- National Key Laboratory of Crop Genetic Improvement, Microelement Research Center, Huazhong Agricultural University, Wuhan, China
Phytohormones play pivotal roles in the response of plants to various biotic and abiotic stresses. Boron (B) is an essential microelement for plants, and Brassica napus (B. napus) is hypersensitive to B deficiency. However, how auxin responds to B deficiency remained a dilemma for many years and little is known about how other phytohormones respond to B deficiency. The identification of B-efficient/inefficient B. napus indicates that breeding might overcome these constraints in the agriculture production. Here, we seek to identify phytohormone-related processes underlying B-deficiency tolerance in B. napus at the physiological and gene expression levels. Our study indicated low-B reduced indole-3-acetic acid (IAA) concentration in both the shoots and roots of B. napus, and affected the expression of the auxin biosynthesis gene BnNIT1 and the efflux gene BnPIN1 in a time-dependent manner. Low-B increased the jasmonates (JAs) and abscisic acid (ABA) concentrations and induced the expression of the ABA biosynthesis gene BnNCED3 and the ABA sensor gene BnPYL4 in the shoot. In two contrasting genotypes, the auxin concentration decreased more drastically in the B-inefficient genotype ‘W10,’ and together the expression of BnNIT1 and BnPIN1 also decreased more significantly in ‘W10’ under long-term B deficiency. While the JAs concentration was considerably higher in this genotype, and the ABA concentration was induced in ‘W10’ compared with the B-efficient genotype ‘QY10.’ Digital gene expression (DGE) profiling confirmed the differential expression of the phytohormone-related genes, indicating more other phyohormone differences involving in gene regulation between ‘QY10’ and ‘W10’ under low-B stress. Additionally, the activity of DR5:GFP was reduced in the root under low-B in Arabidopsis, and the application of exogenous IAA could partly restore the B-defective phenotype in ‘W10.’ Overall, our data suggested that low-B disturbed phytohormone homeostasis in B. napus, which originated from the change of transcriptional regulation of phytohormones-related genes, and the differences between genotypes may partly account for their difference in tolerance (B-efficiency) to low-B.
Introduction
A change in phytohormones is one of the most significantly adaptive strategies used by plants in their response to adverse conditions (Potters et al., 2007). Recently, the roles that phytohormones play in response to limited or excessive nutrients (including nitrogen, phosphorus, potassium, sulfur, and many others) have been well-characterized (Scheible et al., 2004; Liu et al., 2009; Rubio et al., 2009; Kapulnik et al., 2011; Kiba et al., 2011; Romera et al., 2011; Iqbal et al., 2013; Zhang et al., 2014). On one hand, plant mineral nutrition status affects phytohormone metabolism, and interaction. Nitrogen regulated cytokinin (CTK) biosynthesis by triggering the expression of CTK biosynthesis-related genes IPT3 and CYP735A (Kiba et al., 2011) and also facilitated cell-to-cell auxin transport by regulating the abundance of NRT1.1/CHL1 (Ho et al., 2009), known as a dual affinity nitrate transporter and nitrate sensor (Krouk et al., 2010). A low potassium supply can induce the increase in the expression of ethylene (ETH) biosynthesis-related genes, thus boosting up ETH concentration, which was closely correlated with the phenotype of reduced lateral roots when Arabidopsis was exposed to potassium deficiency (Shin and Schachtman, 2004; Jung et al., 2009). OsARF16 was involved in the crosstalk of auxin signaling and iron deficiency signaling transduction pathway (Shen et al., 2015). The genes involved in the first three steps of the jasmonates (JAs) biosynthesis enzymes, i.e., LOX2, AOS, and AOC, were significantly up-regulated in Arabidopsis during potassium starvation and down-regulated after potassium resupply (Armengaud et al., 2004). The JAs metabolism pathway was considered as one of the most significant pathways identified in rice under arsenic stress, although arsenic is not an essential element (Yu et al., 2012). On the other hand, spraying exogenous phytohormones or knockdown of phytohormone biosynthesis/signaling-related genes could alter the expression of some genes implicated in nutrient uptake and transport, which may, to a certain extent, lead to the restoration of phenotypes caused by unfavorable nutrient conditions. It was reported that CTK repressed the phosphate-starvation response through increasing the intracellular phosphate level (Wang et al., 2006). Auxin treatment inhibited the expression of a low-sulfur-responsive gene in Arabidopsis (Dan et al., 2007). Treating maize plants with auxin increased the gene expression of a potassium transporter ZMK1 (Philippar et al., 1999). Arabidopsis mutants, abi2-1, and aba1, which showed low sensitivity or impaired abscisic acid (ABA) biosynthesis, exhibited a reduced accumulation of anthocyanin when grown under low-Pi (inorganic phosphate), whereas not all of the plant defects caused by low-Pi were able to be rescued (Trull et al., 1997; Ciereszko and Kleczkowski, 2002). Exogenous ABA alleviated zinc uptake and accumulation when Populus was exposed to excess zinc (Shi et al., 2015).
