- 1State Key Laboratory of Tree Genetics and Breeding, Key Laboratory of Tree Breeding and Cultivation of the State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
- 2Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 3Research Institute of Wood Industry, Chinese Academy of Forestry, Beijing, China
- 4Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China, University of Chinese Academy of Sciences, Beijing, China
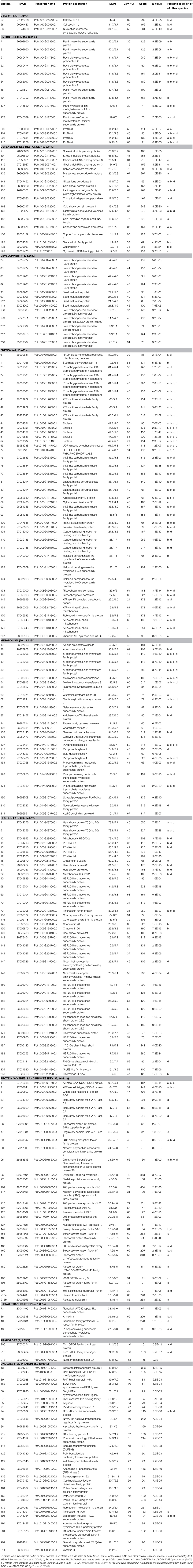
Proteomic analysis was used to generate a map of Populus deltoides CL. “2KEN8” mature pollen proteins. By applying 2-D electrophoresis, we resolved 403 protein spots from mature pollen. Using the matrix-assisted laser desorption/ionization time time-of-flight/time-of-flight tandem mass spectrometry method, we identified 178 distinct proteins from 218 protein spots expressed in mature pollen. Moreover, out of these, 28 proteins were identified as putative allergens. The expression patterns of these putative allergen genes indicate that several of these genes are highly expressed in pollen. In addition, the members of profilin allergen family were analyzed and their expression patterns were compared with their homologous genes in Arabidopsis and rice. Knowledge of these identified allergens has the potential to improve specific diagnosis and allergen immunotherapy treatment for patients with poplar pollen allergy.
Introduction
In spermatophytes, pollen grains are the dispersal agents of sperm cells and are vital for successful sexual reproduction and subsequent seed and fruit production (Sheoran et al., 2007). The development of pollen, microsporogenesis and microgametogenesis involves the coordinated expression of several genes in different tissues of an anther (Mccormick, 2004; Ma, 2005), and pollen grains at maturity contain a large number of transcripts with designated roles in cell wall metabolism, cytoskeleton formation, cell signaling, and vesicle transport (Becker et al., 2003; Honys and Twell, 2003; Pina et al., 2005). It has been reported that pollens are a major cause of Type I allergies due to the presence of several allergens (Nakamura and Teshima, 2013). Manifestation of allergic diseases, spanning from mild rhinitis to anaphylaxis is a major health problem, affecting the quality of life of millions of people all over the world (Sircar et al., 2012). More than 30% of the world population is affected by different kinds of allergy, caused by naturally occurring as well as synthetically produced compounds and their prevalence is increasing daily (Singh and Shahi, 2008).
The proteome is the entire set of proteins expressed by a genome, cell, tissue, or organism (Wilkins et al., 1996). It is highly dynamic and depends on cell cycle, environmental influences and tissue/cell type. Rapid advances in proteomic technologies, along with completion of many plant species genome sequencing projects and availability of comprehensive public sequence databases, have provided tremendous impetus to plant proteomics research (Canovas et al., 2004; Hirano et al., 2004).
In recent years, proteomic techniques that target protein allergens, i.e., allergenomics, emerged as powerful tools for comprehensive allergen analysis (Akagawa et al., 2007; Picariello et al., 2011). At first, proteomic analyses were used to detect novel allergens by identifying proteins following separation by 2-DE and MS (Nakamura and Teshima, 2013). Compared to conventional methods based upon protein isolation processes, proteomics has accelerated identification of numerous allergens in plants. Furthermore, novel allergenomics techniques, which consider the properties (biochemical, structural, reactivity) of the allergens, have been developed (Kitta et al., 2006; Yano and Kuroda, 2008; Shahali et al., 2012). However, up to recent times, very little was known about the molecular basis of poplar pollen allergy, one of the more common causes of allergy symptoms, particularly in spring.
On this basis, the objective of the present study was to identify likely allergenic proteins of Populus pollen. The genus Populus contains approximately 30 species of woody plants, all found in the Northern hemisphere and exhibiting some of the fastest growth rates observed for trees growing in temperate climate (Taylor, 2002). P. deltoides is a poplar species with a high yield, fine wood quality, strong adaptability and disease resistance. Therefore, it is used widely worldwide as an important woody species. However, P. deltoides releases large amounts of pollens in spring and these pollens are surmised to cause the allergenic response in human. Such pollens belong to the most important elicitors of allergy in adults and adolescents (Vieths et al., 2002). Allergy is an adverse reaction to normally harmless substances, such as allergens, by the immune system, and it involves immune response mediated by an increased amount of immunoglobulin E (IgE) or IgG antibodies (Bohle, 2004). Once sensitized, human can become allergic to homologous proteins of other pollens or present in food, via cross-reactivity (Vieths et al., 2002). The inhalation of pollen from several birch trees and grasses is the main cause of primary sensitization in humans (Bartra et al., 2009).
The majority of allergens present in plants belong to four families: pathogenesis-related protein 10 (PR-10 protein, birch allergen Bet v 1 homologs), thaumatin-like proteins (TLP, PR-5 proteins), non-specific lipid transfer proteins (nsLTPs, PR-14 proteins) and profilins (PRF) (Breiteneder and Ebner, 2000). Due to the common structure and properties of such allergens over a wide range of plant species, genera and even families, allergy cross-reactivity has been frequently observed.
By combining two-dimensional gel electrophoresis (2-DE) with matrix-assisted laser desorption/ionization time time-of-flight/time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS/MS), and by using the available databases for P. trichocarpa and other plant species, a comprehensive analysis of P. deltoides CL. “2KEN8” mature pollen proteome was performed in the present work. Many of the proteins identified in this study have predicted roles in defense mechanisms, energy conversion, pollen germination, and pollen tube growth, and possibly in sperm cell formation. To our knowledge, there has been no previous proteomic study to predict the pollen allergens of poplar. Thus, we aimed to identify expressed proteins and the likely allergens in poplar mature pollen.
Materials and Methods
Plant Materials and Pollen Collection
“2KEN8” trees, one of the widely grown high-yield P. deltoids in China were obtained from nursery of Chinese Academy of Forestry, Beijing, China. The flowering branches were cut and then placed in water in a greenhouse. At anthesis, fresh pollen was collected in the morning by shaking the tassel in a plastic bag, while old pollen and anthers were removed from tassels by vigorous shaking the evening of the day before. For RNA isolation and qRT-PCR, four tissues (leaf, stem, root, and pollen) were collected from P. deltoids. Samples were frozen immediately in liquid nitrogen, and stored at −80°C for further analysis. Three biological replicates were performed.
Protein Extraction
The pollen samples (~0.3 g) were ground to a fine powder in a pestle and mortar in liquid nitrogen, and extracted with acetone containing 10% (w/v) TCA (for electrophoresis, Sigma-Aldrich) and 1% (w/v) DTT (biotechnology grade, Amersco). The samples were kept at −20°C for at least 2 h. The samples were centrifuged at 25,000 g for 20 min at 4°C, and the resulting pellets were washed by suspending in acetone containing 1% (w/v) DTT, incubated at −20°C for 2 h, and centrifuged as above. The pellets were suspended again in acetone, sonicated (15 s duration, 3 times with 5 min intervals) on ice at 200 W in 6 mm ultrasonic probe in JY92-II DN sonicator homogenizer (Ningbo Scientz Biotechnology Co, China), and centrifuged at 25,000 g as above. The pellets were vacuum dried and total soluble proteins were extracted by dissolving in isoelectric focusing (IEF) compatible buffer comprising 8 M urea, 20 mM DTT, 4% (w/v) CHAPS (ultrapure bioreagent, Sigma), and 2% (v/v) ampholytes (pH 4–7, GE Healthcare). Solution were vortexed extensively for 1 h at room temperature, centrifuged at 4°C for 20 min at 25,000 g, and the supernatants were collected. The resulting pellets were resolubilized and vortexed for 1 h, centrifuged at 25,000 g (20 min, 4°C), and the supernatants combined with those collected earlier. The resulting protein samples were centrifuged again for 20 min at 25,000 g (4°C). Total soluble protein in the supernatants was estimated with Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA) and used immediately for further analysis or stored at −80°C for later use.
