- Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
Glyoxalase I (GLYI) is a ubiquitous enzyme in all organisms that catalyzes the conversion of the potent cytotoxin methylglyoxal to S-D-lactoylglutathione. Although many reports suggest the importance of GLYI in the plant response to stress, its function in seeds requires further study. Here, we identified a heat-induced GLYI from Brassica napus seeds, BnGLYI, using a comparative proteomics approach. Two-dimensional gel analyses revealed that BnGLYI protein expression upon heat treatment was significantly elevated in thermotolerant seeds but was diminished in heat-sensitive seeds. The BnGLYI-2 and BnGLYI-3 genes from the heat-sensitive and thermotolerant cultivars, respectively, were characterized, and analyzed. Only two amino acid residue variations were found between the amino acid sequences of the two genes. Moreover, overexpressing BnGLYI-3 in yeast cells enhanced tolerance to heat and cold stress and significantly increased GLYI activity compared to overexpressing BnGLYI-2. In addition, BnGLYI-3 transformants showed enhanced superoxide dismutase activities under heat and cold treatment, whereas these activities were diminished for BnGLYI-2 transformants. Taken together, these results indicate that overexpression of the BnGLYI-3 gene imparts thermotolerance and cold tolerance in yeast and that the variations in BnGLYI-3 may play an important role in the responses to temperature stresses.
Introduction
Seeds not only provide the majority of the human food supply but are also an important supply of nutrients in animal husbandry and industrial feedstock; high-quality seeds are of great significance for socioeconomic development (Janmohammadi et al., 2008). Seed quality is determined by factors such as the rate and uniformity of germination, vigor, and storability of a seed lot. Biotic and abiotic factors affect seed vigor, germination, and storability. High temperature is one abiotic factor often incurred in seed storage, which may reduce seed vigor and decelerate or completely inhibit seed germination (Wahid et al., 2007). Tests using artificial aging induced by high temperature are therefore often used to evaluate seed storability (Janmohammadi et al., 2008). In addition, heat stress is a major factor that affects plant development (Hall, 1992; Marcum, 1998; Howarth, 2005). Heat stress, usually in combination with drought stress or other stresses, causes extensive worldwide agricultural losses (Suzuki and Mittler, 2006). Although many studies have investigated the molecular factors related to thermotolerance in plants (Wahid et al., 2007), few studies in seeds have been reported.
Several major tolerance mechanisms, including the activities of ion transporters, osmoprotectants, free-radical scavengers, late embryogenesis abundant proteins, and factors involved in signaling cascades and transcriptional control, are essential for helping a seed counteract damage induced by harsh temperatures (Wang et al., 2004). Previous studies examining seed antioxidant defense have shown that ROS can accumulate due to disruption of the mitochondrial electron transport chain (Leprince et al., 1994) in response to recommencement of metabolism during seed aging. An increasing number of HSPs/chaperones that interact with heat stress-response mechanisms have been identified (Wang et al., 2004). Transgenic Arabidopsis seeds overexpressing NnHSP17.5 in sacred lotus (Nelumbo nucifera Gaertn) exhibited enhanced resistance to accelerated aging treatment (Zhou et al., 2012). Furthermore, other heat-responsive genes have been identified. Recently, a heat-induced NnANN1 gene was identified in N. nucifera seeds and was demonstrated to play an important role in seed thermotolerance (Chu et al., 2012). However, the molecular basis underlying these mechanisms is currently unclear. Considering that the cellular response to abiotic stresses is a multigenic trait resulting from multiple complex metabolic reactions, additional candidate genes contributing to heat tolerance must be explored.
The glyoxalases have been studied for many years in animals and have been proposed to be involved in the regulation of cell division and proliferation, tumor growth, microtubule assembly, and the protection of cells against oxoaldehyde toxicity (Thornalley, 1990). The glyoxalase system is important for detoxification of MG, a cytotoxic metabolite that is primarily produced as a result of carbohydrate and lipid metabolism. Two metalloenzymes, glyoxalase I (GLYI), and GLYII, constitute the glyoxalase system and catalyze a two-step reaction (Thornalley, 1990). First, GLYI catalyzes the conversion of MG to S-D-lactoylglutathione (S-LG) in the presence of GSH. Second, S-LG is hydrolyzed to D-lactate and GSH by GLYII. In this reaction, GLYI is considered as the primary rate-limiting enzyme (Ferguson et al., 1998). However, only a few GLYI genes have been cloned in plants, including Lycopersicon esculentum, Brassica juncea, B. oleracea, Glycine max, Sporobolus stapfianus, Triticum aestivum, Thlaspi caerulescens, Vigna radiata, and Oryza sativa (Espartero et al., 1995; Clugston et al., 1998; Veena et al., 1999; Skipsey et al., 2000; Lin et al., 2010; Hossain et al., 2011; Tuomainen et al., 2011, and Mustafiz et al., 2014). To date, genome projects have revealed that Arabidopsis thaliana and Oryza sativa contain 11 genes encoding GLYI (Mustafiz et al., 2011).
In plants, GLYI showed higher activity in rapidly dividing cells, such as in cell suspensions (soybean), root tips and seedlings (Paulus et al., 1993; Lin et al., 2010). GLYI activity can also be upregulated under stress conditions and in the presence of developmental cues, and this observation suggests that GLYI plays an important role in the stress response and in development (Ramaswamy et al., 1984; Sethi et al., 1988; Deswal et al., 1993; Yadav et al., 2005a,b; Singla-Pareek et al., 2006; Lin et al., 2010; Hossain et al., 2011). Tomato GLYI was upregulated in response to salt stress, and osmotic and phytohormonal stimuli (Espartero et al., 1995). Transgenic tobacco overexpressing the B. juncea GLYI gene (BjGLYI) showed significantly enhanced tolerance to MG and high salt concentrations compared to non-transgenic plants (Veena et al., 1999; Singla-Pareek et al., 2003). In a recent study, tobacco leaves transgenically overexpressing TaGLYI from wheat (T. aestivum L.) showed increased tolerance to ZnCl2 stress compared to control leaves (Lin et al., 2010). Despite the long-standing interest of plant scientists in the function of GLYI in stress responses, the involvement of plant GLYI genes in the thermotolerance of seeds has not yet been described.
Rapeseed (canola, oilseed rape; Brassica napus L., Brassicaceae family), an important oilseed plants worldwide, often encounters high temperature during storage, which leads to decline or deterioration in seed vigor (Nagel and Börner, 2010). Therefore, it is important to identify thermotolerance-related genes in B. napus seeds to improve their storability and thermotolerance. In this study, we analyzed seed tolerance to an extremely high temperature to screen for thermotolerance-related genes. A B. napus GLYI gene, BnGLYI, was identified by comparative proteomic analysis. Two-dimensional gel analyses showed that the protein level of BnGLYI in thermotolerant seeds was significantly increased in response to heat stress. Heterologous expression of BnGLYI in yeast conferred tolerance to severe temperature stress. In conclusion, BnGLYI is a heat-responsive protein that participates in tolerance to heat stress, and the variations in the BnGLYI protein sequence might affect its function. These results shed light on the applicability of a molecular breeding approach to enhance plant thermotolerance and seed storability.
Materials and Methods
Heat Stress Assay in Rapeseeds and Collection of Plant Materials
A heat-tolerant and a heat-sensitive B. napus cultivar, named 3382 and 2682, respectively, were used in this study. The two cultivars were screened by heat stress treatment from many cultivars of B. napus (data unpublished). The heat stress assay was performed using a previously described protocol (Gao et al., 2010; Chu et al., 2012). Briefly, surface-sterilized mature rapeseed seeds were incubated at 37°C for 2 h and then treated at 65°C for 2 h. Immediately after this treatment, the seeds were moved to room temperature and finally incubated at 28°C to assess their ability to germinate. The heat treatments were performed in the dark. Five sets of 100 seeds were used for each genotype. The treated seeds and the control seeds (maintained at 28°C) were simultaneously collected for RNA and protein isolation. The simultaneously harvested seeds were used for either the control or the heat treatment. The leaves from plants grown in a greenhouse were used for DNA extraction according to previously reported methods (Murray and Thompson, 1980).