Boron (B) deficiency is universally acknowledged as a serious agricultural problem worldwide, especially in Japan, China, USA, and Brazil (Tanaka and Fujiwara, 2008). To cope with B deficiency, plants trigger physiological and developmental responses, aiming to strengthen tolerance to B deficiency, which in many cases alters morphology and metabolism and involves changes in phytohormones. A dramatic decrease in IAA exporting from the shoot apex as well as a decline in the IAA concentration in the shoot apex and roots of pea plants have been observed (Li et al., 2001; Wang et al., 2006). Nonetheless, another viewpoint supported that excessive auxin accumulated in bean radicles (Coke and Whittington, 1968), and a similar result was observed in lettuce (Crisp et al., 1976). Under a low-B condition, the auxin efflux gene PIN1 (pin-formed 1) was down-regulated and PIN2 (pin-formed 2) played an important role in the root meristem maintenance in Arabidopsis (Li et al., 2015). The use of Arabidopsis IAA and ETH mutants has provided substantial evidence, which suggested that IAA and ETH were involved in the inhibition of root elongation, and the formation and elongation of root hairs under B deficiency, respectively (Martin-Rejano et al., 2011). Abreu et al. (2014) formulated the hypothesis that signaling mechanisms during cell differentiation and organogenesis were extraordinarily sensitive to B deficiency in the apical root meristem of Arabidopsis, and CTK functioned as the key regulator. B deficiency inhibits root cell elongation and this response is driven by a signaling pathway with the participation of ETH and auxin (Camacho-Cristobal et al., 2015). Using cDNA chips, several differentially expressed genes (DEGs) related to phytohormones were identified in Arabidopsis under B deficiency, including JAs, CTK, IAA, ETH, and ABA (Peng et al., 2012). Although the relationships between B deficiency and phytohormone responses have been reported in numerous studies, the actual auxin response to B deficiency remained a dilemma for many years and how other phytohormones respond to B deficiency remain elusive. In addition, an overall analysis is rare on transcriptional regulation for how phytohormones respond to B deficiency.
Brassica napus (B. napus), in addition to being one of the preeminent sources of cooking oil worldwide (Meyer, 2009), is an agricultural crop that is highly susceptible to B deficiency (Marschner, 1995). The B-efficient (B-deficiency tolerant) genotype ‘QY10’ was identified to cope well with low-B conditions compared with the B-inefficient (B-deficiency sensitive) genotype ‘W10’ (Xu et al., 2002; Zhao et al., 2008). identification of Differential protein abundance and cDNA microarray analysis showed that JAs-, CTK-, IAA-, ETH-, ABA-related genes, and JAs-related proteins were characterized under B deficiency (Wang et al., 2011; Peng et al., 2012), suggesting that B. napus may have evolved sophisticated mechanisms involving changes in phytohormones to enhance the ability to tolerate B deficiency. The physiological variation in the sensitivity of different B. napus genotypes to low-B highlights the possible complex architecture of plant phytohormone responses. The relationship between B and auxin has not yet been elucidated and there is little information on the relationship between B and other phytohormones. Consequently, studies on phytohomone-mediated response to low-B or the effect of low-B on phytohormones and related gene expression in B. napus are crucial for in-depth researches determining the differences in phytohormone responses between B-efficient and B-inefficient genotypes. In this study, we hypothesized: (i) that B deficiency could induce the common and/or distinct phytohormone responses, including concentration and transcriptional regulation levels between B-efficient and B-inefficient B. napus genotypes; and (ii) that the distinct phytohormone responses would be one reason for the severity differences in response to B deficiency between B-efficient and B-inefficient B. napus genotypes. The objectives of this study were to elucidate the changes in B. napus phytohormones in response to B deficiency, at both physiological and molecular levels, as well as the differences in phytohormone responses between B-efficient and B-inefficient B. napus genotypes.
Materials and Methods
Plant Materials and Growth Condition
Two different B. napus genotypes, including B-efficient ‘QY10’ and B-inefficient ‘W10’ (Xu et al., 2002; Zhao et al., 2008), were used to conduct two different B deficiency hydroponic experiments. Plump seeds from the two genotypes were surface-sterilized using 0.5% (w/v) NaClO for 10 min and were rinsed completely in ultrapure water (18.25 MΩ·cm). The seeds were then sown in gauze after being soaked in deionized water for 24 h. After 5 days for germination, uniform seedlings were transplanted into black plastic containers with Hoagland and Arnon solution (Hoagland and Arnon, 1950). Under long-term exposure to low-B, seedlings were first grown in one-quarter-strength nutrient solution containing low-B (0.25 μM H3BO3), and sufficient-B (25 μM H3BO3) was used as control. B concentration was determined according to Asad et al. (1997) and Zhao et al. (2008). The nutrient solution was replaced every 5 days, beginning with one quarter-strength solution, afterwards progressing to one-half-strength and eventually full-strength. The leaves, shoot apexes, root crowns, and roots were harvested separately on the 20th day. When exposed to short-term B-free starvation treatment, seedlings were first grown under sufficient-B condition (10 μM H3BO3) for 12 days, then the roots of half plants were repeatedly rinsed with ultrapure water to remove B on the surface and transferred to B-free solution afterwards, and the rest seedlings transferred to sufficient-B circumstance (10 μM H3BO3) as control. The leaves, shoot apexes and roots were harvested separately at the 0, 3, 6, 12, 24, 72, and 120 h after transferred to B-free condition. For digital gene expression (DGE) profiling experiment, seedlings were grown under low-B (0.25 μM) condition for 10 days when ‘W10’ showed slight B-defective symptoms, then the leaves and roots of ‘W10’ and ‘QY10’ were harvested. All plants were cultivated in an illuminated culture room with a temperature regime of 24/22°C (day/night) and a photoperiod of 14/10 h (day/night) and a light intensity of 300–320 μmol·m−2·s−1. Each treatment was set with four replicates. For the exogenous IAA application experiment, plants were grown under sufficient-B or low-B condition for 10 days, then transferred to either 0.25 or 25 μM B with either 0 or 10 μM IAA (Sigma-Aldrich) for another 10 days, then four plants from each group were harvested. Arabidopsis harboring DR5:GFP (a synthetic auxin-inducible promoter-drived green fluorescent protein) and PIN1::PIN1-GFP seeds were kindly provided by Hongwei Xue and Yingtang Lu, respectively. Arabidopsis plants were grown in MS medium under sufficient-B (30 μM) or low-B condition (0.1 μM) under standard greenhouse conditions with artificial light (16/8-h photoperiod) for 4 or 12 days. The roots were harvested carefully for microscopy analysis.
