- 1Fruit Crop Systems Biology Laboratory, College of Horticulture, Nanjing Agricultural University, Nanjing, China
- 2Institute of Botany, Jiangsu Province and the Chinese Academy of Sciences, Nanjing, China
- 3Department of Agronomy, Food, Natural Resources, Animals and Environment, University of Padova, Legnaro, Italy
- 4Department of Biotechnology, University of Verona, Verona, Italy
WRKY proteins are a class of transcription factors (TFs) involved in the regulation of various physiological processes, including the plant response to biotic and abiotic stresses. Recent studies in Arabidopsis have revealed that some WRKY TFs interact with a class of proteins designed as VQ proteins because of their typical conserved motif (FxxxVQxLTG). So far, no information is available about the genomic organization and the function of VQ motif-containing protein in grapevine (Vitis vinifera L). In the current study, we analyzed the 12X V1 prediction of the nearly homozygous PN40024 genotype identifying up to 18 predicted VQ genes (VvVQ). VvVQs phylogenetic and bioinformatic analyses indicated that the intron-exon structures and motif distribution are highly divergent between different members of the grapevine VQ family. Moreover, the analysis of the V. vinifera cv. Corvina expression atlas revealed a tissue- and stage-specific expression of several members of the family which also showed a significant correlation with WRKY TFs. Grapevine VQ genes also exhibited altered expression in response to drought, powdery mildew infection, salicylic acid (SA) and ethylene (ETH) treatments. The present study represents the first characterization of VQ genes in a grapevine genotype and it is a pivotal foundation for further studies aimed at functionally characterizing this mostly unknown grapevine multigenic family.
Introduction
WRKY proteins are a class of transcription factors (TFs) found almost exclusively in plants, which have been associated to the control of a broad range of biological processes including response to biotic and abiotic stresses (Journot-Catalino et al., 2006; Liu et al., 2006; Xu et al., 2006; Zheng et al., 2006; Kim et al., 2008; Lai et al., 2008; Li et al., 2009), hormone signaling (Chen et al., 2010; Shang et al., 2010), secondary metabolism (Wang et al., 2010b; Suttipanta et al., 2011), and developmental processes (Johnson et al., 2002; Robatzek and Somssich, 2002; Miao et al., 2004; Luo et al., 2005; Jiang and Yu, 2009). In higher plants, genes encoding WRKY TFs are organized in large superfamilies as confirmed by recent genome-wide studies in species such as Arabidopsis (Arabidopsis thaliana; 74 genes), rice (Oryza sativa; 102 genes), and grapevine (Vitis vinifera; 59 genes) (Eulgem et al., 2000; Ramamoorthy et al., 2008; Wang et al., 2014).
One of the common defining features of WRKY TFs is their highly conserved DNA binding region, also called the WRKY domain, which consists of about 60 amino acids at the N-terminus, adjacent to a zinc-finger motif at the C-terminus. The conservation of the WRKY domain is mirrored by a remarkable conservation of its cognate binding site, the W-box (TTGACC/T), which is a common feature in all promoters of WRKY-regulated genes (Rushton et al., 1996; Eulgem et al., 2000).
It has been previously shown that interacting-partners such as co-activators, chromatin re-modelers, and enzymes modifying histones could modulate differential DNA-binding and transcription-regulatory activities of many WRKY TFs. Among these interacting partners is a class of proteins characterized by a short conserved VQ (FxxxVQxLTG) motif and generally designed as VQ proteins. The first VQ protein was identified in Arabidopsis by using a yeast two-hybrid assay with a MAP kinase, MPK4, as bait. The MPK4-interacting protein, designed as MPK Substrate (MKS) was found to be a 222-amino-acid protein member of a plant specific family including proteins from dicots and monocots sharing a VQ motif of unknown function (Andreasson et al., 2005). The VQ protein MKS forms complexes with AtWRKY25 and AtWRKY33, two group I WRKY TFs, involved in the regulation of plant defense responses (Andreasson et al., 2005). In absence of pathogen infection, MPK4 exists in nuclear complexes with WRKY33 through mutual interactions with MKS1 (Andreasson et al., 2005; Qiu et al., 2008). Upon Pseudomonas infection or flagellin treatment, activated MPK4 phosphorylates MKS1 and releases WRKY33, which then targets the expression of PAD3, a gene encoding for a protein involved in the biosynthesis of the phytoalexin camalexin (Andreasson et al., 2005). Subsequently, Lai et al. (2011) identified and functionally characterized other two VQ proteins, namely SIGMA FACTOR BINDING PROTEIN 1 (SIB1) and SIB2, which positively interact with WRKY33 in the plant defense against necrotrophic pathogens. The interaction between VQs and WRKY TFs was also described in the plant response to abiotic environmental stresses. In fact, a study aimed at clarifying the role of WRKY8 in salt tolerance in Arabidopsis showed that the interaction of this TF with the protein VQ9 results in the decrease of the WRKY8-DNA-binding activity and in negative regulation of salinity stress responses (Hu et al., 2013). Finally, another Arabidopsis VQ domain-containing protein, known as HAIKU1 (IKU1, called AtVQ14) was demonstrated to interact with WRKY10 to regulate endosperm growth and seed size (Wang et al., 2010a).
A genome wide screening of the VQ genes in Arabidopsis led to the identification of 34 members. These proteins were found to interact exclusively with group I and II-c WRKY TFs and their overexpression led to altered flowering and growth phenotypes, which supports the hypothesis that this gene family could be involved not only in the plant responses to stresses, but, as observed for WRKYs, also in developmental processes (Cheng et al., 2012). The conserved valine and glutamine residues in the short VQ motif are important for the physical interaction with WRKY C-terminal domains. Recently, a comprehensive analysis of VQ motif containing genes in rice led to the identification of 39 VQ members. These genes, similarly to what observed in Arabidopsis, were shown to be involved in disease resistance and in the plant response to environmental stresses (Kim et al., 2013).
Cultivated grapevine (V. vinifera L.) is an economically important fruit crop. A first genome sequence assembly of a highly homozygous line of V. vinifera Pinot Noir (PN40024) was published in 2007 (Jaillon et al., 2007), followed by a second assembly obtained from a highly heterozygous Pinot Noir line (PN ENTAV 115) (Velasco et al., 2007). These assemblies have been uploaded and improved by the grapevine community during the last few years and represent an invaluable resource for many studies. So far, many gene families and superfamilies have been identified and characterized in grapevine, however, there has been no systematic analysis of VQ genes family. In the current study, a genomic characterization of the VQ family in the 12X V1 prediction of the near homozygous PN40024 genotype led to the identification of 18 predicted VvVQ genes. The genomic sequences and phylogeny of all the VvVQ genes were analyzed, together with their expression profile at different developmental stages and in response to different biotic and abiotic stresses. Moreover, based on coexpression analyses we raised some hypotheses on putative interactions between VQ motif-containing proteins and several WRKY TFs, which has to be confirmed by functional studies. These analyses provide important information about the evolution and function of the grapevine VQ genes and represent the first pivotal foundation for further studies on the role of these genes in the regulation of developmental and defense mechanisms.
Materials and Methods
Identification of Grapevine VQ Gene Family Members
The 12X V1 release of the V. vinifera PN40024 genome containing the most recently uploaded version of gene and protein predictions was retrieved from the CRIBI Biotechnology Center website (http://genomes.cribi.unipd.it/). Arabidopsis and rice genome sequences were obtained from the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) and the Rice Genome Annotation project sites (http://rice.plantbiology.msu.edu/) respectively. The Hidden Markov Model (HMM) of the VQ motif (PF05678) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/). A search of all grapevine VQ protein sequences was carried out with the HMM search tool (HMMER 3.0; http://hmmer.janelia.org/) using the VQ motif HMM profile as a query and default parameters (E < 0.01). All candidate grapevine VQ genes with non-redundant hits were retained and confirmed using the SMART software program (http://smart.embl-heidelberg.de/). Moreover, as a cross check, a Pfam-code protein domain search was performed directly using the query system interface of the CRIBI website (http://genomes.cribi.unipd.it/). Recently, a new prediction (V2) of the PN40024 12X coverage was released (Vitulo et al., 2014); sequences retrieved from the V1 were checked against the V2 in order to identify variants or discrepancies. Length of sequences, isoelectric points, and molecular weights of deduced polypeptides were calculated by the ExPasy website (http://web.expasy.org/protparam/). The protein subcellular location was predicted by the Wolf PSORT program (http://www.genscript.com/psort/wolf_psort.html).
Sequence Alignment and Phylogenetic Analysis
Multiple alignments of grapevine, Arabidopsis and rice VQ domain sequences were performed using MUSCLE with default options (Edgar, 2004). The phylogenetic tree was inferred based on the neighbor-joining (NJ) method with a Kimura 2-parameter model using MEGA 5.0 (Tamura et al., 2011). Bootstrap values were calculated for 1000 iterations. Moreover, an additional phylogenetic tree was constructed based on a MCMC (Bayesian Markov chain Monte Carlo) method with Poisson rate matrix amino-acid model by mean of MrBayes software (Huelsenbeck and Ronquist, 2001).
Motif Search and Exon-Intron Structure Analysis
The predicted VQ coding (CDS) and genomic sequences identified in the V1 prediction of the PN40024 genotype were used to infer the intron/exon structure using the online Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.ch). To assess the structural divergence of grapevine VQ genes, the Multiple Expectation Maximization for Motif Elicitation (MEME) online program (http://meme.sdsc.edu/meme/itro.html) (Bailey et al., 2009) was used to identify motifs in the 18 identified grapevine VQ protein sequences. The optimized parameters of MEME used were the following: maximum number of motifs, 20; minimum motif width, 6; and maximum motif width, 50.
