- Key Laboratory of Rice Genetic Breeding of Anhui Province, Rice Research Institute, Anhui Academy of Agricultural Sciences, Hefei, China
Nitrogen recycling and redistribution are important for the environmental stress response of plants. In non-nitrogen-fixing plants, ureide metabolism is crucial to nitrogen recycling from organic sources. Various studies have suggested that the rate-limiting components of ureide metabolism respond to environmental stresses. However, the underlying regulation mechanism is not well understood. In this report, rice ureidoglycolate amidohydrolase (OsUAH), which is a recently identified enzyme catalyzing the final step of ureide degradation, was identified as low-temperature- (LT) but not abscisic acid- (ABA) regulated. To elucidate the LT regulatory mechanism at the transcriptional level, we isolated and characterized the promoter region of OsUAH (POsUAH). Series deletions revealed that a minimal region between –522 and –420 relative to the transcriptional start site was sufficient for the cold induction of POsUAH. Detailed analyses of this 103-bp fragment indicated that a C-repeat/dehydration-responsive (CRT/DRE) element localized at position –434 was essential for LT-responsive expression. A rice C-repeat-binding factors/DRE-binding proteins 1 (CBFs/DREB1s) subfamily member, OsCBF3, was screened to specifically bind to the CRT/DRE element in the minimal region both in yeast one-hybrid assays and in in vitro gel-shift analysis. Moreover, the promoter could be exclusively trans-activated by the interaction between the CRT/DRE element and OsCBF3 in vivo. These findings may help to elucidate the regulation mechanism of stress-responsive ureide metabolism genes and provide an example of the member-specific manipulation of the CBF/DREB1 subfamily.
Introduction
Metabolic adjustments alter the physiological and developmental reactions of plant stress adaptation (Bohnert et al., 1995; Bohnert and Sheveleva, 1998). Nitrogen recycling and redistribution are important for the environmental stress response (Nicolas et al., 1985; Tahir and Nakata, 2005; Zhu et al., 2007; Maruyama et al., 2009; Guttieri et al., 2013; Zhang et al., 2014). The redistributed nitrogen is transported among cells or organs of a plant in the form of amino acids or ureides (Schubert, 1986). In most plant species, ureides are the intermediates of the nitrogen recycling from the purine nucleotides for remobilization into amino acids. The functions of ureides have been well documented in the source-to-sink transportation of nitrogen-fixing plants, whereas accumulating evidence has led to the hypothesis that ureides might also participate in stress adaptation. Ureide catabolism is involved in nitrogen recycling from stressed and senescent tissues in drought-treated legumes (Alamillo et al., 2010; Díaz-Leal et al., 2014). Allantoin and allantoate, the primary types of ureides, are accumulated by stresses such as drought, cold, and salinity (Brychkova et al., 2008; Alamillo et al., 2010; Kanani et al., 2010; Yobi et al., 2013). The accumulation of rice grain allantoin is positively correlated with seedling tolerance to low-temperature (LT) stress (Wang et al., 2012). Allantoin has been wildly used as a biomarker of oxidative stress in mammalian cells (Esen et al., 2011; Chung and Benzie, 2013; Fukuhara et al., 2013). Although lacking antioxidant activity in vitro, allantoin supplementation could effectively mitigate oxidative damage in plants (Gus’kov et al., 2004; Brychkova et al., 2008). Further evidence has indicated that exogenous allantoin could improve plant resistance to various stresses (Watanabe et al., 2010; Wang et al., 2012). In addition, recent studies in Arabidopsis demonstrated that in vivo, allantoin is crucial to determine the cellular abscisic acid (ABA) level by activating de novo ABA biosynthesis and hydrolysis of the ABA-glucose conjugate (Watanabe et al., 2014a,b).
In plants other than nitrogen-fixing legumes, ureides are converted via the oxidation of purine. Then, ureides are catabolized in a continuous enzymatic reaction to re-assimilate inorganic nitrogen and are finally converted into glycosylate (Werner et al., 2010, 2013; Werner and Witte, 2011). Many ureide generative or degradative enzymes are involved in environmental stresses. For example, in Arabidopsis, tomato, sugarcane, and ryegrass, environmental stresses, including LT, drought, and salinity, coincidently upregulated the expression of xanthine dehydrogenase (XDH), which is rate-limiting enzyme in purine breakdown (Sagi et al., 1998; Nogueira et al., 2003; Hesberg et al., 2004; Yesbergenova et al., 2005). The repression of Arabidopsis XDH leads to increasing stress sensitivity (Brychkova et al., 2008; Watanabe et al., 2010). A key enzyme gene of ureides catabolism, allantoin amidohydrolase (ALN), is upregulated by drought and ABA treatment in some species of common beans (Alamillo et al., 2010; Coleto et al., 2014). Moreover, mutation of the ALN gene of Arabidopsis greatly enhances the tolerance to water deficit by activating stress-response genes genome-wide (Watanabe et al., 2014b).
Despite several genes of ureide metabolism being associated with stress, the involvement of the remaining components, especially those downstream of allantoate degradation, is largely unknown. More importantly, the regulation mechanisms of stress induction are not well understood. Ureidoglycolate amidohydrolase (UAH) is a recently identified ureide catabolic enzyme in Arabidopsis, rice, and soybeans (Werner et al., 2013) that has the ability in vitro to hydrolyze ureidoglycolate into glyoxylate, carbon dioxide, and two molecules of ammonia (Werner et al., 2010). Here, we report the LT-responsive expression of the rice UAH gene (OsUAH, LOC_Os12g40550), which catalyzes the final step of allantoate degradation (Werner et al., 2010). The molecular mechanism of LT induction of OsUAH is investigated. The results obtained here indicate that C-repeat-binding factors/DRE-binding proteins 1 (CBFs/DREB1s) play a critical role in the LT-responsive expression of OsUAH. Our results may enhance the understanding of the regulation of stress-involved ureide metabolism genes.
Materials and Methods
Plant Materials and Growth Conditions
Rice plants (Oryza sativa L. ssp. japonica) were used as a source to isolate promoters and genes and for plant transformation. Mature, non-dormant seeds were sterilized and germinated in 1/2 MS medium under a light/dark cycle of 16 h/8 h at 28°C for at least 10 days. Rice seedlings at the trifoliate stage were transferred to plastic buckets with soil at 30°C during the day and 20°C at night in a greenhouse.
Isolation and Sequence Analysis of the Promoter of OsUAH
According to the genomic sequence of OsUAH (LOC_Os12g40550), the region from 2000 bp upstream to 100 bp downstream of the transcription initiation site for this gene was predicted to be the promoter region (POsUAH, as showed in Supplementary Sequence). POsUAH was PCR-amplified from rice genomic DNA using gene-specific primers. To identify potential functional elements, the full-length sequence of POsUAH was analyzed with the PLACE1 and Plant-PAN2 software packages as previously described (Luo et al., 2013).
Promoter-GUS Chimeric Vector Construction and Generation of Transgenic Rice Plants
The 5′ deletions of POsUAH at positions –1227, –717, –522, –420, and –137 were generated by PCR amplification using different forward primers and a single downstream primer. A HindIII restriction site was introduced into the forward primers, and an EcoRI restriction site was introduced into the reverse primer. The full-length promoter and five deleted derivatives were cut by HindIII and EcoRI and then inserted into the plant transforming binary vector PCAMBIA1391 upstream of the GUS coding sequence. The corresponding plasmids were designated as POsUAH, PTru1, PTru2, PTru3, PTru4, and PTru5 according to the position at the 5′ end.
Site-specific mutation was performed using the Quick Change Site-Directed Mutagenesis Kit (Transgene, China). The pEASY-T plasmid containing the PTru1 fragment was used as the PCR template. The obtained mutated construct was cut by HindIII and EcoRI and ligated into PCAMBIA1391. The corresponding plasmid was designated as PTru1-M.
To construct potential gain-of-function vectors, the sequence of the CaMV 35S promoter from –46 to +1 (mini 35S) was amplified and inserted into PCAMBIA1391 upstream of the GUS coding sequence. The obtained construct was named Pmini and used as a control. A 103-bp fragment that was located in the region from –522 to –420 of POsUAH was obtained by PCR using sequence-specific primers with a HindIII site and an EcoRI site. After digestion, the fragment was inserted into Pmini to obtain the recombinant plasmid P103bp-mini. Full-length POsUAH was also fused to Pmini as a positive control (construct POsUAH-mini). All primer sequences that were used are listed in Supplementary Tables S1–S10.