Determination of Root Architecture
Plant roots were imaged using a scanner after exposure to low-B for 20 days. Total root length, root volume and root surface area were measured using the root image analysis software Win-RHIZO Pro (Regent Instruments, QC, Canada).
Quantification of B Concentration and B Efficiency Coefficient
Samples were prepared from four plants for each replicate. All samples were killed out at 105°C for 30 min, and then oven-dried to constant weight at 65°C. The dry samples were then weighed and ground to fine powder in a carnelian mortar. B was extracted from the dry powder by shaking it in 10 mL of 1 M HCl for shoot and 6 mL for root in a shaker for 2 h. The B concentration was determined by Inductively Coupled Plasma-Optical Emission Spectroscopy (ICP-OES, Varian, Inc. USA). Plant dry matter (DM), B concentration, B content, and B efficiency coefficient (BEC = DM under B deficiency/DM under sufficient B) were calculated for each genotype, respectively.
Digital Gene Expression (DGE) Library Preparation and Sequencing
Leaves and roots of the seedlings with three biological replicates were individually sampled. Total RNA was extracted with Trizol Reagent (Invitrogen, CA, USA) from the young leaves and roots of the two genotypes ‘QY10’ and ‘W10.’ Reverse transcriptions were carried out using M-MLV Reverse Transcriptase (Promega, WI, USA) according to the manufacturer's instructions. Tag libraries from the RNA samples were prepared in parallel, and then were subjected to an Illumina Hiseq 2500 platform, generating 50 bp single-end reads. For differentially expressed genes (DEGs) analysis, the number of expressed tags was calculated and then normalized to FPKM (fragment per kilo base of exon per million fragments mapped) using Cuffdiff 2.0, a software known to robustly perform differential analysis (Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation; significance level at P < 0.05, false discovery rate [FDR] < 0.05).
JA, ABA, and IAA Analysis
Samples were prepared using a slightly modified crude extraction procedure reported by Liu et al. (2012). The standard IAA, JA were purchased from Sigma-Aldrich (St. Louis, MO, USA), and ABA, OPDA (JA biosynthetic precursor 12-oxophytodienoic) and JA-Ile (the isoleucine conjugate of JA) were from OlChemIm (OlChemIm, Olomouc, Czech Republic). The internal standards were 2H6ABA (OlChemIm) for ABA, 10-dihydro-JA (DHJA; OlChemIm) for JA, OPDA and JA-Ile, and D2-IAA (Sigma-Aldrich) for IAA. The modified steps were using internal standards, 30 ng 2H6ABA, 40 ng DHJA, and 50 ng D2-IAA for each sample, and the filtrate was dried then dissolved in 200 μL 50% methanol. Total IAA, JA, OPDA, JA-Ile, and ABA were determined by ultra-fast liquid chromatography-electrospray ionization tandem mass spectrometry (UFLC-ESI-MS).
Quantitative Real Time PCR
Total RNA was extracted and reversely transcribed as above in digital gene expression (DGE) library preparation and sequencing method. Gene sequences were achieved from the available B. napus genome sequence database (http://www.genoscope.cns.fr/brassicanapus/) and degenerate primers were designed by Primer Premier 5.0. Quantitative real time polymerase chain reaction (qRT-PCR) for detecting the relative expression of genes was performed using the SYBR Green Real-Time PCR Master Mix Kit (TOYOBO, Japan) and the CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). All PCR reactions were performed with independently three replicates. The PCR profiles were as follows: 95°C for 1 min, followed by 45 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. The sequences of primers used for qRT-PCR were listed in Supplementary Table 1.
Microscopy
Images of plant roots were observed with a light/fluorescence microscope (Nikon 80I). Root tips of 12-day Arabidopsis DR5:GFP and PIN1::PIN1-GFP were mounted onto microscope slides for observation. Confocal microscopy was performed using a Leica Sp2 Confocal Laser-Scanning Microscope according to the manufacturer's instructions. The 4-day DR5: GFP roots were mounted onto microscope slides with 20 μg mL−1 propidium iodide (PI, Sigma-Aldrich). Ten seedlings were analyzed per treatment. The signal intensity was measured using ImageJ software (W.S. Rasband, National Institutes of Health), and error bars were obtained based on measurements of 10 seedlings per treatment.
