- National Key Laboratory of Plant Molecular Genetics, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
miR319-targeted TCP genes are believed to regulate cell division in leaves and floral organs. However, it remains unknown whether these genes are involved in cell wall development. Here, we report that TCP24 negatively regulates secondary wall thickening in floral organs and roots. The overexpression of the miR319a-resistant version of TCP24 in Arabidopsis disrupted the thickening of secondary cell walls in the anther endothecium, leading to male sterility because of arrested anther dehiscence and pollen release. Several genes linked to secondary cell wall biogenesis and thickening were down-regulated in these transgenic plants. By contrast, the inhibition of TCP24 using the ectopic expression of a TCP24-SRDX repressor fusion protein, or the silencing of TCP genes by miR319a overexpression, increased cell wall lignification and the enhanced secondary cell wall thickening. Our results suggest that TCP24 acts as an important regulator of secondary cell wall thickening and modulates anther endothecium development.
Introduction
Anther dehiscence is a multistage process that involves coordinated programmed events in specific cells, including degeneration of the middle layer and the tapetum, thickening of the endothecium, degradation of septum cells, and breakage of stomium cells (Goldberg et al., 1993; Sanders et al., 1999; Wilson et al., 2011). Secondary wall thickening of the endothecium generates the tensile force necessary to rupture the stomium and in turn, release the pollen grains (Keijzer, 1987; Bonner and Dickinson, 1989). The importance of this process has been demonstrated by genetic analysis. A loss-of-function mutation of MYB26 disrupts secondary thickening of the anther walls, resulting in non-dehiscent anthers (Dawson et al., 1999; Steiner-Lange et al., 2003; Yang et al., 2007). Two NAC domain transcription factors, NST1 and NST2, function redundantly in regulating endothecium wall thickening and act downstream of MYB26. Overexpression of these two genes results in ectopic secondary thickening in various tissues (Mitsuda et al., 2005). Mutations in IRREGULAR XYLEM (IRX) and receptor-like protein kinase 2 (RPK2) genes also lead to the defective secondary wall thickening of the anther (Brown et al., 2005; Mizuno et al., 2007; Hao et al., 2014). Other genes, such as CA2 (carbonic anhydrase 2), AHP4 (Arabidopsis histidine-containing phosphotransfer factor 4), SAF1 (secondary wall thickening-associated F-box 1) and CBSX2 (cystathionine β-synthase domain-containing protein), negatively regulate this process, and the overexpression of these genes in Arabidopsis leads to anther non-dehiscent phenotypes (Jung et al., 2008, 2013; Villarreal et al., 2009; Kim et al., 2012).
The TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family encodes plant-specific transcription factors, which contain a conserved bHLH motif that allows DNA binding and protein–protein interactions (Cubas et al., 1999; Martin-Trillo and Cubas, 2010). The TCP members are grouped into two classes based on sequence homology: class I and class II TCPs (Cubas et al., 1999). It is believed that class I TCP genes promote cell division, while class II genes act antagonistically to inhibit cell division (Li et al., 2005). Duplication and diversification events over millions of years have generated a large family of 24 TCP genes in Arabidopsis of which 11 belong to the class II subfamily (Martin-Trillo and Cubas, 2010). A functional analysis shows that the class II TCP genes regulate several aspects of plant development. Arabidopsis BRANCHED1 (BRC1) and BRC2, both closely related to the TEOSINTE BRANCHED1 from maize (Doebley et al., 1997), are involved in suppressing axillary bud outgrowth (Aguilar-Martinez et al., 2007). TCP2, TCP3, TCP4, TCP10, and TCP24 are the targets of miR319a/JAW. The down-regulation of these genes by overexpression of miR319a in jaw-D mutants generates larger leaves with crinkled surfaces owing to the extended cell proliferation along leaf margins (Palatnik et al., 2003). Conversely, hyper-activation of TCP4 results in decreased cell proliferation, resulting in smaller leaves (Sarvepalli and Nath, 2011). miR319a-targeted genes function redundantly with TCP5, TCP13, and TCP17 to coordinate the maintenance of undifferentiated fates in the shoot apical meristem and the promotion of the differentiated status in leaves (Koyama et al., 2007; Efroni et al., 2008). This coordination is achieved via the negative regulation of CUP-SHAPED COTYLEDON (CUC) genes, and TCP3 can directly activate the expression of miR164A, ASYMMETRIC LEAVES1, INDOLE-3-ACETIC ACID3/SHORT HYPOCOTYL2 (IAA3/SHY2), and At1g29460 to suppress CUC expression (Koyama et al., 2010). miR319a-targeted TCPs can interact with ASYMMETRIC LEAVES2 and repress the expression of BREVIPEDICELLUS and KNAT2 genes by binding to their promoters causing normal leaf development (Li et al., 2012b). It is reported that a transcriptional repressor, TIE1, recruits co-repressors TOPLESS/TOPLESS-related proteins to repress the activities of class II TCP genes (Tao et al., 2013).
Previous studies showed that these CINCINNATA (CIN)-like TCP genes were expressed differentially in various organs, indicating that they might play important roles in many aspects of plant development (Koyama et al., 2007). In this study, we used a reverse genetic approach to investigate the function of TCP24. Overexpression of TCP24 led to non-dehiscent anthers owing to the lack of secondary wall thickening in the endothecium, while fusing it with an EAR motif repressor domain (SRDX) caused enhanced lignin deposition in the anther endothecium, as well as other tissues, suggesting that TCP24 functions as a negative regulator of secondary wall thickening.
Materials and Methods
Plant Materials and Growth Conditions
The wild type and transgenic plants of Arabidopsis thaliana used in this study were of Columbia ecotype (Col-0). Seeds were surface sterilized in 70% ethanol for 1 min, followed by 0.1% HgCl2 for 10 min, then washed five times in sterile distilled water, and plated on solid 1% sugar Murashige and Skoog medium. The plates were sealed with parafilm, incubated at 4°C in the dark for 2 days, and then moved to a growth room at 22°C with 16 h light. Two weeks later, the seedlings were transplanted carefully to peat soil in plastic pots, moved to a growth chamber in the phytotron of Institute of Plant Physiology and Ecology, and grown at 22°C with 16 h of light per day.
Gene Cloning and Transformation
The full length CDS of TCP24 was amplified from cDNA. mTCP24 was generated by site-directed mutagenesis method using the QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene Catalog #210518) with appropriate primers. To construct the plasmid p35S:mTCP24, vector PJP100 (p35S:mTCP2, obtained from Dr. Weigel’s lab) was modified by replacing mTCP2 with mTCP24. The 35S promoter of p35S:mTCP24 was replaced with the TCP24 promoter (2.7 kb fragment upstream from the translational start site) for the construction of pTCP24:mTCP24. p35S:TCP24SRDX was generated in our modified pCAMBIA3301 binary vector by fusing TCP24 with the EAR motif repressor domain SRDX under the control of the 35S promoter. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101 for plant transformation using the floral dip method as described previously (Li et al., 2012a). The transgenic plants of p35S:mTCP24 and pTCP24:mTCP24 were selected on plates containing 1/2 Murashige and Skoog media supplemented with 50 mg/L kanamycin, while p35S:TCP24SRDX plants were selected using 40 mg/L phosphinothricin.
