- Institute of Horticultural Biotechnology, Fujian Agriculture and Forestry University, Fuzhou, China
MicroRNA160 plays a critical role in plant development by negatively regulating the auxin response factors ARF10, -16, and -17. However, the ways in which miR160 expression is regulated at the transcriptional level, and how miR160 interacts with its targets during plant embryo development, remain unknown. Here, we studied the regulatory relationships among endogenous target mimics (eTMs), and miR160 and its targets, and their involvement in hormone signaling and somatic embryogenesis (SE) in Dimocarpus longan. We identified miR160 family members and isolated the miR160 precursor, primary transcript, and promoter. The promoter contained cis-acting elements responsive to stimuli such as light, abscisic acid, salicylic acid (SA) and heat stress. The pri-miR160 was down-regulated in response to SA but up-regulated by gibberellic acid, ethylene, and methyl jasmonate treatment, suggesting that pri-miR160 was associated with hormone transduction. Dlo-miR160a, -a∗ and -d∗ reached expression peaks in torpedo-shaped embryos, globular embryos and cotyledonary embryos, respectively, but were barely detectable in friable-embryogenic callus. This suggests that they have expression-related and functional diversity, especially during the middle and later developmental stages of SE. Four potential eTMs for miR160 were identified. Two of them, glucan endo-1,3-beta- glucosidase-like protein 2-like and calpain-type cysteine protease DEK1, were confirmed to control the corresponding dlo-miR160a∗ expression level. This suggests that they may function to abolish the binding between dlo-miR160a∗ and its targets. These two eTMs also participated in 2,4-D and ABA signal transduction. DlARF10, -16, and -17 targeting by dlo-miR160a was confirmed; their expression levels were higher in friable-embryogenic callus and incomplete compact pro-embryogenic cultures and responded to 2,4-D, suggesting they may play a major role in the early stages of longan SE dependent on 2,4-D. The eTMs, miR160, and ARF10, -16, and -17 exhibited tissue specificity in ‘Sijimi’ longan vegetative and reproductive organs, but were not significant negatively correlated. These results provide insights into the possible role of the eTM-miR160-ARF10-16-17 pathway in longan somatic embryo development.
Background
MicroRNAs, a conserved class of single-stranded non-coding RNAs of approximately 21 nt in length, are derived from pri-miRNAs containing imperfect stem-loop secondary structures (Jones-Rhoades et al., 2006). These hairpin RNAs, which are known as pre-miRNAs genes, are transcribed by RNA polymerase II in plants, processed by DICER- LIKE1 to produce an miRNA:miRNA∗ duplex in the nucleus, and transported to the cytoplasm (Lee et al., 2004; Jones-Rhoades et al., 2006). The mature miRNA recognizes target mRNA sites through perfect or near perfect complementarity, and negatively regulates mRNA expression by cleaving targeted mRNAs (Llave et al., 2002) or repressing translation (Gandikota et al., 2007). miRNAs play vital roles in a range of developmental processes (Carrington and Ambros, 2003; Jones-Rhoades et al., 2006). The expression of miRNA genes is tightly controlled. Recently, a novel mechanism involving eTMs was discovered, whereby miRNA–mRNA duplexes with a large bulge around the cleavage site actually sequester miRNAs, resulting in inhibition of miRNA activity (Franco-Zorrilla et al., 2007). eTMs have been used as a valuable tool to understand the functions of several miRNA families in plants (Wu et al., 2013).
A highly conserved miRNA family, miR160, negatively regulates ARF10 (Liu et al., 2007), -16 (Wang et al., 2005), and -17 (Mallory et al., 2005) of the repressor ARF family, and plays a critical role in maintaining the process of seed germination and the normal developmental programs of embryos, roots, stems, leaves, siliques, and floral organs. An Arabidopsis loss-of-function mutation in miR160a caused various embryonic defects, suggesting that miR160 may control embryo development (Liu et al., 2010b). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean (Turner et al., 2013), and decreases seed dormancy in Arabidopsis (Liu et al., 2013). The over-expression of artificial target mimics for miR160 resulted in plants of a smaller size with serrated and upward-curled leaves (Todesco et al., 2010; Yan et al., 2012). Over-expression of eTMs of miR160 also caused smaller and serrated leaves in transgenic plants (Wu et al., 2013), suggesting that eTMs play a key role in plant development by inhibiting miRNA activity. However, to date, analysis and identification of eTMs for miR160 has rarely been performed in plants (Wu et al., 2013; Shuai et al., 2014).
Dimocarpus longan Lour., also called longan or dragon eye, is a tropical tree of the soapberry family (Sapindaceae) that produces edible fruit. As a traditional medicine, longan fruit is used to enhance memory (Park et al., 2010), promote blood metabolism, relieve insomnia, and prevent amnesia (Yi et al., 2011). Longan seed extracts, which contain high levels of antioxidant polyphenolic compounds (Chung et al., 2010) and polysaccharides (Zheng et al., 2010), are capable of inducing apoptosis in various colorectal cancer cells. Previous studies have shown that longan’s embryo development status influences its seed size, fruit quality, fruit set and yield (Liu et al., 2010a; Lai and Lin, 2013; Lin and Lai, 2013a,b). However, because of the extreme genetic heterozygosity exhibited by this species and the difficulty of sampling early embryos, advances in understanding of the regulation of longan embryo development have been limited.
Somatic embryogenesis, which resembles zygotic embryogenesis, is an essential component of plant cell differentiation and embryo development. The longan SE system has been used widely as a model system to investigate in vitro and in vivo regulation of embryogenesis in plants (Lai et al., 2010; Lin and Lai, 2010). In our previous study, we found that miR160a was barely detectable during the early stages of longan SE, but highly expressed during the heart- and torpedo-shaped embryonic stages (Lin and Lai, 2013b). Previous reports have suggested that larch and Brassica napus miR160s might play a regulatory role during cotyledonary embryo development (Zhang et al., 2012; Huang et al., 2013). However, it is not clear how post-transcriptional regulation of ARF10, -16, and -17 by miR160 regulates plant embryo development or SE, particularly in longan.
The aim of this study was to investigate the regulatory networks of eTMs and miR160-mediated regulation of ARF10, -16, and -17 in longan, especially during SE. Here, we identified and examined the expression of mature miR160 molecules, the miR160 primary transcript and its promoter, eTMs, and homologs of the target genes ARF10, -16, and -17 for miR160. Our results suggest possible general regulation by eTM down-regulation of miR160 cleavage of ARF10, -16, and -17 during SE in longan.
Materials and Methods
Tissue Samples
Synchronized embryogenic cultures at different developmental stages from D. longan ‘Honghezi’ (Fujian, China), consisting of EC, ICpEC, GE, HE, TE, and CE, were generated as detailed in Lai and Chen (1997).
The EC was pre-cultured on Murashige and Skoog (MS) medium containing 2,4-D (1.0 mg/L) for 20 days, and then sub-cultured in 50 mL of liquid MS medium containing 2,4-D (0, 0.5, 1.5, or 2 mg/L), SA (75 μM), MeJA (50 μM), ET (30 ppm), GA3 (35 μM), or ABA (100 μM) in a rotary shaker at 150 rpm for 24 h. These cultures were all kept in the dark.
Dimocarpus longan ‘Sijimi’ samples, including roots, leaves, floral buds, flowers, young fruits, mature fruits, pericarp, pulp and seeds, were collected from the experimental fields of Fujian Academy of Agricultural Science in Putian. All materials from at least six rootstock plants of the same cultivar were combined.
The synchronized embryogenic cultures, EC cultures treated with different hormones, and ‘Sijimi’ samples were collected and stored at -80°C for subsequent analyses.
Database Searching for miR160 Members of Longan
Previously published small RNA data (BioSample accession SAMN04120614, Bio-Project ID PRJNA297248) (Lin and Lai, 2013b), generated using various synchronized embryogenic cultures from the longan ‘Honghezi’ variety, was used to identify miR160 family members. The potential miR160 members were searched using the miRBase server (Release 21: June 2014). Multiple alignment analysis of miR160 members was performed using DNAMAN ver. 6.0 (Lynnon Biosoft Corp, Pointe-Claire, QC, Canada).
Pri-miR160 3′ and 5′ RACE Experiments
To determine whether the miR160 members were present in the D. longan genome, the full-length cDNA sequence of pri-miR160 was cloned by RT-PCR and 3′/5′ RACE PCR from the EC and mixed cDNA. The EC cDNA for RT-PCR of the pre-miR160 conserved regions was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA). The mixed cDNA for pri-miR160 3′/5′ RACE PCR was synthesized from mixed total RNA (EC, ICpEC, GE and HE) using a GeneRacerTM Kit (Invitrogen, Corporation, Carlsbad, CA, USA), following the manufacturer’s instructions.
The amplification of the full-length cDNA of pri-miR160 was carried out by 3′/5′ RACE PCR from the longan mixed cDNA, and its genomic sequence was cloned from the EC DNA template. The sequences of all primers used are provided in Table 1.
To determine the phylogeny of the miR160 gene, we downloaded all precursor sequences of plant miR160s from miRBase release 21. Phylogenetic analysis was performed according to the neighbor-joining method (Saitou and Nei, 1987) using MEGA 5.02 (Kumar et al., 2004), with 1000 bootstrap replicates.
miR160 Promoter Cloning and cis-Acting Element Analysis
To study the molecular mechanism underlying the transcriptional regulation of miR160, genome walking was performed using TAIL-PCR (Takara, Japan) to obtain the miR160 promoter. The promoter was cloned by three nested PCR amplifications from the longan EC DNA template with the same forward primer AP2 and specific reverse primers. The primer sequences can be found in Table 1. Cis-acting element analysis of the miR160 promoter was carried out using the PlantCARE database (Lescot et al., 2002).
Prediction of eTMs for the miR160 Family in D. longan
First, the longan EC transcriptome database (SRA050205) (Lai and Lin, 2013) was used to search for sites in the cDNA sequences that were reverse complements of the miR160 family, and the tool TAPIR was used to search for eTMs according to criteria described previously (Bonnet et al., 2010). The cDNAs that satisfied the above criteria were considered to be target mimic candidates.
Modified 5′ RLM-RACE
To determine the internal cleavage sites in the ARF10, -16, and -17 mRNAs, a modified 5′ RLM-RACE experiment was performed using a GeneRacer kit (Invitrogen). Briefly, total RNAs from equal mixtures of EC, GE, HE, and TE were ligated to a 5′ RACE RNA adapter. The experiment essentially followed the manufacturer’s instructions. The PCR amplification used GeneRacer 5′ primers and gene-specific primers (Table 1). The resulting PCR products were gel-purified, cloned, and sequenced.
Gene Expression Assays
Total RNAs were isolated from the nine synchronized embryogenic cultures described above using the TRIzol Reagent kit (Invitrogen). cDNAs for miRNA and mRNA quantification were synthesized using a One Step PrimeScript® miRNA cDNA Synthesis Kit (Takara Code, D350A), and PrimeScriptTM Perfect Real Time RT Reagent Kit (TaKaRa Code, DR037A), respectively.