Two-dimensional Gel Electrophoresis (2-DE)
2-DE was carried out as previously described (Sheoran et al., 2006). IEF was performed using the Ettan III system (GE Healthcare) and 18-cm Immobiline Dry Strips of 4–7 linear pH gradients (GE Healthcare, OK, USA). The strips were rehydrated overnight in a solution containing 8 M urea, 2% (w/v) CHAPS, 20 mM DTT, 0.002% (w/v) bromophenol blue, 2% (w/v) IPG buffer (pH 4–7), and 600 μg of the protein sample. IEF was carried out by applying a voltage of 250 V for 1 h, followed by an increase to 3500 V over 2 h, and holding at 8000 V until a total of 60 kVh was obtained.
Following IEF, the strips were equilibrated for 15 min in a buffer containing 0.1 M Tris-HCl (pH 8.8), 2% (w/v) SDS (proteomics grade, Amresco), 6 M urea, 30% (v/v) glycerol (biotechnology grade, Amersco) and 0.1 M DTT, and for another 15 min in the same buffer containing 0.25 M iodoacetamide without DTT. The equilibrated strips were applied to vertical SDS–polyacrylamide gels (12.5% resolving 5% stacking) and sealed with 0.5% agarose in SDS buffer (see below for the composition) containing bromophenol blue. Electrophoresis was performed in two gels for 30 min at 10 mA gel−1, and then at 30 mA gel−1 until the dye front reached the bottom of the gels, in an SDS electrophoresis buffer containing 25 mM TRIS base, 192 mM glycine, and 0.1% (w/v) SDS, pH 8.3 in a PROTEAN II XL multi-cell (Bio-Rad, USA).
Gel Staining and Image Analysis
Gels were fixed overnight in 50% (v/v) ethanol with 10% (v/v) orthophosphoric acid, washed with water (1 h), and stained with Colloidal Coomassie Blue G-250 (CCB) as described earlier (Sheoran et al., 2006). Images of the stained gels were captured with a scanner (UMAX Powerlook 2100 XL; UMAX, Taiwan, China) and analyzed with ImageMaster 2D Platinum Software (Version 6.0; Amersham Biosciences, Uppsala, Sweden). Two replicate gels were run for each of three different pooled pollen samples collected from different batches of plants.
MALDI-TOF/TOF MS/MS
Selected spots were excised manually from the 2-DE gels and automatically de-stained, dehydrated, reduced with DTT, alkylated with iodoacetamide, and digested with gold grade trypsin (Mass grade, Promega) using a MassPREP protein digest station (Micromass, Manchester, UK) according to the recommended procedure. Samples were then analyzed by a MALDI-TOF/TOF tandem mass spectrometer ABI 4800 proteomics analyzer (Applied Biosystems, Framingham, MN). For acquisition of mass spectra, 0.4 μl samples were spotted onto a MALDI plate, followed by 0.4 μl matrix solution [0.5 M CHCA in 50% (v/v) ACN (HPLC grade, Fisher) and 0.05% (v/v) TFA (HPLC grade, Merck)]. Mass data were acquired with 4000 Series Explorer Software v3.5 in batch-processing mode of MS/MS. All MS survey scans were acquired over the mass range m/z 800–4000 in the reflection positive-ion mode. The MS peaks were detected on minimum S/N ratio ≥10 and cluster area S/N threshold ≥40 without smoothing and raw spectrum filtering. Peptide precursor ions corresponding to contaminants including keratin and the trypsin autolytic products were excluded in a mass tolerance of 0.5 Da.
Database Search, Annotation, and Allergen Prediction
For protein identification, the acquired MS/MS data were uploaded on the Protein Pilot software (Applied Biosystems, Framingham, MN) and compared against P. trichocarpa genome database (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ptrichocarpa), NCBI non-redundant protein sequence database (NCBI-nr) and Swiss-Prot database. Searches were performed using the following parameters: trypsin as the proteolytic enzyme, allowing for one missed cleavage; carbamidomethylation of cysteine as a fixed modification; oxidation of methionine as a variable modification. Proteins identified with a Mowse score greater than 60 (significant at 95% confidence interval) are reported.
To annotate the identified proteins with Gene Ontology (GO) terms, the sequences were imported into Blast2GO (Conesa et al., 2005), a software package that retrieves GO terms, allowing gene functions to be determined and compared. These GO terms are assigned to query sequences, producing a broad overview of groups of genes catalogs into three ontology vocabularies, biological processes (BP), molecular functions (MF), and cellular components (CC). The output GO terms were then slimmed in REVIGO and treemaps were produced (Supek et al., 2011).
Allergen prediction were realized by using the SDAP-Structural Database of Allergenic Proteins (http://fermi.utmb.edu/SDAP/index.html) under the two conditions of (1) sequence similarity >35% between presently obtained proteins and reported allergen proteins and (2) the presence of at least eight consecutive amino acids in the analyzed protein sequences compared to known allergen proteins (Ivanciuc et al., 2003). Furthermore, predictions for antigenicity were realized using the online software (http://imed.med.ucm.es/Tools/antigenic.pl) based on the algorithm of Kolaskar and Tongaonkar (1990). By these criteria, some of the presently analyzed poplar mature pollen proteins were declared as likely corresponds to allergen related proteins.
Publicly Available Microarray Data Analyses
Microarray data for various tissues were available at NCBI Gene Expression Omnibus (GEO) database (http://www.ncibi.nlm.nih.gow/geo/), notably under the series accession number GSE21481 (for P. trichocarpa). Probe sets corresponding to selected genes were identified using the online Probe Match tool POParray (http://aspendb.uga.edu/poparray). For genes with more than one probe sets, the median of expression values was considered. The expression data were normalized by the Gene Chip Robust Multiarray Analysis (GCRMA) algorithm followed by log transformation and average calculation. Normalized values were extracted for further analyses.
Sequence Alignments and Phylogenetic Analyses
Multiple alignment of profilin protein sequences from poplar, Arabidopsis, and rice were performed using the Clustal X2.1 program (Larkin et al., 2007). Phylogenetic trees were constructed using the neighbor-joining method in the MEGA package V5.2 (Tamura et al., 2011) with bootstrap values from 1000 replicates indicated at each node. Secondary structures of proteins were predicted using the Protein Structure Prediction Server (PRIPRED, http://bioinf.cs.ucl.ac.uk/psipred/).
RNA Isolation and Real-time qRT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with on-column treatment using RNase-free DNase I (Qiagen) to remove any contamination of genomic DNA. First-strand cDNA synthesis was carried out with approximately 1 μg RNA using the SuperScript III reverse transcription kit (Invitrogen) and random primers according to the manufacturer's procedure. Primers with melting temperatures of 58–60°C and amplicon lengths of 100–250 bp were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm). All primer sequences used are listed in Table S1. qRT-PCR was conducted on LightCycler 480 Detection System (Roche, Penzberg, Germany) using SYBR Premix Taq Kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. The PtActin gene was used as internal control.
Results and Discussion
Proteomic Maps of P. deltoides Mature Pollen
After the 2-DE gels were aligned and matched, a total of 403 reproducible protein spots were detected in P. deltoides CL. “2KEN8” mature pollen. These proteins cover the pI range from 4 to 7 and the MW range from 5 to 120 kDa (Figure 1). All detected protein spots were processed by automated in-gel tryptic digestion and MALDI-TOF/TOF MS/MS analysis. After searching various publicly available protein databases, out of the 403 detected protein spots, 218 spots allowed the identification of 178 different proteins. Table 1 lists each of the identified proteins by its PACid number and corresponding P. trichocarpa gene locus, as obtained from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ptrichocarpa). The calculated MW of the identified proteins ranged from 6.9 to 106.0 kDa, and the calculated pI ranged from 4.26 to 8.95, which is close to the experimental data as judged from the location of the spots on the 2-DE gels (Table 1 and Figure 1).
