- Department of Microbiology and Cell Science, University of Florida, Gainesville, FL, USA
Elongator, a six-subunit protein complex, was initially isolated as an interactor of hyperphosphorylated RNA polymerase II in yeast, and was subsequently identified in animals and plants. Elongator has been implicated in multiple cellular activities or biological processes including tRNA modification, histone modification, DNA demethylation or methylation, tubulin acetylation, and exocytosis. Studies in the model plant Arabidopsis thaliana suggest that the structure of Elongator and its functions are highly conserved between plants and yeast. Disruption of the Elongator complex in plants leads to aberrant growth and development, resistance to abiotic stresses, and susceptibility to plant pathogens. The morphological and physiological phenotypes of Arabidopsis Elongator mutants are associated with decreased histone acetylation and/or altered DNA methylation. This review summarizes recent findings related to the epigenetic function of Elongator in plant development and responses to abiotic and biotic stresses.
Introduction
Elongator was first identified as an elongating RNA polymerase II (RNAP II)-associated protein complex in yeast (Otero et al., 1999; Wittschieben et al., 1999), and was later found to be highly conserved in eukaryotes (Versées et al., 2010). This complex consists of six subunits (ELP1–ELP6) with ELP1-3 forming the core subcomplex and ELP4–ELP6 the accessory subcomplex (Li et al., 2001; Winkler et al., 2001). Deletion of any of the six subunits results in almost identical phenotypes, suggesting that all six subunits are required for Elongator’s cellular functions (Krogan and Greenblatt, 2001; Nelissen et al., 2005, 2010; Mehlgarten et al., 2010; Glatt et al., 2012). ELP1 has a nuclear localization sequence essential for Elongator function and WD40 repeats that possibly function together with the WD40-containing subunit ELP2 as scaffolds for complex assembly (Fichtner et al., 2003). ELP3 is the catalytic subunit containing a C-terminal GNAT-type histone acetyltransferase (HAT) domain and an N-terminal iron–sulfur (Fe–S) radical S-adenosylmethionine (SAM) domain (Wittschieben et al., 1999; Chinenov, 2002). ELP4, ELP5, and ELP6 each form a RecA-ATPase-like fold and together assemble into a hexameric ring-shaped structure (Glatt et al., 2012; Lin et al., 2012).
The presence of a conserved HAT domain in ELP3 and the co-purification of Elongator with elongating RNAP II led to the initial assumption that Elongator might facilitate transcription elongation via histone acetylation (Otero et al., 1999; Wittschieben et al., 1999). Indeed, ELP3 is capable of acetylating all four core histones in vitro (Wittschieben et al., 1999), and highly purified holo-Elongator has a dominant preference for lysine-14 of histone H3 and to a small extent for lysine-8 of Histone H4 (Winkler et al., 2002; Li et al., 2009). Consistently, yeast, human, and plant Elongator mutants contain reduced levels of acetylated histone H3 and H4 (Kim et al., 2002; Winkler et al., 2002; Close et al., 2006; Nelissen et al., 2010). Although chromatin immunoprecipitation (ChIP) failed to detect enrichment of Elongator at actively transcribed genomic regions in yeast (Pokholok et al., 2002), RNA immunoprecipitation (RIP) showed that Elongator interacts with nascent mRNA during transcription elongation (Gilbert et al., 2004). Conversely, in humans and plants, ChIP experiments detected association of Elongator with gene promoters and/or coding regions (Kim et al., 2002; Close et al., 2006; Wang et al., 2013). Furthermore, it was recently reported that the ELP4–ELP6 accessory subcomplex assembles into a hexameric ring-shaped structure that is important for recognizing histone H3 (Lin et al., 2012). These results, together with the finding that Elongator facilitates RNAP II transcription through chromatin in an acetyl-CoA-dependent manner (Kim et al., 2002), support that Elongator assists RNAP II during transcription elongation via chromatin remodeling.
ELP3 also contains a putative SAM-binding domain, which was hypothesized to function catalytically in histone demethylation (Chinenov, 2002). In yeast, the radical SAM domain of ELP3 was shown to be a motif required for the structural integrity of Elongator (Greenwood et al., 2009). In contrast, the archaea Methanocaldococcus jannaschii ELP3 SAM-binding motif might have a catalytic role, since it binds and cleaves SAM (Paraskevopoulou et al., 2006). Interestingly, a recent study in mouse indicated that Elongator is required for zygotic paternal genome demethylation, which is mediated by the ELP3 radical SAM domain rather than the HAT domain (Okada et al., 2010).