Quantitative PCR assays of gene expression were performed using a LightCycler 480 qPCR instrument (Roche Applied Science, Switzerland) and SYBR premix (Takara). All reactions were performed in triplicate. The expression levels of the miR160s, their targets (ARF10, -16, -17), and the eTMs were quantified using internal standards (Lin and Lai, 2010, 2013c). Statistical analysis was performed using SPSS 19. Gene names and primer sequences are provided in Table 2.
Results
Analysis of the miR160 Family Members from a Longan Small RNA Dataset
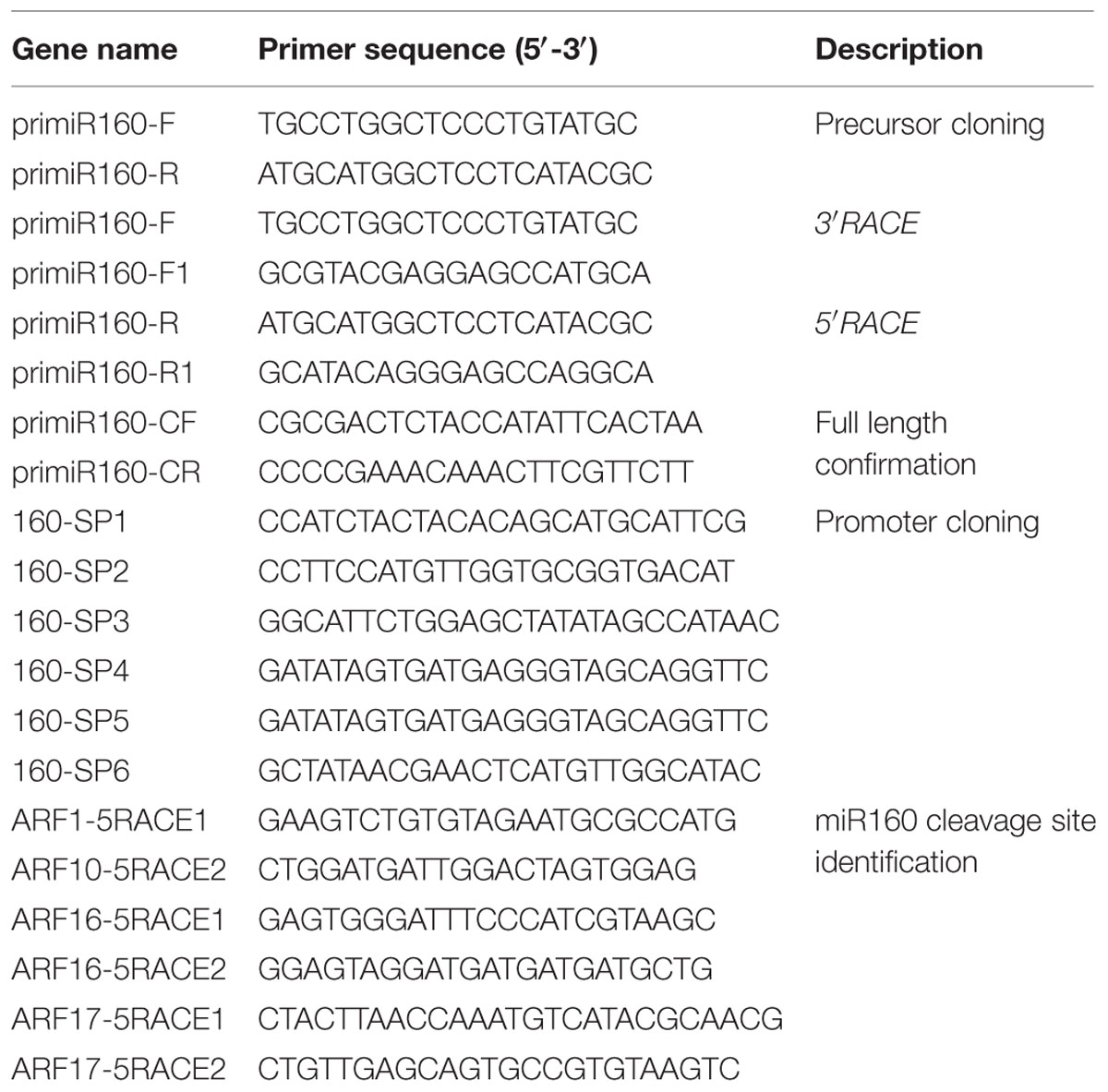
The miR160 family, comprising three members (ath-miR160a–c) in Arabidopsis and six members (osa-miR160a–f) in Oryza sativa, was first identified in plants (Reinhart et al., 2002). By searching a longan small RNA dataset, we found that the miR160 family was represented by 12 members with different read counts (Figures 1A–C). The 12 potential miR160 members were then compared against the miRBase database (Release 21), which revealed that they were distributed into two isoforms. One was identical to ath-miR160a, including ten similar variants; the other was identical to osa-miR160d, including two variants. We named these two isoforms dlo-miR160a and dlo-miR160d, respectively. However, variants similar to miR160b and miR160c were not found. Therefore, there are at least two miR160 isoforms in the D. longan genome. According to the Blast alignment results, of these 12 variants, only two mature miRNAs were excised from the 5′ arm of the hairpin, the others were located in the 3′ arm of the hairpin.
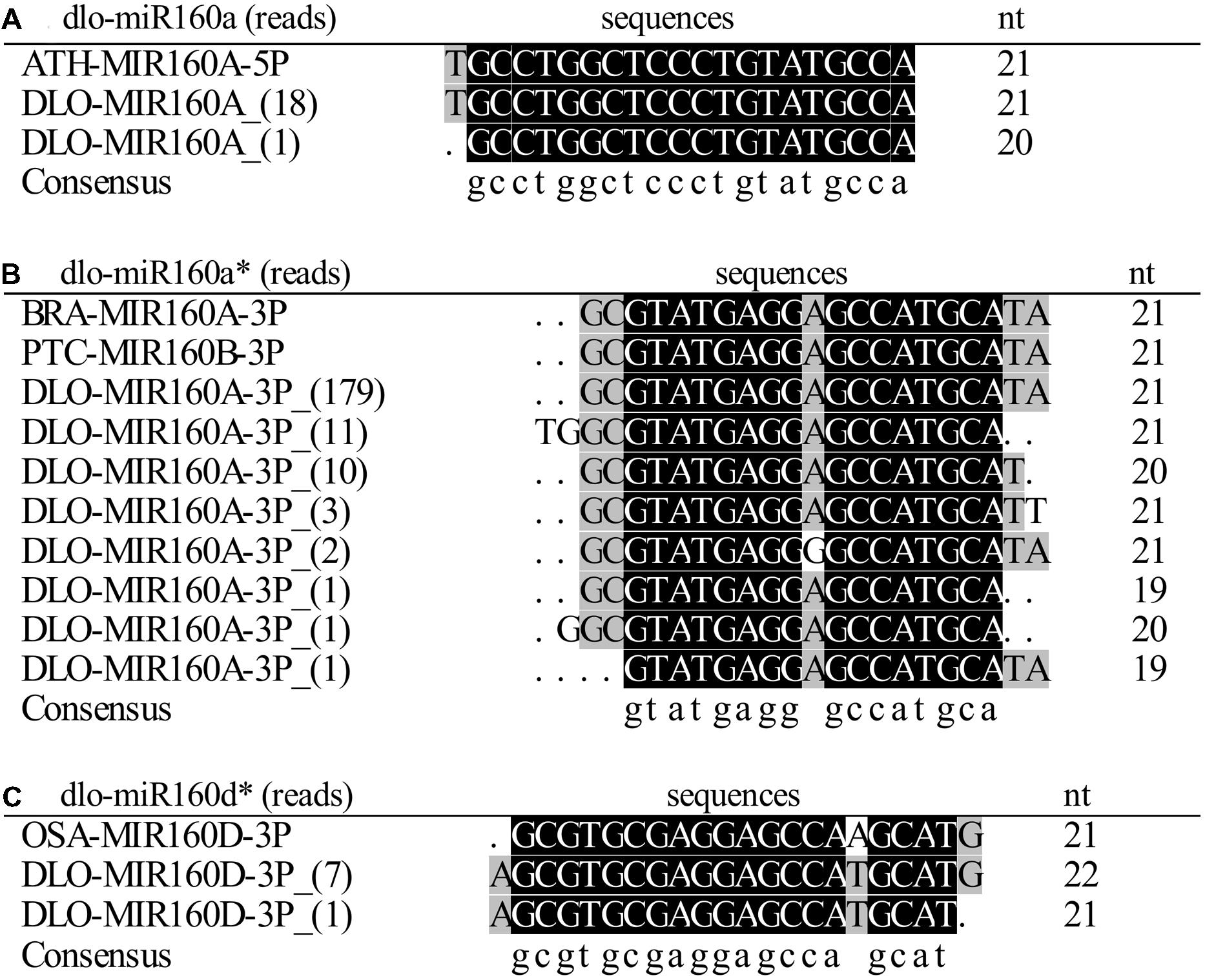
FIGURE 1. Alignment of Dimocarpus longan miR160 members. (A) miR160a; (B) miR160a∗; (C) miR160d∗. ATH, Arabidopsis thaliana (miRBase No. MI0000190); BRA, Brassica rapa (miRBase No. MIMAT0010155); PTC, Populus trichocarpa (miRBase No. MIMAT 0022890); OSA, Oryza sativa (miRBase No. MIMAT0022854); DLO, Dimocarpus longan.
Identification of the miR160 Gene Sequence in Longan
To find the miR160 gene and determine its structure and length in the D. longan genome, the full-length cDNA sequence of pri-miR160 was obtained by 3′/5′-RACE. Two precursors with lengths of 80 and 82 bp were obtained by RT-PCR. Alignment analysis showed that these two precursors were highly homologous except for 13 nucleotide differences (Figure 2A). To determine the hairpin structure of the miR160 precursors and the positions of the miR160 members, secondary structure analysis of the precursors was conducted using the DNAMan6.0 software. The two miR160 precursors both formed the classic stem-loop structure, and minimum free energy (MFE) analysis indicated that the stabilities of their secondary structures were -47.1 and -39.5 kcal/mole, respectively (Figure 2B). By comparing these two precursors and the mature miR160 sequences, dlo-miR160a-5p variants were found that exactly matched the 5′ arms of both precursors, while dlo-miR160a-3p and -d-3p variants were detected in the 3′ arms of the two precursors, respectively. The sequence that exactly matched the dlo-miR160a-3p variants was named pre-miR160a; the other one was identical to dlo-miR160d-3p and was named pre-miR160d (Figure 2B). Phylogenetic analysis (Figure 2C) also suggested that the pre-miR160a sequence in D. longan was homologous to pre-miR160 sequences in Arabidopsis (miR160a), Populus trichocarpa (miR160b, c), and Vitis vinifera (miR160e). Pre-miR160d appeared to be closely related to V. vinifera miR160d.
