- 1Department of Agronomy, Food, Natural Resources, Animals and the Environment, University of Padova, Padova, Italy
- 2Proteomics Center of Padova University, Padova, Italy
- 3Sugarcane Breeding Institute, Coimbatore, India
- 4Department of Bio-Engineering, Birla Institute of Technology, Ranchi, India
- 5Department of Biomedical Sciences, University of Padova, Padova, Italy
Ultraviolet-B radiation acts as an environmental stimulus, but in high doses it has detrimental effects on plant metabolism. Plasma membranes represent a major target for Reactive Oxygen Species (ROS) generated by this harmful radiation. Oxidative reactions occurring in the apoplastic space are counteracted by antioxidative systems mainly involving ascorbate and, to some extent, glutathione. The occurrence of the latter and its exact role in the extracellular space are not well documented, however. In Arabidopsis thaliana, the gamma-glutamyl transferase isoform (GGT1) bound to the cell wall takes part in the so-called gamma-glutamyl cycle for extracellular glutathione degradation and recovery, and may be implicated in redox sensing and balance. In this work, oxidative conditions were imposed with Ultraviolet-B radiation (UV-B) and studied in redox altered ggt1 mutants. The response of ggt1 knockout Arabidopsis leaves to UV-B radiation was assessed by investigating changes in extracellular glutathione and ascorbate content and their redox state, and in apoplastic protein composition. Our results show that, on UV-B exposure, soluble antioxidants respond to the oxidative conditions in both genotypes. Rearrangements occur in their apoplastic protein composition, suggesting an involvement of Hydrogen Peroxide (H2O2), which may ultimately act as a signal. Other important changes relating to hormonal effects, cell wall remodeling, and redox activities are discussed. We argue that oxidative stress conditions imposed by UV-B and disruption of the gamma-glutamyl cycle result in similar stress-induced responses, to some degree at least. Data are available via ProteomeXchange with identifier PXD001807.
Introduction
The apoplast—i.e., the extraprotoplastic matrix of plant cells, including the cell wall—contains a number of enzymatic and non-enzymatic components involved in many physiological processes and is therefore important in the plant cell's response to both abiotic and biotic stress (Dietz, 1997; Agrawal et al., 2010). Being at the interface with the external environment, rapid fluctuations occur in this compartment as a consequence of unfavorable conditions, such as salinity (Hernandez et al., 2001), ozone (Jaspers et al., 2005) drought (Hu et al., 2005), and UV-B radiation (Pristov et al., 2013), with consequent changes in the concentrations and redox state of its components.
Ultraviolet-B radiation (UV-B, 280–315 nm) is a component of the solar electromagnetic spectrum reaching the Earth's surface, which has gained attention in recent years because it has increased as a consequence of ozone layer destruction by anthropogenic emissions.
As a component of the solar radiation reaching the leaf, UV-B also acts as an environmental stimulus for plant growth and development. Recent literature has demonstrated the existence of the UV-B photoreceptor 8 (UVR8), which controls the plant's photomorphogenic response to UV-B radiation. UVR8 promotes a signal cascade that mediates UV-B photomorphogenic responses in order to secure plant acclimation and survival in sunlight (Rizzini et al., 2011).
While it is beneficial at low intensities (Hideg et al., 2013), numerous studies have reported that excess UV-B radiation harms plants by causing oxidative damage to cellular targets (Brosché and Strid, 2003), altering the structure and functions of the leaf epidermis, cell wall, and membranes (Pristov et al., 2013). A common consequence of many types of environmental stress in plants is a greater abundance of some reactive oxygen species (ROS), such as superoxide, hydrogen peroxide (H2O2), hydroxyl radicals and singlet oxygen (Li and van Staden, 1998). Increases in ROS are seen after UV-B exposure too (Noctor et al., 2014), and result in lipid peroxidations and damage to plasma membranes. To prevent these detrimental effects, plant cells deploy an array of non-enzymatic and enzymatic antioxidant systems that act as biochemical barriers to counteract and deactivate ROS.
This complex interplay of several metabolites, enzymes, ROS, antioxidants, and hormones gives rise to signals that are transferred inside the cell through the plasma membrane to activate adaptive and response mechanisms.
A major line of defense in the apoplast is represented by the antioxidant molecule ascorbate and, to a lesser extent, glutathione. While both are involved intracellularly in the Halliwell-Asada pathway for controlling ROS and thereby maintaining the cellular redox state and protecting the cellular components from oxidative threat (Smirnoff and Pallanca, 1996; Schafer and Buettner, 2001; Saruhan et al., 2009; Potters et al., 2010), only ascorbate occurs in high micromolar, or even millimolar quantities in the apoplast (Potters et al., 2010), where it can play a part in redox control. The role of extracellular glutathione in the apoplastic space is controversial because it can only be found in traces under physiological conditions, but it can rise to 2% of the total leaf glutathione under pathogen attack (Vanacker et al., 1998a).
There have been reports, however, of the extracellular enzyme gamma-glutamyl-transferase (GGT; E.C. 2.3.2.2) degrading Glutathione (GSH) (Martin et al., 2007), which means that, like animals (Meister and Anderson, 1983), plants also have a gamma-glutamyl cycle involving intracellular glutathione biosynthesis, extrusion and extracellular degradation, with recovery of the constituent amino acids (Ferretti et al., 2009).
These findings can explain the low levels of glutathione in the extracellular environment on the one hand, but also raise the question of the significance of a gamma-glutamyl cycle in plants. In barley roots, using GGT inhibitors in association with the thiol oxidizing molecule diamide resulted in a net glutathione extrusion and accumulation in the extracellular medium (Ferretti et al., 2009). This leads us to wonder whether a gamma-glutamyl cycle could operate as a redox sensing or redox balancing system.
Another study (Tolin et al., 2013) characterized the leaf proteome of Arabidopsis thaliana ggt1 mutant lines and showed that, even under physiological conditions, a number of antioxidant and defense enzymes were significantly upregulated as a result of impaired extracellular GGT activity. This also implies that GSH turnover involving apoplastic GSH degradation is needed for proper redox sensing and/or a coordinated response to the environment. We speculated that a feedback signal might be missing when the GGT cycle is disrupted, and this would trigger the altered response.
To shed light on these unknown GGT functions in the plant's adaptation to the environment, in this work we investigated the effects of UV-B radiation as an oxidizing stress condition affecting the apoplastic environment in wild type Arabidopsis and a previously-characterized ggt1 knockout mutant line (Destro et al., 2011).
To improve our understanding of protein regulation, it can be helpful to use fractionation (sub-cellular proteomics) to reduce the complexity of the total protein extract and enable the visualization of proteins occurring in low quantities (Brunet et al., 2003).
Since apoplastic proteome analysis can afford a better understanding of the complex network of extracellular proteins involved in plant defense (Agrawal et al., 2010), we investigated the changes occurring in the extracellular proteome as a consequence of the null mutation and/or UV-B treatment by means of Isobaric tags for relative and absolute quantification (iTRAQ) labeling for relative peptide quantification and Liquid Chromatography Mass Spectrometry (LC-MS-MS) analysis. This strategy enables an accurate and sensitive protein quantification, which is essential for the identification of apoplastic proteins in small quantities or small variations in their level of expression.
Following extraction with the extracellular washing fluid (ECWF) technique, we also explored ascorbate and glutathione content and their redox state in the leaf apoplastic fluids.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of A. thaliana and a ggt1 knockout mutant line, both Columbia ecotype (Col-0), were sterilized and incubated at 4°C in the dark for 4 days to synchronize germination and ensure a uniform growth. The ggt1 knockout mutant was established in the mutant collection identified by the Salk Institute (Alonso et al., 2003), and was obtained from the Nottingham A. thaliana Stock Centre (http://nasc.nott.ac.uk; polymorphism SALK_080363). Seeds were sown in soil pots and grown in a greenhouse.
