- 1Institute of Plant Nutritional Physiology and Molecular Biology, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Institute of Materia Medica, Fujian Academy of Medical Sciences, Fuzhou, China
- 5College of Resource and Environmental Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 6The Higher Educational Key Laboratory of Fujian Province for Soil Ecosystem Health and Regulation, Fujian Agriculture and Forestry University, Fuzhou, China
- 7Fujian Key Laboratory for Plant Molecular and Cell Biology, Fujian Agriculture and Forestry University, Fuzhou, China
Magnesium (Mg)-deficiency, which affects crop productivity and quality, widespreadly exists in many agricultural crops, including citrus. However, very limited data are available on Mg-deficiency-responsive microRNAs (miRNAs) in higher plants. Using Illumina sequencing, we isolated 75 (73 known and 2 novel) up- and 71 (64 known and 7 novel) down-regulated miRNAs from Mg-deficient Citrus sinensis leaves. In addition to the remarkable metabolic flexibility as indicated by the great alteration of miRNA expression, the adaptive responses of leaf miRNAs to Mg-deficiency might also involve the following several aspects: (a) up-regulating stress-related genes by down-regulating miR164, miR7812, miR5742, miR3946, and miR5158; (b) enhancing cell transport due to decreased expression of miR3946 and miR5158 and increased expression of miR395, miR1077, miR1160, and miR8019; (c) activating lipid metabolism-related genes by repressing miR158, miR5256, and miR3946; (d) inducing cell wall-related gene expansin 8A by repressing miR779; and (e) down-regulating the expression of genes involved in the maintenance of S, K and Cu by up-regulating miR395 and miR6426. To conclude, we isolated some new known miRNAs (i.e., miR7812, miR8019, miR6218, miR1533, miR6426, miR5256, miR5742, miR5561, miR5158, and miR5818) responsive to nutrient deficiencies and found some candidate miRNAs that might contribute to Mg-deficiency tolerance. Therefore, our results not only provide novel information about the responses of plant to Mg-deficiency, but also are useful for obtaining the key miRNAs for plant Mg-deficiency tolerance.
Introduction
Magnesium (Mg), which serves as a central component of the chlorophyll (Chl) molecule and as a cofactor and allosteric modulator for more than 300 enzymes including ribulose-1,5-bisphosphate carboxylase, ATPase, protein kinases, phosphatases and glutathione synthase (Cakmak and Kirkby, 2008), participates in many physiological and biochemical processes during plant growth and development including photosynthesis (Tang et al., 2012; Yang et al., 2012), respiration (Peng et al., 2015), organic acid metabolism (Yang et al., 2013), carbohydrate partitioning between source and sink organs (Cakmak et al., 1994; Yang et al., 2012), phloem export of sucrose (Cakmak and Kirkby, 2008), and reactive oxygen species (ROS) formation and scavenging (Cakmak and Kirkby, 2008; Yang et al., 2012). Mg-deficiency, which affects crop productivity and quality, widespreadly exists in many agricultural crops, including citrus (Tang et al., 2012; Verbruggen and Hermans, 2013). In China, Mg-deficiency often occurs in citrus orchards, and is responsible for the loss of yield and poor fruit quality (Ling et al., 2009). According to our investigation in 2011, up to 77.4 and 35.6% of “Guanximiyou” pummelo (Citrus grandis) orchards in Pinghe, Zhangzhou, China were deficient in soil exchange Mg and leaf Mg, respectively (Li et al., 2015).
Although the physiological and biochemical responses of plants to Mg-deficiency have been investigated in some detail in various plants (Cakmak and Kirkby, 2008; Hermans et al., 2013; Verbruggen and Hermans, 2013), very limited data are available on the molecular mechanisms of plant tolerance to Mg-deficiency until recently. Peng et al. (2015) isolated 59 up- and 31 down-regulated (19 up- and 12 down-regulated) proteins from Mg-deficient Citrus sinensis leaves (roots). In addition, two studies with Arabidopsis showed that the responses of global transcriptomics to Mg-deficiency were asynchronized, with a less number of differentially expressed genes after 4 or 8 h in leaves and after 28 h or 1 week in roots (Hermans et al., 2010a,b). Although stress-related gene expression programme largely occurs at the transcriptional level, the roles of post-transcriptional gene regulation have been recognized with the discovery of microRNAs (miRNAs) and small-interfering RNAs (siRNAs; Shukla et al., 2008). Approximately 21-nucleotide-long miRNAs generated from non-coding transcripts, one of the most abundant classes of small RNAs, are crucial post-transcriptional regulators of gene expression by repressing translation or directly degrading mRNAs in plants (Jones-Rhoades et al., 2006). MiRNAs have key roles in plant adaptations to nutrient deficiencies (Khraiwesh et al., 2012; Lu et al., 2014, 2015; Zeng et al., 2014; Paul et al., 2015). Plant miR399 and miR827, which are specifically induced by phosphorus (P)-deficiency, play a role in the regulation of P homeostasis by down-regulating their target genes UBC24 and nitrogen (N) limitation adaptation (NLA), respectively (Shukla et al., 2008; Hsieh et al., 2009). Also, many other miRNAs such as miR156, miR159, miR166, miR169, miR395, miR397, miR398, miR408, miR447, miR482, miR1510 and miR2109 are involved in plant response to P-limitation (Valdés-López et al., 2010; Hackenberg et al., 2013; Zhao et al., 2013; Paul et al., 2015).
Recently, Lu et al. (2014, 2015) investigated long-term B-deficiency-responsive miRNAs by Illumina sequencing and obtained 134 (112 known and 22 novel) and 172 (158 known and 14 novel) differentially expressed miRNAs from B-deficient C. sinensis roots and leaves, respectively, demonstrating the possible involvement of miRNAs in the tolerance of citrus plants to B-deficiency. It is worth noting that most of these B-deficiency-responsive miRNAs were identified only from B-deficient leaves or roots, only 22 miRNAs were identified from the both. Obviously, long-term B-deficiency-induced alterations of miRNA expression profiles greatly differed between leaves and roots.
In Arabidopsis, miR857, miR408, miR398, and miR397, which are up-regulated by copper (Cu)-deficiency, have been demonstrated to contribute to plant Cu homeostasis via negatively regulating nonessential Cu protein genes, thus saving Cu for other essential Cu proteins in Cu-deprived plants (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008). Waters et al. (2012) reported that iron (Fe)-deficient Arabidopsis rosettes had more accumulation of Cu accompanied by decreased expression levels of miR397a, miR398a, and miR398b/c, indicating a link between Fe-deficiency and Cu homeostasis.
Many differentially expressed miRNAs (at least 27conserved families) have been isolated from N-deficient soybean, common bean, Arabidopsis and maize (Valdés-López et al., 2010; Liang et al., 2012; Zhao et al., 2012; Zeng et al., 2014). In Arabidopsis, miR169 was greatly repressed and its target genes, NFYA (Nuclear Factor Y, subunit A) family members, were greatly up-regulated by N-deficiency. Transgenic Arabidopsis plants over-expressing miR169a had lower N level, and displayed less tolerance to N-deficiency than the wild type, indicating the possible roles of miR169 in helping plants to deal with N-starvation (Zhao et al., 2011).
MiR395, which targets two sulfur (S) metabolism-related genes [i.e., ATP sulfurylases (APS) and sulfate transporter 2;1 (SULTR2;1)], was induced by S-deprivation. MiR395-over-expressing Arabidopsis exhibited remarkable down-regulation in mRNA levels of its two target genes, and had more accumulation of S in the shoot but not in the root. The aps1-1 sultr2;1 APS4-RNAi mutants displayed similar phenotypes to those of miR395-over-expressing plants. These authors concluded that miR395-mediated regulation of APS and SULTR2;1 might play a crucial role in plant S homeostasis (Liang et al., 2010).
So far, many workers have investigated the roles of plant miRNAs in response to nutrient deficiencies. Most studies, however, have focused on P, B, N, Fe, and S deficiencies. Little is known about Mg-deficiency-responsive miRNAs in higher plants. In this study, we first sequenced two small RNA libraries from Mg-deficient and -sufficient (control) C. sinensis leaves, respectively, using Illumina sequencing in order to identify the Mg-deficiency-responsive miRNAs that might contribute to the tolerance of plants to Mg-deficiency.