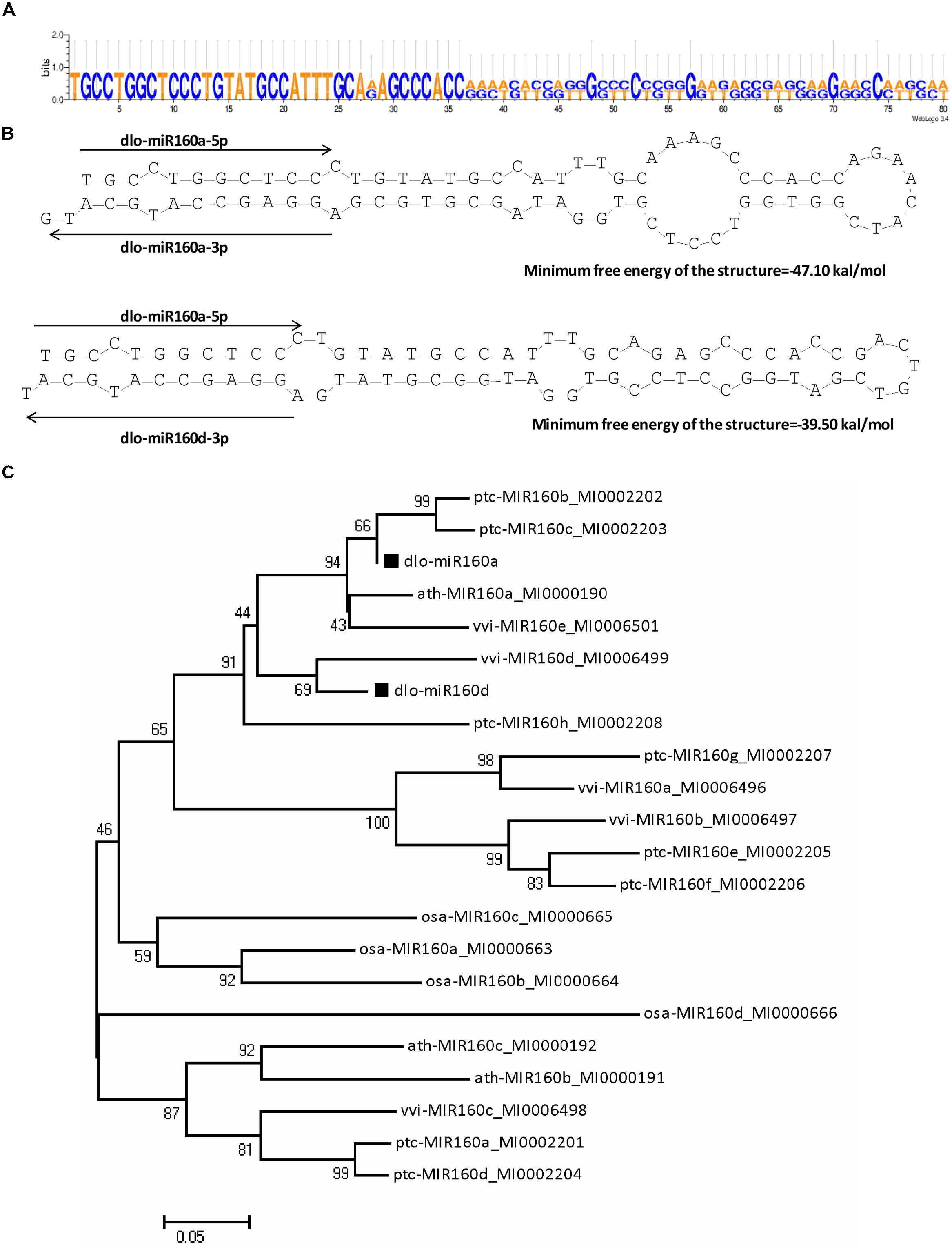
FIGURE 2. Separate primary MIR160 transcripts contain dlo-miR160a/a∗ and -160d∗. (A) Longan miR160 precursors. (B) Hairpin structures of the longan miR160 precursors containing dlo-miR160a/a∗ and -160d∗. (C) Neighbor-joining phylogenetic tree of pre-miR160 sequences in plants. Numbers in the tree represent bootstrap values. ptc, Populus trichocarpa; vvi, Vitis vinifera; dlo, Dimocarpus longan; ath, Arabidopsis thaliana; osa, Oryza sativa.
Next, 5′ and 3′ RACE analyses for the pri-miR160 transcripts were carried out. The full-length sequence was 441 bp (GenBank accession No. KJ372214), with the typical poly(A) tail in the 3′ UTR, and the first nucleotide (transcription initiation site) of the full-length cDNA was A. BLAST analysis of the pri-miR160 showed similarities with existing pri/pre-miR160s in Manihot esculenta, P. trichocarpa, Gossypium hirsutum, and Arabidopsis thaliana. The corresponding pri-miR160 genomic DNA sequence, which comprised 424 bp and contained one 100 bp intron, was also obtained (GenBank accession No. KJ372217). The sequence at the exon–intron junctions was consistent with the eukaryotic GT–AG rule.
Isolation of the miR160 Promoter in Longan
To further understand the regulatory role of the miR160 gene at the transcriptional level, a 957 bp 5′-flanking sequence of the pri-miR160 was cloned from longan EC (GenBank accession No. KJ372217). PlantCARE (Lescot et al., 2002) was used to search for cis-acting elements in the pri-miR160 promoter. The results revealed the presence of conserved TATA and CAAT boxes. Many putative cis-elements associated with responses to light (BOX-4, Box II, G-BOX, GAG-motif, GATA-motif and Sp1), ABA (ABRE), SA (TCA-element) and environmental stresses (heat stress-response element, HSE) were found in the promoter. In addition, an anaerobic response element (ARE) that is essential for anaerobic induction, a Skn-1_motif that is required for endosperm expression, a circadian element involved in circadian control, and four elements of unknown function were also present in the promoter (Figure 3). These results prompted us to carry out further experiments to identify the response of the miR160 gene to various environmental stimuli.

FIGURE 3. 5′ flanking sequence of pri-miR160 in D. longan. The bold A is the transcription start site (TSS). Cis-acting regulatory elements are underlined and shaded in gray; their orientations are shown by arrows.
Transcriptome-wide Prediction of eTMs of the miR160 Family in Longan
To date, an analysis of eTMs for miR160 has not been performed in longan. Here, transcriptome-wide computational prediction of eTMs for miR160 was performed. Four potential eTMs were identified, including an unknown mRNA (Unigene11608), a nitrate transporter family protein (Unigene56365), glucan endo-1,3-beta- glucosidase-like protein 2-like (Unigene64827), and calpain-type cysteine protease DEK1 (Unigene66891) (Figure 4). By contrast, two and eleven non-coding eTMs for miR160 were predicted in Arabidopsis and rice, respectively (Wu et al., 2013). The four potential eTMs all contained a three-nucleotide insertion between the nucleotides interacting with positions 10 and 11 of the corresponding miRNA, and the mimic sites of Unigene11608 and Unigene56365 were on the sense strand of the 3′ UTR and 5′ UTR, respectively, while the other two mimics were located on the antisense strand of the CDS (coding sequence) region (Figure 4). These results suggested that the miR160 family might also be controlled by eTMs in D. longan.

FIGURE 4. Predicted eTMs for the miR160 family in longan. (+), mRNA sense strand; (-), mRNA antisense strand; RPKM, reads per kilobase per million mapped reads; size, unigene length. (A) Unigene11608: an unknown mRNA; (B) Unigene56365: a nitrate transporter family protein; (C) Unigene64827: glucan endo-1,3-beta- glucosidase-like protein 2-like; (D) Unigene66891: calpain-type cysteine protease DEK1.
ARF Transcripts are Cleaved by miR160 During Longan SE
In Arabidopsis, miR160 targets ARF10, -16, and -17, and its regulation of these transcripts appears to be important in seed germination, and shoot and root development (Mallory et al., 2005; Wang et al., 2005; Liu et al., 2007); however, its role in longan SE remains unknown. The mRNA sequences of the D. longan ARFs DlARF10 (KJ410234), -16 (KJ410235), and -17 (KJ410237) were extracted from GenBank to search for possible miR160 cleavage sites. Alignment analysis showed that DlARF10, -16, and -17 shared high amino acid sequence similarities. All contained a conserved family domain and a B3 domain, while DlARF17 was distinguished by its poorly conserved C-terminal domain (Figure 5), which was consistent with the ARFs identified from Arabidopsis (Wang et al., 2005).
FIGURE 5. Cleavage site mapping of dlo-miR160a target genes. The mRNA sequences of DlARF10, -16, and -17 are aligned with dlo-miR160a. Numbers indicate the fraction of cloned PCR products terminating at different positions. The B3 DNA binding domain (B3), Auxin_resp, and AUX_IAA are highlighted in green, red, and blue, respectively, in the ARF10, -16, and -17 proteins. The dlo-miR160a complementary sequence in the DlARF10, -16, and -17 mRNAs and the corresponding amino acid sequences are shown.
Fragments of the DlARF10, -16, and -17 mRNAs were detected from longan embryogenic tissues and were determined to be cleaved by miR160 using a modified form of RLM-RACE. The miR160a target sites in the DlARF10, -16, and -17 mRNAs were located at positions corresponding to the amino acid sequences GMQGAWH, GIQGARH, and GMQGARQ, respectively. The miR160a sequence showed high complementarity to these regions in DlARF10, -16, and -17, with only one or two mismatched bases (Figure 5). To determine the tissues in which miR160a regulates the ARF10, -16, and -17 mRNAs, we searched for cleaved fragments of all three genes in longan EC cDNA and mixed cDNA samples. Fragments of the three genes were not detected in the EC samples, but were found in the mixed cDNA sample, indicating the presence of cleaved products in the mixed cDNA, but not in EC. These results clearly indicated that miR160a cleaved the ARF10, -16, and -17 mRNAs during longan SE, confirming that miR160-ARF10-16-17 signal transduction operates in longan, similarly to Arabidopsis.
Confirmation of an eTM-miR160-ARF10-16-17 Regulatory Pathway During Longan SE
To determine experimentally whether the potential dlo-miR160 family members obtained from the longan small RNA data were indeed expressed, and whether the predicted eTMs function to abolish the binding between miR160 and its targets (ARF10, -16, -17), we investigated their expression patterns during SE in D. longan (Figures 6 and 7).