For the UV-B radiation experiments, plants in the phase of maximum expansion of the rosette (before bolting) were transferred to a climatic cell 2 days before the treatment to enable their acclimation. The growth chamber settings were: 12/12 h light/dark cycle, 21/21°C temperature, 300 μmol m−2 s−1 photosynthetically active radiation, and 60% relative humidity. The UV-B treatment was applied for 8 h at the beginning of the light period. The radiation was provided by two Philips TL40W/12 lamps with an intensity, measured on a level with the plants, of 8.3 kJ m−2 d−1 (UVBBE, biologically effective UV-B). After the 8 h UV-B treatment, leaves were immediately harvested for ECWF and total leaf extraction. Following, both the infiltrate and the leaf extracts were analyzed for ascorbate content by spectrophotometric method, as described, the same day. Aliquots of the extracts were stored in −80° for thiol measurements.
Apoplastic Fluid Extraction
ECWF were extracted by vacuum infiltration according to Lohaus et al. (2001). About 1 g of fresh leaves were cut, rinsed, immersed in infiltration buffer and vacuum-infiltrated for 10 min at 20 kPa. After infiltration, the leaves were blot-dried, weighed and placed vertically in a 5 ml syringe. The syringes were placed in tubes and centrifuged at 200 g, 4°C for 20 min. Apoplastic fluids were collected from the bottom of the tubes. For ascorbate and thiol extraction, 10 μl 0.1N HCl were placed at the bottom of the tubes before centrifugation to prevent oxidation. The composition of the infiltration buffer used for the ascorbate and thiol measurements was: KH2PO4 50 mM, KCl 50 mM, and EDTA 2.5 mM, pH 4.5. For the GGT activity and proteomic analyses, the infiltration buffer contained: KH2PO4 50 mM, KCl 0.2 M, and PMSF 1 mM, pH 6.2.
The contamination level of the extracts obtained with the infiltration/centrifugation technique was assessed by means of malic dehydrogenase activity measurements, and ranged between 1.6 and 2.5% among the replicate extractions (data not shown).
Total Leaf Extraction
Total leaf extraction for the thiol, ascorbate and Dehydroascorbate (DHA) measurements was done using metaphosphoric acid 1.5% and EDTA 1 mM buffer: 1 g of fresh leaves were powdered in a mortar with liquid nitrogen and extracted in a leaves to buffer ratio of 1:4, then centrifuged at 10'000 rpm for 10 min at 4°C. The same extraction procedure was used for total GGT activity, but using the infiltration buffer.
ASC and DHA Determination
Ascorbate and dehydroascorbate were measured by spectrophotometric analysis following the decrease in absorbance at 265 nm according to Hewitt and Dickes (1961).
Chromatographic Low-Molecular-Weight Thiol Assay
To measure total thiol concentration extracts, 50 μL of total leaf extract and ECWF were derivatized with 4-fluoro-7-sulfobenzofurazan ammonium salt fluorophore (SBD-F) (Dojindo, Japan). Low Molecular Weight (LMW) thiols were separated by isocratic High pressure liquid chromatography (HPLC) using the method described elsewhere (Masi et al., 2002) with some modifications. The mobile phase was 75 mM ammonium-formiate, pH 2.9 and 3% methanol (97:3, vol/vol). For oxidized thiol quantification, samples were pre-treated with 2-vinylpyridine according to Griffith (1980), then buffered to basic pH and treated with 2-vinylpyridine for 1 h to protect the free thiol moieties. Afterwards, the samples were washed to remove the resulting complexes, and the remaining unreacted samples (containing the oxidized thiols) were derivatized and analyzed by HPLC.
GGT Activity Measurements
GGT activity was determined spectrophotometrically according to Huseby and Stromme (1974). Leaf extracts were reacted in a mix of solution A (5 mM g-glutamyl-p-nitroanilide 100 mM NaH2PO4, pH 8.0) and solution B (575 mM gly-gly in 100 mM NaH2PO4, pH 8.0) in a ratio of 10:1. Absorbance was recorded for 1 h at 407 nm to measure p-nitroaniline release into the assay medium.
Statistical Analysis
After checking for a normal distribution, data were tested with One-Way ANOVA using the General linear models (GLM) procedure in SAS (SAS 9.2, 2008). Data with a non-normal distribution were submitted to a non-parametric test (Kruskal-Wallis) using XLSTAT (2014 version). In both cases, Bonferroni's test was used to ascertain differences between means. Significance was established at P = 0.05.
Proteomic Analysis
Protein In Situ Digestion
Proteins obtained from infiltration were quantified by bicinchoninic acid spectrophotometric assay; 50 μg of proteins were loaded in a homemade 11% Sodium dodecyl sulfate (SDS) gel and the electrophoretic run was stopped as soon as the protein extracts entered the running gel. Bands were excised and washed several times with 50 mM triethylammonium bicarbonate (TEAB) and dried under vacuum after a short acetonitrile wash. Cysteines were reduced with 10 mM dithiothreitol (in 50 mM TEAB) for 1 h at 56°C, and alkylated with 55 mM iodoacetamide (in 50 mM TEAB) for 45 min at room temperature in the dark. Gel pieces were then washed with alternate steps of TEAB and acetonitrile, and dried. Proteins were digested in situ with sequencing grade modified trypsin (Promega, Madison, WI, USA) at 37°C overnight (12.5 ng·μl−1 trypsin in 50 mM TEAB). Peptides were extracted with three steps of 50% acetonitrile in water. One μg of each sample was withdrawn to check digestion efficiency using LC-MS/MS analysis, and the remaining peptide solution was dried under vacuum.
iTRAQ Labeling and Peptide Fractionation
Peptides were labeled with iTRAQ reagents (ABSciex) according to the manufacturer's instructions. They were labeled with the four iTRAQ tags using a Latin panel strategy: wt UV-B, ggt1 UV-B, wt ctrl, and ggt1 ctrl were labeled respectively with 114, 115, 116, and 117 tags in the first replicate; 115, 116, 117, 114 tags in the second and 116, 117, 114, 115 tags in the third. Prior to mixing the samples in a 1:1:1:1 ratio, 1 μg of each sample was analyzed separately to check label efficiency by LC-MS/MS analysis, setting the iTRAQ labeling as a variable modification in the database search. All the peptides were correctly identified as being iTRAQ-modified at the N-terminus and at each lysine residue. The samples were then pooled and dried under vacuum. The mixture of labeled samples (one per replicate) was suspended in 500 μl of buffer A (10 mM KH2PO4, 25% acetonitrile, pH 2.9) and loaded onto a strong cation exchange cartridge (AB Sciex) for fractionation according to (Tolin et al., 2013). After a washing step with buffer A, the peptides were eluted stepwise with increasing concentrations of KCl in buffer A (25, 50, 100, 200, and 350 mM). The volume of each fraction (500 μl) was reduced under vacuum, and the samples were desalted using C18 cartridges (Sep-Pack, C18, Waters) according to the manufacturer's instructions. The samples were ultimately dried under vacuum and kept at −20°C until MS analysis.
LC-MS/MS Analysis, Database Search, and Protein Quantification
Samples were suspended in H2O/0.1% formic acid and analyzed by LC-MS/MS. The MS analyses were conducted with a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Pittsburgh, CA, USA) coupled online with a nano-HPLC Ultimate 3000 (Dionex - Thermo Fisher Scientific). Samples were loaded in a homemade 10 cm chromatographic column packed into a pico-frit (75 mm id, 10 mm tip, New Objectives) with C18 material (ReproSil, 300 Å, 3 μm). The LC separation and mass spectrometer settings used for the analyses were the same as those described in Tolin et al. (2013), and the method was as described by Köcher et al. (2009).
