- 1Department of Molecular, Cellular and Biomedical Sciences, University of New Hampshire, Durham, NH, USA
- 2Program in Genetics, University of New Hampshire, Durham, NH, USA
Replication protein A (RPA) is a heterotrimeric, single-stranded DNA binding complex required for eukaryotic DNA replication, repair, and recombination. RPA is composed of three subunits, RPA1, RPA2, and RPA3. In contrast to single RPA subunit genes generally found in animals and yeast, plants encode multiple paralogs of RPA subunits, suggesting subfunctionalization. Genetic analysis demonstrates that five Arabidopsis thaliana RPA1 paralogs (RPA1A to RPA1E) have unique and overlapping functions in DNA replication, repair, and meiosis. We hypothesize here that RPA1 subfunctionalities will be reflected in major structural and sequence differences among the paralogs. To address this, we analyzed amino acid and nucleotide sequences of RPA1 paralogs from 25 complete genomes representing a wide spectrum of plants and unicellular green algae. We find here that the plant RPA1 gene family is divided into three general groups termed RPA1A, RPA1B, and RPA1C, which likely arose from two progenitor groups in unicellular green algae. In the family Brassicaceae the RPA1B and RPA1C groups have further expanded to include two unique sub-functional paralogs RPA1D and RPA1E, respectively. In addition, RPA1 groups have unique domains, motifs, cis-elements, gene expression profiles, and pattern of conservation that are consistent with proposed functions in monocot and dicot species, including a novel C-terminal zinc-finger domain found only in plant RPA1C-like sequences. These results allow for improved prediction of RPA1 subunit functions in newly sequenced plant genomes, and potentially provide a unique molecular tool to improve classification of Brassicaceae species.
Introduction
Replication protein A (RPA) is a heterotrimeric, single-stranded DNA (ssDNA) binding protein complex that is highly conserved in eukaryotes (Wold, 1997). It was first identified as an indispensable component for the in vitro replication of simian virus 40 (SV40) DNA (Wobbe et al., 1987). Its primary function is to protect ssDNA from nucleolytic damage and hairpin formation during DNA metabolism. Consistent with its function, RPA plays essential roles in almost all DNA metabolic pathways including DNA replication, transcription, recombination, DNA damage surveillance and recognition, cell-cycle checkpoints, and in all major types of DNA repair including base excision, nucleotide excision, mismatch, and double-strand break repair (Wold, 1997; Iftode et al., 1999; Zou et al., 2006). RPA participates in these diverse DNA metabolic pathways through its ability to interact with DNA and numerous proteins involved in these processes (Iftode et al., 1999; Zou et al., 2006). In both animal and yeast (Saccharomyces cerevisiae) cells, RPA is hyper-phosphorylated upon DNA damage or replication stress by checkpoint kinases including ATM, ATR, and DNA-PK (Binz et al., 2004; Vassin et al., 2004; Liu et al., 2006). The hyper-phosphorylation may switch the function of RPA from DNA replication to DNA repair by inducing changes in the RPA structure and RPA-DNA and RPA–protein interactions, suggesting that animal and yeast cells use a structure-based modulation mechanism of RPA (Binz et al., 2004; Vassin et al., 2004).
Although DNA metabolism and DNA damage responses are highly conserved in eukaryotes, RPA regulation in plants appears different from animals and yeast. For example, RPA hyper-phosphorylation was not detected in rice (Oryza sativa) plants treated with DNA damaging agents (Marwedel et al., 2003). Furthermore, in contrast to the single RPA1, RPA2, and RPA3 subunits of the heterotrimeric RPA complex found in yeast and mammals (except for few mammals with a second RPA2), plants encode multiple RPA1, RPA2, and RPA3 subunits. Rice encodes three RPA1-like genes (RPA1a,b,c), three RPA2-like genes (RPA2-1,-2,-3), and one RPA3-like gene (Ishibashi et al., 2006). Arabidopsis (Arabidopsis thaliana) encodes five RPA1 genes (RPA1-A, B, C, D, and E; Shultz et al., 2007; Aklilu et al., 2014), two RPA2 genes, and two RPA3 genes (Ganpudi and Schroeder, 2011).
Why do plants contain multiple functional RPA1 gene paralogs? Paralogous genes are generated by events such as whole-genome duplication, segmental duplication, and tandem gene duplication. Unlike animals, genome duplication is prominent in plants (Lockton and Gaut, 2005; Cui et al., 2006). For example, Arabidopsis has experienced at least three events of whole genome duplications (Vision et al., 2000; Simillion et al., 2002). There are three possible fates of genes after duplication. These are (i) nonfunctionalization/pseudogenization or loss, where one copy loses its function(s) by degenerative mutations, (ii) subfunctionalization where both copies become partially compromised by mutation accumulation to the point at which their total capacity is reduced to the level of the single-copy ancestral gene, and (iii) neofunctionalization where one copy acquires a novel, beneficial function and become preserved by natural selection, with the other copy retaining the original function (Lynch and Conery, 2000; Zhang, 2003; Moore and Purugganan, 2005; Louis, 2007). The subfunctionalization model also predicts that duplicate genes will share overlapping redundant functions early in the process of functional divergence (Moore and Purugganan, 2005).
Using functional genetic analysis we previously showed that all of the five Arabidopsis RPA1 paralogs are functional (Aklilu et al., 2014). We also reported that these paralogs have undergone subfunctionalization but also share different degrees of overlapping redundant functions. For instance, the rpa1c mutant displays hypersensitivity to DNA double-strand breaks induced by both ionizing radiation and camptothecin, while the rpa1e mutant shows hypersensitivity only to ionizing radiation. Combination of rpa1c and rpa1e results in additive hypersensitivity to a variety of DNA damaging agents. Overall, the results suggest that RPA1C and RPA1E each play unique roles in the repair of DNA damage, with RPA1C playing a leading role in promoting double-strand break repair (Aklilu et al., 2014).
Conversely, the rpa1b rpa1d double mutant has a severely defective growth and developmental phenotype (reduced fitness) under normal growth conditions. However, it displays similar sensitivity as wild-type plants for DNA damaging agents. The growth and developmental abnormalities of the rpa1b rpa1d double mutant are likely a result of defects in DNA replication that generate abnormal cell division suggesting both RPA1B and RPA1D are required for normal DNA replication (Aklilu et al., 2014).
In addition to DNA repair and DNA replication activities, RPA1 proteins play essential roles in progression of meiosis. Of all the five rpa1 single mutants, only the rpa1a single mutant displays an obvious defective meiotic phenotype, observed as lower seed set and reduced crossover formation (Osman et al., 2009; Aklilu et al., 2014). However, the rpa1a rpa1c double mutant is completely sterile, with incomplete synapsis and chromosome fragmentation observed during meiosis (Aklilu et al., 2014). This suggests that both RPA1A and RPA1C play important and perhaps overlapping roles during meiosis.
In order to understand the evolutionary history and identify specific sequence variations responsible for the functional diversification of RPA1 throughout plants, we have analyzed the nucleotide and amino acid sequences, gene conservation, and structural features of all available plant RPA1 paralogs. The sequence and evolutionary analysis together with our new additional genetic analysis reveals that plant RPA1 gene family is divided into three main evolutionary groups (A, B, and C): Group A (including A. thaliana RPA1A) is primarily responsible for meiotic progression. Group B (including A. thaliana RPA1B and RPA1D) is responsible for normal DNA replication. Group C (including A. thaliana RPA1C and RPA1E) is primarily responsible for DNA damage repair. As we show here, these groups have unique coding and regulatory sequences, gene and protein structure, and different level of gene conservation and expression that are consistent with these proposed functions, and highlight how evolution of this gene family occurred into specialized members. These data provide needed sequence structure context for future biochemical studies of RPA function in plants.
Materials and Methods
Plant Materials and Growth
Plant growth conditions and plant lines, including Wild-type (Wt) and all RPA1 single mutant lines used are previously described (Aklilu et al., 2014).
Microscopy
Meiotic chromosome spreads were prepared as described (Armstrong et al., 2001). Slides were mounted with DAPI (2.5 mg/ml) in Vectashield. Chromosomes were visualized with a confocal microscope (Zeiss LSM 510) using the halogen light. Images were then captured using an externally mounted digital camera (Olympus CKX41) and processed with a SPOT microscope imaging software.
Sequences Sources
Promoter, coding, and amino acid sequences were obtained from the National Center for Biotechnology Information (NCBI) and The Arabidopsis Information Resources (TAIR) database. Orthologous RPA1 sequences were identified by employing NCBI BLAST searches. Full length Arabidopsis RPA1 amino acid sequences were used for the BLAST search against the genome of each organism. In addition, sequences identified at each step are used to refine the search. Orthology was confirmed by reverse BLAST. The lowest e-value and maximum percent identity BLAST hit was chosen as the putative ortholog of the respective Arabidopsis RPA1 protein.
Phylogenetic Analysis
Phylogenetic analysis of the RPA1 protein sequences was performed with the MEGA5 software package (Tamura et al., 2011). Amino acid sequences were aligned using ClustalW (default parameters) with in the MEGA5 software package. Trees were constructed using maximum likelihood methods with Jones-Taylor-Thornton (JTT) amino acid substitution model and 1000 bootstrap replicates. Except from bootstrapping and choice of model, all other parameters were left at default settings. Additional neighbor joining (NJ) trees were constructed with the same amino acid substitution model, bootstrap replicates, and NJ tree construction default parameters in MEGA5 to confirm the results of the ML analysis.
Codon Bias/Usage
Frequency of optimal codons (FOP) was calculated based on optimal codons identified for Arabidopsis (Wright et al., 2004) and rice (Liu et al., 2004).
Gene Structure and Protein Domains
Introns in RPA1 genes were identified using the NCBI gene database. RPA1 protein domains and subdomains were identified using the NCBI Conserved Domain Database [CDD] (Marchler-Bauer et al., 2011).
