A. thaliana (Kliebenstein et al., 2001; Windsor et al., 2005).
Methylthioalkylmalate synthases (MAMs) are involved in amino acid chain elongation, and give rise to glucosinolates with diverse chain-lengths during the biosynthesis of methionine-derived glucosinolates in A. thaliana (Kliebenstein et al., 2001). It is thought that MAM genes are derived from isopropylmalate synthase genes (IPMS), which encode the enzymes that catalyze the first step of leucine biosynthesis (De Kraker et al., 2007). MAM genes are often found as clusters of tandem arrays but differentiated genes in the genome. In A. thaliana, the configuration of the MAM cluster comprises three genes in one tandem array, AtMAM2, AtMAM1, and AtMAM3 (AtMAM-L), though there are variations among accessions because of gene deletion or conversion events (Kroymann et al., 2003). AtMAM3 catalyzes the formation of all aliphatic glucosinolates, especially long-chain glucosinolates (6C, 7C, and 8C) in Arabidopsis (Textor et al., 2007). AtMAM2 and AtMAM1 catalyze the formation of short-chain aliphatic glucosinolates (3C and 4C) (Kliebenstein et al., 2001; Kroymann et al., 2003; Textor et al., 2004). In A. lyrata, which produces mainly 3C Met-derived glucosinolates and lower levels of long-chain glucosinolates (Windsor et al., 2005), the MAM cluster contains three directly repeated paralogous sequences (MAMa, MAMb, and MAMc). MAMa controls the first Met chain extension, MAMb is responsible for the long-chain Met-glucosinolates, and the function of MAMc is unclear (Benderoth et al., 2006).
The Brassicaceae is a medium-sized family that contains 338 genera and 3709 species, and includes many economically important crops (Warwick et al., 2006). This family can be split into two major groups: the Aethionema group, and the core group (Franzke et al., 2011). Three major lineages (lineages I, II, and III) have been proposed in the core group, based on the sequences of the chloroplast gene ndhF and supported by subsequent studies (Beilstein et al., 2006; Koch et al., 2007; Couvreur et al., 2010). The core group has undergone three ancient whole-genome duplication (WGD) events (Franzke et al., 2011). These events have played a crucial role in the genetic diversification and species radiation of lineages in Brassicaceae. Furthermore, whole-genome triplication events occurred in Brassica (Br-α), Leavenworthia alabamica (La-α), and Camelina sativa (Cs-α), as determined by analyses of their recently sequenced genomes (Haudry et al., 2013; Slotte et al., 2013; Cheng et al., 2014). These genome duplication events followed by gene losses during diploidization resulted in very complex relationships among the duplicated MAM genes in Brassicaceae (Benderoth et al., 2006; Sonderby et al., 2010). Furthermore, the local tandem duplication (TD) events occurred frequently at the MAM loci after genome duplication, making the relationship even more complicated. Although it is very challenging to clarify the evolutionary history of MAM genes in Brassicaceae, it is important to explore their evolution to deduce the functions of the diversified and duplicated MAM genes in extant Brassicaceae species.
Fortunately, the genomes of 13 crucifer species have been completely or partially sequenced, providing the opportunity to clarify the evolution of MAM genes. The sequenced crucifer species include: (1) five species from lineage I, they are the model plant A. thaliana (Initiative, 2000), A. lyrata (Hu et al., 2011), Capsella rubella (Slotte et al., 2013), L. alabamica (Haudry et al., 2013), and Camelina sativa (Kagale et al., 2014); (2) seven species from lineage II, B. rapa (Wang et al., 2011b), Thellungiella salsuginea (Wu et al., 2012), Schrenkiella parvula (synonym of Thellungiella parvula) (Dassanayake et al., 2011), Thellungiella halophila (Yang et al., 2013b), Sisymbrium irio (Haudry et al., 2013), B. oleracea (Liu et al., 2014), and Raphanus sativus (Kitashiba et al., 2014); (3) Aethionema arabicum, an early branching sister group to the core Brassicaceae group (Haudry et al., 2013). And there is no sequenced lineage III species available to date. Here, we took advantage of the whole genome sequences to investigate the evolution and diversification of MAM genes in Brassicaceae. Our analyses revealed the lineage-specific evolutionary routes that have led to the diversified structure of aliphatic glucosinolates.
Materials and Methods
Sources of Genome Data
B. rapa gene sequences for synteny analyses were obtained from BRAD (V1.5; http://brassicadb.org) (Cheng et al., 2012). Gene and genome data sets for A. thaliana were downloaded from The Arabidopsis Information Resource (TAIR9; http://www.Arabidopsis.org/index.jsp). The genomic dataset for A. lyrata was downloaded from the Joint Genome Initiative database (Gene model 6; http://genome.jgi-psf.org/Araly1/Araly1.home.html) (Hu et al., 2011). S. parvula and T. salsuginea datasets were obtained from Dassanayake et al. (2011) and Wu et al. (2012). Three MAM genes have been annotated in the genomes S. parvula and T. salsuginea, with two of them located at a tandem array. Gene and genome data for T. halophila were obtained from Yang et al. (2013b). The L. alabamica, S. irio and A. arabicum genomic datasets were obtained from Haudry et al. (2013). The C. sativa genomic dataset was obtained from Kagale et al. (2014) with nine syntenic and three non-syntenic MAM genes annotated. B. oleracea genomic was obtained from Liu et al. (2014), which has more than six annotated MAM genes. The R. sativus genomic dataset was obtained from Kitashiba et al. (2014) and contains at least two MAM genes.
Syntenic Ortholog Determination
Multi-syntenic orthologs between A. thaliana and other sequenced Brassicaceae species such as B. rapa, A. lyrata, S. parvula, T. salsuginea, T. halophila, L. alabamica, S. irio, A. arabicum, C. sativa, B. oleracea, and R. sativus were identified with the tool SynOrths and through the following URL: http://brassicadb.org/brad/searchSyntenytPCK.php.
Phylogenetic Analyses and Sequence Features
The full-length sequences of the MAM proteins encoded by genes in the 12 sequenced Brassicaceae species were aligned using Clustal W with default parameters (Larkin et al., 2007). A phylogenetic tree was constructed using the neighbor-joining method with Mega version 5.0 software (Tamura et al., 2011). Support for the topology was estimated from 1000 bootstrap replicates, and nodes occurring in less than 50% of the replicates were collapsed. Gene structures were determined by comparing coding and genomic sequences among MAM genes, based on information obtained from the Gene Structure Display Server (GSDS, Guo et al., 2007).
Motif Identification
MEME version 4.9.1 (Bailey et al., 2009) was used to identify the conserved motifs of syntenic MAM proteins in the sequenced Brassicaceae species. The parameters for the analysis were as follows: number of repetitions, 0 or 1; maximum number of motifs, 9; and optimum motif width, 6–200. The MAST program (Bailey and Gribskov, 1998) was used to search for each of the motifs in MAM sequences. The MEME program was used to extract each motif sequence from the syntenic MAM genes. The motifs were further characterized using the Conserved Domain Search Service (Marchler-Bauer et al., 2011).
Tests for Selective Pressure
Pairwise alignments of each motif in syntenic MAM genes were made using ClustalX2 (Larkin et al., 2007), with the corresponding protein sequences as the alignment guides. Gaps in the alignments were removed. The analysis of synonymous (Ks) and non-synonymous (Ka) substitution rates was carried out using the KaKs Calculator version 1.2 (Zhang et al., 2006). This program implements several candidate models of codon substitution in a maximum likelihood framework. We used the MS method to estimate Ka and Ks values with default parameters.
Results
Identification of MAM Genes in Genomes of Brassicaceae
Shared synteny describes genomic fragments in different species that are inherited from a common ancestor (Lyons et al., 2008). Syntenic genes are orthologs that located at these syntenic fragments, and they often share similar functions. We identified the MAM genes in the 13 sequenced Brassicaceae species according to their gene annotation information and their gene synteny relationship to the model plant species A. thaliana (http://brassicadb.org/brad/searchSyntenytPCK.php). B. rapa contains seven MAM genes including five syntenic and two non-syntenic ones (Wang et al., 2011a). We found one syntenic gene in L. alabamica (LaMAM), two syntenic genes in T. halophila, and four syntenic genes in each of A. arabicum and C. rubella (Table 1). No MAM genes were identified in S. irio. Table 1 lists the information of MAM genes in each genome of the 13 Brassicaceae species.