In Situ Hybridization
The full-length coding sequence of TCP24 was polymerase chain reaction (PCR) amplified and cloned into pBluescript SK. Digoxigenin-labeled sense and antisense probes were synthesized with T7 or T3 RNA polymerase (Roche). Inflorescences from wild type and transgenic plants were pretreated and hybridized as described previously (Liu et al., 2011). Locked nucleic acid (LNA)-modified probe of miR319a was synthesized and labeled with DIG at the 3′ end and used for in situ hybridization.
Real-Time PCR
The total RNA of inflorescences (with opened flowers removed) was extracted using TRIzol Reagent (Invitrogen) and treated with DNase I (TaKaRa) to remove DNA contamination. For cDNA synthesis, ∼4 μg RNA was reverse-transcribed using PrimeScript® Reverse Transcriptase (TaKaRa) with oligo(dT) primers according to the manufacturer’s protocol. A quantitative real-time PCR analysis was performed using the Rotor-Gene 3000 system (Corbett Research, Mortlake, NSW, Australia) using SYBR Premix Ex Taq (Takara). ACTIN mRNA was used as an internal control, and the comparative threshold cycle (2-ΔΔCt) method was used to determine relative transcript levels. Three biological replicates and three technical replicates were performed. The gene specific primers for reverse-transcription PCR were shown in Supplementary Table S1.
Histology
Inflorescences of 5- to 6-week-old wild type and transgenic plants were fixed in formalin/acetic acid/alcohol (FAA) and embedded in paraffin (Sigma). Then, 7 μm sections were stained with 0.05% (w/v) toluidine blue (Sigma) at 37°C for 15 min and then washed with water. For the analysis of semi-thin sections, samples fixed in FAA were embedded in epoxy resin. Then, 2-μm-thick sections were cut with glass knives, affixed to glass slides, and stained in 0.05% (w/v) toluidine blue. The sections were observed under a light microscope (Olympus model BX 51).
To visualize lignin deposition, plant tissues were stained with phloroglucinol-HCl solution (1.25 g of phloroglucinol dissolved in 25 ml of 95% ethanol and 10 ml of concentrated HCl), and observed under a dissecting microscope. To examine the secondary wall thickening in the endothecium, anthers were placed onto glass slides with clearing fluid. The solution was prepared from lactic acid, chloral hydrate, phenol, clove oil and xylene in the ratio 2:2:2:2:1, respectively, by weight (Herr, 1971). The anthers were observed by microscopy using differential interference contrast optics.
Results
Overexpression of TCP24 Disrupts Anther Dehiscence in Arabidopsis
Mutations in single CIN-like TCP genes do not generate visible phenotypes owing to the redundancy among these genes (Koyama et al., 2007). To characterize the role of TCP24 during plant development, we first constructed mTCP24, the miR319a-resistant version of TCP24, which contains nucleotide substitutions in the miR319a-binding region that do not change the encoded amino acid sequence (Figure 1A), as they did for mTCP2, mTCP3, and mTCP4 (Palatnik et al., 2003; Koyama et al., 2007). In the p35S:mTCP24 plants, the rosette leaves were turned slightly downward (Figures 1B,C), the number of branches increased compared with the wild type (Figures 1D,E), and importantly, the flowers were partially or completely sterile (Figure 1F). Under optical microscopy, anther dehiscence was arrested (Figures 1G–J), albeit to different extents between the transgenic lines. Transcripts of TCP24 were elevated in these independent transgenic lines compared with wild type (Figure 1K). Among these transgenic lines, the higher the TCP24 expression was, the higher the male sterility was, indicating a correlation between the expression levels of TCP24 and the severity of the sterile phenotypes (Supplementary Figure S1). The L2 and L23, two completely sterile lines, set seeds when they were pollinated with the wild type pollen, indicating that p35S:mTCP24 did not affect female fertility.
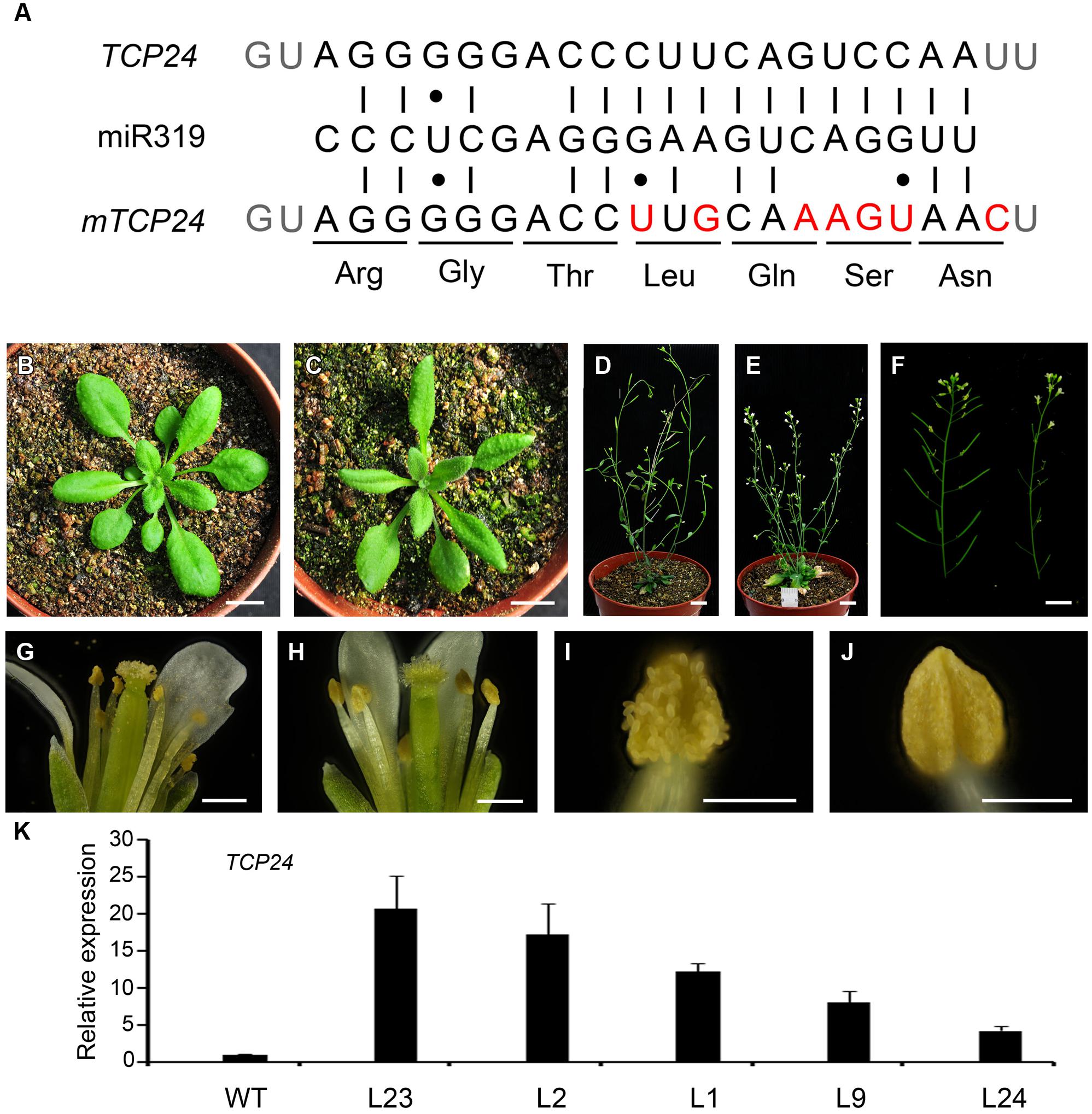
FIGURE 1. Phenotypes of p35S:mTCP24 plants. (A) Diagram of p35S:mTCP24 construct. (B,C) Plants of the wild type (B) and p35S:mTCP24 line (L23; C) at the rosette stage. (D,E) Plants of the wild type (D) and L23 (E) at the inflorescence stage. (F) Inflorescences of the wild type (left) and L23 plants (right). (G,H) Open flowers showing the four whorls of floral organs in the wild type (G) and L23 (H). (I,J) Anthers of the wild type (I) and L23 (J). (K) Real-time PCR analysis of TCP24 expression in the transgenic lines. Scale bars: 1 cm in (B–F); 500 μm in (G,H); 200 μm in (I,J).