Accumulating evidence suggests that Elongator also plays a role in formation of the 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) side chains on uridines at the wobble position in tRNAs (Karlsborn et al., 2015). Yeast Elongator mutants lack tRNA modifications at wobble uridines or thiouridines at position 34 of the anticodon (Huang et al., 2005). Interestingly, elevated levels of two tRNA species rescue the defects of transcription and exocytosis in yeast Elongator mutants (Esberg et al., 2006), and overexpression of tRNALysUUU complements the stress-related phenotypes of the yeast sin3/elp3 mutant cells (Fernández-Vázquez et al., 2013). These results are in line with the recent finding that the hexameric ELP456 accessory subcomplex specifically recognizes tRNA (Glatt et al., 2012). Therefore, it was proposed that the effects of elp mutations on transcription and secretion might be indirect consequences of inappropriate tRNA modifications (Esberg et al., 2006).
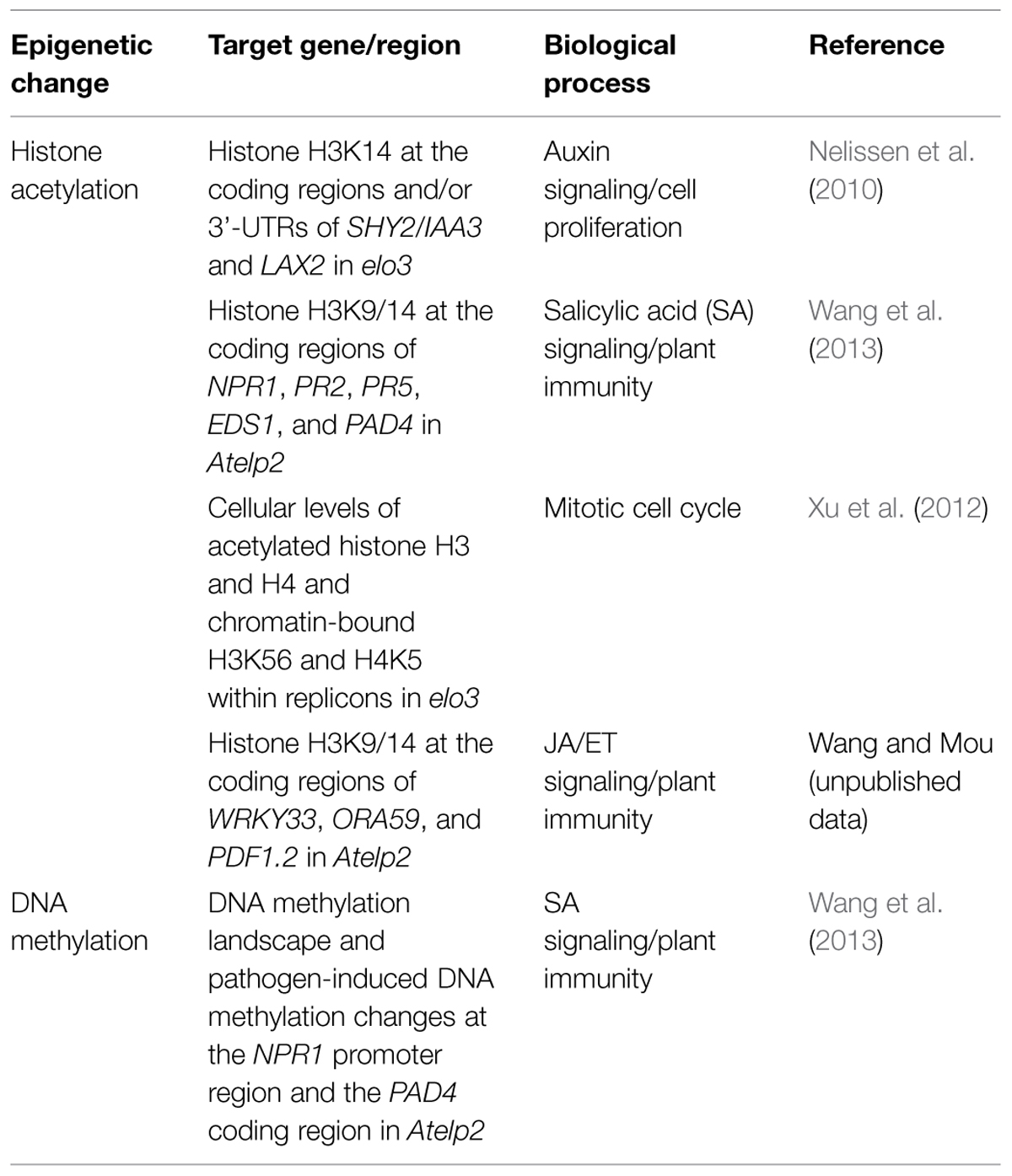
In addition to its functions in histone acetylation and tRNA modification, Elongator has also been implicated in multiple kingdom-specific activities, such as exocytosis in yeast and neuronal development in animals (Rahl et al., 2005; Close et al., 2006). Recent studies performed in the model plant Arabidopsis thaliana have revealed that the structure and function of Elongator are conserved in plants (Figure 1; Table 1; Nelissen et al., 2010; DeFraia and Mou, 2011; Van Lijsebettens et al., 2014; Yan et al., 2014). This review focuses on recent advances in the study of the epigenetic function of Elongator in plant development and responses to biotic and abiotic stresses.
