- 1Plant Genomic Network Research Team, RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 2Core Research for Evolutional Science and Technology, Japan Science and Technology, Kawaguchi, Japan
- 3Kihara Institute for Biological Research, Yokohama City University, Yokohama, Japan
Chromatin regulation is essential to regulate genes and genome activities. In plants, the alteration of histone modification and DNA methylation are coordinated with changes in the expression of stress-responsive genes to adapt to environmental changes. Several chromatin regulators have been shown to be involved in the regulation of stress-responsive gene networks under abiotic stress conditions. Specific histone modification sites and the histone modifiers that regulate key stress-responsive genes have been identified by genetic and biochemical approaches, revealing the importance of chromatin regulation in plant stress responses. Recent studies have also suggested that histone modification plays an important role in plant stress memory. In this review, we summarize recent progress on the regulation and alteration of histone modification (acetylation, methylation, phosphorylation, and SUMOylation) in response to the abiotic stresses, drought, high-salinity, heat, and cold in plants.
Introduction
Past decades, innovation of DNA sequencing technology has brought an evolution of genome biology. Genome projects of many organisms have revealed complexity of genome structure. However, DNA sequence information is not enough to understand the mysterious life phenomenon and how genome is organized. For this, other factors, e.g., chromatin regulation mediated by histone modification and DNA methylation, and RNA-mediated regulation are also involved. Study on such regulation other than genetic information is called as “epigenetics.”
Epigenetic regulation is an important mechanism that is involved in a wide range of biological phenomena, such as genome stability, developmental programming, gene expression, and diseases like cancer through chromatin regulation, small RNA-mediated regulation, and so on (Grewal and Jia, 2007; Feng et al., 2010; Kanherkar et al., 2014). Chromatin regulation mediated by histone modifications and DNA methylation, can be dynamically and statically changed to maintain gene and genome activities (Wolffe, 1998; Zhang and Reinberg, 2001; Kurdistani et al., 2004; Pokholok et al., 2005). Plants perceive stimuli from the surrounding environment and possess sophisticated regulatory networks of genes that can control the accumulation of metabolites, which allows the plants to survive and adapt to environmental changes (Shinozaki and Yamaguchi-Shinozaki, 2007; Urano et al., 2010). Recent studies have reported that histone modifications, such as H3K4me3, H3K9ac, H3K9me2, H3K23ac, H3K27ac, H3K27me3, and H4ac, along with DNA methylation can be correlated with gene expression in response to abiotic stresses, such as water deficit, high-salinity, and temperature shifts (Kim et al., 2008; Luo et al., 2012a). Some histone modifications change rapidly in response to environmental changes, while others change gradually along with changes in gene expression to control physiological homeostasis and development under environmental stresses (Kim et al., 2008, 2012). It still remains unclear which comes first transcriptional changes or chromatin changes, and how they are linked. In this paper, we review recent studies of chromatin regulation in plants, focusing mainly on histone modification and DNA methylation in response to abiotic stresses.
Histone Proteins and Modifiers in Plants
A basic core histone octamer for nucleosomes is composed of histones H2A, H2B, H3, and H4 (Arya and Schlick, 2009; Zhou et al., 2013). Histone variants, such as H2A.Z and CENH3, function in precise and specific regulation of gene activity and genome structure (Deal et al., 2007; Zhang et al., 2008; Yan et al., 2010; Coleman-Derr and Zilberman, 2012).
The N-terminal region of histones is called the histone tail. Histones are enriched with basic amino acid residues such as lysine and arginine. The basic residues in histone tails are covalently modified by methylation, acetylation, phosphorylation, and ubiquitination, and these modifications alter the activity of the genes that are wrapped around the core histones. Histone modification can have different effects depending on which residue is modified and the type of modification. The current understanding of the effect of histone modifications is based mainly on research in yeast. In yeast, mutants in which the modified residue in the histone tail is replaced by a different residue have been used to determine the effects of histone modification (Rundlett et al., 1998; Suka et al., 2001).
In Arabidopsis, histone modification sites were identified by mass-spectrometry and biochemical assays (Earley et al., 2007; Zhang et al., 2007). The results of these studies suggested that post-translational modification sites of histones in Arabidopsis were highly conserved with other eukaryotes. Generally, the acetylation of lysine residues in H3 and H4 N-tails neutralizes the positive charge of the histone tails, which decreases their affinity for DNA and alters the accessibility of transcription factors to the template DNA strand. As a result, histone acetylation tends to induce gene activation (Kuo et al., 1996; Zhang et al., 1998; Shahbazian and Grunstein, 2007). Conversely, the removal of histone acetylation can lead to gene repression and silencing (Kadosh and Struhl, 1998; Rundlett et al., 1998; Chen et al., 2010b; To et al., 2011a). The effects of histone methylation events vary depending on the site of the modification. Although changes in histone modifications can be correlated with gene activity, the molecular mechanisms through which the chemical modifications influence chromosomal structure and the accessibility of transcription factors are still not fully understood. For example, tri-methylation of the fourth lysine of H3 (H3K4me3) is an active mark for gene expression, and tri-methylation in the twenty-seventh lysine of H3 (H3K27me3) is a repressive mark of facultative heterochromatin (Cao et al., 2002; Finnegan and Dennis, 2007; Doyle and Amasino, 2009). These relationships between the alteration of histone modifications and gene activity are highly conserved from yeast to human, and also in plants.
Histone modifiers are also well conserved in angiosperms. For example, the major histone modifiers histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTs), and histone demethylases (HDMs) have been isolated or identified in several plants, including Arabidopsis, tomato, rice, barley, grapevine, Brassica, and Brachypodium. These histone modifiers have been phylogenetically analyzed and classified (Pandey et al., 2002; Chen et al., 2010b; Papaefthimiou et al., 2010; Pontvianne et al., 2010; Aquea et al., 2011; Huang et al., 2011; Aiese-Cigliano et al., 2013), revealing that histone modifiers, such as the HATs, are conserved in plants (Aiese-Cigliano et al., 2013). However, the molecular functions of many of these modifiers have not yet been well characterized.
Histone Modification in Drought Stress Response
Drought stress affects plant growth and survival. Gene regulatory networks associated with the drought stress response in plants have been studied by analyzing drought stress-responsive genes that encode functional and regulatory proteins, such as transcription factors (Shinozaki and Yamaguchi-Shinozaki, 2007; Fujita et al., 2011; Osakabe et al., 2014). The expression of drought stress-responsive genes is positively correlated with the intensity of drought stress (Matsui et al., 2008). The transcriptional responsiveness of drought stress-upregulated genes was found to be correlated with changes in histone modification and nucleosome density (Kim et al., 2008, 2012; To and Kim, 2014). Under strong drought conditions, the histone modifications H3K4me3 and H3K9ac on drought stress-upregulated genes, such as RD20 and RD29A, were more highly enriched than under moderate drought conditions (Kim et al., 2008, 2012; Figure 1). Furthermore, under moderate drought conditions, there was little nucleosome loss from the RD29A region (Kim et al., 2008), while under strong drought conditions, notable nucleosome loss occurred in the same gene region (Kim et al., 2012). These results indicated that epigenetic responsiveness depended on the intensity of the drought stress.