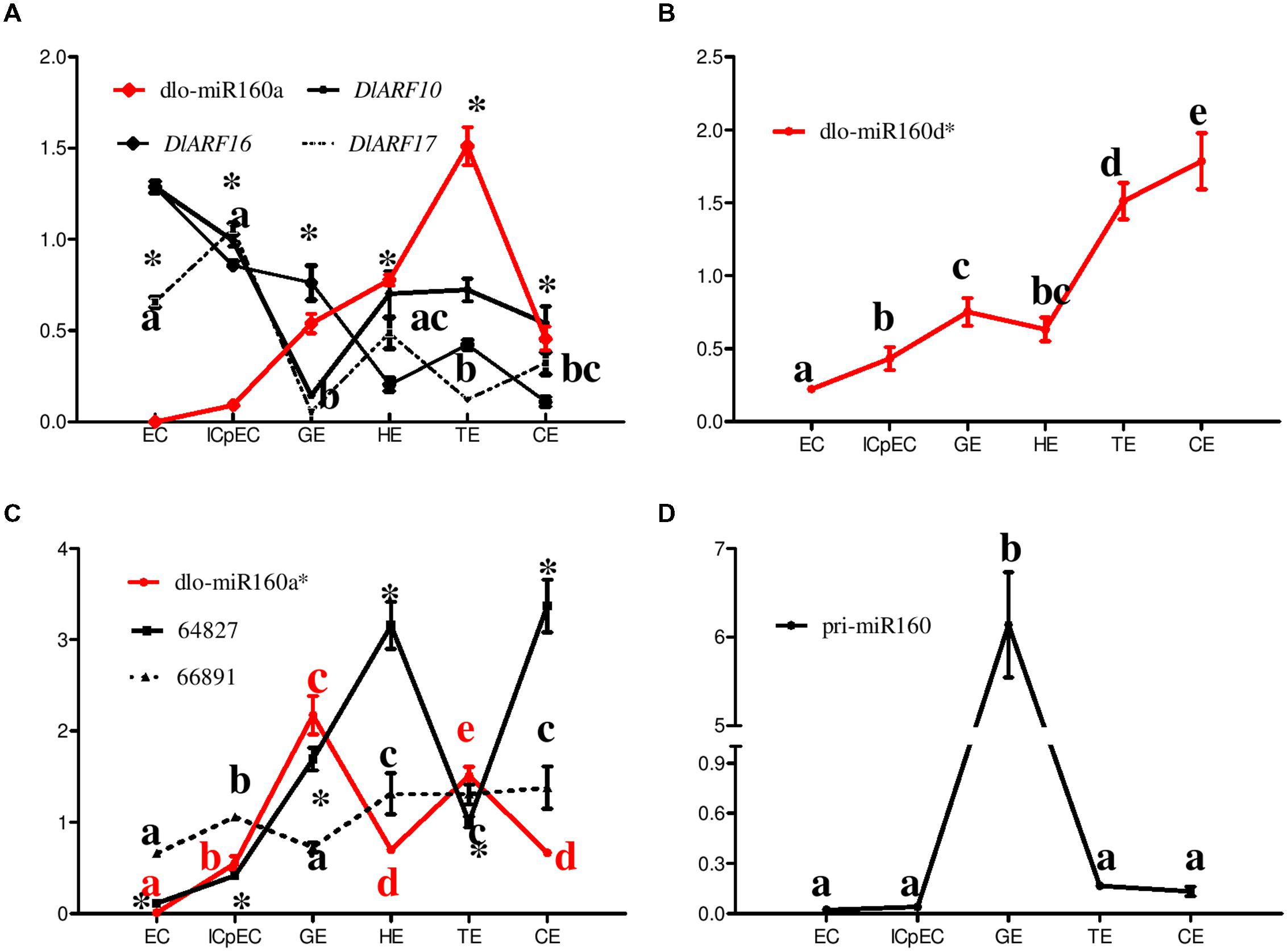
FIGURE 6. Relative expression of eTMs (Unigene64827/66891), the dlo-miR160 family, miR160 targets and pri-miR160 during longan SE. (A–D) The expression levels were normalized to the reference genes DlFSD1a, EF-1a and eIF-4a. Unigene64827: glucan endo-1,3-beta-glucosidase-like protein 2; Unigene66891: calpain-type cysteine protease DEK1 family. Samples: EC, friable-embryogenic callus; ICpEC, incomplete compact pro-embryogenic cultures; GE, globular embryos; HE, heart-shaped embryos; TE, torpedo-shaped embryos; CE, cotyledonary embryos. The y-axes represent the relative expression values; the x-axes represent longan SE samples. Asterisks (∗) and different lowercase letters above the bars indicate a statistically significant difference, and identical lowercase letters denote no significant difference among different embryogenic cultures (P < 0.05).
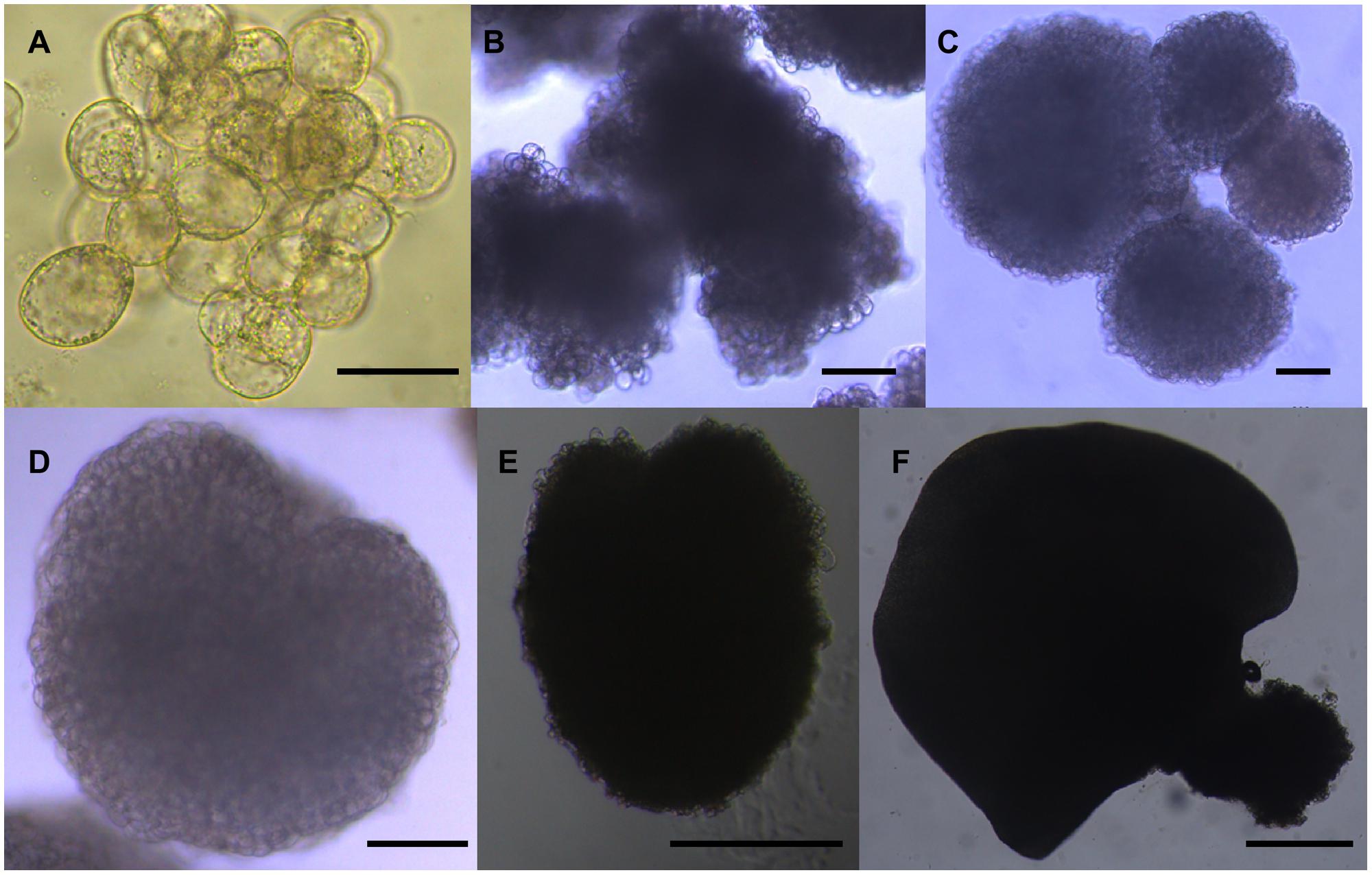
FIGURE 7. Morphology of the six sequential developmental stages of longan SE. (A) Friable-embryogenic callus (EC); (B) incomplete compact pro-embryogenic cultures (ICpEC); (C) globular embryos (GE); (D) heart-shaped embryos (HE); (E) torpedo-shaped embryos (TE); (F) cotyledonary embryos (CE). The bars are 50 μm in (A), 100 μm in (B–D), and 500 μm in (E,F).
The expression levels of dlo-miR160a, -a∗, and -d∗ were investigated during D. longan SE (Figures 6A–C). Members of the dlo-miR160 family exhibited different temporal and spatial expression. Dlo-miR160a and -a∗ both exhibited tissue-specific expression and showed similar expression patterns; both were barely detectable in EC. The abundance of dlo-miR160a increased during the ICpEC, GE and HE stages and reached a peak in the TE stage, while dlo-miR160a∗ showed its highest expression in GE. In addition, dlo-miR160d∗ expression was lowest in EC, but continually increased in ICpEC-GE-TE, reaching its peak in the CE stage. These results showed that different members of the dlo-miR160 family might have different and separate regulatory effects on longan SE. Furthermore, the expression levels of the mature miR160s and pri-miR160 were compared. Pri-miR160 showed a similar expression pattern to dlo-miR160a∗, but had some differences compared with the expression levels of dlo-miR160a and -d∗ (Figure 6D).
To examine whether the four predicted eTMs for dlo-miR160a∗ were indeed transcribed, and to test whether these eTMs control miR160 expression, we validated their expression during longan SE. The unknown mRNA (Unigene11608) and nitrate transporter family protein (Unigene56365) were not detected in the six tissues. However, endo-1,3-beta-glucosidase-like protein 2-like (Unigene64827) and calpain- type cysteine protease DEK1 (Unigene 66891) showed great variation among the tissues (Figure 6C). Both were highly expressed in the HE and CE stages, and weakly expressed in EC, suggesting that they might play potential roles in the late stages of SE. Their expression patterns were largely reciprocal to that of dlo-miR160a∗, especially at the GE to CE stages (Figure 6C). These results showed that the predicted eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain-type cysteine protease DEK1) may indeed function to abolish the binding between dlo-miR160a∗ and its targets.
DlARF10, -16, and -17 targeting by dlo-miR160a members was confirmed by RLM- RACE during longan SE, and their expression levels were further examined in six tissues (Figure 6A). DlARF10 and -16 both reached their peaks in the EC stage, while DlARF17 expression was highest in the ICpEC stage. DlARF10 and -17 expression was lowest in the GE stage, and DlARF16 was expressed weakly in the CE stage. As expected, their expression was mainly inversely proportional to that of dlo-miR160a (Figure 6).
Taken together, these results confirmed the eTM-miR160-ARF10-16-17 regulatory pathway might play a potential role during the SE process in D. longan.
2,4-D Controls the Levels of the eTMs, miR160, and ARF10, -16, and -17
Auxin plays major roles in embryo development. To investigate whether the miR160 level and eTM-miR160-ARF10-16-17 signal transduction were controlled by exogenous auxin, they were analyzed in longan EC grown for 24 h on media containing different concentrations of 2,4-D (an auxinic herbicide). The results showed that the 2,4-D concentration affected the eTM, miR160, and ARF10, -16, and -17 levels (Figure 8).