TCP24 Suppresses Secondary Wall Thickening of the Anther Endothecium
Anther dehiscence requires the degeneration of some tissues and subsequent differentiation of other tissues, including epidermis, stomium, endothecium, and septum (Goldberg et al., 1993). To verify the defects in anther dehiscence in p35S:mTCP24 plants, we examined the anther endothecium of line L23. During stages 9–10 (Sanders et al., 1999) when microspores were formed, L23 anthers were indistinguishable from those of the wild type (Figures 2A,B). At stage 11, the tapetum of the wild type anthers was degenerated and the endothecium thickened, forming bands of striated spring-like structures (Figures 2C,E). However, secondary thickening in L23 anthers was not observed, as fibrous bands were absent in the endothecium although the tapetum was degenerated (Figures 2D,F). Secondary cell wall thickening was necessary to create the shearing force required for anther dehiscence through the stomium (Dawson et al., 1999). At later stages, the septa of L23 anthers were degraded as in the wild type (Figures 2G,H). While the stomium broke in L23, the anthers did not open (Figures 2I,J). Alexander staining showed that the pollen grains in L23 anthers were viable (Supplementary Figure S2). These observations indicate that non-dehiscence in p35S:mTCP24 anthers is due to the defect in secondary wall thickening in the anther endothecium.
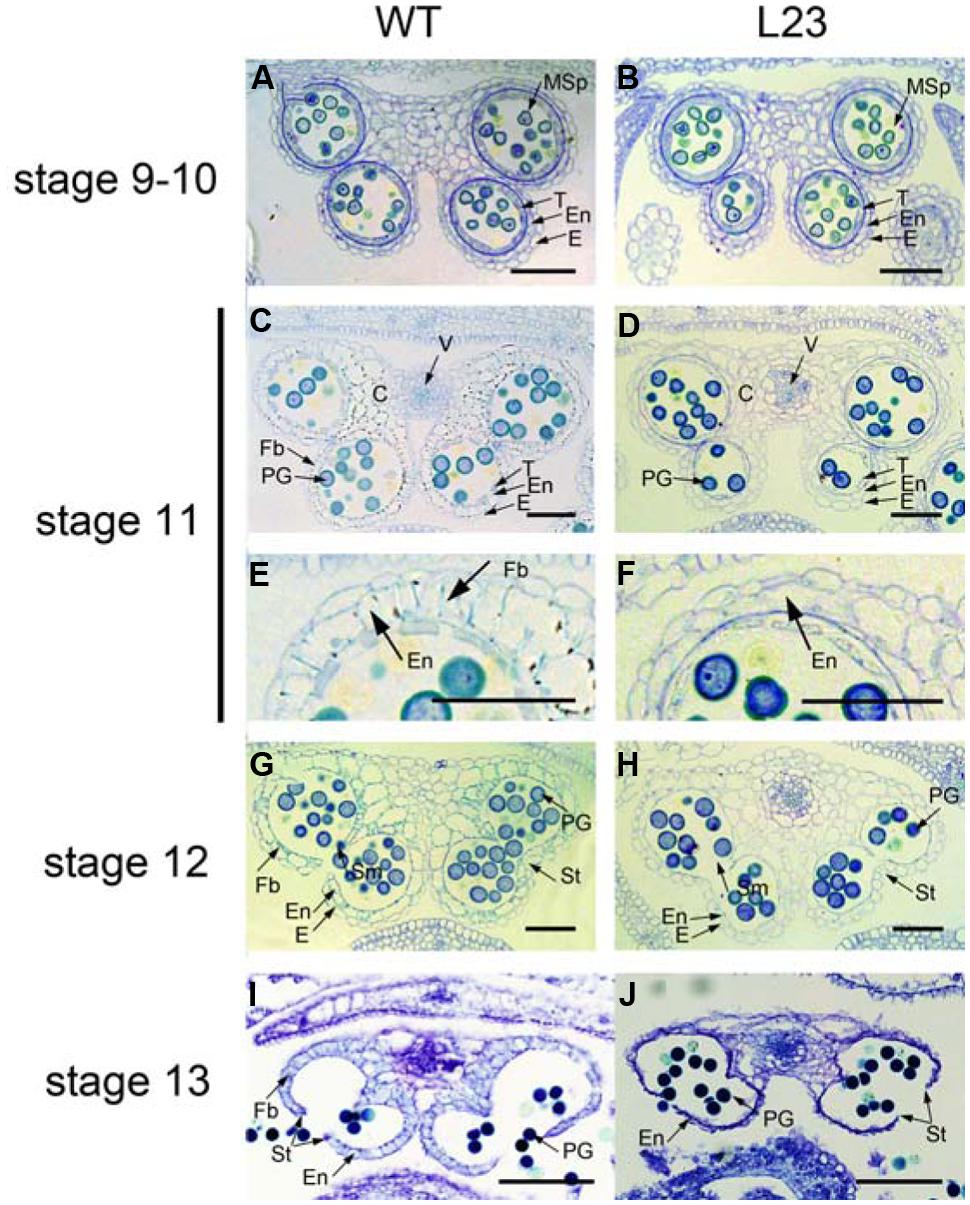
FIGURE 2. Defects of p35S:mTCP24 anthers in secondary cell wall thickening. (A,B) Cross sections of the wild type (A) and L23 (B) anthers at stage 9. (C,D) Cross sections of the wild type (C) and L23 (D) anthers at stage 11. (E,F) Close-up of cross sections of the wild type (E) and L23 (F). (G,H) Cross sections of the wild type (G) and L23 (H) anthers at stage 12. (I,J) Cross sections of the wild type (I) and L23 (J) anthers at stage 13. Anther stages were defined according to Sanders et al. (1999). C, connective; E, epidermis; En, endothecium; Fb, fibrous bands; Msp, microspore; PG, pollen grain; Sm, septum; St, stomium; T, tapetum; V, vascular region. Scale bars: 50 μm in (A–H); 100 μm in (I,J).
We examined the accumulation of lignin, which was the major component of secondary walls according to phloroglucinol staining. The deep red staining of lignified materials by phloroglucinol was clearly observed in the endothecium layer in the wild type anthers (Figure 3A). However, no staining was observed in L23 anthers (Figure 3B). We also treated the anthers with clearing fluid. The thickened cell walls appeared in the wild type endothecium (Figures 3C,E) but were absent in the transgenic plants (Figures 3D,F). These results indicate that TCP24 negatively regulates secondary wall thickening in the anther endothecium.

FIGURE 3. Secondary cell wall thickening of anthers in the transgenic plants with p35S:mTCP24. (A,B) Phloroglucinol staining of wild type (A) and L23 (B) anthers. Lignified material was stained in deep red. (C–F) Microscopic observation of the anthers treated with clearing fluid. (C) The wild type. (D) L23 line. (E,F) Magnification of red box region in (C) and (D). Arrows indicate the thickened endothecium cells. Scale bars: 200 μm in (A,B); 100 μm in (C,D); 10 μm in (E,F).