We analyzed the structures of the MAM genes in these Brassicaceae species. Most MAM genes shared conserved gene structures. Four MAM genes were very short, lost some conserved domains or were heavily differentiated to the other MAM genes. Compared with the majority of MAM genes, LaMAM in lineage I and SpMAM-2 and BoMAM-5 in lineage II had fewer exons, and TsMAM-3 had a different gene structure (Figure 1).
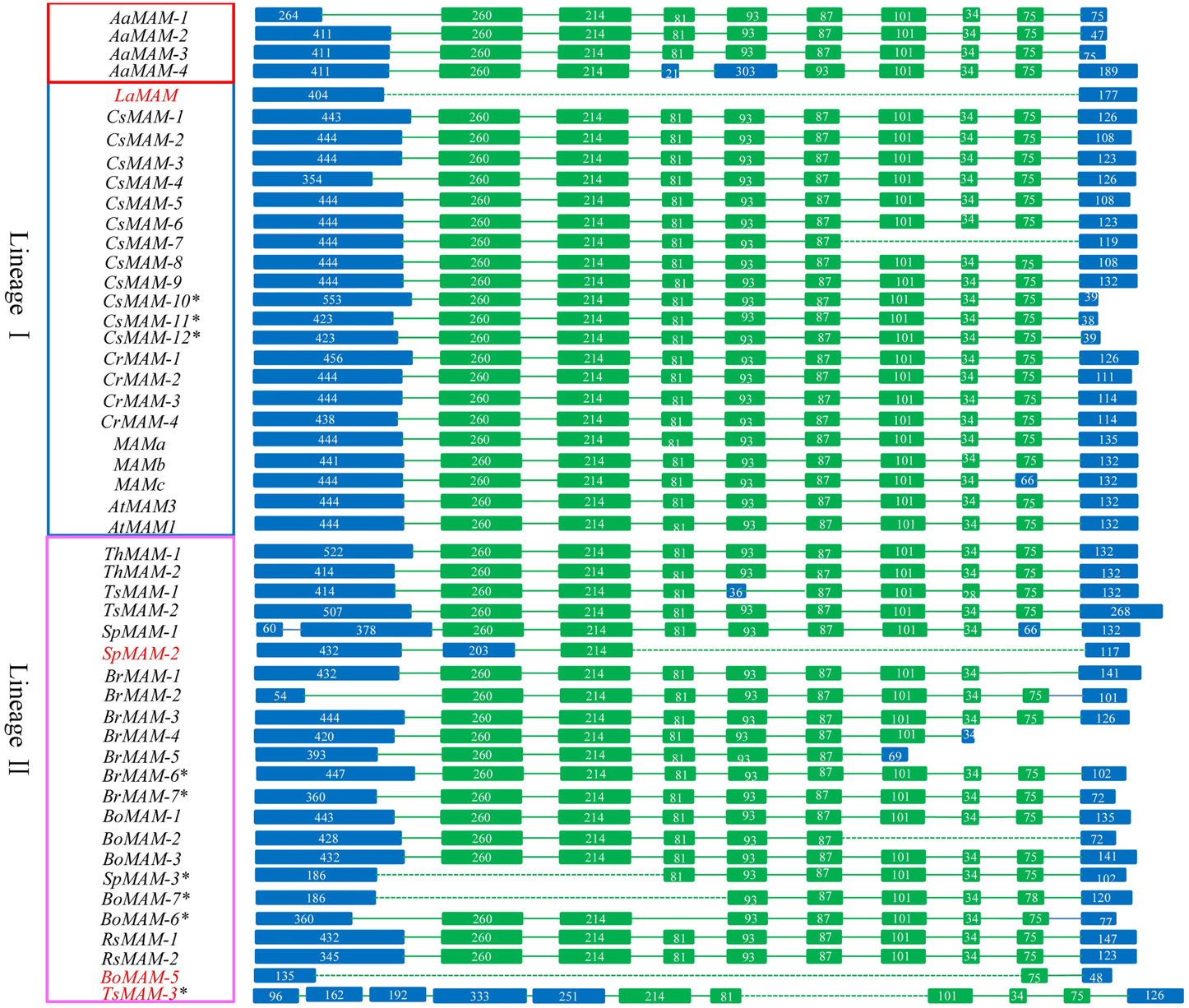
Figure 1. Schematic diagram of structure of MAM genes in 12 sequenced Brassicaceae species (excluding S. irio). Green blocks, conserved exons; blue blocks, variable exons. Most MAMs share the same gene structure, except for LaMAM in lineage I and SpMAM-2, BoMAM-5, and TsMAM-3 in lineage II (gene ID shown in red). Dashed lines indicate absence of exons from the region, compared with corresponding region in A. thaliana. Numbers in blocks indicate exon length. Genes shown in red, blue, and pink blocks (on left) are from A. arabicum, lineage I species, and lineage II species, respectively.
Two Major Groups of MAM Genes Evolved Independently in Core Brassicaceae
Using the neighbor-joining method, we constructed a phylogenetic tree for MAM genes, based on the sequences of MAM proteins in the 12 sequenced species (exclude S. irio). This analysis excluded four short/different genes, LaMAM, SpMAM-2, BoMAM-5, and TsMAM-3, but included IPMS genes (predicted by synteny analyses with AtIPMS1 and AtIPMS2 of Arabidopsis). Rice IPMS served as the outgroup (Figure 2).
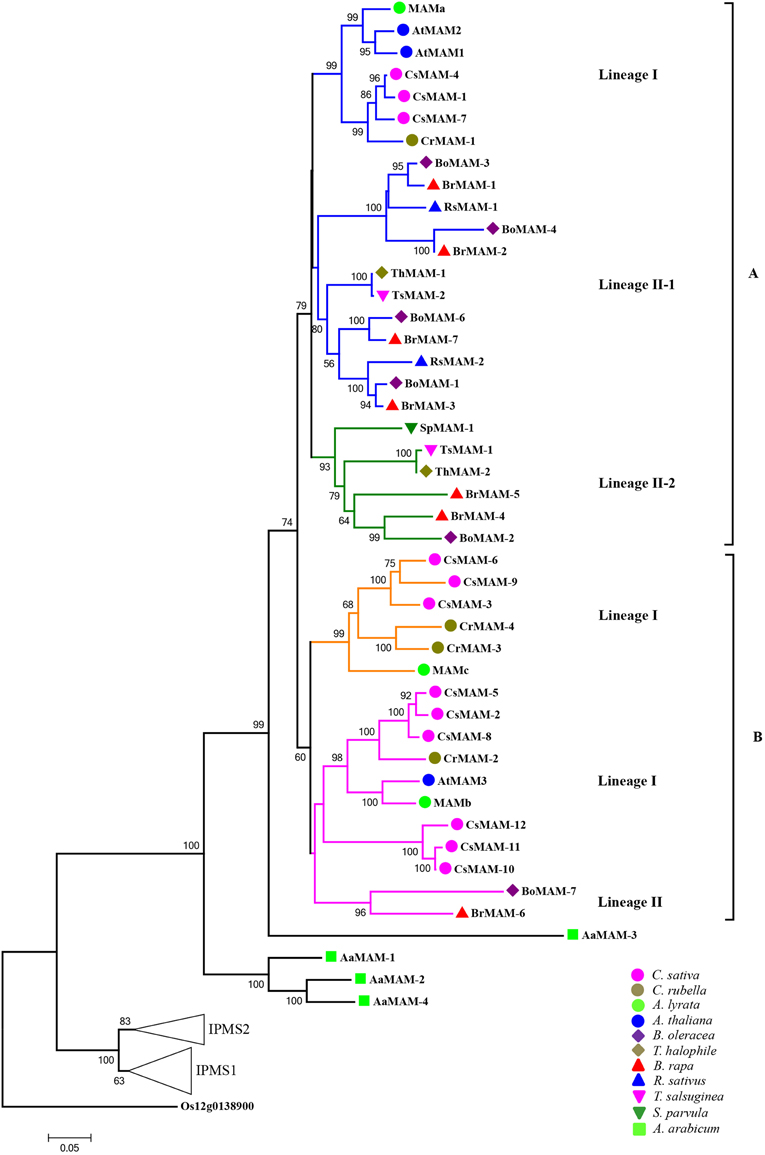
Figure 2. Phylogeny relationships of MAM genes in Brassicaceae, based on protein sequences of MAM genes. Phylogenetic tree was constructed using the full-length sequences of MAM and IPMS proteins encoded by genes in 13 sequenced Brassicaceae species (excluding LaMAM, SpMAM-2, BoMAM-5, and TsMAM-3). The rice gene Os12g0138900 encoded isopropylmalate synthase was used as the outgroup to build the phylogenetic tree. Numbers on branches indicate percentage bootstrap support (1000 replicates). Genes in the core Brassicaceae group formed two major groups (A and B) representing four clades (shown as colored branches). Triangles labeled IPMS1 and IPMS2 represent evolution of IPMS in Brassicaceae (see Supplemental data for more detail).