The binary constructs were introduced into the Agrobacterium tumefaciens strain EHA105. The rice transformation constructs that were used contained the HPT gene under the control of the 35S promoter to enable hygromycin-based plant selection. Embryonic calli from the mature rice seeds (Oryza sativa L. ssp. Japonica) were transformed by co-cultivation, selected with 50 mg/l hygromycin, and used to regenerate transgenic plants as previously described (Duan et al., 2012). The single-copy transgenic lines were screened using the real-time PCR method as described (Yang et al., 2005), and at least four independent T2 lines were selected for further analysis.
Stress Treatments
To assess the expression levels of the OsUAH gene under temperature stress, 10-days-after-germination (DAG) seedlings on agar plates were placed in a growth chamber at constant temperatures of 4 or 42°C under a light/dark cycle of 16 h/8 h. The seedlings were incubated in 1/2 MS solution containing 250 mM NaCl for salt treatment and 100 μM ABA for ABA treatment. For drought stress, the seedlings were dried at 40% relative humidity. Then, the samples were harvested at 0, 4, 8, 12, and 24 h and frozen in liquid nitrogen for RNA extraction.
To analyze the response of POsUAH to LT stress at different temperatures, 10-DAG seedlings on agar plates were placed in growth chambers at 4, 10, and 15°C. The control seedlings were grown under the same conditions but at 30°C. The samples were harvested at 0, 4, 8, 12, and 24 h. Mature plants at 60 DAG were treated for 24 h at 4°C, after which the roots, stems and leaves were collected. To analyze the response to LT stress, transgenic plants of truncation and mutation constructs were treated for 24 h at 4°C as above.
RNA Isolation and qRT-PCR Analysis
The total RNA was extracted from rice using the RNAprep Pure Plant Kit (TIANGEN, China) in accordance with the manufacturer’s instructions. To amplify the corresponding genes, cDNAs were synthesized with random primers using the FastQuant RT Kit (TIANGEN, China) as the template for the qRT-PCR. Real-time quantitative PCR was performed using an ABI PRISM 7500 real-time PCR system (Applied Biosystems, USA) with SYBR Green (TIANGEN, China). The real-time PCR conditions were 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The qRT-PCR reactions were performed in triplicate for each cDNA sample. The ACTIN gene was used as an internal control, and the relative expression levels were determined in accordance with standard protocols (Livak and Schmittgen, 2001). The expression difference were statistically determined by a on-side paired t-test.
Histochemical GUS Staining
The histochemical localization of GUS activity in transgenic plants was performed as previously described (Wu et al., 2003). The samples were incubated in GUS staining solution (50 mM sodium phosphate at pH 7.0, 10 mM Na2-EDTA, 0.1% Triton X-100, 1 mg/ml X-Gluc) at 37°C for 24 h after 15 min of vacuum filtration. After staining, the samples were fixed in 70% ethanol, and photographs were taken under a dissecting microscope.
Generation of Yeast Reporter Strains for One-Hybrid Screening
For the one-hybrid assay, three tandem copies of the fragment from –443 to –418 of POsUAH was synthesized as a bait. In addition, a two-base-substitution fragment and a five-base-substitution fragment at the CRT/DRE element were used as controls. The sequences are shown in Supplementary Table S7. The above three fragments were separately digested with HindIII and SalI and inserted into the plasmid pAbAi. The obtained recombinant bait plasmids were recognized as pAbAi-E1, pAbAi-E1m2, and pAbAi-E1m5. After digestion with BbsI, the linearized bait plasmids were transformed into yeast strains according to the method that is described in the MatchmakerTM Gold Yeast One-Hybrid Library Screening System Kit (Clontech, USA).
To analyze the CRT/DRE-element-binding activity of OsCBFs, the ORFs of the five rice OsCBF transcription factors OsCBF1, OsCBF2, OsCBF3, OsCBF4, and OsDREB1B were cloned from the cDNA of Japonica rice and were inserted into a GAL4 AD backbone. The pGAD-OsCBF plasmids were then transformed into yeast strains that were integrated with a reporter vector via the LiAc yeast transformation method. The empty AD vector was used as a negative control.
Purification of Bacterially Expressed Proteins and Electrophoretic Mobility Shift Assay (EMSA)
The pGEX-4T-1 bacterial expression vector system was used to produce a fusion protein with glutathione S-transferase (GST). To obtain the fused GST-OsCBF3 protein, the recombinant plasmid was transformed into Escherichia coli [Rosetta 2 (DE3) pLysS strain]. The recombinant GST-OsCBF3 protein was induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside for 20 h at 20°C. Protein purification of the GST fusion protein from bacterial extracts was achieved by affinity chromatography with Glutathione Sepharose 4B Resin (GE Healthcare) following the instruction of the manufacturer. Cells carrying the pGEM-4T-1 empty vector were processed as negative controls in an identical manner.
For Electrophoretic Mobility Shift Assay (EMSA), complementary single-stranded oligonucleotides of the E2 probe and its mutants were synthesized, labeled with biotin, and annealed to make probes (Supplementary Table S10). EMSAs were performed using biotin-labeled double-stranded DNA probes with the Light-Shift Chemiluminescent EMSA Kit (Thermo-Fisher Scientific, USA) according to the manufacturer’s instructions.
Trans-activation Experiment with Transgenic Rice
Effector plasmids were constructed with DNA fragments containing OsCBF1, OsCBF2, OsCBF3, OsCBF4, and OsDREB1B coding regions that were under the control of the maize UBI promoter. The effector constructs used PMI as a selected marker gene to enable mannose-based plant selection. The transgenic plants were generated, and the expression of the corresponding CBF genes was examined using qRT-PCR assays. The overexpressing lines were crossed with three independent single-copy PTru1 and PTru1-M reporter lines containing the HPT marker gene as described above. The crossed plants were obtained by hygromycin and mannose double selection.
Results
Identification the LT Induction of OsUAH
To identify the stress response of OsUAH, the transcript levels were monitored in time-course treatments of drought, LT, high temperature (HT), salinity, and ABA. As shown in Figure 1, OsUAH was greatly induced by the cold treatment. The OsUAH transcript began to accumulate after 4 h of cold stress treatment and increased in a time-dependent manner. After 24 h of incubation at 4°C, the mRNA level of OsUAH was 9.14-fold relative to that of the untreated control. Cool stress (10–15°C) is the most frequently abiotic stress during the early growth stage of rice. Similar with the response to cold stress, OsUAH transcripts were also accumulated by the 10 and 15°C incubations (Supplementary Figure S1). In contrast, neither HT, salinity stress nor ABA treatment upregulated the expression of OsUAH at any tested time point (Figure 1). Under water deficit, OsUAH expression did not transcriptionally respond to drought stress in a short period (4 and 8 h air-dry treatment); however, 3.4-fold and 4.2-fold inductions were observed after 12 and 24 h of stress incubation, respectively (Figure 1).
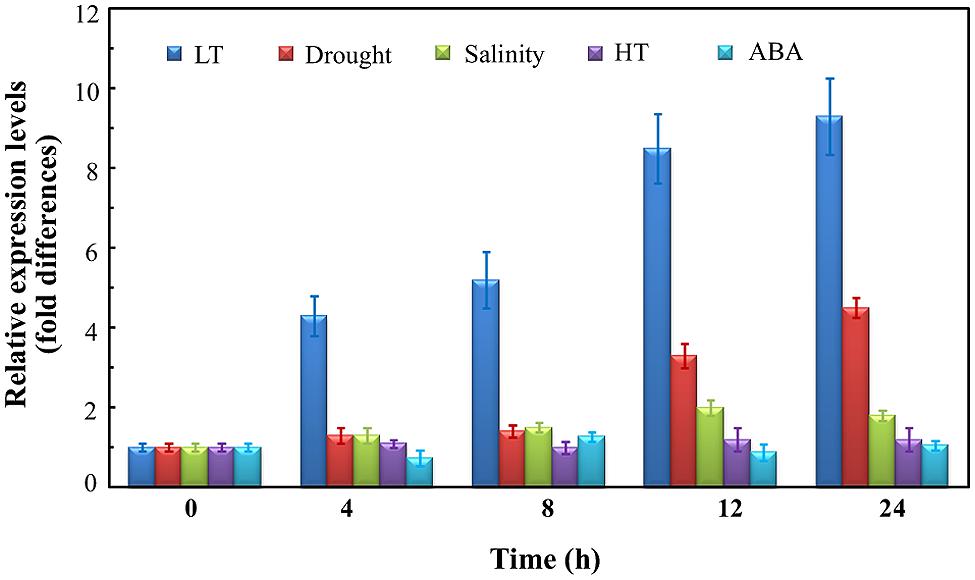
FIGURE 1. Quantitative real-time PCR (qRT-PCR) analysis of the OsUAH transcripts under various stress conditions. Rice seedlings at 10 days-after-germination (DAG) were treated with low-temperature (LT; 4°C), high temperature (HT; 42°C), drought (dried at 40% relative humidity), salt (250 mM NaCl) and abscisic acid (ABA; 100 μM ABA). The samples were collected on a time course for RNA isolation and subsequent qRT-PCR analysis. ACTIN was used as an internal control. The values are the means ± SD of three independent biological experiments.