To exclude the effect of the 35S promoter on ectopic expression, we expressed TCP24 under the control of its native promoter (2.7 kb 5′ upstream of the TCP24 transcriptional start site) (Figure 4A). Among the pTCP24:mTCP24 lines, some had a complete loss of fertility because seed set was not observed, while most showed a reduced fertility compared with the wild type (Figure 4B). In the transgenic line 24-2 which was sterile, there was no pollen on the stigmas, indicating that pollen grain release from the anthers was arrested (Figures 4C,D), and no pollen was observed being released from the anthers (Figures 4F,G). Under the microscope, a few pollen grains were found to be outside the stomium on 24-5 plants (Figures 4E,H). Using phloroglucinol staining, we observed no staining in the anthers of 24-2 plants (Figure 4J) and very weak red staining in the anthers of 24-5 plants (Figures 4I,K). The cell walls of the endothecium were thickened uniformly in the wild type (Figure 4L), but they were not observed in 24-2 anthers (Figure 4M). On the transgenic line 24-5 which had reduced fertility, secondary cell walls occurred in some positions (Figure 4N). These observations confirmed that the overexpression of TCP24 inhibited secondary wall thickening in the anther endothecium.
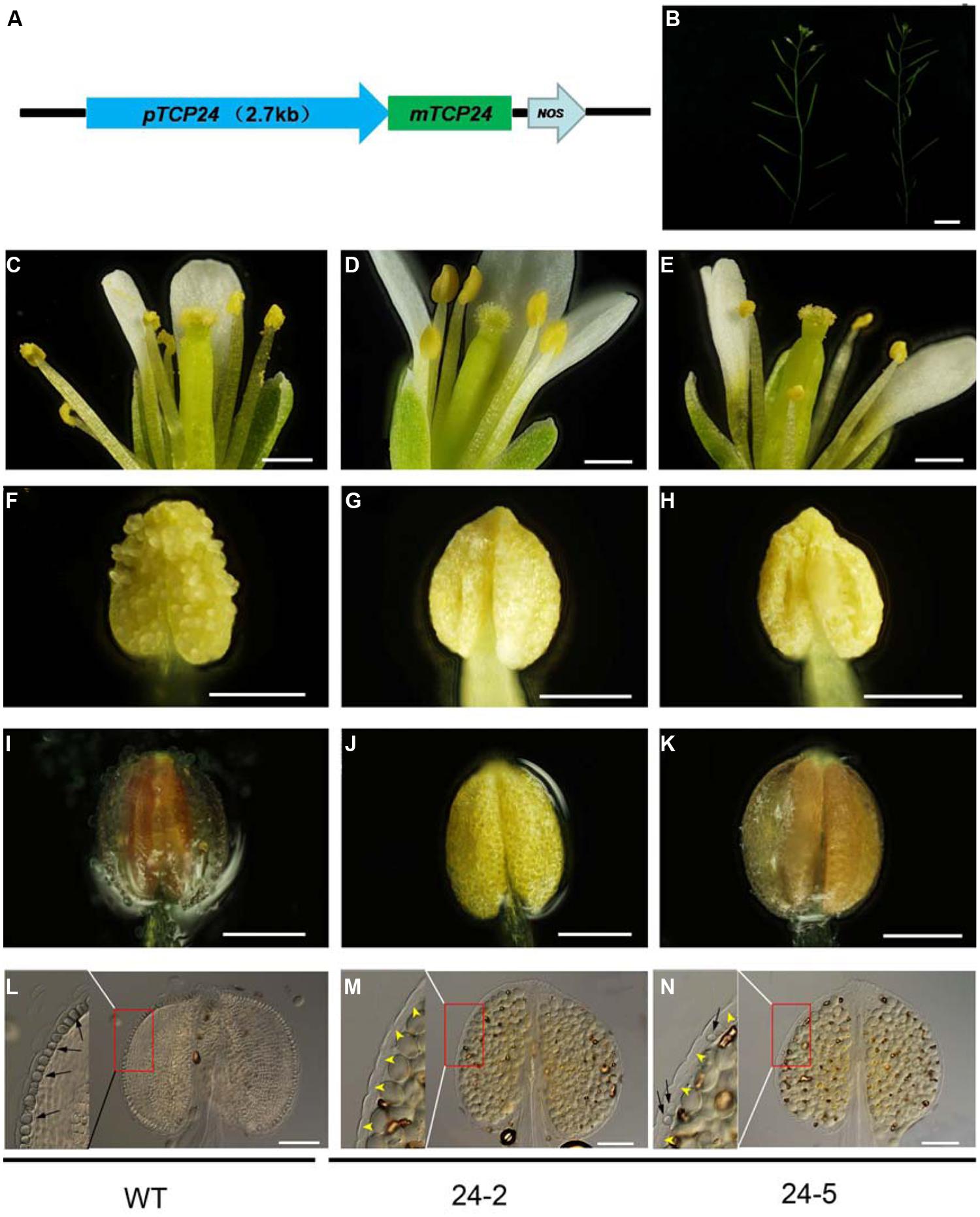
FIGURE 4. The floral organs and anthers of the transgenic plants with pTCP24:mTCP24. (A) Diagram of pTCP24:mTCP24 construct. (B) Inflorescences of the wild type (left) and pTCP24:mTCP24 (24-5; right) plants. (C–E) Open flowers in the wild type (C), 24-2 (D) and 24-5 (E) lines. (F–H) Anthers in the wild type (F), 24-2 (G), and 24-5 (H) lines. (I–K) The wild type (I), 24-2 (J), and 24-5 (K) anthers with phloroglucinol staining. (L–N) The wild type (L), 24-2 (M), and 24-5 (N) anthers treated with clearing fluid. Images in the left of each figures is magnified from the red boxes. Black arrows indicate the thickened endothecium cells, and yellow arrowheads indicate no thickened endothecium cells. Scale bars: 1 cm in (B); 500 μm in (C–E); 200 μm in (F–K); 100 μm in (L–N).
TCP24 Gene Expression Became Weak at the Anther Endothecium Initiation Stage
To examine the temporal and spatial expression of TCP24 during anther development, in situ hybridization was performed using the wild type anthers. The anther development was divided into 14 stages (Sanders et al., 1999). At stage 2, the TCP24 signal was strong in the whole region (Figure 5A). At stage 3, the signal was preferential in the epidermal, parietal layer and sporogenous cells (Figure 5B). At stages 4 to 5, when four clearly defined locules were established, TCP24 was strongly expressed in the epidermis, endothecium, middle layer, tapetum, vascular tissue, and microspore mother cell (Figures 5C–E). At stage 6, the signal became weak, and was clearly localized in the tapetum, microspore, and vascular region (Figures 5F–H). From stage 11, when secondary cell wall thickening begins, the TCP24 signal disappeared in the endothecium, but was still present in the vascular region (Figures 5I–L). The expression domains of miR319a were similar as those of TCP24 (Supplementary Figure S3). To address whether TCP24 overexpression causes the ectopic distribution of TCP24 in endothecium, we detected TCP24 in L23 anthers. The TCP24 expression pattern was the same as that of the wild type at stage 5, although the signal was much stronger than in the wild type (Supplementary Figure S4A). There was no ectopic signal in the endothecium during the secondary wall thickening process (Supplementary Figures S4B–D). These observations suggested that the lack of secondary wall thickening in the anthers of p35S:mTCP24 plants was due to the high level of TCP24 rather than its misexpression.