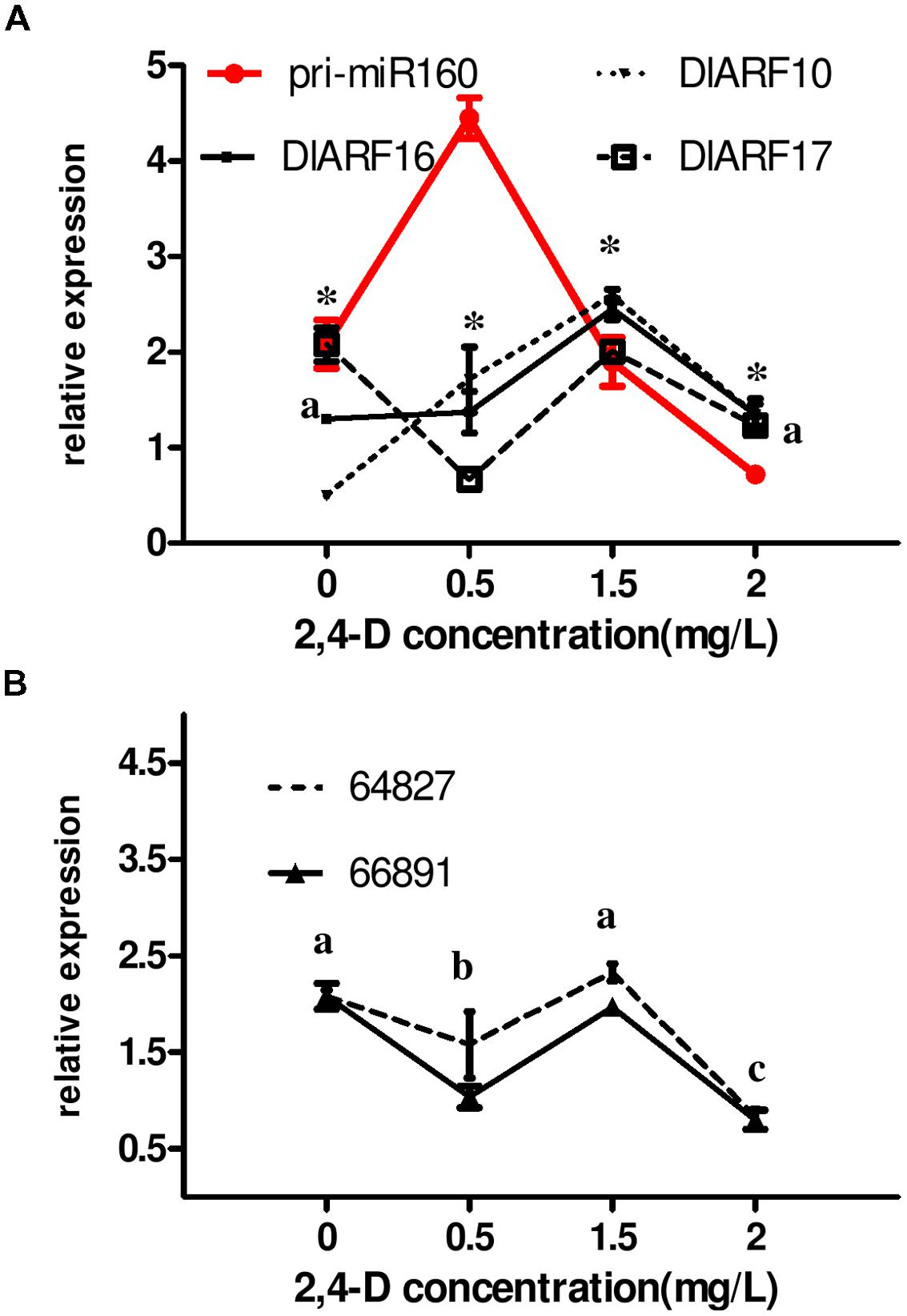
FIGURE 8. Auxin control of eTM, miR160, and ARF10/-16/-17 expression levels. (A,B) The expression levels were normalized to the reference genes DlFSD1a, EF-1a and eIF-4a. Asterisks (∗) and different lowercase letters above the bars indicate a statistically significant difference, and identical lowercase letters denote no significant difference among different embryogenic cultures (P < 0.05).
Pri-miR160 accumulated at 0.5 mg/L 2,4-D after 24 h of treatment compared with auxin-free medium under the same conditions, but decreased significantly at higher concentrations (Figure 8), substantiating the participation of miR160 in the 2,4-D signal pathway. The expression patterns of the eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain-type cysteine protease DEK1) resembled those of DlARF16 and -17 targeted by dlo-miR160a under the tested concentrations of 2,4-D (Figure 8). Both were expressed at lower levels at 0.5 mg/L 2,4-D compared with auxin-free medium, increased at 1.5 mg/L, and decreased at 2.0 mg/L, which was largely reciprocal to pri-miR160 expression. This result demonstrated that the presence of auxin in the medium affected the levels of the eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain-type cysteine protease DEK1), and in turn, miR160- mediated regulation of DlARF16 and -17 in longan EC. However, the level of DlARF10 increased as the 2,4-D concentration increased (0.5–1.5 mg/L), but decreased at 2.0 mg/L, which was not the reverse of pri-miR160. This suggested that DlARF10 responds to 2,4-D in a positive manner, but might be out of miR160’s control.
Taken together, these results clearly demonstrated that 2,4-D controlled the levels of the eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain-type cysteine protease DEK1), and in turn, miR160-mediated regulation of DlARF16 and -17 in longan. However, it is not clear whether auxin affects the eTM levels directly.
ABA Regulates eTM-miR160 Signal Transduction
A previous study showed that miR160-ARF10 plays an important role in ABA-auxin crosstalk in seed germination and post-embryonic developmental programs (Liu et al., 2007). To test whether ABA treatment also affected miR160 expression and eTM-MIR160 signal transduction in longan, longan EC was grown for 24 h on medium containing 100 μM ABA and analyzed. The results showed that ABA treatment affected the eTM and miR160 levels (Figure 9). After ABA treatment, the levels of the eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain-type cysteine protease DEK1) were slightly decreased compared with the control, while pri-miR160 expression was elevated after 24 h. Thus, the increased expression of pri-miR160 in longan EC after ABA treatment might be caused by decreased expression of the eTMs (endo-1,3-beta-glucosidase-like protein 2-like and calpain- type cysteine protease DEK1), or through the ABRE element in its 5′ regulatory region.
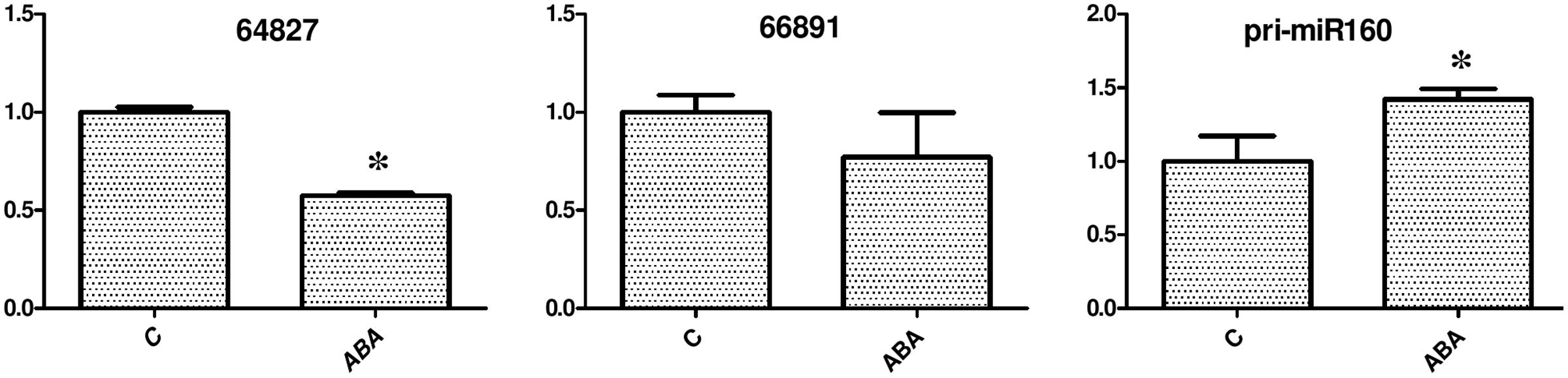
FIGURE 9. Exogenous ABA treatment affects the eTM and miR160 expression levels. The expression levels were normalized to the reference gene EF-1a. Unigene64827: glucan endo-1,3-beta-glucosidase-like protein 2, Unigene66891: calpain-type cysteine protease DEK1 family. An asterisk (∗) indicates a statistically significant difference between the control (C) and hormone treatments (P < 0.05). The y-axes represent relative expression values; the x-axes represent hormone concentration.
Expression Changes of Pri-miR160 in Response to Hormone Treatments
To further analyze the possible functions of miR160 in phytohormone signaling pathways, the expression of pri-miR160 in response to hormonal stimuli was investigated. The pri-miR160 expression level was greatly increased in response to GA3 (35 μM), ET (30 ppm), and MeJA (50 μM) treatments, but was slightly decreased after 24 h of SA (75 μM) treatment (Figure 10). Thus, GA3, ET, and MeJA up-regulate the expression of miR160, while SA down-regulates its expression in longan EC, possibly through related cis-elements in its promoter region. However, only an SA-responsive cis-element (TCA-element) was predicted in the promoter region. These results suggest that miR160 may also mediate these hormone signal interactions.
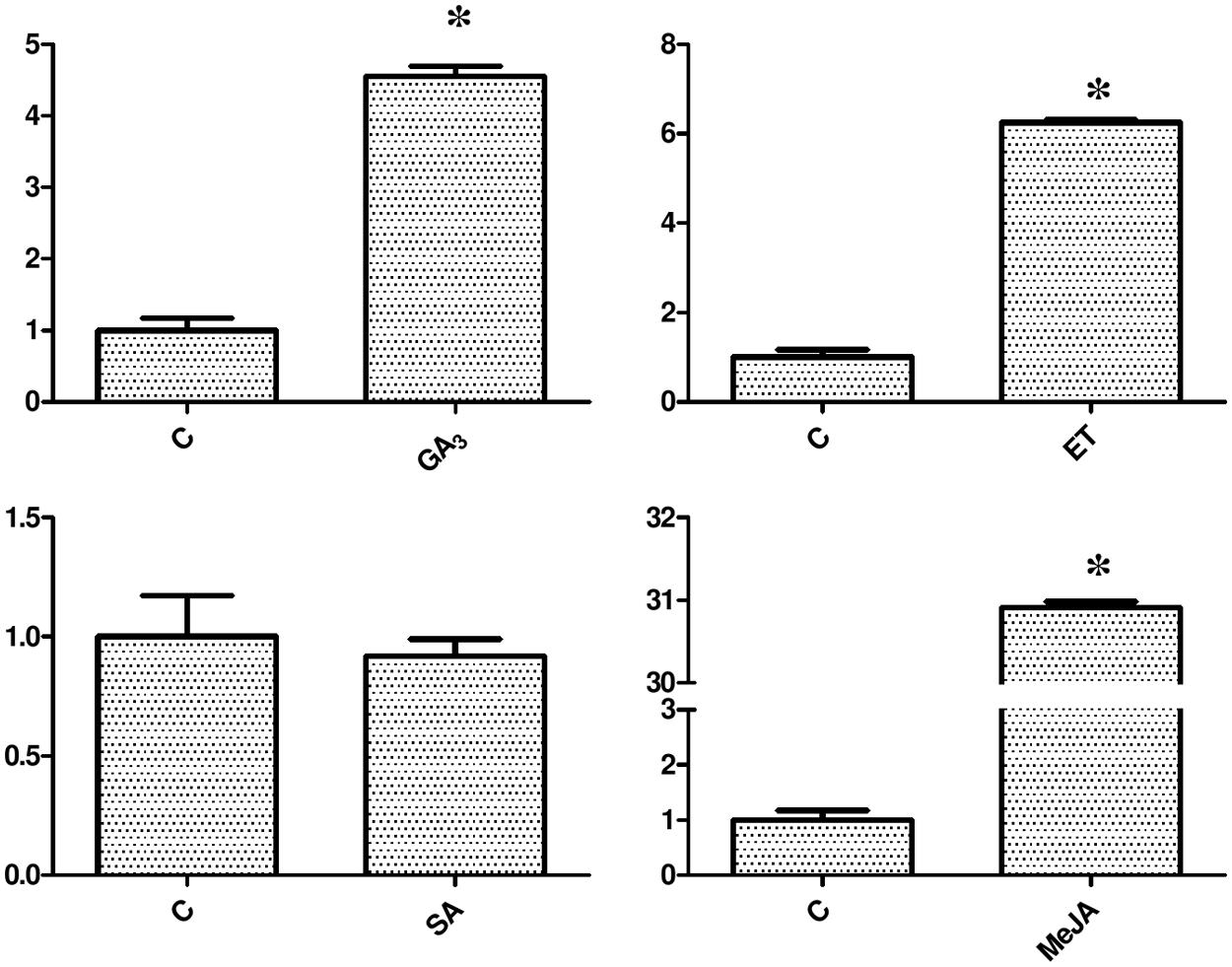
FIGURE 10. Expression analysis of pri-miR160 in response to exogenous hormone treatments. The expression levels were normalized to the reference gene EF-1a. An asterisk (∗) indicates a statistically significant difference between the control (C) and hormone treatments (P < 0.05). The y-axes represent relative expression values; the x-axes represent hormone concentration.