Protein Extraction
Protein was extracted from both the treated and control B. napus seeds via trichloroacetic acid (TCA)/acetone precipitation as described by Damerval et al. (1986) and Gan et al. (2010) with minor modifications. Briefly, the seeds were ground in a precooled mortar in the presence of liquid nitrogen. The powders were precipitated with 10 volumes of precooled acetone (containing 10% TCA and 0.07% dithiothreitol (DTT)), and the mixtures were incubated at –20°C for 1 h. After centrifuging at 20 000 ×g for 30 min and removing the supernatant, the pellet of each sample was rinsed and incubated in ice-cold acetone (containing 0.07% DTT) for 1 h at –20°C. This step was repeated three times. Finally, each pellet was air-dried and resuspended in 500 μl of lysis buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 1 M phenylmethanesulfonyl fluoride (PMSF), 50 mM DTT, 0.5% Triton X-100, and 0.5% ampholine) and then vortexed for 1 h at room temperature. After centrifugation at 20 000 ×g for 30 min, each supernatant was collected in a fresh tube, and the protein concentrations were quantified using Bradford reagent (Aidlab, China) according to the manufacturer’s instructions. The protein samples were stored at –80°C.
Two-Dimensional Gel Electrophoresis
Two-dimensional gel electrophoresis was performed according to Gan et al. (2010, 2013). For total protein analysis, immobilized pH gradient (IPG) strips of 24 cm in length were rehydrated with 450 μl of rehydration solution for 14 h at 20°C, which was loaded with 1 mg of protein (Gan et al., 2010). Isoelectric focusing (IEF) was performed in an Ettan IPGphor IEF system as described by Gan et al. (2010). After IEF, the IPG strips were equilibrated as described by Gan et al. (2013). The second dimension of separation was performed using a 12.5% SDS-PAGE gel in an Ettan DALTsix system according to Gan et al. (2013). The gel was stained with colloidal Coomassie Brilliant Blue G-250 as previously described (Candiano et al., 2004). The experiment was replicated three times.
Data Analysis and Protein Identification
Images (300 dpi, 16-bit grayscale pixel depth) were scanned using UMAX Powerlook 2100XL (UMAX, Taipei, China) and were analyzed using Image Master 2D Platinum Software (Version 5.0, GE Healthcare) as described by Hajduch et al. (2007). Protein spots showing a significant difference in intensity (P < 0.05) of greater than 1.5-fold were selected and manually excised from the gel for protein identification. Mass spectrometry (MS) analysis of selected protein spots was performed by Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China), using the matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) method (Hajduch et al., 2007). The MS/MS data were searched against the ‘plant’ subset of the NCBI non-redundant sequence database using Mascot1 (Matrix Science).
Isolation of GLYI cDNA Clones
To functionally characterize BnGLYI, its coding region was amplified according to the sequence of BjGLYI (Table 1). An aliquot of 5 μl of the cDNA was used as a template to conduct PCR using full-length primers in a PTC-200 Peltier Thermal Cycler (Bio-Rad, USA); the PCR protocol consisted of an initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min, and a final extension of 10 min at 72°C. The desired PCR products were cloned into the pEASY-T1 vector (TransGen Biotech, China), and clones were selected on plates supplemented with IPTG, X-gal, and ampicillin. The clones were identified by PCR and then sequenced. Sequence comparisons were performed using BLAST. The BnGLYI sequences cloned from B. napus 3382 and 2682 were designated as BnGLYI-3 (KT720495) and BnGLYI-2 (KT720496), respectively.
Sequence Analysis
Amino acid (AA) sequence alignment was conducted using the DNAMAN program2 The conserved domains within the GLYI coding nucleotide sequence were searched in the NCBI database3
Quantitative RT-PCR (qRT-PCR) Analysis
Total RNA was extracted from yeast using an RNeasy Mini Kit (QIAGEN, Germany). RNA pellets were dissolved in DEPC-treated water; the presence of RNA was confirmed by agarose gel electrophoresis; and the RNA concentrations were quantified based on the absorbance of the samples at 260 nm. The RNA samples were reverse-transcribed into first-strand cDNA using a ReverTra Ace-α-TM qPCR RT kit (Toyobo, Japan) and stored at –80°C. Expression of the GLYI genes in transgenic yeast was evaluated using LightCycler 480 SYBR Green I Master mix and a LightCycler 480II real-time PCR system (Roche, Switzerland). Expression of the BnGLYI gene was normalized to that of Actin according to the 2 method aaa (Livak and Schmittgen, 2001) using the following formula:
where CT, Actin is the CT value of the reference gene Actin, and CT, target is the CT value of the gene under investigation. The value determined by this formula represents the relative expression of a gene in a sample relative to that in a calibrator. The primers and the amplification conditions used are described in Table 1. Each sample was examined in triplicate.
Ectopic Expression of the BnGLYI Gene in Yeast
The coding sequence of the BnGLYI gene was amplified from cDNA by PCR using pfu DNA polymerase (Tiangen, China). An EcoRI site and a NotI site (underlined in Table 1) were introduced at the 5′-end and the 3′-end of the PCR product, respectively. The EcoRI/NotI fragment (558 bp) was directly cloned into the pEASY-T1 vector (TransGen, China) and was then sub-cloned into the Pichia expression vector pPIC3.5K (Multi-Copy Pichia Expression Kit, Invitrogen, USA) to generate the pPIC3.5K-BnGLYI-2 and pPIC3.5K-BnGLYI-3 plasmids. These plasmids were introduced into Pichia pastoris GS115 cells by electroporation (MicroPulser Electroporation Apparatus, Bio-Rad, USA) according to the manufacturer’s instructions for overexpression of the given fusion protein. Transformants were selected by plating cells on synthetic minimal medium agar lacking histidine, and were then confirmed by PCR using gene-specific primers (Table 1). The cells transformed with empty vector (pPIC3.5K), pPIC3.5K-BnGLYI-2 and pPIC3.5K-BnGLYI-3 were designated as GS3.5K, GSGLYI-2, and GSGLYI-3, respectively.
Heterologous expression of BnGLYI was induced under the transcriptional control of the yeast AOX1 promoter. Expression of BnGLYI in P. pastoris GS115 was induced according to the instructions of the Multi-Copy Pichia Expression Kit as well as the method described by Wang et al. (2013) with slight modification. The transformant expressing the highest abundance of GLYI was selected and used for further studies. Aliquots (1 ml) of cells were harvested at 24, 48, 72, and 96 h and then centrifuged at maximum speed for 2–3 min at room temperature to collect the cells. Total cellular protein samples were electrophoresed via 12% SDS–PAGE, followed by Coomassie blue staining.
Assessment of Yeast Resistance to Temperature Stress
Yeast cultured to the exponential phase in a yeast extract peptone dextrose medium (YPD) was inoculated in buffered glycerol complex medium (BMGY). After induction in buffered methanol complex medium (BMMY), the cultured cells were diluted by 1:10, 000. The diluted cells were treated under heat and cold stress according to a previously reported method (Li et al., 2009) with minor modifications. Three aliquots (100 μl each) were treated at 42°C in a water bath for 2.5, 3.5, and 5 h. Simultaneously, three additional aliquots were frozen at –20°C for 10 days (D) and 13 D. The test samples were cultured on a YPD culture plate at 30°C after the treatments. Three aliquots were directly plated on a YPD plate as a control. The survival rate of the treated cells relative to the control cells is expressed as the percentage of colony-forming units (CFU%). The experiments were repeated three times. Tukey’s test was used to analyze all the data, which were presented as the means ± SD (n = 3), to compare the survival rate between the yeast cells harboring BnGLYI-2 and BnGLYI-3 and the control cells under either normal or stress conditions. A P < 0.05 was considered to indicate a significant difference.
Determination of Enzyme Activities
Glyoxalase I activity in total protein extracts from yeast was measured as described previously (Singla-Pareek et al., 2003; Mustafiz et al., 2010). Each of the measurements was performed using three replicates. The enzymatic production of S-lactoylglutathione (extinction coefficient 3.37 mM–1 cm–1) was measured at 240 nm at 15 s intervals for 2 min. Specific activity of GLYI is expressed as μmol S-lactoylglutathione formed/hr/mg protein (Junaid et al., 2004). The yeast protein was extracted using Y-PER Yeast Protein Extraction Reagent (Thermo Scientific, USA). Bradford reagent (Aidlab, China) was used to determine the protein concentrations. ImageJ software was used to determine the relative expression levels of BnGLYI-2 and BnGLYI-3.
