- 1Southern Crop Protection and Food Research Centre, Agriculture and Agri-Food Canada, London, ON, Canada
- 2Department of Biology, University of Western Ontario, London, ON, Canada
- 3Alberta Glycomics Centre and Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
- 4Department of Biology, University of British Columbia, Kelowna, BC, Canada
The receptor binding domain of the tailspike protein Gp9 from the P22 bacteriophage was recently shown to reduce Salmonella colonization in the chicken gut. In this study, we transiently expressed the receptor binding domain of the Gp9 tailspike protein in Nicotiana benthamiana, and targeted it to the endoplasmic reticulum (ER) or to the chloroplasts. Gp9 was also fused to either an elastin-like polypeptide (ELP) or hydrophobin I tag, which were previously described to improve accumulation levels of recombinant proteins. The highest levels of recombinant protein accumulation occurred when unfused Gp9 was targeted to the ER. Lower levels of chloroplast-targeted Gp9 were also detected. ELP-fused Gp9 was purified and demonstrated to bind to Salmonella enterica serovar Typhimurium in vitro. Upon oral administration of lyophilized leaves expressing Gp9-ELP to newly hatched chickens, we found that this tailspike protein has the potential to be used as a therapeutic to control Salmonella contamination in chickens.
Introduction
The human intestine is a complex microbial ecosystem where hundreds of species of bacteria have adapted to live and grow. A mutualistic relationship has evolved benefiting the health of the host while providing an optimal habitat for microflora to thrive (Guarner and Malagelada, 2003). However, some of these bacteria are pathogenic, causing a wide array of intestinal pathologies. Salmonella enterica is a Gram-negative enteropathogenic bacterium that is widely prevalent and is one of the primary causes of foodborne illness in humans. There are roughly 1.4 million non-typhoidal salmonellosis cases each year in North America, causing approximately 25% of all hospitalizations due to foodborne illness (Mead et al., 1999). S. enterica serotype Typhimurium also referred to as S. Typhimurium, causes gastroenteritis characterized by diarrhea, vomiting, and abdominal pain and is showing the emergence of multidrug-resistant strains (Su et al., 2004; Chen et al., 2013). Poultry and eggs are a major source of infection, but other sources such as vegetables, fruits, nuts, sprouts, leafy greens, roots, and beans have been reported (Rodrigue et al., 1990; Hammack, 2012). In chickens, Salmonella is found throughout the intestinal tract (Fanelli et al., 1971) and the rupturing of intestinal contents during evisceration can readily contaminate poultry meat. For instance, Salmonella has been isolated from 33% of raw chicken breasts sampled from retail grocery stores in Ontario, Canada (Cook et al., 2012).
Antibiotic use has led to the emergence of antibiotic-resistant Salmonella strains. In 2013, 17% of typhoidal Salmonella isolates from Canadians were resistant to ciprofloxacin and 41% of S. Heidelberg infections were resistant to at least one antibiotic1. This growing concern has provoked research into alternative methods for controlling bacterial outbreaks. Considerable research into using bacteriophage therapy to treat or prevent bacterial infections progressed in Eastern Europe and the former Soviet Union during the latter part of the 20th century and could potentially be reconsidered as a viable alternative to antibiotics (Sulakvelidze et al., 2001). Lytic bacteriophages are host-specific, self-replicating, and virtually non-toxic making them attractive alternatives to control bacteria such as Salmonella and bacteriophages have been shown to reduce Salmonella colonization in chickens (Goode et al., 2003; Atterbury et al., 2007). Despite these successes, this therapy is not without drawbacks. Bacteriophages are host-specific requiring diagnosis of the pathogen before the phage is administered (Waseh et al., 2010). Phages can also carry harmful genes and can potentially transfer these genes to the bacteria, increasing virulence (Skurnik and Strauch, 2006). As a result, there has been interest in the use of phage proteins such as endolysins (Roach and Donovan, 2015) as tools for the specific targeting of bacteria and the exploitation of phage receptor binding proteins for use in diagnostics and engineered phage-derived killing machines (Singh et al., 2012; Simpson et al., 2015). Unexpectedly, Waseh et al. (2010) have demonstrated that the P22 phage tailspike protein alone is effective in controlling Salmonella colonization and spread in chickens, presumably through its binding capability. These tailspike proteins are highly stable homotrimers that form the short tail of the bacteriophage and bind to the O-antigenic repeating units on the outer membrane lipopolysaccharide (Baxa et al., 1996). The tailspike protein Gp9 from the P22 bacteriophage can recognize several serovars of Salmonella including S. Typhimurium, S. Paratyphi A, and S. Enteritidis. A shortened version of Gp9 has been shown to agglutinate S. Typhimurium, inhibit bacterial motility and reduce colonization in the chicken gut (Waseh et al., 2010). Therefore, this protein has the potential to act as an effective pre-slaughter feed additive to reduce Salmonella contamination in chickens.
Plant bioreactors have been growing in acceptance as feasible production platforms for therapeutic proteins, as they are highly scalable and can be established with little upfront cost (Fischer et al., 2012). Protein drugs expressed in plant tissue are thought to be protected from digestive enzymes by the plant cell wall (Kwon and Daniell, 2015), and are especially useful for veterinary applications where regulations allow administration of unpurified or partially purified extracts (MacDonald et al., 2015). For example, leaf tissue can be harvested, lyophilized, and orally administered in capsules or suspended in a slurry removing costs associated with protein purification, administration, and cold-storage (Kolotilin et al., 2014). As higher eukaryotic organisms, plants can introduce post-translational modifications required for complex recombinant proteins. Despite these benefits, recombinant protein yield remains a major factor limiting the widespread adoption of plant bioreactors for commercial protein production. Consequently, several approaches are currently being used to increase protein accumulation in plants. Proteins can be targeted to different subcellular compartments such as the ER, the chloroplasts, and the apoplast using signal and transit peptides (Conley et al., 2009b). This is because each subcellular compartment has a unique biochemical environment, protease content, and physical size which influence protein accumulation levels (Streatfield, 2007; Pillay et al., 2014). Additionally, peptide tags can be fused to recombinant protein to increase accumulation. For example, fusion tags such as ELPs and HFBI can increase recombinant protein accumulation levels, and have also been used to purify proteins from plant extracts (Conley et al., 2011).
The goal of this project was to transiently produce the truncated version of Gp9 in Nicotiana benthamiana by targeting the protein to the chloroplasts, to the ER, or to the ER fused with an ELP or HFBI tag. The activity of plant-produced Gp9 was then tested by examining its ability to bind to S. Typhimurium. Lastly, plant tissue containing Gp9 was orally administered to chickens inoculated with S. Typhimurium to determine if this plant produced therapeutic has the potential to limit Salmonella colonization.
Materials and Methods
Gene Cloning
The truncated version (encoding amino acids 109–666) of the endorhamnosidase mutant of gp9 (as described in Waseh et al., 2010) was codon-optimized for plant expression and synthesized by Biobasic Inc. (Markham, ON, Canada). Gp9 was recombined into the previously constructed pCaMGate expression vectors using the LR reaction of Gateway® technology (Thermo Fischer Scientific, Waltham, MA, USA), courtesy of Dr. Andrew Conley of Agriculture and Agri-Food Canada, London Ontario. Recombinant pCAMGate vectors were transformed in Escherichia coli XL1-Blue using the Gene Pulser II system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and PCR screening using gene-specific primers (Forward primer: CGTTAGGTGTAGGTTTTGGTATGGATGGT, Reverse primer: CCGGCAACAGGATTCAATCTTAA) was conducted to screen for positive transformants containing the correct insert. Plasmid DNA was isolated from positive colonies and transformed into electro-competent Agrobacterium tumefaciens EHA105 cells. Electroporated A. tumefaciens cells were spread on yeast extract broth (YEB) plates containing 50 μg/ml kanamycin and 10 μg/ml rifampicin and incubated for 2 days at 28°C.