FIGURE 1. Possible roles of the Elongator complex in plants. The currently available information suggests three possible roles for the Elongator complex in plants. In the nucleus, Elongator is associated with hyperphosphorylated RNA polymerase (RNAP II) and is required for histone acetylation and/or DNA demethylation/methylation at various genetic loci. On the other hand, Elongator may also regulate protein translation through tRNA modification in the cytoplasm.
Identification and Characterization of the Elongator Complex in Plants
In a genetic screen for mutants with abnormal shaped leaves in Arabidopsis, four elongata (elo) mutants, elo1, elo2, elo3, and elo4, were isolated (Berná et al., 1999). The elo4 mutant was later found to be allelic to the drl1-2 (deformed roots and leaves1) mutant, which carries a transposed Ds element in the Arabidopsis homolog of the yeast Elongator-associated protein KTI12 (KILLER TOXIN INSENSITIVE12; Fichtner et al., 2002; Nelissen et al., 2003). Further BLAST search indicated that homologs of all six subunits of the yeast Elongator are also present in the Arabidopsis genome (Nelissen et al., 2003). Indeed, the elo1, elo2, and elo3 mutations were identified in the A. thaliana (At) Elongator subunits AtELP4, AtELP1, and AtELP3, respectively, (Nelissen et al., 2005).
To verify the existence and composition of an Elongator complex in plants, tandem affinity purification (TAP) was performed using Arabidopsis cell suspension cultures overexpressing TAP-tagged ELO3/AtELP3, ELO1/AtELP4, or AtELP5 (Nelissen et al., 2010). All six subunits (ELO2/AtELP1, AtELP2, ELO3/AtELP3, ELO1/AtELP4, AtELP5, and AtELP6) were purified in the TAPs with ELO3/AtELP3, ELO1/AtELP4, or AtELP5 as bait, confirming that the Elongator composition is conserved in plants. Moreover, stoichiometric concentrations of ELO2/AtELP1, AtELP2, and ELO3/AtELP3 were found on gels when ELO2/AtELP1 was used as bait, whereas only stoichiometric concentrations of ELO1/AtELP4, AtELP5, and AtELP6 were detected when using ELO1/AtELP4 or AtELP5 as bait, suggesting that the Arabidopsis holo-Elongator is also composed of two distinct subcomplexes. The structural conservation was further supported by the fact that AtELP6 interacts with both ELO1/AtELP4 and AtELP5 in yeast two-hybrid assays (Nelissen et al., 2010).
ELO3/AtELP3 was shown to colocalize with euchromatin and the phosphorylated form of RNAP II, indicating that plant Elongator is also involved in the process of RNAP II transcription elongation (Nelissen et al., 2010). The functional conservation was further corroborated by results from heterologous complementation experiments. Ectopic expression of ELO2/AtELP1 in a yeast elp1 mutant restores sensitivity toward zymocin, a yeast killer toxin complex (Chen et al., 2006), and ELO2/AtELP1 and ELO3/AtELP3 are able to structurally replace the respective yeast Elongator subunits and functionally restore ochre suppression and γ-toxin sensitivity of a yeast elp1 elp3 double mutant by reconstituting U34 tRNA modifications (Mehlgarten et al., 2010). Moreover, tRNA wobble uridine modifications are compromised in the Arabidopsis elo2/Atelp1, elo3/Atelp3, and drl1 mutants (Chen et al., 2010; Mehlgarten et al., 2010). Therefore, the structure of Elongator and its functions in RNAP II transcription elongation as well as tRNA modification are conserved between yeast and plants.
The Epigenetic Function of Elongator in Plant Growth and Development
As elo/Atelp mutants exhibit abnormal shaped leaves (Berná et al., 1999; Nelissen et al., 2003, 2005), Elongator clearly plays an important role in plant growth and development. At the macroscopic level, elo/Atelp mutants have narrow and elongated leaves, reduced primary roots and lateral root density, abnormal inflorescence phyllotaxis, delayed seedling growth, and reduced apical dominance (Nelissen et al., 2003, 2005, 2010). At the cellular and ultrastructural levels, leaves of elo/Atelp mutants have larger and fewer cells, which show less stacked grana in the chloroplasts, a hypotonic vacuole, and massive presence of Golgi vesicles in the cytoplasm (Falcone et al., 2007).