Chromosomal Locations and Gene Duplication
The chromosomal location of grapevine VQ genes was determined based on the 12X V1 release in the grapevine genome browser at the CRIBI website (http://genomes.cribi.unipd.it/). The distribution of VvVQ genes was drawn using MapInspect software (http://www.plantbreeding.wur.nl/UK/software_mapinspect/). The physical location of Arabidopsis VQ genes was mapped using the Chromosome Map Tool at TAIR (http://arabidopsis.org/jsp/ChromosomeMap/tool.jsp). VQ genes duplication events in Arabidopsis and grapevine were also investigated. Tandem duplicated genes defined as an array of two or more genes that were in the same phylogenetic group and found within a 100 kb genomic window (Yang et al., 2008). The information for segmental duplication was obtained from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). Synteny blocks within the Arabidopsis genome and between grapevine and Arabidopsis genomes were downloaded from the Plant Genome Duplication Database and those containing grapevine VQ genes were identified.
Promoter Analysis
Sequences of 2000 bp upstream the coding regionof each predicted VvVQ gene were downloaded from the grapevine genome website CRIBI (http://genomes.cribi.unipd.it/) and screened for cis-acting elements using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) according to Liu et al. (2009). PlantCARE software was also used for identifying putative plant-specific TF binding sites (TFBSs) in a given DNA sequence.
Public Microarray-Based Expression Analysis of VvVQ Genes
The expression patterns of VvVQ genes, predicted from an analysis of the grapevine genome, was determined in a global V. vinifera cv. Corvina (clone 48) gene expression atlas of different organs at various developmental stages. Microarray data were downloaded from the Gene Expression Omnibus under the series entry GSE36128 (http://www.ncbi.nlm.nih.gov/geo/) (Fasoli et al., 2012) and includes the following tissues: in vitro roots, green stem, buds after budburst (rosette of leaf tips visible), young leaves (leaves collected from shoots with only five leaves), senescing leaves (leaves at the beginning of leaf-fall), berry rachis (from fruit-set to ripening), flowers (50% cap-fall), and berry pericarp (from fruit set to ripe). In addition, berries were also examined which had undergone post-harvesting withering for 1–3 months after harvest. Data were analyzed and graphically represented using MeV (Multi Experiment Viewer) software (Saeed et al., 2006). Data obtained from the V. vinifera cv. Corvina expression atlas were normalized based on the mean expression value of each gene in all tissues/organs analyzed.
Plant Growth and Stress Treatments
Pinot noir 40024 in vitro-grown plants were grown at 25°C under long-day conditions (16 h/8 h light/dark) in a culture room. For the drought treatment, 5-week-old in vitro PN40024 plants were transplanted into pots and acclimated in an environmental chamber at 23 ± 2°C, a 16 h/8 h light/dark photoperiod [100 μmol/(m2• s)], and with a relative humidity of 70–80%. Potted plants were first kept in well-watered conditions until the plants were approximately 40 cm tall with 14 leaves. One group of plants was maintained under well-watered conditions as control, while the other three groups of plants received no water (drought treatment). Samples were harvested at 0, 4, 8, and 12 days after water was withheld and immediately frozen in liquid nitrogen and stored at −70°C until further analysis. Each time point had a corresponding control. For each treatment, leaves from three independent plants, considered as three biological replicates were sampled. For the biotic stress treatment, a local strain of Erysiphe necator was isolated from infected leaves collected from the “Meiguixiang” (Muscat) variety belonging to the Nanjing Agricultural University grape germoplasm resources and maintained on leaves of V. vinifera Pinot noir PN40024 glasshouse-grown plants. Inoculation of grapevine PN40024 leaves were performed by using the compression method pressing infected-leaves on healthy leaves of similar size. PN40024 leaves were collected 0, 4, 12, and 24 h after inoculation and immediately frozen in liquid nitrogen and stored at −70°C until subsequent analysis could take place. For a mock control, similar sized, Pinot noir healthy leaves were pressed onto in vitro leaves of PN40024. For hormone treatments, 5-week old in vitro grapevine plants were sprayed with 5 mM SA or 0.5 g/L ethephon. Plants sprayed with sterile distilled water were collected as control. Samples were harvested at 0, 4, 12, and 24 h after treatment. In these experiments, each sample (treated and untreated) was collected in three biological replicates.
Quantitative RT-PCR Expression Analysis of Grapevine VvVQ Genes
Quantitative RT-PCR was performed on an ABI 7300 Real Time PCR System (Applied Biosystems, USA) using the SYBR-Green (Takara, Japan) method. Designing of the oligonucleotide primers was based on the 3'-untranslated region and the 3'terminal sequences of the predicted coding region of each gene using Beacon designer software (version 2) and are listed in Table S1. Primers were checked using the BLAST tool at NCBI and the dissociation curve for each primer set was analyzed after the PCR reaction to confirm specificity. The specificity of PCR products was also verified by cloning the amplicons into the pMD19-T Vector (Takara). Cloned products were subsequently sequenced and aligned to the grapevine reference genome. Each reaction was carried out in a volume of 20 μl, containing 10 μl SYBR, 8.6 μl ddH2O, 1 μl diluted template (1 μl of the generated first-strand cDNA diluted by 9 μl ddH2O), and 0.2 μl of each gene-specific primers. The following PCR program was used: 94°C for 30 s (pre-denaturation) followed by 40 cycles at 94°C for 30 s (denaturation), 60°C for 20 s (primer annealing) and 72°C for 43 s (extension and obtaining the fluorescent signal). The specificity of the reactions was verified by melting curve analysis. The baseline and threshold cycles (Ct) were automatically determined by the ABI software. Three technical replicates were used for each biological replicate. The actin-101-like gene (VIT_12S0178g00200) whose expression levels remained nearly constant under all experimental conditions (CT was approximately 21 in all samples considered) was used as an internal control. The relative expression for all selected genes was calculated using the formula the 2−ΔΔCt method, where ΔΔCt= (Ct target gene − Ct actin) treatment − (Ct target gene − Ct actin)ck(Yuan et al., 2006). To visualize the relative fold difference, all data were represented by setting the relative expression level of 0-point treatment as 1, “above 1” and “below 1” are considered as up or down regulation during the stress treatment.
Statistical and Correlation Network Analyses
Statistic analyses were performed using Dunnett's two-tailed t-test. Mean values ± SD of at three replicates were presented, and significant differences relative to controls were given at *P ≤ 0.05 and **P ≤ 0.01. Pearson correlation (r) and respective FDR adjusted p-values were calculated between expression values of WRKY genes belonging to group I and II-c and all VQs, by using R programming language (http://cran.r-project.org/). The correlation network analysis (http://www.cytoscape.org/) (Shannon et al., 2003) was performed with VQ and WRKY co-expressed genes that highlight a r > 0.6 and a FDR < 0.05.
Results and Discussion
Identification and Annotation of VVvQ Genes
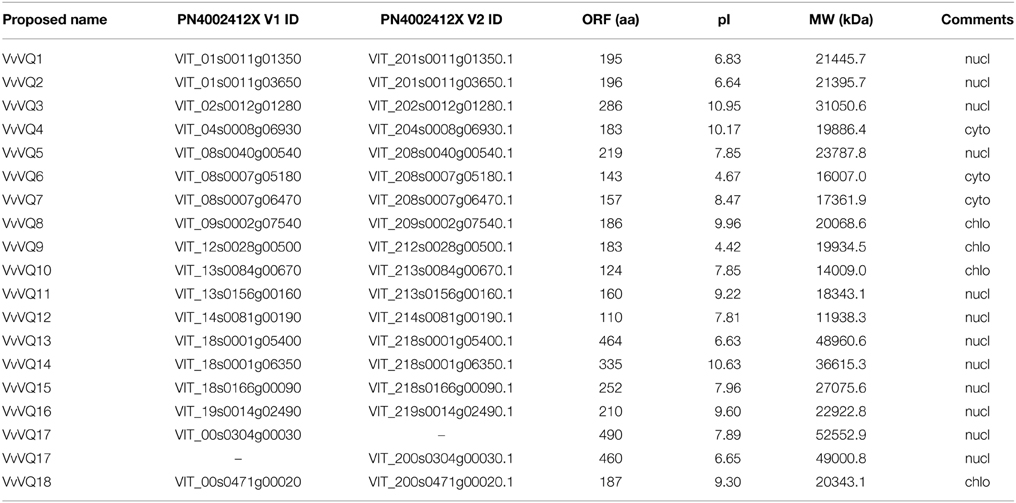
A search for genes encoding VQ proteins in the 12X V1 prediction of the PN40024 grapevine genome (http://genomes.cribi.unipd.it/) was performed, by using the HMM profile of the VQ motif (PF05678) as query. A total of 18 genes putatively encoding VQ proteins were identified in the grapevine reference genome and confirmed by a cross-check using the CRIBI interface for searching prediction based on Pfam codes. 16 VQ genes were located on 10 out of 19 grapevine chromosomes and named from VvVQ1 to VvVQ16 Figure S1). Two additional predicted genes (VIT_00s0304g00030 and VIT_00s0471g00020) were located within scaffolds not anchored to any chromosome (unknown chromosome, chrUn) and were arbitrarilly designated as VvVQ17 and VvVQ18, respectively Figure S1). The compositional characteristics of the grapevine VQ genes, including the deduced protein length (aa), weight (KDa), and isoelectric point (pI) are listed in Table 1. Data indicated that the predicted VvVQ genes encode peptides ranging from 110 (VvVQ12) to 490 (VvVQ17) aa in length. As observed in Arabidopsis, the majority of members encoded small proteins with fewer than 300 aa residues (Cheng et al., 2012). The isoelectric point (pI) of the VQ proteins ranged from 4.42 (VvVQ9) to 10.95 (VvVQ3). Compared to Arabidopsis and rice (39 and 34 VQ genes, respectively) (Cheng et al., 2012; Kim et al., 2013), grapevine has the lowest number of members, even though the size of its genome (475 Mb) is larger than the Arabidopsis (125 Mb) and rice (389 Mb) ones. A possible explanation of the small size of the VQ family in grapevine is the fact that whereas two rounds of genome duplication (α and β) were described in Arabidopsis (Bowers et al., 2003), there was no genome-wide duplication in Vitis after a shared ancient γ triplication (Jaillon et al., 2007; Tang et al., 2008a,b). Moreover, it must be noted that this study was performed on the 12X V1 prediction, which is based on in silico analyses. We cannot rule out the possibility that additional VQ were missed because of gaps in the genome assembly or incomplete/wrong gene predictions.