Materials and Methods
Plant Materials and Mg Treatments
This study was conducted at Fujian Agriculture and Forestry University (FAFU), Fuzhou, China (26°5′ N, 119°14′ E) with an average annual temperature of ca. 20°C and an average annual sunlight hours of ca. 1600 h. Plant culture and Mg treatments were performed according to Peng et al. (2015). Briefly, 15-week-old seedlings of “Xuegan” [Citrus sinensis (L.) Osbeck] in 6 L pots filled with fine river sand were grown in a greenhouse under natural photoperiod at FAFU. Each pot, which contained two seedlings, was irrigated every other day until saturated with nutrient solution containing 2.5 mM KNO3, 2.5 mM Ca(NO3)2, 0.5 mM KH2PO4, 10 μM H3BO3, 2 μM MnCl2, 2 μM ZnSO4, 0.5 μM CuSO4, 0.065 μM (NH4)6Mo7O24, 20 μM Fe-EDTA and 0 mM (Mg-deficiency) or 1 mM (Mg-sufficiency) MgSO4 for 16 weeks. Sulfur concentration was maintained at a constant level by using equivalent moles of Na2SO4 in replace of MgSO4. At the end of the experiment, fully-expanded (about 7 weeks old) leaves from different replicates and treatments were used for all the measurements. Leaves were collected at noon under full sun and immediately frozen in liquid N2, then stored at −80°C until extraction.
Plant Dry Weight (DW) and Leaf Mg Concentration
At the end of the experiment, 9–10 plants per treatment from different pots were collected. Plant DW was measured after being dried at 70°C for 48 h. Leaf Mg concentration was assayed by atomic absorption spectroscopy after digested with 1 N HCl (Kushizaki, 1968).
Leaf SRNA Library Construction and Illumina Sequencing
Equal amounts of frozen Mg-deficient or -sufficient leaves from five plants (one plant per pot) were mixed as a biological replicate. Total RNA was extracted from 0.1 g mixed frozen samples using TRIzol reagent (Invitrogen, Carlsbad, CA) following manufacturer's instructions. Mg-deficient and -sufficient leaf sRNA libraries were constructed according to Lu et al. (2014). Illumina sequencing was performed on a Solexa sequencer at the Beijing Genomics Institute (BGI), Shenzhen, China (Lu et al., 2014).
SRNA Annotation and MiRNA Identification
Both sRNA annotation and miRNA identification were performed according to Lu et al. (2014). Briefly, software developed by the BGI was used to analyze the raw data from the Solexa sequencing. Clean reads were then used to calculate length distribution and common/specific sequences. Thereafter, the clear reads were mapped to C. sinensis genome (JGIversion 1.1, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Csinensis) using SOAP, only perfectly mapped sequences were retained and analyzed further. RRNAs, tRNAs, snRNAs and snoRNAs were removed from the sRNAs sequences through BLASTn search using NCBI Genebank database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi/) and Rfam (12.0) database (http://www.sanger.ac.uk/resources/databases/rfam.html) using following program and parameters: blastall -p blastn -F F -e 0.01. The repeat associated RNA and piRNA were identified using tag2repeat and tag2piRNA (developed by BGI) respectively. SRNA tags were also aligned to exons and introns of mRNA to find the degraded fragments of mRNA. All annotations were summarized using tag2annotation software (developed by BGI) in the following order of preference: rRNA (Genbank > Rfam) > known miRNA > repeat > exon > intron. The remaining sequences were aligned with known plant miRNAs from miRBase 21 (http://www.mirbase.org/) with up to two mismatches. Reads that were not annotated were used to predict novel miRNAs using Mireap (http://sourceforge.net/projects/mireap/), a prediction software developed by the BGI, by exploring the secondary structure, the Dicer cleavage site and the minimum free energy of the unannotated small RNA tags which could be mapped to genome. Parameters were set as follows: minimal miRNA sequence length (18), maximal miRNA sequence length (25), minimal miRNA reference sequence length (20), maximal miRNA reference sequence length (23), maximal copy number of miRNAs on reference (20), maximal free energy allowed for a miRNA precursor (−18 kcal/mol), maximal space between miRNA and miRNA* (300), minimal base pairs of miRNA and miRNA* (16), maximal bulge of miRNA and miRNA* (4), maximal asymmetry of miRNA/miRNA* duplex (4) and flank sequence length of miRNA precursor (20; Lu et al., 2014). In addition, we used MTide (http://bis.zju.edu.cn/MTide; Zhang et al., 2015) and DNAMAN 8 (http://www.lynnon.com/pc/framepc.html) to predict novel miRNA. Only these miRNA candidates that were simultaneously predicted by the three softwares were considered to be novel miRNAs.
Differential Expression Analysis of MiRNAs
Both the fold change between Mg-deficiency and -sufficiency and the P-value were calculated from the normalized expression of TPM (Wang et al., 2011). Normalized expression was calculated by the following formula: Normalized expression = Actual miRNA count/Total count of clean reads*1,000,000. The fold change between B-deficiency and control was calculated as: Fold-change = log2 (B-deficiency/Control). The p-value was calculated by the following formula:
A miRNA was considered differentially expressed when the miRNA had both a P-value of less than 0.01 and a fold change of more than 1.5 (Lu et al., 2014).
Target Prediction of MiRNAs
Target prediction of miRNAs was performed by RNAhybrid based on rules suggested by Allen et al. (2005) and Schwab et al. (2005): (a) no more than four mismatches between sRNA and target (G-U bases count as 0.5 mismatches); (b) no more than two adjacent mismatches in the miRNA/target duplex; (c) no adjacent mismatches in positions 2–12 of the miRNA/target duplex (5′ of miRNA); (d) no mismatches in positions 10–11 of miRNA/target duplex; (e) no more than 2.5 mismatches in positions 1–12 of the of the miRNA/target duplex (5′ of miRNA); and (f) minimum free energy (MFE) of the miRNA/target duplex should be >75% of the MFE of the miRNA bound to it's perfect complement.
Functions of the Potential Targets of the Differentially Expressed MiRNAs
All targets of the differentially expressed miRNAs were mapped to GO terms in the database (http://www.geneontology.org/), and calculated gene numbers for each term. The GO results were expressed as three categories: biological process, molecular function and cellular component (Lu et al., 2014).
Validation of MiRNA Expression by Stem-Loop qRT-PCR
The analysis of miRNA expression was performed using stem-loop qRT-PCR method, stem-loop primers for reverse transcription and primers for qRT-PCR were listed in Table S1. Total RNA was reversetranscribed using Taqman® MicroRNA Reverse Transcription Kit (USA). SYBR® Premix Ex Taq™ II (Takara, Japan) kit was used for qRT-PCR. MiRNA special (forward) primers were designed according to the miRNA sequence but excluded the last six nucleotides at 3′ end of the miRNA. A 5′ extension of several nucleotides, which was chosen randomly and relatively GC-rich, was added to each forward primer to increase the melting temperature (Chen et al., 2005). All the primers were assigned to Primer Software Version 5.0 (PREMIER Biosoft International, USA) to assess their quality. For qRT-PCR, 20 μL reaction solution contained 10 μL ready-to-use SYBR® Premix Ex TaqTM II (Takara, Japan), 0.8 μL 10 μM miRNA forward primer, 0.8 μL 10 μM Uni-miR qPCR primer, 2 μL cDNA template and 6.4 μL dH2O. The cycling conditions were 60 s at 95°C, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s. qRT-PCR was performed on the ABI 7500 Real Time System. Samples for qRT-PCR were run in three biological replicates with two technical replicates. Relative miRNA expression was calculated using ddCt algorithm. For the normalization of miRNA expression, actin (AEK97331.1) was used as an internal standard and the leaves from control plants were used as reference sample, which was set to 1.
qRT-PCR Analysis of MiRNA Target Gene Expression
Total RNA was extracted from frozen Mg-sufficient and -deficient leaves using TRIzol reagent (Invitrogen, Carlsbad, CA) following manufacturer's instructions. The sequences of the F and R primers used were given in Table S2. qRT-PCR analysis of miRNA target gene expression was performed using an ABI 7500 Real Time System according to Lu et al. (2014). Samples for qRT-PCR were run in three biological replicates with two technical replicates.
Experimental Design and Statistical Analysis
There were 20 pot seedlings per treatment in a completely randomized design. Experiments were performed with 3 replicates except for plant DW (n = 9–10) and leaf Mg concentration (n = 5). The unpaired t-test was applied for comparison between two means.
Results
Plant Growth and Leaf Mg Concentration
As shown in Figure 1, 0 mM Mg-treated seedlings had lower plant DW and leaf Mg concentration than 1 mM Mg-treated ones, and leaf Mg concentration was much less than the normal range (Chapman, 1968). Visible Mg-deficient symptoms were observed only in 0 μM Mg-treated leaves (Figure S1). Thus, seedlings treated with 0 mM Mg are considered as Mg-deficient, and those treated with 1 mM Mg are considered as Mg-sufficient (control).
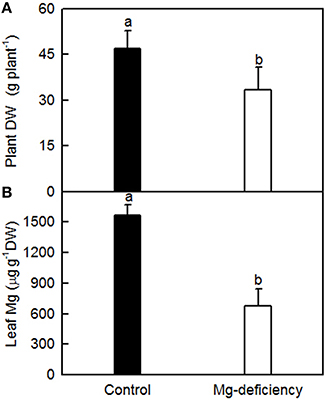
Figure 1. Effects of Mg-deficiency on plant growth (A) and leaf Mg concentration (B). Bars represent mean ± SD (n = 5 for leaf Mg and 9–10 for plant DW). Different letters above the bars indicate a significant difference at P < 0.05.