Sequence Distance and Natural Selection
Analysis of synonymous (dS) and nonsysnonymous (dN) substitution rate were performed with the MEGA5 software package using the Nei-Gojobori substitution model (Tamura et al., 2011). Natural selection (ω) was calculated by dividing dN by dS (dN/dS).
Gene Expression
Level and pattern of RPA1 genes expression at different developmental stages and tissues were retrieved from genevestigator (https://www.genevestigator.com) and Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Genevestigator is an online visualization tool that summarizes results from thousands of high quality transcriptomic experiments often done by cDNA microarrays (Hruz et al., 2008). Arabidopsis eFP browser is also a data visualization tool for exploring degree and location of gene expression (Winter et al., 2007).
Analysis of cis-Elements in Arabidopsis RPA1 Promoter Regions
Promoter elements for all the sequences were analyzed using the PLACE (Plant Cis-acting Regulatory DNA Elements) database (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999).
Arabidopsis RPA1 TAIR Gene Codes
AtRPA1A, AT2G06510; AtRPA1B, AT5G08020; AtRPA1C, AT5G45400; AtRPA1D, AT5G61000; AtRPA1E, AT4G19130.
Results and Discussion
Arabidopsis RPA1A Plays a Leading Role in Meiosis
We previously described the role of RPA1A and RPA1C during meiotic progression employing homozygous mutants rpa1a, rpa1c, and the double mutant rpa1a rpa1c (Aklilu et al., 2014). These results suggest a genetically redundant role for both RPA1A and RPA1C during early stages of meiosis (crossing over and repair). A previous study suggests RPA1A plays a unique role during later stages of meiosis (second-end capture; Osman et al., 2009), but there is currently no evidence to support a role for RPA1C at later stages of meiosis. To determine which if any of these two subunits plays a more predominant role during early (repair) stages of meiosis (and thereby providing additional clarity into the functional classification of these genes), we generated two additional heterozygous mutant combinations, rpa1a (+/−) rpa1c (−/−) and conversely rpa1a (−/−) rpa1c (+/−). By comparing these two combinations, we sought to determine the relative dominance (“gene dosage”) of each in promoting early meiotic repair. As shown in Figures 1A,B, phenotypic analysis of these mutant combinations show that while rpa1a (+/−) rpa1c (−/−) is fully fertile, the rpa1a (−/−) rpa1c (+/−), displays reduced fertility (~92% reduction in fertility) vs. the rpa1a (−/−) single mutant.
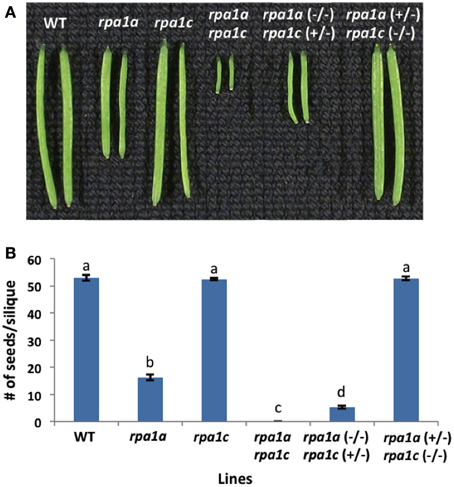
Figure 1. Meiotic defective A. thaliana RPA1 mutant lines. (A) Siliques harvested from ~7 weeks old wild-type (WT) and RPA1 mutant plants. (B) Number of seeds per silique. The mutant plants have the following percentage of fertility reduction: rpa1a: ~70%; rpa1a (−/−) rpa1c (+/−): ~92%; rpa1a rpa1c: 100%. rpa1a (+/−) rpa1c (−/−) has similar fertility level as WT. Data are mean ± SE (n > 10). To analyze statistical difference F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
To evaluate meiotic integrity at the chromosomal level, we prepared chromosomal spreads of pollen-mother cells from WT and mutant lines. In comparison to WT, rpa1a, rpa1c, and rpa1a rpa1c, the rpa1a (+/−) rpa1c (−/−) combination displays normal meiotic progression, while the rpa1a (−/−) rpa1c (+/−) displayed both univalents and fragmented chromosomes during metaphase I (Supplementary Figure S1). In addition, rpa1a (−/−) rpa1c (+/−) combination showed highly fragmented chromosomes and unequal segregation during the subsequent stages anaphase I and II (Supplementary Figure S1). These results show that the rpa1c mutation in the rpa1a background is semi-dominant during meiotic repair, and suggest that RPA1C is either less effective at meiotic repair than is RPA1A, or that RPA1C is only required for repair of a small subset of meiotic double-strand breaks. We therefore propose that RPA1A plays the leading role in both early and late stages of meiotic crossing over, but that RPA1C can either partially or completely fulfill the role of RPA1A during early meiotic stages (DNA repair and initiation of recombination) in its absence. This is consistent with rice rpa1a single mutants that show defective DNA repair during meiosis (Chang et al., 2009). However, it would be useful to determine combined effects of RPA1A and RPA1C deficiency in rice for a more direct comparison of rice and Arabidopsis (monocot vs. dicot) meiotic progression, since rice rpa1c knock-down lines also display meiotic deficiencies (Li et al., 2013).
The Plant RPA1 Gene Family is Composed of Three Distinct Groups
Animal and yeast RPA1 is generally encoded by a single gene (Wold, 1997; Iftode et al., 1999). Genetic studies in rice and Arabidopsis suggest two general groups of RPA1 encoding genes, one responsible for DNA replication and one for DNA repair (Ishibashi et al., 2006; Aklilu et al., 2014). In order to determine conservation of these groups throughout the plant kingdom, we conducted BLAST searches employing known RPA1 genes in Arabidopsis and constructed maximum likelihood phylogenetic trees from identified sequences to determine phylogenetic relationships. Full-length Arabidopsis RPA1 amino acid sequences were employed for BLAST searches against the complete genome of individual plant and algae species found in the NCBI database. In addition sequences identified at each step were used to detect additional homologs. Orthology was confirmed by reverse BLAST. The lowest e-value and maximum percent identity sequence identified was chosen as the putative ortholog of the respective Arabidopsis RPA1 protein (Supplementary Table S1). RPA1 has four conserved domains; DBD-F, DBD-A, DBD-B, and DBD-C. Some of the putative RPA1 orthologs have either a DBD-F or a DBD-C deletion. Human RPA1 containing a DBD-F deletion retains replication activity, while a DBD-C deletion is non-functional (Gomes and Wold, 1996; Haring et al., 2008). To be as inclusive as possible while eliminating potential psuedogenes or unrelated groups, we included those putative sequences that do not contain a DBD-F domain sequence in our analysis (Figures 2, 3).
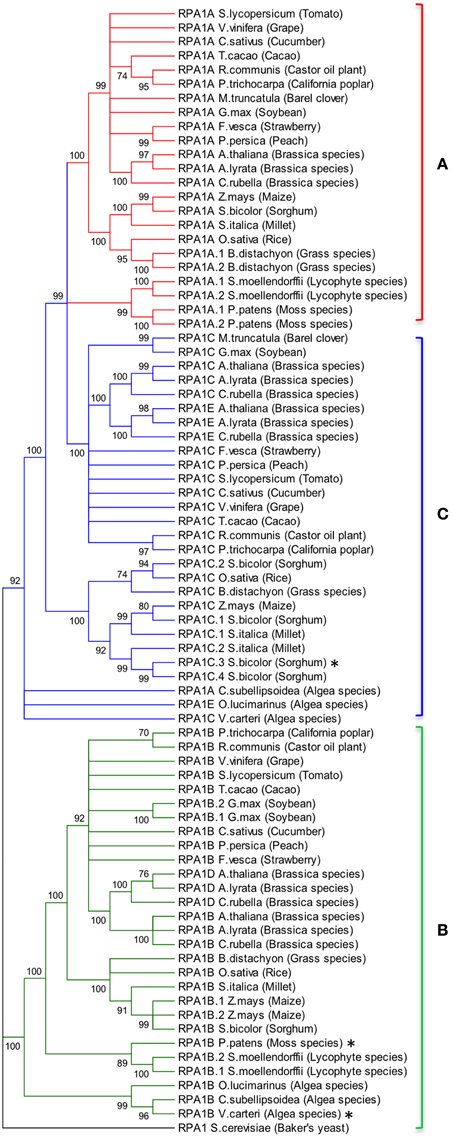
Figure 2. Evolutionary relationships of RPA1 proteins. (A) RPA1A group, (B) RPA1B group, (C) RPA1C group. The evolutionary history was inferred using the Maximum-Likelihood method performed with MEGA5.2 software package. Amino acid sequences were aligned using ClustalW and used to produce phylogenetic trees using the Jones-Taylor-Thornton (JTT) amino acid substitution model. Numbers next to the branches are bootstrap values (1000 replicates). Branches that show less than 70% bootstrap support were collapsed. Some RPA1 sequences denoted by asterisk (*) [sorghum RPA1C-3, P. patens RPA1B, and V. cateri RPA1B] contain DBD-F deletion. Yeast RPA1 was used as an outgroup to root the tree. Except for bootstrapping and choice of model, all other parameters were left at default settings.
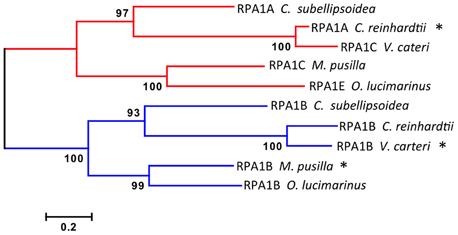
Figure 3. Evolutionary relationships of unicellular green algae RPA1 proteins. The evolutionary history was inferred using the Maximum Likelihood method performed with MEGA5.2 software package. Amino acid sequences were aligned using ClustalW and used to produce phylogenetic trees using the Jones-Taylor-Thornton (JTT) amino acid substitution model. Some RPA1 sequences denoted by asterisk (*) [C. reinhardtii RPA1A, M. pussila RPA1B, and V. cateri RPA1B] contain DBD-F deletion. Numbers next to the branches are bootstrap values (1000 replicates). Scale represents number of amino acid substitutions per site. Except for bootstrapping and choice of model, all other parameters were left at default settings.