Statistical Analysis
Analysis of variance was conducted by SPSS16.0 for Windows software (SPSS 249 Inc, Chicago, IL, USA). The long-term and short-term experiments were conducted separately and the data were dealt separately. Differences among the B-treatments were identified using the least significant difference test (LSD) at the P < 0.05 level. Expression data were normalized with the expression level of the actin by the 2 method (Livak and Schmittgen, 2001).
Results
Variation of Two Genotypes in Response to Low-B Stress
The biomass (dry weight), B content, and B efficiency coefficient (BEC) of two B. napus genotypes, i.e., B-efficient ‘QY10’ and B-inefficient ‘W10,’ were measured and analyzed at the tested B treatments. Under the low-B (0.25 μM) condition, ‘W10’ exhibited typical B-defective symptoms, including curved and thickened leaves, pluriceps shoot apex, short primary root and hypogenetic lateral roots (Figure 1A). In addition, the dry weight of the leaves, stem and root and the B content of ‘W10’ were significantly lower than those of ‘QY10’ (Figures 1B,C). In the presence of sufficient B (25 μM) for 20 days, ‘QY10’ slightly outperformed ‘W10’ (Figure 1A) and accumulated slightly more B (Figure 1D). The BEC of ‘QY10’ reached 0.70, which was markedly higher than that of ‘W10’ (0.40). Analysis of the total root length, root surface area and root volume of the two B. napus genotypes confirmed that ‘W10’ was more sensitive to B deficiency than that of ‘QY10’ (Supplementary Figure 1). These results demonstrated that ‘QY10’ has a greater tolerance (higher B efficiency) to low-B stress than ‘W10.’
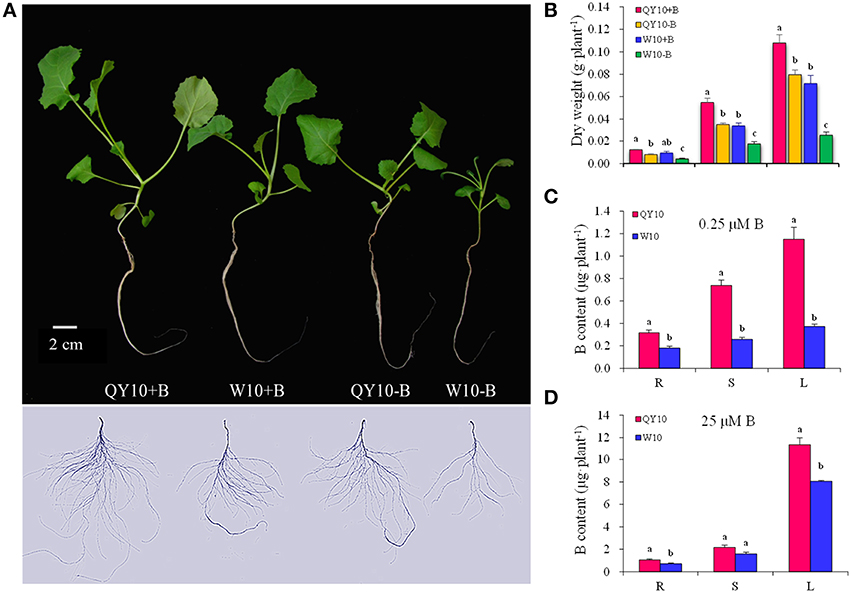
Figure 1. Growth variations of ‘QY10’ and ‘W10.’ (A) Phenotype, (B) dry weight, (C, D) B content of two B. napus cultivars ‘QY10’ (B-efficient) and ‘W10’ (B-inefficient) grown hydroponically under sufficient B (+B, 25 μM B) or low-B (−B, 0.25 μM B) for 20 days. R, root; S, stem; L, leaves. Values represent mean ± SD of four independent replicates, and bars with different letters show significant differences for the same organ among the treatments (ANOVA, LSD, p < 0.05).
Digital Gene Expression (DGE) Profiling of Phytohormone-Related Genes
To assess a comprehensive characterization of different phytohormone transcriptional behaviors under B deficiency, DGE analysis was performed on both the root and leaves of ‘QY10’ and ‘W10.’ Global expression profiling of the genes related to eight phytohormones, including auxin, ABA, ETH, brassinosteroids (BRs), JAs, Gibberellin (GA), CTK and salicylic acid (SA), was conducted. The results showed that among 2,744 phytohormone-related genes, 512 in the root, and 728 in the leaves were indentified to be differentially expressed between ‘QY10’ and ‘W10’ under a 10-day B-deficiency period, accounting for 18.7 and 26.5% of the total, respectively (Supplementary Table 2). Comparative transcriptomic analysis of DEGs related to phytohormones responding to stimuli, together with phytohormone-related transcription factors related to Aux/IAAs, MYBs and ERFs showed that the expression of most of the DEGs in the leaves of ‘QY10’ was lower than that of ‘W10’ (Figure 2A), while the opposite trend was observed in the root (Figure 2B). In general, the expression of Aux/IAAs was more abundant in the root of ‘QY10’ compared with ‘W10,’ while the expression of all ERFs was much higher in the leaf of ‘W10’ compared with ‘QY10’ (Figures 2A,B). The MYB family has been reported to be involved in the response to many phytohormones (Chen Y. H. et al., 2006; Lee et al., 2015). In this study, the expression of MYB34 and MYB47 in response to JA (Chen Y. H. et al., 2006) was significantly higher in both the root and leaves of ‘QY10’ compared with ‘W10’ (Figures 2A,B).