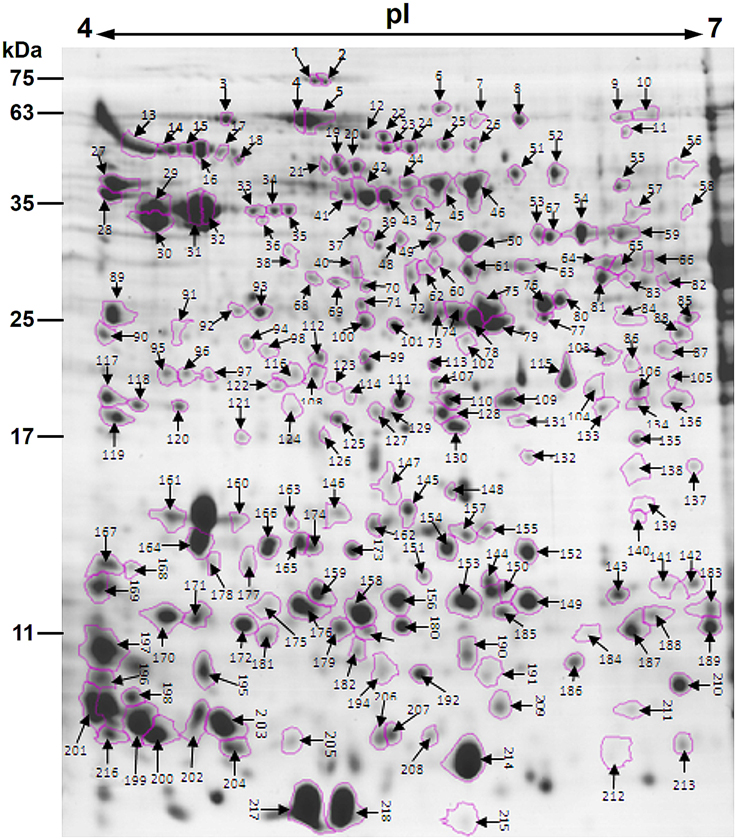
Figure 1. Colloidal Coomassie stained 2-DE gel (pH 4–7) of P. deltoides CL. “2KEN8” mature pollen. Spot numbers indicated on the gel were subjected to MALDI-TOF/TOF MS/MS analysis. Standard molecular weight markers are shown on the left.
Proteomic analyses showed that correlations between identified proteins and spot locations on the gels 2D-might not be one-to-one. In particular, expression of a given gene can give rise to several polypeptides located in different gel spots (Noir et al., 2005; Zou et al., 2009). Consistent with this, the present study showed that 40 proteins were associated with different spots (Table 1). There might be several possibilities to account for different electrophoretic migrations of the same protein in a 2D-gel. One possibility corresponds to post-translational modifications of the proteins, such as phosphorylation, methylation, glycosylation, or acetylation (Jensen, 2004). In general, these modifications do not significantly affect the MW of a protein, but induce a pI shift on the protein spot on the gel (Holmes-Davis et al., 2005; Noir et al., 2005). Another possibility relies on alternative splicing of mRNAs during translation (Brett et al., 2002) or on partial synthesis of proteins during pollen maturation (Loraine et al., 2013).
A previous study identified 135 proteins from Arabidopsis mature pollen proteome (Holmes-Davis et al., 2005), of which 74 were presently detected in P. deltoids mature pollen (Table 1). Another study identified 121 proteins from Arabidopsis mature pollen proteome (Noir et al., 2005), of which 86 were presently detected in P. deltoids mature pollen (Table 1). Zou et al. (2009) by using 2-DE and MALDI-TOF MS identified 189 distinct proteins from Arabidopsis mature pollen and pollen tubes, and 98 of them were detected in the present study (Table 1). In tomato, 133 proteins were identified from tomato pollen (Sheoran et al., 2007), and 56 of them were also detected in P. deltoids mature pollen in our present study. Therefore, despite differences that may be attributed to the methods of protein extraction, choices of immobilized pH gradient (IPG) strips with different pH ranges, spot selection for MS analysis and the different plant species used, our present data support the finding that the P. deltoids mature pollen proteome exhibits common features with mature pollen proteomes from other plant species. However, it is noted that in our study, 89 new proteins were identified that were not reported before in the previous studies mentioned above (Table 1). Most of these newly identified proteins were classified into development and protein fate. Interestingly, a large number of sHsps (spots No. 40, 68, 69, 70, 142, 143, 144, 150, 151, 152, 156, 165, 166, 184, 187, and 188) were identified in P. deltoides mature pollen. It has been reported that sHsp expression is developmentally induced in tobacco pollen. Notably, it appears that different subsets of cytosolic sHsp genes are expressed in a stage-specific fashion suggesting that certain sHsp genes may play specific roles in early stages of pollen development, while others may play a role in later stages, e.g., desiccation tolerance (Volkov et al., 2005). Moreover, we found that spots No. 96, 109, 120, 127, 128, and 134 are proteasome-related proteins, in agreement with the finding that the ubiquitin/proteasome pathway is involved in pollen tube growth (Sheng et al., 2006). Most of the protein fate proteins seem to correspond to Hsps. This presumably reflects a remnant process of late pollen development allowing desiccation tolerance. The majority of identified proteins are preferentially implicated in the determination of protein fate (17.57%) rather than in protein synthesis (14.41%, Table 1). It has been demonstrated that Arabidopsis pollen is charged with stored mRNA and performed translation apparatus enabling rapid activation upon hydration and germination (Honys and Twell, 2003) as it was established for seeds (Rajjou et al., 2004; Galland et al., 2014). Our results support this concept.
Functional Classification of Identified Proteins
To assign functional information to the presently identified proteins, we first classified them into functional groups according to previous proteomic analyses (Holmes-Davis et al., 2005; Noir et al., 2005; Sheoran et al., 2007; Zou et al., 2009). As shown in Table 1, the three major groups of identified proteins in P. deltoides mature pollen were involved in energy regulation (18.47%), protein fate (17.57%), protein synthesis and processing (14.41%), and metabolism (11.71%) in good agreement with previous studies showing that the majority of proteins expressed in mature Arabidopsis pollen are involved in energy and general metabolism (Holmes-Davis et al., 2005; Noir et al., 2005; Sheoran et al., 2006). Furthermore, GO analysis was carried out, which provides a dynamic, controlled vocabulary, and hierarchical relationships for the representation of information on biological process (BP), molecular function (MF), and cellular component (CC), allowing a coherent annotation of genes and their products (Ashburner et al., 2000). For BP, cellular metabolic process (GO:0044237, 107 proteins) was the most represented GO term, followed by macromolecule metabolic process (GO:0043170, 80 proteins) and response to stress (GO:0006950, 67 proteins) (Figure 2A). Regarding MF, proteins with catalytic activity (GO:0003824, 82 proteins) and ion binding (GO:0043167, 38 proteins) were highly represented (Figure 2B). For CC, the most represented category was intracellular (GO:0005622, 132 proteins), cytoplasm (GO:0005737, 126 proteins), and membrane-bounded organelle (GO:0043227, 94 proteins) (Figure 2C).
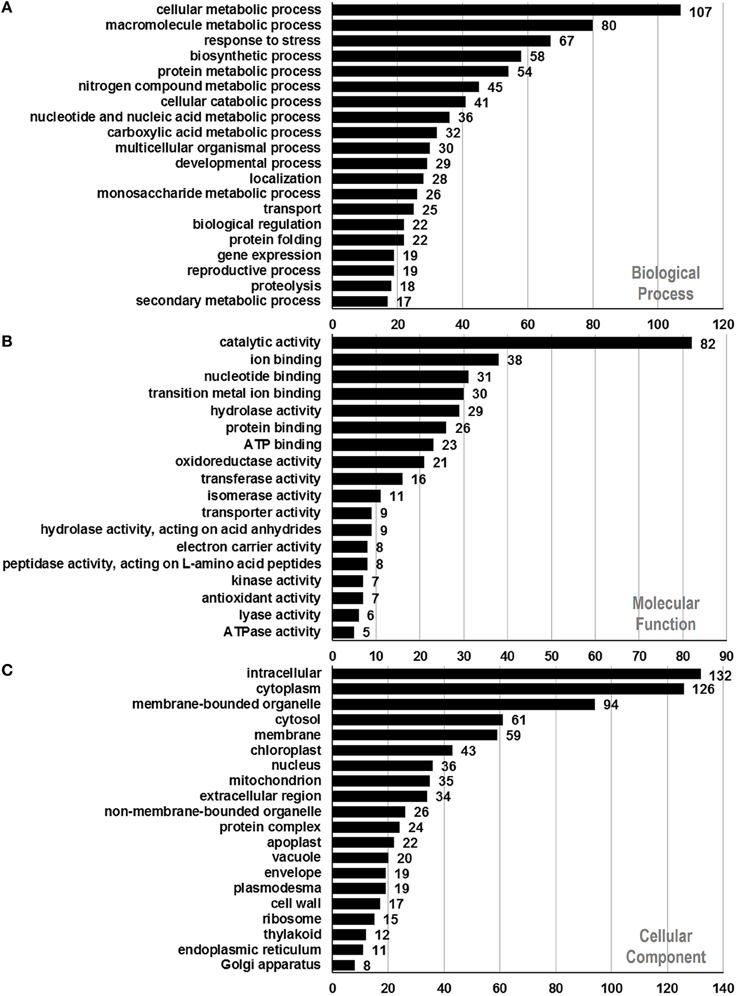
Figure 2. Functional annotation of identified proteins based on GO categorization. Results are summarized for three main GO categories. (A) Biological process, (B) molecular function, and (C) cellular component. The number corresponding to each column indicates the number of proteins in this GO term.