Expression of eTMs, miR160, and ARF10, -16, and -17 in ‘Sijimi’ Tissues
To get more information about the characteristics of the eTM, miR160, and ARF10, -16, and -17 accumulation patterns in other tissues, their expression in D. longan Lour. cv. Sijimi was studied (Figures 11 and 12). ‘Sijimi’ is unique cultivar of longan that blossoms and bears fruit throughout the year. A variety of tissues (Figure 12), including roots (R), mature leaves (L), floral buds (FB), flowers (F), young fruits (YF), mature fruits (MF), pericarp (P1), pulp (P2) and seeds (S), were used to assay RNA accumulation.
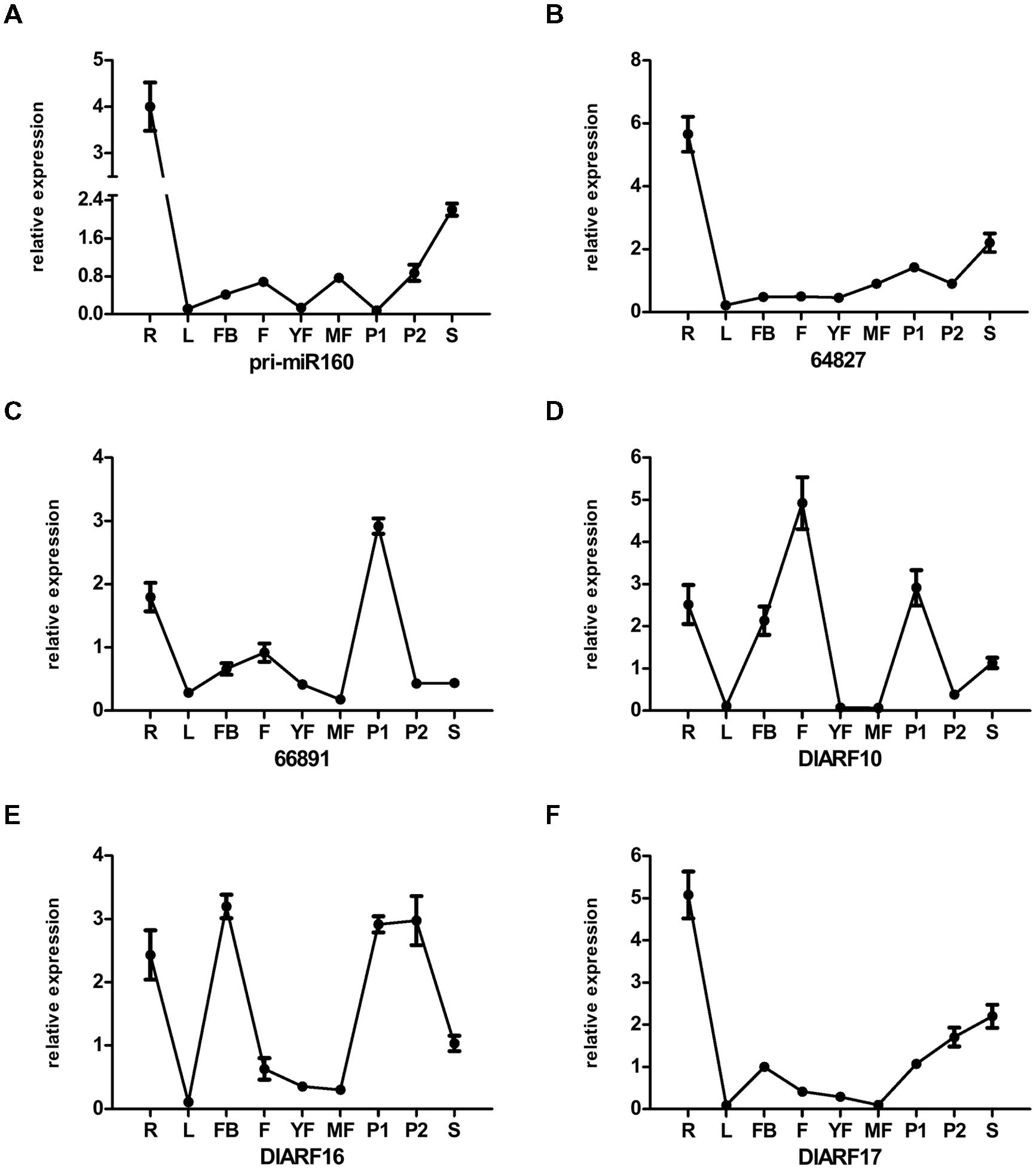
FIGURE 11. Differential expression analysis of eTMs, miR160, and DlARF10, -16, and -17 in vegetative and generative tissues of ‘Sijimi’ longan. (A–F), qPCR analysis of the expression levels of the eTMs, miR160, and DlARF10/-16/-17. Unigene64827: glucan endo-1,3-beta-glucosidase-like protein 2, Unigene66891: calpain-type cysteine protease DEK1 family. The expression levels were normalized to the reference genes DlFSD1a, EF-1a, and eIF-4a. Samples: roots (R), leaves (L), floral buds (FB), flowers (F); young fruit (YF), mature fruit (R), pericarp (P1), pulp (P2), and seeds (S).
FIGURE 12. Morphology of vegetative and generative tissues of ‘Sijimi’ longan. (A) Roots (R); (B) leaves (L); (C) flowers (F) and floral buds (FB); (D), young fruit (YF); (E) mature fruit (R) and pericarp (P1, arrow); (F) pulp (P2, arrow), and seeds (S). Bars are 8 mm.
We examined the expression patterns in all tissues and found that the eTMs, miR160 and ARF10, -16, and -17 exhibited tissue specificity (Figures 11A–F). All the genes were highly expressed in roots, and showed the lowest expression levels in mature leaves, except for calpain-type cysteine protease DEK1 (Unigene 66891), which was least expressed in mature fruits. These observations indicated that eTM-miR160-ARF10- 16-17 signal transduction might affect ‘Sijimi’ root development. In addition, high expression levels were observed for pri-miR160, endo-1,3-beta-glucosidase-like protein 2-like (Unigene 64827) and DlARF17 in seeds, calpain-type cysteine protease DEK1 (Unigene 66891) and DlARF10 and -16 in pulp, DlARF10 in flowers, and DlARF16 in floral buds and pericarp. Furthermore, the expression of pri-miR160 in young fruits and pulp, DlARF10 and -16 in young fruits and mature fruits, and DlARF17 in mature fruits was very low or undetectable. These data suggested eTM-miR160-ARF10-16-17 signal transduction plays an extensive role in ‘Sijimi’ development, but there was no significant negative correlation among the levels of eTM, miR160 and DlARF10, -16, and -17 mRNA accumulation in the organs analyzed.
Discussion
Unique Size and Structure of longan miR160
Plant miRNAs are usually encoded by small gene families of up to 14 members (Parizotto et al., 2004), similar to protein-coding genes. The miR160 family in plants includes three members in Arabidopsis, six members in O. sativa and Zea mays, and seven members in Populus trichocarpa (Jin et al., 2012). In our study, only two members, dlo-miR160a and dlo-miR160d∗, were detected in longan somatic embryos. However, these two members were represented by 12 variants with identical or nearly identical sequences and different read counts. qPCR results further indicated that the different members of the dlo-miR160 family had different expression patterns. A previous study also showed that the multi-copy miR160 family has expression diversification, and may have functional diversity. For example, miR160a regulates embryo development at the very beginning of embryogenesis (Liu et al., 2010b), while miR160c regulates root cap development (Wang et al., 2005), which suggests that the different spatial and temporal expression patterns of the dlo-miR160 family members reflects their functional diversity in longan SE.
The secondary structures of miRNA genes play an integral role in their maturation, regulation and functions (Parizotto et al., 2004; Jiang et al., 2006). miRNA genes have been identified using computational approaches that depend upon the availability of reference or genomic sequences (Megraw et al., 2006; Meng et al., 2009; Branscheid et al., 2011; Han et al., 2014). However, most miRNA genes, especially in non-model plants, have not been investigated, primarily because of a lack of genome-wide datasets. In our study, 5′ and 3′ RACE experiments, together with the data deposited in the GenBank and miRBase databases, were used to obtain the full-length pri-miRNA for miR160. The corresponding pri-miR160a genomic DNA sequence (424 bp, containing one intron) was obtained, which was shorter than the Arabidopsis miR160a gene (2034-bp long; one intron), but longer than the Arabidopsis miR160b gene (378-bp long; no introns) (Szarzynska et al., 2009). This suggests that the pri-miR160 genes from different species and different members vary in size and structure.
The Longan miR160 Promoter is Associated with Hormone Signal Transduction
Many previous studies have focused on elucidating the mechanism of miRNA target gene regulation, but there is limited information on the regulation of miRNA genes themselves. Previous studies have shown that the promoters of most of the known pre-miRNAs share similar characteristics with RNA polymerase II-dependent protein-coding gene promoters (Zhou et al., 2007). The identification of the promoters of miRNA genes is a work in progress; however, most studies have concentrated on A. thaliana (Megraw et al., 2006; Zhou et al., 2007) and O. sativa (Cui et al., 2009; Meng et al., 2009). Here, the pri-miR160 promoter was cloned from longan EC and contained the conserved TATA and CAAT boxes.
