- 1Department of Plant Systems Biology, Vlaams Instituut voor Biotechnologie, Ghent, Belgium
- 2Department of Plant Biotechnology and Genetics, Ghent University, Ghent, Belgium
- 3Department of Medical Protein Research, Vlaams Instituut voor Biotechnologie, Ghent, Belgium
- 4Department of Biochemistry, Ghent University, Ghent, Belgium
A commentary on
A peptide hormone and its receptor protein kinase regulate plant cell expansion
by Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., and Sussman. M. R. (2014). Science 343, 408–411. doi: 10.1126/science.1244454
Small signaling peptides and their receptors play essential roles in plant growth and development (Murphy et al., 2012; Czyzewicz et al., 2013; Matsubayashi, 2014). Arabidopsis thaliana encodes over 600 putative receptor-like kinases and more than 1000 potential secreted peptides (Shiu and Bleecker, 2001; Lease and Walker, 2006), and similar numbers can be expected in other plant species (Shiu et al., 2004; Lehti-Shiu et al., 2009). Taking into account that one peptide can bind or signal through multiple receptors and one receptor can recognize several peptides (Ogawa et al., 2008; Kinoshita et al., 2010; Lee et al., 2011; Shinohara et al., 2012), there is an enormous number of possible peptide ligand-receptor kinase pairs. However, this number of possibilities is in stark contrast with the very few peptide ligand-receptor kinase pairs that have been identified (Butenko et al., 2009; Murphy et al., 2012; Czyzewicz et al., 2013; Endo et al., 2014). Evidently, unambiguous identification of a ligand is crucial to fully understand receptor kinase–mediated signaling pathways (Hirakawa et al., 2008; Jia et al., 2008; Ogawa et al., 2008; Lee et al., 2012; Uchida et al., 2012; Okamoto et al., 2013; Tabata et al., 2014).
The identification of ligand-receptor pairs is technically very challenging, as the genes encoding them regularly belong to gene families with multiple members and are often low expressed, and this only in certain cell types or during specific developmental stages. Therefore, various strategies were followed to identify candidate pairs, such as transcriptional analyses at cellular resolution, microscopic characterization of loss and gain-of-function plants, genetic interaction studies, and biochemical assays demonstrating direct physical interactions (see e.g., Murphy et al., 2012 and Czyzewicz et al., 2013 for more details).
To study the physical interaction of ligands with their receptors, Butenko et al. (2014) recently developed a rapid cellular bioassay that uses the oxidative burst response in Nicotiana benthamiana leaves as readout for activation of (ectopically expressed) receptors by synthetic peptides. However, while a broad range of receptor kinases might be able to activate an oxidative burst, it is likely that this approach cannot be applied to all peptide ligands and/or receptor kinases. In addition, prior knowledge on potential receptor candidates facilitates such studies and there is the requirement for expressing the receptor in N. benthamiana. A similar approach relies on chimeric receptors and monitoring of luciferase activity of known targets in protoplasts transiently expressing signaling components (Albert et al., 2010; Mueller et al., 2012).
Another approach was used by Tabata et al. (2014). Here, overexpression of individual A. thaliana leucine-rich repeat receptor kinases (LRR-RKs) from subfamilies X and XI in tobacco BY-2 cells was combined with photoaffinity labeling of these LRR-RKs by a biologically active small signaling peptide analog derivatized with 125I-labeled photoreactive 4-azidosalicylic acid ([125]ASA). This revealed that two related LRR-RKs of subfamily XI directly and specifically interact with C-TERMINALLY ENCODED PEPTIDE 1 (CEP1). Subsequently, this interaction was confirmed by demonstrating that cepr1 cepr2, a double loss-of-function mutant in the identified CEP1 receptor genes, was insensitive to synthetic CEP1 peptide in a root growth assay. This novel approach represents a major leap forward regarding ligand–receptor pairing, but does not take the ligand-receptor interactions into account that rely on a protein complex status involving co-receptors and/or interacting proteins.