Superoxide dismutase (SOD) activity was assessed by monitoring the inhibition of the photochemical reduction of nitroblue tetrazolium (NBT) according to the method of Beauchamp and Fridovich (1971). One unit of SOD activity was defined as the amount of enzyme required to cause 50% inhibition of the rate of NBT reduction at 560 nm (Beauchamp and Fridovich, 1971). SOD activity is expressed as units per milligram of protein.
Results
Thermotolerance of Oilseed Seeds
To demonstrate the basal thermotolerance of oilseeds, mature seeds were treated at high temperature. When treated at 65°C for 2 h, the germination of 3382 seeds was not evidently affected, as only a slight decrease in the germination rate was observed; however, only approximately 40% (41.7 ± 1.2%) of the 2682 seeds remained alive (Figure 1). With increasing treatment time, the germination rates of 3382 seeds gradually declined. However, the germination rates of 2682 seeds declined sharply, and up to 100% of the 2682 seeds were dead after 6 h at 65°C. The germination rate of 3382 seeds was significantly higher than that of 2682 seeds under heat treatment.
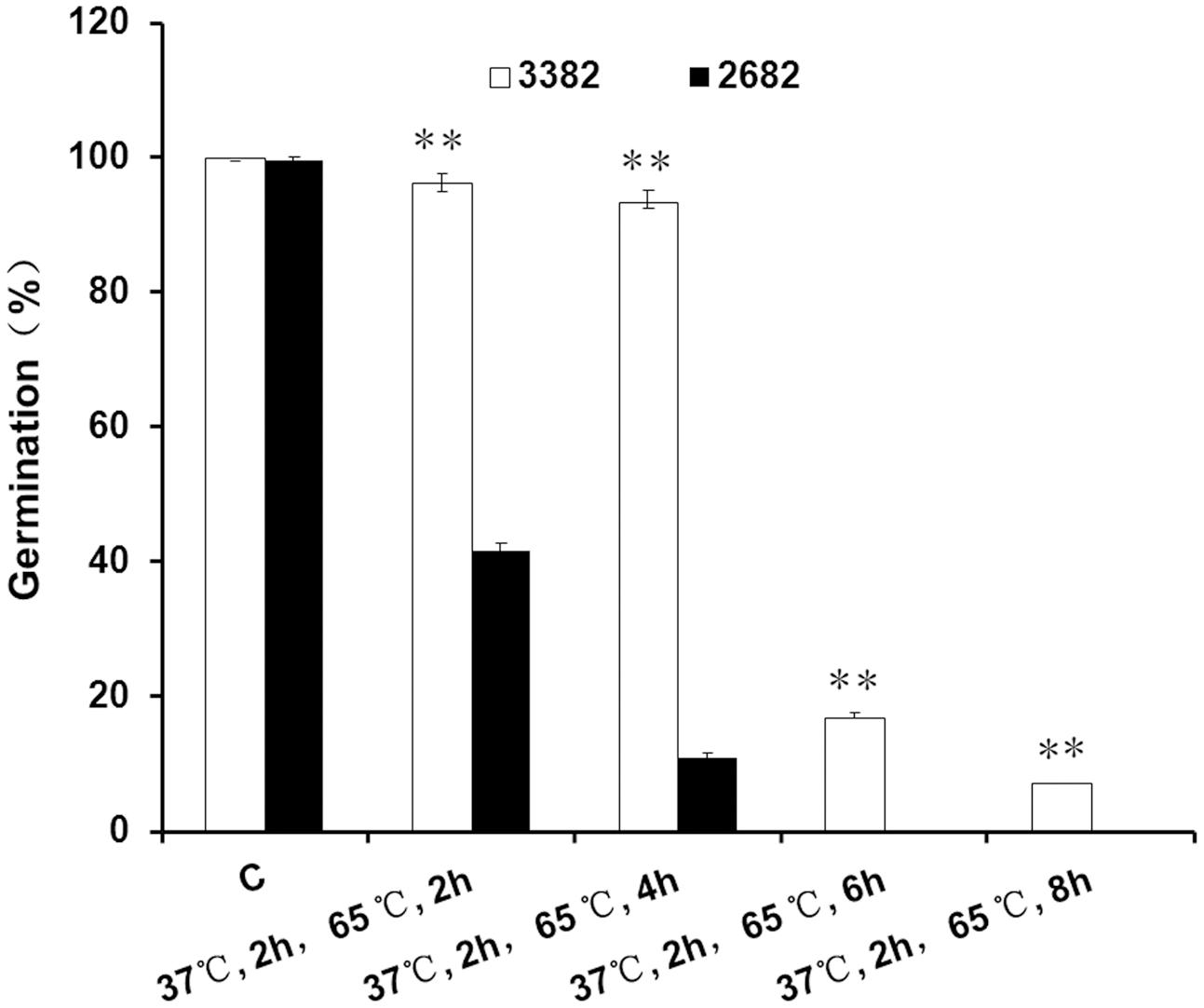
FIGURE 1. Seed thermotolerance assays. Asterisks denote a significant difference compared with 2682 seed germination based on Student’s t-test: ∗∗P < 0.01. The bars represent the means (three replicates) ± SD. C, control.
GLYI is a Heat-Responsive Protein
The objective of this study was to identify novel genes involved in the development of heat tolerance in B. napus. To reveal how plant cells respond to heat stress by modulating protein expression, we performed a two-dimensional gel analysis to screen for heat-responsive proteins. One heat-responsive protein was identified as GLYI (Figures 2A,B, Supplementary Figures S1 and S2). Nine peptides were matched to the predicted GLYI protein sequence with a MASCOT score of 117 (Supplementary Figure S2).
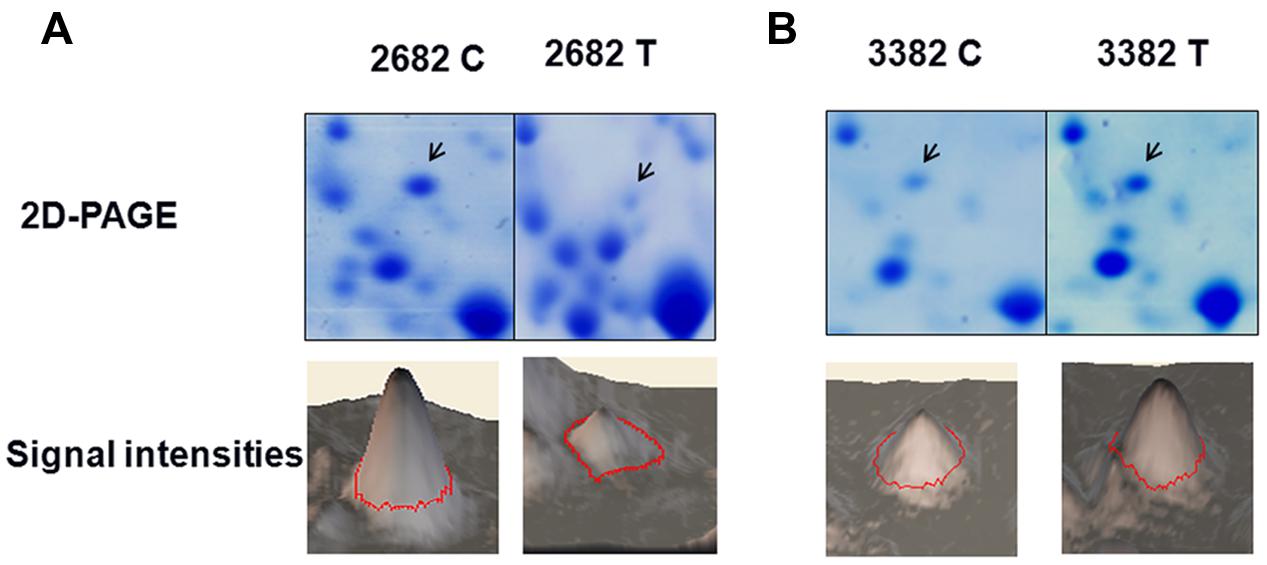
FIGURE 2. Glyoxalase I (GLYI) is a heat-responsive protein. The changes in the level of GLYI protein in response to heat treatment were analyzed by two-dimensional gel electrophoresis. The quantified Coomassie-staining signal intensity of the GLYI spot from each image is, respectively, shown in (A, 2682) and (B, 3382). C, control, T, treatment.