To precisely investigate the LT induction of OsUAH, the sequence of the predicted promoter (POsUAH) with a length of 2100 base pairs (bp) was isolated from the rice (Oryza sativa L. cv. Nipponbare) genome and contains 2000 bp immediately upstream and 100 bp downstream (–2000/+100) of the transcription initiation site. The DNA fragment was inserted into a PCAMBIA 1391 vector, generating a rice transformation construct with POsUAH driving the GUS reporter gene. A total of 28 independent transgenic lines were generated. Among these lines, six single-copy transgenic lines harboring the POsUAH::GUS construct were screened and selected to determine the expression pattern of POsUAH.
The activity of POsUAH was first examined by GUS histochemical staining. Under normal growth conditions, the GUS staining could not be detected in any of transgenic plants, regardless of tissues or developmental stages (Figure 2A). However, after LT treatment, obvious blue staining was observed not only in 5-DAG seedlings, but also in tissues of plants at the booting stage (60 DAG). The LT response of POsUAH was also determined by quantitative reverse real-time PCR (qRT-PCR). The GUS transcript was significantly induced after 4 h of incubation at 4°C (p < 0.05). The expression gradually increased in a time-dependent manner, reaching 8.08-fold at 24 h. The LT responses of the promoter were also examined by the incubation at 10 and 15°C. As shown in Figure 2B, the activities of the promoter could be significantly induced after 4 h of incubations (p < 0.05), after which the activities slowly increased or remained relatively constant (Figure 2B). To further investigate the expression pattern of POsUAH, GUS levels were individually measured in different tissues of 60-DAG plants. The transcripts were markedly increased by LT stress (4°C incubation for 24 h), while the fold-induction level in the roots (11.01-fold) was relatively higher than that in the leaves (5.76-fold) and stems (3.94-fold), (Figure 2C).
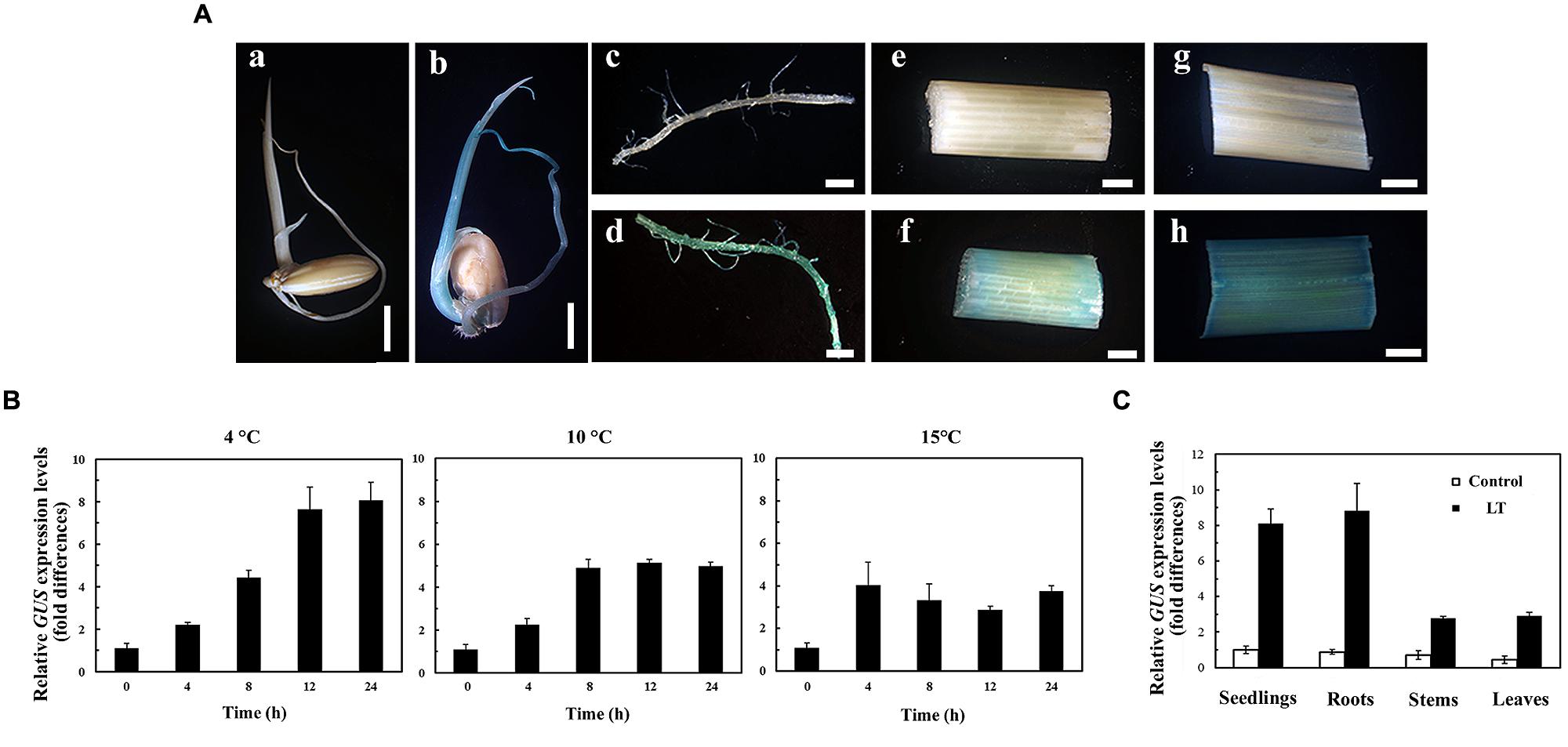
FIGURE 2. The LT response of POsUAH. (A) The GUS staining of the transgenic rice harboring the POsUAH::GUS construct. 5-DAG seedlings (a,b); and roots (c,d), stems (e,f), and leaves (g,h) of 60-DAG plants were incubated under normal growth conditions (a,c,e,g) or LT (4°C) treatment for 24 h (b,d,f,h). (B) Analysis of the promoter activity induction under different LT conditions. Seedlings at 10 DAG were treated with 4, 10, and 15°C for 24 h. GUS transcripts were measured by qRT-PCR. (C) qRT-PCR analysis of GUS induction in different organs of transgenic rice. The 60-DAG plants were treated with LT (4°C) for 24 h, after which the mature leaves, stems and roots were collected. GUS induction was examined with a parallel experiment on 10-DAG seedlings. The GUS mRNAs of the untreated transgenic plants were set to one. The values are the means ± SD of six independent biological experiments.
Identification of the Minimal Promoter Region for LT-Inducible Expression
In an attempt to define the specific regions of POsUAH that are involved in LT-inducible expression, a series of 5′ deletions in the 2100 bp promoter were generated (Figure 3A). After fusion to a GUS reporter gene, five truncation fragments were separately transformed into rice. For each construct, at least four independent single-copy transgenic lines were selected, and the GUS mRNA levels were quantitatively assayed. All of the truncation fragments had similar expression levels as those of the full-length POsUAH under normal growth conditions. Under a LT treatment at 4°C for 24 h, the GUS levels were significantly induced in the constructs containing deletions of the distal part of the full-length sequence to positions –1227 (construct PTru1), –717 (construct PTru2), and –522 (construct PTru3) from the transcription initiation site, while PTru1 had a statistically similar induction fold as that of POsUAH, and the induction levels of PTru2 and PTru3 decreased (Figure 3A). In contrast, the LT inductions were completely lost in the constructs containing deletions to positions –420 (construct PTru4) and –137 (construct PTru5), (Figure 3A), suggesting that the 103-bp fragment between positions –522 and –420 is essential for the LT induction of POsUAH.