The phylogenetic tree showed that all of the A. arabicum MAM genes were clustered with the same A. arabicum MAM gene, AaMAM-3. The rest of the MAM genes in the sequenced species in the core Brassicaceae group originated from AaMAM-3, and formed two major relatively distinct groups, group A and group B. Each group contained one or more MAM genes from the same species, suggesting that duplication and gene diversification had occurred frequently in the subsequent evolution of MAM genes.
Group A included one clade of MAM genes from lineage I species (Figure 2, blue branches) and two clades of MAM genes from lineage II species (Figure 2, blue and green branches). Three previously identified Arabidopsis genes, AtMAM1, AtMAM2, and MAMa, which are responsible for short-chain Met-derived glucosinolate biosynthesis, were all in group A. Three C. sativa genes and one C. rubella gene clustered with the Arabidopsis MAM genes. The other two clades of MAM homologs from lineage II species (lineage II-1 and lineage II-2) clustered together with the lineage I clade, suggesting that they share the same or a similar function as that of Arabidopsis MAM genes, which encode enzymes that biosynthesize short-chain aliphatic glucosinolates. Furthermore, in each lineage II group, the MAM genes in the Brassica genus were more similar to those in the Thellungiella genus than to those in the Arabidopsis genus. This result is consistent with the finding that Brassiceae and Arabidopsis ancestors diverged before the split of Brassiceae-Thellungiella (Cheng et al., 2013).
Group B comprised MAM genes from the same lineage I species (Figure 2, yellow and pink branches) and two non-syntenic MAM genes from B. rapa and B. oleracea (Figure 2, pink branches). AtMAM3 and MAMb (pink clade) encode enzymes that biosynthesize long-chain glucosinolates, suggesting that the other MAM genes in this clade encode enzymes with similar functions. The function of MAMc (Figure 2, yellow clade) remains unclear (Benderoth et al., 2006). All of the MAM and IPMS genes in Brassicaceae clustered together, suggesting that they share a common ancestral gene originating from monocots.
MAM Ancestral Locus is a Tandem Gene Array and Shares Clear Synteny among Brassicaceae species
To assess the contributions of polyploidy and tandem gene duplications to MAM gene diversification, we conducted detailed synteny analyses within these 13 Brassicaceae genomes using the tool SynOrths (Cheng et al., 2012). The MAM region showed conserved synteny across all of the sequenced Brassicaceae genomes (Figure 3), consistent with other studies demonstrating extensive synteny among cruciferous genomes (Rossberg et al., 2001; Boivin et al., 2004; Kuittinen et al., 2004; Cheng et al., 2013).
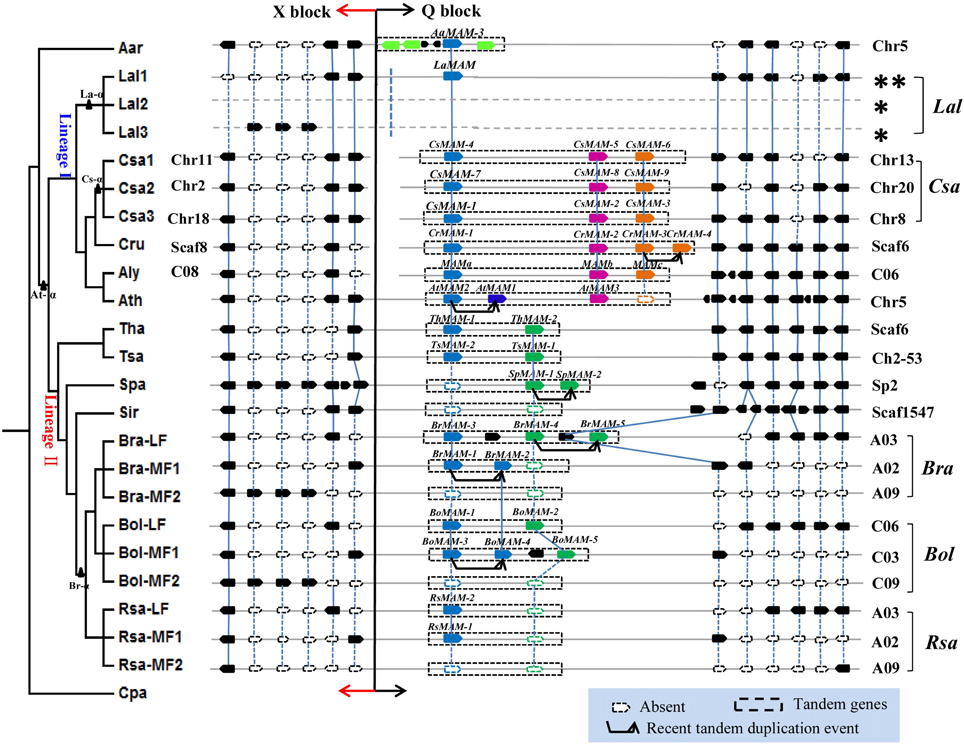
Figure 3. Synteny analyses of MAM region in the family Brassicaceae showing lineage-specific rearrangements, local tandem duplications, and diverse patterns of gene loss. Phylogenetic tree on the left represent the evolutionary relationships among 13 sequenced Brassicaceae species. Black triangles mark whole genome duplication (At-α) and whole genome triplication (La-α, Cs-α, and Br-α) events. MAM region shows conserved synteny across all sequenced Brassicaceae genomes. MAM tandem genes located at the end of Q Block, linked to X Block, are highlighted in colored blocks; blue and green blocks show genes responsible for elongation of short-chain glucosinolates, pink blocks show genes responsible for long-chain glucosinolate biosynthesis, orange blocks show genes whose function is unclear. Genes shown as solid and hollow (white) symbols in MAM region are present and absent, respectively. Chromosomal location of MAM region in each species is shown on the right or left. Single asterisk indicates that sub-genomic location of LaMAM region could not be determined because of scarce syntenic genes in this region. Dashed line at junction between X and Q Blocks and double asterisk on upper right of L. alabamica indicates that it is unclear whether these two blocks are on the same chromosome or not.
Three tandem MAM genes, with the same gene order and orientation, were located in the conserved syntenic region of most lineage I species, except for L. alabamica, which had only one LaMAM. The other two MAM homologs would have been lost from L. alabamica either after its divergence from A. arabicum, or more recently. Based on our phylogenic analysis, these three tandem genes were assumed to be responsible for the different chain lengths of glucosinolates (Figure 3, labeled with different colors). The genomes of species in Brassicaceae comprise 24 genomic blocks (A–X, also known as ancestral karyotypes, AK) (Parkin et al., 2005; Schranz et al., 2006). The X and Q Blocks are located on different chromosomes or scaffolds in C. sativa, C. rubella, and A. lyrata, but on the same chromosome in A. thaliana (Figure 3). These patterns of arrangement suggested that major chromosome rearrangements have occurred near the MAM loci. However, we could not identify whether the Q and X Blocks in L. alabamica were present on the same chromosome or not, because several different scaffolds were involved in this region. Furthermore, apart from the local TD event in A. thaliana that led to AtMAM1 and AtMAM2 (Benderoth et al., 2006), we found another recent TD event in C. rubella that led to the two homologous genes CrMAM-3 and CrMAM-4.