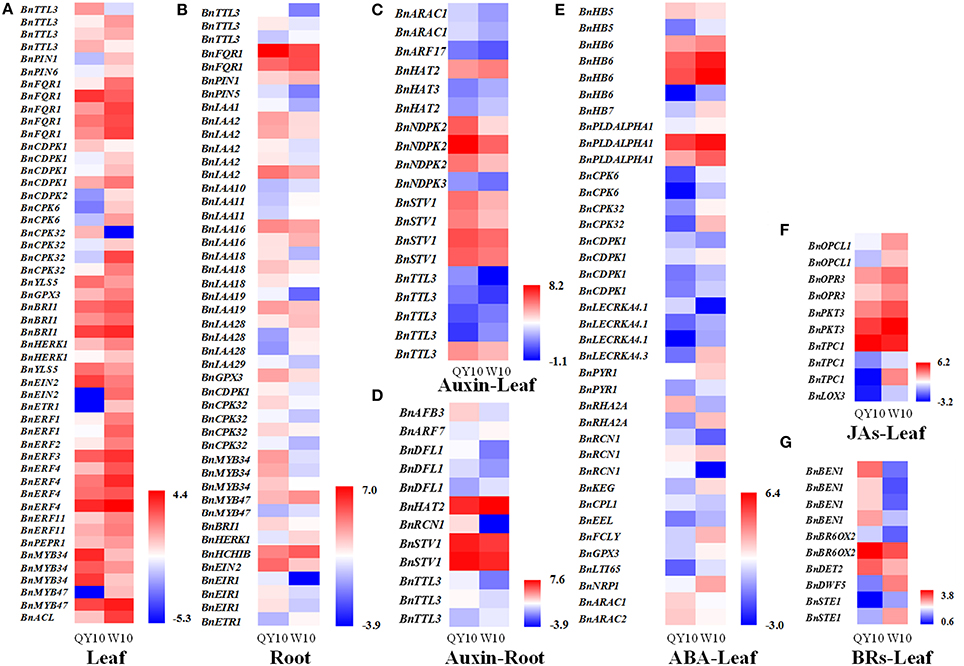
Figure 2. Comparative analysis of phytohormone-related gene expression profiling of ‘QY10’ and ‘W10’ in response to low-B stress. (A,B) Heat map representing the phytohormone-related DEGs responding to stimuli and the transcription factors Aux/IAAs, MYBs, and ERFs, which were differentially expressed in the leaves (A) and root (B) between ‘QY10’ and ‘W10’ under low-B stress. The color from blue to red represents log2 FPKM (Fragments Per Kilobases of exon model per Million mapped reads). (C–G) Heat map representing the DEGs involved in the auxin-mediated signaling pathway in the root (C) and leaves (D), ABA-mediated signaling pathway in the leaves (E), JAs biosynthetic process in the leaves (F), and BRs biosynthetic process in the leaves (G) of ‘QY10’ and ‘W10.’
We further analyzed the expression profiling of the genes related to the phytohormone biosynthetic process and signaling pathway to demonstrate the utility of DGE for understanding the specific functions of these genes in the response of B. napus to B deficiency. The expression of the DEGs involved in the auxin-mediated signaling pathway in the leaves and root of ‘QY10’ was almost higher than those of ‘W10’ (Figures 2C,D). The expression of the DEGs involved in the ABA-mediated signaling pathway in the leaves of ‘W10’ was almost higher than those of ‘QY10’ (Figure 2E). The expression of the DEGs involved in the JAs biosynthetic process in the leaves of ‘QY10’ was lower than those of ‘W10’ (Figure 2F). In contrast to the JAs, the expression of the DEGs involved in the BRs biosynthetic process was higher in the leaves of ‘QY10’ compared with ‘W10’ (Figure 2G). The transcription profiling indicated that the expression of the phytohormone-related genes differed considerably between the B-tolerant and B-sensitive genotypes. To validate this difference, the concentrations of IAA, ABA, and JA, as well as its precursor OPDA or the conjugate JA-Ile, and the related gene expression were measured under B deficiency stress.
Effect of B Deficiency on IAA in ‘QY10’ and ‘W10’
In terms of the long-term low-B stress, as shown in Figure 3, the IAA concentrations in all organs decreased compared with the sufficient B treatment, except in the leaves of ‘QY10.' The decline rate in the shoot apex, root crown and root of ‘QY10’ was 24.5, 20.3, and 26.0%, respectively. However, the rate was much more drastic in ‘W10,’ i.e., 55.4, 71.2, and 43.0%, respectively. Meanwhile, the IAA concentration in the leaves of ‘W10’ decreased by 53.5%. The changes in the expression of the IAA biosynthetic/transport-related genes were similar to the IAA concentration changes under long-term B deficiency. The IAA biosynthesis gene BnNIT1 (NIT1, nitrilase1) was down-regulated by 55.0 and 44.9% in the shoot apexes and by 13.2 and 77.2% in the roots of ‘QY10’ and ‘W10,’ respectively. The auxin-efflux gene BnPIN1 was down-regulated by 15.4 and 55.2% in the shoot apexes and by 96.7 and 94.5% in the roots of ‘QY10’ and ‘W10,’ respectively. Another auxin-efflux gene, BnPIN2, was down-regulated in leaves and the shoot apex, but only in ‘W10’ (Figure 3).