Sequencing and Analysis of Two SRNA Libraries from Mg-Sufficient and -Deficient Citrus Leaves
To isolate Mg-deficiency-responsive miRNAs, two sRNA libraries were constructed from leaves of C. sinensis seedlings submitted to 0 or 1 mM MgSO4 for 16 weeks, respectively. After being sequenced by a Solexa sequencer, we obtained 20,602,570 and 22,513,099 raw reads from Mg-sufficient and -deficient leaf libraries, respectively. These raw reads were cleaned by removing the contaminant reads like adaptors and low quality tags, thus leading to the generation of 20,328,011 (4,024,507) and 22,218,850 (4,480,037) clear reads (unique reads) from Mg-deficient and -deficient libraries, respectively (Table 1). Most of the clear sequences were within the range of 20–24 nt, which accounted for 94 and 96% of the total clear reads from Mg-deficient and -sufficient leaves, respectively. Reads with the length of 24 nt were at the most abundant, followed by the reads with length of 21, 22, 23, and 20 nt (Figure S2), as found for fruits of C. sinensis (Xu et al., 2010) and Poncirus trifoliata (Song et al., 2010), roots and leaves of C. sinensis (Lu et al., 2014, 2015), and flowers of P. trifoliata (Song et al., 2010). Therefore, the data of sRNA libraries obtained in this study are reliable. Compared with the controls, Mg-deficient leaves displayed less 21 and 24 nt clean reads and more 22 and 23 nt clean reads (Figure S2).
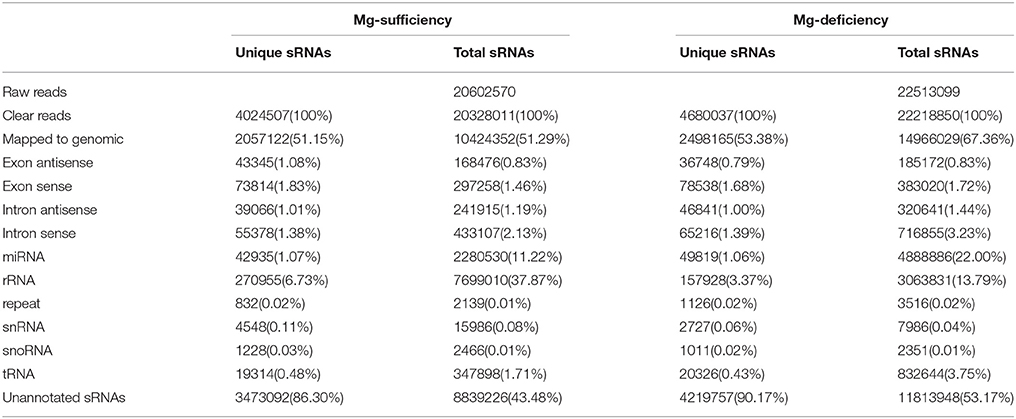
Table 1. Statistical analysis of sRNA sequencing data from Mg-sufficient and -deficient leaves of Citrus sinensis.
As shown in Table 1, 10,424,352 (2,057,122) and 14,966,029 (2,498,165) clean reads (unique reads) from Mg-sufficient and -deficient leaves, respectively were mapped to C. sinensis genome (JGIversion 1.1, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Csinensis) using SOAP (Li et al., 2008). After removal of these annotated reads such as exon, intron, miRNA, rRNA, repeat regions, snRNA, shorn and tRNA, the remained unique read used for the prediction of novel miRNAs for Mg-sufficient and -deficient leaves were 3,473,092 and 4,219,757 reads, respectively.
Isolation of Known and Novel MiRNAs in Citrus Leaves
We isolated 691 known miRNAs from the two libraries constructed from Mg-sufficient and -deficient leaves (Table S3). To compare the abundance of miRNAs in the two libraries, the count of reads was normalized to transcript per million (TPM). In this experiment, known miRNAs with normalized read-count less than ten in the two leaf libraries were not used for further analysis in order to avoid false results caused by the use of low expressed miRNAs (Chen et al., 2012; Lu et al., 2014). After these low expressed miRNAs being excluded, the remained 288 known miRNAs were used for further analysis (Table S4).
As shown in Table 1, the unannotated 3,473,092 and 4,219,757 unique clean reads from Mg-sufficient and -deficient leaf libraries, respectively were used to predict the novel miRNAs. Based on the criteria for annotation of plant miRNAs (Jones-Rhoades et al., 2006; Meyers et al., 2008), we obtained 113 novel miRNAs from the two libraries (Table S5). Like known miRNAs, only 34 novel miRNAs with normalized read-count more than 10 in Mg-sufficient and/or Mg-deficient libraries were used for the expression analysis (Table S6).
Mg-Deficiency-Responsive MiRNAs in Citrus Leaves
A miRNA was considered differentially expressed when it had both a P-value of less than 0.01 and a fold-change of more than 1.5. According to the above criteria, we identified 75 (73 known and 2 novel) up-regulated and 71 (64 known and 7 novel) down-regulated miRNAs from Mg-deficient leaves. The strongest up-regulated known (novel) and down-regulated known (novel) miRNAs were miR5832 with a fold-change of 17.61 (novel_miR_96 with a fold-change of 17.75) and miR4351 with a fold-change of -14.66 (novel_miR_243 with a fold-change of -13.08), respectively (Tables S7, S8).
Validation of Illumina Sequencing Data by qRT-PCR
Since only one mixed sample of Mg-sufficient or -deficient leaf RNA was sequenced, the expression levels of 39 known miRNAs were analyzed using stem-loop qRT-PCR to validate the miRNA expression patterns revealed by Illumina sequencing. The expression levels of all these miRNAs except for miR1222, miR7730, and miR5832 matched with the expression patterns obtained by Illumiona sequencing (Figure 2). Thus, the high-throughput sequencing allowed us to identify the Mg-deficiency-responsive miRNAs in citrus leaves.
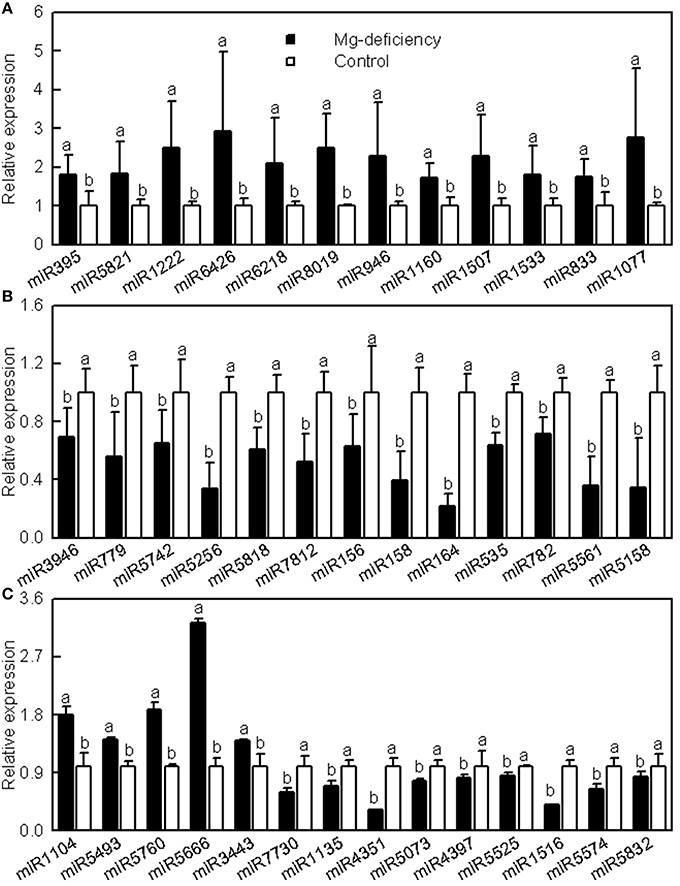
Figure 2. Relative abundances of selected known miRNAs in Mg-deficient and -sufficient (control) leaves revealed by qRT-PCR. Bars represent mean ± SD (n = 3). Significant differences were tested between control and Mg-deficient leaves for the same miRNA. Different letters above the bars indicate a significant difference at P < 0.05. All the values were expressed relative to the control leaves.
Identification of Targets for Mg-Deficiency-Responsive MiRNAs and GO Analysis
We predicted 187 and 24 target genes from the 57 known and 3 novel differentially expressed miRNAs, respectively (Tables S9, S10). Based on the biological process, these target genes for known (novel) were mainly involved in response to stress, regulation of transcription, biological process, protein metabolic process and transport (regulation of transcription and developmental process; Figure 3A). Based on the molecular function, these target genes for the known and novel miRNAs genes were classified into 17 and four categories, respectively, the highest percentage of three categories for known miRNAs were transcription factor activity, nucleic acid binding and other activity (Figure 3B). Based on the cellular component, these target genes for the known and novel miRNAs were associated with 13 and two components, respectively. The most three GO terms for known miRNAs were nucleus, membrane and chloroplast (Figure 3C).