The raw LC-MS/MS files were analyzed using Proteome Discoverer 1.4 (Thermo Fisher Scientific), connected to a Mascot Search Engine server (version 2.2.4, Matrix Science, London, UK). The spectra were searched against a ARATH Uniprot protein database. Enzyme specificity was set to trypsin with two missed cleavages, and peptide and fragment tolerance was set to 10 ppm and 0.6 Da, respectively. Methylthiocysteine, 4-plex iTRAQ at the N-terminus and Lys were set as fixed modifications, while Met oxidation was selected as a variable modification. False discovery rates (FDR) were calculated by the software, based on the search against the corresponding randomized database. Only proteins identified and quantified with at least 2 unique peptides with 99% confidence (FDR 1%) were considered as positive identifications. A 5% FDR was adopted in only two cases (as shown in Table 2 and in Data Sheet 1, Supplementary Material), in which the MS/MS spectra were manually inspected for confirmation. Data were pre-filtered to exclude MS/MS spectra containing less than 5 peaks or with a total ion count below 50. Quantification was done by normalizing the results on the median value of all measured iTRAQ reporter ratios.
Protein expression ratios are given as: wt (UV-B/ctrl), ggt1 (UV-B/ctrl), ctrl (ggt1/wt), and UV-B (ggt1/wt) and they are the mean value of at least 2 biological replicates (see Data Sheet 2, 3, 4, Supplementary Material). To improve the statistical robustness of the data, all proteins were submitted to a two-tailed Z test with a confidence level of p < 0.05. The variations were further restricted to proteins exhibiting an at least ±50% fold change in their expression (1.5 for upregulated and 0.68 for downregulated proteins).
Results
GGT Activity
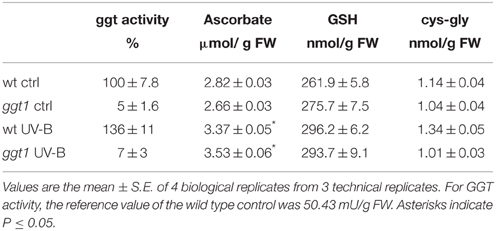
An increase in GGT enzymatic activity was found in wt plants after UV-B irradiation; this increase was greater in total leaf extracts (+35%, Table 1) than in ECWF (+10%, Figure 1A). Activity in the mutant was significantly lower in total leaf extracts and almost undetectable in the ECWF (as was to be expected because GGT1 is the only apoplastic isoform active in leaves), but no significant differences were observed after UV-B exposure (Figure 1A).
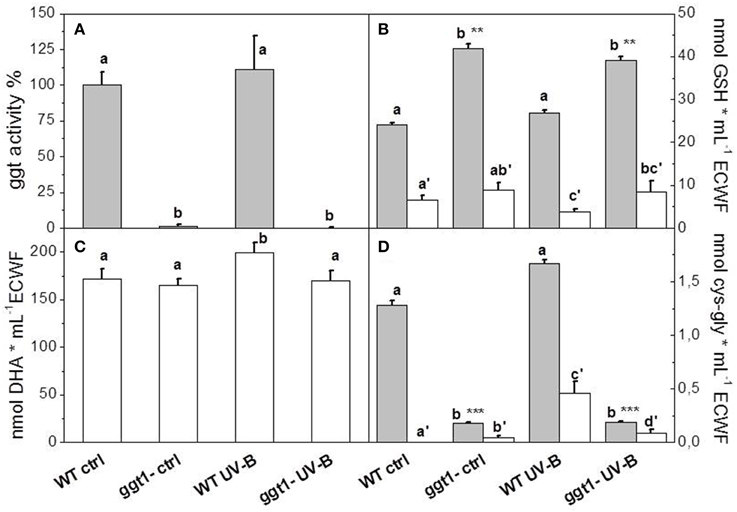
Figure 1. GGT activity (A), glutathione (B), ascorbate (C) and cys-gly (D) in ECWF. Gray bars show total content, white bars oxidized forms. Reported values are the mean ± S.E. of 3 technical replicates, each conducted with at least 4 biological replicates. Different letters indicate significant differences between conditions (P = 0.05*; P = 0.01**; P = 0.001***). For GGT activity, the reference value of the wild type control was 43.05 mU/mL ECWF.
Antioxidant Content (GSH and Ascorbate)
Ascorbate was only found in its reduced form in total leaf extracts, and was increased by UV-B treatment (by approximately 20–30%) in both genotypes (Table 1). We found no reduction in the ascorbate in the apoplastic space, where we could only measure the oxidized form, dehydroascorbate (Figure 1C). We found no significant differences between the genotypes or treatments in the total glutathione or cys-gly content in total leaf extract (Table 1). In ECWF total glutathione content was higher in the ggt1 mutant than in the wild type; and supplementing UV-B radiation did not alter these values (Figure 1B). GSSG was lower in the ECWF from wt leaves under UV-B treatment, whereas oxidized cys-gly increased significantly under the same conditions (Figures 1B,D).
It should be noted that apoplastic glutathione is only a small fraction of total leaf glutathione, so fluctuations in the apoplast are somewhat diluted during the extraction process. For the same reason, variations in the small amount of extracellular DHA may not have been reflected in total leaf extracts.
Proteomic Analysis
In total, 329 proteins were uniquely identified by the LC-MS/MS analyses; 208 were found in at least two biological replicates. Based on the Gene Ontology (GO) assignment for cellular compartmentalization (Uniprot 14, www.uniprot.org), we restricted our analysis to the 118 proteins that were either apoplastic or unlocalized, accounting for approximately 57% of the total.
Should be considered bearing in mind that several truly extracellular proteins have yet to be properly assigned to the apoplast. In fact, it has been reported (Agrawal et al., 2010; Ding et al., 2012) that about 50% of proteins secreted in the apoplast lack a leaderless secretory tag;. There are consequently many unpredicted secretory proteins in plants, and their occurrence is often underestimated or they are even considered improperly as contaminants. Our decision to restrict our assignments according to the Uniprot database was therefore rigorous, but probably led to an underestimation of the truly apoplastic proteins.
The variations considered were further restricted to proteins exhibiting an at least ±50% fold change in expression.
Various information can be drawn from comparisons between the four experimental conditions: (i) the effect of UV-B treatment on each genotype; (ii) differential apoplastic protein composition in ggt1 vs. wt; (iii) possible differences in the behavior of the ggt1 mutant and the wt under UV-B. Comparing the two genotypes, 23 proteins were downregulated and only three were upregulated in ggt1 by comparison with the wt under physiological conditions (Table 2 and Data Sheet 1, Supplementary Material). UV-B treatment resulted in 8 proteins being downregulated in ggt1; and in 12 being downregulated and 11 being upregulated in the wt. When the ggt1 and wt were compared after UV-B treatment, it emerged that 9 proteins were expressed less, and 10 were expressed more in the mutant than in the wild type. A condensed view of all these variations is given in Table 2.
Table 2. Brief overview of expression changes in apoplastic and unlocalized proteins in the four conditions analyzed: wt (UV-B/ctrl), ggt1 (UV-B/ctrl), ctrl (ggt1/wt), and UV-B (ggt1/wt).