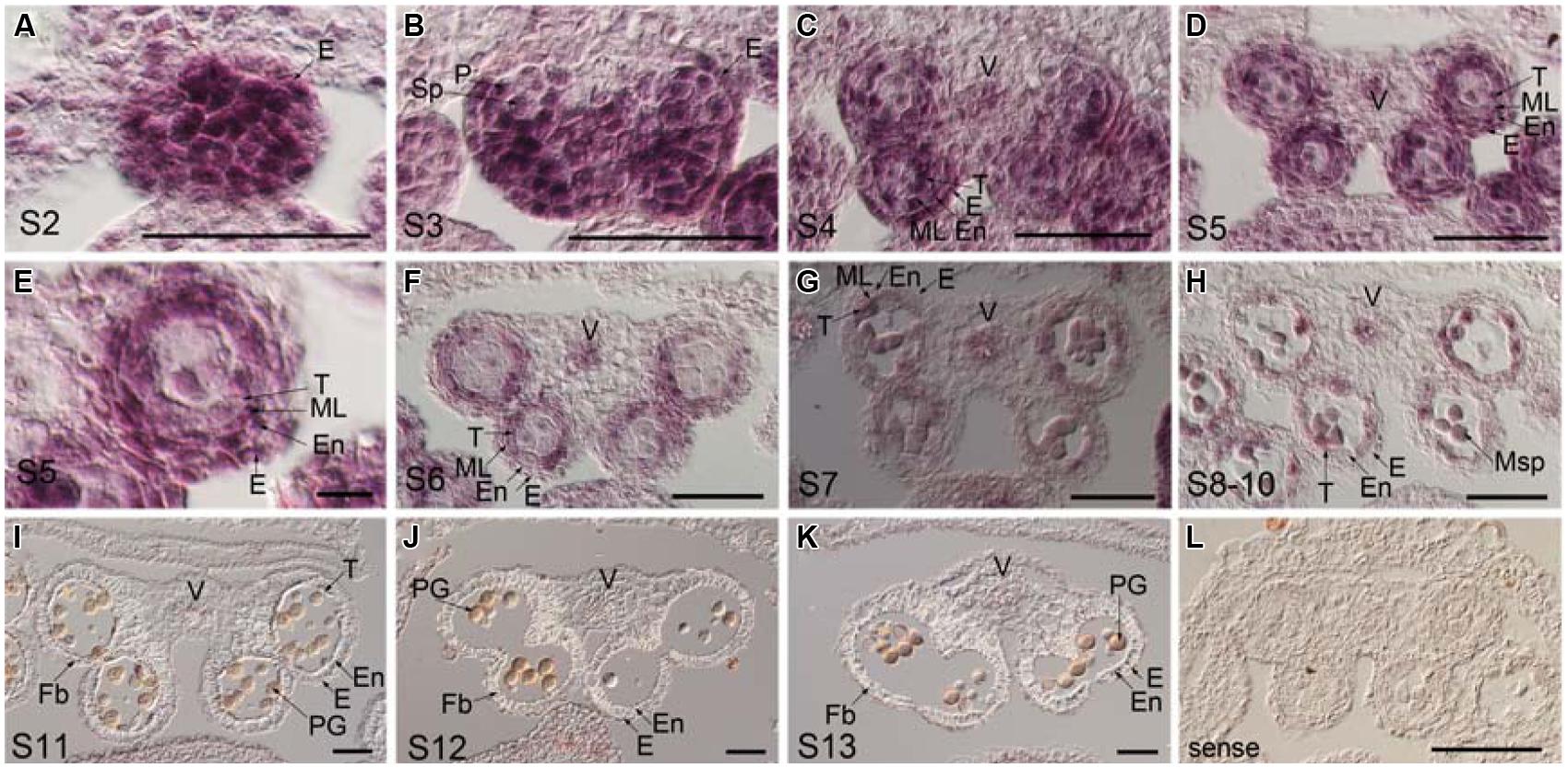
FIGURE 5. Temporal and spatial expression patterns of TCP24 gene during anther development. In situ hybridization using TCP24 probe. (A–K) Transverse sections of anthers at the stages 2 (A), 3 (B), 4 (C), 5 (D,E), and 6 (F), 7 (G), 8–10 (H), 11 (I), 12 (J), and 13 (K) antisense probes. (E) Magnified picture from the top right region in (D). (L) Sense probe of TCP24. E, epidermis; En, endothecium; Fb, fibrous bands; ML, middle layer; Msp, microspore; P, parietal cell; PG, pollen grain; Sp, sporogenous; T, tapetum; V, vascular region. Scale bars: 50 μm in (A–D, F–L); 5 μm in (E).
TCP24 Regulates the Genes Linked to Secondary Cell Wall Thickening
Secondary walls in the anther endothecium are composed of lignin and cellulose. Mutations of the genes involved in these biosynthesis processes cause non-dehiscent anthers (Brown et al., 2005; Thevenin et al., 2011). We examined the expression profiles of the genes involved in the biosynthesis of lignin (C4H, 4CL1, CCoAOMT, and PAL4) and cellulose (IRX1, IRX3, and IRX5) (Boerjan et al., 2003; Somerville, 2006). All of these genes were down-regulated in the flower buds of p35S:mTCP24 plants (Figure 6A). It was reported that mutations in MYB26, NST1, and NST2, as well as the overexpression of AHP4, resulted in the failure of anther dehiscence and that these genes act upstream to regulate secondary wall biosynthesis genes (Dawson et al., 1999; Steiner-Lange et al., 2003; Mitsuda et al., 2005; Yang et al., 2007). We found that TCP24 overexpression significantly reduced the expression of NST1 and NST2, but not MYB26. Unexpectedly, AHP4 was greatly up-regulated (Figure 6B). These results indicate that TCP24 negatively regulates secondary wall biosynthesis genes and is possibly upstream of these genes in the pathways.