Transient Expression in N. benthamiana Plants
Suspensions of A. tumefaciens carrying Gp9 or A. tumefaciens carrying the post-transcriptional gene silencing suppressor p19 from Cymbidium ringspot virus (Silhavy et al., 2002), were incubated overnight at 28°C with shaking at 250 rpm until an optical density at 600 nm (OD600) of 0.5–1.0 was reached. Cultures were then centrifuged at 6000 × g for 30 min and resuspended to an OD600 of 1.0 in Gamborg’s solution containing 3.2 g/l Gamborg’s B5 with vitamins, 20 g/l sucrose, 10 mM MES (pH 5.6), and 200 μM acetosyringone. Cultures were then incubated at room temperature with gentle agitation for 1 h. An equal volume of A. tumefaciens culture containing Gp9 was combined with A. tumefaciens culture carrying p19 and Gamborg’s solution to give a total A. tumefaciens OD600 of approximately 0.67. These suspensions were used to infiltrate 7–8 week-old N. benthamiana plants grown in a growth room under 16 h light/8 h dark conditions at 21–22°C with 55% humidity, and receiving roughly 100 μmol/photons m-2s-1 of light. A 3 ml syringe was used to infiltrate the A. tumefaciens suspensions through the stomata of the abaxial leaf epidermis of N. benthamiana. After infiltration, plants were returned to the growth chamber for up to 6 days.
Tissue Collection and Protein Extraction
Four biological replicates were used in all experiments and consisted of four plants sampled as follows: two leaf disks/leaf (7 mm diameter) were collected from three infiltrated leaves of each plant and pooled. Tissue was flash frozen in liquid nitrogen and stored at -80°C until use. For protein extraction, tissue was homogenized twice in 30 s pulses using a TissueLyser (Qiagen, Venlo, Netherlands) and TSP were extracted in 200 μl of plant extraction buffer (PEB) containing 1X phosphate-buffered saline (PBS), 0.1% (v/v) Tween-20, 2% (w/v) polyvinylpolypyrrolidone (PVPP), 100 mM ascorbic acid, 1 mM ethylenediaminetetraccetic acid (EDTA), 1 mM of phenylmethanesulfonylfluoride (PMSF) and 1 μg/ml leupeptin. TSP concentration for each sample was determined using the Bradford assay (Bradford, 1976). For electrophoresis under non-reducing conditions, protein samples were stored in a 5% (w/v) SDS, 250 mg/ml glycerol, 0.1 mg/ml bromophenol blue, 0.16 M Tris/HCl sample buffer (Seckler et al., 1989) to better visualize protein trimerization. Samples were frozen at -80°C until use.
Western Blotting and Gel Staining
Pooled sample extracts and individual replicates were immunodetected against a standard curve of known amounts of purified Gp9-ELP to accurately quantify protein accumulation levels. Samples were either boiled for ten minutes or not boiled and loaded onto Bio-Rad Mini-Protean® TGXTM Precast 4–20% (w/v) polyacrylamide gradient gels. Separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes and blocked overnight in a 5% (w/v) skim milk powder in TBS-T (Tris-buffered saline-Tween 20) blocking solution. Membranes were incubated with a 1:5000 dilution of mouse anti-c-Myc antibody (Genscript, A00864, Piscataway, NJ, USA) or a 1:8000 dilution of rabbit polyclonal anti-Gp9 antibody (Kropinski et al., 2011). Membranes were washed and incubated with a 1:5000 dilution of goat anti-mouse or anti-rabbit secondary antibody conjugated with horseradish peroxidase (HRP), and visualized with the GE Healthcare Life Sciences (Little Chalfont, UK) ECL Prime Western Blotting Detection Reagent. Recombinant protein was quantified by image densitometry using Totallab TL100 software (Non-linear Dynamics, Durham, NC, USA). For staining of separated proteins, gels were washed for 5 min in water and stained for 1 h with GelCodeTM Blue stain reagent (Thermo Fischer Scientific, Waltham, MA, USA) at room temperature. Gels were then destained using three 5-min washes with water and imaged.
Protein Purification
Gp9-ELP and Gp9-HFBI proteins were purified using a c-Myc tag purification kit from MBL International Corporation MBL (3305, Woburn, MA, USA) according to the manufacturer’s instructions. Purified protein was stored at -80°C until use. The E. coli produced His6-Gp9 was purified as described by Waseh et al. (2010).
Gp9 and Gp9-ELP Adherence to S. Typhimurium
Salmonella enterica serovar Typhimurium (ATCC19585) was purchased from the American Type Culture Collection (Manassas, VA, USA) and grown under aerobic conditions at 37°C on Lysogeny Broth (LB) agar plates. The strain used in the adherence assay was transformed with the pWM1007 plasmid (Miller et al., 2000) which expresses the green fluorescence protein (GFP) and grown on LB supplemented with 25 μg/ml kanamycin.
Five hundred nanograms of the E. coli produced His6-Gp9, BSA, or plant-produced Gp9-ELP were spotted onto hole punch sized pieces of nitrocellulose membranes and then blocked for 1 h in 5% (w/v) skim milk in PBS with 0.05% (v/v) Tween (PBS-T). The membranes were then probed with 108 cfu/ml of GFP-expressing S. Typhimurium in 5% skim milk PBS-T. The disks were washed three times for 5 min in PBS-T and were then placed onto LB agar plates with 25 μg/ml kanamycin and allowed to grow at room temperature overnight, followed by growth at 37°C for 8 h. The disks were imaged with a FujiFilm (Tokyo, Japan) FLA-5000 system using the 473 nm laser at 400V for excitation and LPB (Y510) filter for emission. Fluorescence intensity was measured using the MultiGauge version 3.0 software.
Animal Studies
Animal studies were carried out in accordance with the protocol approved by the Animal Care and Use Committee at the University of Alberta following the procedure described by Waseh et al. (2010). Each group contained 5–8 SPF leghorn chickens (Poultry Research Facility, University of Alberta) that were provided with feed and water ad libitum and were randomly tested for the presence of Salmonella on the day of hatching by plating cloacal swabs onto selective Oxoid Brilliance Salmonella agar (Oxoid, ON, Canada). In all cases no Salmonella colonies were observed after 24 h of incubation at 37°C. Chickens were orally gavaged with 300 μL PBS containing 107 colony forming units (CFUs) of S. Typhimurium the next day and then gavaged with 35 mg lyophilized and powdered leaves resuspended in 300 μl of PBS at 1, 18, and 42 h post-infection. The chickens were culled at 47 h post-infection and the collected cecal contents were serially diluted and plated onto Oxoid Brilliance Salmonella agar. Salmonella CFU were counted after the agar plates were incubated for 24 h at 37°C.
Statistics
Minitab® 17 statistical software (Minitab Ltd., Coventry, UK) was used to perform statistical analysis on the Gp9 accumulation data. A one-way analysis of variance (ANOVA) was performed with a Tukey test on the mean Gp9 accumulation levels for each day of the time course. P < 0.05 was considered significant. A two-tailed Student’s t-test was performed on the data from the chicken experiment. P < 0.05 was once again considered significant.