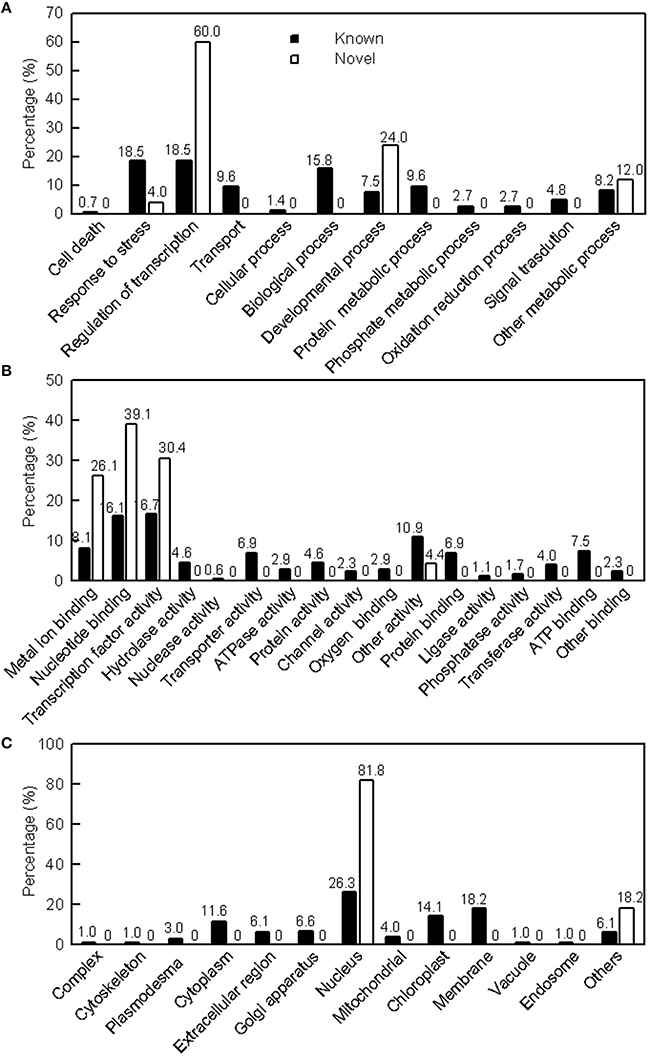
Figure 3. GO of the predicted target genes for 57 (3) differentially expressed known (novel) miRNAs in Citrus sinensis leaves. Categorization of miRNAs target genes was performed according to biological process (A), molecular function (B), and cellular component (C).
qRT-PCR Validation of Target Genes
We used qRT-PCR to assay the transcript levels of 77 genes targeted by 12 down- and 11 up-regulated miRNAs in order to verify the expression of the target genes and how the miRNAs regulate their target genes. As shown in Table 2, 56 (73%) target genes displayed the expected reverse changes in mRNA levels in Mg-deficient leaves with their corresponding miRNAs, demonstrating the possible roles of miRNAs in regulating gene expression under Mg-deficiency by cleaving mRNAs. However, 17 target genes had the similar change trends in transcript levels in Mg-deficient leaves with their corresponding miRNAs. The remaining four target genes were not detected in Mg-sufficient and -deficient C. sinensis leaves.
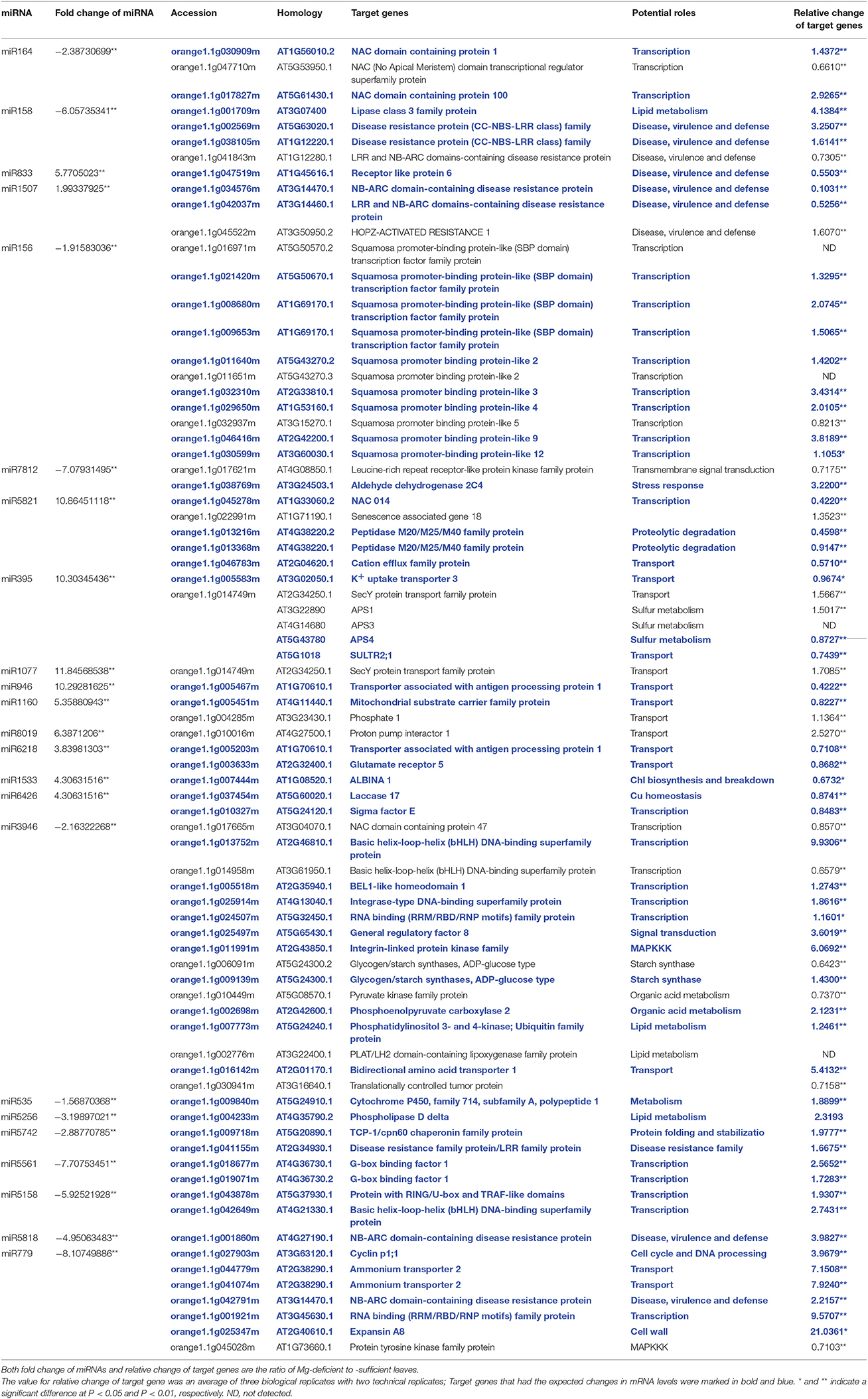
Table 2. qRT-PCR relative expression of experimentally determined or predicted target genes of selected miRNAs.
Discussion
In addition to their involvement in plant growth and development, evidence in Arabidopsis, C. sinensis, barley (Hordeum vulgare), soybean (Glycine max), white lupin (Lupinus albus), common bean (Phaseolus vulgaris), rapeseed (Brassica napus), tomato (Solanum lycopersicum), maize (Zea mays), and wheat (Triticum aestivum) shows that miRNAs play key roles in the adaptations of plants to P, Cu, Fe, N, and B deficiencies (Hsieh et al., 2009; Kong and Yang, 2010; Liang et al., 2010, 2012; Valdés-López et al., 2010; Zhao et al., 2012, 2013; Hackenberg et al., 2013; Lu et al., 2014, 2015; Zeng et al., 2014; Paul et al., 2015). Here, we isolated 137 known and nine novel Mg-deficiency-responsive miRNAs from C. sinensis leaves, respectively (Tables S7, S8), demonstrating the possible involvement of miRNAs in the tolerance of plants to Mg-deficiency.