To facilitate the interpretation of the results, we ran a bioinformatic analysis with Blast2GO, a tool for the functional annotation of sequences and data mining, based on the GO vocabulary (Conesa and Götz, 2008). This made it easy to assess and visualize the relative abundance of functional terms (obtained from the pool of GO terms) in the category of biological processes, based on a score assigned by the Blast2GO algorithm. Within the category of biological processes, the GO terms involved under the four conditions, and either down- or upregulated, are shown in Figures 2, 3, respectively.
Figure 2. GO terms distribution in the biological process of downregulated proteins. Black bars shows wt (UV-B/ctrl), dark gray bars ggt1 (UV-B/ ctrl), light gray is ctrl (ggt1/wt), and white bars is UV-B (ggt1/wt).
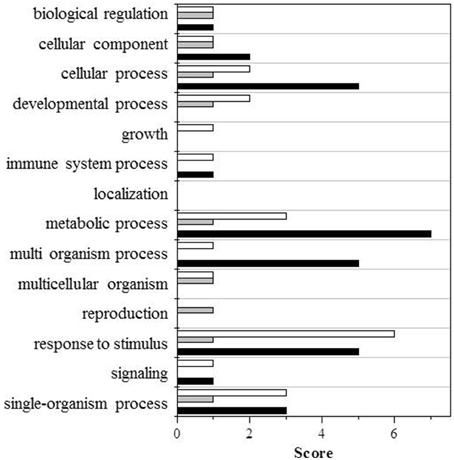
Figure 3. GO terms distribution in the biological process of upregulated proteins. Black bars shows wt (UV-B/ctrl), dark gray bars ggt1 (UV-B/ ctrl), light gray is ctrl (ggt1/wt), and white bars is UV-B (ggt1/wt).
Based on the Blast2GO scores, UV-B in both the wild type and the ggt1 mutant mainly seem to cause a lower expression of proteins in the “metabolic process” and “response to stimulus” categories (Figure 2). Far fewer proteins were upregulated, but the analysis as a whole again showed that the “response to stimulus” and “metabolic process” categories scored highest, but only after the UV-B treatment in both genotypes (Figure 3). Based on the results shown in Table 2, the variations observed were functionally grouped as explained below. For the sake of simplicity, the proteins listed in Table 2 were divided into 4 broad categories, but many of those described here could have been placed in more than one category (depending on whether we considered the protein's biological function or its catalytic activity, for instance).
Pathogenesis and Hormone-Related Proteins
Gibberellins are hormones that can be found in the apoplastic space too (Kramer, 2006). Here, we found the gibberellin-regulated protein Q9LFR3 (At5g14920) upregulated by UV-B treatment in the mutant.
Among the proteins targeted by hormones there is a galactose-binding domain containing protein (At5g25460, Q94F20) with a putative function in response to karrikins, a novel group of plant growth regulators (Nelson et al., 2011). This protein is downregulated in the wild type under UV-B treatment, and in the ggt1 mutant in physiological conditions.
By comparison with the wild type, two pathogen-related proteins are less expressed in the ggt1 mutant, i.e., PR-1 (At2g14610) and PR-5 (At1g75040), reportedly regulated by brassinosteroids (Sävenstrand et al., 2004). Another protein involved in lipid catabolism and response to pathogens is a GDSL esterase/lipase (At3g16370) that is expressed more in the ggt1 mutant than in the wild type under UV-B.
Proteolytic enzymes are directly or indirectly involved in several plant cellular processes, including resistance to pathogens and disease (Xia et al., 2004). In our study, we identified four members of the aspartyl protease family, a class of enzymes acting as endopeptidases to remove aspartic residues from polypeptide chains. One of them (At1g03220) is downregulated in the wild type after UV-B treatment, and in the ggt1 mutant in physiological conditions. This protein and At1g09750 are both upregulated in ggt1 by comparison with the wild type as an effect of UV-B treatment. At5g07030 is downregulated under UV-B in the mutant, and At5g10760 is downregulated in the mutant under control conditions. It could be hard to explain these opposite effects in the expression of members of the same aspartyl protease family, but it is worth noting that these enzymes are reportedly involved in plant defenses and development (Minic et al., 2007).
Cell Wall Remodeling
Up to 90% of plant cell walls consist of three types of polysaccharide: cellulose, hemicelluloses, and pectins. Their composition and structure differ from one species to another, and change as plants develop and with environmental fluctuations (Cosgrove, 1997; Popper and Fry, 2003; Minic et al., 2007). Six proteins belonging to the class of glycoside hydrolases are downregulated by UV-B radiation in the wild type (At2g28470, At5g08380, At4g19410, At5g06870, At4g19810, and At5g08370) while another six proteins are upregulated (At3g14310, At2g45470, At3g57260, At4g30270, At5g20950, and At2g18660).
Comparing the ggt1 mutant with the wild type clearly revealed a constitutive downregulation of proteins related to cell wall remodeling (At4g19410, At4g24780, At5g20950, and At2g43570). One of them is downregulated both in the mutant under physiological conditions and in the wild type after UV-B treatment. An opposite response to UV-B radiation emerged for beta-glucosidase At3g57260, which was higher in the wild type, and lower in ggt1 after the treatment. UV-B radiation resulted in a lower expression in the ggt1 mutant of other cell wall remodeling proteins, namely α-arabinofuranosidase At3g10740, α-xylosidase At1g68560, and the berberine bridge enzyme At5g44400. We also found a lower expression of chitinase At2g43570 in the ggt1 mutant than in the wild type both under physiological conditions and after treatment with UV-B.
Signaling
In this study, we observed changes in four proteins containing leucine-rich repeats, and in two cysteine-rich secretory proteins belonging to a class acting as kinases. Leucine- and cysteine-rich proteins are transmembrane proteins that are reportedly induced by ROS and salicylic acid (Brandes et al., 2009).
All leucine-rich proteins were downregulated in the mutant (At1g33600, At5g12940, At1g33590, and At3g20820) under physiological conditions. It seems particularly interesting that At1g33590 expression was also downregulated in the wild type under UV-B radiation.
The two cysteine-rich repeat secretory proteins, At3g22060 and At5g48540, were both downregulated in the ggt1 mutant under physiological conditions. These proteins are also PM-associated receptor-like kinases, and At3g22060 interacts with one or more unknown PM-localized ABA receptor(s) (Xin et al., 2005), whereas At5g48540 is involved in response to karrikins (Nelson et al., 2011).
The EG45-like domain containing protein (At2g18660) is part of a class of small proteins that act as signaling molecules. In our study, At2g18660 was upregulated in the wild type under UV-B.
GSTs, Redox Regulation and ROS Balance
One protein (At1g31690) involved in response to oxidative stress was downregulated under UV-B in the wild type. This protein is involved in H2O2 metabolism, acting as an oxidase. Also Peroxidase 34 (At3g49120) was downregulated in the ggt1 mutant under control conditions.
Two Glutathione S-Transferases (GSTs) proteins belonging to categories F2 and F9 (At4g02520 and At2g30860, respectively) were upregulated in the wild type plants after UV-B radiation. When the two genotypes were compared after the same treatment, the expression level was lower in the former.
Two proteins in the germin-like family (At5g20630 and At1g02335) were downregulated in ggt1, one in physiological conditions and the other after UV-B treatment. These proteins are involved in defending against biotic and environmental stress.
Superoxide dismutase proteins are reportedly involved in removing superoxide radicals from the apoplast following UV-B radiation (Alscher et al., 2002). In our study, the expression of superoxide dismutase (At4g25100) was upregulated in the wild type after UV-B radiation and in ggt1 under physiological conditions.