To address phylogenetic relationships of all identified sequences, we employed maximum likelihood and neighbor-joining analyses as described in the Materials and Methods section. Shown in Figure 2 is our resulting maximum-likelihood phylogenetic analysis (and is very similar employing neighbor-joining methods) of RPA1 from all identified plant-related sequences. This analysis shows the plant RPA1 family is generally divided into three distinct groups composed of RPA1A (A group), RPA1B (B group), and RPA1C (C group) (Figure 2; Supplementary Figure S2; Supplementary Table S1). RPA1D and RPA1E, which are found only in the Brassicaceae family, are members of the B and C group, respectively. Within each major clade (for example, RPA1B) each taxa are arranged in a sub-clade of dicot, monocot, primitive plant (Physicomitrella patens and Selaginella mollendorffii), and unicellular green algae (V. carteri, O. lucinarinus and C. subellipsoida). The common ancestor of RPA1B of dicots and monocots appears diverged from an RPA1B-like progenitor of primitive plants. This ancestral RPA1B in primitive plants appears to have diverged from an RPA1B-like progenitor in unicellular green algae (Figure 2, Supplementary Figure S2). The evolutionary history for RPA1A and RPA1C appears different from RPA1B. The common ancestor of RPA1A of dicots and monocots appears diverged from RPA1A of primitive plants and RPA1C of dicots. In turn, RPA1A of primitive plants and RPA1C of dicots likely diverged from the common ancestor leading to RPA1C of monocots which itself appears to have diverged from the RPA1A/C progenitor in unicellular green algae (Figure 2, Supplementary Figure S2).
Interestingly, some plants contain additional RPA1 group members. Soybean (Glycin max) and maize (Zea mays) have two RPA1B-like sequences; sorghum (Sorghum bicolor) and millet (Setaria italica) have four and two RPA1C-like sequences, respectively. Members of the Brassicaceae family (A. thaliana, Arabidopsis lyrata, and Capsella rubella) have two additional RPA1 types, RPA1D and RPA1E. In terms of both sequence and functional similarity RPA1D and RPA1E are close paralogs of RPA1B and RPA1C, respectively (Figure 2; Aklilu et al., 2014), suggesting these are duplications of RPA1B and RPA1C that occurred only in the Brassicaceae family. Unlike other plants, barrel clover (Medicago truncatula) has only two types of RPA1 (RPA1A and RPA1C) and it does not appear to have an obvious RPA1B. Given the importance of RPA1B in DNA replication (Aklilu et al., 2014) and the fact that other member of the Fabaceae family (Soybean) has RPA1B, it is possible that lack of this sequence is due to sequencing/annotation errors. Alternatively, barrel clover may actually lack RPA1B since the RPA1A-like sequence from barrel clover contains a subdomain that is mostly found only in the N-terminal domain of RPA1B group (see Discussion of subdomains below). If so, this might suggest that Mt RPA1A evolved to perform dual functions in both meiosis and replication.
From our analysis we find that primitive plant genomes generally contain two types of RPA1 sequences, RPA1A/C-like and RPA1B-like. However, only P. patens and S. mollendorffii from this group have a completed genome sequence (Goodstein et al., 2012). Additional genome sequencing of more species would be needed to determine if RPA1C is present in primitive plants.
Interestingly the RPA1A/C-like progenitor sequences of unicellular green algae are more similar to each other than to their orthologous RPA1A/C counterparts in dicots, monocots, and primitive plants (Figure 2, Supplementary Figure S2). This suggests that the plant RPA1A and RPA1C groups have evolved significantly since divergence from its counterpart in unicellular green algae, or alternatively the algae ancestor has evolved more rapidly since splitting from higher plant lineages. To determine the early evolutionary relationship of RPA1 proteins, we searched for RPA1 orthologs from two additional algal completed genomes (employing the method described above) to construct a maximum likelihood phylogenetic tree focused on algal RPA1 sequences. Interestingly, all five unicellular green algae that represent five different genera have two types of RPA1 sequences, an RPA1B-like group common to all the algae representatives in this study, and an RPA1A/C-like group (Figure 3; Supplementary Table S1; Supplementary Figure S2). This suggests that the first duplication event of RPA1 that led to diversification in plants likely occurred during the evolution of green algae. This is further supported by the fact that red algae generally contain only a single RPA1-like sequence (Supplementary Table S2). We propose a model (described below) for the expansion of RPA1 from green algae to higher plants based upon the phylogenetic relationships and sequence groups described above, in combination with RPA1 domain structure described in the next five sections below.
Structure and Domains of Plant RPA1 Proteins
Eukaryotic RPA1 contains four highly-conserved domains termed the N-terminal domain or DNA Binding Domain F (DBD-F), two structurally similar central DNA Binding Domains (DBD-A and DBD-B), and a C-terminal DNA Binding Domain C (DBD-C) (Wold, 1997; Takashi et al., 2009; Figure 4). Studies in yeasts and animals suggest that the N-terminal domain (DBD-F) is primarily involved in protein-protein interactions, the middle domains (DBD-A and DBD-B) function in ssDNA binding, while the C-terminal domain (DBD-C) is involved in DNA damage recognition during nucleotide excision repair and subunit interaction (Wold, 1997; Lao et al., 2000; Zou et al., 2006).
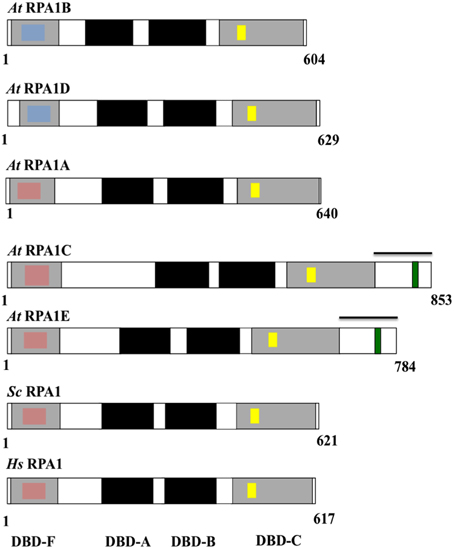
Figure 4. Schematic diagram of the structure and functional domains of RPA1 proteins. At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens. Blue inset boxes represent Generic Binding Surface I. Pink inset boxes represent Binding Surface I (Basic Cleft). Yellow inset boxes represent C5 (in At RPA1B, D), C6 (in At RPA1A, C, E), and C4 (in Sc RPA1and Hs RPA1)—type zinc-finger motifs. Green inset boxes represent CCHC-type zinc-finger motif. Bars above At RPA1C and At RPA1E indicate C-terminal extension region.
To characterize conservation of these domains throughout plant RPA1 sequences as well as relate known functional characteristics to domain structure, we compared all A. thaliana RPA1 sequence domain structure to human and yeast RPA1as summarized in Figure 4. We employed the NCBI Conserved Domain Database [CDD] (Marchler-Bauer et al., 2011) to identify RPA1 structure, domains, and sub-domains. All sequences contain these four conserved domains (DBD-F, DBD-A, DBD-B, DBD-C). However, we find that each group contains unique subdomains or motifs that may contribute to functional differences. In the following sections we describe these subdomains and motifs in detail. With very few exceptions (Sorghum RPA1C and P. patens RPA1B contain DBD-F deletion), the general structure, domains, subdomains, and motifs of the three groups of A. thaliana RPA1 proteins are conserved throughout all plants examined here (Supplementary Figures S3–S6).
RPA1B is Unique Among Other RPA1 Sequences Through Loss of the Binding Surface I Domain
DBD-F of human RPA1 contains a subdomain called “Binding Surface I” (BS-I) or basic cleft (Figure 4, pink inset box) (Jacobs et al., 1999; Marchler-Bauer et al., 2011). Multiple studies demonstrate that the BS-I subdomain in human RPA1 and a similar region in yeast RPA1 is required for DNA repair activity but not required to support DNA replication (Longhese et al., 1994; Umezu et al., 1998; Bochkareva et al., 2005; Haring et al., 2008; Xu et al., 2008). Five highly conserved amino acid residues constitute the protein interaction sites of BS-I; position 1 is generally a polar residue (N/Q/S/T), position 2, 3, and 5 are basic (R/K), and position 4 is hydrophobic (L/V/I) (Supplementary Figure S3A; Supplementary Table S3). In all plants analyzed here, except Sorghum RPA1C-3, sequences phylogenetically predicted to fall into the RPA1A and RPA1C groups generally display these same conserved features as found in animal and fungal RPA1 (Figure 4; Supplementary Figures S3B,C). One notable exception among plant sequences is at position 1 where the conserved polar residue (N/Q/S/T) is replaced by a negatively charged residue (D/E). Interestingly, the predicted plant RPA1B group members do not display the conserved features of BS-I in most or all five positions, being replaced by chemically non-similar residues (Supplementary Figure S3D). For instance, RPA1B sequences generally contain a negatively charged amino acid (D) at position 1 instead of a conserved polar residue. In addition, position 3 in most RPA1B sequences contains a polar residue, while position 5 contains a negatively charged residue (E/D), instead of the conserved basic residues (R/K). Similarly, the hydrophobic residue found at position 4 (L/V/I) is in most cases replaced by a polar residue (T/S). These data suggest that plant RPA1B evolved into a more specialized member of the RPA1 family through loss of this domain (BS-I) to interact primarily with DNA replication-oriented pathways consistent with proposed functions of this group (Aklilu et al., 2014). In unicellular green algae only two of the five RPA1A/C progenitor sequences (O. lucimarinus RPA1E and M. pussila RPA1C) contain BS-I. The other sequences have either an N-terminal (DBD-F) deletion (C. reinhardtii RPA1C) and thus do not contain BS-I, or have N-terminal domain but do not contain BS-I (V. cateri RPA1C and C. subellipsoida RPA1A).