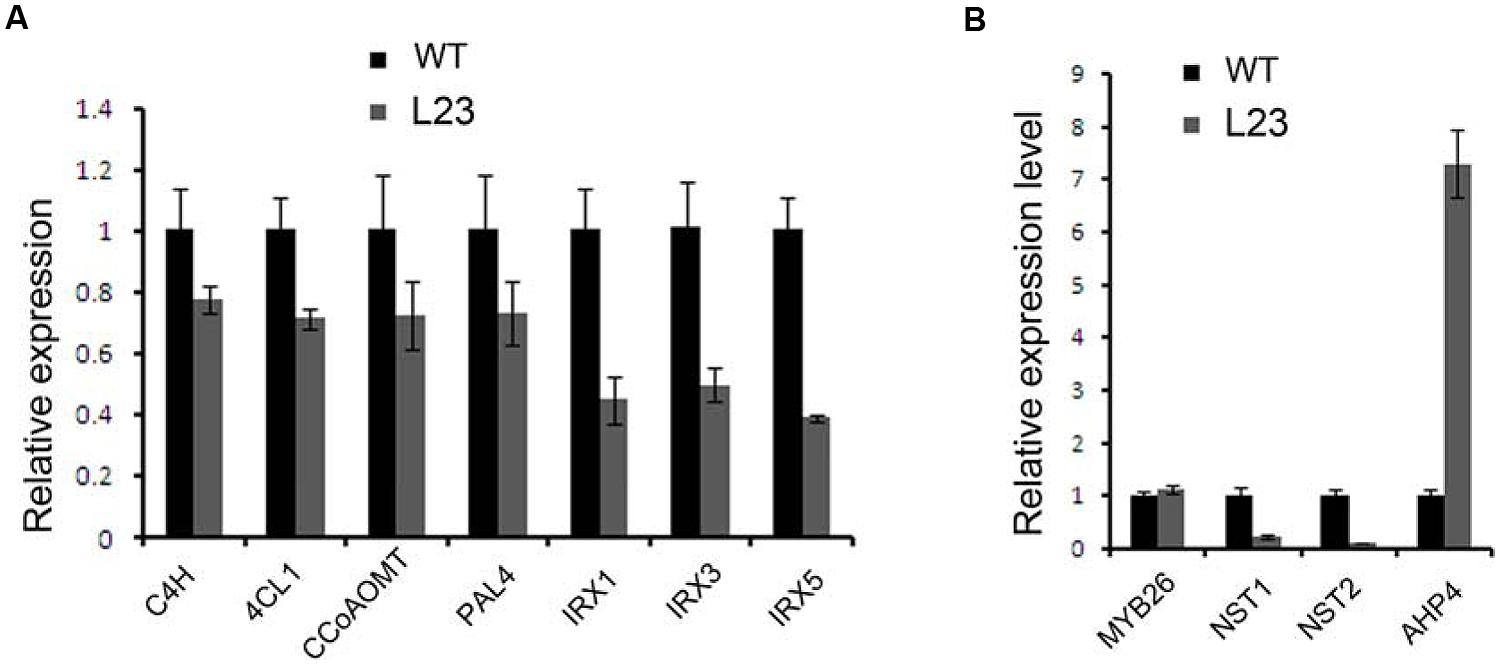
FIGURE 6. Regulation of TCP24 to the genes linked to secondary cell walls. (A) Relative expression levels of genes involved in the biosynthesis of lignin (C4H, 4CL1, CCoAOMT, PAL4) and cellulose (IRX1, IRX3, and IRX5). (B) Relative expression levels of the genes that regulate the secondary wall thickening. Error bars represent SD of three replicates.
TCP24 Silencing is Helpful for Thickening Anther Endothecium Secondary Cell Walls
To investigate whether secondary wall thickening was affected by TCP24, we carefully observed the anthers of jaw-D mutant plants in which miR319a-targeted TCP genes were down-regulated (Palatnik et al., 2003). In the flower buds of jaw-D, these TCP genes were down-regulated (Figure 7A). The epidermal tissues and the anther endothecium were particularly affected in the jaw-D mutant. The periclinal cell walls were thicker than the wild type (Figures 7B,C).
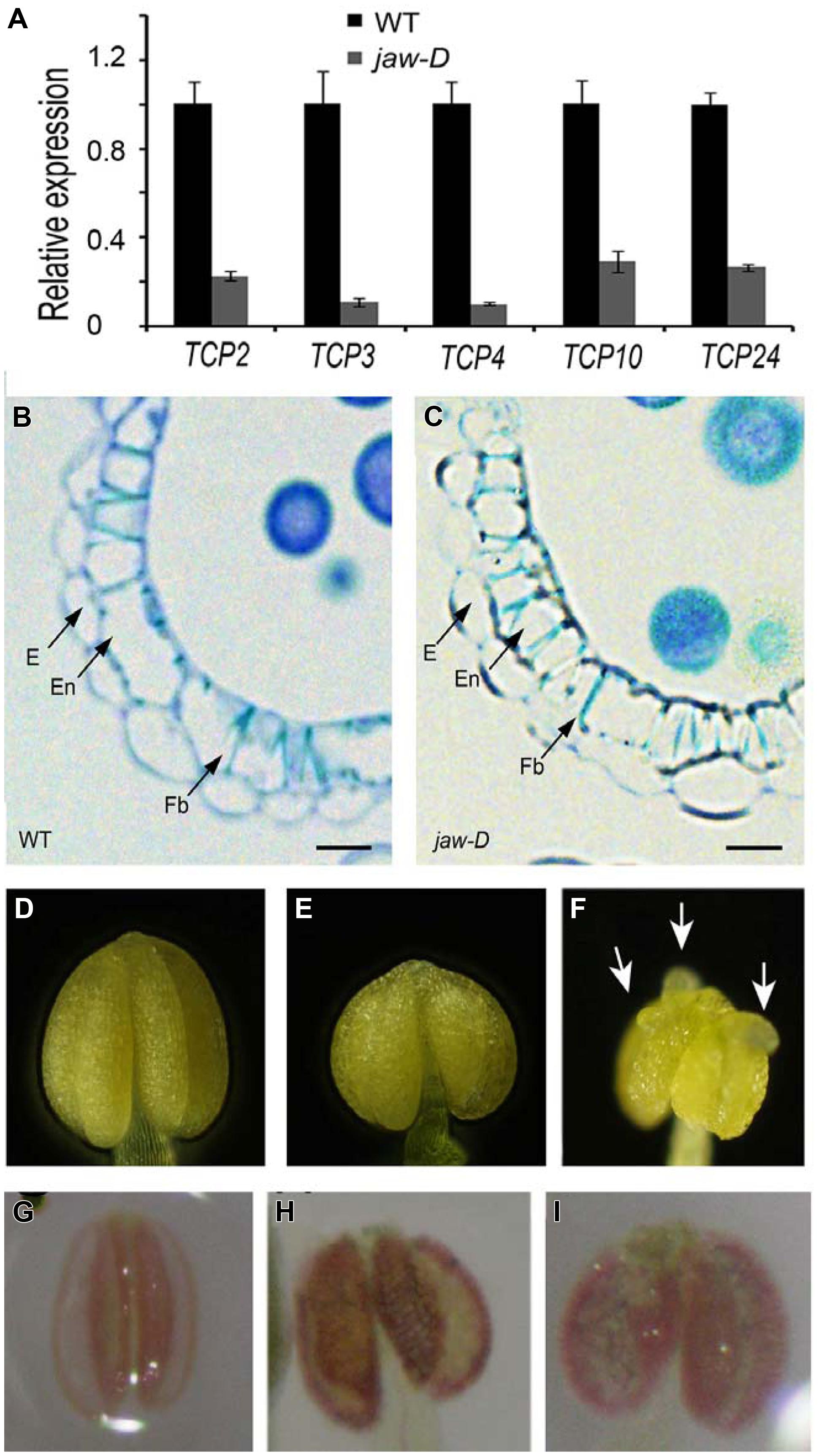
FIGURE 7. Secondary cell wall thickening of anther endothecium in the plants with TCP24 silencing. (A) Relative expression of miR319a-targeted TCP genes in flower buds of jaw-D mutants. (B,C) Transverse sections of the anthers in the wild type (B) and jaw-D (C) plants. (D–F) Anthers of the wild type (D) and the p35S:TCP24SRDX (E,F) plants. Arrows indicate protuberances. (G–I) Phloroglucinol staining of anthers showing secondary cell wall thickening of anther endothecium in the wild type (G) and the p35S:TCP24SRDX (H,I) plants (the strong phenotype in the left and weak phenotype in the right). Scale bars: 10 μm in (B,C).