Since the phytohormone auxin plays a leading role in regulating cell proliferation (Wang and Ruan, 2013), the abnormal phenotypes of elo/Atelp mutants might be due to defects in auxin signaling or distribution. Indeed, a group of auxin-related genes are down-regulated in elo/Atelp mutants (Nelissen et al., 2010). Interestingly, decreased expression of two auxin-related genes, SHY2/IAA3 and LAX2, in the elo3/Atelp3 mutant is correlated with reduced histone H3K14 acetylation at the coding regions and 3′-UTRs (Table 2), suggesting that these two genes might be direct targets for Elongator’s HAT activity during RNAP II transcription elongation. On the other hand, H3K14 acetylation levels in several other auxin-related genes, which are also down-regulated in elo3/Atelp3, are not changed, indicating that Elongator likely targets specific genes for histone acetylation rather than affecting overall histone acetylation levels (Nelissen et al., 2010). Additionally, ethylene (ET) and jasmonic acid (JA) signaling and abiotic stress responses are up-regulated in elo/Atelp plants, which might also contribute to their pleiotropic phenotypes (Chen et al., 2006; Zhou et al., 2009; Nelissen et al., 2010).
It is well known that normal cell proliferation is required for the leaf adaxial–abaxial polarity establishment in Arabidopsis (Yuan et al., 2010). The abnormal leaf polarity formation in elo/Atelp mutants is likely due to the defective cell cycle progression caused by aberrant DNA replication and increased DNA damage (Xu et al., 2012). In yeast, Elongator associates with PCNA (proliferating cell nuclear antigen) and functions in maintenance of genome stability (Li et al., 2009). Elongator also interacts with PCNA in Arabidopsis, and the interaction is required for DNA replication and repair. Moreover, ELO3/AtELP3 is required for cellular histone H3 and H4 acetylation and DNA replication-coupled H3K56 and H4K5 acetylation (Table 2), which are important for DNA replication-coupled chromatin assembly (Xu et al., 2012). Thus, Elongator likely modulates mitotic cell cycle through interacting with PCNA and functioning in histone acetylation.
Elongator may also mediate the establishment of leaf adaxial–abaxial polarity in Arabidopsis by repressing transcription of abaxial-determinant genes and class 1 KNOX genes (Kojima et al., 2011). This negative role of Elongator in gene expression may be an indirect consequence of positively regulated genes.
Recently, the function of a tomato AtELP2-like gene, SlELP2L, was characterized (Zhu et al., 2015). Similar to Atelp mutants, SlELP2L-RNAi transgenic tomato plants display pleiotropic phenotypes, such as delayed seedling development, reduced leaf growth, rapidly senescing leaves and sepals, and dark-green fruits. A number of ET- and ripening-related genes are down-regulated in SlELP2L-silencing plants, whereas several DNA methyltransferase genes are up-regulated. It was therefore proposed that the tomato SlELP2L might regulate plant growth and development by modulating DNA methylation. Additionally, levels of GA and IAA, which have profound effects on plant growth and development (Martí et al., 2006; Zhao, 2010), are reduced in SlELP2L-RNAi plants. Interestingly, some phenotypes of the SlELP2L-RNAi tomato plants are in marked contrast to those of elo/Atelp mutants. For instance, ET signaling is upregulated in elo/Atelp mutants (Nelissen et al., 2010), but down-regulated in SlELP2L-silencing plants (Zhu et al., 2015). Moreover, elo/Atelp mutants accumulate high levels of auxin, whereas SlELP2L-RNAi tomato plants exhibit reduced levels of auxin. These differences suggest that the function of Elongator in different plant species may not be exactly the same.
The Role of Elongator in Plant Responses to Abiotic Stresses
In a genetic screen for Arabidopsis mutants with altered drought sensitivity, a drought-resistant mutant, abo1-1 (ABA-overly sensitive), was identified (Chen et al., 2006). The abo1-1 mutant is hypersensitive to ABA in stomatal closure and seedling growth and carries a mutation in the ELO2/AtELP1 gene. In a separate genetic screen for mutants hypersensitive to ABA in root growth, Atelp2 and Atelp6 were identified (Zhou et al., 2009). It was found that abo1/elo2/Atelp1, Atelp2, elo1/Atelp4, and Atelp6 are all hypersensitive to ABA in seed germination and seedling growth. Similarly, the tomato SlELP2L-silencing plants exhibit an obvious increase in ABA sensitivity during seedling growth (Zhu et al., 2015). On the other hand, only the core subcomplex mutants abo1/elo2/Atelp1 and Atelp2, but not the accessory subcomplex mutants elo1/Atelp4 and Atelp6, exhibit ABA hypersensitivity in stomatal closure (Zhou et al., 2009). The different functions of the two subcomplexes in ABA-mediated stomatal movement appear to conflict with the notion that both core and accessory subcomplexes are essential for the function of the holo-Elongator described to date (Svejstrup, 2007; Van Lijsebettens et al., 2014; Karlsborn et al., 2015).
Disruption of the Arabidopsis Elongator complex results in increased resistance to the oxidative stress caused by CsCl and methyl viologen under light, indicating that Elongator functions as a negative modulator of oxidative stress (Zhou et al., 2009). Consistently, expression of CAT3 (CATALASE 3), which encodes a catalase decomposing hydrogen peroxide in reactive species homeostasis, is up-regulated in elo/Atelp mutants. Elevated expression of CAT3 may contribute to the increased oxidative stress resistance. The elo/Atelp mutants also accumulate high levels of basal and light-induced anthocyanins (Zhou et al., 2009). Since anthocyanins function as antioxidants to protect plants from oxidative stress caused by diverse stressors such as drought, salt, and light (Gould, 2004), the elevated levels of anthocyanins in elo/Atelp mutants might also contribute to the enhanced resistance to oxidative stress.