Results
Gp9 Transient Expression in N. benthamiana
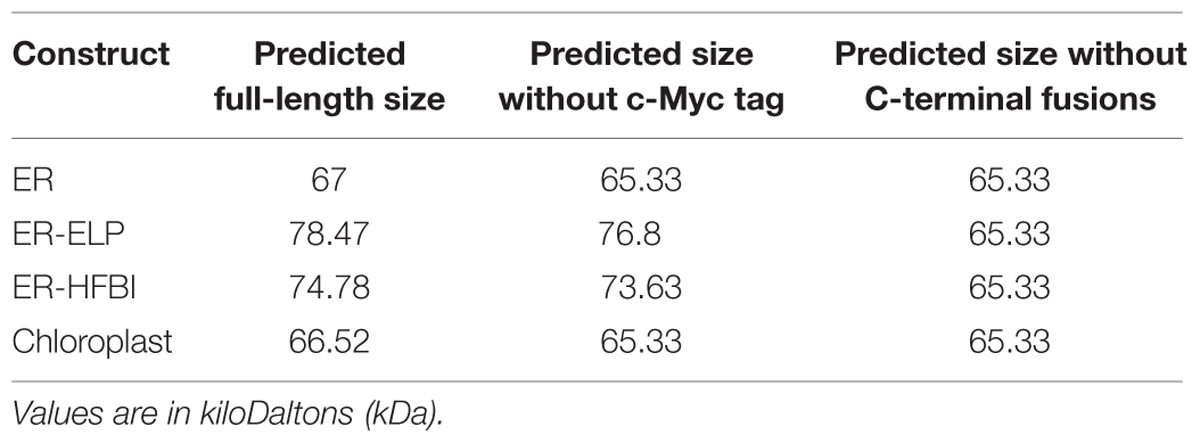
The Gp9 tailspike gene was cloned into pCaMGate expression vectors (Pereira et al., 2014) targeting the ER, the ER fused with an ELP tag (ER-ELP), the ER fused with a HFBI tag (ER-HFBI), or the chloroplasts (Figure 1). Constructs were then agroinfiltrated along with the gene silencing suppressor, p19 (Silhavy et al., 2002), into the leaves of N. benthamiana plants. Plants were monitored over the course of 6 day(s) post-infiltration (dpi). Young, upper leaves infiltrated with the ER-targeted constructs turned a slight yellow–green color but otherwise the phenotypes remained unchanged. Leaf tissue samples were collected from 3 to 6 dpi and analyzed for Gp9 accumulation via Western blot using a c-Myc antibody specific for the C-terminal Myc peptide found in all constructs (Figure 2A). Gp9 targeted to the ER was faintly visible, running higher than predicted (Table 1), and no bands were visible for the chloroplast-targeted Gp9. Conversely, Gp9-ELP and Gp9-HFBI were readily detected, and faint bands were observed for both the ELP and HFBI fused constructs migrating above 150 kDa which could represent potential trimers. Smaller bands ranging in size between 20 and 50 kDa were also observed in the Gp9-ELP and Gp9-HFBI samples which most likely correspond to Gp9 degradation products since they are absent from the p19 negative control lane (Figure 2A). Interestingly, when immunoblots were probed with an anti-Gp9- antibody, a strong band slightly under 75 kDa was observed for ER-targeted Gp9, and a somewhat fainter band was seen in the chloroplast-targeted Gp9 (Figure 2B). Bands potentially representing dimers and trimers were also observed for all four proteins, as well as smaller bands that may represent Gp9 degradation products. It is interesting that the smaller bands observed in the Gp9-ELP and Gp9-HFBI are of a different size when detected with the two antibodies. The protein band migrating between 37.5 and 50 kDa may be a plant protein as it appears faintly in the p19 lane, while the band running between 50 and 75 kDa may represent a degradation product of Gp9. This band is present in all samples, and it may represent the N-terminal portion of the protein detected with the Gp9-specific antibody, while the smaller band running between 20 and 37 kDa on the blot detected with the c-Myc antibody might represent the C-terminal portion of the protein. The low abundance of Gp9 on blots probed with the c-Myc antibody also imply that the c-Myc tag is cleaved off Gp9, or is inaccessible to the antibody.
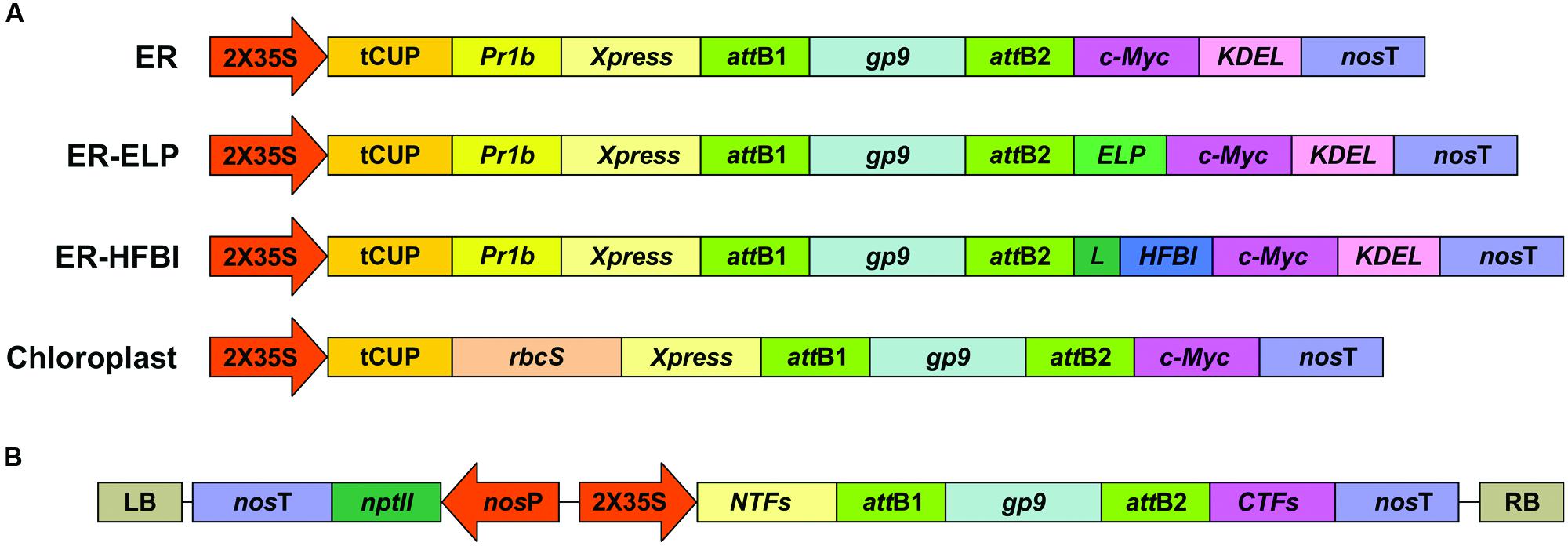
FIGURE 1. Expression cassettes for Gp9 transient expression in N. benthamiana. (A) A schematic representing the gene constructs targeting the ER or the chloroplasts. Two of the cassettes targeting the ER also contain sequences coding for an ELP or a HFBI tag with a linker (L). Expression and targeting elements incorporated into the expression vectors are: the double-enhanced 35S cauliflower mosaic virus promoter (2X35S); tCUP translational enhancer from tobacco; tobacco Pathogenesis-Related-1b signal peptide (PR1b) and the KDEL ER retrieval tetrapeptide; nopaline synthase terminator (nosT); and Xpress and c-Myc tags for detection and purification. A sequence coding for the RuBisCo small subunit transit peptide (rbcS) was used for targeting to the chloroplasts. (B) A zoomed-out schematic of the constructs in the T-DNA sequence. LB and RB are the left and right borders of the T-DNA sequence. nptII codes for neomycin phosphotransferase conferring resistance to kanamycin, driven by a nopaline synthase promoter (nosP) and terminator (nosT). Schematic represents the same constructs in (A) with different N-terminal fusions (NTFs) and C-terminal fusions (CTFs). Schematics are not to scale.