Discussion
GGT Activity and Soluble Antioxidants
Following excess UV-B exposure, plants deploy a wide array of morphological and biochemical defense mechanisms, including soluble antioxidants (Shiu, 2005). The changes observed in this study are consistent with the view that, under UV-B radiation, oxidative conditions in the apoplastic space involve both ascorbate and glutathione, the two main soluble antioxidant molecules in plant cells. Ascorbate in ECWF was found fully oxidized, which is consistent with the view that oxidizing conditions prevail in the apoplastic space (Vanacker et al., 1998a,b; Saruhan et al., 2009).
UV-B radiation induced an increase in apoplastic ascorbate in both genotypes, suggesting that ascorbate is extruded as a means to counteract the artificially-imposed oxidative conditions. While glutathione content was substantially unchanged in total leaf extracts in all the conditions tested, it was altered in the ECWF from mutant leaves, where the effect of the ggt1 null mutation results in a net increase in glutathione content, as a predictable effect of the reduced GGT degradation activity.
Under UV-B, the concurrent decrease in oxidized glutathione and increase in oxidized cys-gly can be interpreted as an enhanced gamma-glutamyl transferase activity; this is supported by the previous finding that GGT1 has a stronger preference for GSSG (Ohkama-Ohtsu et al., 2008).
GGT activity was barely detectable in the mutant, confirming that GGT1 is the main isoform contributing to over 90% of said activity in wild type leaves. Since the GGT2 isoform is not expressed in leaves (Destro et al., 2011) and GGT3 is assumed to be non-functional (Martin et al., 2007), this indicates that the activity found in the mutant represents the contribution of the remaining vacuolar isoform GGT4.
The increased GGT activity following UV-B treatment in the wild type therefore suggests that the rate of the gamma-glutamyl cycle is accelerated by this radiation. The involvement of the vacuolar GGT4 in the degradation of glutathione conjugates, e.g., with lipoperoxides and/or other damaged molecules, might be implicated too, but this seems unlikely since no significant increase in GGT activity was apparent in the mutant under the same conditions.
Collectively, these novel findings thus imply that the gamma-glutamyl cycle is accelerated under oxidative conditions imposed by ultraviolet-B radiation, and support the conviction that it is involved in oxidative stress sensing and/or response.
Apoplastic Proteome Readjustments
Proteome analysis has proved a powerful tool for deciphering cell metabolism under different perturbations and has been found useful in apoplastic studies too (Agrawal et al., 2010). Apoplastic proteins establish a constitutive systemic defense network, with only a few of them changing under environmental and/or biotic stress (Delaunois et al., 2014).
Two main approaches are currently adopted in plant physiology studies: the application of stress conditions, and the use of mutants. These alternative and converging strategies may provide tools for deciphering metabolism. In this work, oxidative conditions were imposed with UV-B and studied in redox-altered ggt1 mutants. Subcellular fractionation and apoplastic proteome analysis were then used to arrive at a better understanding of the rearrangements in the extracellular compartment.
The experimental design adopted here could consequently help to describe and compare the effects of UV-B treatment on the two genotypes, and the differences in apoplastic proteome composition between the mutant and wild type leaves under control conditions.
In both genotypes, UV-B treatment caused a downregulation of different kinds of protein related to cell wall biosynthesis, response to stress and proteolysis. It prompted an upregulation, but only in the wild type, of other proteins involved in cell wall remodeling and two glutathione S-transferases, GST-F2 and GST-F9. No proteins were found upregulated in the ggt1 mutant after UV-B (Table 2).
The hormonal changes occurring in the ggt1 mutant, with or without exposure to UV-B radiation, were not considered in the experimental setup, and were beyond the scope of this work. Several proteins seen here to change in expression could be targets for hormones, however. For instance, one protein whose expression was stimulated by UV-B is reportedly a gibberellin-regulated protein (At5g14920), suggesting that gibberellins could be implicated in the response. The expression of a galactose-binding domain containing protein (At5g25460), which is stimulated by karrikins (a novel group of plant growth regulators (Nelson et al., 2011), was also higher in the mutant than in the wild type after UV-B. Interplay with hormones may also concern two PRPs (PR-1 and PR-5), whose expression was lower in the ggt1 mutant under physiological conditions. A previous study (Sävenstrand et al., 2004) had found their expression strongly reduced in brassinosteroid metabolism mutants. It would be interesting to see whether the brassinosteroid pathway is altered in ggt1 mutants too.
Broadly speaking, cell wall modifying proteins such as glycosyl hydrolases (GHs), peroxidases, esterases, transglycosylases, and lyases, are involved in the construction, remodeling or turnover of cell wall components (Cosgrove, 1997; Stolle-Smits et al., 1999; Obel et al., 2002; Reiter, 2002). Some of them may have other functions too, e.g., in the glycosylation state of target proteins (Kang et al., 2008), which in turn could be involved in signaling processes (Minic et al., 2007).
Taking a broader look at the changes found in this category suggests that UV-B affects the expression of some proteins in the wild type (e.g., pectine-acetylesterase and its inhibitor, xyloglucan endotransglucosylase, beta 1,3-glucanase, beta-galactosidase, alpha-galactosidase and a polygalacturonase inhibitor) and others in the ggt1 mutant (alpha-xylosidase, myrosinase, alpha-arabinofuranosidase, and a berberine bridge enzyme), confirming the view that the cell walls are the target of this radiation. Notably, these remodeling processes are affected in the ggt1 mutant not only by UV-B treatment, but also under physiological conditions. Since cell wall structure is reportedly altered during development and by exposure to stress (Potters et al., 2009), our findings could be explained by the existence of a stress-like condition in the mutant, where some signals mimic the oxidative state induced by UV-B in the wild type.
Myrosinase, a protein in the class of glycoside hydrolases, was less expressed under UV-B in the ggt1 mutant, and this could have ecophysiological consequences because in Brassicaceae myrosinases play a part in growth, development, and defenses against microbes, as well as deterring insects and herbivores (Rodman, 1991). The two germin-like proteins that were downregulated in the ggt1 mutant could also be consistent with alterations in the defense systems against biotic and environmental stress.
The expression of some other proteins was altered in opposite ways (up- or downregulated) after UV-B exposure, depending on the genotype considered: for instance, the stress-responsive glucan endo-1,3-beta-glucosidase was upregulated in the wild type, but downregulated in ggt1.
A group of proteins involved in response to stimuli, i.e., the leucine-rich and cysteine-rich proteins, was downregulated in ggt1 in physiological conditions. Leucine-rich proteins contain a leucine-rich repeat (LRR) motif that has revealed a central role in recognizing different pathogen-associated molecules in the innate host defense of plants and animals (Gunawardena et al., 2011).
In this study, we also identified 4 aspartyl proteases that were altered under our experimental conditions: this may mean that members of this category of enzymes related to plant defenses are sensitive to redox variations. Aspartyl proteases are important for plant development. They have been implicated in the ABA-dependent responsiveness to drought-induced stress (Yao et al., 2012), and in Arabidopsis a gene encoding the aspartyl protease protein was found downregulated by cold and high-salinity stress (Seki et al., 2002).
The EG45-like domain containing protein 2 (At2g18660) was upregulated in the wild type under UV-B. This protein belongs to the category of plant natriuretic peptides (PNPs), a novel class of small proteins showing homology with the N-terminus of expansins, though they are significantly shorter and lack the wall-binding domain (Ludidi et al., 2002). Previous studies found PNPs upregulated under saline and osmotic conditions (Rafudeen et al., 2003), but the effects of UV-B on this class of peptidic signaling molecules had not been reported before.
Among the variations in apoplastic enzyme expression found in the present study, some that particularly attracted our attention are closely related to ROS metabolism.