Two types of MAM genes with minor sequence variations (Figure 2, lineage II-1, lineage II-2) were located in the conserved syntenic region of most lineage II species. This syntenic region in lineage II species differed from the MAM loci of lineage I species. SpMAM-1 from S. parvula clustered with TsMAM-1 and ThMAM-2, but not with TsMAM-2 and ThMAM-1 (Figure 2), indicating that the ancestral type (blue) had been lost, and that SpMAM-1 gained the function of the MAM ancestral gene. In S. irio, there were no MAM genes in this syntenic region. The phylogenetic tree indicated that SpMAM-1, BrMAM-1, BrMAM-4 and BoMAM-3 had undergone recent TD events, giving rise to more MAM homologous genes in S. parvula, B. rapa, and B. oleracea, respectively, to allow adaptation to environmental changes.
Lineage-Specific Evolutionary Patterns of MAM Loci
Based on the phylogeny and synteny analyses across the sequenced Brassicaceae species, we proposed a lineage-specific evolution pattern for the syntenic MAM loci (Figure 4). The evolution of MAM loci could be divided into two independent lineage-specific routes after the divergence of A. arabicum; a lineage I-specific route and a lineage II-specific route.
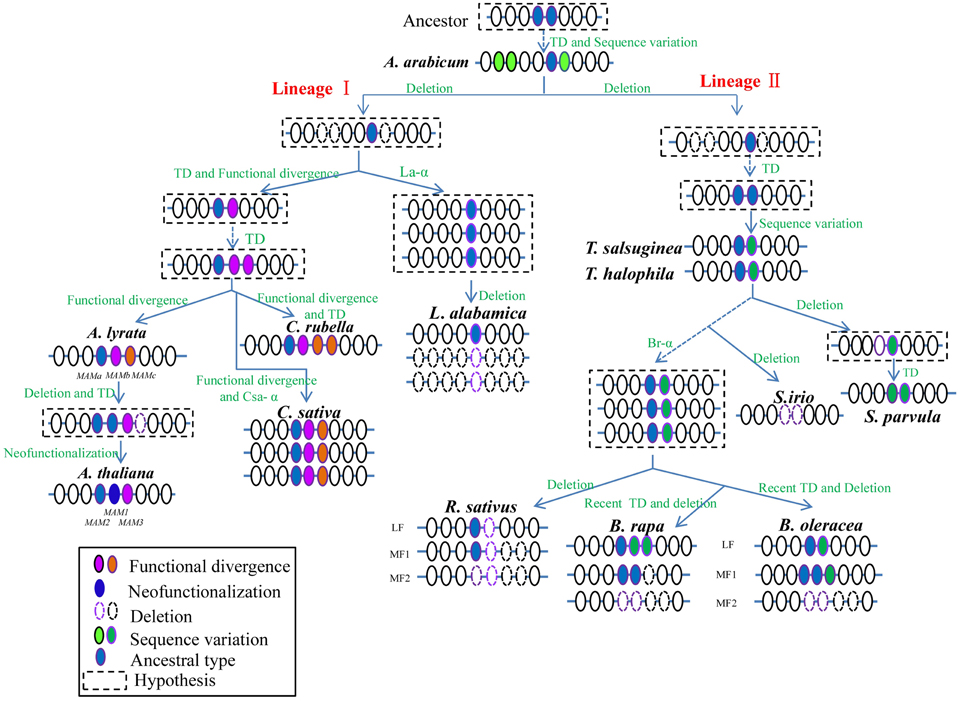
Figure 4. Putative model for MAM loci lineage-specific evolution in Brassicaceae. After the divergence of A. arabicum, the evolution of MAM loci proceeded via two independent lineage-specific routes: the lineage I-specific evolutionary route, and the lineage II-specific route. Colored solid circles represent MAM genes, white circles show their flanking genes. Changes in colors of MAM genes and dashed lines on flanking genes indicate specific evolutionary events. Genes within dashed lines indicate presumed process during evolution. TD, tandem duplication.
Before the separation of A. arabicum and the core Brassicaceae group, an ancient ancestral locus with one or two MAM homologs should have evolved to give rise to multiple AaMAM genes through several TD events. Then, AaMAM-3 diverged from the multiple copies, forming the ancestral MAM gene (Figure 4, blue circles) of the core Brassicaceae species. The other three AaMAM homologs (Figure 4, green circles) were possibly lost from the core Brassicaceae species. The lineage-specific evolution of MAM loci in the core Brassicaceae group was assumed to proceed alongside the split of the three major lineages (lineages I, II, and III), possibly driven by At-α.
During the formation of the MAM loci in lineage I, the ancestral gene was first amplified by a TD event, followed by functional divergence to give rise to two distinct MAM genes (Figure 4, blue and pink). Then, the duplicated gene (Figure 4, pink circles) underwent another round of TD and functional divergence events to generate three MAM genes in A. lyrata (MAMa, MAMb, and MAMc) and C. rubella. Subsequently, in C. rubella, one CrMAM gene (Figure 4, orange circles) underwent another local TD event, leading to four CrMAM genes, possibly encoding proteins with three different functions. This TD may have occurred soon after the split or more recently. In A. thaliana, AtMAM1, and AtMAM2 evolved from a MAMa duplication event. AtMAM2 retained the original function, while AtMAM1 acquired a new function (neofunctionalization) (Benderoth et al., 2006). AtMAM3 originated from MAMb, and MAMc was lost. C. sativa underwent an extra whole genome triplication (WGT) event (Cs-α). In the well-preserved hexaploid genome of C. sativa, three clusters with a total of nine syntenic CsMAM genes were retained. However, in L. alabamica, which also underwent a WGT event (La-α), only one syntenic LaMAM gene was retained because of extensive gene losses during rediploidization.
Compared with the lineage I route, the lineage II route represents a different evolutionary process for MAM loci. After the first amplification of the ancestral gene, two copies (Figure 4, blue circles) were retained until the speciation of T. halophila and T. salsuginea, when minor sequence variations arose in the duplicated gene, but were not substantial enough to lead to functional divergence (Figure 4, green circles). When S. parvula diverged, the ancestral gene (Figure 4, blue circles) tended to degenerate and be lost, but the duplicated gene (Figure 4, green circles) underwent a local TD event to produce two SpMAM homologs. However, in S. irio, the two syntenic MAM copies were completely lost, either when it diverged from Thellungiella or more recently. Subsequently, when the Brassica genus split out, B. rapa, B. oleracea, and R. sativus retained various copies of MAM genes that were generated from WGT (Br-α) followed by biased gene lost. The fact that there are two pairs of duplicates (BrMAM-1/BrMAM-2 and BrMAM-4/BrMAM-5) in the LF and MF1 subgenomes of B. rapa, and one pair (BoMAM-3/BoMAM-4) in the LF subgenome of B. oleracea further suggested that recent TD events occurred after Br-α.
Different Structures of Proteins Encoded by Syntenic MAM Genes
To assess whether the sequence divergence among duplicates changed the proteins structures of syntenic MAM genes, we analyzed nine conserved motif patterns in 41 MAM proteins in the 12 Brassicaceae species using the MEME tool (Bailey et al., 2009) (Figure 5). Motifs 1, 2, 3, and 4 were identified to belong to the DRE_TIM_metallolyase super family [cl18962] conserved domain, and motifs 5, 6, 7, 8, and 9 were to belong to the PLN03228 (methylthioalkylmalate synthase) conserved domain reported by the Conserved Domain Search Service (Marchler-Bauer et al., 2011).