Characterization of GLYI cDNA Clones from B. napus
Based on the sequences of BjGLYI (GenBank accession no. Y13239) and A. thaliana AtGLYI (AT1G08110.1), full-length cDNAs of BnGLYI, encoding proteins sharing 99 and 100% AA identity with the BjGLYI sequence, respectively, were cloned from the 3382 and 2682 cultivars. The open reading frame (ORF) of BnGLYI cDNA consisted of 558 bp encoding a protein of 185 amino acids. The calculated molecular mass and pI were 20.8 kDa and 5.42, respectively, and these values were consistent with the apparent molecular mass and pI of the target protein spot on the two-dimensional gels.
Proteins containing a glyoxalase domain (PF00903) and having a putative lactoylglutathione lyase function have been classified as GLYI proteins (Mustafiz et al., 2011). In the two deduced protein sequences, two glyoxalase domains were found (AA 29–171, E-value 18.e-25) based on Pfam 23.04, and this result showed that the gene corresponding to both proteins was BnGLYI. Multi-alignment analysis of the deduced AA sequences from the BnGLYI gene revealed that the BnGLYI gene exhibited significantly high similarity to other GLYI genes (Figure 3A). For example, the BnGLYI-3 protein (from B. napus 3382) shared 79, 94, 99, 98, and 99% AA identities with GLYI from Solanum lycopersicum (Accession no. Z48183), A. thaliana (Accession no. AT1G08110.1), B. juncea (Accession no. Y13239), Arachis hypogaea (Accession no. DQ989209.2), and B. napus 2682 (BnGLYI-2), respectively (Figure 3A). These results suggested that this protein has been evolutionarily conserved. AA residues probably involved in metal and GSH binding sites were also conserved among the GLYI sequences (Figure 3B). Moreover, we found only two variations (Pro/Ala at position 9 and Ala/Val at position 93) in the AA sequence between BnGLYI-2 and BnGLYI-3. The variation at position 93 may affect the putative metal binding sites and this position may be an active site based on the analysis presented in Figure 3B.
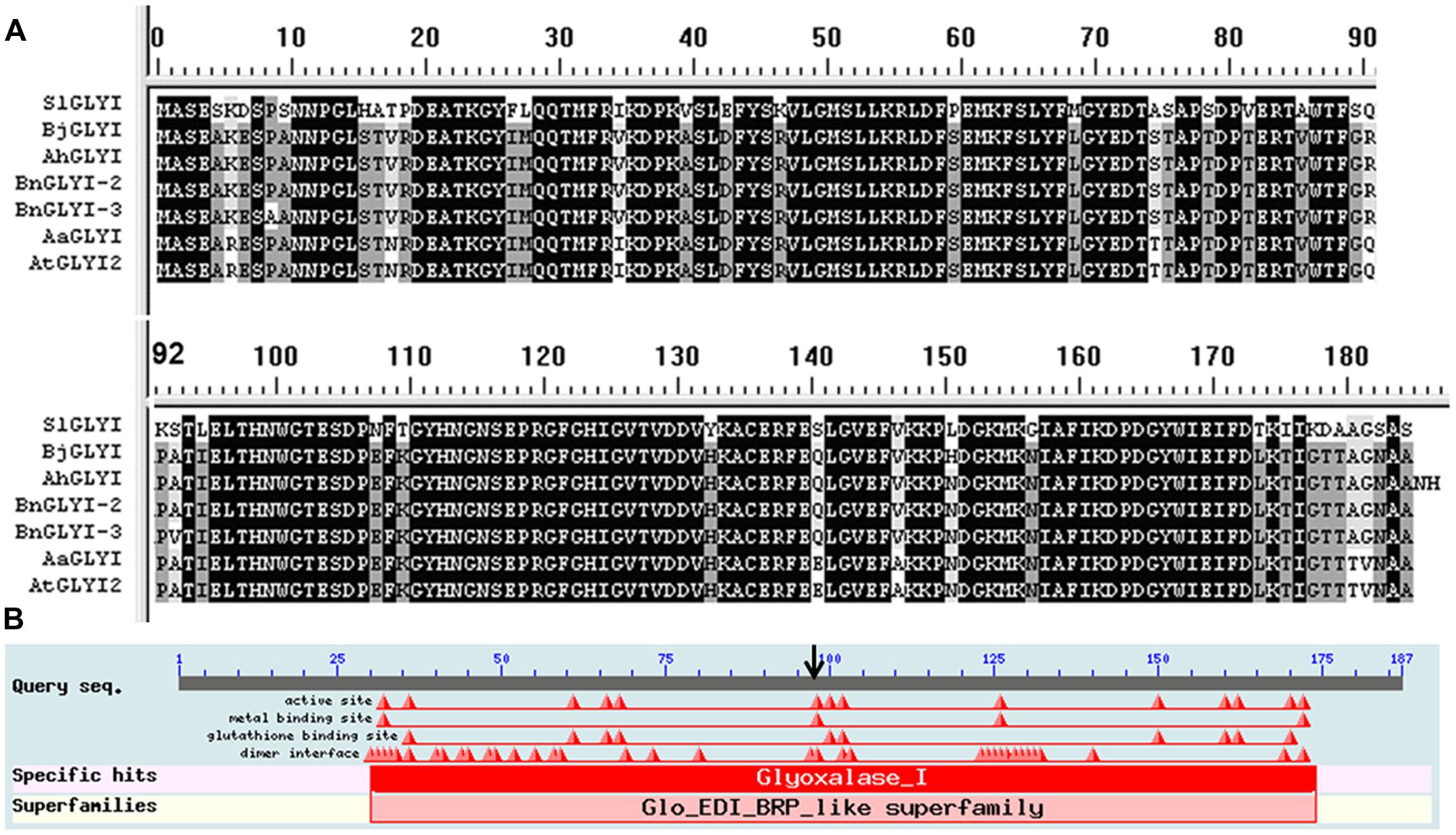
FIGURE 3. Sequence alignment and conserved domain analysis of BnGLYI. (A) Multiple sequence alignment of the deduced AA sequence of the GLYI cDNA clone from B. napus with previously reported GLYI sequences. The sequences were aligned using the DNAMAN program. SlGLYI: from Solanum lycopersicum (Accession no. Z48183), AtGLYI2: from Arabidopsis thaliana (Accession no. AT1G08110), BjGLYI: from Brassica juncea (Accession no. Y13239), AhGLYI: from Arachis hypogaea (Accession no. DQ989209.2), BnGLYI-3: from B. napus 3382 (Accession no. KT720495); BnGLYI-2: from B. napus 2682 (Accession no. KT720496). (B) Conserved domain analysis of BnGLYI. GLYI has a requirement for bound metal ions for catalysis.
Overexpressing BnGLYI-3 in P. pastoris Enhances Tolerance to Temperature Stresses
To determine whether the variations between the two BnGLYI protein sequences affected the function of the protein, we overexpressed the two BnGLYI genes in P. pastoris cells. Each full-length BnGLYI coding sequence was subcloned into the pPIC3.5K vector, and the resulting plasmids were transformed into yeast. The qRT-PCR results showed that the transformants harboring BnGLYI-2 and BnGLYI-3 displayed significantly increased BnGLYI expression, but that BnGLYI expression was not detected in the transformants carrying the empty vector or in the wild-type cells (Supplementary Figure S3). In addition, the soluble proteins extracted from P. pastoris cells were analyzed by SDS-PAGE. A distinct band with a molecular mass of ∗21 kDa was visible in the protein extract obtained from the GSGLYI-2 and GSGLYI-3 cells (Figure 4A). Moreover, the GSGLYI-2 and GSGLYI-3 cells exhibited a similar BnGLYI protein level for the same amount of total protein (Figure 4A). These results indicated that the two sequence variations did not affect BnGLYI protein stability. Serial dilutions (10–4, 10–5, 10–6) of cell cultures plated on a YPD plate showed that the transformants had similar growth rates to GS3.5K and GS115 cells when incubated at 30°C (data not shown).