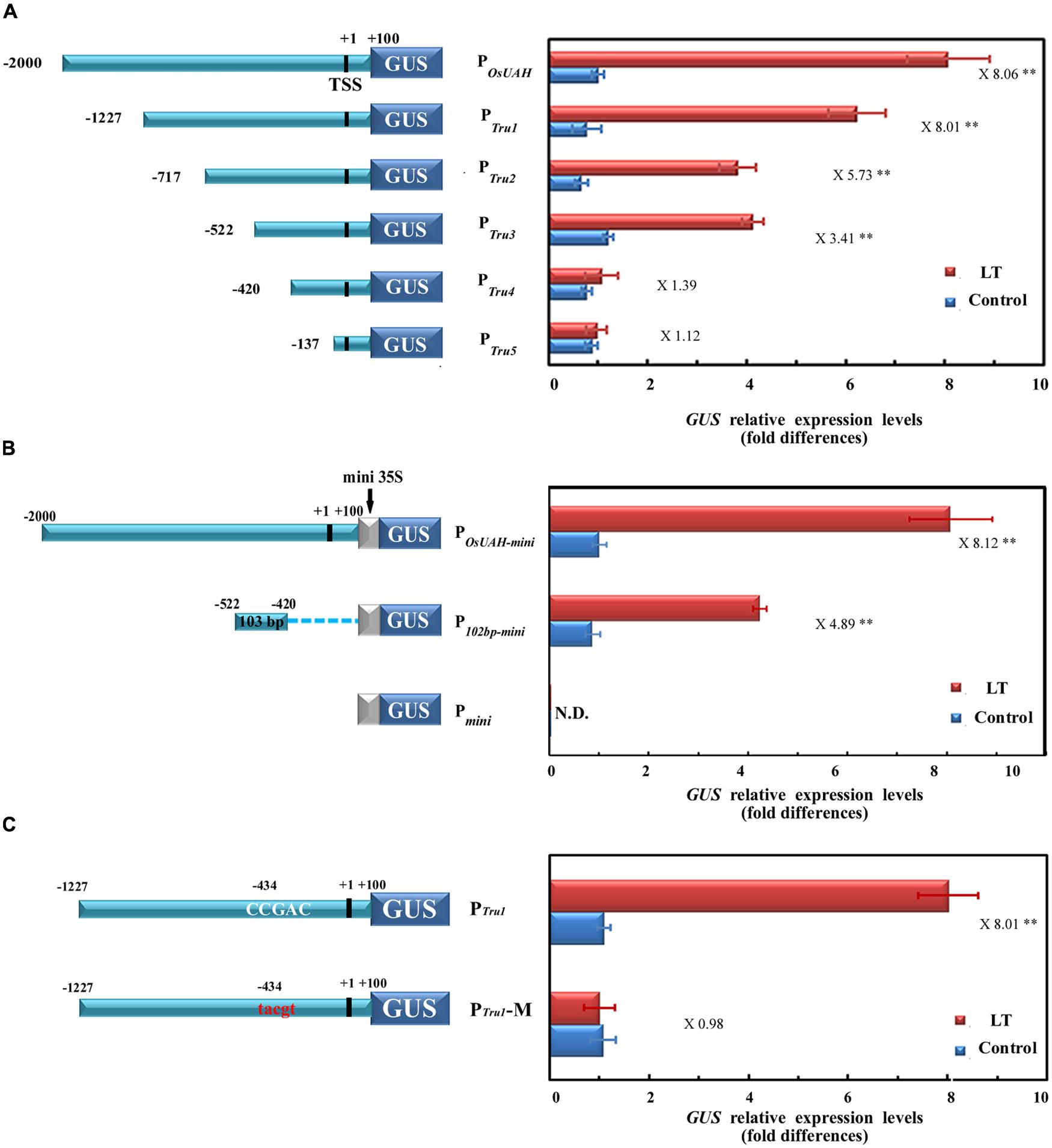
FIGURE 3. Activity analysis of POsUAH for determining the minimal region of LT induction. (A) Deletion analysis of POsUAH. Left, schematic diagrams of serial 5′ deletion constructs of POsUAH. The locations of the 5′ ends of the five fragments of POsUAH are indicated. LT stress was applied by incubating 10-DAG seedling at 4°C for 24 h. The activities were then compared by analyzing GUS transcript abundance in corresponding transgenic populations. The GUS mRNAs of untreated full-length POsUAH transgenic plants were averaged and set to one. (B) Identification of the 103-bp region that was responsible for the LT response. The 103-bp sequence between position –522 and –420 was linked at the 5′ region upstream of the mini35S promoter region as the P103bp-mini construct. Full-length POsUAH was also fused to the mini35S to generate POsUAH-mini as a positive control, and the empty Pmini construct was used as a negative control. After generating transgenic plants, their LT induction abilities were examined by determining GUS transcriptional expression. The GUS mRNAs of untreated POsUAH-mini transgenic plants were averaged and set to one. (C) Identification of the cis-element that is responsible for the LT response. Left, Schematic diagrams of the PTru1 construct and the mutated PTru1 (PTru1-M) containing a mutation on the core C-repeat/dehydration-responsive (CRT/DRE) element (from the conserved CCGAC to the irrelevant 5-bp sequence TGCGA). Right, the corresponding promoter activity as determined in the stably transformed transgenic rice plants. The GUS mRNAs of untreated PTru1 transgenic plants were averaged and set to one. The ratio of the promoter activity with LT stress to that without LT stress (induction ratio) is shown at the far right. TSS: transcriptional start site. The relative expression levels were averaged from at least four independent biological experiments. The significant difference between the LT-stress and non-stress conditions was analyzed using a one-side paired t-test (∗∗p < 0.01).
To confirm that the region between positions –522 and –420 plays a crucial role in POsUAH, the 103-bp fragment was separated and fused to a mini35S promoter (construct P103bp-mini) and then linked to the binary vector to drive GUS. Meanwhile, full-length POsUAH was also fused to the mini35S promoter as a positive control (construct POsUAH-mini). The constructs, as well as the empty mini35S::GUS vector (Pmini), were stably transformed into rice. As shown in Figure 3B, both POsUAH-mini and the P103bp-mini were still able to be induced by LT treatment, although the fold-induction level of POsUAH-mini (8.1-fold) was relatively higher than the level of P103bp-mini (4.89-fold). In addition, the expression of Pmini was not altered by LT stress. These results indicate that the 103-bp minimal region is sufficient to activate the LT-induced expression and should contain cis-elements, which are responsible for the LT stress response.
To further identify the element(s) that are responsible for LT induction, a sequence analysis was performed on the 103-bp fragment between positions –522 and –420. A CCGAC element, which is the core sequence of the C-repeat/dehydration-responsive (CRT/DRE) element, was located at position –434, closely associated with the expression of LT or drought stresses. To determine whether the CRT/DRE element is involved in LT induction, the CCGAC sequence was substituted in situ in the construct PTru1 with an irrelevant 5-bp sequence, TGCGA, generating the construct PTru1-M. Under normal conditions, the activity of PTru1-M remained the same as that of the non-mutated PTru1 in the corresponding transgenic plants. However, the GUS mRNA accumulations by LT treatment were fully abolished in all of the tested PTru1-M transgenic rice compared to the 8.01-fold induction in plants harboring PTru1 (Figure 3C). These results suggest that this element is essential to the LT induction of OsUAH.