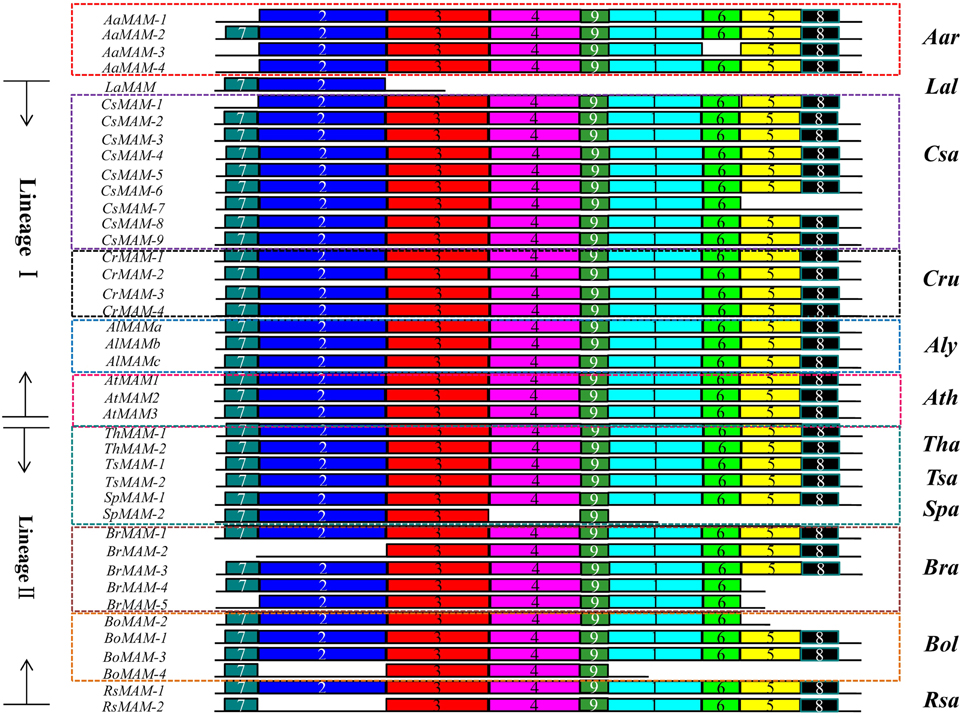
Figure 5. Motif structures of syntenic MAM genes in Brassicaceae. MAM genes in lineage I encode proteins with nine intact motifs (except for LaMAM, CsMAM-1, and CsMAM-7). In lineage II species, seven genes in four species encode proteins with that lack some motifs, indicating that the proteins encoded by genes at the MAM loci underwent rapid functional divergence. Different colored boxes with numbers show protein motifs.
The MAM genes in lineage I species encode proteins with more conservative structures than those of proteins encoded by genes in A. arabicum and lineage II species. One MAM genes of A. arabicum encode protein with intact motifs, while the others lack one or two motifs. In lineage I, apart from LaMAM, CsMAM-1, and CsMAM-7, the other MAM genes all encode proteins with nine intact motifs. In lineage II, especially in the Brassica genus, three-fifths of the syntenic MAM genes of B. rapa and half of the MAM genes of B. oleracea and R. sativus encode proteins with incomplete motifs, suggesting that local TD and Br-α events led to major divergences in protein structure. The lost motifs were either [cl18962] or PLN03228 motifs for different MAM gene copies. Because enzymes with these motifs catalyze the first committed step in leucine biosynthesis or methionine biosynthesis, the lost motifs may result in lower enzyme activity, or inactive enzymes.
Purifying Selection Events were Detected and Even More Frequent than Positive Selection
Next, we inferred whether selection acted on the nine conserved motifs during the lineage-specific evolution of MAMs. We calculated the non-synonymous/synonymous substitution ratio (Ka/Ks) for each motif of above syntenic MAM genes using the tool KaKs Calculator (Zhang et al., 2006). Motifs 1, 2, and 5 showed an overall excess, while motifs 3, 4, 6, and 8 showed more than 95% excess of synonymous changes relative to non-synonymous changes (Ka/Ks<1) (Supplementary Table 3), indicating strong purifying selection. However, Ka/Ks analysis for motifs 7 and 9 of the overall syntenic MAM genes showed that more than 10% of the sequences had undergone rapid non-synonymous to synonymous substitutions (Ka/Ks>1), indicating positive selection. The excess of non-synonymous changes in these two motifs may reflect general variations in the 3D structural framework that differentiate the activities or function of MAM enzymes.
To test this hypothesis, we calculated the Ka/Ks ratio of motif 7 and motif 9 respectively of syntenic MAM genes of lineages I and lineage II species. We found that more than 80% ancestral-like genes (Figure 3, blue blocks) showed excess synonymous changes (Ka/Ks<1) (Table 2), indicating the general conservation of these two motifs. That is, the activity of the enzymes encoded by these genes was conserved. However, in motif 7 (Table 2), more than 80% of the lineage I ancestral-like genes vs. MAMb, 70% of the lineage II genes vs. MAMb, and 50% of the lineage II ancestral-like genes vs. AtMAM3 showed a Ka/Ks of greater than 1, indicating an excess of non-synonymous changes. This result showed that positive selection had driven motif 7 of MAMb and AtMAM3 (Figure 3, pink blocks) distinguished from the ancestral-like genes (Figure 3, blue blocks).
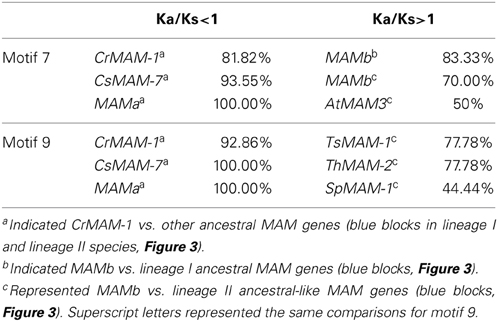
Table 2. Statistical analysis of Ka/Ks for motif 7 and motif 9 of syntenic MAM genes responsible for short- and long-chain glucosinolate biosynthesis.
For species from lineage II, the split out of Thellungiella is prior to the origin of Brassica genus (Haudry et al., 2013). We calculated the Ka/Ks ratio for three relatively ancient MAM genes of Thellungiella vs. the other lineage II MAM genes to test the selection pressure. We found for the motif 9 (Table 2), more than 75%, 75%, and 44% of the ancestral-like genes in lineage II vs. TsMAM-1, ThMAM-2, and SpMAM-1, respectively, had a Ka/Ks greater than 1. This indicated that there was strong positive selection driving the divergence between the ancestral-like genes and their tandem duplicates in lineage II species (Figure 3, green blocks).
Discussion
The MAMs encoded by the MAM gene cluster are central to the diversification of Met-derived aliphatic glucosinolates in crucifer species. We took advantage of the completely or partially sequenced genomes of 13 Brassicaceae species (A. thaliana, A. lyrata, C. rubella, L. alabamica, C. sativa, B. rapa, T. salsuginea, S. parvula, T. halophila, S. irio, B. oleracea, R. sativus, and A. arabicum (Initiative, 2000; Dassanayake et al., 2011; Hu et al., 2011; Wang et al., 2011b; Wu et al., 2012; Haudry et al., 2013; Slotte et al., 2013; Yang et al., 2013b; Kagale et al., 2014; Kitashiba et al., 2014; Liu et al., 2014) to investigate the evolution and diversification of MAM genes at specific MAM loci in Brassicaceae. We proposed that the syntenic loci of MAM gene, which underwent frequent tandem duplications, evolved via two independent lineage-specific routes after their divergence from A. arabicum. Our analyses indicate that positive selection has driven the diversification of MAM genes involved in ailphatic glucosinolates. These findings will help further study of the function of MAM genes in Brassicaceae species.
A lineage I-Specific Chromosome Rearrangement Occurred Near MAM Loci
Genome polyploidization is an evolutionary process that plays a key role in generating the diversity of plant species and providing abundant genetic materials for the evolution or expansion of gene families (Hittinger and Carroll, 2007; Spillane et al., 2007). The chromosomal constitution of each organism is reflected by its karyotype. Each species has a particular number of chromosomes with unique sizes and shapes (Schubert and Lysak, 2011). The genomes of species in Brassicaceae comprise 24 genomic blocks (A–X, also known as ancestral karyotypes, AK) (Parkin et al., 2005; Schranz et al., 2006), which can be observed in the recently sequenced genomes of A. lyrata, S. parvula, and B. rapa (Dassanayake et al., 2011; Hu et al., 2011; Wang et al., 2011b), as well as in the genome of the model plant A. thaliana (Initiative, 2000), etc.