In this General Commentary, we would like to highlight an original proteomics-driven approach overcoming the above-described limitations, including the need for constructs, which was employed by the Sussman lab in their quest for the receptor of the secreted RAPID ALKALINIZATION FACTOR (RALF) peptide (Haruta et al., 2014) (Figure 1). Specifically, the phosphorylation status of plasma membrane proteins—in their natural in planta environment—was studied in response to treatment with recombinant, biologically active RALF peptide. To obtain quantitative phosphoprotein profiles, 15N metabolic labeling of A. thaliana seedlings was combined with mass spectrometry–based phosphoproteomics. This strategy allowed the identification of five plasma membrane proteins that displayed a RALF-induced change in phosphorylation pattern, of which two receptor kinases. These can be considered as putative RALF receptors, given that ligand binding likely results in an immediate change in phosphostatus, as illustrated through phosphorylation of the brassinosteroid receptor BRI1 upon ligand binding (Wang et al., 2005). Among these was the FERONIA (FER) receptor kinase, which, following necessary biochemical and functional characterization, was indeed confirmed as a RALF receptor. It is hypothesized that, upon recognition of the peptide ligand, phosphorylation at the C-terminus activates the kinase and initiates a RALF-induced signaling cascade. Additionally, the phosphoproteome analysis pinpointed a plasma membrane H+-ATPase, AHA2, as a potential downstream protein and/or putative FER substrate. Their findings support a model where RALF recognition by the FER kinase affects proton transport, and consequently cell elongation and plant development (see also Murphy and De Smet, 2014). This powerful and straightforward approach hence identified RALF and FER as peptide–receptor pair and shed light on the molecular mechanism that regulates cell elongation. However, it should be taken into account that depending on the experimental set-up, the selected treatments, and the developmental and physiological growth stage, this approach might not give the complete picture. This is exemplified by the fer4 knockout mutant, which is not completely insensitive to RALF at higher concentrations, suggesting there are likely other RALF receptors. Nevertheless, this is a powerful approach that allows identifying receptors for selected ligands and/or universal downstream responses, without (transient) expression of components.
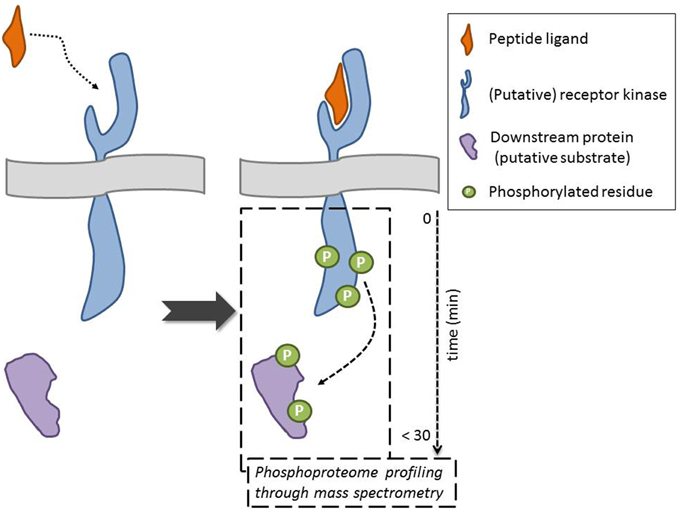
Figure 1. Mass spectrometry-based approaches point to putative receptor kinase for orphan ligand and to downstream components and/or potential substrates following ligand binding through evaluation of changes in the phosphorylation status of proteins. The ideal scenario, where indeed the peptide interacts with the receptor, is depicted (dotted arrow). An alternative outcome is, for example, that the peptide indirectly effects the phosphorylation status of a membrane-associated receptor kinase. The phosphorylation of the downstream protein can be direct (receptor kinase substrate) or indirect (requiring intermediate kinases) (dashed arrow).
In conclusion, enormous progress has been made in the matching of peptide ligand–receptor kinase pairs in the last years, with mass spectrometry-driven phosphoprotein profiling as a promising strategy that can be applied to identify receptors for orphan ligands and to progressively close the gap in the plant peptide–receptor field.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
ES is a Postdoctoral Research Fellows of the Fund for Scientific Research (FWO)-Flanders (Belgium).