Superoxide anion formation is reportedly triggered by ultraviolet-B radiation (Alscher et al., 2002). It seems noteworthy that no plant superoxide dismutase (SOD) identified to date contains a signal peptide, but extracellular SOD activity in stressed or pathogen-infected plants has been reported in many works (Hernandez et al., 2001; Karpinska et al., 2001; Kaffarnik et al., 2009; Pechanova et al., 2010). SODs produce H2O2, which is degraded to H2O by ascorbate peroxidase. By removing superoxide anions, SODs may limit the duration of the oxidative burst to an early event in plant defense (Pristov et al., 2013; Scheler et al., 2013).
In this study, a superoxide dismutase (At4g25100) was found upregulated by UV-B radiation in wild type leaves: this can be interpreted as the need to improve scavenging activity to remove excess superoxide anions. Although its localization is not reported in the official databases, its occurrence in the apoplast was noted in other studies too (Kwon et al., 2005; Ding et al., 2012). Higher SOD levels combined with lower levels of the putative copper amine oxidase (At1g31690) may result in higher H2O2 levels. An increased GST expression could also result in its scavenging, however, so while it seems clear that ROS metabolism is affected by UV-B treatment in the wild type, it is hard to draw any conclusions on H2O2 levels, and further experimentation is needed to validate our hypothesis.
The ROS scavenging scenario in the ggt1 mutant is undoubtedly more complex. The above-mentioned apoplastic SOD is upregulated under physiological conditions, and a peroxidase is downregulated. These effects may be interpreted as readjustments in the redox-altered ggt1 background. Such readjustments may be needed to sustain a higher H2O2 level, which could act as a signal.
Taken together, these effects may result in higher H2O2 levels in the mutant under physiological conditions (schematically shown in Figure 4), whereas the rise in H2O2 in the wild type is a direct consequence of oxidative stress conditions induced by UV-B radiation.
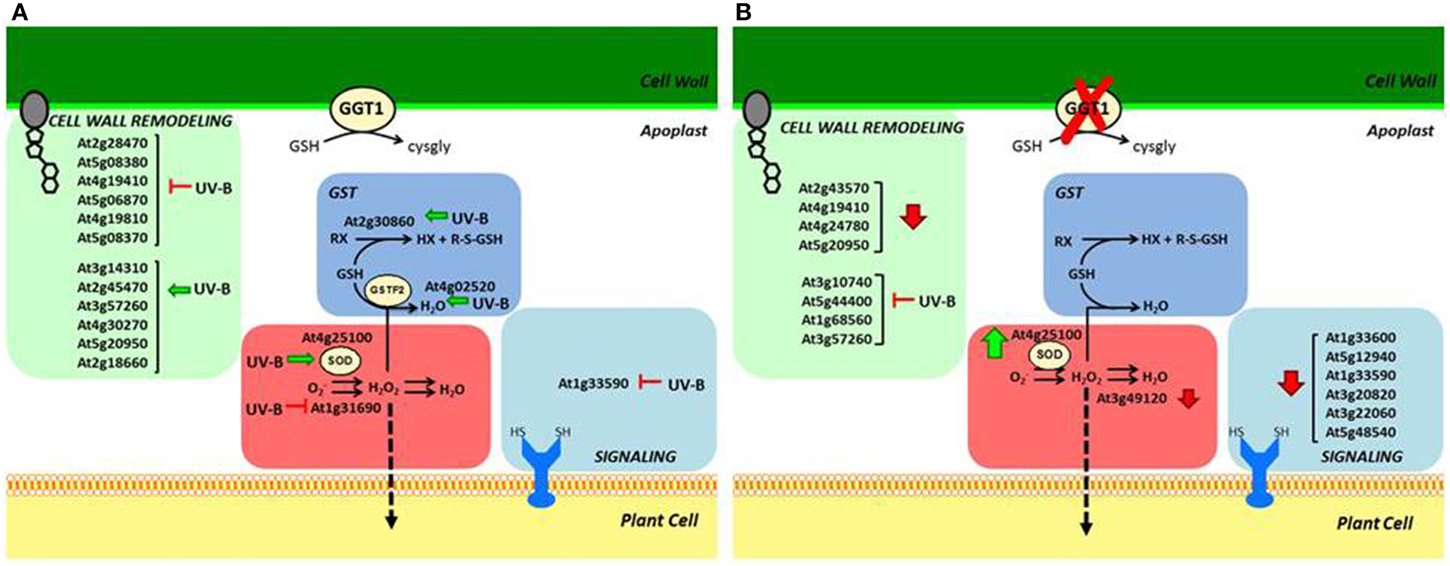
Figure 4. Schematic overview of apoplastic proteome variations in: (A) wild type, induced by UV-B; (B) ggt1 genotype due to the mutation and/or to UV-B treatment. Vertical arrows refer to stimulation (↑) or repression (↓) caused by the mutation; horizontal arrows indicate repression (⊢) or stimulation (←) caused by UV-B treatment.
As a signaling molecule, H2O2 may cross membranes in a process facilitated by aquaporins (Bienert et al., 2006), reaching internal cell compartments and the nucleus, where it can activate defense gene expression (Mullineaux et al., 2006). If this assumption holds true, it might explain the “constitutive alert response” effect observed in a previous proteomic analysis of total leaf extracts from ggt1 mutant leaves (Tolin et al., 2013).
Future research is therefore needed to ascertain the level of ROS, and especially H2O2, in the apoplast of ggt1 mutants, and the possible involvement of hormones (e.g., brassinosteroids and gibberellins) in the response. Both H2O2 and hormones are signals arising in the apoplast that can be transferred intracellularly and evoke the cell's responses. For this signal transduction function we could also consider four leucine-rich and two cysteine-rich proteins belonging to the superfamily of receptor-like kinases (RLKs), which are associated with the plasma membrane and contain redox-sensitive thiols, which were found at lower level in the ggt1 mutant. Disrupting of the gamma-glutamyl cycle could result in an altered signal perception pathway.
While hormonal and redox readjustments seem to be implicated in the modified metabolism of ggt1 mutants, it remains to be seen how silencing the gamma-glutamyl transferase activity and consequently impairing the gamma-glutamyl cycle may lead to the effects reported here. Further experiments are needed to clarify the link between the gamma-glutamyl cycle and apoplastic redox events.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Renato Millioni from the Proteomics Center of Padova University for his kind support, and Frances Coburn for text and style improvement. The authors also wish to thank the “Cassa di Risparmio di Padova e Rovigo” (CARIPARO) Foundation for funding the acquisition of the LTQ-Orbitrap XL mass spectrometer. DP was granted a University of Padova post-doctoral fellowship for Indian researchers; SMMM was supported by an Erasmus Mundus - EMMA Fellowship. LEB was granted a “Fondazione A. Gini” fellowship. MP was funded by a University of Padova research fellowship. MF was granted a fellowship for doctoral students from the CARIPARO Foundation. This work was funded by the University of Padova - MURST ex-60%.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00128/abstract.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001807.
Abbreviations
UV-B, Ultraviolet-B radiation; ROS, Reactive Oxygen Species; GGT1, gamma-glutamyl transferase 1 isoform; H2O2, Hydrogen Peroxide; iTRAQ, Isobaric tags for relative and absolute quantification; UVR8, UV-B photoreceptor 8; GSH, Glutathione; LC-MS-MS, Liquid Chromatography Mass Spectrometry; ECWF, Extracellular washing fluid; DHA, Dehydroascorbate; SBD-F, 4-fluoro-7-sulfobenzofurazan ammonium salt fluorophore; LMW, Low Molecular Weight; HPLC, High pressure liquid chromatography; ANOVA, Analysis of variance; GLM, General linear models; SDS, Sodium dodecyl sulfate; TEAB, Triethyl ammonium bicarbonate; FDR, False discovery rates; FW, Formula Weight; GSSG, Glutathione disulfide/Oxidized GSH; SOD, Superoxide dismutase; PNPs, Plant natriuretic peptides; GSTs, Glutathione S-Transferases; GHs, Glycosyl hydrolases; PRPs, Pathogenesis-related proteins.