RPA1B Sequences Contain a Conserved N-Terminal (DBD-F) Nucleic Acid Binding Surface Termed Generic Binding Surface I
OB-fold domains are known for their ssDNA-binding activity. However, no clear conservation of amino acid residues that directly interact with nucleic acids has been identified among the domains (Theobald et al., 2003). For example, except for two aromatic residues that stack with DNA bases, there is no conservation among the rest of the 6–9 amino acids that are predicted to contact ssDNA and are found in the two main ssDNA-binding OB fold domains (DBD-A and DBD-B) of human RPA1 (Bochkarev et al., 1997). Nevertheless, alignments of multiple OB fold domains reveal a pattern of hydrophobic residues, mostly flanked by polar or charged residues, in alternating amino acid positions that are conserved for short stretches of sequence around the site of ssDNA-binding (Theobald et al., 2003). These patterns are generally referred to as Generic Binding Surface I (GBS-I), and are found in DBD-A, DBD-B, and DBD-C of eukaryotic RPA1.
Analysis of plant RPA1 sequences employing NCBI CDD reveals that an additional GBS-I domain is present in DBD-F of RPA1B group sequences [Figure 4 (blue inset box); Supplementary Figure S4A]. This domain displays the classic pattern of hydrophobic residues flanked by polar or charged residues found in all other GBS-I domains, although the conservation of amino acid identity is unique among this particular domain in plant DBD-F. Interestingly, some plant RPA1A- and RPA1C-like sequences also contain this GBS-I in DBD-F (Supplementary Figure S4C) and these special cases are discussed below. In unicellular green algae only three of the five RPA1B-like sequences (RPA1B of O. lucimarinus, C. reinhardtii, and C. subellipsoida) contain GBS-I. The other two sequences (RPA1B of M. pussila and V. cateri) do not contain GBS-I as they have a truncated N-terminal domain (DBD-F). Interestingly, however, RPA1A of C. subellipsoida does contain a GBS-I domain in DBD-F. In general, this domain structure is unique to plant and green algae, and not found in animals and fungi.
Human RPA binds to ssDNA in two modes. The first mode has an occluded size of 8–10 nucleotides (Blackwell and Borowiec, 1994) and accomplished by the two major ssDNA-binding domains, DBD-A and DBD-B (Bochkareva et al., 2001; Hass et al., 2012). The second binding mode has an occluded binding size of ~30 nucleotides (Kim et al., 1992, 1994) and involves DBD-A, -B, and -C of RPA1 and DBD-D of RPA2 (Bastin-Shanower and Brill, 2001; Cai et al., 2007). DBD-F has a weak ssDNA-binding affinity (Daughdrill et al., 2001; Bochkareva et al., 2005) and is not involved in either binding modes (Cai et al., 2007). The ability of RPA binding to short nucleotides (8–10 nt) is required for DNA repair function, but not DNA replication (Hass et al., 2012). Conversely, it is proposed that the ability of RPA to bind to long stretches of ssDNA (~30 nt) is required for its function in DNA replication rather than in DNA repair, as there is extensive DNA unwinding and exposure of long ssDNA intermediates during DNA replication vs. DNA repair (Hass et al., 2012). Accordingly, our result suggests that in plants, DBD-F of the RPA1B group may participate in ssDNA-binding (as it contains GBS-I) so that together with other DBDs (DBD-A, DBD-B, DBD-C, and DBD-D in RPA2) it may result in an occluded size of more than 30 nucleotides.
The presence of GBS I in DBD-F of barrel clover and Brachypodium distachyon RPA1A-like sequences, and millet and sorghum RPA1C-like sequences (Supplementary Figure S4C) suggests these sequences could participate in DNA replication activities. For example, since we were unable to identify a predicted RPA1B sequence in barrel clover, it is possible the RPA1A-like sequence might have gained the GBS I subdomain to function in place of RPA1B. B. distachyon has two RPA1A (RPA1A-1 and RPA1A-2), perhaps allowing the paralogs to accumulate small gradual mutations and thereby gain new domains and functions as purifying selection is weaker on duplicate genes (Castillo-Davis et al., 2004). This same explanation can be argued for RPA1C-2 and RPA1C-4 of millet and sorghum, respectively.
Plant RPA1 Sequences Contain Unique Zinc-Finger Motifs Within the DBD-C C-Terminal Domain
RPA1 sequences from yeasts and animals contain a C4-type (C-X2-C-X13-C-X2-C) zinc-finger motif (ZFM) within the DBD-C domain (Wold, 1997). This motif is required for DNA replication (Lin et al., 1996, 1998; Walther et al., 1999), DNA damage recognition (Lao et al., 2000), and for proper structural formation of RPA complexes (Lin et al., 1996). Employing NCBI CDD protein domain analysis we find that indeed all plant RPA1 sequences also contain ZFM sequences. However, RPA1A and RPA1C contain a C6-type (C-X3-C-X8-C-X13-C-X2-C-X6-C) ZFM, while RPA1B contains a C5-type (C-X2-C-X13-C-X2-C-X6-C) ZFM. Conservation of these domains among all plant species analyzed is high, with only a few exceptions seen in sorghum RPA1C-3 and RPAC-4, rice RPA1C, and in the Lycophyte S. moellendorffii RPA1B-like sequences (Supplementary Figure S5). RPA1A/C-like progenitor sequences of unicellular green algae contain a C-4 type ZFM (Supplementary Figure S5E). However, in M. pussila RPA1C and O. lucimarinus RPA1E, the fourth residue of the conserved motif is histidine (H) instead of cysteine (C). Interestingly, RPA1B-like sequences of unicellular green algae do not contain a conserved C4-type ZFM in DBD-C (Supplementary Figure S5F).
ZFMs are known for their role in protein-DNA and protein-protein interactions (Krishna et al., 2003; Gamsjaeger et al., 2007). The human RPA1 ZFM interacts with both normal and damaged ssDNA with low and high affinity, respectively, thereby contributing to optimal ssDNA-binding activity (Dong et al., 1999; Walther et al., 1999; Lao et al., 2000). Human RPA1 that contains a mutation in either the two cysteine amino acids of the ZFM, or a complete deletion of the motif, retains ssDNA binding activity, heterotrimeric complex formation, and DNA repair promoting activities, but does not support DNA replication (Lin et al., 1996, 1998; Walther et al., 1999). The latter replication defect may be due to a failure to load polymerase δ at replication forks during the elongation step of DNA replication (Lin et al., 1998), or due to improper heterotrimeric complex formation, as the ZFM generally affects the structure of the RPA complex (Dong et al., 1999; Walther et al., 1999; Bochkareva et al., 2000). In plants, it is possible that the different types of ZFMs in the RPA1A/1C and RPA1B group may contribute to the functional specificity of these proteins in DNA metabolism through direct and specific protein-DNA and/or protein-protein interactions. The ZFMs may also contribute to the formation of different types of RPA heterotrimeric complexes. In support of this, Arabidopsis RPA1A preferentially forms a complex with RPA2B, and RPA1B preferentially forms a complex with RPA2A (Eschbach and Kobbe, 2014). Also in rice, each of the three RPA1 subunits preferentially forms a complex with a specific RPA2 subunit (Ishibashi et al., 2006).
RPA1C Group has a C-Terminal Extension Region that Contains a Glycine/Serine-Rich Domain Interspersed by a CCHC-Type ZFM
In contrast to other RPA1 sequences examined here, plant RPA1C-like sequences (including RPA1E in Brassicaceae) contain a unique C-terminal extension region that contains an average amino acid sequence length of ~176 in RPA1C and ~119 in RPA1E-like sequences (Figure 4; Supplementary Table S4). CLUSTAL alignments of these C-terminal extensions (beginning at the end of the putative DBD-C domain in each case to the end of the predicted sequence), NCBI CDD domain searches, and manual comparisons were employed to identify common sequence domains or regions. Common among all of these sequences is the presence of at least one zinc-knuckle CCHC-type (CX2CX4HX4C) ZFM. All Brassicaceae members display one ZFM in RPA1C, and one ZFM in RPA1E C-terminal extensions, are roughly at the same position within the alignments (both C-terminal only, and the full-length sequence alignments), and share high sequence identity. Interestingly, in plants that do not have the RPA1C/RPA1E paradigm (non-Brassicaceae), the RPA1C sequence(s) contain multiple ZFM that fall into two clusters (termed here C1 and C2) each with unique sequence identity (Supplementary Figure S6). In most cases (tomato, cucumber, and maize for example) there are two unique ZFM separated by ~30 amino acids (C1 followed by C2). In other cases, such as rice for example, there is a C1 followed by multiple (three in this case) C2s. The fact that there are two unique cluster types of the ZFM suggests either (1) that these motifs arose independently through acquisition of ZFM-like sequences from two independent sources (genes) in the ancestral gene, or (2) were duplicated from the same ancestral gene but evolved early into two unique ZFM sequences. In the case of Brassicaceae, a likely scenario is that the ancestral RPA1C that gave rise to current RPA1C and RPA1E contained both C1 and C2, but in each case lost C2. If so, this would suggest C2 is only necessary in the presence of C1 to carry out multiple DNA repair-related functions. In unicellular green algae only one of the RPA1A/C progenitor sequences (M. pussila RPA1C) contains CCHC-type ZFM in the C-terminal extension region. The other sequences either do not contain a C-terminal extension region (O. lucimarinus RPA1E) or contain a C-terminal extension region that does not have CCHC-ZFM (V. cateri RPA1C).