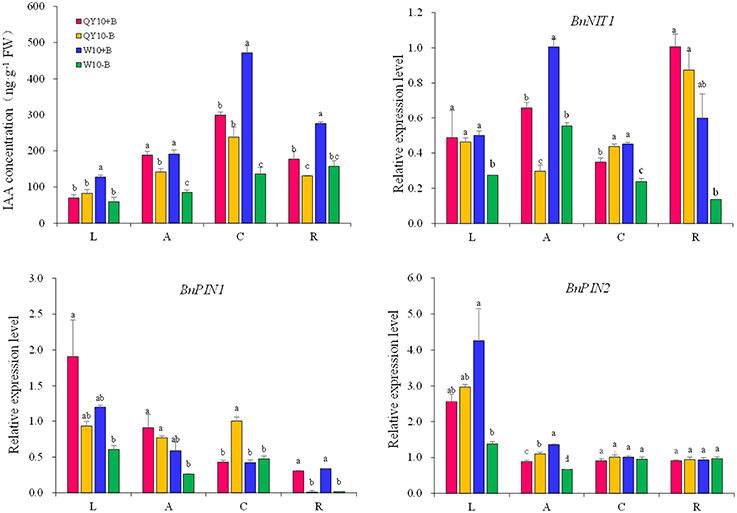
Figure 3. IAA concentration and expression of IAA-related genes under long-term low-B stress. BnNIT1: auxin biosynthesis-related gene. BnPIN1 and BnPIN2: auxin efflux genes. Plants were grown hydroponically under sufficient B (+B, 25 μM B) or low-B (−B, 0.25 μM B) for 20 days. L, leaves; A, shoot apex; C, root crown; R, root. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same organ among the treatments (ANOVA, LSD, p < 0.05).
As the removal of B could cause some early B-defective reactions (Goldbach et al., 2000), we carried out a short-term (5-day) B-free starvation experiment to corroborate the response of auxin to low-B. There was no obvious change in the IAA concentration in the three organs of the two genotypes within a 12-h period of B-free starvation compared with the B-sufficient condition. Compared with the B-sufficient condition, the IAA concentration in the shoot apex and the root of ‘W10’ decreased by 42.3 and 58.3%, respectively, from 0 h to the 5th day, while that of ‘QY10’ remained in a dynamic equilibrium (Figure 4A). These results demonstrated that IAA mediated B deficiency stress but did not instantaneously respond to B deficiency, which is in agreement with previous report by Xiong et al. (2001). Curiously, the expression of BnNIT1 decreased at 3 and 12 h, especially in ‘W10,’ and then increased on the 5th day. The expression of BnPIN1 was induced at 3 and 12 h, and then recovered to the level measured in the sufficient B treatment by the 5th day. The expression of BnPIN2 was similar to that of BnPIN1 (Figure 4B). These results pointed to the likelihood that the change in IAA in response to low-B stress was involved in the regulation of IAA biosynthetic/transport-related genes expression, which was induced by the B level in a complicated time-dependent way in B. napus.
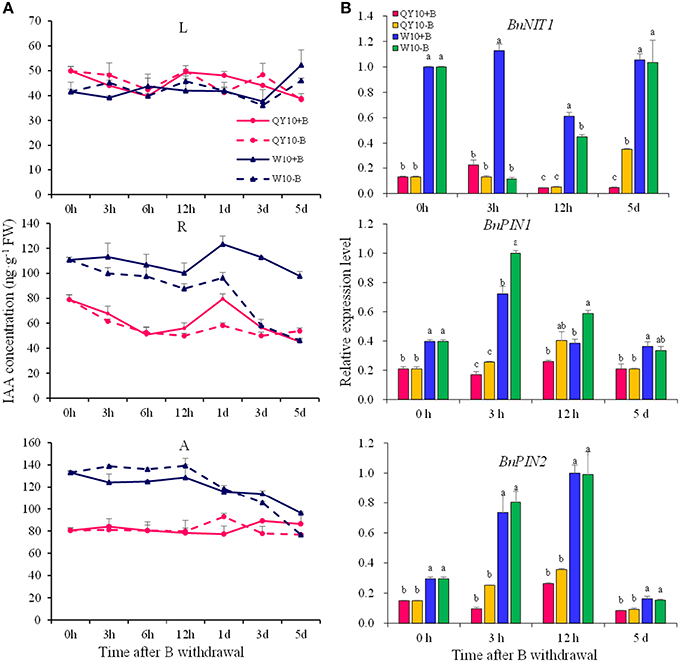
Figure 4. IAA concentration and related-gene expression of ‘QY10’ and ‘W10’ under B-free starvation. (A) IAA concentration. (B) IAA related-gene expression in root. After been grown hydroponically under sufficient B (+B, 25 μM B) for 12 days, half of the plants were transplanted to B-free solution (−B) and then sampled at different time points, and the other plants, which served as the control, were grown continuously with sufficient B. L, leaves; R, root; A, shoot apex. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same time among the treatments (ANOVA, LSD, p < 0.05).