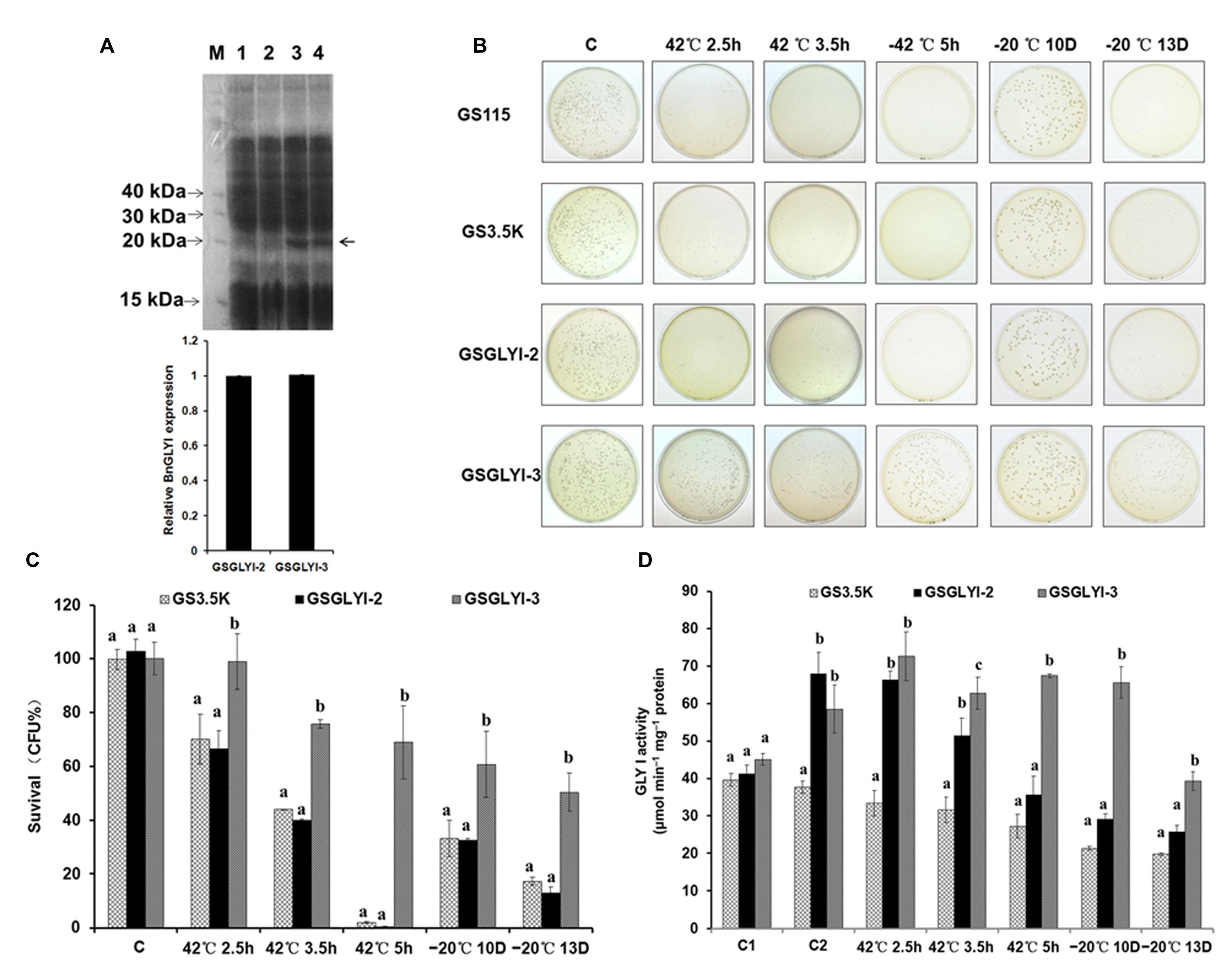
FIGURE 4. Analysis of BnGLYI expression in Pichia pastoris. (A) SDS-PAGE analysis of P. pastoris proteins. M: protein marker (KDa); 1: GS115; 2: GS3.5K; 3: GSGLYI-2; 4: GSGLYI-3. The relative BnGLYI expression levels were calculated after normalization to the total protein content. The expression levels of GSGLYI-2 were normalized to 1.0, and the data were analyzed using ImageJ software. The bars represent the means (three replicates) ± SD. (B) Growth of P. pastoris cells stressed at 42°C for 2.5, 3.5, and 5 h or stressed at –20°C for 10 D and 13 D. Diluted culture samples were treated at 42°C in a water bath and were then plated on YPD plates that were incubated at 30°C. (C) Viability of P. pastoris transformants carrying pPIC3.5K-BnGLYI-2 (GSGLYI-2), pPIC3.5K-BnGLYI-3 (GSGLYI-3), or pPIC3.5K (GS3.5K) that were stressed as described in (B). Cell viability is plotted as the percentage of CFUs relative to the number of CFUs in their respective controls. The data are presented as the means ± SD of three independent assays; different letters indicate significant differences (P < 0.05) under each stress condition, and values followed by the same letter are not significantly different according to Tukey’s test. (D) GLYI activity in crude soluble protein extracted from P. pastoris transformed with pPIC3.5K (GS3.5K), pPIC3.5K-BnGLYI-2 (GSGLYI-2), or pPIC3.5K-BnGLYI-3 (GSGLYI-3) under different temperature stress conditions. C, control; C1, non-induced cells cultured under the control conditions; C2, induced cells cultured under the control conditions.
A thermotolerance assay was conducted on yeast cells transformed with the recombinant plasmids pPIC3.5k-BnGLY-2 and pPIC3.5k-BnGLY-3, and cells harboring the empty pPIC3.5K vector were used as a control. Compared with the GS3.5K cells, the GSGLYI-3 cells showed increased tolerance to high temperatures; however, the GSGLYI-2 transformant did not show any thermotolerance under heat stress (Figures 4B,C). Within the first 2.5 h of heat treatment, loss of cell viability was observed. The survival rates of GS3.5K, GSGLYI-2, and GSGLYI-3 cells were approximately 70.2, 66.7, and 99.1%, respectively. The viability of GSGLYI-3 cells was significantly different from that of GS3.5K and GSGLYI-2 cells (P < 0.05). After 3.5 h of heat treatment at 42°C, 40.0 and 75.9% of the cells overexpressing BnGLYI-3 and BnGLYI-2, respectively, had survived. After 5 h of heat treatment at 42°C, 70.1% of the GSGLYI-3 cells remained alive, but most of the GS3.5K and GSGLYI-2 cells were dead (Figures 4B,C). Taken together, the survival rate of the BnGLYI-3-transformed cells was significantly higher than that of the BnGLYI-2 and empty vector transformants. However, the survival rate of GSGLYI-2 cells was not significantly different from that of GS3.5K cells.
To investigate the role of the BnGLYI protein in the response to cold shock, the GSGLYI-2 and GSGLYI-3 transformants as well as the empty vector control transformants were treated at –20°C. After being frozen for 10 D, approximately 60% of the GSGLYI-3 cells and 30% of the GSGLYI-2 and GS3.5K cells remained alive. After being frozen for 13 D, clearly more surviving cells (approximately 50%) remained in the GSGLYI-3 cell lines than in the GSGLYI-2 and control GS3.5K cells. Statistical analysis revealed that the survival rate of the GSGLYI-3 cells was 50.5±13.1%, whereas the survival rate of GSGLYI-2 and GS3.5K was approximately 17.4±1.5 and 12.9±2.5%, respectively (Figures 4B,C). These results indicated that overexpressing the BnGLYI-3 gene significantly enhanced the freeze tolerance of the host cells.