References
Albert, M., Jehle, A. K., Mueller, K., Eisele, C., Lipschis, M., and Felix, G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285, 19035–19042. doi: 10.1074/jbc.M110.124800
Butenko, M. A., Vie, A. K., Brembu, T., Aalen, R. B., and Bones, A. M. (2009). Plant peptides in signalling: looking for new partners. Trends Plant Sci. 14, 255–263. doi: 10.1016/j.tplants.2009.02.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Butenko, M. A., Wildhagen, M., Albert, M., Jehle, A., Kalbacher, H., Aalen, R. B., et al. (2014). Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26, 1838–1847. doi: 10.1105/tpc.113.120071
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Czyzewicz, N., Yue, K., Beeckman, T., and De Smet, I. (2013). Message in a bottle: small signalling peptide outputs during growth and development. J. Exp. Bot. 64, 5281–5296. doi: 10.1093/jxb/ert283
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Endo, S., Betsuyaku, S., and Fukuda, H. (2014). Endogenous peptide ligand-receptor systems for diverse signaling networks in plants. Curr. Opin. Plant Biol. 21C, 140–146. doi: 10.1016/j.pbi.2014.07.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., and Sussman, M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. doi: 10.1126/science.1244454
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. doi: 10.1073/pnas.0808444105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jia, G., Liu, X., Owen, H. A., and Zhao, D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 2220–2225. doi: 10.1073/pnas.0708795105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kinoshita, A., Betsuyaku, S., Osakabe, Y., Mizuno, S., Nagawa, S., Stahl, Y., et al. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920. doi: 10.1242/dev.048199
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lease, K. A., and Walker, J. C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142, 831–838. doi: 10.1104/pp.106.086041
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, H., Chah, O. K., and Sheen, J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473, 376–379. doi: 10.1038/nature09958
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, J. S., Kuroha, T., Hnilova, M., Khatayevich, D., Kanaoka, M. M., Mcabee, J. M., et al. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126–136. doi: 10.1101/gad.179895.111
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lehti-Shiu, M. D., Zou, C., Hanada, K., and Shiu, S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26. doi: 10.1104/pp.108.134353
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matsubayashi, Y. (2014). Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. doi: 10.1146/annurev-arplant-050312-120122
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mueller, K., Bittel, P., Chinchilla, D., Jehle, A. K., Albert, M., Boller, T., et al. (2012). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24, 2213–2224. doi: 10.1105/tpc.112.096073
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Murphy, E., and De Smet, I. (2014). Understanding the RALF family: a tale of many species. Trends Plant Sci. 19, 664–671. doi: 10.1016/j.tplants.2014.06.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Murphy, E., Smith, S., and De Smet, I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24, 3198–3217. doi: 10.1105/tpc.112.099010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ogawa, M., Shinohara, H., Sakagami, Y., and Matsubayashi, Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. doi: 10.1126/science.1150083
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Okamoto, S., Shinohara, H., Mori, T., Matsubayashi, Y., and Kawaguchi, M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4:2191. doi: 10.1038/ncomms3191
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shinohara, H., Moriyama, Y., Ohyama, K., and Matsubayashi, Y. (2012). Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70, 845–854. doi: 10.1111/j.1365-313X.2012.04934.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shiu, S. H., and Bleecker, A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE 2001:re22. doi: 10.1126/stke.2001.113.re22
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tabata, R., Sumida, K., Yoshii, T., Ohyama, K., Shinohara, H., and Matsubayashi, Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. doi: 10.1126/science.1257800
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Uchida, N., Lee, J. S., Horst, R. J., Lai, H. H., Kajita, R., Kakimoto, T., et al. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. U.S.A. 109, 6337–6342. doi: 10.1073/pnas.1117537109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, X., Goshe, M. B., Soderblom, E. J., Phinney, B. S., Kuchar, J. A., Li, J., et al. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17, 1685–1703. doi: 10.1105/tpc.105.031393
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: peptide ligand, receptor kinase, phosphorylation, mass spectrometry, phosphoproteomics
Citation: Stes E, Gevaert K and De Smet I (2015) Phosphoproteomics-based peptide ligand-receptor kinase pairing. Commentary on: “A peptide hormone and its receptor protein kinase regulate plant cell expansion”. Front. Plant Sci. 6:224. doi: 10.3389/fpls.2015.00224
Received: 17 December 2014; Accepted: 21 March 2015;
Published: 09 April 2015.
Edited by:
Sabine Lüthje, University of Hamburg, GermanyCopyright © 2015 Stes, Gevaert and De Smet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ive De Smet,aXZzbWVAcHNiLnZpYi11Z2VudC5iZQ==
 Elisabeth Stes
Elisabeth Stes Kris Gevaert
Kris Gevaert Ive De Smet
Ive De Smet