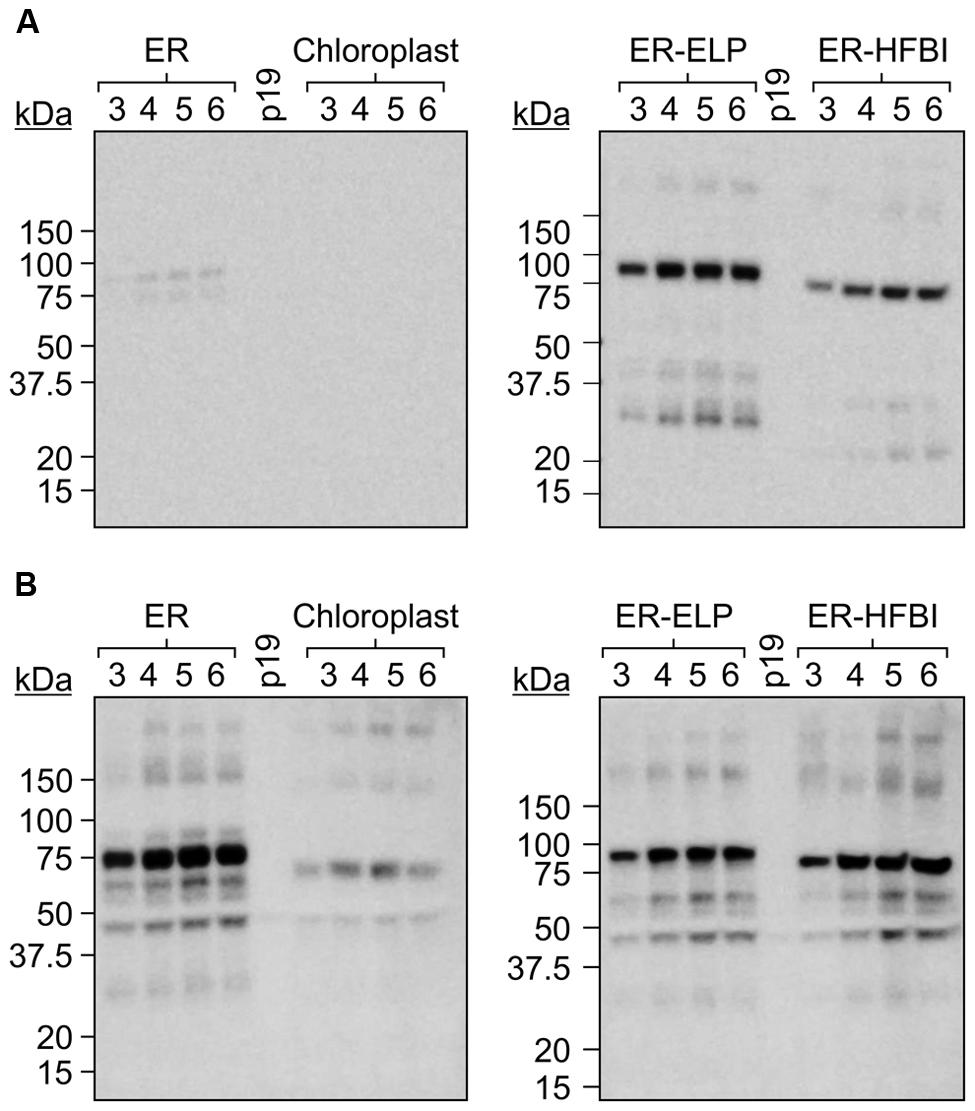
FIGURE 2. A time course of Gp9 accumulation levels in Nicotiana benthamiana. (A) Immunoblots of TSP extracted from N. benthamiana leaf tissue infiltrated with gp9 constructs targeting the ER, the chloroplasts, or the ER fused to an ELP tag (ER-ELP) or fused to a hydrophobin tag (ER-HFBI). Four plants were infiltrated with each construct. Tissue was sampled from infiltrated leaves from 3 to 6 dpi. Equal volumes of TSP from each of the four replicates were pooled, and 20 μg was loaded per well. Immunoblots were probed with (A) an anti-c-Myc antibody or (B) an anti-Gp9 antibody. p19: TSP from plants infiltrated with p19 serving as a negative control.
Gp9 Accumulation in N. benthamiana
Since more protein was detected using the anti-Gp9-antibody, this antibody was used to accurately quantify Gp9 accumulation via densitometry analysis. For the purpose of protein quantitation, only bands representing the full length Gp9 monomer were quantified. Immunoblots using individual replicates revealed that Gp9 accumulation increases from the 3rd dpi, peaks on day 4 or 5, and subsequently decreases (Figure 3). ER-targeted Gp9 accumulated in significantly higher amounts than the other proteins reaching on average of 1.64 ± 0.09% of TSP on day 5. When recombinant protein accumulation is calculated in terms of micrograms of Gp9 per gram of FLW, Gp9 accumulates to an average of 235.01 ± 21.12 μg/g of FLW on day 4 (Figure 3B). The presence of either the ELP or HFBI tag appears to significantly decrease Gp9 accumulation on 3–5 dpi (P < 0.05). Gp9-ELP accumulates on average to 0.98 ± 0.05% of TSP on day 5 or 135.86 ± 25.56 μg/g of FLW on day 4, roughly 0.6 times or 1.7 times less, respectively, than when unfused. The presence of the HFBI tag caused Gp9 to accumulate in even lower amounts on average to 0.79 ± 0.09% of TSP (0.48 times less) or 116.49 ± 16.29 μg/g of FLW (two times less) on day 4. Gp9 accumulated significantly less when targeted to the chloroplasts compared to ER-targeted proteins on days 4 and 5 (P < 0.05). Accumulation reached an average of 0.21 ± 0.01% of TSP or 28.45 ± 1.65 μg/g of FLW on the 5th dpi.
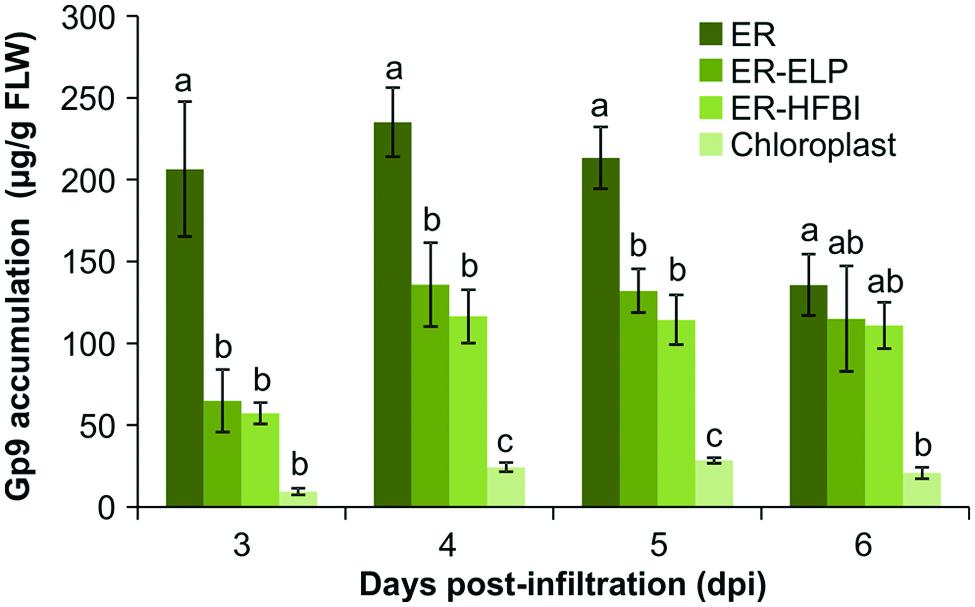
FIGURE 3. Quantification of Gp9 accumulation in N. benthamiana over 3, 4, 5, and 6 dpi. Quantification was performed on TSP extracts from infiltrated N. benthamiana tissue using a standard curve of known amounts of purified Gp9-ELP. Gp9 was targeted to the ER, the chloroplasts, or the ER fused to an ELP tag (ER-ELP) or fused to a hydrophobin tag (ER-HFBI). Immunoblots were probed with an anti-Gp9 antibody. Accumulation levels of Gp9 are shown in μg per g of FLW. Error bars represent the standard error of the mean value of four biological replicates. Treatments which do not share a letter are significantly different (P < 0.05) as determined by a one way ANOVA and the Tukey test.