In the long-term low-B stress and short-term B-free starvation experiments, ‘W10’ had a higher IAA concentration than ‘QY10’ under the B-sufficient condition (Figures 3, 4A). Meanwhile, the expression of the auxin biosynthesis gene BnNIT1 was much higher in the root of ‘W10’ than in the root of ‘QY10,’ suggesting that ‘W10’ has a higher IAA demand (Figure 4B). This may partly account for the more severe symptoms exhibited by ‘W10’ under B deficiency.
Activity of DR5:GFP and the PIN1::PIN1-GFP Protein in Arabidopsis Root
Auxin accumulation in the root mediated by auxin biosynthesis and transport is important for the onset of cell division in root meristems (Blilou et al., 2005; Zhu et al., 2015). Our assays afore-mentioned showed that low-B stress not only decreased auxin concentration but also changed its biosynthesis gene BnNIT1 and the efflux gene BnPIN1 mRNA abundance in the root in a time-dependent manner, and the decline was rather dramatic under long-term B deficiency. Thus, we tested Arabidopsis transgenic plants harboring DR5:GFP and PIN1::PIN1-GFP to investigate auxin distribution and flow in the root tip. Under the B-sufficient condition, DR5:GFP was mainly expressed in the root tips, specifically in the quiescent center (QC; Figure 5A), which is in accordance with previous report by Tanaka et al. (2006), and it was richly expressed when the seedlings were cultivated for 12 days (Figure 5B). The fluorescence intensity in the QC decreased to 69.2 and 59.2% after 4 and 12 days, respectively, under the low-B condition (Figures 5A,B). With respect to the 18-h B deficiency, there was no significant difference in the fluorescence intensity, except for a slight increase (Figures 5A,C). PIN1::PIN1-GFP fluorescence intensity decreased to 71.3% in the root subjected to low-B stress for 12 days, compared with that treated with sufficient B (Supplementary Figure 2), which implies that the relatively long-term B deficiency decreased the acropetal transport of auxin. These results indicate that auxin plays a vital role in root growth by decreasing the auxin concentration in the QC under B deficiency.
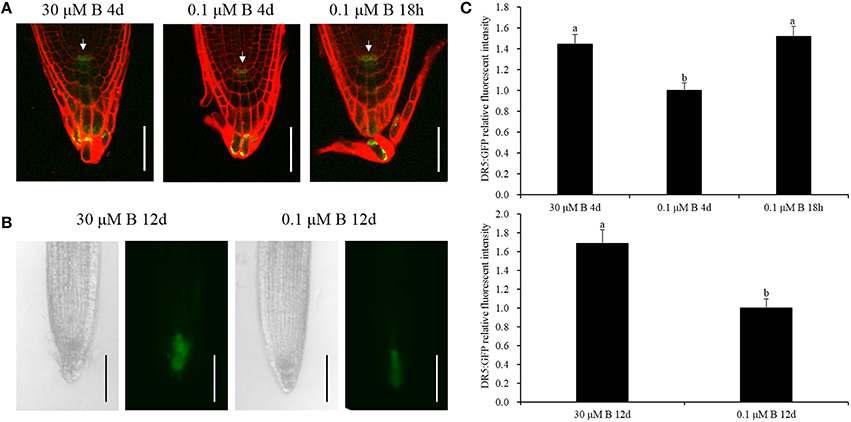
Figure 5. GFP fluorescence of the DR5:GFP protein in the root tips. (A) Transgenic Arabidopsis thaliana plants were grown in MS media containing 30 μM B for 4 days, 0.1 μM B for 4 days, or 30 μM B for 3 days and 6 h and then transferred to 0.1 μM B for 18 h. The cell wall was stained with PI (red). White arrows represent the quiescent center (QC). Scale bar = 50 μm. (B) Transgenic Arabidopsis thaliana plants were grown in MS media containing 30 or 0.1μM B for 12 days. Scale bar = 200 μm. (C) Quantification of the DR5: GFP fluorescence intensities in the quiescent center zones in (A,B). Total of 10 seedlings were imaged for each treatment, and representative photos were shown. Values represent the mean ± SD of 10 independent replicates, and bars with different letters show significant differences (ANOVA, LSD, p < 0.05).
Effect of Exogenous IAA on B Deficiency in ‘QY10’ and ‘W10’
The effects of exogenous IAA application on the ‘QY10’ and ‘W10’ phenotypes under sufficient- and low-B conditions are shown in Figure 6A. The shoot dry weight of IAA-treated ‘W10’ was significantly higher than that of ‘W10’ without exogenous IAA application (increased by 211.5%) under B deficiency. However, the shoot dry weights in IAA-treated B-deficient ‘QY10,’ B-sufficient ‘QY10’ and ‘W10’ were significantly lower compared with the plants without exogenous IAA application. The root dry weight increased except for ‘QY10’ under sufficient B. (Figure 6B). In addition, auxin significantly affected the number of axillary buds. There were an average of 4 axillary buds per plant in the QY10+B, QY10−B, and W10+B treatments (data not shown), compared with 7 under B deficiency (W10−B), which was reduced to 4.25 in the W10−B+IAA treatment (Figure 6C). The exogenous IAA experiment suggested that under B-sufficient conditions, plants with a suitable phytohormone status would suffer from a hyper-auxin toxicity when supplied with exogenous IAA. However, under B-deficient conditions, the B deficiency-sensitive genotype ‘W10’ with reduced IAA concentration was partly restored by exogenous IAA application.