The Epigenetic Function of Elongator in Plant Responses to Biotic Stresses
Salicylic acid (SA) is a key defense signal molecule against biotrophic and hemibiotrophic pathogens in plants (Vlot et al., 2009). SA accumulation occurs after pathogen infection, which is essential for activation of both local and systemic acquired resistance (SAR; Durrant and Dong, 2004). The transcription coactivator NPR1 [NONEXPRESSOR OF PATHOGENESIS-RELATED (PR) GENES] is a master regulator of SA-mediated defense responses (Durrant and Dong, 2004). Mutations in the NPR1 gene significantly compromise basal immunity, effector-triggered immunity (ETI), and SAR (Durrant and Dong, 2004). These mutations also lead to SA hyperaccumulation during pathogen infection (Shah et al., 1997; Wildermuth et al., 2001) and failure of seedling development on Murashige and Skoog (MS) medium containing high concentrations of SA (Cao et al., 1997). In a genetic screen for gns (green npr1 seedling on SA medium) mutants, we found that mutations in AtELP2 and ELO3/AtELP3 restore SA tolerance to npr1 on half-strength MS medium containing 0.5 mM SA and also suppress npr1-mediated SA hyperaccumulation (DeFraia et al., 2010, 2013). Since high levels of SA trigger production of reactive oxygen species and subsequent cellular damage (Rao et al., 1997), Elongator may facilitate SA toxicity by suppressing the expression of antioxidant genes such as CAT3 (Zhou et al., 2009).
Mutations in AtELP2 and ELO3/AtELP3 also compromise plant immunity (DeFraia et al., 2010, 2013). While gns1/Atelp2 and gns2/Atelp3 are more susceptible to both virulent and avirulent Pseudomonas syringae, SAR induction in these mutants is normal. Interestingly, simultaneous removal of AtELP2 and NPR1 completely abolishes resistance to two different ETI-inducing pathogens, P. syringae pv. tomato (Pst) DC3000/avrRpt2 and Pst DC3000/avrRps4. Microarray analysis revealed that the gns1/Atelp2 mutation has a broader impact than npr1 on Pst DC3000/avrRpt2-induced transcriptome changes, indicating that AtELP2 is a more general modulator of transcription than NPR1. Furthermore, Pst DC3000/avrRpt2-induced expression of a group of SA pathway defense genes is delayed and/or decreased in the gns1/Atelp2 mutant compared to wild type. Similarly, both gns1/Atelp2 and gns2/Atelp3 are significantly more susceptible than wild type to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola and B. cinerea-induced expression of the JA/ET pathway defense genes OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59) and PLANT DEFENSIN1.2 (PDF1.2) is delayed and/or decreased in gns1/Atelp2 plants (Wang and Mou, unpublished data). These results together suggest that AtELP2 is an accelerator of plant defense responses (DeFraia et al., 2010).
Genome-wide DNA methylation analysis revealed that the gns1/Atelp2 mutation increases total number of methylcytosines, decreases average methylation levels of methylcytosines, and alters methylation levels of specific cytosines (Wang et al., 2013). Further analysis showed that AtELP2 is required for pathogen-induced dynamic changes in DNA methylation levels of two major defense genes NPR1 and PAD4 (Table 2). On the other hand, histone acetylation assay indicated that histone H3K9/14ac levels in the coding regions of several defense genes, including NPR1, PAD4, EDS1, PR2, PR5, WRK33, ORA59, and PDF1.2, are significantly reduced in gns1/Atelp2 (Table 2). The reduced histone H3K9/14ac levels are correlated with delayed and/or decreased induction of these defense genes in gns1/Atelp2, suggesting a role for AtELP2 in histone acetylation (Wang et al., 2013; Wang and Mou, unpublished data). Consistently, mutants of the Arabidopsis Elongator catalytic subunit (ELO3/AtELP3) lacking the conserved residues in either the HAT domain or the radical SAM domain (Figure 2), fail to complement gns2/Atelp3 mutant phenotypes (DeFraia et al., 2013), indicating that both domains are required for Elongator to function in Arabidopsis. Therefore, Elongator likely plays an epigenetic role in response to pathogen infections.