This analysis also revealed that the C-terminal extension regions in RPA1C sequences are glycine- and serine-rich (Supplementary Figures S6A,B). Glycine-rich regions are found in RNA-Binding Proteins (RBPs) and they may be enriched by additional polar (hydrophilic) residues, such as serine, arginine, asparagine, glutamine, and tyrosine. However, the function of these additional polar residues is poorly understood (Rogelj et al., 2011). Glycine-rich domains interspersed by CCHC-type ZFMs are found in RNA-binding plant proteins involved in post-transcriptional regulation of gene expression under various stress conditions (Karlson et al., 2002; Karlson and Imai, 2003; Kim et al., 2005, 2007; Kim and Kang, 2006). Therefore, it is possible that RPA1C may also bind to RNA and play a role in post-transcriptional regulation. Furthermore, CCHC-type ZFMs are predicted to bind to both normal GT-rich ssDNA (Rajavashisth et al., 1989; Tzfati et al., 1995; Kim et al., 2005) and damaged ssDNA [e.g., Arabidopsis DDB2 (Ly et al., 2013), yeast RAD18 (Jones et al., 1988), and human PARP-1 (Langelier et al., 2011)]. This suggests that besides binding to RNA and ssDNA and thereby playing regulatory role, the CCHC-type zinc-finger motif may also play a role in DNA damage recognition, as suggested from genetic analysis (Aklilu et al., 2014).
In summary, the protein structure analysis result is consistent with genetic and phylogenetic data that plant RPA1 proteins fall into three distinct groups with unique functions (RPA1A, RPA1B, and RPA1C). Based on these data we propose that duplication of RPA1 in unicellular green algae led to two main progenitor groups in primitive plants, and later diverged into three groups in higher plants with specialized functions (Figure 5).
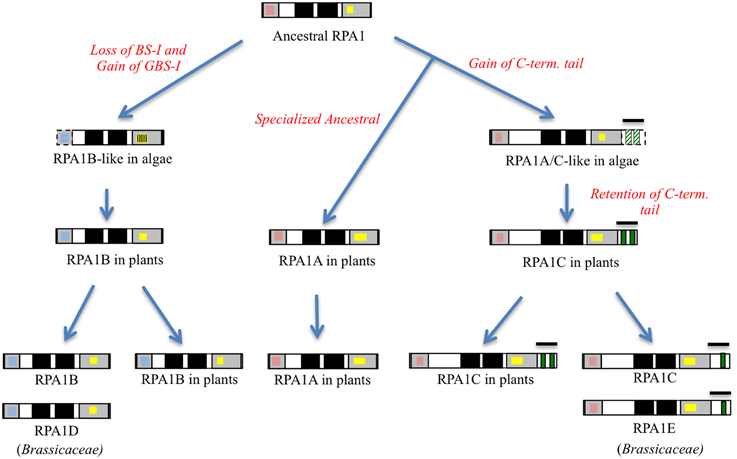
Figure 5. Proposed model of RPA1 evolution in plants and algae. Gray boxes from right to left are DBD-F and DBD-C, respectively, dark boxes from right to left are DBD-A and DBD- B, respectively, red boxes are Binding Surface I (BS-I), blue boxes are Generic Binding Surface I (GBS-I), yellow boxes are C4/C5/C6—type zinc-finger motifs (ZFM), green boxes are CCHC-type ZFM. Line bars indicate C-terminal extension (tail) region, broken lines and strips indicate poorly conserved domains.
The RPA1B Group has Higher Levels of Gene Expression, Intron Frequency, and Optimal Codon Usage
Depending on their functional specificity and biochemical role, genes in general and duplicated genes in particular can have different temporal, spatial, and levels of gene expression (Chiapello et al., 1998; Casneuf et al., 2006; Hyun et al., 2014). As discussed above, genetic analysis of rpa1 mutants suggests specialized functions for individual RPA1 subunits. Based on this we predict unique gene expression patterns of RPA1 genes that are consistent with their proposed function. For example, we might expect to see higher basal expression of individual RPA1 members if they participate in “housekeeping” type functions, such as normal DNA replication activity. To this end, we examined expression patterns and levels of the Arabidopsis, soybean, rice, and P. patens RPA1 paralogs (Figure 6) from Genevestigator, an online gene expression visualization tool that summarizes results from thousands of high quality transcriptomic experiments, typically employing cDNA microarrays (Hruz et al., 2008). In general the RPA1B group displays higher basal expression levels in most developmental stages in comparison with the RPA1A and RPA1C groups of all four included plant species, in both shoots and roots (Figures 6A,B). In contrast, members of the RPA1C group and to a lesser extent the RPA1A group are induced by DNA damage (Culligan et al., 2006). The higher overall level of gene expression of the RPA1B group suggests a requirement throughout the developmental growth of the plant, and is consistent with the proposed function of RPA1B in normal DNA replication functions (Aklilu et al., 2014).

Figure 6. Level and pattern of RPA1 expression at different developmental stages and root zones. (A) A. thaliana; (1) Germinated seed, (2) Seedling, (3) Young rosette leaf, (4) Developing rosette leaf, (5) Bolting, (6) Young flower, (7) Developed flower, (8) Flowers and siliques, (9) Mature siliques, (10) Senescence. (B) AtRPA1 expression in different root zones of 7-day-old Arabidopsis seedlings. (C) Soybean; (1) Germination, (2) Main shoot growth, (3) Flowering, (4) Fruit formation, (5) Bean development. (D) Rice; (1) Germination, (2) Seedling, (3) Tillering stage, (4) Stem elongation stage, (5) Booting stage, (6) Heading stage, (7) Flowering stage, (8) Milk stage, (9) Dough stage. (E) Psychomitrela patens; (1) Germination [protenema development], (2) Gametophore growth, (3) Gametangia development. Data were collected from genevestigator (A,C–E; Hruz et al., 2008) and Arabidopsis eFP Browser (B; Winter et al., 2007). Error bars indicate standard error.
In A. thaliana, RPA1B and RPA1D display ~10-fold more introns vs. RPA1A, RPA1C, and RPA1E (Supplementary Table S1). To determine if this paradigm is consistent throughout plants, we established intron frequencies in all three major groups of RPA1 paralogs (A, B, and C) as predicted by our phylogenetic analysis above, employing NCBI sequence databases. As shown in Figure 7, RPA1B-like genes from 20 plant species contain ~six-fold more introns on average than either group RPA1A, or group RPA1C. However, in unicellular green algae, there is no clear pattern of intron frequency between the B-like group and A/C-like group (Supplementary Table S1). In many eukaryotes, intron frequency is generally proportional to gene expression levels (Callis et al., 1987; Brinster et al., 1988; Duncker et al., 1997; Juneau et al., 2006; Shabalina et al., 2010), and mRNA stability (Le Hir et al., 2003; Niu and Yang, 2011). For example, highly expressed genes in both Arabidopsis and rice show increased intron frequency (number of introns per kilobase of coding sequence) vs. lower expressed genes (Ren et al., 2006). Therefore, these data are consistent with higher gene expression levels of RPA1B group members vs. RPA1A or RPA1C members.
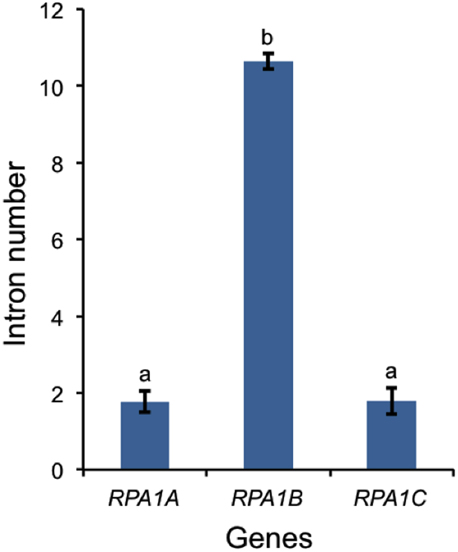
Figure 7. Number of introns in plant RPA1 genes. Twenty plants (Supplementary Table S1) are included in the analysis. Data are mean ± SE. To analyze statistical difference F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
Codon preference (optimal codons) is determined by tRNA abundance and gene expression level and occurs with unequal frequencies (Ikemura, 1985; Duret and Mouchiroud, 1999). Highly expressed genes in both prokaryotes and eukaryotes have increased frequency of codons that match abundant tRNAs (Ikemura, 1985; Bulmer, 1988; Wright et al., 2004). Codon bias may reflect a selective pressure to enhance translational efficiency for highly expressed genes (Bulmer, 1988; Marais and Duret, 2001; Wright et al., 2004; Sablok et al., 2013). Since the RPA1B group displays higher overall gene expression levels vs. RPA1A and RPA1C, we further hypothesized that RPA1 genes may reflect this through a bias in optimal codon usage. In order to test this we calculated the frequency of optimal codons (FOP) for Arabidopsis and rice RPA1 genes, since these species have the most complete data set of developmental stages from Genevestigator (Hruz et al., 2008) and optimal codons for highly expressed genes have been identified in these organisms (Liu et al., 2004; Wright et al., 2004). FOP is calculated as the number of occurrences of optimal codons divided by the total number of codons (Ikemura, 1985). As shown in Figure 8, we find that the RPA1B group has the highest number of optimal codons (FOP = 0.54) followed by RPA1A (FOP = 0.45) and RPA1C (FOP = 0.41). FOP-values for the Arabidopsis RPA1D (0.45) and RPA1E (0.42) are not included in the F-test as they are only found in Brassicacea. This suggests that RPA1B group is likely under the control of translational selection due to a demand for its higher translational protein product during DNA replication.