The expression level of miR164 was decreased in Mg-deficient leaves (Table 2), as found for transient low nitrate-stressed maize leaves (Xu et al., 2011) and water stressed cassava (Manihot esculenta) leaves (Phookaew et al., 2014). As expected, the expression of its target genes (NAC domain containing protein 1 and NAC domain containing protein 100) was induced in Mg-deficient leaves (Table 2). Transgenic rice over-expressing NAC1 and NAC6 displayed higher drought and salt tolerance (Hu et al., 2006; Nakashima et al., 2007), and SINAC4-RNAi tomato plants were more sensitive to drought and salt stress (Zhu et al., 2014). Therefore, Mg-deficiency-induced down-regulation of leaf miR164 might play a role in the tolerance of plants to Mg-deficiency by enhancing the expression of NAC. However, the expression of NAC domain containing protein 47 targeted by miR3946 and NAC 014 targeted by miR5821 was down-regulated in Mg-deficient leaves. Xu et al. (2011) observed that the expression level of miR164 in maize leaves increased in response to chronic N limitation, concluding that miR164 might function in remobilizing the N from old to new leaves via boost senescence due to decreased expression of NAC under N limitation.
The expression level of miR158 was lower in Mg-deficient leaves than in controls (Table 2). Similar results have been obtained on N-deficient Arabidopsis seedlings (Liang et al., 2012), B-deficient C. sinensis roots (Lu et al., 2014) and leaves (Lu et al., 2015). The observed lower expression level of miR158 indicated that its target genes might be induced in these leaves. In fact, the expression of all target genes (i.e., one lipase class 3 family protein and two disease resistance protein (CC-NBS-LRR class) family) were up-regulated in Mg-deficient citrus leaves except for LRR and NB-ARC domains-containing disease resistance. Also, the expression levels of HOPZ-ACTIVATED RESISTANCE 1 targeted by miR1507, disease resistance family protein/LRR family protein targeted by miR5742, and NB-ARC domain-containing disease resistance protein (AT4G27190.1 and AT3G14470.1) targeted by miR5818 and miR779 were increased in Mg-deficient leaves (Table 2). These implied that the disease resistance might be enhanced in Mg-deficient leaves. This agrees with our results that Mg-deficient citrus leaves had higher concentrations of K and Ca (Xu, 2015), which play a role in plant disease resistance (Amtmann et al., 2008; Huber and Jones, 2013). However, the expression levels of receptor like proteins 6 targeted by miR833, LRR and NB-ARC domains-containing disease resistance protein and NB-ARC domain-containing disease resistance protein targeted by miR1507 were decreased in Mg-deficient leaves (Table 2).
MiR156, which targets a series of squamosa promoter binding protein-like (SPL) genes, determines plastochron length by regulating SPL levels (Wang et al., 2008). As shown in Table 2, most of the target genes showed expected reverse changes in mRNA levels in Mg-deficient leaves compared with miR156. Transgenic Arabidopsis over-expressing miR156h displayed enhanced rate of leaf initiation (Stief et al., 2014). A similar effect was detected in the spl9 spl15 double mutant (Wang et al., 2008). The observed down-regulation of miR156 in Mg-deficient leaves indicated that the rate of leaf initiation might be decreased in Mg-deficient seedlings, thus decreasing leaf number and leaf DW (Peng et al., 2015). Study showed that increased miR156 activity resulted in high concentration of anthocyanins, while decreased miR156 activity led to the accumulation of flavonols (Gou et al., 2011). Therefore, Mg-deficient citrus leaves might have less accumulation of anthocyanin and more accumulation of flavonols due to decreased abundance of miR156 (Table 2).
MiR7812 was repressed and its target gene aldehyde dehydrogenase (ALDH) 2C4 was induced in Mg-deficient leaves (Table 2). Aldehyde dehydrogenases, which catalyze the oxidation of aldehydes arising from reactions of ROS with lipids and proteins to carboxylic acids, function in the detoxification of aldehydes generated in plants exposed to abiotic stress. Over-expression of ALDH3I1 and ALDH7B4 in Arabidopsis increased tolerance to abiotic stresses and protected plants against lipid peroxidation and oxidative stress (Kotchoni et al., 2006). Transgenic Arabidopsis over-expressing a stress-inducible ALDH from Arabidopsis displayed enhanced stress tolerance, which was correlated with decreased accumulation of lipid peroxidation-derived reactive aldehydes compared to wild-type plants (Sunkar et al., 2003). Heat shock proteins (HSPs)/chaperones play crucial roles in protecting plants against stress by reestablishing normal protein conformation and thus cellular homeostasis. Our finding that Mg-deficiency increased leaf expression of TCP-1/cpn60 chaperonin family protein targeted by 5742 (Table 2) agrees with the report that the abundances of several HSPs were increased in Mg-deficient C. sinensis leaves (Peng et al., 2015).
Our finding that miR395 was induced in Mg-deficient leaves (Table 2) agrees with the report that miR395 in Arabidopsis leaves was enhanced by S-deficiency. MiR395 targets APS1, APS2, APS4 and sulfate transporter 2;1 (SULTR2;1). Their transcripts were decreased in transgenic Arabidopsis over-expressing miR395 accompanied by increased accumulation of S in the shoot but not in the root. MiR395 might play a role in the regulation of plant S accumulation and allocation by targeting APS and SULTR2;1 (Liang et al., 2010). As expected, the expression of APS4 and SULTR2;1 was down-regulated in Mg-deficient leaves (Table 2). Therefore, Mg-deficiency-induced up-regulation of leaf miR395 might contribute to the homeostasis of S in plants, which agrees with our data that Mg-deficiency did not significantly affect S concentration in C. sinensis roots, stems and leaves (Xu, 2015). However, the expression of APS1 was up-regulated in Mg-deficient leaves (Table 2). Also, K+ uptake transporter 3 (KUP3), a target gene of miR395 was inhibited in Mg-deficient leaves (Table 2). Kim et al. (1998) showed that AtKUP3 transcripts increased in K+-starved Arabidopsis roots. Because K concentration was higher in Mg-deficient C. sinensis roots, stems and leaves than in controls (Xu, 2015), the down-regulation of KUP3 might provide an adaptive strategy of plants to Mg-deficiency by lowering K uptake, thus maintaining nutrient balance. Thus, it is reasonable to assume that miR395 played a role in the maintenance of S and K homeostasis.
Study showed that Arabidopsis phosphate 1 (PHO1; At3g23430) played a role in the regulation of P homeostasis through the phosphate (Pi) loading to the xylem (Wang et al., 2004). The pho1 mutant of Arabidopsis had ca. 95% less Pi and 50–75% less total P in shoots than wild-type plants (Poirier et al., 1991). As shown in Table 2, the expression levels of both miR1160 and its target gene PHO1 were enhanced in Mg-deficient leaves. The up-regulation of PHO1 might be advantageous to alleviating Mg-deficiency-induced decrease in leaf P level, since Mg-deficient C. sinensis plants accumulated less P in roots, stems and leaves than controls (Xu, 2015).
Both leaf miR8019 and its target gene: proton pump interactor 1 (PPI1) were induced by Mg-deficiency (Table 2). Anzi et al. (2008) showed that PPI1 stimulated in vitro activity of plasma membrane (PM) H+-ATPase. Thus, its activity might be enhanced in these leaves. This agrees with the previous reports that Fe-deficiency strongly increased PM H+-ATPase activity in cucumber roots (Dell'Orto et al., 2000), and that P-deficient soybean roots had increased PM H+-ATPase activity (Shen et al., 2006), because the concentrations of P and Fe were lower in Mg-deficient roots, stems and leaves than in controls (Xu, 2015).
MiR1533 was up-regulated in Mg-deficient leaves and its target gene: ALBINA 1 encoding the CHLD subunit of the Mg-chelatase involved in Chl biosynthesis, was down-regulated in these leaves (Table 2). This implied that Chl biosynthesis might be impaired, thus decreasing leaf Chl concentration and accelerating leaf senescence. This agrees with our data that senescence associated gene 18 targeted by 5821 was up-regulated in Mg-deficient leaves (Table 2) and previous reports that Mg-deficient citrus leaves had lower Chl concentration (Tang et al., 2012; Yang et al., 2012).
Leaf miR6426 was up-regulated and its target genes: laccase 17 and sigma factor E (SIGE, SIG5) were down-regulated by Mg-deficiency (Table 2). The down-regulation of laccase 17 might be advantageous to the maintenance of Cu homeostasis (Abdel-Ghany and Pilon, 2008), because Cu concentration was lower in Mg-deficient leaves than in controls (Xu, 2015). Studies showed that laccase down-regulation caused an increase in total phenolic content in poplar (Ranocha et al., 2002), and that Mg and Cu concentrations were negatively correlated with total phenols in beech (Fagus sylvatica) leaves (Påhlsson, 1989). Thus, it is reasonable to assume that Mg-deficient citrus had higher concentration of total phenols. Kanamaru and Tanaka (2004) demonstrated that SIG5 was induced by various stresses and might contribute to the repair of damaged photosystem II (PSII) in higher plants. The down-regulation of SIG5 indicated that PSII might be damaged in Mg-deficient leaves (Tang et al., 2012; Yang et al., 2012).