To exclude the redundant effects of the other TCP genes, we created p35S:TCP24SRDX plants by fusing TCP24 with the SRDX repression domain. This approach converted transcription factors into dominant repressors, even in the presence of redundant genes (Hiratsu et al., 2003), and has been extensively used to study the functions of TCP genes (Koyama et al., 2007, 2010; Guo et al., 2010; Kieffer et al., 2011; Uberti-Manassero et al., 2012). A total of 48 independent transgenic lines were obtained. Their anthers were wider than the wild type (Figures 7D,E) and some had protuberances on their surface (Figure 7F). Phloroglucinol staining showed that lignification was enhanced and the endothecium layers were much thicker in the anthers of p35S:TCP24SRDX plants compared with the wild type (Figures 7G–I). This result indicates that the posttranscriptional silencing of TCP24 promotes the thickening of secondary cell walls in the anther endothecium.
Besides the anther endothecium, secondary wall thickening was observed in the other tissues using phloroglucinol staining. In the wild type roots, lignified secondary wall thickening was observed in vascular bundles but not in the parenchymatous cells (Figure 8A) as observed (Herve et al., 2009). In p35S:TCP24SRDX roots, however, it was seen in the parenchymatous cells as well (Figure 8B). In vascular bundles of the mature sepals and petals the transgenic plants exhibited stronger signals of lignified secondary wall thickening compared with the wild type plants (Figures 8C,D). These results indicate that TCP24 repression influences the ectopic thickening of the secondary walls in various tissues.
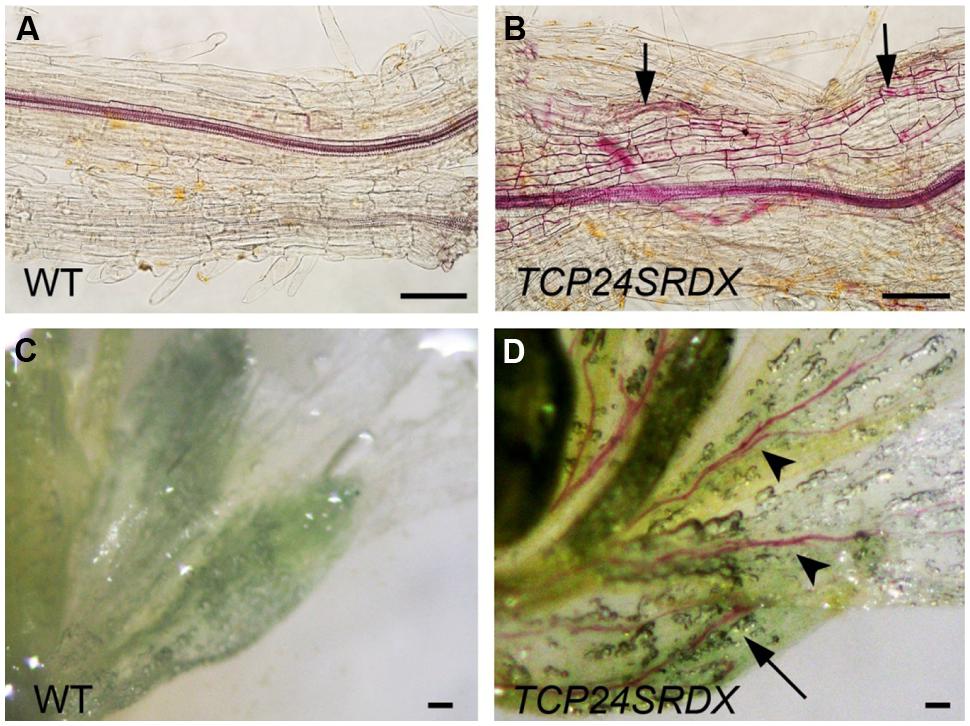
FIGURE 8. Ectopic thickening of secondary walls in p35S:TCP24SRDX plants. The tissues were stained with phloroglucinol staining to detect the lignified secondary wall thickening. (A,B) Roots of the wild type (A) and p35S:TCP24SRDX plants (B). Arrows in (B) indicate ectopic deposition of secondary walls. (C,D) Flowers of the wild type (C) and p35S:TCP24SRDX plants (D). Arrow and arrowheads in (D) indicate enhanced deposition of secondary walls in sepals (arrow) and petals (arrowheads). Scale bars: 100 μm.
Discussion
miR319a-targeted TCP genes may play important roles in controlling cell division and differentiation during leaf development (Palatnik et al., 2003; Koyama et al., 2007, 2010; Ori et al., 2007; Efroni et al., 2008; Li et al., 2012b). In this study, we found that miR319a-targeted TCP24 negatively regulates secondary cell wall thickening in the anther endothecium. This result suggests that miR319a-targeted TCP genes are multifunctional in their regulation of cell development. It has also been reported that TCP4 can bind to the LOX2 promoter, regulating leaf senescence by controlling the expression of jasmonic acid biosynthesis genes (Schommer et al., 2008). The proper level of active TCP4 is critical for petal and stamen development (Nag et al., 2009), and TCP2 and TCP3 interact with components of the core circadian clock (Giraud et al., 2010). Additionally, TCP3 interacts with R2R3-MYB proteins and participates in the flavonoid biosynthesis pathway (Li and Zachgo, 2013).
In transgenic plants containing p35S:mTCP24, secondary cell wall thickening does not occur in the anther endothecium. Overexpression of TCP24 under its native promoter also exhibits a similar phenotype. However, silencing TCP24 by enhanced miR319a expression or the repression of TCP24 using the SRDX repressor domain causes increased lignification and the deposition of secondary cell walls in the anther endothecium. Apparently, TCP24 represses the secondary cell wall thickening in the anther endothecium. In the wild type anthers, TCP24 strongly expresses in the endothecium when this cell layer is formed, and the expression weakens and eventually disappears when secondary wall thickening occurs. Clearly, TCP24 acts as a repressor of secondary wall thickening at the early stage of endothecium development.
Secondary wall thickening in the anther endothecium is important for anther dehiscence. Several processes, such as degeneration of the tapetum, septum, and breakage of stomium cells that affect dehiscence, occur normally in the transgenic plants. Pollen grains are fertile but they remain locked into the non-dehiscent anthers. Microscopic observation and histological staining suggest that this defect is due to the lack of secondary wall thickening in the anther endothecium. The importance of this process has been verified in several studies (Dawson et al., 1999; Steiner-Lange et al., 2003; Mitsuda et al., 2005; Yang et al., 2007; Jung et al., 2008; Kim et al., 2012).
A few genes have been linked to secondary thickening in anther endothecium. MYB26, NST1, and NST2 positively control secondary thickening by regulating the expression of secondary wall biogenesis genes, and AHP4 negatively regulates this process (Steiner-Lange et al., 2003; Mitsuda et al., 2005; Yang et al., 2007). Meanwhile, mutations in the genes that encode secondary wall biogenesis, such as IRX, 4CL3, CCR, and CAD, also result in failed secondary thickening, resulting in the non-dehiscent phenotype (Brown et al., 2005; Gui et al., 2011; Thevenin et al., 2011; Hao et al., 2014). Other mechanisms exist that can be illustrated by studying the function of mitochondrial gamma CA2 and CBSX2. p35S:CA2 plants cause a dramatic decrease in the reactive oxygen species production in anthers, which may impair H2O2-dependent lignin polymerization and deposition in the anther endothecium, resulting in a lack of secondary wall thickening in the endothecium (Villarreal et al., 2009). CBSX2 modulates the H2O2 status and may be linked to the jasmonic acid response, which in turn controls secondary wall thickening of the anther endothelial cells (Jung et al., 2013). In this study, we demonstrate that several genes linked to secondary wall biogenesis are down-regulated in TCP24-overexpressing plants. NST1 and NST2 are down-regulated and AHP4 is up-regulated in the transgenic plants. These results indicate that TCP24 acts upstream of the genes that promote secondary wall thickening. It has been reported that MYB26 is an upstream regulator of NST1 and NST2 (Yang et al., 2007). However, MYB26 transcripts are not changed in TCP24 overexpressing plants. We speculate that TCP24 functions in a MYB26-independent manner.