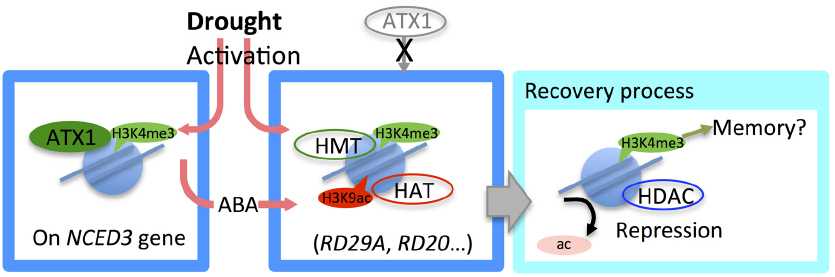
Figure 1. Epigenetic regulation in the drought stress response. Under drought stress, NCED3, a key gene in the abscisic acid (ABA) biosynthesis pathway, is activated with an increase in H3K4me3 marked by the histone methyltransferase ATX1. Active histone marks, such as H3K4me3 and H3K9ac were enriched on many drought-responsive genes. During recovery from drought stress, H3K9ac is rapidly removed, while H3K4me3 is removed more slowly, indicating that H3K4me3 may be involved in epigenetic memory.
To regulate gene activity, an epigenetic mode shift is required for precise transcriptional regulation. To fully repress the expression of stress-upregulated genes and to reset chromatin status under non-drought conditions, aggressive histone deacetylation and nucleosome replacement are required (Kim et al., 2012). During recovery from drought stress, H3K9ac rapidly decreased on the drought stress-upregulated genes, RD29A, RD20, and AtGOLS2 (Figure 1). Simultaneously, RNA polymerase II was shown to be rapidly removed from those gene regions. These results suggested that histone deacetylation was promoted to remove the H3K9ac in conjunction with transcriptional repression (Figure 1). H3K4me3 was removed more slowly than H3K9ac; its enrichment decreased gradually with rehydration treatment (Figure 1). Surprisingly, the results of our previous study suggested that nucleosome density on drought stress-inducible genes behaved in an oscillation-like manner during recovery from dehydration (Kim et al., 2012), suggesting that nucleosome replacement is required to remove H3K4me3 and to reset the chromatin status.
It has been reported that drought stress response is memorized via histone modification on several drought stress-upregulated genes (Ding et al., 2012). H3K4me3 is a good marker of gene activation and stress memory (Oh et al., 2008; Zhang et al., 2009; Ding et al., 2011, 2012; Kim et al., 2012; Figure 1). H3K4me3 enrichment was found to be correlated with transcriptionally active gene regions based on a genome-wide analysis using ChIP-seq (van Dijk et al., 2010). Interestingly, in Arabidopsis, genome-wide H3K4me3 enrichment peaked 300 bp downstream from the transcriptional start site, similar to the H3K4me3 distribution pattern found in human cells (Barski et al., 2007). In contrast, H3K4me3 was distributed broadly on many dehydration/ABA (abscisic acid) inducible genes (van Dijk et al., 2010).
H3K4me3 modification by the HMT Arabidopsis trithorax-like 1 (ATX1) is involved in the activation of NCED3, which encodes a key enzyme in the ABA biosynthesis pathway, under drought stress conditions (Ding et al., 2011; Figure 1). ATX1 binding to the NCED3 region increased under drought treatment. The atx1 mutant plant had remarkably lower enrichments of RNA pol II and H3K4me3 under drought stress conditions compared with the wild type. Transcript levels of several drought stress and ABA-upregulated genes, such as RD29A and RD29B, were reduced during drought treatment in the atx1 mutant. By training with multiple drought treatments, the enriched H3K4me3 levels in the RD29B and RAB18 gene regions were maintained after rehydration, and Ser5P RNA pol II, the activated form of RNA pol II, was found to be stalled on these trained gene regions (Ding et al., 2012). The H3K4me3 level on RD29B was lower in the atx1 mutant than in the wild-type plant after drought stress (Ding et al., 2012). Thus, ATX1-modified H3K4me3 may be considered to have an important role in regulation of the drought-responsive gene network via NCED3; however, ATX1 does not seem to have a critical impact on drought stress-memory in Arabidopsis.
Histone acetylation status has also been correlated with drought stress and ABA responses in plants. For example, in response to drought stress, the histone acetylation level increased on the drought-responsive genes, such as RD20, RD29A, and RD29B (Kim et al., 2008) and H3K9ac was enriched rapidly in these gene regions. Interestingly, specific patterns of histone acetylation were found on each drought-responsive gene, suggesting that differences in histone modification may contribute to the responsiveness of these genes.
In rice, drought stress significantly induced four HAT genes (OsHAC703, OsHAG703, OsHAF701, and OsHAM701; Fang et al., 2014) and enhanced acetylation of H3K9, H3K18, H3K27, and H4K5 under drought stress conditions was found by western blotting analysis. In barley (Hordeum vulgare L.), the expression of all three GNAT-MYST family HAT genes (HvMYST, HvELP3, and HvGCN5) was induced by ABA treatment (Papaefthimiou et al., 2010). Phylogenetically, OsHAM701 and OsHAG703 belong to the HvMYST and HvELP3 clades, respectively. Thus, the epigenetic response to drought stress may be partially conserved between rice and barley.
The HD2-type HDACs, HD2A (HDT1), HD2B (HDT2), HD2C (HDT3), and HD2D (HDT4) belong to a plant-specific HDAC family in Arabidopsis, rice, barley, and tomato (Dangl et al., 2001; Pandey et al., 2002; Wu et al., 2003; Demetriou et al., 2009). AtHD2C-overexpressing Arabidopsis plants showed ABA insensitivity, reduced transpiration, and enhanced tolerance to drought and salt stresses (Sridha and Wu, 2006). In barley, the expression of HD2 genes responded to stress-related plant hormones such as ABA, jasmonic acid (JA), and salicylic acid (SA; Demetriou et al., 2009). These results suggested that HD2 genes play roles in resistance to abiotic and biotic stresses in monocot and dicot plants.
Histone Modification in Salt Stress Response
Understanding the involvement of histone modifications in salt stress responses has gradually progressed and three histone modifications, acetylation, methylation, and phosphorylation, have been shown to influence the salinity stress response in plants.
Histone acetylation is generally correlated with gene activation, and is controlled by antagonistic actions between the HAT and HDAC proteins. In maize roots, the upregulation of cell wall-related genes, such as ZmEXPB2 and ZmXET1, has been associated with increased H3K9 acetylation in the promoter and coding regions, which is thought to be necessary for high salinity response. It has been speculated that the upregulation of these genes might be mediated by two HAT genes (ZmHATB and ZmGCN5), because their mRNA expression was found to increase under salt stress conditions (Li et al., 2014). In Arabidopsis, a mutant for the transcriptional adaptor ADA2b, which modulates HAT activity, showed hypersensitivity to salt (Kaldis et al., 2011), suggesting that HATs play a pivotal role in salinity tolerance. However, Arabidopsis mutants for HDAC proteins such as HD2C, histone deacetylase 6 (HDA6), and histone deacetylase 19 (HDA19), which should increase histone acetylation, showed hypersensitivity to salt (Chen and Wu, 2010; Chen et al., 2010a; Luo et al., 2012b).