Mg-deficient leaves had lower expression level of miR3946 and its target genes: glycogen/starch synthases (orange1.1g009139m) involved in starch biosynthesis and phosphoenolpyruvate carboxylase (PEPC) 2 involved in organic acid metabolism were up-regulated in these leaves (Table 2). This agrees with our reports that Mg-deficient C. sinensis leaves had higher or similar concentrations of starch, glucose, fructose and sucrose, higher activities of pyruvate kinase (PK) and PEPC, and enhanced organic acid metabolism and respiration, which was considered to be an adaptive response to Mg-deficiency by providing energy to maintain the basic metabolic processes in Mg-deficient leaves with lower photosynthetic rate (Yang et al., 2012, 2013; Peng et al., 2015). Mg-deficiency-induced up-regulation of phosphatidylinositol 3- and 4- kinase involved in lipid metabolism agrees with our report that the abundances of two protein species involved in lipid metabolism were enhanced in Mg-deficient C. sinensis leaves, thus contributing to the tolerance of plants to Mg-deficiency (Peng et al., 2015). Similarly, the expression level of the lipid metabolism-related gene, lipase class 3 family protein targeted by miR158 was up-regulated in Mg-deficient leaves due to decreased expression levels of their miRNAs (Table 2). Therefore, lipid metabolism might be up-regulated in Mg-deficient leaves. BAT1, a bidirectional amino acid transporter in Arabidopsis could be involved in amino acid export from the phloem into sink tissues (Dündar and Bush, 2009). Recently, Ladwig et al. (2012) showed that SIAR1, encoding a BAT from Arabidopsis, played an important role in organic N allocation and particularly in amino acid homeostasis in developing siliques. Mutant alleles of SIAR1 displayed more accumulation of anthocyanins and lower concentration of amino acids in the early stages of silique development. The up-regulation of BAT1 agrees with the report that Mg-deficient spinach leaves displayed more accumulation of amino acids (Fischer et al., 1988). The basic helix-loop-helix (bHLH) proteins, a large superfamily of transcription factors (TFs) involved in DNA binding, play key roles in plant development and environmental responses (Hudson and Hudson, 2015). Huang et al. (2013) suggested that a bHLH of P. trifoliata might play a key role in cold tolerance via positively regulating peroxidase-mediated ROS scavenging. Vorwieger et al. (2007) reported that two Arabidopsis bHLH TF were strongly induced by Fe-deficiency. Long et al. (2010) observed that POPEYE encoding a bHLH TF was induced by Fe-deficiency, concluding that POPEYE might play a crucial role in Arabidopsis Fe homeostasis. The up-regulation of bHLH DNA-binding superfamily protein genes (AT2G46810.1 and AT4G21330.1) targeted by miR3946 and miR5158 in Mg-deficient leaves (Table 2) agrees with the report that Mg-deficiency decreased root, stem and leaf concentration of Fe (Xu, 2015).
The expression level of miR535 was decreased in Mg-deficient leaves (Table 2), as obtained on drought potato leaves (Zhang et al., 2014) and Xanthomonas axonopodis pv. manihotis inoculated cassava leaves (Pérez-Quintero et al., 2012). MiR535 was predicted to target genes encoding cytochrome P450, family 714, subfamily A, polypeptide 1 in citrus (Table 2), disease resistance family protein, pectinesterase family protein, zinc ion binding, MLP-LIKE PROTEIN 423 and leucine-rich repeat transmembrane protein kinase, putative in cassava (Pérez-Quintero et al., 2012) and MYB domain-containing protein in potato (Zhang et al., 2014), which are involved in the regulation of various physiological processes. Thus, miR535 might play a role in the tolerance of plants to (a)biotic stresses.
As shown in Table 2, leaf miR5561 was repressed and its target gene G-box binding factor 1 (GBF1) was induced by Mg-deficiency. Smykowski et al. (2010) observed that GBF1 negatively regulated the expression of catalase2, and that gbf1 Arabidopsis mutants had a delayed senescence phenotype and postponed expression of senescence-associated genes. Mg-deficiency-induced up-regulation of GBF1 agrees with our above inference that Mg-deficiency accelerated leaf senescence, and that our report that Mg-deficient C. sinensis and C. grandis leaves had lower catalase (CAT) activity compared with controls (Yang et al., 2012).
MiR779 was down-regulated and all its target genes except for protein tyrosine kinase family protein were induced in Mg-deficient leaves (Table 2). Expansins are essential for cell enlargement and cell wall loosening during many developmental processes in plants. Choi et al. (2003) showed that expansins participated in enhancing plant growth by mediating cell wall loosening. Evidence shows the involvement of expansions in plant tolerance to abiotic stresses including dehydration (Dai et al., 2012), heat (Xu et al., 2014) and salt (Lü et al., 2013). Thus, the up-regulation of expansin A8 might have a positive role in plant Mg-deficiency tolerance. Through the interaction existing between Mg2+ and in the absorption process, levels would increase in Mg-deficient plants, thus leading to toxicity, which could be reversed by increasing Mg supply (Lasa et al., 2000). Ammonium transporter 1;1 (AMT1;1) transgenic rice had enhanced N use efficiency, growth and yield under optimal and suboptimal conditions (Ranathunge et al., 2014). Thus, the up-regulation of AMT2 might be an adaptive response of plants to Mg-deficiency. Similarly, the expression levels of the other transport-related genes targeted by miR395 and miR1077 (SecY protein transport family protein), miR1160 (PHO1), miR8019 (PPI1), and miR3946 (BAT1) were up-regulated in Mg-deficient leaves (Table 2). This agrees with our report that only two up-regulated protein species involved in protein transport were detected in Mg-deficient leaves, and that transport of proteins might be enhanced in Mg-deficient leaves (Peng et al., 2015). By contrast, the expression levels of miR5821, miR395, miR946, miR1160, and miR6218 were increased in Mg-deficient leaves, and their some target genes related to transport [i.e., cation efflux family protein, K+ uptake transporter 3 (KUT3), transporter associated with antigen processing protein 1, mitochondrial substrate carrier family protein and glutamate receptor 5 (GLR5)] were down-regulated in these leaves. Therefore, the transport of some substances might be down-regulated in Mg-deficient leaves.
Conclusions
Using Illumina sequencing, we isolated 691 known and 113 novel miRNAs from Mg-deficient and -sufficient citrus leaves. A miRNA was considered differentially expressed when it had both a fold-change of more than 1.5 and a P-value of less than 0.01. Based on the two criteria, we obtained 75 (73 known and 2 novel) up-regulated and 71 (64 known and 7 novel) down-regulated miRNAs from Mg-deficient leaves. This indicated that C. sinensis leaves owned remarkable metabolic plasticity, which might contribute to Mg-deficiency tolerance of plants. As shown in Figure 4, a possible model for the responses of leaf miRNAs to Mg-deficiency was proposed via integrating the present findings with the data available on the previous reports. The adaptive responses of leaf miRNAs to Mg-deficiency might include following several aspects: (a) inducing stress-related genes by repressing miR164, miR7812, miR5742, miR3946, and miR5158; (b) up-regulating transport-related genes; (c) increasing the expression of genes related to lipid metabolism by inhibiting miR158, miR5256, and miR3946 expression; (d) activating cell wall-related gene expansis 8A by down-regulating miR779; and (e) down-regulating the expression of genes involved in the maintenance of S, K and Cu by up-regulating miR395 and miR6426. To sum up, we identified some new known miRNAs (i.e., miR7812, miR8019, miR6218, miR1533, miR6426, miR5256, miR5742, miR5561, miR5158, and miR5818) responsive to nutrient deficiencies and obtained some candidate miRNAs that might contribute to Mg-deficiency tolerance of C. sinensis plants. Further study is needed to elucidate the roles of these candidate miRNAs in responses to Mg-deficiency, which will be useful to us for obtaining the key miRNAs for plant Mg-deficiency tolerance.
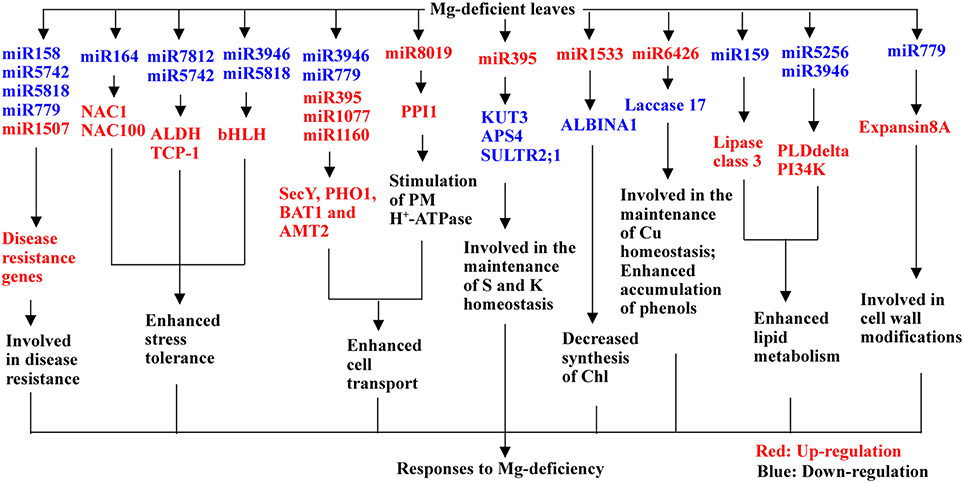
Figure 4. A potential model for responses of C. sinensis leaf miRNAs to Mg-deficiency. AMT2, ammonium transporter 2; BAT1, bidirectional amino acid transporter 1; PI34K, phosphatidylinositol 3- and 4- kinase; PLD delta, phospholipase D delta.