OsCBF3 Specifically Binds to the CRT/DRE Element in the Minimal Region of POsUAH
The interaction between the CRT/DRE elements with the CBF transcript factors has been identified in various plants (Sakuma et al., 2002; Qin et al., 2004). In the rice genome, 10 putative CBF/DREB homologs (OsDREB1A – OsDREB1J) have been identified. Five of these homologs, OsDREB1C/OsCBF1 (LOC_Os06g03670), OsDREB1F/OsCBF2 (LOC_Os01g73770), OsDREB1A/OsCBF3 (LOC_Os09g35030), OsDREB1D/OsCBF4 (LOC_Os06g06970), and OsDREB1B (LOC_Os09g35010), are induced by LT stress (Dubouzet et al., 2003; Liu et al., 2007; Wang et al., 2008). To determine the specific rice CBF factor that could directly target the core CRT/DRE element in the minimal region of POsUAH, a yeast one-hybrid assay was used. Three tandem copies of the 25-bp sequence surrounding the CRT/DRE element were synthesized as E1 bait. E1 was fused in front of the reporter gene AUR1-C, an antibiotic resistance gene that confers Aureobasidin A (AbA) resistance in yeast. Meanwhile, five OsCBF/DREB1 genes were separately cloned and fused to a GAL4 activation domain (AD) as preys. After being co-transformed with the promoter and individual CBF, only the yeast cells harboring OsCBF3 and E1 could grow on the 100 ng/ml SD/-Leu/AbA medium, while the co-transformant with other rice CBFs could not survive under AbA selection. These observations indicate that OsCBF3 is the only candidate binding to POsUAH among the tested rice CBF members in yeast (Figure 4A).
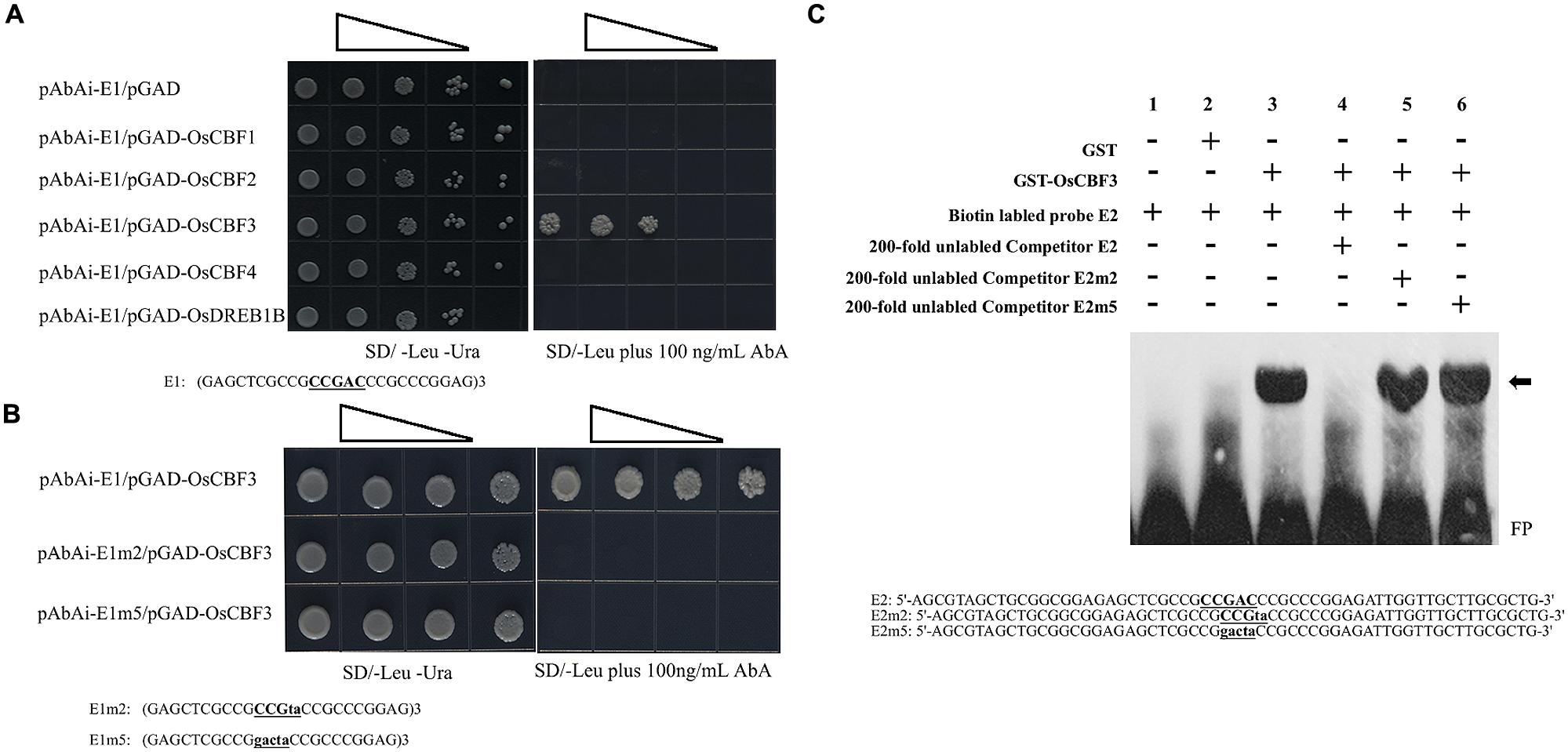
FIGURE 4. The core CRT/DRE element specifically interacted with OsCBF3. (A) Yeast one-hybrid analysis of the binding of rice CBFs to POsUAH. Upper, OsCBF3 specifically interacted with POsUAH. Yeast cells were co-transformed with a bait vector containing three consecutive repeats of the 25-bp POsUAH fragment surrounding the core CRT/DRE element (underlined) and fused to an AUR1-C reporter gene (pAbAi-E1) and with a prey vector containing a CBF transcription factor coding sequence that was fused to a GAL4 activation domain (pGAD-CBF). The binding of the GAL4 activation domain (pGAD empty vector) to the POsUAH fragment (pAbAi-E1) was used as a negative control. Lower, OsCBF3 interacted with the CRT/DRE element in POsUAH. The bait vector carrying the corresponding fragment and the prey pGAD-OsCBF3 vector were co-transfected into yeast cells. The fragments: E1, Three copies of the POsUAH fragment; E2m, two-base mutation of the CRT/DRE element in E1; and E5m, five-base mutation of the CRT/DRE element in E1. The cells were grown in liquid media to an OD600 of 0.1 (10-1) and diluted in a 10× dilution series (10-2 to 10-4/10-3). Of each dilution, 6 μL was spotted onto media that selected for both of the plasmids (SD–Ura–Leu) and selecting for interaction (SD–Leu) and that was supplemented with 100 ng/ml Aureobasidin A (AbA) to suppress the background growth and test the strength of the interaction. (B) The Electrophoretic Mobility Shift Assay (EMSA) assays indicate that the glutathione S-transferase (GST)-OsCBF3 fusion protein but not GST can specifically bind to the CRT/DRE element (underlined) in the 59-bp POsUAH sequence. The probes were incubated with recombinant proteins in 20-μL reactions. (C) Lane 1, 20 fmol of the labeled wild-type probe (E2) alone. Lane 2, 20 fmol of labeled E2 probe with 1 μg of the recombinant GST protein. Lane 3, 20 fmol of the labeled E2 probe with 1 μg of the recombinant GST-OsCBF3 protein. Lane 4, 20 fmol of the labeled E2 probe with 1 μg of the recombinant GST-OsCBF3 protein and 2 pmol of the unlabeled competitor E2 probe. Lane 5, 20 fmol of the labeled E2 probe with 1 μg of the recombinant GST-OsCBF3 protein and 2 pmol of the unlabeled competitor probe with the two-base mutation on the CRT/DRE element (E2m2). Lane 6, 20 fmol of the labeled E2 probe with 1 μg of the recombinant GST-OsCBF3 protein and 2 pmol of the unlabeled competitor probe with the five-base mutation on the CRT/DRE element (E2m5). The mutation sites are labeled with lower case letters. The arrow indicates the up-shifted bands. FP, Free probe. GST is from the expression vector pGEM-4T-1.
To explore whether the element is the binding site of OsCBF3 in the promoter, two site-directed mutations were performed on the core sequence of E1, generating a two-base substitution (E1m2: CCGta) and a five-base substitution (E1m5: gacta). The AD-OsCBF3 yeast cells harboring baits with either E1m2 or E1m5 could not grow on the leucine dropout medium that was supplemented with 100 ng/ml AbA. In contrast, the growth of yeast cells with OsCBF3 and wild-type E1 was not inhibited by AbA (Figure 4B). These results suggest that the core CRT/DRE element is the specific binding site of OsCBF3 in the POsUAH fragment.