Furthermore, we measured GLYI activity in the crude protein samples extracted from GSGLYI-2, GSGLYI-3, and GS3.5K cells. As shown in Figure 4D, GS3.5K, GSGLYI-2, and GSGLYI-3 cells exhibited similar GLYI activity under normal conditions without induction. After induction, the GSGLYI-3 and GSGLYI-2 cells displayed approximately 55 and 80% higher activity, respectively, than the GS3.5K cells. GSGLYI-3 cells showed significantly higher GLYI activity than the GS3.5K and GSGLYI-2 cells after treatment at 42°C for 3.5 and 5 h, as well as after treatment at –20°C (Figure 4D). GSGLYI-3 cells showed approximately 1.4-fold and 0.9-fold higher GLYI activity than GS3.5K and GSGLYI-2 cells, respectively, after the 5-h treatment. In addition, GSGLYI-3 cells displayed approximately twofold and 1.3-fold higher GLYI activity than GS3.5K and GSGLYI-2 cells, respectively, after 10 D of treatment at –20°C. These results suggested that overexpressing GSGLYI-3 may improve heat and cold stress tolerance in P. pastoris.
SOD Activity in the Transformed Cells
To further explore the physiological traits by which the GSGLYI-3 cells exhibited enhanced tolerance to temperature stress, the activity of the antioxidant enzyme SOD was examined (Figure 5). SOD showed different activity patterns among the different cells under heat and cold stress. SOD activity in GSGLYI-3, GSGLYI-2 and GS3.5K cells was similar under heat and cold stress treatments. However, compared to the same cells under control conditions, SOD activity was decreased by 8.6% in GSGLYI-2 cells treated at 42°C for 5 h, but was increased by 2.1 and 57.1% in GS3.5K and GSGLYI-3 cells, respectively (Figure 5). After treatment at –20°C for 13 D, SOD activity was decreased by 56.5 and 28.9% in GS3.5K and GSGLYI-2 cells, respectively, but was increased by 6.4% in GSGLYI-3 cells relative to the levels in the corresponding control-treated cells (Figure 5).
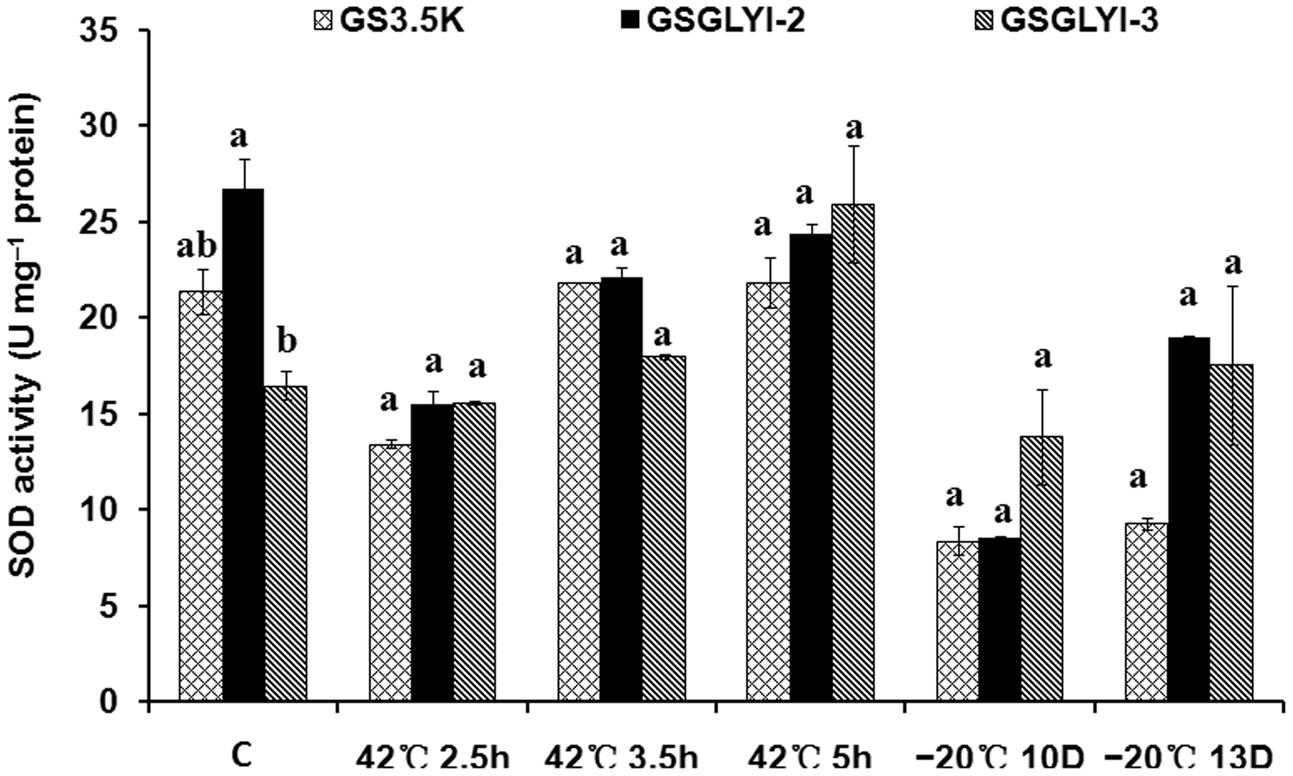
FIGURE 5. Superoxide dismutase activity in transformed P. pastoris cells under temperature stress treatments. The lowercase letters indicate significant differences among the transformants under the same treatment (Tukey’s multiple comparisons test, P < 0.05). Variants possessing the same letter are not statistically significant. C, control.
Discussion
In the present study, two BnGLYI genes were isolated from B. napus seeds via a proteomic analysis of thermotolerant (3382) and thermosensitive (2682) seeds. The coding sequences of BnGLYI-3 and BnGLYI-2 were cloned and characterized from the thermotolerant and thermosensitive B. napus cultivars, respectively. A previous study found that the BjGLYI sequence showed significant homology with other GLYI sequences from plant and microbial species at the nucleotide and AA levels (Veena et al., 1999). In our study, the AA sequences of BnGLYI exhibited up to 79–98% identity to the AA sequences of GLYI from other seed plants. The GLYI coding sequence encodes for a 185 AA protein that contains zinc and GSH binding domains similar to the other reported GLYI proteins. Most interestingly, we found only two variations (Pro/Ala at position 9 and Ala/Val at position 93) between the AA sequences of BnGLYI-2 and BnGLYI-3.
There are 11 members of the GLYI family in both Arabidopsis and rice (Mustafiz et al., 2011). B. napus is an allotetraploid that may harbor many GLYI genes (at least two). After the release of the B. napus genomic sequence data5, BLAST searches were performed against the B. napus database. The results showed that the top hit was the assembled CDS of accession no. GSBRNA2T00156270001 using the AA sequence of BjGLYI (the peptides detected by MALDI-TOF MS analysis) as a query. When the corresponding CDS sequence was used as a query, the top hit was also GSBRNA2T00156270001. These results indicated that we cloned the gene identified by two-dimensional gel analysis. To fully understand the relationship between the two BnGLYI variants and resistance to temperature stress, we expressed the two BnGLYI genes in P. pastoris, a relatively simple system, and assessed their resistance to stresses in vitro. The results indicated that cells expressing the BnGLYI-3 gene were significantly resistant to heat and cold stress compared with cells harboring an empty vector or expressing BnGLYI-2. Thus, these results demonstrated that the Pro9Ala and Ala93Vla variants may be responsible for the different levels of resistance to thermal stress between the two transformants.
Although glyoxalases have been highly conserved throughout evolution, they exhibit a significant structural variation in terms of active site position and number (Mustafiz et al., 2014). In the case of yeast (possessing a monomeric molecular structure), the AA residue mutation E163Q/E318Q of GLYI substantially reduced its activity, and this result confirmed that the yeast GLYI contains two active sites (Frickel et al., 2001). Human glyoxalase I is a homodimeric protein (Ridderström et al., 1998). It has been proposed that residue 172 in the human enzyme, a glutamate, acts as the base for catalysis of the reaction (Ridderström et al., 1998; Cameron et al., 1999). Others have argued that residue 157 of human GLYI has no direct role in catalysis, but is rather involved in forming the substrate-binding site (Ridderström et al., 1997). Met157 was changed into Ala, His or Glu, which resulted in either sharply decreased activity or the same high specific activity as wild-type GLYI (Ridderström et al., 1997). In the present study, of the two variations, only residue 93 was in the metal binding site, according to the structure of GLYI. Converting Ala93 to Val93 caused the cells to lose their resistance to heat and cold stress. This variation might lead to structural changes that could be related to the catalytic activity of GLYI.