To gain further insight into functional classification of proteins present in the P. deltoides mature pollen proteome, the GO terms along with their P-values were further summarized independently by the REVIGO reduction analysis tool that condenses the GO description by removing redundant terms (Supek et al., 2011). The results of these further reductions are visualized in Figure 3. For categories based on BP, the translational elongation and hexose metabolism processes were main GO terms in P. deltoides mature pollen. For categories based on CC, the identified proteins were mainly related with mitochondrial function.
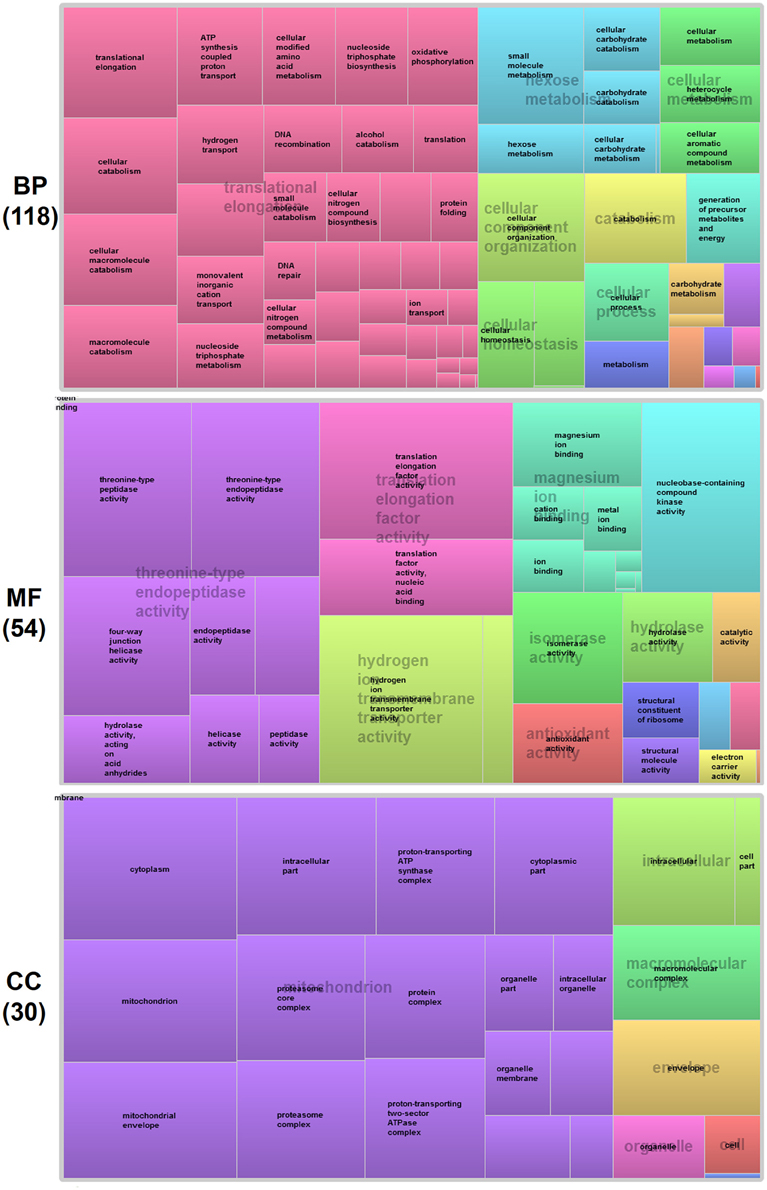
Figure 3. GO treemaps for the identified proteins in P. deltoides pollen. GO terms for proteins identified in P. deltoides pollen are shown. The box size correlates to the -log10 P-value of the GO-term. Boxes with the same color can be grouped together and correspond to the same upper-hierarchy GO-term which is found in the middle of each box.
Prediction of Allergens in P. deltoides Mature Pollen
Pollen grains are known to contain a number of proteins that can act as allergens (Mohapatra and Knox, 1996). Poplar trees release large amounts of pollen in spring that might cause the allergenic response. To date, many sequences and structures of allergenic proteins have been determined. Most of them can be grouped into a few families (Aalberse, 2000; Breiteneder and Ebner, 2000), suggesting that they share common characteristics that contribute to their ability to bind IgE and trigger an allergic reaction (Ipsen and Løwenstein, 1997; Sicherer, 2001).
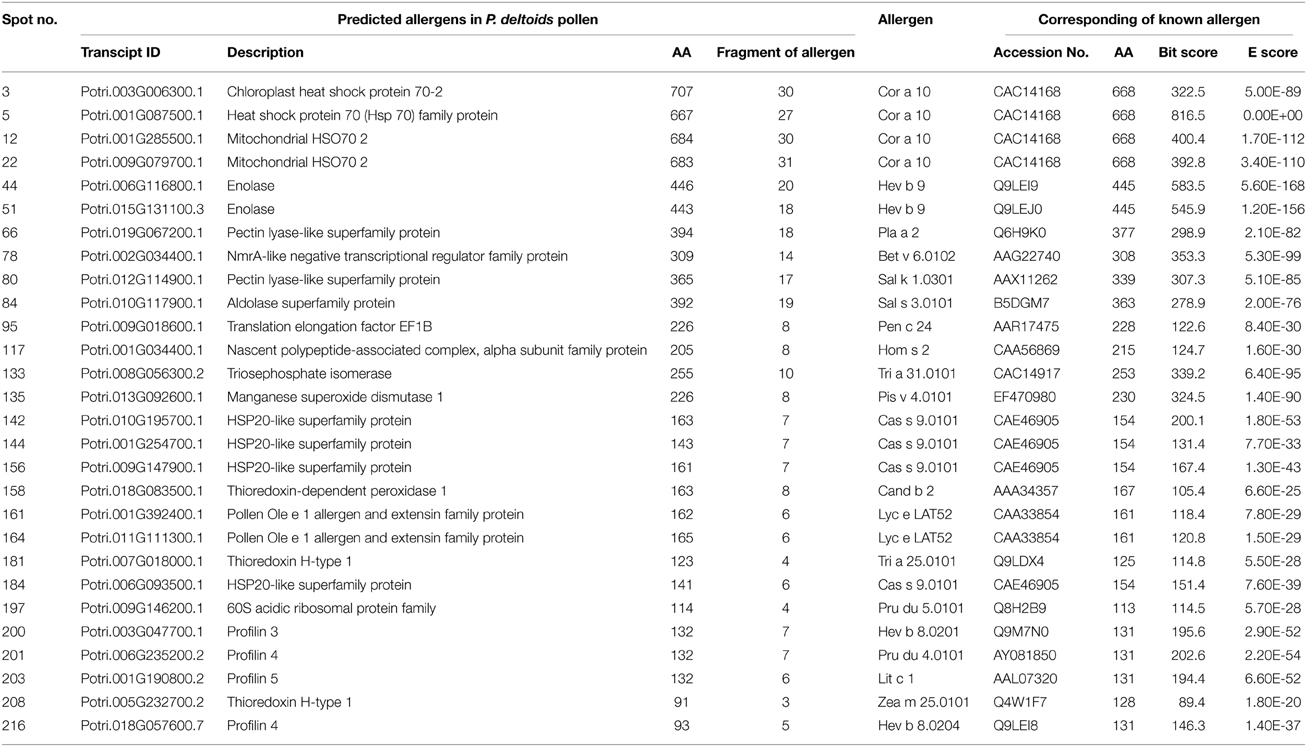
To identify the likely allergen proteins present in P. deltoides mature pollen, the currently identified proteins were searched through SDAP, a web server that provides rapid, cross-referenced access to the sequences, structures and IgE epitopes of allergenic proteins (Ivanciuc et al., 2003). In this way, 28 mature pollen poplar proteins were predicted as being candidate allergens (Table 2). Then the potential antigenic peptides were determined using the method of Kolaskar and Tongaonkar (1990). Here, predictions are based on a table that reflects the occurrence of amino acid residues in experimentally known segmental epitopes. Segments are only reported if they have a minimum size of eight residues. From this search, the predicted antigenic peptides in the 28 predicted P. deltoides antigen proteins are shown in Figure 4 and Figure S1.