The miR160 family has the highest number of identified ARF binding sites (AuxREs) in cassava (Perez-Quintero et al., 2012), G. max (miR160d and miR160f) (Han et al., 2014), and rice (osamiR160f). However, ARF binding sites were not found in the longan miR160 promoter. Previous experiments showed that indole acetic acid (IAA) treatment did not influence the auxin-related rice homologous miRNAs significantly, such as osa-miR160, osa-miR164, and osa-miR167 (Meng et al., 2009). Here, exogenous 2,4-D treatment significantly influenced pri-miR160 expression, suggesting that ARF elements may be present outside of the 960 bp promoter region. Indeed, AuxREs in the 3′ regulatory region of miR160a are required for auxin regulation of miR160a expression in Arabidopsis (Liu et al., 2010b). However, AuxREs were not found in the 3′ regulatory region of pri-miR160.
Other cis-regulatory elements associated with plant hormones, such as ABA (ABRE) and SA (TCA-element), necessary for plant responses to various abiotic stresses (Kruszka et al., 2012), were detected in longan. In Arabidopsis, miR160, miR393 and miR397b were up-regulated, whereas miR169 and miR398 were down-regulated, in response to ABA stress. In another study, negative regulation of ARF10 by miR160 modulated the response to ABA stress during germination (Liu et al., 2007). Our qPCR results showed that ABA up-regulated the expression of miR160, which suggests that miR160 is involved in ABA signaling in longan. Furthermore, pri-miR160 was down-regulated in response to SA treatment, implying that miR160 is also involved in SA signaling in longan. GA3, ET, and MeJA also up-regulated the expression level of pri-miR160, but corresponding cis-regulatory elements were not identified in the 960 bp longan miR160 promoter region. However, gibberellin response factor elements were found in the putative promoter regions of miR160f, miR160g, and miR160h in G. max (Han et al., 2014), and auxin-, MeJA-, ET-, GA3-, and SA-responsive elements were found in the promoter regions of miR160s in cassava (Pinweha et al., 2015). Taken together, these results clearly demonstrated that the pri-miR160 was down-regulated in response to SA (75 μM), but up-regulated by MeJA (50 μM), ET (30 ppm), GA3 (35 μM), or ABA (100 μM), suggesting that miR160s are widely involved in hormone signal transduction in longan. However, it is not clear whether these hormones affect the morphogenesis of somatic embryos; this requires further experimental validation.
The other types of cis-regulatory elements in the upstream region of the miR160 gene in longan were defense and stress responsive, such as the anaerobic-response element (ARE) and HSE, which were also found in the cassava miR160 promoter (Pinweha et al., 2015). Recently, reports showed that miR160 plays a role in the defense and stress responses of plants (Jin et al., 2012; Pinweha et al., 2015). miR160s were highly induced after infection with Pseudomonas syringae pv. tomato (Fahlgren et al., 2007). These results suggest that miR160 might play a role in defense and stress in plants.
In conclusion, these results form the basis for further experiments to identify the function of miR160 in responses to various hormone and environmental stimuli.
eTMs Down-regulated miR160 Cleavage of DlARF10, -16, and -17 mRNAs During Longan SE
Target mimicry, a new regulatory mechanism for miRNA functions, was first studied in Arabidopsis. eTMs for miR160 have been identified in Arabidopsis (Wu et al., 2013), rice (Wu et al., 2013), and P. trichocarpa (Shuai et al., 2014), and are functional target mimics that cause smaller and serrated leaves in transgenic plants (Wu et al., 2013). However, previous studies focused mainly on eTMs derived from intergenic non-coding RNA genes. In our study, four potential eTMs derived from annotated genes were initially identified for miR160a∗ in longan; however, only two eTMs: glucan endo-1,3-beta-glucosidase-like protein 2-like (Unigene64827) and calpain-type cysteine protease DEK1 (Unigene 66891), were detected during longan SE by qPCR. Their expression patterns were inversely proportional to that of the corresponding dlo-miR160a∗, especially at the GE to CE stages. Moreover, their expression was down-regulated while dlo-miR160a∗ was up-regulated by auxin and ABA treatment. These results further proved that Unigene64827 and Unigene 66891 inhibited the expression level of dlo-miR160a∗ and participated in auxin and ABA signal transduction during longan SE. Unigene64827 encodes a homolog of glucan endo-1,3-beta-glucosidase-like protein 2, which is related to cell wall synthesis (Meitzel et al., 2011). Unigene66891 encodes a protein similar to calpain-type cysteine protease DEK1, which is required for epidermal development in the embryo (Johnson et al., 2005). These results suggest that they play a potential role in cell wall synthesis and epidermal development in the embryo by regulating dlo-miR160a∗, especially at the GE to CE stages. This hypothesis requires further experimental validation.
miR160 and its target ARFs are conserved in dicots and monocots (Rhoades et al., 2002). Here, DlARF10, -16, and -17 targeting by dlo-miR160a was confirmed, and their expression levels were mostly the reverse of that of dlo-miR160a. In addition, dlo-miR160a was up-regulated while DlARF16 and -17 were down-regulated by 2,4-D treatment. These results were consistent with earlier reports (Liu et al., 2010b), which suggested that the increased expression of dlo-miR160a induced by 2,4-D treatment down-regulated the expression of ARF16 and -17, further confirming that miR160-ARF16-17 auxin signal transduction operates in longan similarly to Arabidopsis. By contrast, DlARF10 was up-regulated after 2,4-D treatment, suggesting it may not be controlled by dlo-miR160a under auxin treatment.
Auxin response factor proteins can either activate or repress transcription, based on their protein structures and transient expression experiments. ARFs 5–8 and ARF19 contain Gln-Leu-Ser–rich middle regions that may function as transcriptional activators, whereas ARFs 1–4, 9–18, and 20–22 possibly act as repressors, with Pro-Ser-Thr–rich middle regions (Tiwari et al., 2003; Mallory et al., 2005). In our study, DlARF10, -16, and -17 expression was stronger in whole embryos at the EC and ICpEC stages, and gradually decreased during somatic embryo development, suggesting they might function as repressors to establish and maintain auxin signals during embryogenesis (Mallory et al., 2005; Liu et al., 2007, 2010b).
miR160-resistant mutants have been used successfully to characterize the biological functions of ARF10 (Liu et al., 2007), -16 (Wang et al., 2005) and -17 (Mallory et al., 2005), revealing that down-regulation of these genes is essential to maintain the normal developmental programs of the roots, leaves, flowers and siliques. In our study, neither pri-miR160 nor ARF10, -16, or -17 exhibited tissue specificity in ‘Sijimi’ longan. They all showed high expression in roots and were not detected in mature leaves. By contrast, miR160c expression was low or undetectable in juvenile leaves and became high in older leaves, as assessed by GUS staining of ProMIR160c:GUS plants (Wang et al., 2005). miR160a is expressed in all organs, especially in reproductive organs (Liu et al., 2010b), suggesting that miR160 regulation of ARF10, -16, and -17 signal transduction has an extensive role in ‘Sijimi’ development. However, there were no remarkable negative correlations among the levels of miR160 and the DlARF10, -16, and -17 mRNAs in the analyzed tissues.
Conclusion
In summary, we performed dlo-miR160 family member identification, primary transcript and promoter cloning; target mimic prediction, miRNA target identification, and qPCR verification. According to our experiments as well as the negative feedback model, we propose a possible “eTM–miR160–target” regulatory loop during longan SE (Figure 13). The eTMs glucan endo-1,3-beta-glucosidase-like protein 2-like (Unigene64827) and calpain-type cysteine protease DEK1 (Unigene 66891) down- regulation of miR160’s mediation of ARF10, -16, and -17 cleavage is associated with SE, and responds to 2,4-D and ABA signaling transduction in longan. In addition, the expression of pri-miR160 is controlled by GA3, ET, MeJA, and SA.
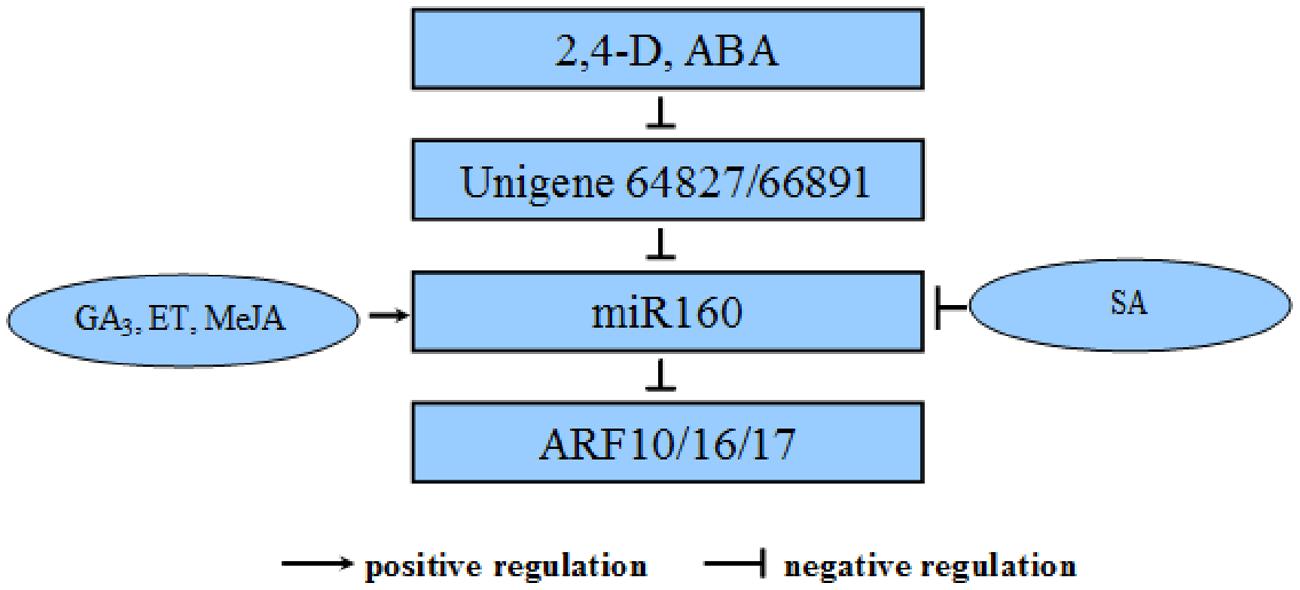
FIGURE 13. Negative regulatory relationships among the eTMs, miR160, and its targets involved in SE in D. longan in relation to hormone metabolism. The eTMs Unigene64827 and Unigene66891 belong to the glucan endo-1,3-beta-glucosidase- like protein 2 and calpain-type cysteine protease DEK1 families, respectively. They down-regulate miR160 by mediating cleavage of ARF10, -16, and -17, which are involved in SE in D. longan, and respond to 2,4-D and ABA signal transduction. In addition, the expression of pri-miR160 is controlled by exogenous GA3, ET, MeJA and SA.