The glyoxalase system, a ubiquitous detoxification system, is considered to play an important role in the removal of MG, a cytotoxic compound whose abundance increases during various stresses (Yadav et al., 2005a). In plants, this pathway is considered to be associated with tolerance to various abiotic stresses. GLYI in B. juncea was reported to confer resistance to salt stress (Veena et al., 1999; Singla-Pareek et al., 2003). In onions, upregulation of GLYI activity was observed in response to various stresses (Hossain and Fujita, 2009). In addition to MG detoxification, the physiological significance of the glyoxalase system is the maintenance of redox homeostasis via the regeneration of GSH. In our study, overexpressing BnGLYI-3 gene from the heat-tolerant B. napus cultivar in yeast conferred resistance to heat and cold stress and significantly elevated GLYI activity compared to the control. These results suggest the protective role of enhanced BnGLYI-3 expression. Moreover, reduced GLYI activity results in MG accumulation (Shamsi et al., 2000). Thus, we speculated that an increase in activity of BnGLYI-3 could protect plants from heat and cold damage to a certain extent.
Furthermore, abiotic stress can stimulate ROS production and induce oxidative stress in plants (Hoque et al., 2012). During oxidative stress, the activity of antioxidant enzymes such as SOD generally increases in plants, and this elevated antioxidant enzyme activity correlates with increased stress tolerance (Gill and Tuteja, 2010). In a previous study, overexpression of a B. campestris HSP70 in tobacco clearly conferred enhanced tolerance to heat stress, and transgenic tobacco plants exhibited significantly higher SOD activity than wild-type plants. These results indicated that elevated SOD activity helps to alleviate the damage to the membrane system caused by heat stress (Wang et al., 2015). In our study, compared with the same cells under control condition, the SOD activity of GSGLYI-2 cells was decreased, but that of GSGLYI-3 cells was significantly increased after treatment at 42°C for 5 h. After treatment at –20°C for 13 D, GS3.5K and GSGLYI-2 cells exhibited decreased SOD activity, but GSGLYI-3 cells exhibited increased SOD activity. These results suggested thatGSGLYI-3 cells exhibit less accumulation of ROS and more effective growth than GS3.5K and GSGLYI-2 cells under temperature stress.
Conclusion
We have identified a BnGLYI gene, and the overexpression of a variant of this gene in yeast conferred thermotolerance. BnGLYI may also possess a similar function in B. napus seeds. Additionally, the BnGLYI activity level may be affected by the AA variation at position 93. The results of our study may shed light on the mechanisms by which plants cope with heat stress.
Author Contributions
Designed the experiments: GY and XW. Conducted the experiments: GY, XL, and XX. Analyzed the data: GY, XL, LW, and XX. Contributed materials/reagents/analysis tools: GY, XL, XX, GG, FL, JQ, JL, BC, KX, LW, and XW. Wrote the paper: GY.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank American Journal Experts for providing language assistance. We are grateful to Dr. LZ Chen and WH Ma (College of Plant Science and Technology, Huazhong Agricultural University) as well as YM Gong (Oil Crops Research Institute, Chinese Academy of Agricultural Sciences) for providing suggestions concerning protein expression. The National Natural Science Foundation of China (31100911) and the National Science and Technology Pillar Program during the Twelfth Five-Year Plan Period (2013BAD01B01 and 2013BAD01B03) funded the study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00150
Abbreviations
ROS, reactive oxygen species; GSH, glutathione (reduced); MG, methylglyoxal; GLYI, glyoxalase I; GLYII, glyoxalase II; BjGLYI, B. juncea GLYI gene; BnGLYI, B. napus GLYI gene.
Footnotes
- ^ http://www.matrixscience.com
- ^ http://www.lynnon.com.
- ^ http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
- ^ http://pfam.sanger.ac.uk/
- ^ http://BrassicaDB.org/brad/
References
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Cameron, A. D., Ridderström, M., Olin, B., Kavarana, M. J., Creighton, D. J., and Mannervik, B. (1999). Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue. Biochemistry 38, 13480–13490. doi: 10.1021/bi990696c
Candiano, G., Bruschi, M., Musante, L., Santucci, L., Ghiggeri, G. M., Carnemolla, B., et al. (2004). Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333. doi: 10.1002/elps.200305844
Chu, P., Chen, H., Zhou, Y., Li, Y., Ding, Y., Jiang, L., et al. (2012). Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta 235, 1271–1288. doi: 10.1007/s00425-011-1573-y
Clugston, S. L., Daub, E., and Honek, J. F. (1998). Identification of glyoxalase I sequences in Brassica oleracea and Sporobolus stapfianus: evidence for gene duplication events. J. Mol. Evol. 47, 230–234. doi: 10.1007/PL00006380
Damerval, C., Vienne, D. D., Zivy, M., and Thiellement, H. (1986). Technical improvement in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling protein. Electrophoresis 7, 53–54. doi: 10.1002/elps.1150070108
Deswal, R., Chakaravarty, T. N., and Sopory, S. K. (1993). The glyoxalase system in higher plants: regulation in growth and differentiation. Biochem. Soc. Trans. 21, 527–530. doi: 10.1042/bst0210527
Espartero, J., Sánchez-Aguayo, I., and Pardo, J. M. (1995). Molecular characterization of glyoxalase-I from a higher plant; up regulation by stress. Plant Mol. Biol. 29, 1223–1233. doi: 10.1007/BF00020464
Ferguson, G. P., Tötemeyer, S., MacLean, M. J., and Booth, I. R. (1998). Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170, 209–218. doi: 10.1007/s002030050635
Frickel, E. M., Jemth, P., Widersten, M., and Mannervik, B. (2001). Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 276, 1845–1849. doi: 10.1074/jbc.M005760200
Gan, L., Li, D. Y., Zang, X., Fu, C. H., Yu, L. J., and Li, M. T. (2010). Construction of protein two-dimensional polyacrylamide gel electrophoresis system for Brassica napus. Acta Agron. Sin. 36, 612–619. doi: 10.1016/S1875-2780(09)60046-8
Gan, L., Zhang, C. Y., Wang, X. D., Wang, H., Long, Y., Yin, Y. T., et al. (2013). Proteomic and comparative genomic analysis of two Brassica napus lines differing in oil content. J. Proteome Res. 12, 4965–4978. doi: 10.1021/pr4005635
Gao, G. Z., Wu, X. M., Lv, X. D., Chen, B. Y., Xu, K., and Yan, G. X. (2010). Genotype differences of seed viability in rapeseed during storage at different temperature. Chin. J. Oil Crop Sci. 32, 495–499.
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hajduch, M., Casteel, J. E., Tang, S., Hearne, L. B., Knapp, S., and Thelen, J. J. (2007). Proteomic analysis of near-isogenic sunflower varieties differing in seed oil traits. J. Proteome Res. 6, 3232–3241. doi: 10.1021/pr070149a
Hoque, T. S., Uraji, M., Ye, W. X., Hossain, M. A., Nakamura, Y., and Murata, Y. (2012). Methylglyoxal-induced stomatal closure accompanied by peroxidase-mediated ROS production in Arabidopsis. J. Plant Physiol. 169, 979–986. doi: 10.1016/j.jplph.2012.02.007
Hossain, M. A., and Fujita, M. (2009). Purification of glyoxalase I from onion bulbs and molecular cloning of its cDNA. Biosci. Biotechnol. Biochem. 73, 2007–2013. doi: 10.1271/bbb.90194
Hossain, M. A., Hasanuzzaman, M., and Fujita, M. (2011). Coordinate induction of antioxidant defense and glyoxalase system by exogenous proline and glycinebetaine is correlated with salt tolerance in mung bean. Front. Agric. China 5:1–14. doi: 10.1007/s11703-010-1070-2
Howarth, C. J. (2005). Genetic Improvements of Tolerance to High Temperature. New York, NY: Howarth Press.