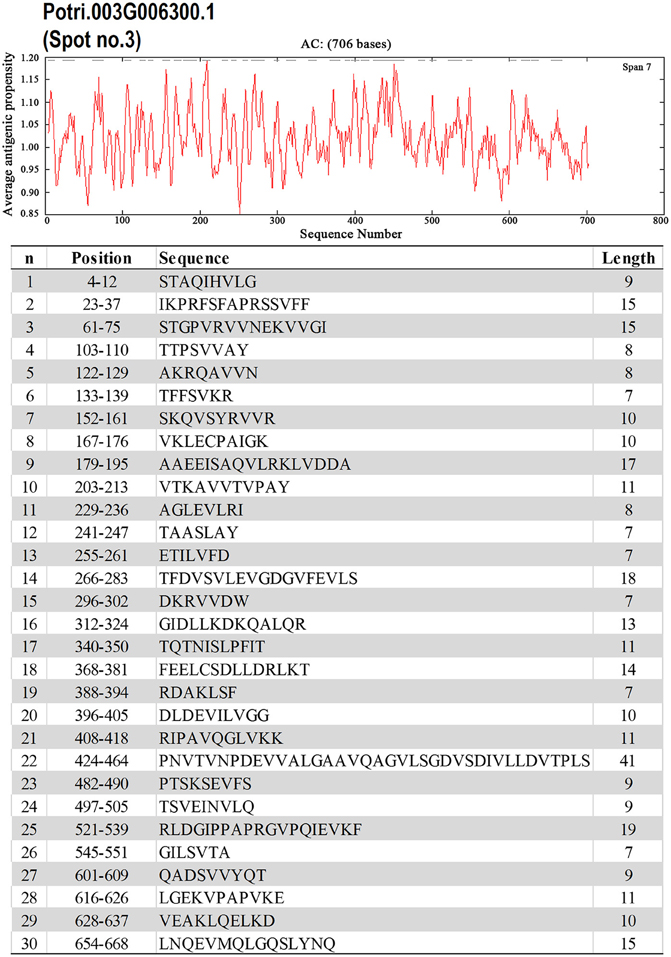
Figure 4. One example (spot No. 3) of determined antigenic peptides in predicted antigen proteins. Predictions are based on a table that reflects the occurrence of amino acid residues in experimentally known segmental epitopes (Kolaskar and Tongaonkar, 1990; Ivanciuc et al., 2003). Total prediction data of 28 candidate antigen proteins were shown in Figure S1.
In P. deltoids pollen, four small Hsps (spots No. 142, 144, 156, and 184) and four Hsp70 (spots No. 3, 5, 12, and 22) were identified as corresponding to allergenic molecules (Table 2). It has reported that class I small heat shock protein (Hsp) is one of allergens in soybean (Gagnon et al., 2010). Hsp70 proteins have been demonstrated to bind to human IgE from patients sensitized to penicillium (Shen et al., 1997), cystic echinococcosis (Ortona et al., 2003), and to corn and wheat dust (Chiung et al., 2000). Spots No. 161 and 164 correspond to pollen Ole e 1 allergen, which was first purified from Olea europea (Lauzurica et al., 1988) and named as Ole e 1 according to the IUIS nomenclature (Marsh et al., 1986). This protein is surmised to control pregermination and pollen tube emergence and guidance (de Dios Alché et al., 2004). In addition, thioredoxin proteins (spots No. 181 and 208) and profilin (spots No. 200, 201, 203, and 216) were identified in our study, consistent with the finding that these two types of proteins have previously been reported as being allergens in wheat, maize, and other plant species (Kleber-Janke et al., 1999; Weichel et al., 2006; Villalta and Asero, 2010).
Expression Patterns of the Predicted Allergen Genes across Various Tissues
Whole genome microarray proved to be a useful means for studying gene expression profiles in poplar (Zhang et al., 2013). To examine whether the predicted pollen-allergen genes presently characterized are expressed in poplar and to study their expression patterns, a comprehensive analysis was conducted based on an Affymetrix microarray data (GSE21481). The expression patterns of the 28 predicted poplar allergen genes across various tissues are shown in Figure 5. Noticeably, two genes corresponding to spots No. 161 and 164 (pollen Ole e 1 allergen proteins) were highly expressed in male catkin, thereby suggesting their specific expression in pollen. It is known that Ole e I is expressed in pollen wall and tapetum but not in petals, roots, or leaves (de Dios Alché et al., 1999). Muschietti et al. (1994) reported that antisense repression of LAT52, a homolog of Ole e I, was associated with abnormal pollen function, consistent with a role of this protein in pollen hydration and/or pollen germination. While only three (spots No. 66, 161, and 164) of pollen allergen genes were highly expressed in male catkin. These genes might play important roles in not only reproductive but also vegetative development. Thus, our data contribute to the identification of new pollen allergenic genes.
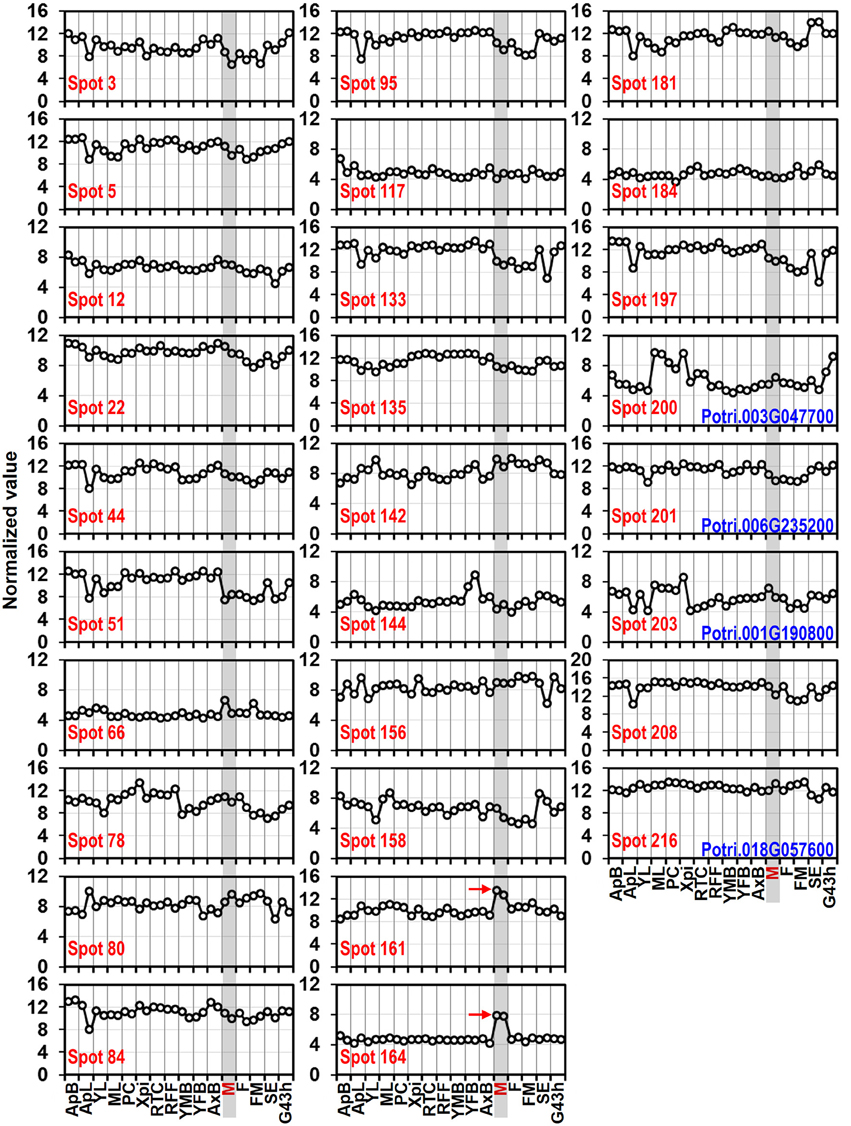
Figure 5. Expression profiles of genes coding 28 predicted allergen related proteins across different tissues. The Affymetrix microarray data were obtained from NCBI Gene Expression Omnibus (GEO) database under the series accession number GSE21481. ApB, Shoot apex; ApL, Young leaves at apex; YL, Young leaves plastochron #2; ML, Mature leaves plastochron #5; PC, Phloem and Cortex; XPi, Developing xylem and pith; RTC, Roots from tissue culture; RFF, Roots from field trees; YMB, Male floral bud initials; YFB, Female floral bud initials; AxB, Axillary buds; M, Male catkin - 3 stages pooled; F, Female catkin post-pollination; FM, Mature catkin before seed release; SE, Seed; G43 h, Seedling 43 h post-imbibition. The spots No. 200, 201, 203, and 216 correspond to profilin (see Table 1).