Gp9-ELP Purification and Characterization
Gp9 polypeptides are found in monomeric, dimeric, and trimeric intermediates before forming the stable, native trimer (Benton et al., 2002). The trimeric intermediate species or protrimer consists of associated subunits which have not completely folded forming a transient, less-stable precursor to the trimer (Goldenberg and King, 1982). We successfully purified Gp9-ELP using a c-Myc-tag purification kit (Figure 4A) and decided to investigate if purified Gp9-ELP is present in either of these states by avoiding complete denaturation (unheated sample) and by avoiding reducing agents such as dithiothreitol (DTT) in the sample buffer to keep the disulfide bonds oxidized (unheated, no DTT). While most of the Gp9-ELP was found in the monomeric form when it was reduced and denatured, there was very little monomer present when heat denaturation was omitted. Instead, an intense band was observed below 250 kDa, which corresponds to the expected size for the 215 kDa trimer (Figures 4A,B). Higher banding was observed when samples were electrophoresed under non-reducing conditions (Figure 4B). Gp9 contains eight cysteine residues and has disulfide bonds while existing as a protrimer, despite the fact that all are reduced during conversion to the native trimeric state (Robinson and King, 1997). Consequently, bands running above 250 kDa could represent the protrimer intermediate running slower than the trimer. Generally, an oxidizing environment is needed for trimer folding due to the presence of the disulfide bonds in the protrimer, yet increasing concentrations of DTT increases the conversion of protrimer to trimer (Robinson and King, 1997). Even larger bands were visualized and could represent higher-order multimers or aggregates (Speed et al., 1997).
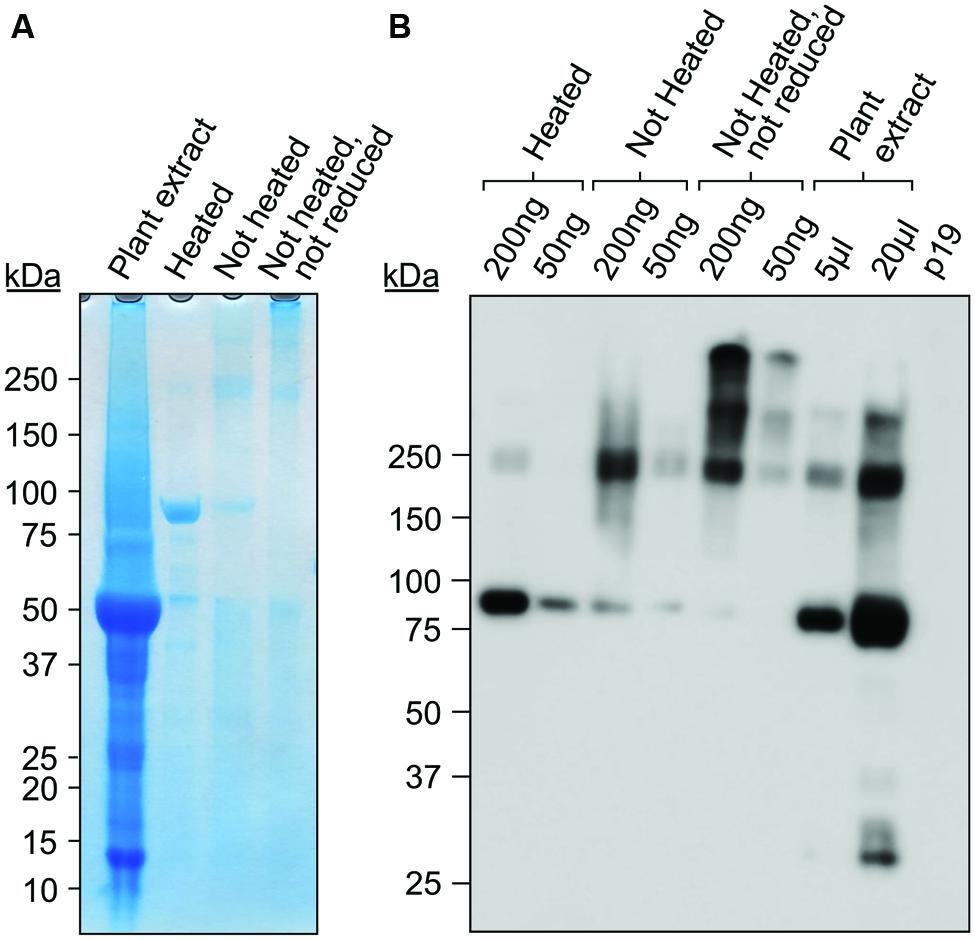
FIGURE 4. Purified Gp9-ELP is found in higher molecular weight protein complexes. Purified Gp9-ELP was electrophoresed under the following conditions: in sample buffer containing DTT and heat-denatured by boiling for 10 min, in sample buffer containing DTT with no heat denaturation, or in sample buffer without DTT and no heat denaturation. The plant extract containing Gp9-ELP was also loaded as a positive control in sample buffer containing DTT and heat-denatured by boiling for 10 min. Twenty microliters of extract from plants infiltrated with p19 were used as a negative control. Gels were either coomassie stained (A) or immunodetected (B) using an anti-c-Myc antibody.
Our results suggest that Gp9 is present in the stable trimeric form when purified and is expected to remain functional to some degree.
Gp9-ELP Binding to S. Typhimurium
To determine if the plant produced Gp9 can bind to S. Typhimurium, 500 ng of purified Gp9-ELP was spotted onto a nitrocellulose membrane, blocked and then probed for 1 h with 108 cfu/ml of GFP expressing S. Typhimurium. BSA and E. coli-produced Gp9 were used as negative and positive controls, respectively. Gp9-ELP was able to bind S. Typhimurium at similar levels as the E. coli-produced Gp9 as measured by fluorescence (Figure 5) suggesting that the plant-produced protein is functional.
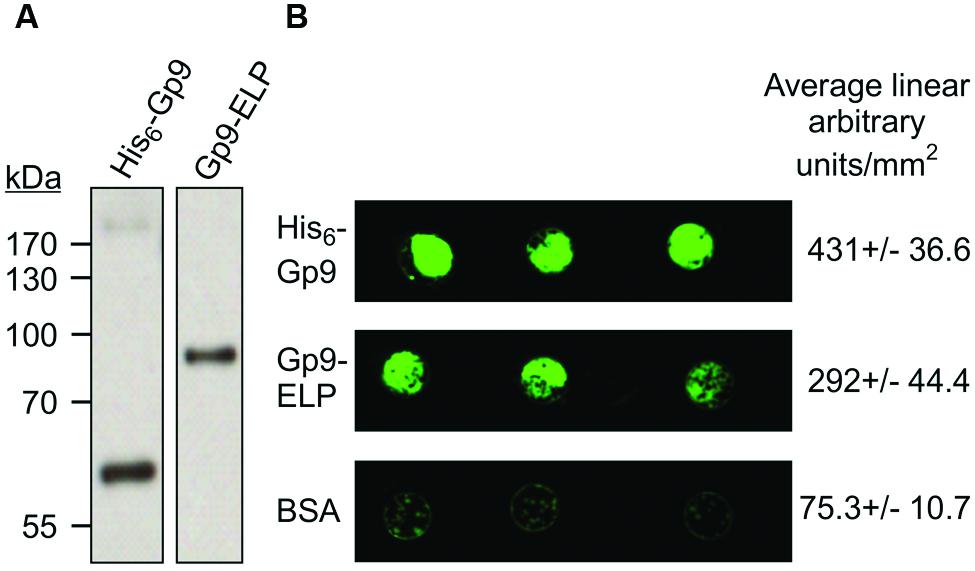
FIGURE 5. S. Typhimurium binding activity to plant produced Gp9-ELP. (A) The Escherichia coli produced His6-Gp9 (62 kDa) and the plant-produced Gp9-ELP (82 kDa) both react with anti-Gp9 antibodies. (B) Either 500 ng of His6-Gp9, BSA, or Gp9-ELP protein were spotted onto nitrocellulose and then probed with 108 cfu/ml GFP-expressing S. Typhimurium and allowed to grow overnight. Two biological replicates were done, each with three technical replicates in each. One representative image and densitometry analysis is shown.