Based on Wang et al. (2014), also the number of grapevine WRKYs is lower when compared with Arabidopsis and rice. Similar sizes of the group I and IIc WRKYs and VQ multigenic families were reported in Arabidopsis (Cheng et al., 2012), but despite that, interaction partnership between the WRKY and VQ proteins was found not to be highly specific. In fact, yeast two-hybrid assays showed that a single VQ protein is able to interact with multiple WRKYs, and different VQ proteins have partially overlapping pools of interacting WRKY partners. Thus, there might not be a biunivocal relation between a given WRKY TF and a VQ protein.
Multiple Sequence Alignment and Structural Analysis
The deduced VQ domains of the 18 VvVQ proteins identified in the grapevine reference genome span across 57 aa in length and were aligned by means of MUSCLE software as reported in Figure S2. The core amino acid sequence, which was found to be conserved within all grapevine VQ proteins, is FXXXVQX(L/V/F)TG. No alternative variants, such as the VH core sequence in rice (Kim et al., 2013) were found in grapevine. An unrooted tree was constructed with the 18, 39, and 34 VQ-domainsequences found in grapevine, rice, and Arabidopsis, respectively, using the NJ method (Figure 1). Based on the rice VQ domain classification and the phylogenetic tree (Kim et al., 2013), the grapevine VQ domains were classified into seven subgroups even though this subdivision has to be considered with the due prudence considering the low bootstrap values obtained likely due to the divergent sequences of the protein family as proposed by Cheng et al. (2012) analysing the VQ family in Arabidopsis. It is not surprising that the phylogenetic tree used produced with a different Bayesian-based alghoritm (Figure S3) gave a different clusterization resect to what obtained with the NJ method in this analysis and in the one performed by Kim et al. (2013). As shown in Figure 1, only one VvVQ protein was located in group VI. In group II, V, and VI, the VQ domains of the genes in the grapevine and Arabidosis clustered together, while the genes of the rice VQ domains clustered by themselves. This further confirmed that grapevine has a close evolutionary relationship with Arabidopsis. Figure 2 illustrated the pattern of motifs detected within each protein, and the exon-intron structure of the grapevine VQ genes. A total number of 20 different motifs, ranging from 6 to 50 amino acids in length, were detected using MEME 4.3.0 software (http://meme.sdsc.edu/meme/intro.html) (Bailey et al., 2009) (Figure 2A, Table 2). As illustrated in Figure 2A, motif 1 is the VQ motif widely distributed in all 18 proteins. The differences in the type and number of motifs within the VvVQ proteins represent the structural basis for the diversity in gene function.
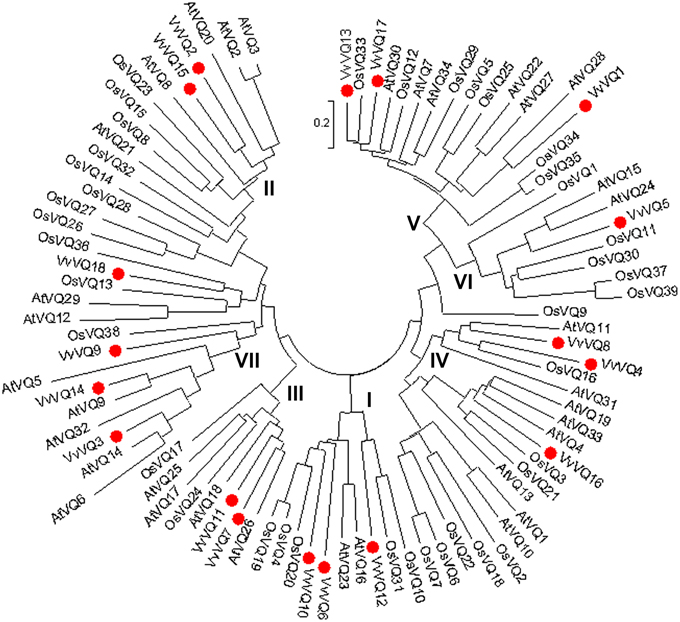
Figure 1. Phylogenetic analysis of Arabidopsis, rice, and grapevine VQ proteins. The unrooted tree, based on the core VQ domains in the three species, was constructed using the Neighbor-Joining (NJ) method (bootstrap 1000 replicates) using MEGA5 software.
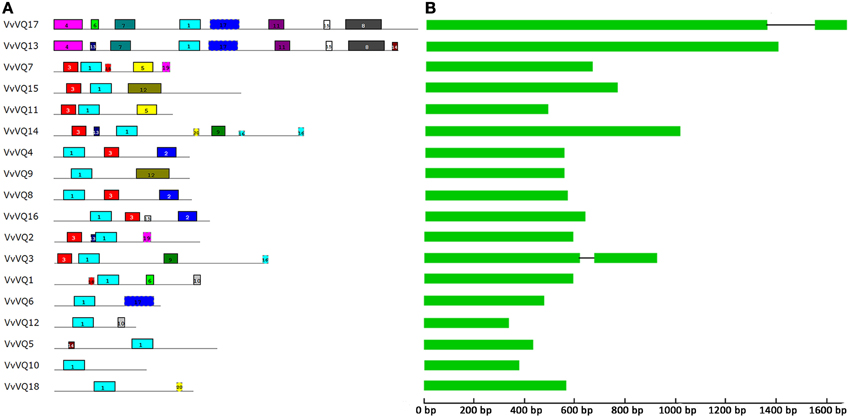
Figure 2. Structural analysis of grapevine VQ genes. (A) The motif composition within each VvVQ protein. The motifs, numbered 1–20, are displayed in different colored boxes. The sequence information for each motif is provided in Table 2. (B) Exon/intron structures of grapevine VQ genes. Exons were drawn to scale and are representred by boxes. Solid lines connecting two exons represent introns.

Table 2. Multilevel Consensus Sequence for each motif of VvVQ proteins as predicted by MEME program.
Sequence comparisons of grapevine full-length cDNAs and genomic DNA revealed the number and position of exons and introns for each of the individual VQ genes. With the exception of two VvVQ genes (VvVQ3 and VvVQ17), the majority of VQ genes in grapevine lack introns (Figure 2B), which is consistent with Arabidopsis and rice. A screening of the V2 prediction of the PN40024 genome, recently released by CRIBI Biotechnology Center (Vitulo et al., 2014) (http://genomes.cribi.unipd.it/grape/) indicates some discrepancies when compared with the V1 prediction. In fact, VvVQ17, which is predicted to contain an intron in the V1 release, does not appear to contain any intron in the V2. A comparison with the corresponding sequence in the PN ENTAV 115 genome (Velasco et al., 2007) confirmed that the right prediction is the most recent one (V2). Thus, among the 18 VvVQ genes, VvVQ3 remains the only one presenting an intronic fragment in its gene structure. Although, the analysis of the exon-intron structure of VQ genes was performed on both the V1 and V2 version of the PN40024 genome and checked also in the PN ENTAV one, it is worth to point out that the analysis of gene structure was based on in silico predictions, and thus could carry imprecisions and errors before a good quality and reliable gene annotation will be improved in grapevine.
Chromosomal Locations and Gene Duplication
The chromosomal position of the 18 grapevine VvVQ genes and of 34 AtVQ genes was depicted in Figures S1, S4, respectively. The distribution of VQ genes among the chromosomes in the two species appeared to be different. As shown in Figure S4, Arabidopsis chromosome 1 (chr1) contained the highest density (32.35% of VQ genes) whereas chr4 and chr5 contained the lowest density (11.76% each) of VQ genes. Based on the plant genome duplication database, there were 10 segmental duplication events between chromosomes in Arabidopsis (Figure S4). In addition to these segmental duplications, it appeared that chr1 had undergone five tandem-duplication events (Yang et al., 2008). In relation to VQ gene expansion in Arabidopsis, segmental duplication was predominant, although tandem duplication was also involved. As previously indicated, 16 VvVQ genes were mapped to 10 different grapevine chromosomes, whereas two of them (VvVQ17 and VvVQ18) were situated on unanchored contigs (chromosome unknown). Most of the grapevine chromosomes had only one single VQ member, although the number varied from 1 to 3. Unlike rice and Arabidopsis, no gene duplication events were observed in the VQ genes of grapevine. Syntenic regions between grapevine and Arabidopsis genomes were downloaded from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) and syntenic blocks containing 4 VQ genes in grapevine and 5 VQ genes in Arabidopsis were identified (Table S2 and Figure S5). Three pairs of VQ genes (VvVQ3-AtVQ32, VvVQ11-AtVQ15, and VvVQ15-AtVQ3) between grapevine and Arabidopsis exhibited VQ gene correspondences. Each pair of genes clustered within the same clade in the phylogenetic tree, indicating that these genes were derived from a common ancestor. There was also one case (VvVQ6-AtVQ17/AtVQ25) where a single grapevine gene corresponded to multiple Arabidopsis VQ genes. Genomic comparisons represent a quick way to transfer knowledge acquired in one taxon to other taxa (Lyons et al., 2008). Thus, the abundance of information about gene function in Arabidopsis allows one to infer a potential gene function for orthologous genes in the syntenic region of other species, such as grapevine species.