In Arabidopsis, a HDAC complex, which included histone deacetylase complex1 (HDC1), HDA6, HDA19, and Arabidopsis Swi-Indipendent3 (AtSin3) was reported (Perrella et al., 2013). HDACs often form functional complexes of multiple proteins to regulate gene activity in eukaryotes (Carrozza et al., 2005a,b; Roguev and Krogan, 2007; Yang and Seto, 2008; Chen et al., 2012). HDA6, HDA19, and AtSin3 are considered as homologs of yeast (Saccharomyces cerevisiae) ScRPD3 deacetylase and ScSin3 proteins. Likewise, HDC1 was found to be an Arabidopsis homolog of ScRXT3. Interestingly, it was reported that the yeast RPD3L complex, comprising ScRPD3, ScSin3, and ScRXT3, responded to heat stress in yeast (Ruiz-Roig et al., 2010). Moreover, the yeast RPD3μ complex that consists of ScRPD3, ScSNT2, and ScECM5 may mediate an oxidative stress response in yeast (McDaniel and Strahl, 2013). These reports of yeast RPD3 complexes indicate that the Arabidopsis HDAC complex involving HDC1, HDA6, and HDA19 may function in the plant’s abiotic stress responses. Indeed, an hdc1 mutant of Arabidopsis induced the expression of ABA1, ABA3, and RAB18, while overexpression of HDC1 led to repressed expression of ABA1, RAB18, and RD29A under high-salinity conditions (Perrella et al., 2013). These results indicated that histone modifier complexes involving HATs and HDACs might be required to fine-tune the histone acetylation status for plant adaptation in response to high-salinity stress (Figure 2, top).
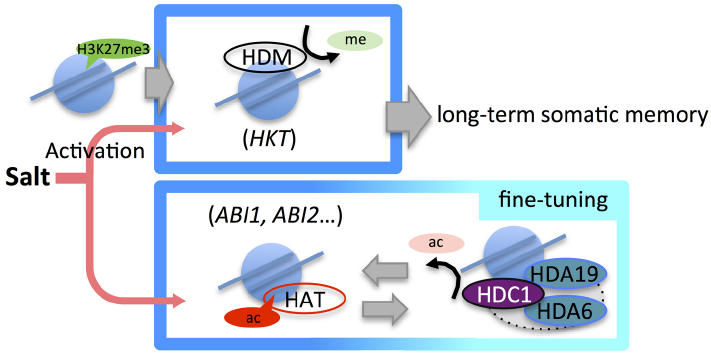
Figure 2. Epigenetic regulation in the high-salinity stress response. Salt stress induces the activation of high-salinity stress-responsive genes with changes in several histone modification marks. Treatment with low concentrations of salt prior to a second stress (priming) induces reduction of H3K27me3 in HKT, which controls Na+ partitioning. Priming-induced low H3K27me3 levels in HKT established long-term memory for rapid response to a second stress (top). Under salt stress, the expression of genes such as ABI1 and ABI2, which are associated with the abscisic acid (ABA) signaling pathway, is fine-tuned by epigenetic regulation mechanisms involving histone acetylation (bottom).
Most reports into the salt stress response in plants have addressed how changes in histone acetylation states are connected to changes in gene expression. Indeed, it has often been the case that histone methylation was surveyed to confirm the interaction with acetylation in the stress response. However, an interesting study by Sani et al. (2013) aimed to decipher the epigenetic action of long-term somatic memory for salinity response. They showed that Na+-pretreated plants showed more drought tolerance than non-treated control plants after a Na+ stress-free period. A change in the H3K27me3 level around the HKT1 gene, which encodes a high-affinity K(+) transporter, induced by mild salt stress was found to be a candidate for explaining the physiological effects caused by the priming treatment (Sani et al., 2013). Many studies have contributed to the current understanding of the core salt-tolerance mechanisms in plants (Deinlein et al., 2014 and references therein). In plant salinity tolerance, maintaining low Na+ levels is important to mitigate ionic Na+ stress. The sodium transporter HKT1 controls root–shoot Na+ partitioning and plays a major role in salt tolerance (Apse et al., 1999; Horie et al., 2009). Sani et al. (2013) identified the shortening and fractionation of H3K27me3 islands after priming treatment by whole-genome ChIP-seq, but found that H3K4me2, H3K4me3, and H3K9me2 islands rarely changed. In the chromatin transition, a long-lasting loss of H3K27me3 was found to occur, implying a release from gene repression in the island and resulting in a rapid and transient increase in the HKT1 mRNA level (Figure 2, bottom; Sani et al., 2013). It is still unclear which genes contribute to enhanced drought stress tolerance in response to mild salt stress priming; however, Sani et al. (2013) have reported that the shortening and fractionation of H3K27me3 in a large number of genes including HKT1 play an important role in somatic memory caused by salt response.
In tobacco BY-2 and Arabidopsis T87 culture cells, rapid transient upregulation of histone H3 Ser-10 phosphorylation occurs, and H3 phosphoacetylation and histone H4 acetylation follow immediately. Interestingly, the onset and persistence of these H3 histone modifications differed between cold and high salinity stress responses (Sokol et al., 2007), suggesting that histone phosphorylation along with other histone modification might be controlled selectively by each stress type, although H3 Ser-10 phosphorylation by itself occurred on a massive scale during active cell division (Paulson and Taylor, 1982).
Histone Modification in Heat Stress Response
Histone variant deposition and histone modifications through acetylation and/or SUMOylation are considered to be involved in the thermal stress response. SUMO (small ubiquitin-related modifier) was identified as a reversible post-translational modifier that plays an important role in the regulation of protein interactions in eukaryotes. Recent studies have revealed that the occupancy of each histone variant of a core histone, in particular H2A and H3, plays important roles in not only gene expression, but also in the repair of DNA breaks and the assembly of chromosome centromeres in eukaryotes (Mizuguchi et al., 2004; Lu et al., 2009; Choi et al., 2013). In Arabidopsis, it has been suggested that H2A.Z deposition in gene bodies promotes variability in the levels and patterns of gene expression (Zilberman et al., 2008; Lu et al., 2009; Coleman-Derr and Zilberman, 2012). In addition to the regulation of gene expression, a genetic screen revealed that H2A.Z contributed to the thermosensory response via its nucleosome occupancy. A screen of Arabidopsis mutants deficient in temperature sensing under ambient temperatures (12–27°C) identified ARP6 (actin-related protein 6) as a regulator of the coordinated changes in gene expression in response to ambient temperature changes (Kumar and Wigge, 2010). ARP6 encodes a subunit of the SWR1 complex (March-Díaz and Reyes, 2009) that is necessary for inserting the alternative histone H2A.Z into nucleosomes replacing the core histone H2A, and could be involved in temperature sensing (Deal and Henikoff, 2010; Kumar and Wigge, 2010). In Brachypodium, the impairment of H2A.Z deposition reduced grain yield under heat stress conditions (Boden et al., 2013). Together, these studies suggest that H2A.Z deposition plays an important role in thermal stress responses. However, other modification such as H3K56 acetylation facilitated by histone chaperon AtASF1A/B is associated with nucleosome loss and causes the accumulation of RNA polymerase II (Weng et al., 2014), and the activation of transcription factors (Sidaway-Lee et al., 2014) also operate in response to heat stress. Because histone acetylation was shown to be necessary for H2A.Z deposition in yeast (Watanabe et al., 2013), histone modification may control the deposition of H2A.Z and possibly other histone variants in the heat stress response in plants. Further analyses are needed to uncover what kinds of histone modification and histone deposition contribute to the heat stress response in plants (Figure 3A).