To further confirm that OsCBF3 binds to POsUAH at the CRT/DRE element, purified full-length OsCBF3 protein was obtained using a GST- fusion purification system and used to perform Electrophoretic Mobility Shift Assay (EMSA). As shown in Figure 4C, the GST-OsCBF3 protein bound to a 59-bp E2 probe from the minimal region of POsUAH that contained the CRT/DRE element. A competition EMSA was performed in parallel with wild-type and mutant unlabeled E2 probes. Figure 4C showed that excessive wild-type E2 probe could compete with the labeled probe, but the same amount of unlabeled mutant E2 probe with the two-base substitution (E2m2) or five-base substitution (E2m5) on the core sequence of E2 did not, suggesting that OsCBF3 protein could bind specifically to POsUAH at the CRT/DRE element in vitro.
OsCBF3 Trans-activates the Expression of POsUAH in a CRT/DRE Element-Dependent Manner
To detect the in vivo interactions between CBFs and POsUAH, promoter trans-activation assays were performed. The constructs are schematically represented in Figure 5A. Because the truncation fragment at position –1227 (PTru1) has a similar expression pattern as that of the full-length promoter POsUAH, PTru1 and the above-described PTru1-M containing the mutated CRT/DRE element were used as reporters. Five LT-responsive rice CBF members were overexpressed by a maize ubiquitin promoter as effectors. The transgenic plants containing the effector were regenerated and crossed with the single-copy reporter plant lines. The crossed plants harboring PTru1 and OsCBF3 resulted in an 11.47-fold increase in GUS mRNA accumulation compared to the background level of the cross of PTru1 and an empty-effector vector, whereas the co-expression of other CBF members exhibited the same GUS levels as that of the empty-effector vector (Figure 5B). These results indicate that PTru1 only could be activated by OsCBF3, which is consistent with the results of the previous binding activity assay in yeast. None of the effectors could induce the expression of the mutated PTru1-M reporter, suggesting that the trans-activation of POsUAH by OsCBF3 depends on the CRT/DRE element.
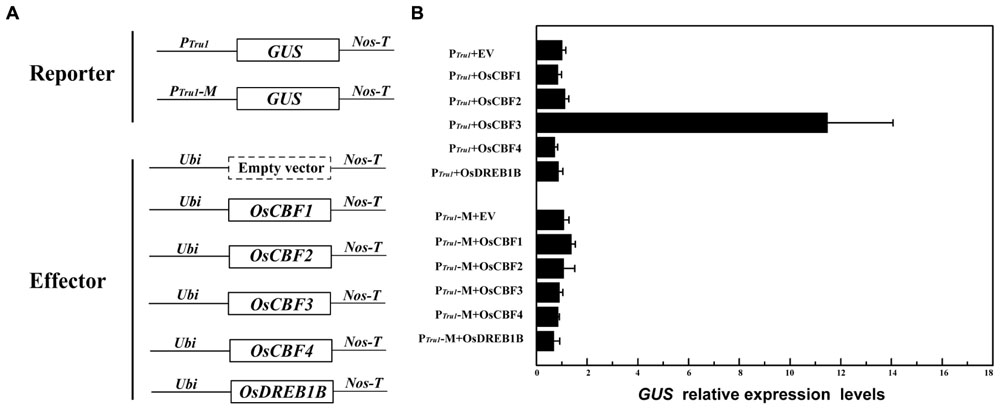
FIGURE 5. OsCBF3 specifically trans-activated POsUAH in planta. (A) Schematic diagrams of the vectors. The CBFs were driven by a maize ubiquitin promoter as effectors, and the truncation promoter PTru1 and the CRT/DRE element mutated derivate PTru1-M (described in Figure 3) were fused to drive GUS as reporters. (B) qRT-PCR analysis of GUS expression. The GUS mRNAs of the crossed transgenic plants containing PTru1 plus the empty effector vector were set to one. The relative expression levels were averaged from three independent biological experiments.
Discussion
Nitrogen supplementation plays a critical role in the utilization of absorbed light energy and photosynthetic carbon metabolism (Kato et al., 2003; Huang et al., 2004). Adequate nitrogen supplementation mitigates the damages of abiotic stresses (Huang et al., 2004; Waraich et al., 2011), while a high risk of photo-oxidative damage is expected in the nitrogen-deficient leaves under stress (Verhoeven et al., 1997). Purine catabolism is important for nitrogen recycling. In this study, we identified OsUAH, the catalyzer of the final step of the ureide-degrading reactions of purine ring catabolism (Werner et al., 2010), as an LT-responsive gene. This result has been confirmed by independent transcriptome analyses (Maruyama et al., 2011; Zhang et al., 2012; Shaik and Ramakrishna, 2013). In addition to previous evidences of the LT induction of other catabolic genes, our results suggest that ureide degradation might be critical for nitrogen redistribution in the LT adaption of plants. Furthermore, our results indicate the OsUAH has a relatively higher induction level in the roots than in photosynthetic tissues, suggesting a potential nitrogen redistribution pattern in response to stress. Various studies have emphasized that nitrogen metabolism genes are regulated by LT stress (Cui et al., 2005; Pageau et al., 2006; Zhu et al., 2007; Robinson and Parkin, 2008; Maruyama et al., 2009; Zhang et al., 2012), while the underlying mechanisms have seldom been reported. The hydrolase genes of purine catabolism, e.g., XDH and ALN, are regulated by ABA (Hesberg et al., 2004; Alamillo et al., 2010). However, we found that the expression of OsUAH was not induced by exogenous ABA (Figure 1), and our data indicated the LT-induction of OsUAH was associated with a CRT/DRE element of POsUAH. The evidence from yeast assays and the in vitro interaction analysis demonstrate that this element physically binds to the CBF transcriptional factor OsCBF3. Further assays demonstrated that POsUAH activity could be upregulated by overexpressing OsCBF3 in vivo. These data suggest that the regulation of OsUAH is involved in a CBF-related LT-response pathway. In addition to LT stress, OsUAH is also induced by drought stress, which is consistent with previous transcriptional profile studies (Degenkolbe et al., 2009; Gao et al., 2009). Because OsCBF3 is a transcription activator of cold- and drought-responsive gene expression, the induction of OsUAH could be explained by the interaction between the CRT/DRE element of POsUAH and OsCBF3. In most plants, the CBF pathway of the drought and LT stress responses is ABA-independent (Shinozaki et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2006). Therefore, our results suggest the OsUAH might respond to environmental stress in a distinct way compared to other ureide metabolism genes.
C-repeat-binding factor transcriptional factors are involved in the activation of LT-responsive genes by interacting with CRT/DRE elements in the promoter. Most LT-responsive genes have multiple CRT/DRE elements in their promoters. However, the element that is located at –434 bp is an unique copy in the POsUAH sequence. Our assays identified a synthetic promoter with a 103-bp fragment containing an element that exhibited similar LT-induction activity as that of POsUAH, whereas the deletion or mutation of this element thoroughly abolished this induction, indicating that this element is sufficient to determine the LT response of POsUAH. In addition, the deletion between –1227 and –717 of POsUAH significantly decreased LT induction level. However, the bioinformatic screening did not detect any potential stress response element in this region. Therefore, it is likely that an enhancer exists in the area that facilitates the intensity of the promoter.