The Effect of Oral Administration of Gp9-ELP on Salmonella Colonization
Due to the demonstrated binding activity of Gp9-ELP, we investigated the effects of orally administering plant tissue containing Gp9-ELP on Salmonella colonization in chicks. The chicks were orally gavaged with 35 mg of resuspended plant tissue containing 7.7 μg Gp9-ELP 1, 18 and 42 h after gavaging with S. Typhimurium. Chicks were culled 47 h after bacterial infection and the Salmonella CFU in the cecal contents were enumerated (Figure 6). Gp9-ELP showed approximately 1-log reduction in S. Typhimurium colonization compared to the untreated control birds (P = 0.058). These results are promising considering plant leaves were fed directly to the birds without further purification, and each dose contained much less Gp9-ELP compared with the Waseh et al. (2010) study where a dose consisted of 30 μg of purified Gp9.
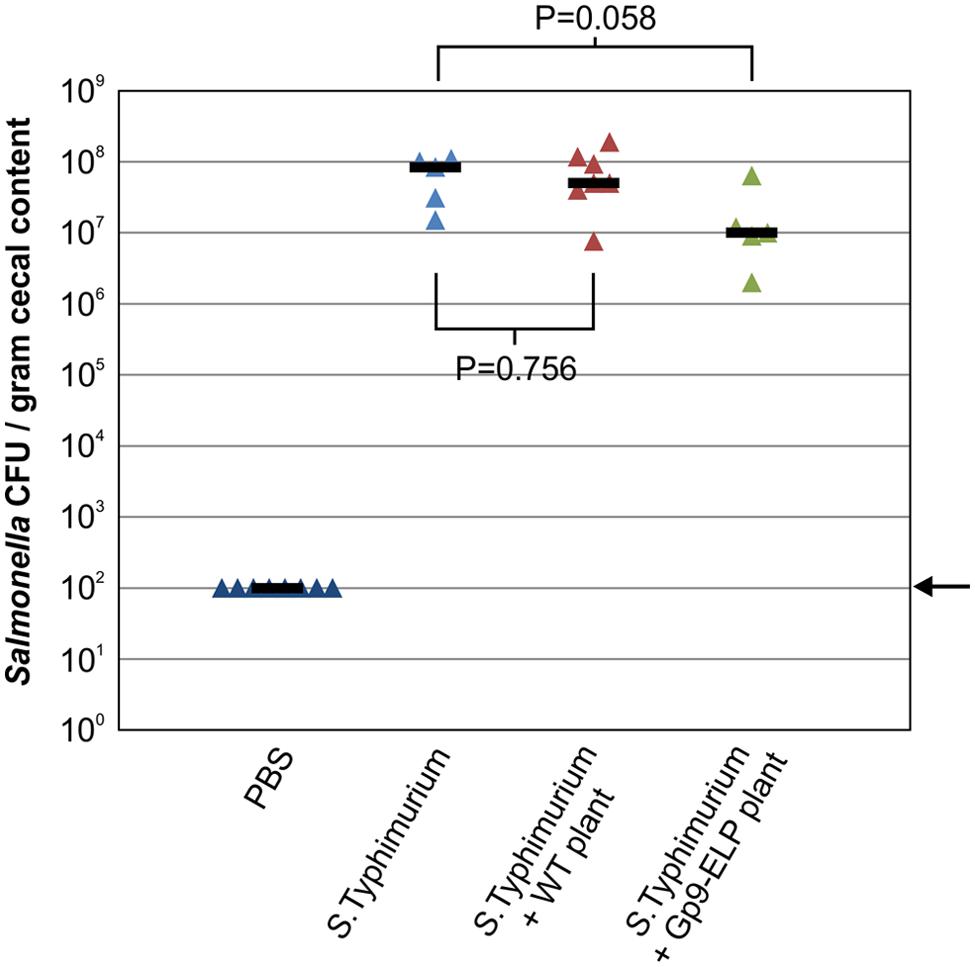
FIGURE 6. Treatment of S. Typhimurium infected chickens with Gp9-ELP. SPF leghorn chickens were orally gavaged with PBS or S. Typhimurium as indicated. One hour later (and at 18 and 42 h), two of the groups were orally gavaged with N. benthamiana extracts with or without Gp9-ELP, while the control groups received PBS. Chickens were fed 7.7 μg of Gp9-ELP per dose. Salmonella colonization levels of the chicken ceca were determined 47 h post-infection. The median is shown for each group and P-values were determined for the plant-fed groups in comparison to the S. Typhimurium group. The arrow represents the detection limit of the experiment.
Discussion
This study demonstrated that the receptor binding domain of the P22 bacteriophage tailspike protein, Gp9, could be transiently expressed in N. benthamiana at reasonable levels and is able to bind to S. Typhimurium. The results presented here suggest that the correct choice of antibody for protein detection is essential to accurately quantify recombinant protein accumulation. When probing with an antibody specific to the C-terminal Myc tag, Gp9 appeared to accumulate to the highest levels when fused to an ELP or HFBI tag, supporting the hypothesis of this study that these fusion tags would increase accumulation levels as previously reported (Patel et al., 2007; Conley et al., 2009a; Joensuu et al., 2010). These results were expected as ELP and HFBI tags promote the formation and distribution of protein bodies (PBs) which are thought to protect recombinant protein from hydrolysis and protease cleavage (Conley et al., 2011; Saberianfar et al., 2015).
However, probing with a Gp9-specific antibody gave contrasting results. When the Gp9-specific antibody was used for immunodetection, unfused ER-targeted Gp9 displayed about 50% more accumulation than the ELP and HFBI fusions. This result was unexpected, and suggests that Gp9 is stable in N. benthamiana and the additional ELP or HFBI tags do not increase protein stability further. Indeed, ELP tags have negligible effects on already high accumulating proteins such as GFP (Conley et al., 2009a). It appears that the addition of an ELP or HFBI tag actually decreases Gp9 accumulation when targeted for retrieval to the ER of N. benthamiana and this should be investigated in future studies. These results also imply that the c-Myc tag is partially cleaved or inaccessible causing us to underestimate Gp9 accumulation levels when detecting with an anti-c-Myc tag antibody. It is possible that the ELP and HFBI tags protect the c-Myc tag from cleavage, possibly through protein body formation, and therefore the fusion constructs were able to be detected with the c-Myc antibody. However, when probing with the Gp9 antibody the ER and chloroplast constructs produce a slightly higher banding pattern than expected if the c-Myc is indeed cleaved off (Figure 2; Table 1). It can be postulated then that these proteins may fold into a conformation that makes the c-Myc tag inaccessible. Regardless, as many recombinant proteins are designed to have tags for easy detection, these results are a cautionary note on the limits of short tags, and that perhaps both N- and C-terminal tags might be used to improve chances of accurate quantitation, while protein-specific antibodies are most probably the best option if available. Our constructs also contain an N-terminal Xpress tag (Figure 1), however, commercially available antibodies have not detected any recombinant protein in crude extracts (unpublished).
Gp9 also accumulated in the chloroplasts, but to a lower level than when targeted to the secretory pathway. Generally, chloroplast targeting is a promising strategy for increasing recombinant protein accumulation (Hyunjong et al., 2006). Nevertheless, some proteins do not accumulate well when targeted to these organelles. For example, one study described lower accumulation levels of zeolin, a chimeric storage protein, when targeted to the chloroplasts compared to the ER. A visualized degradation pattern of zeolin provided evidence for protein degradation in the chloroplasts (Bellucci et al., 2007). On the other hand, Oey et al. (2009) were able to express a phage lysin protein in tobacco chloroplasts with accumulation levels reaching 70% of TSP. This protein was produced by transforming the plastid genome and the gene was codon-optimized for plastid expression. The phage lytic protein was shown to be extremely stable in the plastids, which is understandable as phages have evolved substantial resistance to bacterial proteases. By analogy, the Gp9 should be stable in the plastids, and therefore it is possible that the protein is unable to efficiently translocate into the chloroplasts. Therefore, future work could focus on transforming the plastid genome of N. tabacum for high level, stable expression of Gp9 without having to rely on chloroplast protein import.