Data Access
RNAseq are submitted to Gene Expression Omnibus (GEO) under accession number GSE75758 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75758).
Author Contributions
CM carried out most of the experiments and drafted the manuscript; YQ participated in the design of the study. WL participated in data analysis. LY directed the study; YL participated in qRT-PCR analysis; PG participated in data analysis; XY determined leaf Mg concentration. LC designed and directed the study and revised the manuscript. All authors edited the manuscript.
Funding
Our work was funded by the earmarked fund for China Agriculture Research System (No. CARS-27).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00201
References
Abdel-Ghany, S. E., and Pilon, M. (2008). MicroRNA-mdiated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/jbc.M801406200
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Amtmann, A., Troufflard, S., and Armengaud, P. (2008). The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 133, 682–691. doi: 10.1111/j.1399-3054.2008.01075.x
Anzi, C., Pelucchi, P., Vazzola, V., Murgia, I., Gomarasca, S., Piccoli, M. B., et al. (2008). The proton pump interactor (PPI) gene family of Arabidopsis thaliana: expression pattern of PPI1 and characterisation of knockout mutants for PPI1 and 2. Plant Biol. 10, 237–249. doi: 10.1111/j.1438-8677.2007.00022.x
Cakmak, I., Hengeler, C., and Marschner, H. (1994). Partitioning of shoot and root dry matter and carbohydrates in bean plants suffering from phosphorus, potassium and magnesium deficiency. J. Exp. Bot. 45, 1245–1250. doi: 10.1093/jxb/45.9.1245
Cakmak, I., and Kirkby, E. A. (2008). Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol. Plant. 133, 692–704. doi: 10.1111/j.1399-3054.2007.01042.x
Chapman, H. D. (1968). “The mineral nutrition of citrus,” in The Citrus Industry, Vol. 2, eds W. Reuther, H. J. Webber, and L. D. Batchelor (Berkeley, CA: Division of Agricultural Sciences, University of California), 127–189.
Chen, J. C., Jiang, C. Z., and Reid, M. S. (2005). Silencing a prohibitin alters plant development and senescence. Plant J. 44, 16–24. doi: 10.1111/j.1365-313X.2005.02505.x
Chen, L., Wang, T., Zhao, M., Tian, Q., and Zhang, W. H. (2012). Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235, 375–386. doi: 10.1007/s00425-011-1514-9
Choi, D., Lee, Y., Cho, H. T., and Kende, H. (2003). Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15, 1386–1398. doi: 10.1105/tpc.011965
Dai, F., Zhang, C., Jiang, X., Kang, M., Yin, X., Lü, P., et al. (2012). RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 160, 2064–2082. doi: 10.1104/pp.112.207720
Dell'Orto, M., Santi, S., De Nisi, P., Cesco, S., Varanini, Z., Zocchi, G., et al. (2000). Development of Fe-deficiency responses in cucumber (Cucumis sativus L.) roots: involvement of plasma membrane H+-ATPase activity. J. Exp. Bot. 51, 695–701. doi: 10.1093/jexbot/51.345.695
Dündar, E., and Bush, D. R. (2009). BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta 229, 1047–1056. doi: 10.1007/s00425-009-0892-8
Fischer, E. S., Lohaus, G., Heineke, D., and Heldt, H. W. (1988). Magnesium deficiency results in accumulation of carbohydrates and amino acids in source and sink leaves of spinach. Physiol. Plant. 102, 16–20. doi: 10.1034/j.1399-3054.1998.1020103.x
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., and Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Hackenberg, M., Huang, P. J., Huang, C. Y., Shi, B. J., Gustafson, P., and Langridge, P. (2013). A comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions. DNA Res. 20, 109–125. doi: 10.1093/dnares/dss037
Hermans, C., Conn, S. J., Chen, J., Xiao, Q., and Verbruggen, N. (2013). An update on magnesium homeostasis mechanisms in plants. Metallomics 5, 1170–1183. doi: 10.1039/c3mt20223b
Hermans, C., Vuylsteke, M., Coppens, F., Craciun, A., Inzé, D., and Verbruggen, N. (2010a). Early transcriptomic changes induced by magnesium deficiency in Arabidopsis thaliana reveal the alteration of circadian clock gene expression in roots and the triggering of abscisic acid-responsive genes. New Phytol. 187, 119–131. doi: 10.1111/j.1469-8137.2010.03258.x
Hermans, C., Vuylsteke, M., Coppens, F., Cristescu, S. M., Harren, F. J., Inzé, D., et al. (2010b). Systems analysis of the responses to long-term magnesium deficiency and restoration in Arabidopsis thaliana. New Phytol. 187, 132–144. doi: 10.1111/j.1469-8137.2010.03257.x
Hsieh, L. C., Lin, S. I., Shih, A. C. C., Chen, J. W., Lin, W. Y., Tseng, C. Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151, 2120–2132. doi: 10.1104/pp.109.147280
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, X. S., Wang, W., Zhang, Q., and Liu, J. H. (2013). A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 162, 1178–1194. doi: 10.1104/pp.112.210740
Huber, D. M., and Jones, J. B. (2013). The role of magnesium in plant disease. Plant Soil 368, 73–85. doi: 10.1007/s11104-012-1476-0
Hudson, K. A., and Hudson, M. E. (2015). A classification of basic helix-loop-helix transcription factors of soybean. Int. J. Genomics 2015, 603182. doi: 10.1155/2015/603182
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kanamaru, K., and Tanaka, K. (2004). Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci. Biotech. Biochem. 68, 2215–2223. doi: 10.1271/bbb.68.2215
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Gene Regul. Mech. 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kim, E. J., Kwak, J. M., Uozumi, N., and Schroeder, J. I. (1998). AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10, 51–62. doi: 10.1105/tpc.10.1.51
Kong, W. W., and Yang, Z. M. (2010). Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol. Biochem. 48, 153–159. doi: 10.1016/j.plaphy.2009.12.008
Kotchoni, S. O., Kuhns, C., Ditzer, A., Kirch, H. H., and Bartels, D. (2006). Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 29, 1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x
Kushizaki, M. (1968). An extraction procedure of plant materials for the rapid determination of Mn, Cu, Zn and Mg by the atomic absorption analysis. J. Sci. Soil Manure Japan 39, 489–490.
Ladwig, F., Stahl, M., Ludewig, U., Hirner, A. A., Hammes, U. Z., Stadler, R., et al. (2012). Siliques Are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol. 158, 1643–1655. doi: 10.1104/pp.111.192583
Lasa, B. S., Frechilla, S., Aleu, M., González-Moro, B., Lamsfus, C., and Aparicio-Tejo, P. M. (2000). Effects of low and high levels of magnesium on the response of sunflower plants grown with ammonium and nitrate. Plant Soil 225, 167–174. doi: 10.1023/A:1026568329860
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Li, Y., Han, M. Q., Lin, F., Ten, Y., Lin, J., Zhu, D. H., et al. (2015). Soil chemical properties, ‘Guanximiyou’ pummelo leaf mineral nutrient status and fruit quality in the southern region of Fujian province, China. J. Soil Sci. Plant Nutr. 15, 615–628. doi: 10.4067/s0718-95162015005000029
Liang, G., He, H., and Yu, D. (2012). Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 7:e48951. doi: 10.1371/journal.pone.0048951
Liang, G., Yang, F., and Yu, D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62, 1046–1057. doi: 10.1111/j.1365-313x.2010.04216.x
Ling, L. L., Peng, L. Z., Cao, L., Jiang, C. L., Chun, C. P., Zhang, G. Y., et al. (2009). Effect of magnesium deficiency on photosynthesis characteristic of Beibei 447 Jinchen orange. J. Fruit Sci. 26, 275–280.