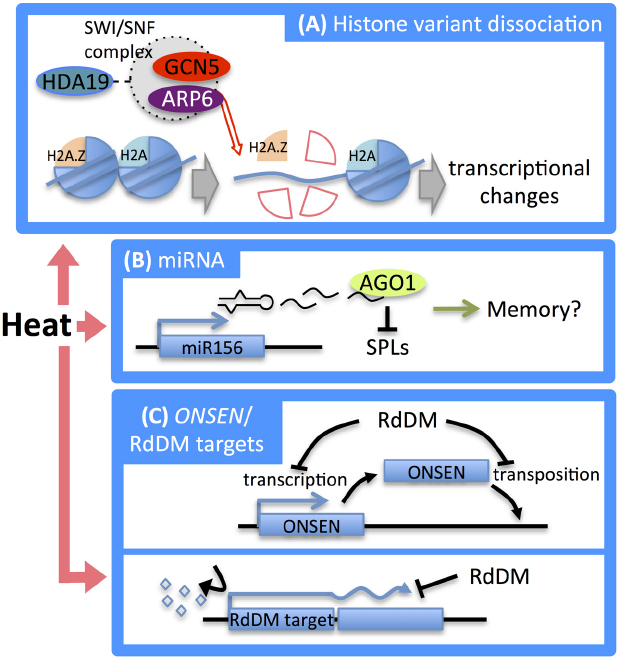
Figure 3. Epigenetic regulation in the heat stress response. (A) At high temperatures, H2A.Z is dissociated by the SWI/SNF chromatin remodeling complex which contains ARP6. This dissociation causes transcriptional changes in downstream genes. (B) After heat stress treatment, the expression of the microRNA miR156, which targets SPL genes, is increased. ago1 mutants of Arabidopsis show defects in heat acclimation, implying involvement of miR156-mediated post-transcriptional regulation of SPL genes in heat stress memory. (C) Heat stress induced activation of the ONSEN retrotransposon. RdDM suppresses transcriptional and transpositional activation of ONSEN (top). Heat stress-induced DNA methylation causes aberrant transcription of genes that are located proximate to RdDM targets (bottom). Induction of DNA methylation restores normal transcription (bottom).
In animals, SUMOylation is an important regulatory mechanism for the control of transcriptional repression mediated by histone modifiers such as HDACs (Kirsh et al., 2002; Geiss-Friedlander and Melchior, 2007). In Arabidopsis, the understanding of SUMOylation under heat stress conditions has progressed and a variety of chromatin modifier and components such as H2B, GCN5, HDA19, and the deubiquitinating enzyme UBP26, which removes ubiquinone bound to H2B, have been found to be SUMOylated (Miller et al., 2010). Interestingly, heat stress treatment (37°C for 30 min) was reported to decrease the SUMOylation status of H2B and increase the status of the GCN5 HAT (Miller et al., 2010, 2013).
There is growing evidence that epigenetic mechanisms contribute to stress memory in plants. Epigenetic memory for stress responses has been summarized in excellent reviews (Chinnusamy and Zhu, 2009; Mirouze and Paszkowski, 2011). A recent study indicated that the microRNA miR156 is involved in memory of heat stress (Stief et al., 2014). Arabidopsis plants that are pre-treated with moderate heat stress (priming) were found to acquire thermotolerance (heat stress memory) to severe heat stress. Hypomorphic mutants of AGO1, a component required for microRNA-mediated post-transcriptional gene silencing, were defective in maintaining the acquired thermotolerance. During priming heat stress, the amount of miR156 increased, while the SPL (squamosal-promoter binding-like) transcription factor genes, which are master regulators of developmental transitions and targeted by miR156, were downregulated. An acquired adaptation to recurring heat stress was reported to be achieved by miR156-mediated post-transcriptional regulation of SPL genes (Stief et al., 2014; Figure 3B).
Histone Modification in Cold Stress Response
In Arabidopsis, vernalization, which is the best characterized pathway involved in epigenetic regulation induced by environmental stresses, can be achieved by long-term exposure to cold temperatures (Song et al., 2012a). Short term exposure to non-freezing low temperatures enhances freezing tolerance, and this process is known as cold acclimation. Both of these processes are low temperature responses; however, they are mediated by independent pathways (Bond et al., 2011).
Low temperature treatment induces genome-wide transcriptional changes. In Arabidopsis, it has been estimated that the transcription of 3 to 20% of the genes change in response to cold stress (Chinnusamy et al., 2007; Matsui et al., 2008). Some epigenetic regulators are transcriptionally upregulated under cold stress conditions, suggesting that their upregulation may cause epigenetic and transcriptional changes of the target genes. In Arabidopsis, the expression of HDA6 was induced by long-term low temperature treatments (Figure 4), and a mutation in this gene resulted in sensitivity to freezing stress (To et al., 2011b). In maize, the expression of HDACs was upregulated during cold acclimation, and global deacetylation at H3 and H4 was observed (Hu et al., 2011). Moreover, during cold stress treatment, heterochromatic tandem repeats were selectively unsilenced, then H3K9ac was accumulated and DNA methylation and H3K9me2 were reduced in the unsilenced regions (Hu et al., 2012; Figure 4).
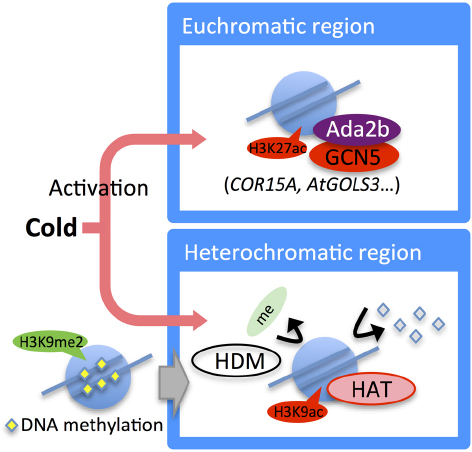
Figure 4. Epigenetic regulation in the cold stress response. Expression of the HDA6 gene is induced by long-term cold treatment. Under cold stress, the histone acetylation level in cold-responsive genes (e.g., COR15A and AtGolS3) is increased by Ada2b and GCN5. In heterochromatic regions, H3K9me2 is removed and converted to acetylation mark. DNA methylation also decreased.
Local epigenetic changes in cold-responsive genes were also induced. In Arabidopsis, H3K27me3 on the cold-responsive genes, COR15A, and AtGolS3, decreased gradually in both a histone occupancy-dependent and -independent manner during cold stress treatment (Kwon et al., 2009). In maize, histone acetylation in cold-responsive genes such as ZmDREB1 and ZmCOR413 increased (Hu et al., 2011). Histone acetylation of OsDREB1b was induced by cold stress in rice (Roy et al., 2014). Thus, epigenetic changes induced by cold stress are likely to contribute to the acquirement of stress tolerance through changes in the expression of cold-responsive genes (Figure 4).
Several epigenetic regulators that are involved in the cold stress response were characterized by forward and reverse genetic studies. ADA2b, a transcriptional activator of HATs (Stockinger et al., 2001), was reported to interact with GCN5, an Arabidopsis HAT, and enhance the HAT activity of GCN5. In ada2b and gcn5 mutants of Arabidopsis, the induction of COR (cold-regulated) transcripts by low temperature was delayed, and the final mRNA expression levels were reduced. The ada2b mutants showed increased freezing tolerance, indicating that ADA2b may function in the repression of freezing tolerance through histone acetylation (Vlachonasios et al., 2003).
Under cold stress, HOS1 (high expression of osmotically responsive gene 1), a RING finger E3 ligase, is a negative regulator of cold-responsive genes such as CBFs/DREB1s and COR/RD/KIN, and the ICE1 transcription factor (Dong et al., 2006). A recent study showed that HOS1 regulated chromatin status in the FLC locus by dissociating HDA6 from FLC chromatin (Jung et al., 2013).