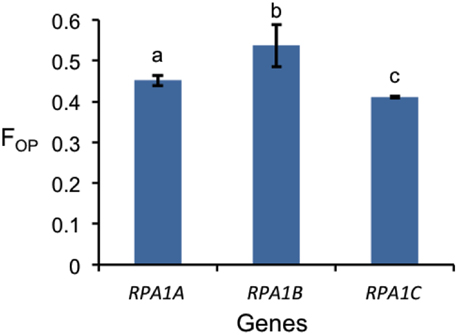
Figure 8. Frequency of optimal codon (FOP) values for RPA1 genes of Arabidopsis and rice. Data are mean ± SE. To analyze statistical difference F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
RPA1A and RPA1B are More Conserved than RPA1C
Highly expressed genes are usually under a high degree of selective constraint and thus display a higher degree of conservation (Pál et al., 2001; Drummond et al., 2005). As described above, plant RPA1 groups have unique expression patterns (Figure 6). Accordingly, we hypothesize unique sequence conservation of RPA1 subunits that is consistent with their expression level. To test this, we analyzed and compared the type and degree of natural selection applied on each RPA1 group. Natural selection is measured by the ratio of non-synonymous (dN) and synonymous (dS) nucleotide substitution rates (ω or dN/dS) and its value ranges from zero (0) to infinity (∞). While a value of < 1 is a sign of purifying selection (where sequence conservation is preferred), a value of >1 is a sign of positive selection (where change in sequence is preferred). We carried out the analysis by dividing plants into two groups. (1) The Brassicacea group (A. thaliana and A. lyrata), RPA1 from this group is analyzed separately because it has five paralogs, and (2) Other dicot group that contains only three RPA1 paralogs (Tomato, Cucumber, Strawberry, Castor oil plant, Grape, Cacao, Peach, and California Poplar). Plants that have only two RPA1 paralogs or more than three, as a result of lineage specific duplication, were not included in the analysis since these cases would affect sequence conservation and selection, and comparison of orthologs would lead to unreliable comparisons. dN and dS analyses employed RPA1 coding sequence comparisons to a common “outgroup” orthologous RPA1 (pair-wise comparison). To this end, RPA1 of C. rubella and rice were used as an “outgroup” to the first and second group, respectively.
All RPA1 are under purifying selection, as they all have ω-values < 1 (Figure 9). However, the degree of selection pressure or conservation is stronger on RPA1B followed by RPA1A and RPA1C. This can be due to the difference in gene expression level and consistent with the high transcript accumulation of RPA1B group vs. the RPA1A/C group (Figure 6). However, gene expression alone does not explain the sequence conservation difference seen among the groups as, for example, there is no clear expression difference between the RPA1A group and RPA1C group (Figure 6). In this case, functional differences may play a role. While RPA1A is primarily responsible for the highly conserved meiotic DNA recombination process, RPA1C likely functions in many types of DNA repair pathways and interacts with various proteins found in each pathway. Proteins involved in many biochemical pathways that employ various interaction partners tend to be more conserved (Krylov et al., 2003). However, since some repair pathways and associated proteins are relatively less conserved across plants (Singh et al., 2010), lineage specific co-evolution of RPA1C may result in less conservation of the protein across species.
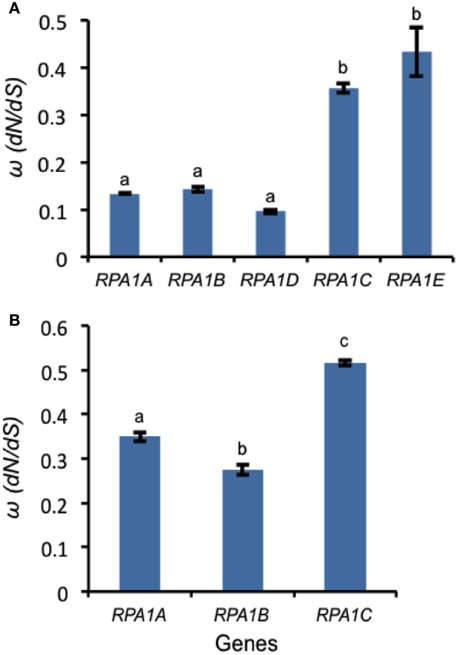
Figure 9. Natural selection (ω) values for RPA1 genes. (A) ω-values for Arabidopsis and A. lyrata RPA1 genes. C. rubella RPA1 genes were used as a reference for ortholog pairwise sequence distance measurement (dN and dS). (B) ω-values for RPA1 genes of eight plants (Tomato, Cucumber, Strawberry, Castor oil plant, Grape, Cacao, Peach, and California Poplar). Rice RPA1 genes were used as a reference for ortholog pairwise sequence distance measurement (dN and dS). dN and dS analyses were conducted in MEGA5 using the Nei-Gojobori model. Data are mean ± SE. To analyze statistical difference F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
Arabidopsis RPA1 Paralogs Contain Unique cis-Acting Element Composition Associated with Biological Function
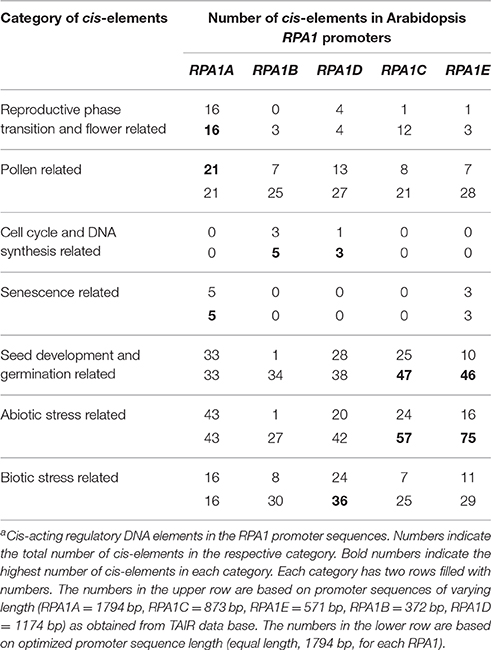
As discussed above, RPA1 members display unique gene expression patterns consistent with their proposed function in meiosis, DNA replication and repair. Accordingly, we hypothesize that respective RPA1 paralog members should display cis-acting elements that regulate their spatial, temporal and induced expression pattern consistent with proposed activities. To identify cis-acting regulatory elements, sequences were analyzed using PLACE (Plant cis-acting regulatory DNA elements) database (Higo et al., 1999). For this analysis we used two sets of promoter sequences. The first set includes predicted promoter sequences upstream of transcriptional start site for each paralogs retrieved from TAIR (The Arabidopsis Information Resource) database. Since the predicted promoter sequence of each paralog is different in length, we also analyzed a second set with an equal length (1794 bp) of promoter sequence for each paralog (Table 1). This is based on the predicted promoter sequence length of RPA1A that contains the largest promoter region of the group. A complete list of cis-acting elements along with their description is presented in Supplementary Table S5.
As is summarized in Table 1, RPA1A promoter sequences are enriched in cis- elements related to reproductive phase transition, flower, and senescence vs. other RPA1B and RPA1C group members. In addition, these sequences appear enriched in pollen-related cis-acting elements vs. other groups, but only when limited to predicted promoter sequences (Table 1). These data are consistent with a proposed leading role for RPA1A during meiosis, and coincides with RPA1A expression up-regulation immediately after reproductive phase transition, and during flowering and pollen formation stage (Figure 6A). Interestingly, RPA1A promoter enrichment of cis- acting elements related to senescence is well correlated to its late-developmental stage (senescence) transcriptional up-regulation (Figure 6A), suggesting a potential role for RPA1A in regulation of senescence, but this would need to be tested biologically.
Cis-elements related to DNA synthesis and cell cycle regulation were only identified in the promoters of RPA1B and RPA1D, consistent with their proposed primary role in DNA replication (Aklilu et al., 2014). Interestingly, RPA1D appears to be uniquely enriched in cis-elements related to biotic stress responses.
Lastly, both RPA1C and RPA1E display enrichment of cis-elements related to seed development and germination and abiotic stress (Table 1). Obviously, abiotic stress encompasses a wide spectrum of insults to tissues (Waterworth et al., 2011; Roy, 2014), but includes agents that damage DNA, such as UV light or ionizing radiation for example. RPA1C and RPA1E are predicted to play a leading role in DNA repair activities (Aklilu et al., 2014) and are induced by ionizing radiation (Culligan et al., 2006), suggesting perhaps many of these abiotic stress-related elements are directly involved in the transcriptional response to DNA damage. Interestingly, DNA repair pathways are known to regulate seed quality and longevity by repairing DNA damage accumulated during storage and imbibition (Cheah and Osborne, 1978; Dandoy et al., 1987; Waterworth et al., 2010, 2015; Chen et al., 2012; Bueso et al., 2014). The observed enrichment of seed related cis-elements in the promoters of both RPA1C and RPA1E raises the question of whether RPA could play a role in seed quality and longevity.
Summary
We describe in this study the evolution of plant RPA1 family and the unique sequence variations that can be used to categorize RPA1 sequences into three general groups in plants, RPA1A, RPA1B, and RPA1C. Our data suggest that these three groups of plant RPA sequences evolved from two groups of green algal RPA1 progenitor sequences, RPA1B-like and RPA1A/C-like sequences, by losing and gaining unique motifs, domains, and subdomains (Figure 5).
The unique sequence variations that exist within each group of plant RPA1 can be employed to predict biochemical function of RPA1 genes from newly sequenced genomes. These predictions will be valuable in future biochemical characterization studies of RPA complexes, which are ultimately necessary to accurately determine RPA functions in plants. Since the expansion of RPA1B and RPA1C into RPA1D and RPA1E respectively, appears to have occurred early and specifically in the Brassicacea family, these sequences could also prove useful in determining phylogenetic relationships within this family.
Author Contributions
BA designed and carried out the experiments and wrote the paper. KC designed experiments and wrote the paper.