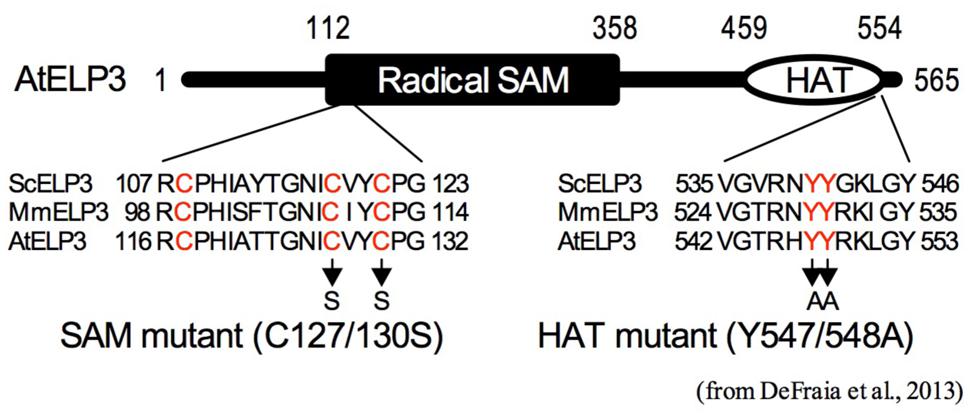
FIGURE 2. Schematic representation of the histone acetyltransferase (HAT) and radical S-adenosylmethionine (SAM) domains of AtELP3. The radical SAM and HAT domains of AtELP3 are aligned with those of the yeast (ScELP3) and mouse ELP3 (MmELP3). Only sequences that are part of the alignment are shown. The conserved amino acid residues are labeled in red. Arrows indicate the mutations created in the SAM and HAT mutants (DeFraia et al., 2013).
Conclusions and Perspectives
Studies in Arabidopsis indicate that the structure of the Elongator complex and its functions are highly conserved between plants and yeast (Otero et al., 1999; Winkler et al., 2002; Nelissen et al., 2010). Disruption of the Elongator complex in Arabidopsis leads to pleiotropic growth and defense phenotypes, which could be attributed to delayed and/or decreased expression of some genes involved in these processes (Nelissen et al., 2005; Zhou et al., 2009; DeFraia et al., 2010; Xu et al., 2012). The delayed and/or decreased gene expression in Atelp mutants is associated with reduced histone acetylation and/or altered DNA methylation (Nelissen et al., 2010; Wang et al., 2013). Although the intrinsic relationship between histone acetylation and DNA demethylation/methylation in Arabidopsis Elongator mutants remains to be determined, these results suggest that Elongator may epigenetically modulate plant development and responses to abiotic and biotic stresses.
However, a direct role of Elongator in histone acetylation and DNA demethylation or methylation in plants has not been established. Moreover, the enzymatic activities (HAT and DNA demethylase or methyltransferase) of the catalytic subunit ELP3 in plants have not been tested. Without such information, it would be difficult to fully appreciate the epigenetic function of Elongator in plants. Additionally, recent evidence suggests that herbivore and pathogen attack of plants generates defense phenotypes across generations and such transgenerational memory appears to be associated with DNA methylation, histone modifications, and small RNAs (Holeski et al., 2012; Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012). Since Elongator modulates histone acetylation and DNA methylation (Nelissen et al., 2010; Xu et al., 2012; Wang et al., 2013), it would be interesting to test whether Elongator is also involved in transgenerational defense induction and epigenetic inheritance in plants.
Accumulating evidence suggests that the yeast Elongator may primarily function in tRNA modification (Karlsborn et al., 2015). Studies in Arabidopsis also indicate that Elongator may play a role in tRNA modification in plants (Chen et al., 2010; Mehlgarten et al., 2010). However, the connection between Elongator’s function in tRNA modification and plant development and responses to abiotic and biotic stresses still remains elusive. It would be very interesting to test whether overexpression of certain types of hypomodified tRNAs (tRNALysUUU, tRNAGlnUUG, and tRNAGluUUC) could rescue some of the plant Elongator mutant phenotypes. Such experiments would help resolve the mystery of the multitasking role of Elongator in plants.
Author Contributions
YD and ZM wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by a grant from the National Science Foundation (IOS-0842716) awarded to ZM.
References
Berná, G., Robles, P., and Micol, J. L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742.