The expression of HOS15, a WD40-repeat protein, was found to be induced by abiotic stresses including cold stress (Zhu et al., 2008). In a hos15 mutant of Arabidopsis, transcripts of stress-regulated genes, such as RD29A, were reported to accumulate. Moreover, hos15 mutants showed hypersensitivity to freezing stress. HOS15 shares sequence similarity with human TBL1 (transducin-beta like protein 1), which is a component of the SMRT/N-CoR repressor complex that associates with HDACs. It has been reported that HOS15 is involved in cold tolerance through the regulation of deacetylation at histone H4 (Zhu et al., 2008).
Compared with vernalization, the understanding of epigenetic regulation underlying the cold acclimation mechanism is limited. Further research is needed to increase the knowledge about this process.
Regulation Mechanism of DNA Methylation in Plants
DNA methylation is another conserved epigenetic modification in eukaryotic organisms. Although the cytosines in CG sites are methylated in differentiated mammalian cells, cytosines in all contexts (CG, CHG, and CHH, where H indicates A, T, or C) can be methylated in plants. While DNA methylation is considered to be a recessive mark, recent studies have revealed that CG methylation can occur within the coding regions of transcribed genes (gene body methylation). Non-CG methylation is found exclusively in heterochromatin. DNA methylation status is regulated by de novo methylation, maintenance methylation, and active demethylation.
In plants, de novo DNA methylation was shown to be induced by a 24-nt short interfering (si) RNA-mediated pathway called RNA-directed DNA methylation (RdDM; Matzke and Mosher, 2014). In RdDM, the plant-specific RNA polymerases Pol IV and Pol V have important functions. These polymerases are evolutionarily related to Pol II but have different roles in RdDM, namely siRNA biogenesis (Pol IV) and recruitment of the silencing complex (Pol V). In brief, 24-nt siRNA is generated from Pol IV transcripts through processes mediated by RNA interference (RNAi) machineries such as RDR2 (RNA-dependent RNA polymerase 2) and DCL3 (Dicer-like 3), Then, the siRNA is incorporated into AGO4 (Argonaute 4). Chromatin association of Pol IV partially depends on SHH1 (sawadee homeodomain homolog 1), which can recognize unmethylated K4 and methylated K9 of histone H3 (Law et al., 2013), while Pol V transcribe genes from target loci and the siRNA-AGO4 complex is recruited to the transcript in a sequence-dependent manner. For transcription by Pol V, the so-called DDR complex, which is composed of DRD1 (defective in RNA-directed DNA methylation), DMS3 (defective in meristem silencing 3), and RDM1 (RNA-directed DNA methylation 1), is required (Kanno et al., 2004, 2008; Gao et al., 2010; Law et al., 2010). Finally, Dnmt3 class de novo methylase DRM2 (domains rearranged methylase 2) is recruited to target loci where it adds a methyl group to cytosines (Cao and Jacobsen, 2002). Several other effectors required for RdDM are discussed in a recent review by Matzke and Mosher (2014).
Induced DNA methylation is retained by maintenance methylases. CG methylation is maintained by conserved Dnmt1 class methylase MET1 (methyltransferase 1), while the plant-specific methylase CMT3 (chromomethylase 3) is required for maintenance of non-CG methylation, especially CHG methylation. Essential to this process is histone H3K9 dimethylation (H3K9me2), which is mediated by KYP/SUVH4, and CMT3 and KYP form self-reinforcement mechanism between DNA methylation and histone methylation (Jackson et al., 2002; Johnson et al., 2007). Maintenance of CHH methylation is regulated by both RdDM and the recently characterized CMT2. In addition to these DNA methyltransferases, chromatin remodeling factor DDM1 (decrease in DNA methylation 1) is also required (Vongs et al., 1993; Jeddeloh et al., 1999; Zemach et al., 2013). DDM1 is necessary to recruit maintenance methylases, such as CMT2, CMT3, and MET1 to linker histone H1 binding regions (Zemach et al., 2013).
DNA methylation is reversible. Active DNA demethylation was found to be achieved through a base excision–repair pathway (Zhu, 2009). The Arabidopsis genome encodes four DNA demethylase genes. DME (demeter) was shown to be preferentially expressed in central cells of female gametophytes, where it induces maternal allele-specific DNA demethylation of imprinted genes such as MEA (medea) and FWA (flowering Wageningen; Choi et al., 2002; Kinoshita et al., 2004). Recent studies revealed that DME was also expressed in the vegetative cells of pollen, but not in sperm cells, and that it played important roles in the establishment of epigenetic status in gametophytes (Schoft et al., 2011; Ibarra et al., 2012). Other DNA demethylases, ROS1 (repressor of silencing 1), DML2 (demeter-like 2), and DML3, are expressed in a wide range of organs (Penterman et al., 2007). Forward and reverse genetic screening revealed other factors that were required for active demethylation; for example, ROS3, an RNA binding protein, ZDP (zinc finger DNA 3′ phosphoesterase), a DNA 3′ phosphatase, and a HAT IDM1 (increase in DNA methylation 1) that can bind with methylated DNA and unmethylated histone H3K4 (Zheng et al., 2009; Martínez-Macías et al., 2012; Qian et al., 2012).
Although interaction between DNA methylation and histone modification, especially between non-CG methylation and H3K9me2, has been implicated, the current understanding of DNA methylation regulation in response to stress is limited compared to the better understanding of histone regulation. Further research will help clarify the underlying mechanisms that connect histone modification, DNA methylation, and stress responses.
DNA Methylation in Abiotic Stress Response
The involvement of DNA methylation in the stress response is still poorly understood; however, recent studies have implicated the involvement of this epigenetic modification in the stress response. Global changes in DNA methylation, including hyper- and hypo-methylation, in response to abiotic stress have been reported in several plant species (Boyko et al., 2010; Bilichak et al., 2012; Karan et al., 2012; Wang et al., 2014) and alterations of DNA methylation in some stress-responsive genes have also been reported. Oxidative stress induced demethylation and transcriptional activation of NtGPDL (glycerophosphodiesterase-like protein) in Nicotiana tabacum (Choi and Sano, 2007). In soybean, salinity stress treatment induced the reduction of DNA methylation and transcriptional activation in genes that encoded salt stress-responsive transcription factors (Song et al., 2012b). Induction of DNA methylation by abiotic stress treatments has also been reported. In a low relative humidity stress condition, the numbers of stomata on the leaf epidermis were found to decrease in Arabidopsis. DNA methylation and transcriptional suppression in two positive regulator genes for stomatal development, SPCH (speechless) and FAMA, were reported to be induced by low relative humidity stress in Arabidopsis (Tricker et al., 2012). Thus, DNA methylation may play an important role in the transcriptional regulation of stress-responsive genes.
The system that regulates DNA methylation also functions as a genomic defense mechanism against abiotic stresses. The siRNA-mediated RdDM pathway restricts transcriptional activity and retrotransposition of the copia-type retrotransposon ONSEN triggered by heat stress (Ito et al., 2011; Figure 3C, top). ONSEN is transcriptionally reactivated by heat treatment, and during the recovery period after heat treatment the number of transcripts was found to diminish gradually. Although ONSEN is reactivated by heat stress, transposition has not been observed in wild-type plants. Interestingly, transcriptional reactivation was enhanced and transposition occurred in Arabidopsis mutants of RdDM components such as nrpd1 (largest subunit of Pol IV), nrpd2 (common second largest subunit of Pol IV and Pol V), and rdr2 (Ito et al., 2011), indicating that the RdDM pathway restricts transcriptional reactivation and retrotransposition of ONSEN induced by heat treatment (Figure 3C, top).