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell, 22, 2219–2236. doi: 10.1105/tpc.110.074096
Lü, P., Kang, M., Jiang, X., Dai, F., Gao, J., and Zhang, C. (2013). RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237, 1547–1559. doi: 10.1007/s00425-013-1867-3
Lu, Y. B., Qi, Y. P., Yang, L. T., Guo, P., Li, Y., and Chen, L. S. (2015). Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biol. 15:271. doi: 10.1186/s12870-015-0642-y
Lu, Y. B., Yang, L. T., Qi, Y. P., Li, Y., Li, Z., Chen, Y. B., et al. (2014). Identification of boron-deficiency-responsive microRNAs in Citrus sinensis roots by Illumina sequencing. BMC Plant Biol. 14:123. doi: 10.1186/1471-2229-14-123
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190. doi: 10.1105/tpc.108.064311
Nakashima, K., Tran, L. S., Nguyen, D. V., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Påhlsson, B. A. M. (1989). Mineral nutrients, carbohydrates and phenolic compounds in leaves of beech (Fagus sylvatica L.) in southern Sweden as related to environmental factors. Tree Physiol. 5, 485–495. doi: 10.1093/treephys/5.4.485
Paul, S., Datta, S. K., and Datta, K. (2015). MiRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 6:232. doi: 10.3389/fpls.2015.00232
Peng, H. Y., Qi, Y. P., Lee, J., Yang, L. T., Guo, P., Jiang, H. X., et al. (2015). Proteomic analysis of Citrus sinensis roots and leaves in response to long-term magnesium-deficiency. BMC Genomics 16:253. doi: 10.1186/s12864-015-1462-z
Pérez-Quintero, A. L., Quintero, A., Urrego, O., Vanegas, P., and Lopez, C. (2012). Bioinformatic identification of cassava miRNAs differentially expressed in response to infection by Xanthomonas axonopodis pv. manihotis. BMC Plant Biol. 12:29. doi: 10.1186/1471-2229-12-29
Phookaew, P., Netrphan, S., Sojikul, P., and Narangajavana, J. (2014). Involvement of miR164- and miR167-mediated target gene expressions in responses to water deficit in cassava. Biol. Plant. 58, 469–478. doi: 10.1007/s10535-014-0410-0
Poirier, Y., Thoma, S., Somerville, C., and Schiefelbein, J. (1991). A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97, 1087–1093. doi: 10.1104/pp.97.3.1087
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced permeability show superior growth and higher yield under optimal and suboptimal conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Ranocha, P., Chabannes, M., Chamayou, S., Danoun, S., Jauneau, A., Boudet, A. M., et al. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 129, 145–155. doi: 10.1104/pp.010988
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. doi: 10.1016/j.devcel.2005.01.018
Shen, H., Chen, J., Wang, Z., Yang, C., Sasaki, T., Yamamoto, Y., et al. (2006). Root plasma membrane H+-ATPase is involved in the adaptation of soybean to phosphorus starvation. J. Exp. Bot. 57, 1353–1362. doi: 10.1093/jxb/erj111
Shukla, L. I., Chinnusamy, V., and Sunkar, R. (2008). The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim. Biophys. Acta 1779, 743–748. doi: 10.1016/j.bbagrm.2008.04.004
Smykowski, A., Zimmermann, P., and Zentgraf, U. (2010). G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 153, 1321–1331. doi: 10.1104/pp.110.157180
Song, C., Wang, C., Zhang, C., Korir, N. K., Yu, H., Ma, Z., et al. (2010). Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431. doi: 10.1186/1471-2164-11-431
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Bäurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Sunkar, R., Bartels, D., and Kirch, H. H. (2003). Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 35, 452–464. doi: 10.1046/j.1365-313X.2003.01819.x
Tang, N., Li, Y., and Chen, L. S. (2012). Magnesium deficiency-induced impairment of photosynthesis in leaves of fruiting Citrus reticulata trees accompanied by up-regulation of antioxidant metabolism to avoid photooxidative damage. J. Plant Nutr. Soil Sci. 175, 784–793. doi: 10.1002/jpln.201100329
Valdés-López, O., Yang, S. S., Aparicio-Fabre, R., Graham, P. H., Reyes, J. L., Vance, C. P., et al. (2010). MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 187, 805–818. doi: 10.1111/j.1469-8137.2010.03320.x
Verbruggen, N., and Hermans, C. (2013). Physiological and molecular responses to magnesium nutritional imbalance in plants. Plant Soil 368, 87–99. doi: 10.1007/s11104-013-1589-0
Vorwieger, A., Gryczka, C., Czihal, A., Douchkov, D., Tiedemann, J., Mock, H. P., et al. (2007). Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226, 147–158. doi: 10.1007/s00425-006-0476-9
Wang, J. W., Schwab, R., Czech, B., Mica, E., and Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. doi: 10.1105/tpc.108.058180
Wang, T., Chen, L., Zhao, M., Tian, Q., and Zhang, W. H. (2011). Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high throughout sequencing. BMC Genomics 12:367. doi: 10.1186/1471-2164-12-367
Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. doi: 10.1104/pp.103.037945
Waters, B. M., Mclnturf, S. A., and Stein, R. J. (2012). Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 63, 5903–5918. doi: 10.1093/jxb/ers239
Xu, J. (2015). Effects of Magnesium Deficiency on Citrus Elements and Root and Leaf Anatomical Structures. Master Thesis, Fujian Agriculture and Forestry University, Fuzhou.
Xu, Q., Liu, Y., Zhu, A., Wu, X., Ye, J., Yu, K., et al. (2010). Discovery and comparative and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11:246. doi: 10.1186/1471-2164-11-246
Xu, Q., Xu, X., Shi, Y., Xu, J., and Huang, B. (2014). Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress. PLoS ONE 9:e100792. doi: 10.1371/journal.pone.0100792
Xu, Z., Zhong, S., Li, X., Li, W., Rothstein, S. J., Zhang, S., et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 6:e28009. doi: 10.1371/journal.pone.0028009
Yamasaki, H., Abdel-Ghany, S. E., Cohu, C. M., Kobayashi, Y., Shikanai, T., and Pilon, M. (2007). Regulation of copper homeostasis by microRNA in Arabidopsis. J. Biol. Chem. 282, 16369–16378. doi: 10.1074/jbc.M700138200
Yang, G. H., Yang, L. T., Jiang, H. X., Wang, P., and Chen, L. S. (2012). Physiological impacts of magnesium-deficiency in citrus seedlings: photosynthesis, antioxidant system and carbohydrates. Trees Struct. Funct. 26, 1237–1250. doi: 10.1007/s00468-012-0699-2
Yang, L. T., Yang, G. H., You, X., Zhou, C. P., Lu, Y. B., and Chen, L. S. (2013). Magnesium deficiency induced changes in organic acid metabolism of Citrus sinensis roots and leaves. Biol. Plant. 57, 481–486. doi: 10.1007/s10535-013-0313-5
Zeng, H., Wang, G., Hu, X., Wang, H., Du, L., and Zhu, Y. (2014). Role of microRNAs in plant responses to nutrient stress. Plant Soil 374, 1005–1021. doi: 10.1007/s11104-013-1907-6
Zhang, N., Yang, J., Wang, Z., Wen, Y., Wang, J., He, W., et al. (2014). Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS ONE 9:e95489. doi: 10.1371/journal.pone.0095489
Zhang, Z., Jiang, L., Wang, J., Gu, P., and Chen, M. (2015). MTide: an integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics 31, 290–291. doi: 10.1093/bioinformatics/btu633
Zhao, M., Ding, H., Zhu, J. K., Zhang, F., and Li, W. X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190, 906–915. doi: 10.1111/j.1469-8137.2011.03647.x
Zhao, M., Tai, H., Sun, S., Zhang, F., Xu, Y., and Li, W. X. (2012). Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS ONE 7:e29669. doi: 10.1371/journal.pone.0029669
Zhao, X., Liu, X., Guo, C., Gu, J., and Xiao, K. (2013). Identification and characterization of microRNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J. Plant Biochem. Biotech. 22, 113–123. doi: 10.1007/s13562-012-0117-2
Keywords: Mg-deficiency, Citrus sinensis, Illumina sequencing, leaves, microRNA
Citation: Ma C-L, Qi Y-P, Liang W-W, Yang L-T, Lu Y-B, Guo P, Ye X and Chen L-S (2016) MicroRNA Regulatory Mechanisms on Citrus sinensis leaves to Magnesium-Deficiency. Front. Plant Sci. 7:201. doi: 10.3389/fpls.2016.00201
Received: 25 September 2015; Accepted: 05 February 2016;
Published: 04 March 2016.
Edited by:
Anna Maria Mastrangelo, Centro di Ricerca per la Cerealicoltura, ItalyReviewed by:
Lijun Chai, Huazhong Agricultural University, ChinaWen-Wu Guo, Huazhong Agricultural University, China
Copyright © 2016 Ma, Qi, Liang, Yang, Lu, Guo, Ye and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Song Chen, bGlzb25nY2hlbjIwMDJAaG90bWFpbC5jb20=
 Cui-Lan Ma1,2,3
Cui-Lan Ma1,2,3 Peng Guo
Peng Guo Xin Ye
Xin Ye Li-Song Chen
Li-Song Chen