To verify the expression patterns of the presently characterized predicted allergen genes, qRT-PCR analysis was performed on four tissues for 16 genes (Figure 6). Microarray data in Figure 5 show that the genes corresponding to spot No. 66 (pectin lyase-like family), spot No. 80 (pectin lyase-like family), spot No. 161 (extension family), and spot No. 164 (extension family) were highly expressed in male catkin. Our present qRT-PCR results indicate that the four genes were high expressed in pollen (Figure 6). Moreover, two genes corresponding to spot No. 78 (NmrA-like negative transcriptional regulator family protein) and spot No. 158 (thioredoxin-dependent peroxidase) also exhibited high abundance in pollen. One likely reason is the pollen we used in qRT-PCR was purer than the male catkin used in the microarray analysis in tissue level. In general, the present qRT-PCR results were in good agreement with the microarray data sets analyzed in this study.
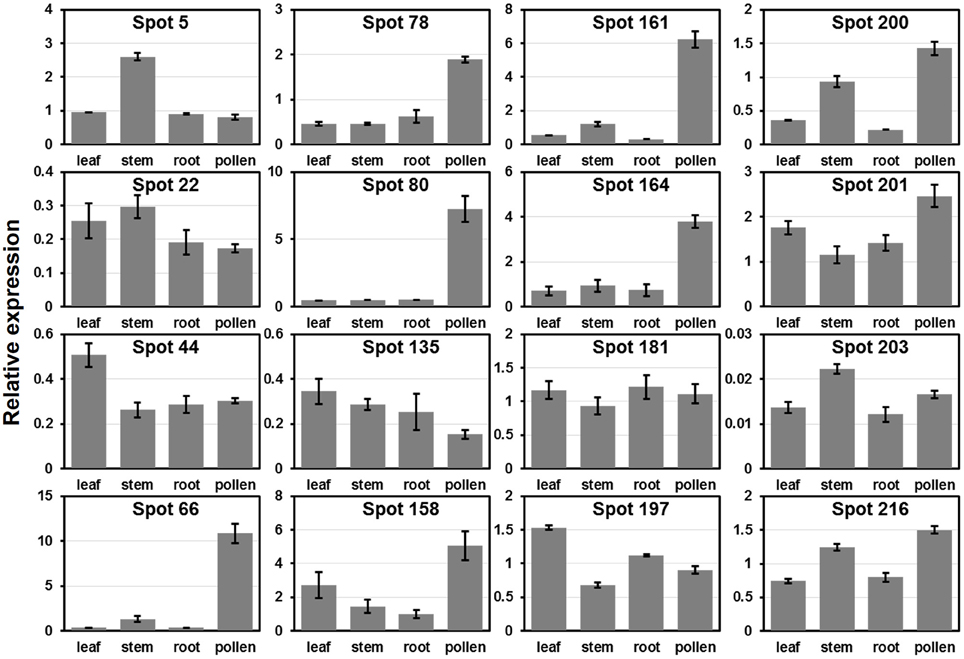
Figure 6. Expression analysis of allergen related genes in different tissues using qRT-PCR. The relative mRNA abundance of 16 allergen related genes presently described were quantified in four tissues (leaf, stem, root, and pollen). The average expression of each gene was calculated relatively to the reference gene PtActin. Relative expression represents log2 expression values. All primer sequences are listed in Table S1. The spots No. 200, 201, 203, and 216 correspond to identified profilin (see Table 1).
Profilin Gene Family and Its Expression Patterns in Poplar
Profilin was first identified as an allergen in birch pollen Bet v2 (Valenta et al., 1991). These proteins probably function as important mediators of membrane-cytoskeleton communication (Machesky and Poland, 1993). Profilins specifically bind to several ligands, that is, actin, phosphatidylinositol-4,5-bisphosphate (PIP2), and poly-L-proline. These characteristics enable them to participate in the regulation of actin polymerization and to interact with the PIP2 pathway of signal transduction (Vieths et al., 2002). Plant profilins exhibit conserved amino acid sequences, share IgE-reactive epitopes, and correspond to highly cross-reactive allergens (Sankian et al., 2005). In the present study, four profilin proteins were identified in P. deltoides mature pollen (Tables 1, 2). To identify all profilin genes in poplar, we performed a BLASTp search against P. trichocarpa genome using profilin protein sequences in Arabidopsis and rice as queries. After confirmation of the protein secondary structure, a total of four profilin genes in poplar were identified. It is noted that all members of this poplar profilin gene family were identified in our proteomic study, suggesting that these profilin proteins are present in high abundance in poplar pollen.
To examine the evolutionary relationships of profilin proteins, we constructed a phylogenetic tree by neighbor-joining method using the full-length profilin proteins in poplar, Arabidopsis, and rice. The phylogenetic tree and amino acid sequences alignment indicate that profilin proteins were highly conserved across three species (Figures 7A,C). We then analyzed the gene structure in the coding sequence of the profilin gene family. The profilin genes in the three species have three exons with conserved length, except one Arabidopsis profilin gene (At5g56600) of which first exon is longer than for the others (Figure 7B). Similar to known profilin structures, poplar profilins consist of three helices and seven β-strands (Figure 7D). In birch pollen profilin, the seven β-strands appear as two orthogonal β-sheets, the first sheet being formed by β1, β2, β4, β5, and β6 strands, while the second sheet is formed by β3 and β7 strands (Fedorov et al., 1997).
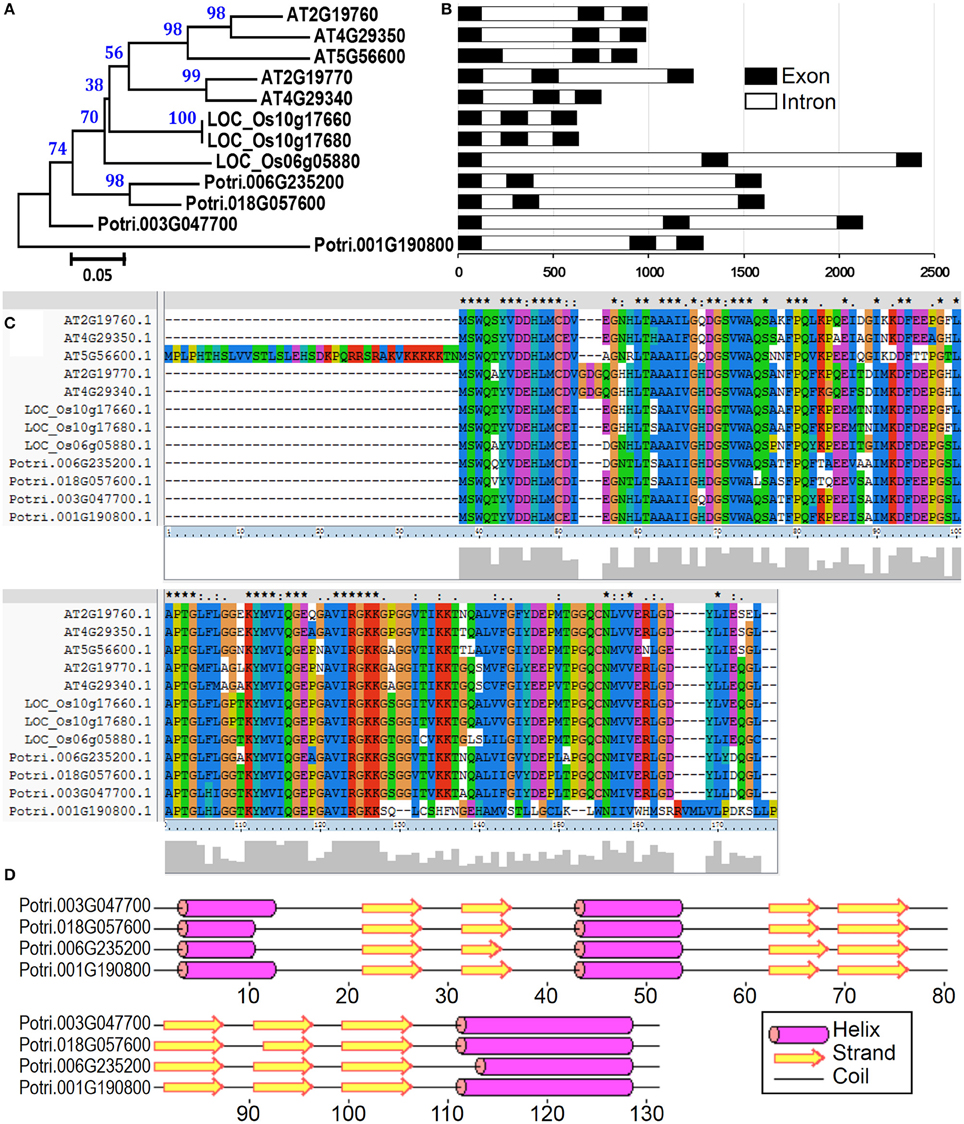
Figure 7. The profilin gene family from poplar, Arabidopsis, and rice. (A) A multiple alignment of full-length profilin protein sequences from three species was executed using Clustal X2.1 and a phylogenetic tree was constructed by the neighbor-joining (NJ) method with 1000 bootstrap replicates. Bootstrap support values are shown on each node. (B) Gene structures of profilin genes from three species. Exons and introns are represented with black and white boxes. (C) Amino acid sequence alignment of profilin proteins in three species. (D) Secondary structure prediction of profilin proteins in poplar.