Identification of cis-elements in the Promoter Regions of VvVQ Genes
In order to obtain useful information related to the regulatory mechanism of VvVQ gene expression, the putative transcription factor binding sites (TFBSs) in the 2000bp DNA sequences upstream the start codon of VvVQ genes were identified using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) online software. Four categories of cis-acting elements were found to be the most represented in the promoter region of VvVQ genes in addition to the basic TATA and CAAT boxes (Table S3). The first class was constituted by “Light responsive elements,” such as the G-box (Menkens et al., 1995), I-box (Manzara et al., 1991), Box I, Sp1, and Box 4 (Lois et al., 1989), detected in most of VvVQ promoters. The second category of cis-acting element in the VvVQ promoter regions was constituted by hormone-responsive elements, such as ABRE (Simpson et al., 2003), P-box (Kim et al., 1992), and the TCA-element (Pastuglia et al., 1997). The CGTCA-motif (present in 77.8%, 14 out of 18 promoter sequences) and TGACG-motif (present in 83.3%, 15 out of 18 promoter sequences) are both cis-acting regulatory elements involved in methyl jasmonate (MeJA)-responsiveness (Rouster et al., 1997). The TCA-element (present in 77.8% of members, 14 out of 18 promoter sequences) is a cis-acting element involved in salicylic acid (SA) responsiveness. Both MeJA and SA are thought to play key roles in plant defense signaling (Gaffney et al., 1993; Xu et al., 1994). Therefore, it appears that most of the VvVQ genes may be involved in pathogen resistance. The ABRE element (72.2%, 13 of 18 promoter sequences) also accounts for a large proportion of hormone-related cis-acting elements in grapevine, suggesting that abscisic acid (ABA) may also regulate the expression of some VvVQ genes. The third major class of cis-acting elements consisted of TFBS related to external environmental stresses. 15 out of 18 VvVQ promoter sequences were found to contain a heat shock element HSE (83.3%) (Freitas and Bertolini, 2004), MBS, a cis element involved in drought response was detected in 10 out of 18 promoter sequences (55.6%) (Yamaguchi-Shinozaki and Shinozaki, 1993), and Box-W1, a fungal-elicitor-responsive element was also represented in 83.3% of sequences (15 out of 18 promoter sequences) (Rushton et al., 1996). The fourth most abundant class of cis-acting elements consisted of elements involved in various aspects of plant development. The Skn-1 motif which is involved in endosperm expression (Washida et al., 1999) is present in all VvVQ genes, with the exception of VvVQ13 and VvVQ17. The mean number of copies (2,72) of this cis-acting element in the VvVQ gene family was higher than that of other types of cis-acting elements except for the TATA-box, CAAT-box, 5'UTR Py-rich stretch (Daraselia et al., 1996) and the unnamed-4, which indicate that most VvVQ genes could be involved in the process of endosperm development. The diversity in the types of cis-acting elements found in the upstream region of VvVQ genes indicates that these genes may have a wide range of functional roles, even thoughit is worth noting that the low number of plant cis-element currently annotated on databases represents a limitation in interpreting genomic data.
VQ proteins are likely to play an important role in plant growth, development, and response to environmental conditions acting as cofactors of group I and IIc WRKY TFs, as already proposed in Arabidopsis (Cheng et al., 2012). The average frequency of the W-boxes (TTGAC, WRKY-binding sites) in the 1.5-kb promoters regions of the 34 Arabidopsis VQ genes is approximately 3.8, which is substantially higher than the statistically expected frequencies (Dong et al., 2003). This suggests a complex regulatory mechanism in which the WRKY TFs regulate the expression of the VQ genes, that are in turn necessary as cofactors of the WRKY proteins (Cheng et al., 2012). In our analysis, W-box sequenceswere found in 14 out of 18 VvVQ genes, but the average frequency of W-boxes in the promoter of grapevine VQ genes was only 1.2. Based on a recent study (Corso, personal communication), the average frequency of TTGAC W-box in 10000 randomly chosen 2 Kb promoter regions is approximately 1.9. This comparison suggests a lower level of transcriptional interaction between WRKYs and VQ genes in grapevine compared to Arabidopsis. Nevertheless, this observation must be considered with due caution, considering the analysis was performed in one third of the gene predictions and in the promoter region longer than those ones considered in the present study.
Expression Analysis of VvVQ Genes in Various Grapevine Tissues at Different Developmental Stages
Using an expression atlas of V. vinifera cv. Corvina (Fasoli et al., 2012), it was possible to investigate patterns of expression of all the predicted coding members of the VvVQ gene family in various tissues and at different developmental stages. All VvVQ genes had corresponding probes on the NimbleGen array. Figure 3 reports a graphical representation of the expression pattern of each VvVQ gene where both genes and samples were ordered based on a hierarchical clustering analysis. HCL tree based on gene leaves indicated a first clear difference between a group of VvVQ genes (VvVQ8, −6, −14, −5, −16, −3, −17, and −13) characterized by a marked modulation of expression in different tissues analyzed and the remaining genes, which showed much lower variability in terms of gene induction and/or repression. Among these were VvVQ9 and VvVQ11, which did not show any variation in expression within all samples analyzed. This observation could be explained by the fact that both VvVQ9 and VvVQ11 have different roles in other developmental processes, tissues, or stress responses than those analyzed, or alternatively by a technical problem with the process of hybridizations in the array. VvVQ18 is a particularly interesting gene because it is the only one whose expression appears to be limited to one specific organ: roots. To our knowledge, there is no information about a possible role of VQ proteins in root development. Nevertheless, it is well documented that there is a role for WRKY TFs in trichome development (Johnson et al., 2002). In a recent study aimed at characterizing the WRKY gene family in grapevine, Wang et al. (2014) identified several VvWRKY genes expressed in roots and, among them, VvWRKY53 appeared exclusively expressed in this organ. VvWRKY53 belongs to the group II-b and, based on previous Y2H assays performed by Cheng et al. (2012), it is not supposed to interact with VQ proteins. In any case, the possibility of an interaction between VQ proteins and WRKY TF belonging to groups other than I and II-c cannot be excluded a priori without any functional analysis. Other WRKYs highly expressed in roots are good candidates to interact with VvVQ18, such as VvWRKY45 and VvWRKY16, both belonging to group I, and VvWRKY43 which belongs to group II-c.
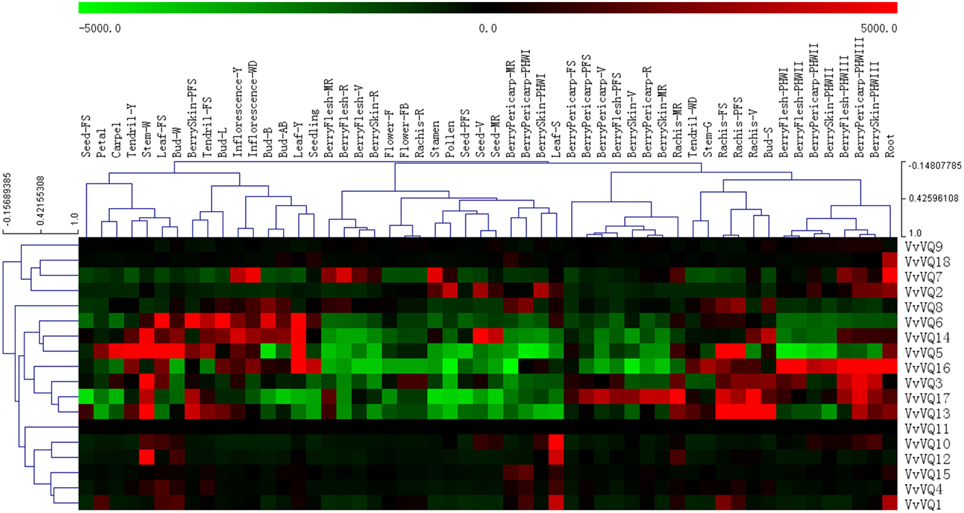
Figure 3. Expression profile of 18 grapevine VQ genes in various tissues and at different developmental stages. Expression data were normalized based on the mean expression value of each gene in all tissues/organs. Different organs/tissues are displayed vertically above each column. Gene names are displayed to the right of each row. Black boxes indicate a low variation with respect to the overall mean value of expression, green boxes indicate a fold decrease, and red boxes indicate a fold increase with respect to the mean value. Samples and genes were hierarchically clustered based on the average Pearson's distance. Abbreviations indicating developmental stages are as follows: FS, fruit set; PFS, post fruit set; V, véraison; MR, mid- ripening; R, ripening; Bud-L, latent bud; Bud-W, winter bud; Bud-S, bud swell; Bud-B, Bud burst; Bud-AB, bud after burst; Inflorescence-Y, young inflorescence with single flowers separated; Inflorescence-WD, well developed inflorescence; Flower-FB, flowering begins; Flower-F, flowering; Tendril-Y, young tendril; Tendril-WD, well developed tendril; Tendril-FS, mature tendril; Leaf-Y, young leaf; Leaf-FS, mature leaf; Leaf-S, senescing leaf; Stem-G, green stem; and Stem-W, woody stem.