Janmohammadi, M., Fallahnezhad, Y., Golsha, M., and Mohammadi, H. (2008). Controlled ageing for storability assessment and predicting seedling early growth of canola cultivars (Brassica napus L.). J. Agric. Biol. Sci. 3, 22–26.
Junaid, M. A., Kowal, D., Barua, M., Pullarkat, P. S., Sklower, B. S., and Pullarkat, R. K. (2004). Proteomic studies identified a single nucleotide polymorphism in glyoxalase I as autism susceptibility factor. Am. J. Med. Genet. A 131, 11–17. doi: 10.1104/pp.105.062257
Leprince, O., Atherton, N. M., Deltour, R., and Hendry, G. A. F. (1994). The involvement of respiration in free radical processes during loss of desiccation tolerance in germinating Zea mays L (An electron paramagnetic resonance study). Plant Physiol. 104, 1333–1339. doi: 10.1104/pp.104.4.1333
Li, D. D., Tai, F. J., Zhang, Z. T., Li, Y., Zheng, Y., and Wu, Y. F. (2009). A cotton gene encodes a tonoplast aquaporin that is involved in cell tolerance to cold stress. Gene 438, 26–32. doi: 10.1016/j.gene.2009.02.023
Lin, F., Xu, J., Shi, J., Li, H., and Li, B. (2010). Molecular cloning and characterization of a novel glyoxalase I gene TaGLY I in wheat (Triticum aestivum L.). Mol. Biol. Rep. 37, 729–735. doi: 10.1007/s11033-009-9578-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marcum, K. B. (1998). Cell membrane thermostability and whole plant heat tolerance of Kentucky bluegrass. Crop Sci. 38, 1214–1218. doi: 10.2135/cropsci1998.0011183X003800050017x
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Mustafiz, A., Ghosh, A., Tripathi, A. K., Kaur, C., Ganguly, A. K., Bhavesh, N. S., et al. (2014). A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78, 951–963. doi: 10.1111/tpj.12521
Mustafiz, A., Sahoo, K. K., Singla-Pareek, S. L., and Sopory, S. K. (2010). Metabolic engineering of glyoxalase pathway for enhancing stress tolerance in plants. Methods Mol. Biol. 639, 95–118. doi: 10.1007/978-1-60761-702-0_6
Mustafiz, A., Singh, A. K., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11, 293–305. doi: 10.1007/s10142-010-0203-2
Nagel, M., and Börner, A. (2010). The longevity of crop seeds stored under ambient conditions. Seed Sci. Res. 20, 1–12. doi: 10.1017/S0960258509990213
Paulus, C., Köllner, B., and Jacobsen, H. J. (1993). Physiological and biochemical characterization of glyoxalase I, a general marker for cell proliferation, from a soybean cell suspension. Planta 189, 561–566. doi: 10.1007/BF00198220
Ramaswamy, O., Pal, S., Guha-Mukherjee, S., and Sopory, S. K. (1984). Correlation of glyoxalase I activity with cell proliferation in Datura callus culture. Plant Cell Rep. 3, 121–124. doi: 10.1007/BF02441015
Ridderström, M., Cameron, A. D., Jones, T. A., and Mannervik, B. (1997). Mutagenesis of residue 157 in the active site of human glyoxalase I. Biochem. J. 328, 231–235. doi: 10.1042/bj3280231
Ridderström, M., Cameron, A. D., Jones, T. A., and Mannervik, B. (1998). Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I. J. Biol. Chem. 273, 21623–21628. doi: 10.1074/jbc.273.34.21623
Sethi, U., Basu, A., and Guha-Mukherjee, S. (1988). Control of cell proliferation and differentiation by regulating polyamine biosynthesis in cultures of Brassica and its correlation with glyoxalase-I activity. Plant Sci. 56, 167–175. doi: 10.1016/0168-9452(88)90031-3
Shamsi, F. A., Sharkey, E., Creighton, D., and Nagaraj, R. H. (2000). Maillard reactions in lens proteins: methylglyoxal-mediated modi?cations in the rat lens. Exp. Eye Res. 70, 369–380. doi: 10.1006/exer.1999.0800
Singla-Pareek, S. L., Reddy, M. K., and Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U.S.A. 100, 14672–14677. doi: 10.1073/pnas.2034667100
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., and Sopory, S. K. (2006). Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 140, 613–623. doi: 10.1104/pp.105.073734
Skipsey, M., Andrews, C. J., Townson, J. K., Jepson, I., and Edwards, R. (2000). Cloning and characterization of glyoxalase I from soybean. Arch. Biochem. Biophys. 374, 261–268. doi: 10.1006/abbi.1999.1596
Suzuki, N., and Mittler, R. (2006). Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol. Plant. 126, 45–51. doi: 10.1111/j.1399-3054.2005.00582.x
Thornalley, P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269, 1–11. doi: 10.1042/bj2690001
Tuomainen, M., Ahonen, V., Kärenlampi, S. O., Schat, H., Paasela, T., and Švanys, A. (2011). Characterization of the glyoxalase 1 gene TcGLX1 in the metal hyperaccumulator plant Thlaspi caerulescens. Planta 233, 1173–1184. doi: 10.1007/s00425-011-1370-7
Veena, Reddy, V. S., and Sopory, S. K. (1999). Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 17, 385–395. doi: 10.1046/j.1365-313X.1999.00390.x
Wahid, A., Gelani, S., Ashraf, M., and Foolad, M. R. (2007). Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223. doi: 10.1016/j.envexpbot.2007.05.011
Wang, J. Q., Yin, X., Wu, M. C., Zhang, H. M., Gao, S. J., Wei, J. T., et al. (2013). Expression of a family 10 xylanase gene from Aspergillus usamii E001 in Pichia pastoris and characterization of the recombinant enzyme. J. Ind. Microbiol. Biotechnol. 40, 75–83. doi: 10.1007/s10295-012-1201-2
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, X. R., Yan, B., Shi, M., Zhou, W., Zekria, D., Wang, H. Z., et al. (2015). Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma doi: 10.1007/s00709-015-0867-5 [Epub ahead of print].
Yadav, S. K., Singla-Pareek, S. L., Ray, M., Reddy, M. K., and Sopory, S. K. (2005a). Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 337, 61–67. doi: 10.1016/j.bbrc.2005.08.263
Yadav, S. K., Singla-Pareek, S. L., Reddy, M. K., and Sopory, S. K. (2005b). Methylglyoxal detoxification by glyoxalase system: a survival strategy during environmental stresses. Physiol. Mol. Biol. Plants 11, 1–11.
Zhang, T., Ge, C., Deng, L., Tan, T., and Wang, F. (2015). C4-dicarboxylic acid production by overexpressing the reductive TCA pathway. FEMS Microbiol. Lett. 362, fnv052. doi: 10.1093/femsle/fnv052
Zhou, Y., Chen, H., Chu, P., Li, Y., Tan, B., Ding, Y., et al. (2012). NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 31, 379–389. doi: 10.1007/s00299-011-1173-0
Keywords: Glyoxalase I gene, seed thermotolerance, proteomics, Brassica napus L., yeast
Citation: Yan G, Lv X, Gao G, Li F, Li J, Qiao J, Xu K, Chen B, Wang L, Xiao X and Wu X (2016) Identification and Characterization of a Glyoxalase I Gene in a Rapeseed Cultivar with Seed Thermotolerance. Front. Plant Sci. 7:150. doi: 10.3389/fpls.2016.00150
Received: 23 September 2015; Accepted: 28 January 2016;
Published: 16 February 2016.
Edited by:
Richard S. Winder, Natural Resources Canada, CanadaReviewed by:
Biswapriya Biswavas Misra, University of Florida, USAElisabetta Mazzucotelli, Consiglio per la Ricerca e Sperimentazione in Agricoltura, Italy
Copyright © 2016 Yan, Lv, Gao, Li, Li, Qiao, Xu, Chen, Wang, Xiao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Wu, d3V4bUBvaWxjcm9wcy5jbg==
 Guixin Yan
Guixin Yan Xiaodan Lv
Xiaodan Lv Jiangwei Qiao
Jiangwei Qiao Biyun Chen
Biyun Chen Xiaoming Wu
Xiaoming Wu