We then compared the expression patterns of profilin genes in poplar, Arabidopsis, and rice. As shown in Figure 6, two poplar genes (corresponding to spots No. 200 and 201) out of the four poplar profilin genes (corresponding to spots No. 200, 201, 203, and 216) exhibit high expression level in pollen. In Arabidopsis, two profilins (At2g19770 and At4g29340) are highly expressed in flowers and floral organs (Figure S2A). In contrast in rice only one profilin (LOC_Os19g17680) is highly expressed in anther (Figure S2B). The similar expression patterns of these genes across plant species (poplar, Arabidopsis, and rice, Figure 6 and Figure S2) suggest that they kept some conserved functions during plant evolution.
Conclusion
In conclusion, the present study analyzed the proteome of P. deltoides mature pollen for the first time. A total of 403 protein spots were isolated by 2-DE, and 178 distinct proteins were identified from 218 protein spots using MALDI-TOF/TOF MS/MS analysis. Furthermore, 28 proteins were identified as putative allergens and their expression patterns across various tissues were analyzed. The expression patterns across various tissues showed that several of allergenic genes are highly expressed in pollen. Moreover, the members of profilin allergen family were analyzed and their expression patterns were compared with the homologous in Arabidopsis and rice. The similar expression profiles of profilins across different plant species support their conserved functions during plant evolution.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Special Fund for Forestry Scientific Research in the Public Interest [201304103-4] and National High-tech R&D Program of China [2011AA100201] to JH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00548
Figure S1. The determined antigenic peptides in 28 predicted antigen proteins. Predictions are based on a table that reflects the occurrence of amino acid residues in experimentally known segmental epitopes (Kolaskar and Tongaonkar, 1990; Ivanciuc et al., 2003).
Figure S2. Expression patterns of Profilin genes in various tissues of Arabidopsis (A) and rice (B). The microarray data was obtained from AtGenExpress Visualization Tool (AVT, http://jsp.weigelworld.org/expviz/) for Arabidopsis and the Rice Expression Profile Database (RiceXPro, http://ricexpro.dna.affrc.go.jp/) for rice.
Table S1. Sequences of qRT-PCR primers.
References
Aalberse, R. C. (2000). Structural biology of allergens. J. Allergy Clin. Immunol. 106, 228–238. doi: 10.1067/mai.2000.108434
Akagawa, M., Handoyo, T., Ishii, T., Kumazawa, S., Morita, N., and Suyama, K. (2007). Proteomic analysis of wheat flour allergens. J. Agric. Food Chem. 55, 6863–6870. doi: 10.1021/jf070843a
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bartra, J., Sastre, J., Del Cuvillo, A., Montoro, J., Jauregui, I., Davila, I., et al. (2009). From pollinosis to digestive allergy. J. Investig. Allergol. Clin. Immunol. 19(Suppl. 1), 3–10.
Becker, J. D., Boavida, L. C., Carneiro, J., Haury, M., and Feijó, J. A. (2003). Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133, 713–725. doi: 10.1104/pp.103.028241
Bohle, B. (2004). T lymphocytes and food allergy. Mol. Nutr. Food Res. 48, 424–433. doi: 10.1002/mnfr.200400003
Breiteneder, H., and Ebner, C. (2000). Molecular and biochemical classification of plant-derived food allergens. J. Allergy Clin. Immunol. 106, 27–36. doi: 10.1067/mai.2000.106929
Brett, D., Pospisil, H., Valcárcel, J., Reich, J., and Bork, P. (2002). Alternative splicing and genome complexity. Nat. Genet. 30, 29–30. doi: 10.1038/ng803
Canovas, F. M., Dumas-Gaudot, E., Recorbet, G., Jorrin, J., Mock, H. P., and Rossignol, M. (2004). Plant proteome analysis. Proteomics 4, 285–298. doi: 10.1002/pmic.200300602
Chiung, Y. M., Lin, B. L., Yeh, C. H., and Lin, C. Y. (2000). Heat shock protein (hsp 70) -related epitopes are common allergenic determinants for barley and corn antigens. Electrophoresis 21, 297–300. doi: 10.1002/(SICI)1522-2683(20000101)21:2<297::AID-ELPS297>3.0.CO;2-U
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
de Dios Alché, J., Castro, A. J., Olmedilla, A., Fernandez, M. D. C., Rodríguez, R., Villalba, M., et al. (1999). The major olive pollen allergen (Ole e I) shows both gametophytic and sporophytic expression during anther development, and its synthesis and storage takes place in the RER. J. Cell Sci. 112, 2501–2509.
de Dios Alché, J., M'rani-Alaoui, M., Castro, A. J., and Rodríguez-García, M. I. (2004). Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 45, 1149–1157. doi: 10.1093/pcp/pch127
Fedorov, A. A., Ball, T., Mahoney, N. M., Valenta, R., and Almo, S. C. (1997). The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin. Structure 5, 33–45. doi: 10.1016/S0969-2126(97)00164-0
Gagnon, C., Poysa, V., Cober, E. R., and Gleddie, S. (2010). Soybean allergens affecting North American patients identified by 2D gels and mass spectrometry. Food Anal. Method 3, 363–374. doi: 10.1007/s12161-009-9090-3
Galland, M., Huguet, R., Arc, E., Cueff, G., Job, D., and Rajjou, L. (2014). Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell Proteomics 13, 252–268. doi: 10.1074/mcp.M113.032227
Hirano, H., Islam, N., and Kawasaki, H. (2004). Technical aspects of functional proteomics in plants. Phytochemistry 65, 1487–1498. doi: 10.1016/j.phytochem.2004.05.019
Holmes-Davis, R., Tanaka, C. K., Vensel, W. H., Hurkman, W. J., and Mccormick, S. (2005). Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5, 4864–4884. doi: 10.1002/pmic.200402011
Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652. doi: 10.1104/pp.103.020925
Ipsen, H., and Løwenstein, H. (1997). Basic features of crossreactivity in tree and grass pollen allergy. Clin. Rev. Allergy Immunol. 15, 389–396. doi: 10.1007/BF02737734
Ivanciuc, O., Schein, C. H., and Braun, W. (2003). SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 31, 359–362. doi: 10.1093/nar/gkg010
Jensen, O. N. (2004). Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33–41. doi: 10.1016/j.cbpa.2003.12.009
Kitta, K., Ohnishi-Kameyama, M., Moriyama, T., Ogawa, T., and Kawamoto, S. (2006). Detection of low-molecular weight allergens resolved on two-dimensional electrophoresis with acid-urea polyacrylamide gel. Anal. Biochem. 351, 290–297. doi: 10.1016/j.ab.2005.12.030
Kleber-Janke, T., Crameri, R., Appenzeller, U., Schlaak, M., and Becker, W.-M. (1999). Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int. Arch. Allergy Immunol. 119, 265–274. doi: 10.1159/000024203
Kolaskar, A., and Tongaonkar, P. C. (1990). A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 276, 172–174. doi: 10.1016/0014-5793(90)80535-Q
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lauzurica, P., Gurbindo, C., Maruri, N., Galocha, B., Diaz, R., González, J., et al. (1988). Olive (Olea europea) pollen allergens-I immunochemical characterization by immunoblotting, crie and immunodetection by a monoclonal antibody. Mol. Immunol. 25, 329–335. doi: 10.1016/0161-5890(88)90027-2
Loraine, A. E., McCormick, S., Estrada, A., Patel, K., and Qin, P. (2013). RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 162, 1092–1109. doi: 10.1104/pp.112.211441
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Machesky, L. M., and Poland, T. D. (1993). Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 3, 381–385. doi: 10.1016/0962-8924(93)90087-H
Marsh, D., Goodfriend, L., King, T., Lowenstein, H., and Platts-Mills, T. (1986). Allergen nomenclature. Bull. World Health Organ. 64, 767.