RNA-directed DNA methylation is required for basal tolerance against heat stress (Popova et al., 2013). The nrpd2 mutants showed hypersensitivity against heat stress and aberrant read-through transcription induced by heat treatment was diminished during the recovery process after heat stress in wild-type plants, but not in nrpd2 mutants (Popova et al., 2013). Such misregulated genes harbor RdDM target sequences, such as transposon remnants, in proximal regions. Heat treatment was found to remove CHH methylation at the transposon remnants in wild-type plants and to induce aberrant transcription of RdDM targets and their nearby genes (Popova et al., 2013). Induction of CHH demethylation on RdDM targets during the recovery period diminished such aberrant transcription in wild-type plants; however, a hypomethylated status continued and misregulation of gene expression was not restored in nrpd2 mutants. These results indicated that the altered heat responsiveness in nrpd2 mutants may be caused by defective epigenetic regulation of nearby RdDM targets (Popova et al., 2013; Figure 3C,bottom).
Conclusions and Perspectives
It is currently understood that the regulation of abiotic stress-responsive genes is related to chromatin alterations. This new understanding has introduced new research aspects to the study of plant abiotic stress responses. It has been shown that dynamic epigenetic changes occur in response to abiotic stresses, and the epigenetic response and memory of gene activation in response to abiotic stresses have become major topics of interest. However, the entire correlation network between abiotic stress responses and epigenetic information, such as the stress-responsive epigenetic modifiers, the targeted stress-responsive genes, and the specific histone modification sites, remain unclear. Investigating the direct effects of histone modification in plants is difficult, because plant genomes harbor multiple copies of histone genes. ChIP assays have proven valuable in helping to identify the histone modifications responsible for epigenetic regulation; however, the results obtained by these assays provide only direct/indirect candidate residues that may be targets for modification. Identifying the functions of histone modification in plant epigenetic studies directly is still a challenge. Generation of mutants of the histone modifiers and of mutated amino acid residues in the histone N-tail by CRISPR/Cas system (Cong et al., 2013) will be an important contribution to these studies, because they will help to determine the superiority between transcriptional changes and epigenetic alterations. Moreover, there is a possibility that tissue-specific alterations are necessary for plants to acquire stress tolerance (Baxter et al., 2010); therefore, because it is difficult to detect tissue-specific alterations of histone modifications by ChIP experiments with the current resolution, the results of ChIP experiments should be evaluated cautiously. Reliable and higher resolution chromatin studies are undoubtedly required to reveal key epigenetic information involved in abiotic stress responses. In this review, we have shown that it is essential to establish experimental materials and efficient technologies for future intensive and acute analyses in plant epigenetics. When the master regulators of epigenetic regulation and novel epigenetic regulation mechanisms in the stress response are discovered, in-depth studies on plant abiotic stress responses will progress rapidly.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST), grants from RIKEN, Japan [to MS], Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Innovative Areas 25119724 [to MS], and 26113719 and (C) 24570065 [to J-MK]), and the Japan Society for the Promotion of Science for Young Scientists [to KS].
References
Aiese-Cigliano, R., Sanseverino, W., Cremona, G., Ercolano, M. R., Conicella, C., and Consiglio, F. M. (2013). Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14:57. doi: 10.1186/1471-2164-14-57
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Apse M, P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aquea, F., Vega, A., Timmermann, T., Poupin, M. J., and Arce-Johnson, P. (2011). Genome-wide analysis of the SET DOMAIN GROUP family in Grapevine. Plant Cell Rep. 30, 1087–1097. doi: 10.1007/s00299-011-1015-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arya, G., and Schlick, T. (2009). A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. 113, 4045–4059. doi: 10.1021/jp810375d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution profiling of histone methylation in the human genome. Cell 129, 823–837. doi: 10.1016/j.cell.2007.05.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baxter, I., Brazelton, J. N., Yu, D., Huang, Y. S., Lahner, B., Yakubova, E., et al. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6:e1001193. doi: 10.1371/journal.pgen.1001193
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bilichak, A., Ilnystkyy, Y., Hollunder, J., and Kovalchuk, I. (2012). The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7:e30515. doi: 10.1371/journal.pone.0030515
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boden, S. A., Kavanová, M., Finnegan, E. J., and Wigge, P. A. (2013). Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 14, R65. doi: 10.1186/gb-2013-14-6-r65
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bond, D. M., Dennis, E. S., and Finnegan, E. J. (2011). The low temperature response pathways for cold acclimation and vernalization are independent. Plant Cell Environ. 34, 1737–1748. doi: 10.1111/j.1365-3040.2011.02370.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boyko, A., Blevins, T., Yao, Y., Golubov, A., Bilichak, A., Ilnytskyy, Y., et al. (2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5:e9514. doi: 10.1371/journal.pone.0009514
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., et al. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043. doi: 10.1126/science.1076997
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, X., and Jacobsen, S. E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144. doi: 10.1016/S0960-9822(02)00925-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carrozza, M. J., Florens, L., Swanson, S. K., Shia, W. J., Anderson, S., Yates, J., et al. (2005a). Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731, 77–87. doi: 10.1016/j.bbaexp.2005.09.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carrozza, M. J., Li, B., Florens, L., Suganuma, T., Swanson, S. K., Lee, K. K., et al. (2005b). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592. doi: 10.1016/j.cell.2005.10.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010a). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, M., Lv, S., and Meng, Y. (2010b). Epigenetic performers in plants. Develop. Growth Differ. 52, 555–566. doi: 10.1111/j.1440-169X.2010.01192.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, L. T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, X. F., Kuryan, B., Kitada, T., Tran, N., Li, J. Y., Kurdistani, S., et al. (2012). The Rpd3 core complex is a chromatin stabilization module. Curr. Biol. 22, 56–63. doi: 10.1016/j.cub.2011.11.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chinnusamy, V., and Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Choi, C. S., and Sano, H. (2007). Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genomics 277, 589–600. doi: 10.1007/s00438-007-0209-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Choi, K., Zhao, X., Kelly, K. A., Venn, O., Higgins, J. D., Yelina, N. E., et al. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336. doi: 10.1038/ng.2766
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. doi: 10.1016/S0092-8674(02)00807-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodiles regulates responsive genes. PLoS Genet. 8:e1002988. doi: 10.1371/journal.pgen.1002988
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cong, L., Ran, F. A., Lin, S., Barretto, R., Habib, N., Hsu, P. D., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dangl, M., Brosch, G., Haas, H., Loidl, P., and Lusser, A. (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213, 280–285. doi: 10.1007/s004250000506
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deal, R. B., and Henikoff, S. (2010). Gene regulation: a chromatin thermostat. Nature 463, 887–888. doi: 10.1038/463887a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deal, R. B., Topp, C. N., McKinney, E. C., and Meagher, R. B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19, 74–83. doi: 10.1105/tpc.106.048447
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Demetriou, K., Kapazoglou, A., Tondelli, A., Francia, T., Stanca, M. A., Bladenopoulos, K., et al. (2009). Epigenetic chromatin modifiers in barley: I. cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol. Plant. 136, 358–368. doi: 10.1111/j.1399-3054.2009.01236.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, Y., Avramova, Z., and Fromm, M. (2011). The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 3, 740. doi: 10.1038/ncomms1732
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doyle, M. R., and Amasino, R. M. (2009). A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151, 1688–1697. doi: 10.1104/pp.109.145581
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., and Pikaard, C. S. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627. doi: 10.1126/science.1190614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fang, H., Liu, X., Thorn, G., Duan, J., and Tian, L. (2014). Expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 443, 400–405. doi: 10.1016/j.bbrc.2013.11.102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Finnegan, E. J., and Dennis, E. S. (2007). Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983. doi: 10.1016/j.cub.2007.10.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, Z., Liu, H. L., Daxinger, L., Pontes, O., He, X., Qian, W., et al. (2010). An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465, 106–109. doi: 10.1038/nature09025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geiss-Friedlander, R., and Melchior, F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956. doi: 10.1038/nrm2293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grewal, S. I., and Jia, S. (2007). Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46. doi: 10.1038/nrg2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horie, T., Hauser, F., and Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668. doi: 10.1016/j.tplants.2009.08.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Zhang, L., He, S., Huang, M., Tan, J., Zhao, L., et al. (2012). Cold stress selectively unsilences tandem repeats in heterochromatin associated with accumulation of H3K9ac. Plant Cell Environ. 35, 2130–2142. doi: 10.1111/j.1365-3040.2012.02541.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Zhang, L., Zhao, L., Li, J., He, S., Zhou, K., et al. (2011). Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 6:e22132. doi: 10.1371/journal.pone.0022132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, Y., Liu, C., Shen, W. H., and Ruan, Y. (2011). Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol. 11:175. doi: 10.1186/1471-2229-11-175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T. F., Uzawa, R., Rodrigues, J. A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. doi: 10.1126/science.1224839
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119, doi: 10.1038/nature09861
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jackson, J. P., Lindroth, A. M., Cao, X., and Jacobsen, S. E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. doi: 10.1038/nature731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jeddeloh, J. A., Stokes, T. L., and Richards, E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. doi: 10.1038/8803
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Johnson, L. M., Bostick, M., Zhang, X., Kraft, E., Henderson, I., Callis, J., et al. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384. doi: 10.1016/j.cub.2007.01.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jung, J. H., Park, J. H., Lee, S., To, T. K., Kim, J. M., Seki, M., et al. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25, 4378–4390. doi: 10.1105/tpc.113.118364
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kadosh, D., and Struhl, K. (1998). Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18, 5121–5127.