Together with VvVQ18, other VQ genes showed a quite specific expression in several tissues. Among these was VvVQ1, which similarly to VvVQ18 was induced in roots, but also in senescing leaves (Leaf-S) and rachis prior to véraison (Rachis-FS and -PFS). VvVQ10 and -12 transcripts accumulated mainly in senescing leaves, even if limited to VvVQ12, a remarkable induction was also detected in winter stem (Stem -W). No other VvVQs (apart from VvVQ2, −4, and −6 which showed a low induction) were strongly induced in senescing leaves. A role for VQ proteins in senescence has not been described before. Nevertheless, several WRKY proteins are known to regulate developmental processes such as leaf senescence (Robatzek and Somssich, 2002; Miao et al., 2004) and, considering the role of VQ proteins as WRKY co-factors, the hypothesis that some members of this gene family are involved also in these processes is far than unlikely. Leaf senescence is an active and highly regulated process that involves an integrated response of leaf cells to age information and other internal and environmental signals and is accompanied by a decreased expression of genes related to photosynthesis and protein synthesis and an increase in the expression of hundreds of senescence-associated genes (Espinoza et al., 2007). Among these are a number of pathogenesis-related (PR) genes and it was proposed that the senescence program might have incorporated features of the pathogen-defense response to protect the senescing leaf against opportunistic pathogens (Quirino et al., 2000). This would be in agreement with the well-documented role of several VQ proteins, in combination with WRKY TFs in the response to biotic stresses. Alternatively, the induction of VvVQ genes in senescing leaves may simply be a consequence of changes in the levels of various phytohormones, including SA and ETH, which are known to play an important role in regulating leaf senescence. This was also true in the present study and in a previous one (Cheng et al., 2012). SA- and ETH- responsive elements were found in the promoter regions of VvVQ10 and VvVQ12, respectively.
Among those genes showing a higher level of variation within the Corvina expression atlas were VvVQ2 and VvVQ5, which were highly expressed in pollen and carpel tissues, respectively. Some other VvVQ genes showed tissue specific expression pattern: 4 VvVQs (VvVQ5, −6, −14, and −16) had higher levels of expression in early leaf developmental stages (Y), even if a remarkable induction was also detectable in other tissues. Some VvVQ genes were more strictly associated with berry development. VvVQ16, for example, exhibited higher expression levels in berry flesh, berry pericarp, and berry skin during post-harvest withering (PHWII, PHWIII), while VvVQ17 had higher expression in berry pericarp and berry skin at the earliest stages of development before ripening commenced (PF, V, MR) and post-harvest withering (PHWIII). Again, as postulated about the senescence process, many WRKY TFs were described to have a role in berry developmental processes and it is not surprising to find several VvVQ genes thatare expressed in developing berries. Based on the analysis performed by Wang et al. (2014), a strong induction of three WRKY TFs, namely VvWRKY14, −19, and −52 was detected in developing berries. Interestingly, these WRKY TFs all belong to the group II-c, which is one of the two WRKY subfamilies able to interact with VQ proteins (Cheng et al., 2012).
To better understand VvVQ genes expression patterns, genes that showed differential expression during various tissues development were analyzed. The expression levels of each tissue at the first development stage were normalized (Figure S6). Genes listed above or below the specified tissue or stage of development indicated that they were up-regulated or down-regulated, respectively. The data indicated that VvVQ genes were expressed in a wide array of tissues and/or at specific stages of development. Up regulation of VvVQ genes mainly occurred in maturity and post-harvest stages of the berry pericarp, bud dormancy, and during stem and leaf development. Down regulation of many VvVQ genes was also evident in post-harvest berry pericarp tissues, berry skin (maturity), and during rachis development.
Network Co-expression Analysis between VvVQs and Group I and II-c WRKY TFs
Previous studies have shown that 14 of the 34 Arabidopsis VQ motif-containing proteins can interact physiologically with WRKY proteins and that most of them have more than one WRKY partner (Andreasson et al., 2005; Wang et al., 2010a; Lai et al., 2011; Cheng et al., 2012; Hu et al., 2013). To gain information about hypothetical interactions between WRKY TFs and VQ proteins in grapevines, we performed a correlation analysis of expression of both VvVQs and VvWRKYs in the Corvina expression atlas. In order to simplify the network patterns, only the WRKY TFs belonging to group I and II-c, previously demonstrated to interact with the VQs in Arabidopsis (Cheng et al., 2012), were considered in the analysis. Two distinct datasets were used, one including the expression data in all tissues analyzed in the Corvina atlas (Fasoli et al., 2012) except the post-harvest withering, and the other one encompassing only berry pericarp, flesh and skin that were subjected to the PHW process. As reported by Vannozzi et al. (2012), the process of post-harvest berry drying (berry withering) involves harvesting of ripe grapevines, allowing them to dry over a period of 3 months in a naturally ventilated room. Its primary purpose is to alter berry quality characteristics and increase the concentration of simple sugars in the production of dessert and fortified wines typical of the Valpolicella region in Italy. However, the drying of harvested grapevines in this way results in a loss of over 30% of their weight through evaporation during this post-harvest treatment (Zamboni et al., 2008) and the speed of water loss induced by withering or other post-harvest techniques imposes a significant water stress on the berries (Corso et al., 2013). In the light of this information, the PHW process can be considered more as a “stress treatment” than a natural condition and the analysis restricted to these stages allowed us to compare the expression of both WRKYs and VQs under this “stress” condition and to assess putative transcriptional relationships. A first general observation was that no similar gene clusters (i.e., co-expressed VQ and WRKY genes) were found in the two considered correlation networks, highlighting strong specificity of VQs and WRKYs expression upon water stress in grapevine. These results were not unexpected, considering that several recent studies suggested a pivotal role of WRKYs in regulating osmotic stress responses in several species, such as Arabidopsis, rice (Rushton et al., 2012; Tripathi et al., 2014) and grapevine (Corso, personal communication). PHW correlation networks (Figure 4) divided VQ and WRKY genes into two main clusters characterized by high co-expression between WRKY and VQ genes (r > 0.89). In Figure 4A, Cluster 1 was characterized by the presence of co-expressed transcripts that showed very high expression values, whereas cluster 2 was composed of genes showing a significantly lower expression. In addition, a third cluster formed only by VvVQ10 and VvWRKY03 was also found. It is noteworthy that in PHW conditions, negative correlations were observed only for VvWRKY52 with VvVQ3 (r = −0.96), VvVQ13 (r = −0.90), VvVQ16 (r = −0.93) and VvVQ17 (r = −0.96). As reported by Chi et al. (2013) a single WRKY protein belonging to Group I or II-c can potentially interact with several VQ proteins differentially affecting DNA-binding specificity or other properties of the interacting WRKY. Moreover, this interaction can also be linked to a negative regulation of the TF activities (Qiu et al., 2008). The co-expression of VvWRKY52 and above mentioned VvVQ transcripts poses the basis for further examinations aimed at determining if a real physical interaction exists between candidate partners.
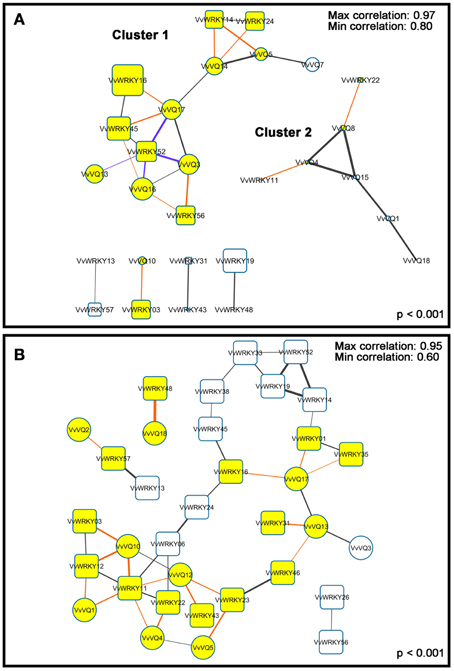
Figure 4. Co-expression network of grapevine VQ motif-containing genes and WRKY genes according to a p < 0.001. Thicker lines indicate more correlated genes. Blue and red lines, indicate negative and positive co-expression between a VQ motif-containing gene and a WRKY gene. VQ-WRKY correlated genes are showed in yellow. (A) Correlation network of PHW berries (Fasoli et al., 2012). The correlation coefficients of co-expression is between 0.8 and 0.97. Larger squares indicate more expressed genes. (B) Correlation network of all tissues described by Fasoli et al. (2012) except for PHW berries. The correlation coefficients of co-expression is between 0.6 and 0.95.