Like the full length Gp9, shortened versions lacking the amino acid N-terminus domain maintain the stability and enzymatic activity of the full length parent proteins (Miller et al., 1998). To the best of our knowledge, this is the first study to transiently express this truncated protein in plants. Previously, truncated tailspikes have been expressed in E. coli, isolated and shown to reduce Salmonella colonization in the chicken GI tract. It is believed that the tailspikes bind to Salmonella and retard its motility and binding capability (Waseh et al., 2010). Consequently, plant-produced Gp9 has the potential to serve as a therapeutic to control Salmonella. These proteins can potentially be added to chicken or other livestock feed, reducing contamination and reducing infections in humans. While the plant produced Gp9 did not yield statistically significant results in the chicken experiment, the results are indeed promising. We purified Gp9-ELP using a Myc column to show that Gp9-ELP exists as a trimer and remains active by binding to S. Typhimurium. However, it is still possible that the ELP tag could alter Gp9 conformation in vivo, influencing activity and future work could focus on unfused Gp9. In vitro, ELPs have been reported to reduce protein activity compared to non-fused proteins (Shamji et al., 2007; Kaldis et al., 2013). It is possible that the physiological temperature of chickens could influence the solubility of Gp9-ELP. This is because ELPs undergo a reversible phase transition from soluble to insoluble aggregates when heated past a certain transition temperature. However, this transition temperature can be manipulated by the altering the number and/or the residue composition of the peptide repeats in order to adjust the solubility of Gp9-ELP, thus allowing for better therapeutic efficacy at physiological temperatures (Hassouneh et al., 2012). Furthermore, it is likely that the 1 log reduction in Salmonella colonization observed in this study compared to the 2 log reduction in the Waseh et al. (2010) study is due to the lower Gp9 dose administered to the birds (30 μg of purified His6-Gp9 versus 7.7 μg Gp9-ELP in 35 mg of lyophilized plant tissue per dose) as well as the fact that the Gp9-ELP protein is contained within plant cells and thus the chicken’s gut proteases need to digest through the plant cell wall in order to release the protein. In the previous study, it was shown that if the first dose was delivered at 18 h instead of 1 h post-infection, the benefits of the treatment were less. In our study, the protein would be released from the plant tissue sometime after the treatment is administered, so it is possible that this delay is partially responsible for the lower reduction in infection. Future studies will compare administration of higher levels of Gp9 protein and providing the feed throughout the experiment in order to more closely resemble the natural conditions in which this protein would be used.
One of our goals in this study was to demonstrate that Gp9 could be directly administered while in minimally processed plant tissue, without having to rely on taxing purification and formulation techniques. It can be postulated that purified Gp9 may better reduce Salmonella colonization in chickens as previously reported (Waseh et al., 2010). While a direct comparison between purified and non-purified Gp9-ELP was not done in this study, we have shown that wild type plant tissue alone is not significantly influencing S. Typhiumurium colonization in chicks. Therefore, while future work should mainly focus on increasing Gp9 accumulation in N. benthamiana, this study serves as another successful example of engineering plant factories for the production of a functional therapeutic.
Author Contributions
SM and DS designed the research, performed the experiments, SM wrote the manuscript. RM, CS, and MD conceived the study, participated in its design and edited the manuscript.
Abbreviations
BSA, bovine serum albumin; CFU, colony-forming units; ELP, synthetic elastin-like polypeptide; ER, endoplasmic reticulum; FLW, fresh leaf weight; GI tract, gastro-intestinal tract; HFBI, hydrophobin I from Trichoderma reesei; S. Typhimurium, Salmonella enterica serovar Typhimurium; S. Paratyphi, Salmonella enterica serovar Paratyphi; S. Enteritidis, Salmonella enterica serovar Enteriditis; S. Heidelberg, Salmonella Heidelberg; TSP, total soluble protein(s).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Angelo Kaldis, Hong Zhu, Bernadette Beadle, and Cory Wenzel for their technical support and Alex Molnar for help with the figures. This work was funded by Agriculture and Agri-Food Canada’s A-base grant #1107, and the Natural Sciences and Engineering Research Council of Canada Strategic Grant #397260. CMS is an Alberta Innovates Technology Futures iCORE Strategic Chair in Bacterial Glycomics.
Footnotes
- ^http://healthycanadians.gc.ca/alt/pdf/publications/drugs-products-medicaments-produits/antibiotic-resistance-antibiotique/antimicrobial-surveillance-antimicrobioresistance-eng.pdf
References
Atterbury, R. J., Van Bergen, M. A., Ortiz, F., Lovell, M. A., Harris, J. A., De Boer, A., et al. (2007). Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73, 4543–4549. doi: 10.1128/AEM.00049-07
Baxa, U., Steinbacher, S., Miller, S., Weintraub, A., Huber, R., and Seckler, R. (1996). Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 71, 2040–2048. doi: 10.1016/S0006-3495(96)79402-X
Bellucci, M., De Marchis, F., Nicoletti, I., and Arcioni, S. (2007). Zeolin is a recombinant storage protein with different solubility and stability properties according to its localization in the endoplasmic reticulum or in the chloroplast. J. Biotechnol. 131, 97–105. doi: 10.1016/j.jbiotec.2007.06.004
Benton, C. B., King, J., and Clark, P. L. (2002). Characterization of the protrimer intermediate in the folding pathway of the interdigitated beta-helix tailspike protein. Biochemistry 41, 5093–5103. doi: 10.1021/bi025167e
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chen, H. M., Wang, Y., Su, L. H., and Chiu, C. H. (2013). Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr. Neonatol. 54, 147–152. doi: 10.1016/j.pedneo.2013.01.010
Conley, A. J., Joensuu, J. J., Jevnikar, A. M., Menassa, R., and Brandle, J. E. (2009a). Optimization of elastin-like polypeptide fusions for expression and purification of recombinant proteins in plants. Biotechnol. Bioeng. 103, 562–573. doi: 10.1002/bit.22278
Conley, A. J., Joensuu, J. J., Menassa, R., and Brandle, J. E. (2009b). Induction of protein body formation in plant leaves by elastin-like polypeptide fusions. BMC Biol. 7:48. doi: 10.1186/1741-7007-7-48
Conley, A. J., Joensuu, J. J., Richman, A., and Menassa, R. (2011). Protein body-inducing fusions for high-level production and purification of recombinant proteins in plants. Plant Biotechnol. J. 9, 419–433. doi: 10.1111/j.1467-7652.2011.00596.x
Cook, A., Odumeru, J., Lee, S., and Pollari, F. (2012). Campylobacter, Salmonella, Listeria monocytogenes, verotoxigenic Escherichia coli, and Escherichia coli prevalence, enumeration, and subtypes on retail chicken breasts with and without skin. J. Food Prot. 75, 34–40. doi: 10.4315/0362-028X.JFP-11-206
Fanelli, M. J., Sadler, W. W., Franti, C. E., and Brownell, J. R. (1971). Localization of Salmonellae within the intestinal tract of chickens. Avian Dis. 15, 366–375. doi: 10.2307/1588708
Fischer, R., Schillberg, S., Hellwig, S., Twyman, R. M., and Drossard, J. (2012). GMP issues for recombinant plant-derived pharmaceutical proteins. Biotechnol. Adv. 30, 434–439. doi: 10.1016/j.biotechadv.2011.08.007
Goldenberg, D., and King, J. (1982). Trimeric intermediate in the in vivo folding and subunit assembly of the tail spike endorhamnosidase of bacteriophage P22. Proc. Natl. Acad. Sci. U.S.A. 79, 3403–3407. doi: 10.1073/pnas.79.11.3403
Goode, D., Allen, V. M., and Barrow, P. A. (2003). Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69, 5032–5036. doi: 10.1128/AEM.69.8.5032-5036.2003
Guarner, F., and Malagelada, J. R. (2003). Gut flora in health and disease. Lancet 361, 512–519. doi: 10.1016/S0140-6736(03)12489-0
Hammack, T. (2012). “Salmonella species,” in Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins, 2nd Edn, eds K. Lampel, S. Al-Khaldi, and S. Cahill (Silver Spring, MD: Food and Drug Administration).