In all of the sequenced genomes of Brassicaceae, the MAM loci are located at the end of Q Block, linked to X Block. Strikingly, the Q and X Blocks are distributed on different chromosomes in C. sativa, C. rubella, and A. lyrata, but on the same chromosome in A. thaliana (Chr5) and in lineage II species. This arrangement is proof of a large-scale chromosomal rearrangement event that occurred in lineage I species before the divergence of A. lyrata and A. thaliana approximately 10 million years ago (Hu et al., 2011), but after their split from A. arabicum. The lineage-I-specific genome rearrangement can be used to reconstruct ancient karyotypes and to detect very old polyploidization events in other lineage I species (Semon and Wolfe, 2007). Even though a large chromosomal-scale rearrangement event occurred during speciation, the MAM loci were not deleted, but were retained with highly conserved syntenic arrangements (Figure 3). Their retention and high degree of conservation are consistent with the importance of the enzymes encoded by genes at the MAM loci in secondary metabolite biosynthesis.
Diversified Functions of MAM Loci may Result in Different Met-Derived Aliphatic Glucosinolates
Glucosinolates are secondary metabolites that are well known for their role in resistance to insects and pathogens, as well as for their cancer-prevention properties. There are large differences among glucosinolate profiles because of differences in their amino acid precursors. Our analyses of the MAM loci in the 13 sequenced species indicate that there were two independent lineage-specific patterns of evolution.
In the early diverged sister A. arabicum, the four identified AaMAM genes could explain the abundance of 3-methylsulfonylpropyl, 4-methylsulfonylbutyl, and 8-methylsulfinyloctyl aliphatic glucosinolates, produced via one, two, and six rounds of carbon-chain extension, respectively (Al-Shammary, 1987). Additionally, the distinct glucosinolate profiles suggest these AaMAM genes could have gained diverse functions to biosynthesize glucosinolates with different chain-length. However, these four AaMAM genes could not be distinguished from MAM1 or MAM3 in our phylogenetic analyses. However, one gene (AaMAM-3) that differed from the others was identified as the ancestor of those in the other Brassicaceae species. In future research, it will be useful to experimentally test their activities in vivo or in vitro to determine which ones are responsible for short chain elongation or long chain elongation, respectively.
In the model plant A. thaliana and its congener A. lyrata, the natural variations in MAMs were shown to determine the glucosinolate phenotypes (Kroymann et al., 2001, 2003; Textor et al., 2004; Benderoth et al., 2006; Heidel et al., 2006). In C. sativa, which retains a highly undifferentiated hexaploid genome structure, 12 MAM genes were identified; 3 were derived from MAMa, 3 from MAMc, and 6 from MAMb. The large-scale expansion of CsMAM genes, especially those originating from MAMb, could contribute to the large quantities of long-chain aliphatic glucosinolates such as glucoarabin (7C), glucocamelinin (10C), and 11-(methylsulfinyl)-undecylglucosinolate (11C) in C. sativa seeds (Berhow et al., 2013). In C. rubella, the four annotated syntenic MAM genes are responsible for the biosynthesis of different chain-length glucosinolates; therefore, glucosinolates with various chain-lengths should be detectable in the organs of this species.
The six sequenced species in lineage II contain abundant short-chain glucosinolates and trace amounts long-chain glucosinolates. This is consistent with the presence of syntenic MAM genes encoding enzymes responsible for short-chain glucosinolate biosynthesis. For example, in Thellungiella, two syntenic tandem MAM genes with a MAMa function (Figure 2) were annotated in T. salsuginea, S. parvula, and T. halophila (Table 1). The presence of these tandem MAM genes can account for the three abundant short-chain glucosinolates, allylglucosinolate (3C), 3-ethylsulphinylpropylglucosinolate (3C), and 3-methylthiopropylpropylglucosinolate (3C) identified in Thellungiella flowers, siliques, and seeds (Pang et al., 2009, 2012). The other long-chain glucosinolate (10MSD) present at trace levels in T. salsuginea and S. parvula should be biosynthesized by enzymes encoded by the non-syntenic genes TsMAM-3 and SpMAM-3. However, in T. halophila, the two syntenic MAM genes are thought to encode enzymes involved in short-chain glucosinolate biosynthesis. Therefore, we proposed that the T. halophila genome should contain another non-syntenic MAM gene encoding an enzyme for long-chain aliphatic glucosinolate biosynthesis. In Brassica, which experienced an additional WGT event compared with the model plant A. thaliana, five syntenic MAM genes sharing MAMa functions encode biosynthetic enzymes for the most abundant short-chain glucosinolates. These genes are involved in the biosynthesis of gluconapin (4C), glucobrassicanapin (5C), and progoitrin (4C) in B. rapa (Padilla et al., 2007; Lou et al., 2008; Kim et al., 2010) and in the biosynthesis of the major aliphatic glucosinolates sinigrin (3C) and glucoiberin (3C) in B. oleracea (Cartea et al., 2008). In R. sativus, two syntenic MAM genes were identified that could be responsible for the biosynthesis of the three main short-chain glucosinolates: glucosisaustricin (2C), glucosisymbrin (3C), and glucoraphenin (4C) (Ediage et al., 2011).
Some lineage I species (e.g., Lepidium sativum, Cardamine hirsuta, and Rorippa islandica) and lineage II species (e.g., S. irio) lack Met-derived glucosinolates but are rich in Val-, Ile-, and Leu-derived glucosinolates or aromatic glucosinolates (Franzke et al., 2011). The results of our synteny analyses suggest that the MAM genes of S. irio were lost after its divergence from Thellungiella, resulting in the lack of aliphatic glucosinolates. Therefore, we speculate that MAM genes should also have been lost from L. sativum, C. hirsute, and R. islandica when the genetic backgrounds were altered or as a result of environmental adaptation.
Positive Selection Drove the Diversification of MAM Loci in Core Brassicaceae
Based on the results obtained here, we propose a scenario for the evolutionary history of the MAM loci (Figure 4) in the family Brassicaceae. In this scenario, all MAM genes in the core Brassicaceae group evolved from a shared ancestor with A. arabicum (AaMAM-3) but were subjected to lineage-specific evolutionary processes by positive selection.
In lineage I, before the speciation of A. lyrata and C. rubella, two MAM genes with different functions were generated in the ancestor by amplification and functional differentiation. Subsequently, the duplicated gene (e.g., that in A. lyrata, C. rubella, and C. sativa) or the ancient gene (e.g., that in A. thaliana) underwent further rounds of TD and functional divergence, giving rise to three MAM genes with distinct functions. During the lineage I evolutionary process, the most conservative motifs (motifs 1, 2, and 5) underwent strong purifying selection, allowing the proteins encoded by the ancestral MAM genes (Figure 3, blue blocks) to share the same or similar enzyme activity. However, between the ancestral genes and the duplicates, motif 9 exhibited an excess of non-synonymous relative to synonymous changes. This indicated that strong positive selection had forced the diversification of the function of enzymes encoded by MAM genes. Additionally, the Ka/Ks ratios of motif 7 were greater than 1 in the following pairs: AtMAM3 vs. CsMAM-3, AtMAM2 vs. CsMAM-3, CsMAM-7 vs. CsMAM-3 as well as AtMAM1 vs. CsMAM-9, AtMAM2 vs. CsMAM-9, CsMAM-1 vs. CsMAM-9, and CsMAM-7 vs. CsMAM-9. This result indicated that positive selection had driven the above genes (Figure 3, orange blocks) to diverge from the other MAM genes (Figure 3, blue and pink blocks). We found that three motifs (motifs 3, 7, and 9) of AtMAM1 vs. AtMAM2 had excess non-synonymous changes (data not shown), indicating strong positive selection of the gene AtMAM1, consistent with the results of a previous report (Benderoth et al., 2006).