Among VQs in the PHW subset, the gene with the higher average expression was VvVQ16 (intensity: 9998), which was anti-correlated with VvWRKY52 as described above, and co-expressed with VvWRKY56 and VvWRKY45, orthologues of AtWKY20 and AtWKY58, respectively (Figure 4). Interestingly, two different studies demonstrated that the Arabidopsis orthologues of VvWRKY52 (AtWRKY75) and of VvWRKY45 (AtWRKY58) are positive and negative regulators of plant responses and defense mechanisms to biotic stress (Balbi and Devoto, 2008; Encinas-Villarejo et al., 2009). In withering, VvWRKY52 was repressed and VvWRKY45 was induced, supporting the hypothesis of their involvement in response to stress and indicating that in grapevine diverse mechanisms are related to the kind of stress imposed, with the induction of different WRKYs TFs upon biotic or abiotic stresses.
In recent years, one of the most studied WRKY TFs has been AtWRKY33, which was reported to be significantly induced by osmotic and oxidative stresses (Jiang and Deyholos, 2009). VvWRKY24 (average expression: 4471) and VvWRKY16 (average expression: 23064), both orthologues of AtWRKY33 (Wang et al., 2014) were strongly correlated (r = 0.91) with VvVQ14 and VvVQ17, respectively, showing an increased expression from PHWI to PHWIII (Figure 3). As observed for VvWRKY52 and correlated VvVQ transcripts, a functional analysis is mandatory to validate a cooperation of VvVQ14/17 andVvWRKY25/16 inmediating the abiotic stress responses in Vitis. The other dataset (Figure 4B) contains all of the grapevine organs and tissues described by Fasoli et al. (2012) except PHW berries. Correlation network analysis conducted on these data showed significant correlations (FDR < 0.05 and r > 0.6) for 10 VQ and 24 WRKY transcripts. Among these, VvVQ18 and VvWRKY48 were the genes showing the highest correlation values (r = 0.95). These genes are both characterized by a high and specific expression in roots as previously described (Figure 3). Another interesting gene was VvVQ10, that is mainly expressed in senescing leaves and co-expressed with VvWRKY03, −11, and −12 of group II-c (Wang et al., 2014). Phylogenetic analysis revealed that VvWRKY03 is similar to AtWRKY75, while VvWRKY11 and −12 shows sequence similarity with AtWRKY51. Guo et al. (2004) suggested that AtWRKY75 and −51 are senescence-specific genes and, only for the first one, is leaf senescence significantly delayed in related Arabidopsis antisense lines.
Interestingly, some water stress-induced WRKY genes were co-expressed with one another (Figure 4). Despite accumulating data suggesting that some WRKY proteins belonging to groups II-a, -b, -d and III function in the same pathway in response to different biotic and abiotic stresses (Chi et al., 2013), it is possible that these interactions may occur also between group I and II-c WRKYs TFs. Thus, further studies could also focus on determining whether different combinations of WRKY proteins cooperate with VQ motif-containing proteins in grapevine berries upon abiotic stresses.
Expression Profiles of Grapevine VQ Genes in Response to Different Stress and Exogenous Hormone Treatments
Stress Treatments
Increasing evidence indicates that VQ genes are associated with plant response to environmental stimuli and that the expression of VQ genes is enhanced by drought, pathogen inoculation, and treatment with hormones (Cheng et al., 2012; Kim et al., 2013). It has been reported that 22 rice VQ genes are up-regulated in response to drought treatment (Kim et al., 2013). The Arabidopsis VQ9 protein has been shown to act as a repressor of WRKY8 to maintain a balance in the activation of WRKY8-mediated signaling pathways that are involved in salinity stress tolerance (Hu et al., 2013). In our expression analysis, we identified few VvVQ genes, VvVQ3, −16, and −17, strictly related to dehydration and linked to the withering process (Figure 3). Anyway, it's worthwhile to note that, although involving a dehydration process, post-withering represents a very particular condition and is limited to berry tissues. In order to analyse more in detail the response of VvVQ genes to drought stress, the expression levels of all the 18 VQ genes were analyzed in leaves of Pinot Noir 40024 plants subjected to water deprivation. Transcript accumulation was measured by quantitative RT-PCR and is expressed as fold-change (FC) in Figure 5. The majority of VvVQ genes were found to be responsive to water stress. For four VQs (VvVQ4, −7, −11, and −13), a significant induction was detected 4 days after the imposition of the stress, 13 VvVQs were highly expressed from the 8th day and only VvVQ14 showed an induction after the 12th day. VvVQ1 and −6 were the most responsive genes, with FC values higher than 20, while VvVQ4 was the only gene very weakly modulated by the stress and with a FC < 2 and thus it can be considered not responsive to the water stress. VvVQ8, −9, and −14 were the only genes shown to gradually increase their expression and showed a peak at 12 day with VvVQ8 being the only one reaching FC values higher than 10. The up-regulation of the VvVQ3, −16, and −17 under water stress conditions, their high expression levels in PHW berry tissues observed in the Corvina atlas (Figure 3) and the presence of MBS cis-elements involved in drought response in the promoter regions strongly suggest a role of these VQs in the response to water stress. As in Wang et al. (2014), the expression of some VvWRKY genes is induced by water deficit stress, such as VvWRKY03, VvWRKY16, and VvWRKY52. In the correlation network analysis on berry tissues undergoing the withering process, (Figure 4A) these genes were linked to VvVQ10, −13, −16, −3, and −17. However, the induction of both VvWRKY52 and VvVQ13, −16, −3, and −17 detected by qRT-PCR analysis is in contrast with the negative correlation observed in the network analysis. As reported in Table S3, the majority of VvVQ genes showing induction upon water stress presents at least one drought responsive MBS element within the first 2 Kb upstream the start codon. These data are in agreement with previous expression analyses performed in rice (Kim et al., 2013), where most VQ genes were demonstrated to be involved in the response to water stress.
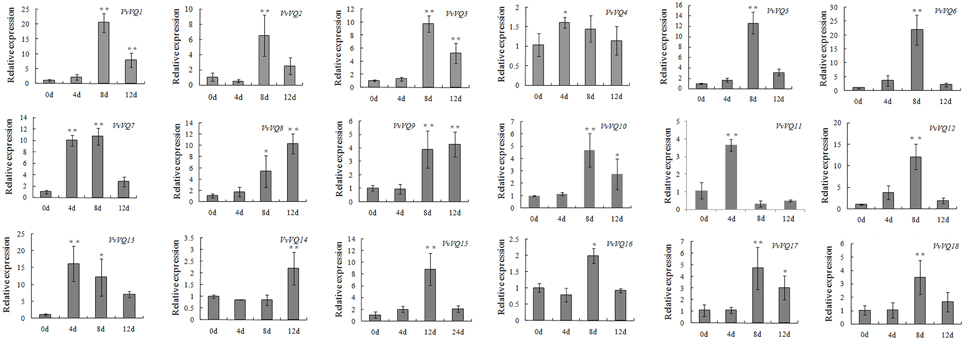
Figure 5. Expression patterns of grapevine VQ genes in response to a drought treatment. Quantitative RT-PCR was used to determine the level of expression of VQ genes in response to a drought treatment. The Y axis represents the level of relative gene expression while the X axis represents the time course over which samples were collected after water was withheld. The name of each VvVQ gene is provided in each graph. The values of VvVQ gene expression were normalized using the expression of a grapevine actin gene. Data points represent the mean of three independent biological replicates. The error bars indicate standard deviation. * indicates that the level of expression is significantly different from the value of the control (*P < 0.05, **P < 0.01).
As demonstrated for the WRKY TFs, many VQ proteins of Arabidopsis and rice are involved in the plant response to biotic stresses (Cheng et al., 2012; Kim et al., 2013). In order to investigate the response of the whole VvVQ family in grapevine upon biotic stress, the expression patterns of all VvVQ transcripts was monitored by qRT-PCR also upon powdery mildew infection. As shown in Figure 6, 5 out of 18 genes (VvVQ3, −5, −6, −11, and −12) were significantly up-regulated upon powdery mildew infection. VvVQ12 is the most responsive gene, increasing its expression across all profiles and reaching a peak at 24 h after inoculation with the FC value of approximately 15 in respect to the mock-inoculated samples. VvVQ3 showed an up regulation at 4 h after inoculation and maintained the same level through the whole profile. The transcript abundance of VvVQ5 in inoculated leaves peaked at 12 h after infection, whereas VvVQ6 and VvVQ11 were induced only at 4 h. The inoculation treatment with powdery mildew also caused an increase in the transcription levels of VvVQ16 at 4 h, however, the expression level then dropped below the level of expression observed prior to inoculation. In contrast, the transcript abundance of VvVQ17 and VvVQ18 was initially decreased up to 12 h after inoculation, but then increased substantially at 24 h (Figure 6). The expression of VvVQ1, −10, and −14, despite having different patterns, showed a clear down-regulation upon infection. These analyses indicated that VvVQ TFs were implicated in the powdery mildew response, possibly through interaction with WRKY proteins. Involvement of both VQ and WRKY proteins in plant defense was already reported, showing that the Arabidopsis VQ proteins SIB1 and SIB2 act as activators of WRKY33, which in turn is involved in the plant defense against necrotrophic pathogens (Lai et al., 2011). Moreover, MKS1, a VQ protein that acts as a substrate for MAPK4 (MPK4) in Arabidopsis, interacts with WRKY25 and WRKY33 and thus may contribute to MAPK-regulated defense activation by coupling the kinase to specific WRKY TFs (Andreasson et al., 2005).