Cao, H., Glazebrook, J., Clark, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, P., Jäger, G., and Zheng, B. (2010). Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 10:201. doi: 10.1186/1471-2229-10-201
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Z., Zhang, H., Jablonowski, D., Zhou, X., Ren, X., Hong, X., et al. (2006). Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 26, 6902–6912. doi: 10.1128/MCB.00433-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chinenov, Y. (2002). A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem. Sci. 27, 115–117. doi: 10.1016/S0968-0004(02)02058-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Close, P., Hawkes, N., Cornez, I., Creppe, C., Lambert, C. A., Rogister, B., et al. (2006). Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22, 521–531. doi: 10.1016/j.molcel.2006.04.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Crane, Y. M., and Gelvin, S. B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. U.S.A. 104, 15156–15161. doi: 10.1073/pnas.0706986104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
DeFraia, C. T., and Mou, Z. (2011). The role of the Elongator complex in plants. Plant Signal. Behav. 6, 19–22. doi: 10.4161/psb.6.1.14040
DeFraia, C. T., Wang, Y., Yao, J., and Mou, Z. (2013). Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 13:102. doi: 10.1186/1471-2229-13-102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
DeFraia, C. T., Zhang, X., and Mou, Z. (2010). Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 64, 511–523. doi: 10.1111/j.1365-313X.2010.04345.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Esberg, A., Huang, B., Johansson, M. J., and Byström, A. S. (2006). Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148. doi: 10.1016/j.molcel.2006.07.031
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Falcone, A., Nelissen, H., Fleury, D., Van Lijsebettens, M., and Bitonti, M. B. (2007). Cytological investigations of the Arabidopsis thaliana elo1 mutant give new insights into leaf lateral growth and Elongator function. Ann. Bot. 100, 261–270. doi: 10.1093/aob/mcm102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fernández-Vázquez, J., Vargas-Perez, I., Sanso, M., Buhne, K., Carmona, M., Paulo, E., et al. (2013). Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9:e1003647. doi: 10.1371/journal.pgen.1003647
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fichtner, L., Frohloff, F., Bürkner, K., Larsen, M., Breunig, K. D., and Schaffrath, R. (2002). Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 43, 783–791. doi: 10.1046/j.1365-2958.2002.02794.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fichtner, L., Jablonowski, D., Schierhorn, A., Kitamoto, H. K., Stark, M. J., and Schaffrath, R. (2003). Elongator’s toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol. Microbiol. 49, 1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x
Gilbert, C., Kristjuhan, A., Winkler, G. S., and Svejstrup, J. Q. (2004). Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14, 457–464. doi: 10.1016/S1097-2765(04)00239-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Glatt, S., Letoquart, J., Faux, C., Taylor, N. M., Seraphin, B., and Müller, C. W. (2012). The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 19, 314–320. doi: 10.1038/nsmb.2234
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gould, K. S. (2004). Nature’s swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 5, 314–320. doi: 10.1155/S1110724304406147
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Greenwood, C., Selth, L. A., Dirac-Svejstrup, A. B., and Svejstrup, J. Q. (2009). An iron-sulfur cluster domain in Elp3 important for the structural integrity of Elongator. J. Biol. Chem. 284, 141–149. doi: 10.1074/jbc.M805312200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Holeski, L. M., Jander, G., and Agrawal, A. A. (2012). Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 27, 618–626. doi: 10.1016/j.tree.2012.07.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huang, B., Johansson, M. J., and Bystrom, A. S. (2005). An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11, 424–436. doi: 10.1261/rna.7247705
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karlsborn, T., Tükenmez, H., Mahmud, A. K., Xu, F., Xu, H., and Byström, A. S. (2015). Elongator, a conserved complex required for wobble uridine modifications in Eukaryotes. RNA Biol. 11, 1519–1528. doi: 10.4161/15476286.2014.992276
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, J. H., Lane, W. S., and Reinberg, D. (2002). Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. U.S.A. 99, 1241–1246. doi: 10.1073/pnas.251672198
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kojima, S., Iwasaki, M., Takahashi, H., Imai, T., Matsumura, Y., Fleury, D., et al. (2011). Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52, 1259–1273. doi: 10.1093/pcp/pcr083
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Krogan, N. J., and Greenblatt, J. F. (2001). Characterization of a six subunit holo-Elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, Q., Fazly, A. M., Zhou, H., Huang, S., Zhang, Z., and Stillman, B. (2009). The Elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 5:e1000684. doi: 10.1371/journal.pgen.1000684
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, Y., Takagi, Y., Jiang, Y., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., et al. (2001). A multiprotein complex that interacts with RNA polymerase II Elongator. J. Biol. Chem. 276, 29628–29631. doi: 10.1074/jbc.C100274200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, Z., Zhao, W., Diao, W., Xie, X., Wang, Z., Zhang, J., et al. (2012). Crystal structure of the Elongator subcomplex Elp4-6. J. Biol. Chem. 287, 21501–21508. doi: 10.1074/jbc.M112.341560
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Martí, E., Gisbert, C., Bishop, G. J., Dixon, M. S., and García-Martínez, J. L. (2006). Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 57, 2037–2047. doi: 10.1093/jxb/erj154
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mehlgarten, C., Jablonowski, D., Wrackmeyer, U., Tschitschmann, S., Sondermann, D., Jäger, G., et al. (2010). Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 76, 1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nelissen, H., Clarke, J. H., De Block, M., De Block, S., Vanderhaeghen, R., Zielinski, R. E., et al. (2003). DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15, 639–654. doi: 10.1105/tpc.007062