References
Agrawal, G. K., Jwa, N. S., Lebrun, M. H., Job, D., and Rakwal, R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10, 799–827. doi: 10.1002/pmic.200900514
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Alscher, R. G., Erturk, N., and Health, L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jexbot/53.372.1331
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bienert, G. P., Schjoerring, J. K., and Jahn, T. P. (2006). Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 1758, 994–1004. doi: 10.1016/j.bbamem.2006.02.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brandes, N., Schmitt, S., and Jakob, U. (2009). Thiol-based redox switches in eukaryotic proteins. Antiox. Redox Signal. 11, 997–1014. doi: 10.1089/ars.2008.2285
Brosché, M., and Strid, Å. (2003). Molecular events following perception of ultraviolet-B radiation by plants. Physiol. Plant. 117, 1–10. doi: 10.1034/j.1399-3054.2003.1170101.x
Brunet, S., Thibault, P., Gagnon, E., Kearney, P., Bergeron, J. J., and Desjardins, M. (2003). Organelle proteomics: looking at less to see more. Trends Cell Biol. 13, 629–638. doi: 10.1016/j.tcb.2003.10.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. doi: 10.1155/2008/619832
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cosgrove, D. J. (1997). Assembly and enlargement of the primary cell wall in plants. Ann. Rev. Cell Dev. Biol. 13, 171–201.
Delaunois, B., Jeandet, P., Clément, C., Baillieul, F., Dorey, S., and Cordelier, S. (2014). Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant. Sci. 5:249. doi: 10.3389/fpls.2014.00249
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Destro, T., Prasad, D., Martignago, D., Bernet, I. L., Trentin, A. R., Renu, I. K., et al. (2011). Compensatory expression and substrate inducibility of γ-glutamyl transferase GGT2 isoform in Arabidopsis thaliana. J. Exp. Bot. 62, 805–814. doi: 10.1093/jxb/erq316
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dietz, K. J. (1997). Functions and responses of the leaf apoplast under stress. Prog. Bot. 58, 221–254. doi: 10.1007/978-3-642-60458-4_9
Ding, Y., Wang, J., Wang, J., Stierhof, Y. D., Robinson, D. G., and Jiang, L. (2012). Unconventional protein secretion. Trends Plant Sci. 17, 606–615. doi: 10.1016/j.tplants.2012.06.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferretti, M., Destro, T., Tosatto, S. C., La Rocca, N., Rascio, N., and Masi, A. (2009). Gamma-glutamyl transferase in the cell wall participates in extracellular glutathione salvage from the root apoplast. New Phytol. 181, 115–126. doi: 10.1111/j.1469-8137.2008.02653.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Griffith, O. W. (1980). Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 106, 207–212. doi: 10.1016/0003-2697(80)90139-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gunawardena, H. P., Huang, Y., Kenjale, R., Wang, H., Xie, L., and Chen, X. (2011). Unambiguous Characterization of Site-specific Phosphorylation of Leucine-rich Repeat Fli-I-interacting Protein 2 (LRRFIP2) in Toll-like Receptor 4 (TLR4)-mediated Signaling. J. Biol. Chem. 286, 10897–10910. doi: 10.1074/jbc.M110.168179
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hernandez, J. A., Ferrer, M. A., Jimenez, A., Barcelo, A. R., and Sevilla, F. (2001). Antioxidant systems and O2.-/ H2O2 production in the apoplast of Pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 127, 817–831. doi: 10.1104/pp.010188
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hewitt, E. J., and Dickes, G. J. (1961). Spectrophotometric measurements on ascorbic acid and their use for the estimation of ascorbic acid and dehydroascorbic acid in plant tissues. Biochem. J. 78, 384–391.
Hideg, E., Jansen, M. A. K., and Strid, A. (2013). UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 18, 107–115. doi: 10.1016/j.tplants.2012.09.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hu, J. F., Li, G. F., Gao, Z. H., Chen, L., Ren, H. B., and Jia, W. S. (2005). Regulation of water deficit-induced abscisic acid accumulation by apoplastic ascorbic acid in maize seedlings. J. Integr. Plant Biol. 47, 1335–1344. doi: 10.1111/j.1744-7909.2005.00165.x
Huseby, N. E., and Stromme, J. H. (1974). Practical points regarding routine determination of γ-glutamyl transferase (γ-GT) in serum with a kinetic method at 37°C. Scand. J. Clin. Lab. Invest. 34, 357–363. doi: 10.3109/00365517409049892
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jaspers, P., Kollist, H., Langebartels, C., and Kangasjarvi, J. (2005). “Plant responses to ozone,” in Antioxidants and Reactive Oxygen Species in Plants, ed N. Smirnoff (Oxford: Blackwell Publishing), 268–292
Kaffarnik, F. A., Jones, A. M., Rathjen, J. P., and Peck, S. C. (2009). Effector proteins of the bacterial pathogen pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol. Cell. Proteomics 8, 145–156. doi: 10.1074/mcp.M800043-MCP200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kang, J. S., Frank, J., Kang, C. H., Kajiura, H., Vikram, M., Ueda, A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938. doi: 10.1073/pnas.0800237105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karpinska, B., Karlsson, M., Schinkel, H., Streller, S., Su, K., Melzer, M., et al. (2001). A novel superoxide dismutase with a high isoelectric point in higher plants. Expression, regulation, and protein localization. Plant Physiol. 126, 1668–1677. doi: 10.1104/pp.126.4.1668
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Köcher, T., Pichler, P., Schutzbier, M., Stingl, C., Kaul, A., Teucher, N., et al. (2009). High precision quantitative proteomics using iTRAQ on an LTQ orbitrap: a new mass spectrometric method combining the benefits of all. J. Proteome Res. 8, 4743–4752. doi: 10.1021/pr900451u
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kramer, E. M. (2006). How far can a molecule of weak acid travel in the apoplast or xylem? Plant Physiol. 141, 1233–1236. doi: 10.1104/pp.106.083790
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kwon, H. K., Yokoyama, R., and Nishitani, K. (2005). A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 46, 843–857. doi: 10.1093/pcp/pci089
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, L., and van Staden, J. (1998). Effects of plant growth regulators on the antioxidant system in callus of two maize cultivars subjected to water stress. Plant Growth Regul. 24, 55–66. doi: 10.1023/A:1005954532397
Lohaus, G., Pennewiss, K., Sattelmacher, B., Hussmann, M., and Muehling, K. H. (2001). Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol. Plant. 111, 457–465. doi: 10.1034/j.1399-3054.2001.1110405.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ludidi, N. N., Heazlewood, J. L., Seoighe, C., Irving, H. R., and Gehring, C. A. J. (2002). Expansin-like molecules: novel functions derived from common domains. Mol. Evol. 54, 587–594. doi: 10.1007/s00239-001-0055-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martin, M. N., Saladores, P. H., Lambert, E., Hudson, A. O., and Leustek, T. (2007). Localization of members of the gamma-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 144, 1715–1732. doi: 10.1104/pp.106.094409
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Masi, A., Ghisi, R., and Ferretti, M. (2002). Measuring low-molecular-weight thiols by detecting the fluorescence of fheir SBD-derivatives: application to studies of diurnal and UV-B induced changes in Zea Mays L. J. Plant Physiol. 159, 499–507. doi: 10.1078/0176-1617-00655
Minic, Z., Jamet, E., Négroni, L., Arsene der Garabedian, P., Zivy, M., and Jouanin, L. (2007). A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 58, 2503–2512. doi: 10.1093/jxb/erm082