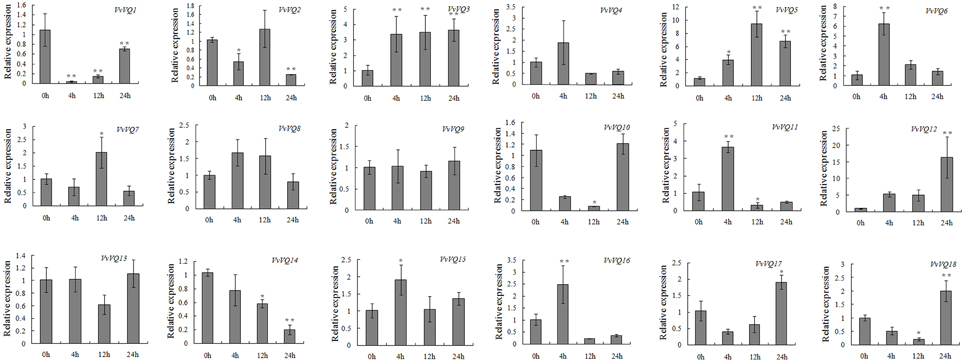
Figure 6. Expression patterns of grapevine VQ genes in response to inoculation with Erysiphe necator (powdery mildew). Quantitative RT-PCR was used to determine the level of expression of VQ genes in response to inoculation with powdery mildew. Gene names are indicated in each graph. The values of VvVQ gene expression were normalized using the expression of a grapevine actin gene. Data points represent the mean of three independent biological replicates. The error bars indicate standard deviation. * indicates that the level of expression is significantly different from the value of the control (*P < 0.05, **P < 0.01).
Hormone Treatments
SA and ETH mediate plant responses to biotic stress (Broekaert et al., 2006; Loake and Grant, 2007). To investigate their effect on the expression of the VvVQ genes, in the current study, quantitative RT-PCR was conducted on the 18 VvVQ genes in plants treated with these two hormones. As shown in Figure 7, the expression profiles after SA treatment appeared modulated for most of the VvVQ genes with the exception of VvVQ3 and VvVQ16, which showed unaltered transcript levels. In particular, VvVQ2, −5, −6, −10, and −18 were strongly up-regulated even if by varying degrees throughout the treatment. Whereas, VvVQ4 and VvVQ9, were significantly down-regulated. VvVQ7 and VvVQ15 were significantly induced at 4 h after SA treatment and then declined to the normal level.
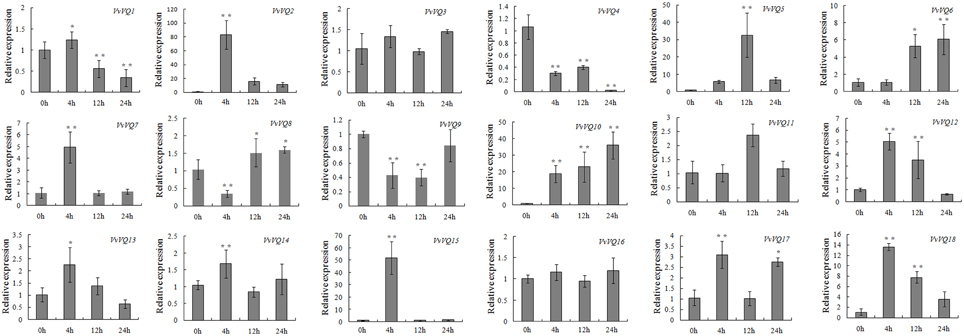
Figure 7. Expression patterns of grapevine VQ genes in response to salicylic acid (SA) treatment. Quantitative RT-PCR was used to determine the level of expression of VQ genes in response to the application of salicylic acid (SA). Gene names are indicated in each graph. The values of VvVQ gene expression were normalized using the expression of a grapevine actin gene. Data points represent the mean of three independent biological replicates. The error bars indicate standard deviation. * indicates that the level of expression is significantly different from the value of the control (*P < 0.05, **P < 0.01).
Analysis of transcript abundance in grapevine leaves treated exogenously with ETH, reported in Figure 8, indicated that VvVQ2, −12, and −18 were the only VvVQ genes exhibiting higher levels of expression than that of the control, suggesting that in general the VvVQs were induced more by SA than ETH. For VvVQ4 and VvVQ7, the expression patterns showed a very slight drop only at 4, −12 h after treatment, whereas the levels of VvVQ13, −16, −15, and -17 remained unchanged. The expression of VvVQ1, −5, −8, and−14 were significantly repressed throughout the experiment.
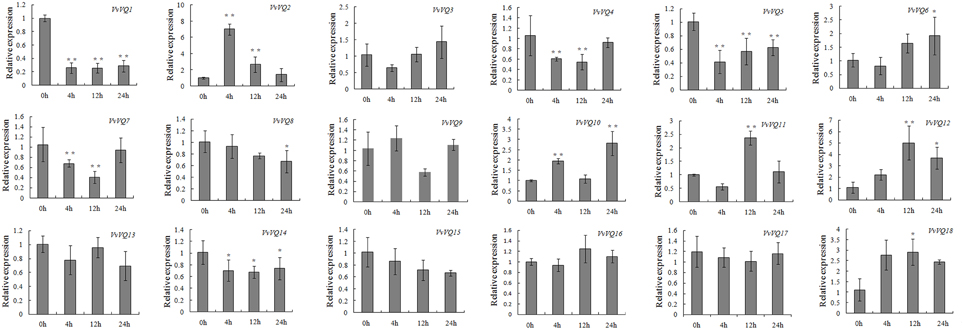
Figure 8. Expression patterns of grapevine VQ genes in response to ethylene (ETH) treatment. Quantitative RT-PCR was used to determine the level of expression of VQ genes in response to an application of ETH. Gene names are indicated in each graph. The values of VvVQ gene expression were normalized using the expression of a grapevine actin gene. Data points represent the mean of three independent biological replicates. The error bars indicate standard deviation. * indicates that the level of expression is significantly different from the value of the control (*P < 0.05, **P < 0.01).
In general, 11 VvVQ genes presented similar expression patterns in both SA and ETH experiments, even if with different FCs. Among these, VvVQ3, −9, and −16 were not modulated, indicating that they are probably not responsive to these hormones. Interestingly, VvVQ5, −7, −8, −13, −15, −17, and −18 showed different responses to the hormones, being down-regulated or not modulated by the ETH treatment, but strongly induced by SA. Similar responses to SA and ETH hormones were also observed for many VvWRKY genes by Guo et al. (2014). Interestingly, among them was VvWRKY44 of subgroup II-c (corresponding to VvWRKY43) (Wang et al., 2014), whose expression was induced by both SA and ETH (Guo et al., 2014), and highly correlated with VvVQ12 in our analysis. VvVQ12 was also up-regulated by both treatments (Figures 7, 8); it is conceivable that VvWRKY43 and VvVQ12 proteins cooperate in the response to SA and ETH. Our expression analysis of the VvVQ gene family after abiotic, biotic stresses and treatments by hormones together with the correlation analysis of gene expression provide useful information about the hypothetical roles of VvVQ genes in the regulation of developmental and defense mechanisms and about possible interactions with the WRKY proteins.
Conclusions
Based on studies with Arabidopsis, it appears that plant VQ proteins primarily function as cofactors of WRKY proteins and play critical roles in regulating WRKY-mediated gene expression (Cheng et al., 2012). In the present study, the grapevine VQ gene family was examined and 18 VvVQ genes were identified. The phylogeny, compositional structure, and cellular location (based on homology with VQ genes in other species) of VvVQ genes were determined, providing a general overview of this multigenic family in grapevine. The expression of VvVQ genes in different tissues and different developmental stages was examined by using publicly available microarray data. Expression data, together with the correlation network analysis carried out with highly correlated VQ and WRKY genes, highlighted that VvWRKY52 and VQ3, −13, −16, and −17 were strongly anti-correlated in PHW conditions, suggesting that further analyses need to be conducted to determine whether the proteins encoded by these genes interact one another. Moreover, these data led to the hypotesis that VvVQ14 and VvVQ17 may cooperate with VvWRKY25 and VvWRKY16 to mediate abiotic stress responses in grapevines. In this study, qRT-PCR was used to analyse the expression of VvVQ transcripts in response to drought, powdery mildew inoculation, and treatment with SA and ETH. Tissue-specific and developmental-specific patterns of VQ gene expressions were observed in grapevine, as was the up- and down-regulation of specific VvVQ genes in response to biotic and abiotic stress treatments, and hormone treatments. Collectively, the data generated in this study of VQ genes in grapevine provided a useful resource for future functional studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Priority Academic Program Developmentof Modern Horticultural Science in the Jiangsu Province.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00417/abstract
Table S1. List of primers used to quantify VvVQ genes expression by qRT-PCR.
Table S2. Syntenic blocks of VQ genes between grapevine and Arabidopsis genomes.
Table S3. Promoter analysis of the grapevine VQ gene family.
Figure S1. Chromosomal location of VQ genes in grapevine. The chromosomal location of grapevine VQ genes was determined based on the 12X V1 release of the PN40024 grapevine genome (http://genomes.cribi.unipd.it/). The distribution of VvVQ genes was drawn using MapInspect software.
Figure S2. Alignment of VQ domain amino acid sequences of grapevine VQ proteins. Multiple alignment of grapevine VQ domain amino acid sequences was performed using Muscle with default options.
Figure S3. Phylogenetic tree of grapevine, Arabidopsis and rice VQ proteins obtained using a Bayesian-base method.
Figure S4. Chromosomal location of VQ genes in Arabidopsis indicating tandem and segmental duplications. Diagram of the five chromosomes of Arabidopsis depicting the location and distribution of 34VQ genes. The segmental duplication genes were connected with a blue line. Tandem duplication genes were indicated with a pink vertical line.