Funding
This work was supported by the National Science Foundation [grant number MCB-0818603]; National Science Foundation ADVANCE [grant number UNH147493] to KC; and the Dissertation Year Fellowship from the University of New Hampshire graduate school to BA. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2654. This work was supported by the USDA National Institute of Food and Agriculture HATCH Project NH00543.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Vaughn S. Cooper for helpful comments and critical reading during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00033
Abbreviations
RPA, Replication Protein A; DSB, Double Strand Break; DBD, DNA Binding Domain; ATM, Ataxia Telangiectasia Mutated; ATR, Ataxia Telangiectasia Rad3-Related; ssDNA, single-stranded DNA.
References
Aklilu, B. B., Soderquist, R. S., and Culligan, K. M. (2014). Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 42, 3104–3118. doi: 10.1093/nar/gkt1292
Armstrong, S. J., Franklin, F. C., and Jones, G. H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114(Pt 23), 4207–4217. Available online at: http://jcs.biologists.org/content/114/23/4207.article-info
Bastin-Shanower, S. A., and Brill, S. J. (2001). Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 276, 36446–36453. doi: 10.1074/jbc.M104386200
Binz, S. K., Sheehan, A. M., and Wold, M. S. (2004). Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 3, 1015–1024. doi: 10.1016/j.dnarep.2004.03.028
Blackwell, L. J., and Borowiec, J. A. (1994). Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol. 14, 3993–4001. doi: 10.1128/MCB.14.6.3993
Bochkarev, A., Pfuetzner, R. A., Edwards, A. M., and Frappier, L. (1997). Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385, 176–181. doi: 10.1038/385176a0
Bochkareva, E., Belegu, V., Korolev, S., and Bochkarev, A. (2001). Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 20, 612–618. doi: 10.1093/emboj/20.3.612
Bochkareva, E., Kaustov, L., Ayed, A., Yi, G. S., Lu, Y., Pineda-Lucena, A., et al. (2005). Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. U.S.A. 102, 15412–15417. doi: 10.1073/pnas.0504614102
Bochkareva, E., Korolev, S., and Bochkarev, A. (2000). The role for zinc in replication protein A. J. Biol. Chem. 275, 27332–27338. doi: 10.1074/jbc.m000620200
Brinster, R. L., Allen, J. M., Behringer, R. R., Gelinas, R. E., and Palmiter, R. D. (1988). Introns increase transcriptional efficiency in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 85, 836–840. doi: 10.1073/pnas.85.3.836
Bueso, E., Ibañez, C., Sayas, E., Muñoz-Bertomeu, J., Gonzalez-Guzmán, M., Rodriguez, P. L., et al. (2014). A forward genetic approach in Arabidopsis thaliana identifies a RING-type ubiquitin ligase as a novel determinant of seed longevity. Plant Sci. 215–216, 110–116. doi: 10.1016/j.plantsci.2013.11.004
Bulmer, M. (1988). Are codon usage patterns in unicellular organisms determined by selection-mutation balance. Evol. Biol. 1, 15. doi: 10.1046/j.1420-9101.1988.1010015.x
Cai, L., Roginskaya, M., Qu, Y., Yang, Z., Xu, Y., and Zou, Y. (2007). Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry 46, 8226–8233. doi: 10.1021/bi7004976
Callis, J., Fromm, M., and Walbot, V. (1987). Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200. doi: 10.1101/gad.1.10.1183
Casneuf, T., De Bodt, S., Raes, J., Maere, S., and Van de Peer, Y. (2006). Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 7:R13. doi: 10.1186/gb-2006-7-2-r13
Castillo-Davis, C. I., Hartl, D. L., and Achaz, G. (2004). cis-Regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 14, 1530–1536. doi: 10.1101/gr.2662504
Chang, Y., Gong, L., Yuan, W., Li, X., Chen, G., Li, X., et al. (2009). Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol. 151, 2162–2173. doi: 10.1104/pp.109.142877
Cheah, K. S., and Osborne, D. J. (1978). DNA lesions occur with loss of viability in embryos of ageing rye seed. Nature 272, 593–599. doi: 10.1038/272593a0
Chen, H., Chu, P., Zhou, Y., Li, Y., Liu, J., Ding, Y., et al. (2012). Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J. Exp. Bot. 63, 4107–4121. doi: 10.1093/jxb/ers093
Chiapello, H., Lisacek, F., Caboche, M., and Hénaut, A. (1998). Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209, GC1–GC38. doi: 10.1016/S0378-1119(97)00671-9
Cui, L., Wall, P. K., Leebens-Mack, J. H., Lindsay, B. G., Soltis, D. E., Doyle, J. J., et al. (2006). Widespread genome duplications throughout the history of flowering plants. Genome Res. 16, 738–749. doi: 10.1101/gr.4825606
Culligan, K. M., Robertson, C. E., Foreman, J., Doerner, P., and Britt, A. B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961. doi: 10.1111/j.1365-313X.2006.02931.x
Dandoy, E., Schnys, R., Deltour, R., and Verly, W. G. (1987). Appearance and repair of apurinic/apyrimidinic sites in DNA during early germination of Zea mays. Mutat. Res. Fund. Mol. Mech. Muta. 181, 57–60. doi: 10.1016/0027-5107(87)90287-9
Daughdrill, G. W., Ackerman, J., Isern, N. G., Botuyan, M. V., Arrowsmith, C., Wold, M. S., et al. (2001). The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 29, 3270–3276. doi: 10.1093/nar/29.15.3270
Dong, J., Park, J. S., and Lee, S. H. (1999). In vitro analysis of the zinc-finger motif in human replication protein A. Biochem. J. 337(Pt 2), 311–317.
Drummond, D. A., Bloom, J. D., Adami, C., Wilke, C. O., and Arnold, F. H. (2005). Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U.S.A. 102, 14338–14343. doi: 10.1073/pnas.0504070102
Duncker, B. P., Davies, P. L., and Walker, V. K. (1997). Introns boost transgene expression in Drosophila melanogaster. Mol. Gen. Genet. 254, 291–296. doi: 10.1007/s004380050418
Duret, L., and Mouchiroud, D. (1999). Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 4482–4487. doi: 10.1073/pnas.96.8.4482
Eschbach, V., and Kobbe, D. (2014). Different replication protein A complexes of Arabidopsis thaliana have different DNA-binding properties as a function of heterotrimer composition. Plant Cell Physiol. 55, 1460–1472. doi: 10.1093/pcp/pcu076
Gamsjaeger, R., Liew, C. K., Loughlin, F. E., Crossley, M., and Mackay, J. P. (2007). Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70. doi: 10.1016/j.tibs.2006.12.007
Ganpudi, A. L., and Schroeder, D. F. (2011). “UV damaged DNA repair & tolerance in plants,” in Selected Topics in DNA Repair, ed C. Chen (San Diego, CA: InTech), 73–96.
Gomes, X. V., and Wold, M. S. (1996). Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry 35, 10558–10568. doi: 10.1021/bi9607517
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Haring, S. J., Mason, A. C., Binz, S. K., and Wold, M. S. (2008). Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J. Biol. Chem. 283, 19095–19111. doi: 10.1074/jbc.M800881200
Hass, C. S., Lam, K., and Wold, M. S. (2012). Repair-specific functions of replication protein A. J. Biol. Chem. 287, 3908–3918. doi: 10.1074/jbc.M111.287441
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. doi: 10.1155/2008/420747
Hyun, T. K., Eom, S. H., Han, X., and Kim, J. S. (2014). Evolution and expression analysis of the soybean glutamate decarboxylase gene family. J. Biosci. 39, 899–907. doi: 10.1007/s12038-014-9484-2
Iftode, C., Daniely, Y., and Borowiec, J. A. (1999). Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180. doi: 10.1080/10409239991209255
Ikemura, T. (1985). Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2, 13–34.