For species Thellungiella and Brassica from lineage II, TD and minor sequence variations could have occurred frequently to generate two types of MAM genes, which encoded products with the same or similar functions, under strong positive selection. Besides motifs 7 and 9, which provide positive selection pressure for the lineage II homologs, in motif 8, the pairwise analysis of the Ka/Ks for CsMAM-1 vs. ThMAM-2 (Ka/Ks = 1.92) and for CsMAM-1 vs. TsMAM-1(Ka/Ks = 1.92) showed that there were strong positive selection pressures forcing the formation of the MAM loci in lineage II species. In this study, we found that various gene duplications (e.g., WGD and TD), functional divergence, and positive selection of MAM loci, via two different lineage-specific evolutionary routes, have contributed to the diversification of glucosinolates.
Previous studies have reported that different glucosinolates are involved in resistance responses to different herbivores and pathogens (Kroymann et al., 2003; Clay et al., 2009) and in increasing plant fitness (Manzaneda et al., 2010). Some specific glucosinolates (e.g., glucoraphanin, glucoraphenin, 4C) show powerful cancer-preventive properties (Hecht, 2000). MAM genes play an essential role in the diversity of aliphatic glucosinolates. In this context, understanding the evolution of the MAM genes will help us to understand the specific functions of MAM genes in the recently sequenced Brassicaceae species. For example, based on our analyses of the phylogenetic and syntenic relationships, we can predict which genes are related to long-chain glucosinolate biosynthesis and which are related to short-chain glucosinolate biosynthesis in C. sativa, and then predict its glucosinolates profile. In B. napus, the main glucosinolate in the leaves and seeds was identified as alkenyl glucosinolate (3C); this information was used to enhance the quality of rapeseed (Mithen, 1992; Parkin et al., 1994). Consistent with that study, our results predicted that BnMAM genes encode enzymes catalyzing short-chain glucosinolate biosynthesis. Indeed, in the recently published B. napus genome, seven syntenic BnMAM genes were annotated on the recent allopolyploid genome (Chalhoub et al., 2014). Our results suggest that these seven genes encode enzymes involved in short-chain glucosinolate biosynthesis.
Thus, understanding the evolution of MAM genes is not only helpful for answering questions about the patterns of conservation and divergence of MAM genes and the forces driving their evolution, but also for predicting the function of MAM genes and the glucosinolate profiles in Brassicaceae species. Therefore, this information will be useful for altering the glucosinolate profiles of Brassicaceae crops.
Author Contributions
Xiaowu Wang, Feng Cheng, and Wencai Yang designed the research. Jifang Zhang performed the research and analyzed the data. Xiaobo Wang, Feng Cheng, Jian Wu, and Jianli Liang contributed new computational tools and data. Jifang Zhang and Xiaowu Wang wrote the article.
Conflict of Interest Statement
The Associate Editor Tiegang Lu and Review Editor Xiao Han declare that, despite being affiliated to the same institution as authors Jifang Zhang, Xiaobo Wang, Feng Cheng, Jian Wu, Jianli Liang and Xiaowu Wang, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bo Liu and Chao Sun for their helpful suggestions. This work was supported by the National Program Fund for Key Basic Research Projects (The 973 Program: 2013CB127000 and 2012CB113900) and the National High Technology R&D Program of China (2012AA100101). Research was carried out in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00018/abstract
References
Al-Shammary, I. K. (1987). Distribution and chemotaxonomic significance of glucosinolates in certain Middle-Eastern cruciferae. Biochem. Syst. Ecol. 15, 559–569. doi: 10.1016/0305-1978(87)90106-2
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beilstein, M. A., Al-Shehbaz, I. A., and Kellogg, E. A. (2006). Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93, 607–619. doi: 10.3732/ajb.93.4.607
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Benderoth, M., Textor, S., Windsor, A. J., Mitchell-Olds, T., Gershenzon, J., and Kroymann, J. (2006). Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 103, 9118–9123. doi: 10.1073/pnas.0601738103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berhow, M. A., Polat, U., Glinski, J. A., Glensk, M., Vaughn, S. F., Isbell, T., et al. (2013). Optimized analysis and quantification of glucosinolates from Camelina sativa seeds by reverse-phase liquid chromatography. Ind. Crops Prod. 43, 119–125. doi: 10.1016/j.indcrop.2012.07.018
Boivin, K., Acarkan, A., Mbulu, R. S., Clarenz, O., and Schmidt, R. (2004). The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135, 735–744. doi: 10.1104/pp.104.040030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cartea, M. E., Velasco, P., Obregón, S., Padilla, G., and de Haro, A. (2008). Seasonal variation in glucosinolate content in Brassica oleracea crops grown in northwestern Spain. Phytochemistry 69, 403–410. doi: 10.1016/j.phytochem.2007.08.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wincker, P., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, F., Mandakova, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, F., Wu, J., Fang, L., and Wang, X. (2012). Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 3:198. doi: 10.3389/fpls.2012.00198
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, F., Wu, J., and Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Hortic. Res. 1:14024. doi: 10.1038/hortres.2014.24
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Couvreur, T. L., Franzke, A., Al-Shehbaz, I. A., Bakker, F. T., Koch, M. A., and Mummenhoff, K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27, 55–71. doi: 10.1093/molbev/msp202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dassanayake, M., Oh, D. H., Haas, J. S., Hernandez, A., Hong, H., Ali, S., et al. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918. doi: 10.1038/ng.889
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Kraker, J. W., Luck, K., Textor, S., Tokuhisa, J. G., and Gershenzon, J. (2007). Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 143, 970–986. doi: 10.1104/pp.106.085555
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ediage, E. N., Di Mavungu, J. D., Scippo, M. L., Schneider, Y. J., Larondelle, Y., Callebaut, A., et al. (2011). Screening, identification and quantification of glucosinolates in black radish (Raphanus sativus L. niger) based dietary supplements using liquid chromatography coupled with a photodiode array and liquid chromatography-mass spectrometry. J. Chromatogr. A 1218, 4395–4405. doi: 10.1016/j.chroma.2011.05.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., and Mummenhoff, K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16, 108–116. doi: 10.1016/j.tplants.2010.11.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, T. L, and Gribskov, M. (1998). Combining evidence using p-values: application to sequence homology searches. Bioinformatics 12, 48–54.