Hassouneh, W., MacEwan, S. R., and Chilkoti, A. (2012). Fusions of elastin-like polypeptides to pharmaceutical proteins. Methods Enzymol. 502, 215–237. doi: 10.1016/B978-0-12-416039-2.00024-0
Hyunjong, B., Lee, D. S., and Hwang, I. (2006). Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J. Exp. Bot. 57, 161–169. doi: 10.1093/jxb/erj019
Joensuu, J. J., Conley, A. J., Lienemann, M., Brandle, J. E., Linder, M. B., and Menassa, R. (2010). Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol. 152, 622–633. doi: 10.1104/pp.109.149021
Kaldis, A., Ahmad, A., Reid, A., McGarvey, B., Brandle, J., Shengwu et al. (2013). High-level production of human interleukin-10 fusions in tobacco cell suspension cultures. Plant Biotechnol. 11, 535–545. doi: 10.1111/pbi.12041
Kolotilin, I., Topp, E., Cox, E., Devriendt, B., Conrad, U., Joensuu, J., et al. (2014). Plant-based solutions for veterinary immunotherapeutics and prophylactics. Vet. Res. 45, 117. doi: 10.1186/s13567-014-0117-4
Kropinski, A. M., Arutyunov, D., Foss, M., Cunningham, A., Ding, W., Singh, A., et al. (2011). Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673. Appl. Environ. Microbiol. 77, 8265–8271. doi: 10.1128/AEM.05562-11
Kwon, K. C., and Daniell, H. (2015). Low-cost oral delivery of protein drugs bioencapsulated in plant cells. Plant Biotechnol. J. 13, 1017–1022. doi: 10.1111/pbi.12462
MacDonald, J., Doshi, K., Dussault, M., Hall, J. C., Holbrook, L., Jones, G., et al. (2015). Bringing plant-based veterinary vaccines to market: managing regulatory and commercial hurdles. Biotechnol. Adv. 33, 1572–1581. doi: 10.1016/j.biotechadv.2015.07.007
Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., et al. (1999). Food-related illness and death in the United States. Emerg. Infect. Dis. 5, 607–625. doi: 10.3201/eid0505.990502
Miller, S., Schuler, B., and Seckler, R. (1998). Phage P22 tailspike protein: removal of head-binding domain unmasks effects of folding mutations on native-state thermal stability. Protein Sci. 7, 2223–2232. doi: 10.1002/pro.5560071021
Miller, W. G., Bates, A. H., Horn, S. T., Brandl, M. T., Wachtel, M. R., and Mandrell, R. E. (2000). Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66, 5426–5436. doi: 10.1128/AEM.66.12.5426-5436.2000
Oey, M., Lohse, M., Kreikemeyer, B., and Bock, R. (2009). Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445. doi: 10.1111/j.1365-313X.2008.03702.x
Patel, J., Zhu, H., Menassa, R., Gyenis, L., Richman, A., and Brandle, J. (2007). Elastin-like polypeptide fusions enhance the accumulation of recombinant proteins in tobacco leaves. Transgenic Res. 16, 239–249. doi: 10.1007/s11248-006-9026-2
Pereira, E. O., Kolotilin, I., Conley, A. J., and Menassa, R. (2014). Production and characterization of in planta transiently produced polygalacturanase from Aspergillus niger and its fusions with hydrophobin or ELP tags. BMC Biotechnol. 14:59. doi: 10.1186/1472-6750-14-59
Pillay, P., Schluter, U., van Wyk, S., Kunert, K. J., and Vorster, B. J. (2014). Proteolysis of recombinant proteins in bioengineered plant cells. Bioengineered 5, 15–20. doi: 10.4161/bioe.25158
Roach, D. R., and Donovan, D. M. (2015). Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 5, e1062590. doi: 10.1080/21597081.2015.1062590
Robinson, A. S., and King, J. (1997). Disulphide-bonded intermediate on the folding and assembly pathway of a non-disulphide bonded protein. Nat. Struct. Biol. 4, 450–455. doi: 10.1038/nsb0697-450
Rodrigue, D. C., Tauxe, R. V., and Rowe, B. (1990). International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105, 21–27. doi: 10.1017/S0950268800047609
Saberianfar, R., Joensuu, J. J., Conley, A. J., and Menassa, R. (2015). Protein body formation in leaves of Nicotiana benthamiana: a concentration-dependent mechanism influenced by the presence of fusion tags. Plant Biotechnol. J. 13, 927–937. doi: 10.1111/pbi.12329
Seckler, R., Fuchs, A., King, J., and Jaenicke, R. (1989). Reconstitution of the thermostable trimeric phage P22 tailspike protein from denatured chains in vitro. J. Biol. Chem. 264, 11750–11753.
Shamji, M. F., Betre, H., Kraus, V. B., Chen, J., Chilkoti, A., Pichika, R., et al. (2007). Development and characterization of a fusion protein between thermally responsive elastin-like polypeptide and interleukin-1 receptor antagonist. Arthritis Rheum. 56, 3650–3661. doi: 10.1002/art.22952
Silhavy, D., Molnar, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., et al. (2002). A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21, 3070–3080. doi: 10.1093/emboj/cdf312
Simpson, D. J., Sacher, J. C., and Szymanski, C. M. (2015). Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 34, 69–77. doi: 10.1016/j.sbi.2015.07.006
Singh, A., Arutyunov, D., Szymanski, C. M., and Evoy, S. (2012). Bacteriophage based probes for pathogen detection. Analyst 137, 3405–3421. doi: 10.1039/c2an35371g
Skurnik, M., and Strauch, E. (2006). Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296, 5–14. doi: 10.1016/j.ijmm.2005.09.002
Speed, M. A., Morshead, T., Wang, D. I., and King, J. (1997). Conformation of P22 tailspike folding and aggregation intermediates probed by monoclonal antibodies. Protein Sci. 6, 99–108. doi: 10.1002/pro.5560060111
Streatfield, S. J. (2007). Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol. J. 5, 2–15. doi: 10.1111/j.1467-7652.2006.00216.x
Su, L. H., Chiu, C. H., Chu, C., and Ou, J. T. (2004). Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin. Infect. Dis. 39, 546–551. doi: 10.1086/422726
Sulakvelidze, A., Alavidze, Z., and Morris, J. G. Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Keywords: bacteriophage tailspike protein, chickens, Nicotiana benthamiana, plant biotechnology, Salmonella, transient transformation
Citation: Miletic S, Simpson DJ, Szymanski CM, Deyholos MK and Menassa R (2016) A Plant-Produced Bacteriophage Tailspike Protein for the Control of Salmonella. Front. Plant Sci. 6:1221. doi: 10.3389/fpls.2015.01221
Received: 15 October 2015; Accepted: 18 December 2015;
Published: 08 January 2016.
Edited by:
Edward Rybicki, University of Cape Town, South AfricaReviewed by:
Biswapriya Biswavas Misra, University of Florida, USAInga Isabel Hitzeroth, University of Cape Town, South Africa
Anatoli Giritch, Nomad Bioscience GmbH, Germany
Copyright © 2016 Miletic, Simpson, Szymanski, Deyholos and Menassa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rima Menassa, cmltYS5tZW5hc3NhQGFnci5nYy5jYQ==
 Sean Miletic
Sean Miletic David J. Simpson
David J. Simpson Christine M. Szymanski
Christine M. Szymanski Michael K. Deyholos
Michael K. Deyholos Rima Menassa
Rima Menassa