The deregulation of TCP24 causes defects not only in the anther endothecium, but also in roots and flower tissues. It has been reported that TCP proteins form homo- and heterodimers, and the latter bind DNA more efficiently than the former (Kosugi and Ohashi, 2002; Danisman et al., 2012). TCP4 can recognize the GGACCA motif, while TCP3 can activate downstream gene expression by directly binding the GGnCCC motif in the respective promoter (Schommer et al., 2008; Koyama et al., 2010). TCP24 may interact with other proteins and bind to the corresponding motifs in the targeted genes to execute its function. Further work is necessary to elucidate the molecular mechanisms of secondary cell wall thickening in plants.
Our results expand the classical roles of TCP24 in cell division. TCP24 regulates the genes that encode the enzymes responsible for secondary cell wall biogenesis, which modify cell walls. It will be interesting to determine whether TCP24 is involved in the relationship between cell division and cell wall development. Further studies on the TCP genes will provide insights into gene regulation pathways in cell differentiation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Basic Research Program (“973” Program) of China (Grant no. 2012CB113903), National High Technology Research and Development Program (“863”Program) of China (Grant no. 2012AA100104), and Natural Science Foundation of China (Grant no. 31200235).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00436
References
Aguilar-Martinez, J. A., Poza-Carrion, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bonner, L. J., and Dickinson, H. G. (1989). Anther dehiscence in lycopersicon-esculentum mill. New Phytol. 113, 97–115. doi: 10.1111/j.1469-8137.1989.tb02399.x
Brown, D. M., Zeef, L. A., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Danisman, S., Van Der Wal, F., Dhondt, S., Waites, R., De Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Dawson, J., Sozen, E., Vizir, I., Van Waeyenberge, S., Wilson, Z. A., and Mulligan, B. J. (1999). Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144, 213–222. doi: 10.1046/j.1469-8137.1999.00507.x
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Goldberg, R. B., Beals, T. P., and Sanders, P. M. (1993). Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229. doi: 10.1105/tpc.5.10.1217
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., and Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
Hao, Z., Avci, U., Tan, L., Zhu, X., Glushka, J., Pattathil, S., et al. (2014). Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 5:357 doi: 10.3389/fpls.2014.00357
Herr J. M. (1971). A new clearing squash technique for the study of ovule development in angiosperms. Am. J. Bot. 58, 785–790. doi: 10.2307/2441475
Herve, C., Dabos, P., Bardet, C., Jauneau, A., Auriac, M. C., Ramboer, A., et al. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149, 1462–1477. doi: 10.1104/pp.108.126136
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313X.2003.01759.x
Jung, K. W., Kim, Y. Y., Yoo, K. S., Ok, S. H., Cui, M. H., Jeong, B. C., et al. (2013). A cystathionine-beta-synthase domain-containing protein, CBSX2, regulates endothecial secondary cell wall thickening in anther development. Plant Cell Physiol. 54, 195–208. doi: 10.1093/pcp/pcs166
Jung, K. W., Oh, S. I., Kim, Y. Y., Yoo, K. S., Cui, M. H., and Shin, J. S. (2008). Arabidopsis histidine-containing phosphotransfer factor 4 (AHP4) negatively regulates secondary wall thickening of the anther endothecium during flowering. Mol. Cells 25, 294–300.
Keijzer, C. J. (1987). The processes of anther dehiscence and pollen dispersal. 1. the opening mechanism of longitudinally dehiscing anthers. New Phytol. 105, 487–498. doi: 10.1111/j.1469-8137.1987.tb00886.x
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kim, Y. Y., Jung, K. W., Jeung, J. U., and Shin, J. S. (2012). A novel F-box protein represses endothecial secondary wall thickening for anther dehiscence in Arabidopsis thaliana. J. Plant Physiol. 169, 212–216. doi: 10.1016/j.jplph.2011.09.006
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Koyama, T., Furutani, M., Tasaka, M., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Li, C., Potuschak, T., Colon-Carmona, A., Gutierrez, R. A., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, S. X., Yang, X., Wu, F. J., and He, Y. K. (2012a). HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J. Exp. Bot. 63, 2787–2798. doi: 10.1093/jxb/err465
Li, Z., Li, B., Shen, W. H., Huang, H., and Dong, A. (2012b). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. doi: 10.1111/j.1365-313X.2012.04973.x
Li, S., and Zachgo, S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913. doi: 10.1111/tpj.12348
Liu, Z. Y., Jia, L. G., Wang, H., and He, Y. K. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62, 4367–4381. doi: 10.1093/Jxb/Err167
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mizuno, S., Osakabe, Y., Maruyama, K., Ito, T., Osakabe, K., Sato, T., et al. (2007). Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50, 751–766. doi: 10.1111/j.1365-313X.2007.03083.x
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. doi: 10.1038/Ng2036
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Sanders, P. M., Bui, A. Q., Weterings, K., Mcintire, K. N., Hsu, Y. C., Lee, P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. doi: 10.1007/s004970050158
Sarvepalli, K., and Nath, U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67, 595–607. doi: 10.1111/j.1365-313X.2011.04616.x
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Somerville, C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22, 53–78. doi: 10.1146/annurev.cellbio.22.022206.160206
Steiner-Lange, S., Unte, U. S., Eckstein, L., Yang, C. Y., Wilson, Z. A., Schmelzer, E., et al. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34, 519–528. doi: 10.1046/j.1365-313X.2003.01745.x
Tao, Q., Guo, D., Wei, B., Zhang, F., Pang, C., Jiang, H., et al. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25, 421–437. doi: 10.1105/tpc.113.109223
Thevenin, J., Pollet, B., Letarnec, B., Saulnier, L., Gissot, L., Maia-Grondard, A., et al. (2011). The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 4, 70–82. doi: 10.1093/mp/ssq045
Uberti-Manassero, N. G., Lucero, L. E., Viola, I. L., Vegetti, A. C., and Gonzalez, D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823. doi: 10.1093/jxb/err305
Villarreal, F., Martin, V., Colaneri, A., Gonzalez-Schain, N., Perales, M., Martin, M., et al. (2009). Ectopic expression of mitochondrial gamma carbonic anhydrase 2 causes male sterility by anther indehiscence. Plant Mol. Biol. 70, 471–485. doi: 10.1007/s11103-009-9484-z
Wilson, Z. A., Song, J., Taylor, B., and Yang, C. Y. (2011). The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649. doi: 10.1093/Jxb/Err014
Keywords: Arabidopsis, TCP24, male sterility, anther dehiscence, secondary wall thickening, SRDX
Citation: Wang H, Mao Y, Yang J and He Y (2015) TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 6:436. doi: 10.3389/fpls.2015.00436
Received: 26 February 2015; Accepted: 27 May 2015;
Published: 24 June 2015.
Edited by:
Ravishankar Palanivelu, University of Arizona, USAReviewed by:
Derek William Richard White, AgResearch Limited, New ZealandSimona Masiero, Università degli Studi di Milano, Italy
Copyright © 2015 Wang, Mao, Yang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuke He, National Key Laboratory of Plant Molecular Genetics, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Fenglin Road 300, Shanghai 200032, China,eWtoZUBzaWJzLmFjLmNu
 Han Wang
Han Wang Yuke He
Yuke He