Ishibashi, T., Kimura, S., and Sakaguchi, K. (2006). A higher plant has three different types of RPA heterotrimeric complex. J. Biochem. 139, 99–104. doi: 10.1093/jb/mvj014
Jacobs, D. M., Lipton, A. S., Isern, N. G., Daughdrill, G. W., Lowry, D. F., Gomes, X., et al. (1999). Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR 14, 321–331. doi: 10.1023/A:1008373009786
Jones, J. S., Weber, S., and Prakash, L. (1988). The Saccharomyces cerevisiae RAD18 gene encodes a protein that contains potential zinc finger domains for nucleic acid binding and a putative nucleotide binding sequence. Nucleic Acids Res. 16, 7119–7131. doi: 10.1093/nar/16.14.7119
Juneau, K., Miranda, M., Hillenmeyer, M. E., Nislow, C., and Davis, R. W. (2006). Introns regulate RNA and protein abundance in yeast. Genetics 174, 511–518. doi: 10.1534/genetics.106.058560
Karlson, D., and Imai, R. (2003). Conservation of the cold shock domain protein family in plants. Plant Physiol. 131, 12–15. doi: 10.1104/pp.014472
Karlson, D., Nakaminami, K., Toyomasu, T., and Imai, R. (2002). A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 277, 35248–35256. doi: 10.1074/jbc.M205774200
Kim, C., Paulus, B. F., and Wold, M. S. (1994). Interactions of human replication protein A with oligonucleotides. Biochemistry 33, 14197–14206. doi: 10.1021/bi00251a031
Kim, C., Snyder, R. O., and Wold, M. S. (1992). Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12, 3050–3059. doi: 10.1128/MCB.12.7.3050
Kim, Y. O., and Kang, H. (2006). The role of a zinc finger-containing glycine-rich RNA-binding protein during the cold adaptation process in Arabidopsis thaliana. Plant Cell Physiol. 47, 793–798. doi: 10.1093/pcp/pcj047
Kim, Y. O., Kim, J. S., and Kang, H. (2005). Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42, 890–900. doi: 10.1111/j.1365-313X.2005.02420.x
Kim, Y. O., Pan, S., Jung, C. H., and Kang, H. (2007). A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 48, 1170–1181. doi: 10.1093/pcp/pcm087
Krishna, S. S., Majumdar, I., and Grishin, N. V. (2003). Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31, 532–550. doi: 10.1093/nar/gkg161
Krylov, D. M., Wolf, Y. I., Rogozin, I. B., and Koonin, E. V. (2003). Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13, 2229–2235. doi: 10.1101/gr.1589103
Langelier, M. F., Planck, J. L., Roy, S., and Pascal, J. M. (2011). Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 286, 10690–10701. doi: 10.1074/jbc.m110.202507
Lao, Y., Gomes, X. V., Ren, Y., Taylor, J. S., and Wold, M. S. (2000). Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry 39, 850–859. doi: 10.1021/bi991704s
Le Hir, H., Nott, A., and Moore, M. J. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220. doi: 10.1016/S0968-0004(03)00052-5
Li, X., Chang, Y., Xin, X., Zhu, C., Li, X., Higgins, J. D., et al. (2013). Replication protein A2c coupled with replication protein A1c regulates crossover formation during meiosis in rice. Plant Cell 25, 3885–3899. doi: 10.1105/tpc.113.118042
Lin, Y. L., Chen, C., Keshav, K. F., Winchester, E., and Dutta, A. (1996). Dissection of functional domains of the human DNA replication protein complex replication protein A. J. Biol. Chem. 271, 17190–17198. doi: 10.1074/jbc.271.29.17190
Lin, Y. L., Shivji, M. K., Chen, C., Kolodner, R., Wood, R. D., and Dutta, A. (1998). The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J. Biol. Chem. 273, 1453–1461. doi: 10.1074/jbc.273.3.1453
Liu, J. S., Kuo, S. R., and Melendy, T. (2006). DNA damage-induced RPA focalization is independent of gamma-H2AX and RPA hyper-phosphorylation. J. Cell. Biochem. 99, 1452–1462. doi: 10.1002/jcb.21066
Liu, Q., Feng, Y., Zhao, X., Dong, H., and Xue, Q. (2004). Synonymous codon usage bias in Oryza sativa. Plant Sci. 167, 101–105. doi: 10.1016/j.plantsci.2004.03.003
Lockton, S., and Gaut, B. S. (2005). Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21, 60–65. doi: 10.1016/j.tig.2004.11.013
Longhese, M. P., Plevani, P., and Lucchini, G. (1994). Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 14, 7884–7890. doi: 10.1128/MCB.14.12.7884
Louis, E. J. (2007). Evolutionary genetics: making the most of redundancy. Nature 449, 673–674. doi: 10.1038/449673a
Ly, V., Hatherell, A., Kim, E., Chan, A., Belmonte, M. F., and Schroeder, D. F. (2013). Interactions between Arabidopsis DNA repair genes UVH6, DDB1A, and DDB2 during abiotic stress tolerance and floral development. Plant Sci. 213, 88–97. doi: 10.1016/j.plantsci.2013.09.004
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Marais, G., and Duret, L. (2001). Synonymous codon usage, accuracy of translation, and gene length in Caenorhabditis elegans. J. Mol. Evol. 52, 275–280. doi: 10.1007/s002390010155
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Marwedel, T., Ishibashi, T., Lorbiecke, R., Jacob, S., Sakaguchi, K., and Sauter, M. (2003). Plant-specific regulation of replication protein A2 (OsRPA2) from rice during the cell cycle and in response to ultraviolet light exposure. Planta 217, 457–465. doi: 10.1007/s00425-003-1001-z
Moore, R. C., and Purugganan, M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. doi: 10.1016/j.pbi.2004.12.001
Niu, D. K., and Yang, Y. F. (2011). Why eukaryotic cells use introns to enhance gene expression: splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biol. Direct 6:24. doi: 10.1186/1745-6150-6-24
Osman, K., Sanchez-Moran, E., Mann, S. C., Jones, G. H., and Franklin, F. C. (2009). Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 28, 394–404. doi: 10.1038/emboj.2008.295
Pál, C., Papp, B., and Hurst, L. D. (2001). Highly expressed genes in yeast evolve slowly. Genetics 158, 927–931. Available online at: http://genetics.org/content/158/2/927.long
Rajavashisth, T. B., Taylor, A. K., Andalibi, A., Svenson, K. L., and Lusis, A. J. (1989). Identification of a zinc finger protein that binds to the sterol regulatory element. Science 245, 640–643. doi: 10.1126/science.2562787
Ren, X. Y., Vorst, O., Fiers, M. W., Stiekema, W. J., and Nap, J. P. (2006). In plants, highly expressed genes are the least compact. Trends Genet. 22, 528–532. doi: 10.1016/j.tig.2006.08.008
Rogelj, B., Godin, K. S., Shaw, C. E., and Ule, J. (2011). “The functions of glycine-rich regions in TDP-43, FUS and related RNA-binding proteins,” in RNA Binding Proteins, ed Z. J. Lorkovic (Austin, TX: 'Landes Bioscience and Springer Science+Business Media), 1–17.
Roy, S. (2014). Maintenance of genome stability in plants: repairing DNA double strand breaks and chromatin structure stability. Front. Plant Sci. 5:487. doi: 10.3389/fpls.2014.00487
Sablok, G., Wu, X., Kuo, J., Nayak, K. C., Baev, V., Varotto, C., et al. (2013). Combinational effect of mutational bias and translational selection for translation efficiency in tomato (Solanum lycopersicum) cv. Micro-Tom. Genomics 101, 290–295. doi: 10.1016/j.ygeno.2013.02.008
Shabalina, S. A., Ogurtsov, A. Y., Spiridonov, A. N., Novichkov, P. S., Spiridonov, N. A., and Koonin, E. V. (2010). Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 27, 1745–1749. doi: 10.1093/molbev/msq086
Shultz, R. W., Tatineni, V. M., Hanley-Bowdoin, L., and Thompson, W. F. (2007). Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 144, 1697–1714. doi: 10.1104/pp.107.101105
Simillion, C., Vandepoele, K., Van Montagu, M. C., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 13627–13632. doi: 10.1073/pnas.212522399
Singh, S. K., Roy, S., Choudhury, S. R., and Sengupta, D. N. (2010). DNA repair and recombination in higher plants: insights from comparative genomics of Arabidopsis and rice. BMC Genomics 11:443. doi: 10.1186/1471-2164-11-443
Takashi, Y., Kobayashi, Y., Tanaka, K., and Tamura, K. (2009). Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis. Plant Cell Physiol. 50, 1965–1976. doi: 10.1093/pcp/pcp140
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Theobald, D. L., Mitton-Fry, R. M., and Wuttke, D. S. (2003). Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32, 115–133. doi: 10.1146/annurev.biophys.32.110601.142506
Tzfati, Y., Abeliovich, H., Avrahami, D., and Shlomai, J. (1995). Universal minicircle sequence binding protein, a CCHC-type zinc finger protein that binds the universal minicircle sequence of trypanosomatids. Purification and characterization. J. Biol. Chem. 270, 21339–21345. doi: 10.1074/jbc.270.36.21339
Umezu, K., Sugawara, N., Chen, C., Haber, J. E., and Kolodner, R. D. (1998). Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148, 989–1005.
Vassin, V. M., Wold, M. S., and Borowiec, J. A. (2004). Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 24, 1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Walther, A. P., Gomes, X. V., Lao, Y., Lee, C. G., and Wold, M. S. (1999). Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry 38, 3963–3973. doi: 10.1021/bi982370u
Waterworth, W. M., Bray, C. M., and West, C. E. (2015). The importance of safeguarding genome integrity in germination and seed longevity. J. Exp. Bot. 66, 3549–3558. doi: 10.1093/jxb/erv080
Waterworth, W. M., Drury, G. E., Bray, C. M., and West, C. E. (2011). Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 192, 805–822. doi: 10.1111/j.1469-8137.2011.03926.x
Waterworth, W. M., Masnavi, G., Bhardwaj, R. M., Jiang, Q., Bray, C. M., and West, C. E. (2010). A plant DNA ligase is an important determinant of seed longevity. Plant J. 63, 848–860. doi: 10.1111/j.1365-313X.2010.04285.x
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. doi: 10.1371/journal.pone.0000718
Wobbe, C. R., Weissbach, L., Borowiec, J. A., Dean, F. B., Murakami, Y., Bullock, P., et al. (1987). Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl. Acad. Sci. U.S.A. 84, 1834–1838. doi: 10.1073/pnas.84.7.1834
Wold, M. S. (1997). Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92. doi: 10.1146/annurev.biochem.66.1.61
Wright, S. I., Yau, C. B., Looseley, M., and Meyers, B. C. (2004). Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol. Biol. Evol. 21, 1719–1726. doi: 10.1093/molbev/msh191
Xu, X., Vaithiyalingam, S., Glick, G. G., Mordes, D. A., Chazin, W. J., and Cortez, D. (2008). The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 28, 7345–7353. doi: 10.1128/MCB.01079-08
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Keywords: RPA, replication, meiosis, DNA repair
Citation: Aklilu BB and Culligan KM (2016) Molecular Evolution and Functional Diversification of Replication Protein A1 in Plants. Front. Plant Sci. 7:33. doi: 10.3389/fpls.2016.00033
Received: 21 August 2015; Accepted: 10 January 2016;
Published: 29 January 2016.
Edited by:
Jan Dvorak, University of California, Davis, USAReviewed by:
Crisanto Gutierrez, Consejo Superior de Investigaciones Cientificas, SpainChung-Ju Rachel Wang, Academia Sinica, Taiwan
Copyright © 2016 Aklilu and Culligan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin M. Culligan, ay5jdWxsaWdhbkB1bmguZWR1
 Behailu B. Aklilu
Behailu B. Aklilu Kevin M. Culligan
Kevin M. Culligan