C-repeat-binding factor/DREB1 is a small subfamily of the APETALA2/Ethylene response factor (AP2/EREBP) family of transcription factors. In Arabidopsis, this subfamily contains six members (Nakano et al., 2006; Lata and Prasad, 2011). Extensive studies have demonstrated that three tandem-distributing CBF genes, CBF1, CBF2, and CBF3 (also known as DREB1B, DREB1C, and DREB1A, respectively), play a central role in the transcriptional regulation of LT responsive genes. Five rice CBFs were identified as induced by cold stress, and overexpressing any of them can result in the enhancement of plant cold tolerance (Dubouzet et al., 2003; Ito et al., 2006; Liu et al., 2007; Qin et al., 2007; Wang et al., 2008). CBFs have highly conserved functional domains. In most cases, the cold-induced CBFs have redundant activities in plant development and stress adaption as identified in gain-of-function experiments by activating the same gene clusters (Gilmour et al., 2004; Zhou et al., 2010; Lata and Prasad, 2011). However, little evidence had suggested that each individual CBF might regulate different targets and thus separately contribute to transcriptome alteration in response to cold. In Arabidopsis, CBF1 and CBF3 RNAi transgenic plants impair the induction of cold-responsive genes; in contrast, the cold tolerance of the loss-of-function mutant of CBF2, cbf2, is enhanced by upregulating CBF1 and CBF3 (Novillo et al., 2004), indicating that the functional differentiation despite the gain-of-function modification leads to exactly same phenotype (Gilmour et al., 2004). In this study, we first demonstrated that POsUAH exclusively bound to OsCBF3 in yeast. Then, we found that OsCBF3 could activate POsUAH by interacting with the CRT/DRE element. Meanwhile, the other four rice CBFs did not affect the promoter activity, although their expression was successfully enhanced similar to OsCBF3 (Supplementary Figure S2). These results suggest that rice CBFs also regulate different objectives, similar to Arabidopsis homologs, and that their function does not fully overlap. Although the overexpression of rice CBFs exhibited a similar stress adaption phenotype (Dubouzet et al., 2003; Ito et al., 2006; Wang et al., 2008), it was reasonable to expect a diversity of function when individual CBF mutant of rice was generated. Furthermore, the rice CBF family has a more complicated structure than that of Arabidopsis or other identified dicot plants. In this study, the activity of POsUAH was not detected before or after LT induction in a transient expression system of tobacco leaves. However, POsUAH could be activated via the co-agroinjection of OsCBF3 in tobacco (Supplementary Figure S3). These results indicate that tobacco may lack functional homologs of OsCBF3, suggesting that the OsCBF3-specific trans-activated regulation might be unique between species.
Funding
This work was supported by the National Natural Science Foundation of China [No. 31501239 to R-YQ, No. 31401454 to HL]; and the Creative Foundation of The Anhui Agricultural Academy of Sciences [13C0101 and 14A0101 to P-CW, 14B0113 to R-YQ].
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01011
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Alamillo, J. M., Díaz-Leal, J. L., Sánchez-Moran, M. V., and Pineda, M. (2010). Molecular analysis of ureide accumulation under drought stress in Phaseolus vulgaris L. Plant Cell Environ. 33, 1828–1837. doi: 10.1111/j.1365-3040.2010.02187.x
Bohnert, H. J., Nelson, D. E., and Jensen, R. G. (1995). Adaptations to environmental stresses. Plant Cell 7, 1099–1111. doi: 10.1105/tpc.7.7.1099
Bohnert, H. J., and Sheveleva, E. (1998). Plant stress adaptations — making metabolism move. Curr. Opin. Plant Biol. 1, 267–274. doi: 10.1016/S1369-5266(98)80115-5
Brychkova, G., Alikulov, Z., Fluhr, R., and Sagi, M. (2008). A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 54, 496–509. doi: 10.1111/j.1365-313X.2008.03440.x
Chung, W.-Y., and Benzie, I. F. F. (2013). Plasma allantoin measurement by isocratic liquid chromatography with tandem mass spectrometry: method evaluation and application in oxidative stress biomonitoring. Clin. Chim. Acta 424, 237–244. doi: 10.1016/j.cca.2013.06.015
Coleto, I., Pineda, M., Rodiño, A. P., De Ron, A. M., and Alamillo, J. M. (2014). Comparison of inhibition of N2 fixation and ureide accumulation under water deficit in four common bean genotypes of contrasting drought tolerance. Ann. Bot. 113, 1071–1082. doi: 10.1093/aob/mcu029
Cui, S., Huang, F., Wang, J., Ma, X., Cheng, Y., and Liu, J. (2005). A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5, 3162–3172. doi: 10.1002/pmic.200401148
Degenkolbe, T., Do, P., Zuther, E., Repsilber, D., Walther, D., Hincha, D., et al. (2009). Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Mol. Biol. 69, 133–153. doi: 10.1007/s11103-008-9412-7
Díaz-Leal, J. L., Torralbo, F., Antonio Quiles, F., Pineda, M., and Alamillo, J. M. (2014). Molecular and functional characterization of allantoate amidohydrolase from Phaseolus vulgaris. Physiol. Plant. 152, 43–58. doi: 10.1111/ppl.12157
Duan, Y., Zhai, C., Li, H., Li, J., Mei, W., Gui, H., et al. (2012). An efficient and high-throughput protocol for Agrobacterium-mediated transformation based on phosphomannose isomerase positive selection in Japonica rice (Oryza sativa L.). Plant Cell Rep. 31, 1611–1624. doi: 10.1007/s00299-012-1275-3
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought–, high-salt– and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Esen, A. M., Akcakoyun, M., Esen, O., Acar, G., Emiroglu, Y., Pala, S., et al. (2011). Uric acid as a marker of oxidative stress in dilatation of the ascending aorta. Am. J. Hypertens. 24, 149–154. doi: 10.1038/ajh.2010.219
Fukuhara, K., Ohno, A., Ota, Y., Senoo, Y., Maekawa, K., Okuda, H., et al. (2013). NMR-based metabolomics of urine in a mouse model of Alzheimer’s disease: identification of oxidative stress biomarkers. J. Clin. Biochem. Nutr. 52, 133–138. doi: 10.3164/jcbn.12-118
Gao, F., Zhang, H., Wang, H., Gao, H., and Li, Z. (2009). Comparative transcriptional profiling under drought stress between upland and lowland rice (Oryza sativa L.) using cDNA-AFLP. Chin. Sci. Bull. 54, 3555–3571. doi: 10.1007/s11434-009-0524-5
Gilmour, S., Fowler, S., and Thomashow, M. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Gus’kov, E. P., Prokof’ev, V. N., Kletskii, M. E., Kornienko, I. V., Gapurenko, O. A., Olekhnovich, L. P., et al. (2004). Allantoin as a vitamin. Dokl. Biochem. Biophys. 398, 320–324. doi: 10.1023/B:DOBI.0000046649.11374.8d
Guttieri, M., Stein, R., and Waters, B. (2013). Nutrient partitioning and grain yield of TaNAM-RNAi wheat under abiotic stress. Plant Soil 371, 573–591. doi: 10.1007/s11104-013-1713-1
Hesberg, C., Hänsch, R., Mendel, R. R., and Bittner, F. (2004). Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J. Biol. Chem. 279, 13547–13554. doi: 10.1074/jbc.M312929200
Huang, Z. A., Jiang, D. A., Yang, Y., Sun, J. W., and Jin, S. H. (2004). Effects of nitrogen deficiency on gas exchange, chlorophyll fluorescence, and antioxidant enzymes in leaves of rice plants. Photosynthetica 42, 357–364. doi: 10.1023/B:PHOT.0000046153.08935.4c
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Kanani, H., Dutta, B., and Klapa, M. (2010). Individual vs. combinatorial effect of elevated CO2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Syst. Biol. 4:177. doi: 10.1186/1752-0509-4-177
Kato, M. C., Hikosaka, K., Hirotsu, N., Makino, A., and Hirose, T. (2003). The excess light energy that is neither utilized in photosynthesis nor dissipated by photoprotective mechanisms determines the rate of photoinactivation in photosystem II. Plant Cell Physiol. 44, 318–325. doi: 10.1093/pcp/pcg045
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Liu, J.-G., Zhang, Z., Qin, Q.-L., Peng, R.-H., Xiong, A.-S., Chen, J.-M., et al. (2007). Isolated and characterization of a cDNA encoding ethylene-responsive element binding protein (EREBP)/AP2-type protein, RCBF2, in Oryza sativa L. Biotechnol. Lett. 29, 165–173. doi: 10.1007/s10529-006-9214-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Q., Li, Y., Gu, H., Zhao, L., Gu, X., and Li, W. (2013). The promoter of soybean photoreceptor GmPLP1 gene enhances gene expression under plant growth regulator and light stresses. Plant Cell Tissue Organ Cult. 114, 109–119. doi: 10.1007/s11240-013-0310-6
Maruyama, K., Takeda, M., Kidokoro, S., Yamada, K., Sakuma, Y., Urano, K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2011). Identification of cis-acting promoter elements in cold– and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nicolas, M. E., Simpson, R. J., Lambers, H., and Dalling, M. J. (1985). Effects of drought on partitioning of nitrogen in two wheat varieties differing in drought-tolerance. Ann. Bot. 55, 743–754.