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). [GSDS: a gene structure display server]. Yi Chuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halkier, B. A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haudry, A., Platts, A. E., Vello, E., Hoen, D. R., Leclercq, M., Williamson, R. J., et al. (2013). An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45, 891–898. doi: 10.1038/ng.2684
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayes, J. D., Kelleher, M. O., and Eggleston, I. M. (2008). The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 47, 73–88. doi: 10.1007/s00394-008-2009-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hecht, S. S. (2000). Inhibition of carcinogenesis by isothiocyanates. Drug Metab. Rev. 32, 395–411. doi: 10.1081/DMR-100102342
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heidel, A. J., Clauss, M. J., Kroymann, J., Savolainen, O., and Mitchell-Olds, T. (2006). Natural variation in MAM within and between populations of Arabidopsis lyrata determines glucosinolate phenotype. Genetics 173, 1629–1636. doi: 10.1534/genetics.106.056986
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hittinger, C. T., and Carroll, S. B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–U671. doi: 10.1038/nature06151
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, T. T., Pattyn, P., Bakker, E. G., Cao, J., Cheng, J. F., Clark, R. M., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481. doi: 10.1038/ng.807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Initiative, A. G. (2000). Analyses of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kagale, S., Koh, C., Nixon, J., Bollina, V., Clarke, W. E., Tuteja, R., et al. (2014). The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5:3706. doi: 10.1038/ncomms4706
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. K., Chu, S. M., Kim, S. J., Lee, D. J., Lee, S. Y., Lim, S. H., et al. (2010). Variation of glucosinolates in vegetable crops of Brassica rapa L. ssp. pekinensis. Food Chem. 119, 423–428. doi: 10.1016/j.foodchem.2009.08.051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kitashiba, H., Li, F., Hirakawa, H., Kawanabe, T., Zou, Z., Hasegawa, Y., et al. (2014). Draft Sequences of the Radish (Raphanus sativus L.) Genome. DNA Res. 21, 481–490. doi: 10.1093/dnares/dsu014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kliebenstein, D. J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., et al. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825. doi: 10.1104/pp.126.2.811
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koch, M. A., Dobes, C., Kiefer, C., Schmickl, R., Klimes, L., and Lysak, M. A. (2007). Supernetwork identifies multiple events of plastid trnF(GAA) pseudogene evolution in the Brassicaceae. Mol. Biol. Evol. 24, 63–73. doi: 10.1093/molbev/msl130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kroymann, J., Donnerhacke, S., Schnabelrauch, D., and Mitchell-Olds, T. (2003). Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. U.S.A. 100(Suppl. 2), 14587–14592. doi: 10.1073/pnas.1734046100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kroymann, J., Textor, S., Tokuhisa, J. G., Falk, K. L., Bartram, S., Gershenzon, J., et al. (2001). A gene controlling variation in arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127, 1077–1088. doi: 10.1104/pp.010416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuittinen, H., De Haan, A. A., Vogl, C., Oikarinen, S., Leppala, J., Koch, M., et al. (2004). Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168, 1575–1584. doi: 10.1534/genetics.103.022343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lou, P., Zhao, J., He, H., Hanhart, C., Del Carpio, D. P., Verkerk, R., et al. (2008). Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New Phytol. 179, 1017–1032. doi: 10.1111/j.1469-8137.2008.02530.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Manzaneda, A. J., Prasad, K. V., and Mitchell-Olds, T. (2010). Variation and fitness costs for tolerance to different types of herbivore damage in Boechera stricta genotypes with contrasting glucosinolate structures. New Phytol. 188, 464–477. doi: 10.1111/j.1469-8137.2010.03385.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marchler-Bauer, A., Lu, S. N., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., Deweese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rossberg, M., Theres, K., Acarkan, A., Herrero, R., Schmitt, T., Schumacher, K., et al. (2001). Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and capsella genomes. Plant Cell 13:11. doi: 10.1105/tpc.13.4.979
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mithen, R. (1992). Leaf glucosinolate profiles and their relationship to pest and disease resistance in oilseed rape. Euphytica 63, 71–83. doi: 10.1007/BF00023913
Nakajima, M., Yoshida, R., Shimada, N., Yamazaki, H., and Yokoi, T. (2001). Inhibition and inactivation of human cytochrome P450 isoforms by phenethyl isothiocyanate. Drug Metab. Dispos. 29, 1110–1113. Available online at: http://dmd.aspetjournals.org/content/29/8/1110.full.pdf+html
Padilla, G., Cartea, M. E., Velasco, P., De Haro, A., and Ordas, A. (2007). Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 68, 536–545. doi: 10.1016/j.phytochem.2006.11.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pang, Q., Chen, S., Li, L., and Yan, X. (2009). Characterization of glucosinolate–myrosinase system in developing salt cress Thellungiella halophila. Physiol. Plant 136, 1–9. doi: 10.1111/j.1399-3054.2009.01211.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pang, Q., Guo, J., Chen, S., Chen, Y., Zhang, L., Fei, M., et al. (2012). Effect of salt treatment on the glucosinolate-myrosinase system in Thellungiella salsuginea. Plant Soil 355, 363–374. doi: 10.1007/s11104-011-1108-0
Parkin, I., Magrath, R., Keith, D., Sharpe, A., Mithen, R., and Lydiate, D. (1994). Genetics of aliphatic glucosinolates. II.Hydroxylation of alkenyl glucosinolates in Brassica napus. Heredity 72:5. doi: 10.1038/hdy.1994.82
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parkin, I. A., Gulden, S. M., Sharpe, A. G., Lukens, L., Trick, M., Osborn, T. C., et al. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171, 765–781. doi: 10.1534/genetics.105.042093
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schonhof, I., Krumbein, A., and Bruckner, B. (2004). Genotypic effects on glucosinolates and sensory properties of broccoli and cauliflower. Nahrung 48, 25–33. doi: 10.1002/food.200300329
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schranz, M. E., Lysak, M. A., and Mitchell-Olds, T. (2006). The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. doi: 10.1016/j.tplants.2006.09.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schubert, I., and Lysak, M. A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27, 207–216. doi: 10.1016/j.tig.2011.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Semon, M., and Wolfe, K. H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17, 505–512. doi: 10.1016/j.gde.2007.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slotte, T., Hazzouri, K. M., Agren, J. A., Koenig, D., Maumus, F., Guo, Y. L., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835. doi: 10.1038/ng.2669
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sonderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spillane, C., Schmid, K. J., Laoueille-Duprat, S., Pien, S., Escobar-Restrepo, J. M., Baroux, C., et al. (2007). Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448, 349–U348. doi: 10.1038/nature05984
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Textor, S., Bartram, S., Kroymann, J., Falk, K. L., Hick, A., Pickett, J. A., et al. (2004). Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218, 1026–1035. doi: 10.1007/s00425-003-1184-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Textor, S., De Kraker, J. W., Hause, B., Gershenzon, J., and Tokuhisa, J. G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in arabidopsis. Plant Physiol. 144, 60–71. doi: 10.1104/pp.106.091579
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Wu, J., Sun, S. L., Liu, B., Cheng, F., Sun, R. F., et al. (2011a). Glucosinolate biosynthetic genes in Brassica rapa. Gene 487, 135–142. doi: 10.1016/j.gene.2011.07.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X. W., Wang, H. Z., Wang, J., Sun, R. F., Wu, J., Liu, S. Y., et al. (2011b). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, U1035–U1157. doi: 10.1038/ng.919
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warwick, S. I., Francis, A., and Al-Shehbaz, I. A. (2006). Brassicaceae: Species checklist and database on CD-Rom. Plant Syst. Evol. 259, 249–258. doi: 10.1007/s00606-006-0422-0
Windsor, A. J., Reichelt, M., Figuth, A., Svatos, A., Kroymann, J., Kliebenstein, D. J., et al. (2005). Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae). Phytochemistry 66, 1321–1333. doi: 10.1016/j.phytochem.2005.04.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wittstock, U., Kliebenstein, D. J., Lambrix, V., Reichelt, M., and Gershenzon, J. (2003). Chapter five glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores. Recent Adv. phytochemistry 37, 101–125. doi: 10.1016/S0079-9920(03)80020-5
Wu, H. J., Zhang, Z., Wang, J. Y., Oh, D. H., Dassanayake, M., Liu, B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224. doi: 10.1073/pnas.1209954109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, R., Jarvis, D. E., Chen, H., Beilstein, M. A., Grimwood, J., Jenkins, J., et al. (2013a). The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46. doi: 10.3389/fpls.2013.00046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, R. L., Jarvis, D. E., Chen, H., Beilstein, M. A., Grimwood, J., Jenkins, J., et al. (2013b). The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46. doi: 10.3389/fpls.2013.00046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z., Li, J., Zhao, X.-Q., Wang, J., Wong, G. K.-S., and Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinformat. 4, 259–263. doi: 10.1016/s1672-0229(07)60007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: glucosinolates, MAM genes, syntenic, evolution, Brassicaceae
Citation: Zhang J, Wang X, Cheng F, Wu J, Liang J, Yang W and Wang X (2015) Lineage-specific evolution of Methylthioalkylmalate synthases (MAMs) involved in glucosinolates biosynthesis. Front. Plant Sci. 6:18. doi: 10.3389/fpls.2015.00018
Received: 26 October 2014; Accepted: 09 January 2015;
Published online: 03 February 2015.
Edited by:
Tiegang Lu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Dongying Gao, University of Gerogia, USAXiao Han, Chinese Academy of Agricultural Sciences, China
Copyright © 2015 Zhang, Wang, Cheng, Wu, Liang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowu Wang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Zhongguancun Nandajie No.12, Haidian District, Beijing 100081, China e-mail:d2FuZ3hpYW93dUBjYWFzLmNu
 Jifang Zhang
Jifang Zhang Xiaobo Wang1
Xiaobo Wang1 Feng Cheng
Feng Cheng Jian Wu
Jian Wu Xiaowu Wang
Xiaowu Wang