Author Contributions
YL conceived the study, participated in its design, carried out the experimental work and wrote the manuscript. ZL conceived the study, participated in its design and coordination, and helped to draft the manuscript. QT, LL, RL, MY, DZ, YC, and ZZ prepared the materials. All authors read and approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was funded by Research Funds for the Doctoral Program of Higher Education of the Chinese Ministry of Education (20123515120008), the National Natural Science Foundation of China (31201614 and 31272149), the Natural Science Funds for Distinguished Young Scholars in Fujian Province (2015J06004), the Fujian Agriculture and Forestry University Funds for Distinguished Youth Fund (xjq201405), and the Outstanding Youth Fund of the Fujian Provincial Department of Education Teacher Education of Young and Middle-aged Teachers in Science and Technology Research (Science and Technology Class A) (JA14099).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
2,4-D, 2,4-dichlorophenoxyacetic acid; ABA, abscisic acid; ARF, auxin response factor; CE, cotyledonary embryos; EC, friable embryogenic callus; ET, ethylene; eTMs, endogenous target mimics; GA3, gibberellic acid; GE, globular embryos; HE, heart- shaped embryos; ICpEC, incomplete compact pro-embryogenic cultures; MeJA, methyl jasmonate; miRNAs, microRNAs; pre-miRNA, precursor miRNA; pri-miRNAs, primary miRNA transcripts; qPCR, real-time quantitative PCR; RLM-RACE, RNA ligase-mediated rapid amplification of cDNA ends; SA, salicylic acid; SE, somatic embryogenesis; TE, torpedo-shaped embryos; UTR, untranslated region.
References
Bonnet, E., He, Y., Billiau, K., and Van De Peer, Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566–1568. doi: 10.1093/bioinformatics/btq233
Branscheid, A., Devers, E. A., May, P., and Krajinski, F. (2011). Distribution pattern of small RNA and degradome reads provides information on miRNA gene structure and regulation. Plant Signal. Behav. 6, 1609–1611. doi: 10.4161/psb.6.10.17305
Carrington, J. C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. doi: 10.1126/science.1085242
Chung, Y. C., Lin, C. C., Chou, C. C., and Hsu, C. P. (2010). The effect of Longan seed polyphenols on colorectal carcinoma cells. Eur. J. Clin. Invest. 40, 713–721. doi: 10.1111/j.1365-2362.2010.02322.x
Cui, X., Xu, S. M., Mu, D. S., and Yang, Z. M. (2009). Genomic analysis of rice microRNA promoters and clusters. Gene 431, 61–66. doi: 10.1016/j.gene.2008.11.016
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gandikota, M., Birkenbihl, R. P., Hohmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313X.2006.02983.x
Han, Y. Q., Hu, Z., Zheng, D. F., and Gao, Y. M. (2014). Analysis of promoters of microRNAs from a Glycine max degradome library. J. Zhejiang Univ. Sci. B 15, 125–132. doi: 10.1631/jzus.B1300179
Huang, D., Koh, C., Feurtado, J. A., Tsang, E. W., and Cutler, A. J. (2013). MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 14:140. doi: 10.1186/1471-2164-14-140
Jiang, D., Yin, C., Yu, A., Zhou, X., Liang, W., Yuan, Z., et al. (2006). Duplication and expression analysis of multicopy miRNA gene family members in Arabidopsis and rice. Cell Res. 16, 507–518. doi: 10.1038/sj.cr.7310062
Jin, W., Wu, F., Xiao, L., Liang, G., Zhen, Y., Guo, Z., et al. (2012). Microarray-based analysis of tomato miRNA regulated by Botrytis cinerea. J. Plant Growth Regul. 31, 38–46. doi: 10.1007/s00344-011-9217-9
Johnson, K. L., Degnan, K. A., Ross Walker, J., and Ingram, G. C. (2005). AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44, 114–127. doi: 10.1111/j.1365-313X.2005.02514.x
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kruszka, K., Pieczynski, M., Windels, D., Bielewicz, D., Jarmolowski, A., Szweykowska-Kulinska, Z., et al. (2012). Role of microRNAs and other sRNAs of plants in their changing environments. J. Plant Physiol. 169, 1664–1672. doi: 10.1016/j.jplph.2012.03.009
Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163. doi: 10.1093/bib/5.2.150
Lai, Z., and Chen, C. (1997). Somatic embryogenesis of high frequency from longan embryogenic calli. J. Fujian. Agric. Univ. 26, 271–276.
Lai, Z. X., He, Y., Chen, Y. T., Cai, Y. Q., Lai, C. C., Lin, Y. L., et al. (2010). Molecular biology and proteomics during somatic embryogenesis in Dimocarpus longan Lour. Acta Hortic. 863, 95–102. doi: 10.17660/ActaHortic.2010.863.10
Lai, Z., and Lin, Y. (2013). Analysis of the global transcriptome of longan (Dimocarpus longan Lour.) embryogenic callus using Illumina paired-end sequencing. BMC Genomics 14:561. doi: 10.1186/1471-2164-14-561
Lee, Y. S., Nakahara, K., Pham, J. W., Kim, K., He, Z., Sontheimer, E. J., et al. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81. doi: 10.1016/S0092-8674(04)00261-2
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lin, Y. L., and Lai, Z. X. (2010). Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 178, 359–365. doi: 10.1016/j.plantsci.2010.02.005
Lin, Y.-L., and Lai, Z.-X. (2013a). Superoxide dismutase multigene family in longan somatic embryos: a comparison of CuZn-SOD, Fe-SOD, and Mn-SOD gene structure, splicing, phylogeny, and expression. Mol. Breed. 32, 595–615. doi: 10.1007/s11032-013-9892-2
Lin, Y., and Lai, Z. (2013b). Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.). PLoS ONE 8:e60337. doi: 10.1371/journal.pone.0060337
Lin, Y. L., and Lai, Z. X. (2013c). Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol. Biochem. 66, 20–25. doi: 10.1016/j.plaphy.2013.02.002
Liu, H., Liu, Y. Z., Zheng, S. Q., Jiang, J. M., Wang, P., and Chen, W. (2010a). Comparative proteomic analysis of longan (Dimocarpus longan Lour.) seed abortion. Planta 231, 847–860. doi: 10.1007/s00425-009-1093-1
Liu, X., Huang, J., Wang, Y., Khanna, K., Xie, Z., Owen, H. A., et al. (2010b). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62, 416–428. doi: 10.1111/j.1365-313X.2010.04164.x
Liu, P. P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H., and Carrington, J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146. doi: 10.1111/j.1365-313X.2007.03218.x
Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q., Yang, H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 15485–15490. doi: 10.1073/pnas.1304651110
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Mallory, A. C., Bartel, D. P., and Bartel, B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17, 1360–1375. doi: 10.1105/tpc.105.031716
Megraw, M., Baev, V., Rusinov, V., Jensen, S. T., Kalantidis, K., and Hatzigeorgiou, A. G. (2006). MicroRNA promoter element discovery in Arabidopsis. RNA 12, 1612–1619. doi: 10.1261/rna.130506
Meitzel, T., Radchuk, R., Nunes-Nesi, A., Fernie, A. R., Link, W., Weschke, W., et al. (2011). Hybrid embryos of Vicia faba develop enhanced sink strength, which is established during early development. Plant J. 65, 517–531. doi: 10.1111/j.1365-313X.2010.04450.x
Meng, Y., Huang, F., Shi, Q., Cao, J., Chen, D., Zhang, J., et al. (2009). Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta 230, 883–898. doi: 10.1007/s00425-009-0994-3
Parizotto, E. A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242. doi: 10.1101/gad.307804
Park, S. J., Park, D. H., Kim, D. H., Lee, S., Yoon, B. H., Jung, W. Y., et al. (2010). The memory-enhancing effects of Euphoria longan fruit extract in mice. J. Ethnopharmacol. 128, 160–165. doi: 10.1016/j.jep.2010.01.001
Perez-Quintero, A. L., Quintero, A., Urrego, O., Vanegas, P., and Lopez, C. (2012). Bioinformatic identification of cassava miRNAs differentially expressed in response to infection by Xanthomonas axonopodis pv. manihotis. BMC Plant Biol. 12:29. doi: 10.1186/1471-2229-12-29
Pinweha, N., Asvarak, T., Viboonjun, U., and Narangajavana, J. (2015). Involvement of miR160/miR393 and their targets in cassava responses to anthracnose disease. J. Plant Physiol. 174, 26–35. doi: 10.1016/j.jplph.2014.09.006
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Shuai, P., Liang, D., Tang, S., Zhang, Z., Ye, C. Y., Su, Y., et al. (2014). Genome-wide identification and functional prediction of novel and drought- responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 65, 4975–4983. doi: 10.1093/jxb/eru256
Szarzynska, B., Sobkowiak, L., Pant, B. D., Balazadeh, S., Scheible, W. R., Mueller- Roeber, B., et al. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37, 3083–3093. doi: 10.1093/nar/gkp189
Tiwari, S. B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. doi: 10.1105/tpc.008417
Todesco, M., Rubio-Somoza, I., Paz-Ares, J., and Weigel, D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6:e1001031. doi: 10.1371/journal.pgen.1001031
Turner, M., Nizampatnam, N. R., Baron, M., Coppin, S., Damodaran, S., Adhikari, S., et al. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162, 2042–2055. doi: 10.1104/pp.113.220699
Wang, J. W., Wang, L. J., Mao, Y. B., Cai, W. J., Xue, H. W., and Chen, X. Y. (2005). Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216. doi: 10.1105/tpc.105.033076
Wu, H. J., Wang, Z. M., Wang, M., and Wang, X. J. (2013). Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161, 1875–1884. doi: 10.1104/pp.113.215962
Yan, J., Gu, Y., Jia, X., Kang, W., Pan, S., Tang, X., et al. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yi, Y., Liao, S. T., Zhang, M. W., Shi, J., Zhang, R. F., Deng, Y. Y., et al. (2011). Physicochemical characteristics and immunomodulatory activities of three polysaccharide-protein complexes of longan pulp. Molecules 16, 6148–6164. doi: 10.3390/molecules16076148
Zhang, J., Zhang, S., Han, S., Wu, T., Li, X., Li, W., et al. (2012). Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236, 647–657. doi: 10.1007/s00425-012-1643-9
Zheng, S. Q., Jiang, F., Gao, H. Y., and Zheng, J. G. (2010). Preliminary observations on the antifatigue effects of longan (Dimocarpus longan Lour.) seed polysaccharides. Phytother. Res. 24, 622–624. doi: 10.1002/ptr.2963
Keywords: Dimocarpus longan, somatic embryogenesis, microRNA160, endogenous target mimics, auxin response factors, gene regulation
Citation: Lin Y, Lai Z, Tian Q, Lin L, Lai R, Yang M, Zhang D, Chen Y and Zhang Z (2015) Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 6:956. doi: 10.3389/fpls.2015.00956
Received: 05 August 2015; Accepted: 20 October 2015;
Published: 05 November 2015.
Edited by:
Agnieszka Ludwików, Adam Mickiewicz University, PolandReviewed by:
Biswapriya Biswavas Misra, University of Florida, USATaras P. Pasternak, University of Freiburg, Germany
Copyright © 2015 Lin, Lai, Tian, Lin, Lai, Yang, Zhang, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongxiong Lai, bGFpengwMUAxNjMuY29t
 Yuling Lin
Yuling Lin Zhongxiong Lai
Zhongxiong Lai