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mullineaux, P. M., Karpinski, S., and Baker, N. R. (2006). Spatial dependence for hydrogen peroxide-directed signalling in light-stressed plants. Plant Physiol. 141, 346–350. doi: 10.1104/pp.106.078162
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nelson, D. C., Scaffidi, A., Dun, E. A., Waters, M. T., Flematti, G. R., Dixon, K. W., et al. (2011). F-Box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902. doi: 10.1073/pnas.1100987108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Obel, N., Porchia, A. C., and Scheller, H. V. (2002). Dynamic changes in cell wall polysaccharides during wheat seedling development. Phytochemistry 60, 603–610. doi: 10.1016/S0031-9422(02)00148-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ohkama-Ohtsu, N., Oikawa, A., Zhao, P., Xiang, C., Saito, K., and Oliver, D. J. (2008). A gamma-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol. 148, 1603–1613. doi: 10.1104/pp.108.125716
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pechanova, O., Hsu, C. Y., Adams, J. P., Pechan, T., Vandervelde, L., Drnevich, J., et al. (2010). Apoplast proteome reveals that extracellular matrix contributes to multistress response in Poplar. BMC Genomics 11:674. doi: 10.1186/1471-2164-11-674
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Popper, Z. A., and Fry, S. C. (2003). Primary cell wall composition of Bryophytes and Charophytes. Ann. Bot. 91, 1–12. doi: 10.1093/aob/mcg013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Potters, G., Horemans, N., and Jansen, M. K. (2010). The cellular redox state in plant stress biology a charging concept. Plant Physiol. Biochem. 48, 292–300. doi: 10.1016/j.plaphy.2009.12.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Potters, G., Pasternak, T. P., Guisez, Y., and Jansen, M. A. (2009). Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ. 32, 158–169. doi: 10.1111/j.1365-3040.2008.01908.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pristov, J. B., Jovanović, S. V., Mitrović, A., and Spasojević, I. (2013). UV-irradiation provokes generation of superoxide on cell wall polygalacturonic acid. Physiol. Plant. 148, 574–581. doi: 10.1111/j.1399-3054.2012.12001.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rafudeen, S., Gxaba, G., Makgoke, G., Bradley, G., Pironcheva, G., Raitt, L., et al. (2003). A role for plant natriuretic peptide immuno-analogues in NaCl- and drought-stress responses. Physiol. Plant. 199, 554–562. doi: 10.1046/j.1399-3054.2003.00201.x
Reiter, W. D. (2002). Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5, 536–542. doi: 10.1016/S1369-5266(02)00306-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rizzini, L., Favory, J. J., Cloix, C., Faggionato, D., O'Hara, A., Kaiserli, E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rodman, J. E. (1991). A taxonomic analysis of glucosinolate-producing plants. Part 1: Phenetics, Syst. Bot. 16, 598–618. doi: 10.2307/2418864
Saruhan, N., Terzi, R., Saglam, A., and Kadioglu, A. (2009). The relationship between leaf rolling and ascorbate-glutathione cycle enzymes in apoplastic and symplastic areas of Ctenanthe Setosa subjected to drought stress. Biol. Res. 42, 315–326. doi: 10.4067/S0716-97602009000300006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sävenstrand, H., Brosché, M., and Strid, A. (2004). Ultraviolet-B signalling: Arabidopsis brassinosteroid mutants are defective in UV-B regulated defence gene expression. Plant Physiol. Biochem. 42, 687–694. doi: 10.1016/j.plaphy.2004.06.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schafer, F. Q., and Buettner, G. R. (2001). Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212. doi: 10.1016/S0891-5849(01)00480-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Scheler, C., Durner, J., and Astier, J. (2013). Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 16, 534–539. doi: 10.1016/j.pbi.2013.06.020
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Seki, M., Narusaka, M., Ishida, J., Nanjo, T., Fujita, M., Oono, Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high -salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. doi: 10.1046/j.1365-313X.2002.01359.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shiu, C.-T. (2005). Ultraviolet-B-induced oxidative stress and responses of the ascorbate-glutathione cycle in a marine macroalga Ulva Fasciata. J. Exp. Bot. 56, 2851–2865. doi: 10.1093/jxb/eri277
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smirnoff, N., and Pallanca, J. E. (1996). Ascorbate metabolism in relation to oxidative stress. Biochem. Soc. Trans. 24, 472–478.
Stolle-Smits, T., Beekhuizen, J. G., Kok, M. T., Pijnenburg, M., Recourt, K., Derksen, J., et al. (1999). Changes in cell wall polysaccharides of green bean pods during development. Plant Physiol. 121, 363–372. doi: 10.1104/pp.121.2.363
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tolin, S., Arrigoni, G., Trentin, A. R., Veljovic-Jovanovic, S., Pivato, M., Zechman, B., et al. (2013). Biochemical and quantitative proteomics investigations in Arabidopsis ggt1 mutant leaves reveal a role for the gamma-glutamyl cycle in plant's adaptation to environment. Proteomics 13, 2031–2045. doi: 10.1002/pmic.201200479
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vanacker, H., Carver, T. L. W., and Foyer, C. H. (1998a). Pathogen-Induced changes in the antioxidant status of the apoplast in barley leaves. Plant Physiol. 117, 1103–1114. doi: 10.1104/pp.117.3.1103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vanacker, H., Harbinson, J., Ruisch, J., Carver, T. L. W., and Foyer, C. H. (1998b). Antioxidant defences of the apoplast. Protoplasma 205, 129–140. doi: 10.1007/BF01279303
Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., et al. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23, 980–988. doi: 10.1038/sj.emboj.7600086
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xin, Z., Zhao, Y., and Zheng, Z. L. (2005). Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 139, 1350–1365. doi: 10.1104/pp.105.068064
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yao, X., Xiong, W., Ye, T., and Wu, Y. (2012). Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J. Exp. Bot. 63, 2579–2593. doi: 10.1093/jxb/err433
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: glutathione, gamma-glutamyl-transferase, oxidative stress, iTRAQ labeling, apoplast, ultraviolet-B radiation
Citation: Trentin AR, Pivato M, Mehdi SMM, Barnabas LE, Giaretta S, Fabrega-Prats M, Prasad D, Arrigoni G and Masi A (2015) Proteome readjustments in the apoplastic space of Arabidopsis thaliana ggt1 mutant leaves exposed to UV-B radiation. Front. Plant Sci. 6:128. doi: 10.3389/fpls.2015.00128
Received: 31 October 2014; Accepted: 17 February 2015;
Published: 24 March 2015.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Ganesh Kumar Agrawal, Research Laboratory for Biotechnology and Biochemistry (RLABB), NepalIris Finkemeier, Max Planck Institute for Plant Breeding Research, Germany
Copyright © 2015 Trentin, Pivato, Mehdi, Barnabas, Giaretta, Fabrega-Prats, Prasad, Arrigoni and Masi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Masi, Department of Agronomy, Food, Natural Resources, Animals and the Environment, University of Padova, Viale dell'Università 16, Legnaro (PD), 35020, ItalyYW50b25pby5tYXNpQHVuaXBkLml0
 Anna Rita Trentin1
Anna Rita Trentin1 Syed M. M. Mehdi
Syed M. M. Mehdi Dinesh Prasad
Dinesh Prasad Giorgio Arrigoni
Giorgio Arrigoni Antonio Masi
Antonio Masi