Figure S5. Syntenic blocks of VQ genes (indicated in red) between the grapevine and Arabidopsis genomes. Arrows represent gene loci in the grapevine and Arabidopsis chromosomes. Blue arrows represent duplicated loci in a syntenic block. The data were obtained from the Plant Genome Duplication Database, and syntenic blocks containing grapevine VQ genes were illustrated.
Figure S6. Differential expression of VvVQ genes in various tissues and at different developmental stages based on microarray analysis. The expression levels of the first development stage of each tissue were set at a value of “1” to determine up and down regulation at subsequent stages of development. Tissues containing only one phase were not considered. The genes above and below the colored tissue bar indicated the up-regulated and down-regulated genes, respectively, at this developmental stage. Genes were considered to be differentially expressed if the fold change (up- or down- regulated) was greater than two.
Abbreviations
VvVQ, Grapevine VQ; SA, Salicylic acid; ETH, Ethylene; TF, Transcription factor; qRT-PCR, Quantitative reverse transcription polymerase chain reaction; MEME, Multiple Expectation Maximization for Motif Elicitation; aa, amino acid/s.
References
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Balbi, V., and Devoto, A. (2008). Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 177, 301–318. doi: 10.1111/j.1469-8137.2007.02292.x
Bowers, J. E., Chapman, B. A., Rong, J. K., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Broekaert, W. F., Delaure, S. L., De Bolle, M. F., and Cammue, B. P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. doi: 10.1146/annurev.phyto.44.070505.143440
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., and Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. doi: 10.1186/1471-2229-10-281
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J., Chen, J. Y., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J.-Q., et al. (2013). Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant. 6, 287–300. doi: 10.1093/mp/sst026
Corso, M., Ziliotto, F., Rizzini, F. M., Teo, G., Cargnello, G., and Bonghi, C. (2013). Sensorial, biochemical and molecular changes in Raboso Piave grape berries applying “Double Maturation Raisonnée” and late harvest techniques. Plant Sci. 208, 50–57. doi: 10.1016/j.plantsci.2013.03.010
Daraselia, N. D., Tarchevskaya, S., and Narita, J. O. (1996). The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol. 112, 727–733. doi: 10.1104/pp.112.2.727
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Encinas-Villarejo, S., Maldonado, A. M., Amil-Ruiz, F., De Los Santos, B., Romero, F., Pliego-Alfaro, F., et al. (2009). Evidence for a positive regulatory role of strawberry (Fragaria×ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J. Exp. Bot. 60, 3043–3065. doi: 10.1093/jxb/erp152
Espinoza, C., Medina, C., Somerville, S., and Arce-Johnson, P. (2007). Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J. Exp. Bot. 58, 3197–3212. doi: 10.1093/jxb/erm165
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fasoli, M., Dal Santo, S., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.1105/tpc.112.100230
Freitas, F. Z., and Bertolini, M. C. (2004). Genomic organization of the Neurospora crassa gsn gene: possible involvement of the STRE and HSE elements in the modulation of transcription during heat shock. Mol. Genet. Genomics. 272, 550–561. doi: 10.1007/s00438-004-1086-5
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. doi: 10.1111/j.1365-3040.2003.01158.x
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Jiang, Y., and Deyholos, M. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kim, D. Y., Kwon, S. I., Choi, C., Lee, H., Ahn, I., Park, S. R., et al. (2013). Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 529, 208–214. doi: 10.1016/j.gene.2013.08.023
Kim, J. K., Cao, J., and Wu, R. (1992). Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pI alpha-amylase gene. Mol. Gen. Genet. 232, 383–393. doi: 10.1007/BF00266241
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Lai, Z., Li, Y., Wang, F., Cheng, Y., Fan, B., Yu, J. Q., et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841. doi: 10.1105/tpc.111.090571
Lai, Z., Vinod, K., Zheng, Z., Fan, B., and Chen, Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8:68. doi: 10.1186/1471-2229-8-68
Li, S., Fu, Q., Huang, W., and Yu, D. (2009). Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 28, 683–693. doi: 10.1007/s00299-008-0666-y
Liu, Q., Wang, H., Zhang, Z., Wu, J., Feng, Y., and Zhu, Z. (2009). Divergence in function and expression of the NOD26-like intrinsic proteins in plants. BMC Genomics 10:313. doi: 10.1186/1471-2164-10-313
Liu, X., Wang, X., Pang, Y., Liang, J., Liu, S., Sun, X., et al. (2006). [Molecular cloning and characterization of a novel WRKY gene from Brassica chinensis]. Mol. Biol. (Mosk) 40, 816–824. doi: 10.1134/s0026893306050074
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence–the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
Lois, R., Dietrich, A., Hahlbrock, K., and Schulz, W. (1989). A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 8, 1641–1648.
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Manzara, T., Carrasco, P., and Gruissem, W. (1991). Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell 3, 1305–1316. doi: 10.1105/tpc.3.12.1305
Menkens, A. E., Schindler, U., and Cashmore, A. R. (1995). The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. doi: 10.1016/S0968-0004(00)89118-5
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-005-2142-1
Pastuglia, M., Roby, D., Dumas, C., and Cock, J. M. (1997). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9, 49–60. doi: 10.1105/tpc.9.1.49
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Quirino, B. F., Noh, Y.-S., Himelblau, E., and Amasino, R. M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5, 278–282. doi: 10.1016/S1360-1385(00)01655-1
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. doi: 10.1093/pcp/pcn061
Robatzek, S., and Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Rouster, J., Leah, R., Mundy, J., and Cameron-Mills, V. (1997). Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 11, 513–523. doi: 10.1046/j.1365-313X.1997.11030513.x
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P. J., Torres, J. T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I. E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700.
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 microarray software suite. Methods Enzymol. 411, 134–193. doi: 10.1016/s0076-6879(06)11009-5
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Simpson, S. D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. doi: 10.1046/j.1365-313X.2003.01624.x
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, H., Wang, X., Bowers, J. E., Ming, R., Alam, M., and Paterson, A. H. (2008a). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954. doi: 10.1101/gr.080978.108
Tang, H. B., Bowers, J. E., Wang, X. Y., Ming, R., Alam, M., and Paterson, A. H. (2008b). Perspective - Synteny and collinearity in plant genomes. Science 320, 486–488. doi: 10.1126/science.1153917
Tripathi, P., Rabara, R., and Rushton, P. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. doi: 10.1007/s00425-013-1985-y
Vannozzi, A., Dry, I. B., Fasoli, M., Zenoni, S., and Lucchin, M. (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 12:130. doi: 10.1186/1471-2229-12-130
Velasco, R., Zharkikh, A., Troggio, M., Cartwright, D. A., Cestaro, A., Pruss, D., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2:e1326. doi: 10.1371/journal.pone.0001326
Vitulo, N., Forcato, C., Carpinelli, E. C., Telatin, A., Campagna, D., D'Angelo, M., et al. (2014). A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14:99. doi: 10.1186/1471-2229-14-99
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010a). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010b). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U.S.A. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Wang, M., Vannozzi, A., Wang, G., Liang, Y.-H., Tornielli, G. B., Zenoni, S., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticult. Res. 1:16. doi: 10.1038/hortres.2014.16
Washida, H., Wu, C. Y., Suzuki, A., Yamanouchi, U., Akihama, T., Harada, K., et al. (1999). Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol. Biol. 40, 1–12. doi: 10.1023/A:1026459229671
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Xu, Y., Chang, P., Liu, D., Narasimhan, M. L., Raghothama, K. G., Hasegawa, P. M., et al. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6, 1077–1085. doi: 10.1105/tpc.6.8.1077
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol. 101, 1119–1120. doi: 10.1104/pp.101.3.1119
Yang, Z., Wang, X., Gu, S., Hu, Z., Xu, H., and Xu, C. (2008). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407, 1–11. doi: 10.1016/j.gene.2007.02.034
Yuan, J. S., Reed, A., Chen, F., and Stewart, C. N. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85
Zamboni, A., Minoia, L., Ferrarini, A., Tornielli, G. B., Zago, E., Delledonne, M., et al. (2008). Molecular analysis of post-harvest withering in grape by AFLP transcriptional profiling. J. Exp. Bot. 59, 4145–4159. doi: 10.1093/jxb/ern256
Keywords: grapevine, VQ gene family, gene expression, drought treatment, biotic stress, WRKY protein, co-expression network
Citation: Wang M, Vannozzi A, Wang G, Zhong Y, Corso M, Cavallini E and Cheng Z-M (2015) A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 6:417. doi: 10.3389/fpls.2015.00417
Received: 19 January 2015; Accepted: 23 May 2015;
Published: 12 June 2015.
Edited by:
Ariel Orellana, Universidad Andres Bello, ChileReviewed by:
Dario Cantu, University of California, Davis, USAPatricio Hinrichsen, Instituto de Investigaciones Agropecuarias, Chile
Copyright © 2015 Wang, Vannozzi, Wang, Zhong, Corso, Cavallini and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong-Ming (Max) Cheng, Fruit Crop Systems Biology Laboratory, Nanjing Agricultural University, Weigang 1, Nanjing 210095, China,em1jQG5qYXUuZWR1LmNu
†Present Address: Massimiliano Corso, Laboratory of Plant Physiology and Molecular Genetics, Université Libre de Bruxelles, Brussels, Belgium
 Min Wang1,2
Min Wang1,2 Alessandro Vannozzi
Alessandro Vannozzi Massimiliano Corso
Massimiliano Corso Zong-Ming (Max) Cheng
Zong-Ming (Max) Cheng