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nelissen, H., De Groeve, S., Fleury, D., Neyt, P., Bruno, L., Bitonti, M. B., et al. (2010). Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. U.S.A. 107, 1678–1683. doi: 10.1073/pnas.0913559107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nelissen, H., Fleury, D., Bruno, L., Robles, P., De Veylder, L., Traas, J., et al. (2005). The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. U.S.A. 102, 7754–7759. doi: 10.1073/pnas.0502600102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Okada, Y., Yamagata, K., Hong, K., Wakayama, T., and Zhang, Y. (2010). A role for the Elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558. doi: 10.1038/nature08732
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Otero, G., Fellows, J., Li, Y., De Bizemont, T., Dirac, A. M., Gustafsson, C. M., et al. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118. doi: 10.1016/S1097-2765(00)80179-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paraskevopoulou, C., Fairhurst, S. A., Lowe, D. J., Brick, P., and Onesti, S. (2006). The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 59, 795–806. doi: 10.1111/j.1365-2958.2005.04989.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pokholok, D. K., Hannett, N. M., and Young, R. A. (2002). Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809. doi: 10.1016/S1097-2765(02)00502-6
Rahl, P. B., Chen, C. Z., and Collins, R. N. (2005). Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17, 841–853. doi: 10.1016/j.molcel.2005.02.018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rao, M. V., Paliyath, G., Ormrod, D. P., Murr, M. P., and Watkins, C. B. (1997). Influence of salicylic acid on H2O2 production, oxidative stress and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 115, 137–149. doi: 10.1104/pp.115.1.137
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rasmann, S., De Vos, M., Casteel, C. L., Tian, D., Halitschke, R., Sun, J. Y., et al. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158, 854–863. doi: 10.1104/pp.111.187831
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slaughter, A., Daniel, X., Flors, V., Luna, E., Hohn, B., and Mauch-Mani, B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158, 835–843. doi: 10.1104/ppp.111.191593
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Svejstrup, J. Q. (2007). Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336. doi: 10.1016/j.ceb.2007.04.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Lijsebettens, M., Dürr, J., Woloszynska, M., and Grasser, K. D. (2014). Elongator and SPT4/SPT5 complexes as proxy to study RNA polymerase II transcript elongation control of plant development. Proteomics 14, 2109–2114. doi: 10.1002/pmic.201400024
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Versées, W., de Groeve, S., and van Lijsebettens, M. (2010). Elongator, a conserved multitasking complex? Mol. Microbiol. 76, 1065–1069. doi: 10.1111/j.1365-2958.2010.07162.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, L., and Ruan, Y. L. (2013). Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 4:163. doi: 10.3389/fpls.2013.00163
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, Y., An, C., Zhang, X., Yao, J., Zhang, Y., Sun, Y., et al. (2013). The Arabidopsis Elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell 25, 762–776. doi: 10.1105/tpc.113.109116
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Winkler, G. S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J. Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 3517–3522. doi: 10.1073/pnas.022042899
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Winkler, G. S., Petrakis, T. G., Ethelberg, S., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., et al. (2001). RNA polymerase II Elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743–32749. doi: 10.1074/jbc.M105303200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wittschieben, B. O., Otero, G., De Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., et al. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128. doi: 10.1016/S1097-2765(00)80194-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xu, D., Huang, W., Li, Y., Wang, H., Huang, H., and Cui, X. (2012). Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J. 69, 792–808. doi: 10.1111/j.1365-313X.2011.04831.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yan, X., Jin, X., Wang, Y., Zheng, B., and Chen, P. (2014). Recent advances in the roles of the Elongator complex in plant physiology and tRNA modification: a review. J. Integr. Agric. 13, 1640–1650. doi: 10.1016/S2095-3119(13)60524-9
Yuan, Z., Luo, D., Li, G., Yao, X., Wang, H., Zeng, M., et al. (2010). Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J. 64, 331–342. doi: 10.1111/j.1365-313X.2010.04326.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, X., Hua, D., Chen, Z., Zhou, Z., and Gong, Z. (2009). Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60, 79–90. doi: 10.1111/j.1365-313X.2009.03931.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhu, M., Li, Y., Chen, G., Ren, L., Xie, Q., Zhao, Z., et al. (2015). Silencing SlELP2L, a tomato Elongator complex protein 2-like gene, inhibits leaf growth, accelerates leaf, sepal senescence, and produces dark-green fruit. Sci. Rep. 5:7693. doi: 10.1038/srep07693
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Elongator, tRNA modification, histone acetylation, DNA methylation, plant development, abiotic stress, plant immunity
Citation: Ding Y and Mou Z (2015) Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front. Plant Sci. 6:296. doi: 10.3389/fpls.2015.00296
Received: 24 March 2015; Accepted: 13 April 2015;
Published online: 29 April 2015
Edited by:
Mahmoud W. Yaish, Sultan Qaboos University, OmanReviewed by:
Mieke Van Lijsebettens, Flanders Institute Biotechnology, BelgiumPeng Chen, HuaZhong Agricultural University, China
Copyright © 2015 Ding and Mou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonglin Mou, Department of Microbiology and Cell Science, University of Florida, Museum Road, Building 981, Gainesville, FL 32611, USA,emhsbW91QHVmbC5lZHU=
 Yezhang Ding
Yezhang Ding Zhonglin Mou
Zhonglin Mou