Mccormick, S. (2004). Control of male gametophyte development. Plant Cell 16, S142–S153. doi: 10.1105/tpc.016659
Mohapatra, S. S., and Knox, R. B. (1996). Pollen Biotechnology, Gene Expression and Allergen Characterization. London: Chapman & Hall.
Muschietti, J., Dircks, L., Vancanneyt, G., and Mccormick, S. (1994). LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6, 321–338. doi: 10.1046/j.1365-313X.1994.06030321.x
Nakamura, R., and Teshima, R. (2013). Proteomics-based allergen analysis in plants. J. Proteomics 93, 40–49. doi: 10.1016/j.jprot.2013.03.018
Noir, S., Bräutigam, A., Colby, T., Schmidt, J., and Panstruga, R. (2005). A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem. Biophys. Res. Commun. 337, 1257–1266. doi: 10.1016/j.bbrc.2005.09.185
Ortona, E., Margutti, P., Delunardo, F., Vaccari, S., Rigano, R., Profumo, E., et al. (2003). Molecular and immunological characterization of the C-terminal region of a new Echinococcus granulosus heat shock protein 70. Parasite Immunol. 25, 119–126. doi: 10.1046/j.1365-3024.2003.00617.x
Picariello, G., Mamone, G., Addeo, F., and Ferranti, P. (2011). The frontiers of mass spectrometry-based techniques in food allergenomics. J. Chromatogr. A 1218, 7386–7398. doi: 10.1016/j.chroma.2011.06.033
Pina, C., Pinto, F., Feijó, J. A., and Becker, J. D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138, 744–756. doi: 10.1104/pp.104.057935
Rajjou, L., Gallardo, K., Debeaujon, I., Vandekerckhove, J., Job, C., and Job, D. (2004). The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134, 1598–1613. doi: 10.1104/pp.103.036293
Sankian, M., Varasteh, A., Pazouki, N., and Mahmoudi, M. (2005). Sequence homology: a poor predictive value for profilins cross-reactivity. Clin. Mol. Allergy 3, 13. doi: 10.1186/1476-7961-3-13
Shahali, Y., Sutra, J. P., Haddad, I., Vinh, J., Guilloux, L., Peltre, G., et al. (2012). Proteomics of cypress pollen allergens using double and triple one-dimensional electrophoresis. Electrophoresis 33, 462–469. doi: 10.1002/elps.201100324
Shen, H. D., Au, L. C., Lin, W. L., Liaw, S. F., Tsai, J. J., and Han, S. H. (1997). Molecular cloning and expression of a Penicillium citrinum allergen with sequence homology and antigenic crossreactivity to a hsp 70 human heat shock protein. Clin. Exp. Allergy 27, 682–690. doi: 10.1111/j.1365-2222.1997.tb01197.x
Sheng, X., Hu, Z., Lü, H., Wang, X., Baluška, F., Šamaj, J., and Lin, J. (2006). Roles of the ubiquitin/proteasome pathway in pollen tube growth with emphasis on MG132-induced alterations in ultrastructure, cytoskeleton, and cell wall components. Plant Physiol. 141, 1578–1590. doi: 10.1104/pp.106.081703
Sheoran, I. S., Ross, A. R., Olson, D. J., and Sawhney, V. K. (2007). Proteomic analysis of tomato (Lycopersicon esculentum) pollen. J. Exp. Bot. 58, 3525–3535. doi: 10.1093/jxb/erm199
Sheoran, I. S., Sproule, K. A., Olson, D. J., Ross, A. R., and Sawhney, V. K. (2006). Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex. Plant Reprod. 19, 185–196. doi: 10.1007/s00497-006-0035-3
Sicherer, S. H. (2001). Clinical implications of cross-reactive food allergens. J. Allergy Clin. Immunol. 108, 881–890. doi: 10.1067/mai.2001.118515
Singh, A. B., and Shahi, S. (2008). Aeroallergens in clinical practice of allergy in India-ARIA Asia Pacific Workshop report. Asian Pac. J. Allergy Immunol. 26, 245–256.
Sircar, G., Chakrabarti, H. S., Saha, B., and Gupta-Bhattacharya, S. (2012). Identification of aero-allergens from Rhizopus oryzae: an immunoproteomic approach. J. Proteomics 77, 455–468. doi: 10.1016/j.jprot.2012.09.023
Supek, F., Bosnjak, M., Skunca, N., and Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. doi: 10.1371/journal.pone.0021800
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Taylor, G. (2002). Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 90, 681–689. doi: 10.1093/aob/mcf255
Valenta, R., Duchene, M., Pettenburger, K., Sillaber, C., Valent, P., Bettelheim, P., et al. (1991). Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 253, 557–560. doi: 10.1126/science.1857985
Vieths, S., Scheurer, S., and Ballmer-Weber, B. (2002). Current understanding of cross−reactivity of food allergens and pollen. Ann. N.Y. Acad. Sci. 964, 47–68. doi: 10.1111/j.1749-6632.2002.tb04132.x
Villalta, D., and Asero, R. (2010). Sensitization to the pollen pan-allergen profilin. Is the detection of immunoglobulin E to multiple homologous proteins from different sources clinically useful? J. Invest. Allergy Clin. 20, 591.
Volkov, R. A., Panchuk, I. I., and Schöffl, F. (2005). Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol. Biol. 57, 487–502. doi: 10.1007/s11103-005-0339-y
Weichel, M., Glaser, A. G., Ballmer-Weber, B. K., Schmid-Grendelmeier, P., and Crameri, R. (2006). Wheat and maize thioredoxins: a novel cross-reactive cereal allergen family related to baker's asthma. J. Allergy Clin. Immunol. 117, 676–681. doi: 10.1016/j.jaci.2005.11.040
Wilkins, M. R., Pasquali, C., Appel, R. D., Ou, K., Golaz, O., Sanchez, J.-C., et al. (1996). From proteins to proteomes: large scale protein identification by two dimensional electrophoresis and amino acid analysis. Biotechnolgy 14, 61–65. doi: 10.1038/nbt0196-61
Yano, H., and Kuroda, S. (2008). Introduction of the disulfide proteome: application of a technique for the analysis of plant storage proteins as well as allergens. J. Proteome Res. 7, 3071–3079. doi: 10.1021/pr8003453
Zhang, J., Li, J., Liu, B., Zhang, L., Chen, J., and Lu, M. (2013). Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics 14:532. doi: 10.1186/1471-2164-14-532
Keywords: allergen, MALDI-TOF/TOF MS/MS, pollen, Populus deltoids, profilin, proteomics, two-dimensional gel electrophoresis
Citation: Zhang J, Wu L-S, Fan W, Zhang X-L, Jia H-X, Li Y, Yin Y-F, Hu J-J and Lu M-Z (2015) Proteomic analysis and candidate allergenic proteins in Populus deltoides CL. “2KEN8” mature pollen. Front. Plant Sci. 6:548. doi: 10.3389/fpls.2015.00548
Received: 21 April 2015; Accepted: 06 July 2015;
Published: 29 July 2015.
Edited by:
Dominique Job, Centre National de la Recherche Scientifique, FranceReviewed by:
Curtis G. Wilkerson, Michigan State University, USAZhuo Renying, The Research Institute of Subtropical Forestry, China
Copyright © 2015 Zhang, Wu, Fan, Zhang, Jia, Li, Yin, Hu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Jun Hu, State Key Laboratory of Tree Genetics and Breeding, Key Laboratory of Tree Breeding and Cultivation of the State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing 100091, China,SHVqakBjYWYuYWMuY24=
†These authors have contributed equally to this work.
 Jin Zhang
Jin Zhang Li-Shuan Wu1,4†
Li-Shuan Wu1,4† Jian-Jun Hu
Jian-Jun Hu Meng-Zhu Lu
Meng-Zhu Lu