Nogueira, F. T. S., De Rosa, V. E., Menossi, M., Ulian, E. C., and Arruda, P. (2003). RNA expression profiles and data mining of sugarcane response to low temperature. Plant Physiol. 132, 1811–1824. doi: 10.1104/pp.102.017483
Novillo, F., Alonso, J. M., Ecker, J. R., and Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 3985–3990. doi: 10.1073/pnas.0303029101
Pageau, K., Reisdorf-Cren, M., Morot-Gaudry, J.-F., and Masclaux-Daubresse, C. (2006). The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. J. Exp. Bot. 57, 547–557. doi: 10.1093/jxb/erj035
Qin, F., Sakuma, Y., Li, J., Liu, Q., Li, Y.-Q., Shinozaki, K., et al. (2004). Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 45, 1042–1052. doi: 10.1093/pcp/pch118
Qin, Q.-L., Liu, J.-G., Zhang, Z., Peng, R.-H., Xiong, A.-S., Yao, Q.-H., et al. (2007). Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza Sativa L. Mol. Breed. 19, 329–340. doi: 10.1007/s11032-006-9065-7
Robinson, S., and Parkin, I. (2008). Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genomics 9:434. doi: 10.1186/1471-2164-9-434
Sagi, M., Omarov, R. T., and Lips, S. H. (1998). The Mo-hydroxylases xanthine dehydrogenase and aldehyde oxidase in ryegrass as affected by nitrogen and salinity. Plant Sci. 135, 125–135. doi: 10.1016/S0168-9452(98)00075-2
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration– and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Schubert, K. R. (1986). Products of biological nitrogen fixation in higher plants: synthesis, transport, and metabolism. Annu. Rev. Plant Physiol. 37, 539–574. doi: 10.1146/annurev.pp.37.060186.002543
Shaik, R., and Ramakrishna, W. (2013). Machine learning approaches distinguish multiple stress conditions using stress responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol. 64, 481–495. doi: 10.1104/pp.113.225862
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Tahir, I. S. A., and Nakata, N. (2005). Remobilization of nitrogen and carbohydrate from stems of bread wheat in response to heat stress during grain filling. J. Agron. Crop Sci. 191, 106–115. doi: 10.1111/j.1439-037X.2004.00127.x
Verhoeven, A. S., Demmig-Adams, B., and Adams Iii, W. W. (1997). Enhanced employment of the xanthophyll cycle and thermal energy dissipation in spinach exposed to high light and N stress. Plant Physiol. 113, 817–824. doi: 10.1104/pp.113.3.817
Wang, P., Kong, C.-H., Sun, B., and Xu, X.-H. (2012). Distribution and function of allantoin (5-Ureidohydantoin) in rice grains. J. Agric. Food Chem. 60, 2793–2798. doi: 10.1021/jf2051043
Wang, Q., Guan, Y., Wu, Y., Chen, H., Chen, F., and Chu, C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67, 589–602. doi: 10.1007/s11103-008-9340-6
Waraich, E. A., Ahmad, R., Ashraf, M. Y., Saifullah, and Ahmad, M. (2011). Improving agricultural water use efficiency by nutrient management in crop plants. Acta Agric. Scand. B Soil Plant Sci. 61, 291–304. doi: 10.1080/09064710.2010.491954
Watanabe, S., Kounosu, Y., Shimada, H., and Sakamoto, A. (2014a). Arabidopsis xanthine dehydrogenase mutants defective in purine degradation show a compromised protective response to drought and oxidative stress. Plant Biotechnol. 31, 173–178. doi: 10.5511/plantbiotechnology.14.0117a
Watanabe, S., Matsumoto, M., Hakomori, Y., Takagi, H., Shimada, H., and Sakamoto, A. (2014b). The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 37, 1022–1036. doi: 10.1111/pce.12218
Watanabe, S., Nakagawa, A., Izumi, S., Shimada, H., and Sakamoto, A. (2010). RNA interference-mediated suppression of xanthine dehydrogenase reveals the role of purine metabolism in drought tolerance in Arabidopsis. FEBS Lett. 584, 1181–1186. doi: 10.1016/j.febslet.2010.02.023
Werner, A. K., Medina-Escobar, N., Zulawski, M., Sparkes, I. A., Cao, F.-Q., and Witte, C.-P. (2013). The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase in Arabidopsis. soybean, and rice. Plant Physiol. 163, 672–681. doi: 10.1104/pp.113.224261
Werner, A. K., Romeis, T., and Witte, C.-P. (2010). Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 6, 19–21. doi: 10.1038/nchembio.265
Werner, A. K., and Witte, C.-P. (2011). The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci. 16, 381–387. doi: 10.1016/j.tplants.2011.03.012
Wu, C., Li, X., Yuan, W., Chen, G., Kilian, A., Li, J., et al. (2003). Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35, 418–427. doi: 10.1046/j.1365-313X.2003.01808.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, L., Ding, J., Zhang, C., Jia, J., Weng, H., Liu, W., et al. (2005). Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep. 23, 759–763. doi: 10.1007/s00299-004-0881-0
Yesbergenova, Z., Yang, G., Oron, E., Soffer, D., Fluhr, R., and Sagi, M. (2005). The plant Mo-hydroxylases aldehyde oxidase and xanthine dehydrogenase have distinct reactive oxygen species signatures and are induced by drought and abscisic acid. Plant J. 42, 862–876. doi: 10.1111/j.1365-313X.2005.02422.x
Yobi, A., Wone, B. W. M., Xu, W., Alexander, D. C., Guo, L., Ryals, J. A., et al. (2013). Metabolomic profiling in Selaginella lepidophylla at various hydration states provides new insights into the mechanistic basis of desiccation tolerance. Mol. Plant 6, 369–385. doi: 10.1093/mp/sss155
Zhang, T., Zhao, X., Wang, W., Pan, Y., Huang, L., Liu, X., et al. (2012). Comparative Transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 7:e43274. doi: 10.1371/journal.pone.0043274
Zhang, Z.-H., Song, H.-X., Liu, Q., Rong, X.-M., Peng, J.-W., Xie, G.-X., et al. (2014). Distribution characters of absorption nitrogen in oilseed rape (Brassica napus L.) at different growth stages. J. Plant Nutr. 37, 1648–1660. doi: 10.1080/01904167.2014.888747
Zhou, M. Q., Shen, C., Wu, L. H., Tang, K. X., and Lin, J. (2010). CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 31, 186–192. doi: 10.3109/07388551.2010.505910
Keywords: CRT/DRE element, low temperature stress, OsCBF3, transcriptional regulation, ureidoglycolate amidohydrolase
Citation: Li J, Qin R-Y, Li H, Xu R-F, Yang Y-C, Ni D-H, Ma H, Li L, Wei P-C and Yang J-B (2015) Low-Temperature-Induced Expression of Rice Ureidoglycolate Amidohydrolase is Mediated by a C-Repeat/Dehydration-Responsive Element that Specifically Interacts with Rice C-Repeat-Binding Factor 3. Front. Plant Sci. 6:1011. doi: 10.3389/fpls.2015.01011
Received: 17 August 2015; Accepted: 02 November 2015;
Published: 13 November 2015.
Edited by:
Michael Wisniewski, United States Department of Agriculture – Agricultural Research Service, USAReviewed by:
Rajeev Arora, Iowa State University, USATimothy Artlip, United States Department of Agriculture – Agricultural Research Service, USA
Copyright © 2015 Li, Qin, Li, Xu, Yang, Ni, Ma, Li, Wei and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng-Cheng Wei, d2VpcGVuZ2NoZW5nQGdtYWlsLmNvbQ==; Jian-Bo Yang, eWppYW5ib0AyNjMubmV0
 Juan Li
Juan Li Rui-Ying Qin
Rui-Ying Qin Hao Li
Hao Li Rong-Fang Xu
Rong-Fang Xu Ya-Chun Yang
Ya-Chun Yang Da-Hu Ni
Da-Hu Ni Hui Ma
Hui Ma Li Li
Li Li Peng-Cheng Wei
Peng-Cheng Wei Jian-Bo Yang
Jian-Bo Yang