Kaldis, A., Tsementzi, D., Tanriverdi, O., and Vlachonasios, K. E. (2011). Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 233, 749–762. doi: 10.1007/s00425-010-1337-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanherkar, R. R., Bhatia-Dey, N., and Csoka, A. B. (2014). Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2:49. doi: 10.3389/fcell.2014.00049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanno, T., Bucher, E., Daxinger, L., Huettel, B., Böhmdorfer, G., Gregor, W., et al. (2008). A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40, 670–675. doi: 10.1038/ng.119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanno, T., Mette, M. F., Kreil, D. P., Aufsatz, W., Matzke, M., and Matzke, A. J. (2004). Involvement of putative SNF2 chromatin remodeling factor DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805. doi: 10.1016/j.cub.2004.04.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karan, R., DeLeon, T., Biradar, H., and Subudhi, P. K. (2012). Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 7:e40203. doi: 10.1371/journal.pone.0040203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S. E., et al. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303, 521–523. doi: 10.1126/science.1089835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirsh, O., Seeler, J. S., Pichler, A., Gast, A., Müller, S., Miska, E., et al. (2002). The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21, 2682–2691. doi: 10.1093/emboj/21.11.2682
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuo, M. H., Brownell, J. E., Sobel, R. E., Ranalli, T. A., Cook, R. G., Edmondson, D. G., et al. (1996). Transcription-linked acetylation by Gcn5p of histone H3 and H4 at specific lysines. Nature 383, 269–272. doi: 10.1038/383269a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurdistani, S. K., Tavazoie, S., and Grunstein, M. (2004). Mapping global histone acetylation patterns to gene expression. Cell 117, 721–733. doi: 10.1016/j.cell.2004.05.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kwon, C. S., Lee, D., Choi, G., and Chung, W. I. (2009). Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 60, 112–121. doi: 10.1111/j.1365-313X.2009.03938.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Law, J. A., Ausin, I., Johnson, L. M., Vashisht, A. A., Zhu, J. K., Wohlschegel, J. A., et al. (2010). A protein complex required for Polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr. Biol. 20, 951–956. doi: 10.1016/j.cub.2010.03.062
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Law, J. A., Du, J., Hale, C. J., Feng, S., Krajewski, K., Palanca, A. M., et al. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389. doi: 10.1038/nature12178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., Yan, S., Zhao, L., Tan, J., Zhang, Q., Gao, F., et al. (2014). Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 14:105. doi: 10.1186/1471-2229-14-105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, P. Y., Lévesque, N., and Kobor, M. S. (2009). NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem. Cell Biol. 87, 799–815. doi: 10.1139/O09-062
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, M., Liu, X., Singh, P., Cui, Y., Zimmerli, L., and Wu, K. (2012a). Chromatin modifications and remodeling in plant abiotic stress responses. Biochem. Biophys. Acta 1819, 129–136. doi: 10.1016/j.bbagrm.2011.06.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, M., Wang, Y. Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012b). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
March-Díaz, R., and Reyes, J. C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2, 565–577. doi: 10.1093/mp/ssp019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martínez-Macías, M. I., Qian, W., Miki, D., Ponted, O., Liu, Y., Tang, K., et al. (2012). A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45, 357–370. doi: 10.1016/j.molcel.2011.11.034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsui, A., Ishida, J., Morosawa, T., Mochuzuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McDaniel, S. L., and Strahl, B. D. (2013). Stress-free with Rpd3: a unique chromatin complex mediates the response to oxidative stress. Mol. Cell. Biol. 33, 3726–3727. doi: 10.1128/MCB.01000-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, M. J., Barrett-Wilt, G. A., Hua, Z., and Vierstra, R. D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517. doi: 10.1073/pnas.1004181107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, M. J., Scalf, M., Rytz, T. C., Hubler, S. L., Smith, L. M., and Vierstra, R. D. (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12, 449–463. doi: 10.1074/mcp.M112.025056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mirouze, M., and Paszkowski, J. (2011). Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14, 267–274. doi: 10.1016/j.pbi.2011.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. doi: 10.1126/science.1090701
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oh, S., Park, S., and van Nocker, S. (2008). Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 4:e1000077. doi: 10.1371/journal.pgen.1000077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandey, R., Muller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Papaefthimiou, D., Likotrafiti, E., Kapazoglou, A., Bladenopoulos, K., and Tsaftaris, A. (2010). Epigenetic chromatin modifiers in barley: III. Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA. Plant Phyiol. Biochem. 48, 98–107. doi: 10.1016/j.plaphy.2010.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paulson, J. R., and Taylor, S. S. (1982). Phosphorylation of histones 1 and 3 and nonhistone high mobility group 14 by an endogenous kinase in HeLa metaphase chromosomes. J. Biol. Chem. 257, 6064–6072.
Penterman, J., Zilberman, D., Huh, J. H., Ballinger, T., Henikoff, S., and Fischer, R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757. doi: 10.1073/pnas.0701861104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Grossele, V., Verduyn, C., et al. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pokholok, D. K., Harbison, C. T., Levine, S., Cole, M., Hannett, N. M., Lee, T. I., et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527. doi: 10.1016/j.cell.2005.06.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pontvianne, E., Blevins, T., and Pikaard, C. S. (2010). Arabidopsis histone lysine methyltransferases. Adv. Bot. Res. 53, 1–22. doi: 10.1016/S0065-2296(10)53001-5
Popova, O. V., Dihn, H. Q., Aufsatz, W., and Jonak, C. (2013). The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 6, 396–410. doi: 10.1093/mp/sst023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qian, W., Miki, D., Zhang, H., Liu, Y., Zhang, X., Tang, K., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336, 1445–1448. doi: 10.1126/science.1219416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roguev, A., and Krogan, N. J. (2007). SIN-fully silent: HDAC complex in fission yeast. Nat. Struct. Mol. Biol. 14, 358–359. doi: 10.1038/nsmb0507-358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roy, D., Paul, A., Roy, A., Ghosh, R., Ganguly, P., and Chaudhuri, S. (2014). Differential acetylation of histone H3 at the regulatory region of OsDREB1b facilitates chromatin remodeling and transcription activation during cold stress. PLoS ONE 9:e100343. doi: 10.1371/journal.pone.0100343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ruiz-Roig, C., Vieitez, C., Posas, F., and de Nadal, E. (2010). The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 76, 1049–1062. doi: 10.1111/j.1365-2958.2010.07167.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rundlett, S. E., Carmen, A, A., Suka, N., Turner, B. M., and Grunstein, M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835. doi: 10.1038/33952
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sani, E., Herzyk, P., Perrella, G., Colot, V., and Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14, R59. doi: 10.1186/gb-2013-14-6-r59
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schoft, V. K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., et al. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. U.S.A. 108, 8042–8047. doi: 10.1073/pnas.1105117108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sidaway-Lee, K., Costa, M. J., Rand, D. A., Finkenstadt, B., and Penfield, S. (2014). Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol. 15, R45. doi: 10.1186/gb-2014-15-3-r45
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sokol, A., Kwiatkowska, A., Jerzmanowski, A., and Prymakowska-Bosak, M. (2007). Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227, 245–254. doi: 10.1007/s00425-007-0612-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, J., Angel, A., Howard, M., and Dean, C. (2012a). Vernalization—a cold-induced epigenetic switch. J. Cell Sci. 125, 3723–3731. doi: 10.1242/jcs.084764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, Y., Ji, D., Li, S., Wang, P., Li, Q., and Xiang, F. (2012b). The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 7:e41274. doi: 10.1371/journal.pone.0041274
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sridha, S., and Wu, K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. doi: 10.1111/j.1365-313X.2006.02678.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Baurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stockinger, E. J., Mao, Y., Regier, M. K., Triezenberg, S. J., and Thomashow, M. F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acid Res. 29, 1524–1533. doi: 10.1093/nar/29.7.1524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suka, N., Suka, Y., Carmen, A. A., Wu, J., and Grunstein, M. (2001). Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8, 473–479. doi: 10.1016/S1097-2765(01)00301-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
To, T. K., and Kim, J. M. (2014). Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 4:548. doi: 10.3389/fpls.2013.00548
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
To, T. K., Kim, J. M., Matsui, A., Kurihara, Y., Morosawa, T., Ishida, J., et al. (2011a). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7:e1002055. doi: 10.1371/journal.pgen.1002055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
To, K. T., Nakaminami, K., Kim, J. M., Morosawa, T., Ishida, J., Tanaka, M., et al. (2011b). Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 406, 414–419. doi: 10.1016/j.bbrc.2011.02.058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tricker, P. J., Gibbings, J. G., Rodríguez-López, C. M., Hadley, P., and Wilkinson, M. J. (2012). Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J. Exp. Bot. 63, 3799–3813. doi: 10.1093/jxb/ers076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Urano, K., Kurihara, Y., Seki, M., and Shinozaki, K. (2010). ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13, 132–138. doi: 10.1016/j.pbi.2009.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J. M., Liu, R., Yang, J., et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10:238. doi: 10.1186/1471-2229-10-238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vongs, A., Kakutani, T., Martienssen, R. A., and Richards, E. J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. doi: 10.1126/science.8316832
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M., Qin, L., Xie, C., Li, W., Yuan, J., Kong, L., et al. (2014). Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol. 55, 1354–1365. doi: 10.1093/pcp/pcu059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watanabe, S., Radman-Livaja, M., Rando, O. J., and Peterson, C. L. (2013). A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340, 195–199. doi: 10.1126/science.1229758
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weng, M., Yang, Y., Feng, H., Pan, Z., Shen, W. H., Zhu, Y., et al. (2014). Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ. 37, 2128–2138. doi: 10.1111/pce.12299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wolffe, A. P. (1998). Packing principle: how DNA methylation and histone acetylation control the transcriptional activity of chromatin. J. Exp. Zool. 282, 239–244. doi: 10.1002/(SICI)1097-010X(199809/10)282:1/2<239::AID-JEZ25>3.0.CO;2-N
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, K., Tian, L., Zhou, C., Brown, D., and Miki, B. (2003). Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34, 241–247. doi: 10.1046/j.1365-313X.2003.01714.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yan, H., Kikuchi, S., Neumann, P., Zhang, W., Wu, Y., Chen, F., et al. (2010). Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation pattern associated with genes and centromeres in rice. Plant J. 63, 353–365. doi: 10.1111/j.1365-313X.2010.04246.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, X. J., and Seto, E. (2008). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218. doi: 10.1038/nrm2346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zemach, A., Kim, M. Y., Hsieh, P. H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., et al. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205. doi: 10.1016/j.cell.2013.02.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, W., Bone, J. R., Edomondson, D. G., Turner, B. M., and Roth, S. Y. (1998). Essential and redundant functions of histone acetylation revealed mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17, 3155–3167. doi: 10.1093/emboj/17.11.3155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, K., Sridhar, V. V., Zhu, J., Kapoor, A., and Zhu, J. K. (2007). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2:e1210. doi: 10.1371/journal.pone.0001210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, W., Lee, H. R., Koo, D. H., and Jiang, J. (2008). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in CENH3-associated chromatin in Arabidopsis and maize. Plant Cell 20, 25–34. doi: 10.1105/tpc.107.057083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di-, and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62. doi: 10.1186/gb-2009-10-6-r62
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. doi: 10.1101/gad.927301
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zheng, X., Pontes, O., Zhu, J., Miki, D., Zhang, F., Li, W. X., et al. (2009). ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 455, 1259–1262. doi: 10.1038/nature07305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, B. R., Feng, H., Kato, H., Dai, L., Yang, Y., Zhou, Y., et al. (2013). Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 110, 19390–19395. doi: 10.1073/pnas.1314905110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, J., Jeong, J. C., Zhu, Y., Sokolchik, I., Miyazaki, S., Zhu, J. K., et al. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. U.S.A. 105, 4945–4950. doi: 10.1073/pnas.0801029105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, J. K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166. doi: 10.1146/annurev-genet-102108-134205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zilberman, D., Coleman-Derr, D., Ballinger, T., and Henikoff, S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129. doi: 10.1038/nature07324
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: abiotic stress response, DNA methylation, epigenetic regulation, histone modification, stress memory
Citation: Kim J-M, Sasaki T, Ueda M, Sako K and Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Received: 31 October 2014; Accepted: 11 February 2015;
Published online: 02 March 2015.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Igor Kovalchuk, University of Lethbridge, CanadaZoya Avramova, University of Nebraska–Lincoln, USA
Copyright © 2015 Kim, Sasaki, Ueda, Sako and Seki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Motoaki Seki, Plant Genomic Network Research Team, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan e-mail:bW90b2FraS5zZWtpQHJpa2VuLmpw
 Jong-Myong Kim
Jong-Myong Kim