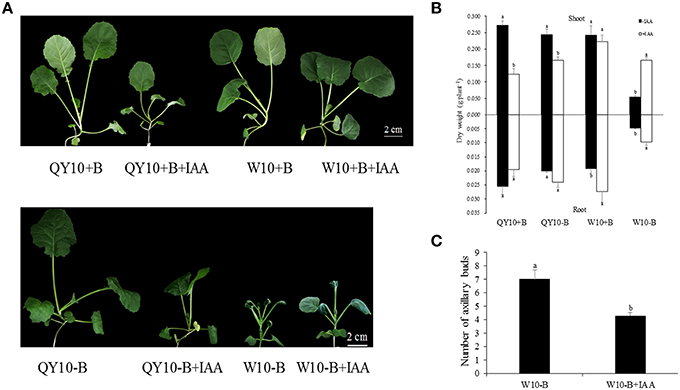
Figure 6. Growth variations of ‘QY10’ and ‘W10’ under sufficient and low-B conditions with or without IAA. (A) Phenotype, (B) dry weight, and (C) number of axillary buds of ‘QY10’ and ‘W10.’ Plants were grown hydroponically under sufficient B (+B, 25 μM B) or low-B (−B, 0.25 μM B) for 10 days, and then 10 μM IAA (+IAA) was supplied for another 10 days. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same organ and material between the treatments (ANOVA, LSD, p < 0.05).
Effect of B Deficiency on JAs in ‘QY10’ and ‘W10’
In this study, JAs were found to be the most abundant phytohormones among the three phytohormones tested in B. napus, especially in the leaves and root crown. Under long-term low-B conditions, the JA concentration in the leaves and the OPDA concentration in the leaves, shoot apex and root increased significantly in ‘QY10,’ whereas the JA concentration in the root crown and the OPDA concentration in the root crown and root of ‘W10’ decreased significantly (Figure 7). The expression of the JA biosynthesis genes BnAOC1 and BnLOX4 (AOC1, allene oxide cyclase1; LOX4, lipoxygenase 4) indicated an alteration trend under long-term B deficiency that was similar to the change in the JAs concentration in the leaves. Both BnAOC1 and BnLOX4 were up-regulated in the leaves of ‘W10’ under B deficiency, while the expression of BnLOX4 in the leaves of ‘QY10’ was slightly down-regulated (Figure 7).
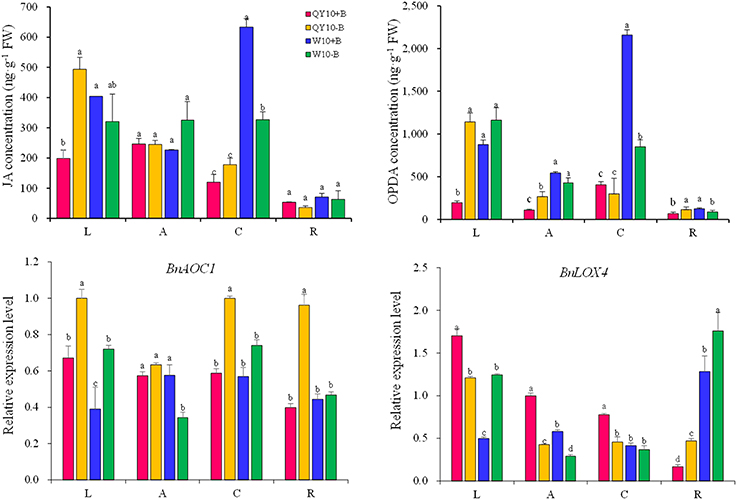
Figure 7. JA and OPDA concentrations and expression of JAs biosynthesis genes under long-term low-B stress. BnAOC1, BnLOX4: JA biosynthesis-related genes. Plants were grown hydroponically under sufficient B (+B, 25 μM B) or low-B (−B, 0.25 μM B) for 20 days. L, leaves; A, shoot apex; C, root crown; R, root. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same organ among the treatments (ANOVA, LSD, p < 0.05).
Under short-term B-free starvation, an apparent pattern in the concentration of JAs was observed. For both ‘QY10’ and ‘W10,’ the JA concentration in the leaves, shoot apexes and roots peaked at 12 h (9:00 p.m.), followed by an attenuation at 1 day to a level that was parallel to that observed at 0 h (9:00 a.m.), regardless of whether the plants were under the B-sufficient condition or the B withdrawal treatment (Figures 8A,B). When the B-free starvation continued to the 5th day, the JAs concentration increased drastically. Compared with ‘QY10,’ the change in the JAs concentration under B-free starvation was more dramatic in the B-inefficient genotype ‘W10.’ On the 5th day, the respective JA concentration in the leaves, shoot apex and root increased by 107.2, 27.8, and 121.9% in ‘QY10’ compared with 507.1, 143.8, and 92.5% in ‘W10.’In addition, the respective JA-Ile concentration in the leaves, shoot apex and root increased by 58.1, 319.3, and 16.1% in ‘QY10’ compared with 15.1, 261.5, and 50.6% in ‘W10’ (Figures 8A,B). Compared with the B-sufficient condition, the expression of the BnAOC1 and BnLOX4 genes for JAs biosynthesis was down-regulated from 3 h to 5 days under short-term B-free starvation. However, these genes were significantly up-regulated on the 5th day in ‘W10’ (Figure 8C). Collectively, our data suggested that low-B stress increased the JA concentration in a time- and organ-specific manner.
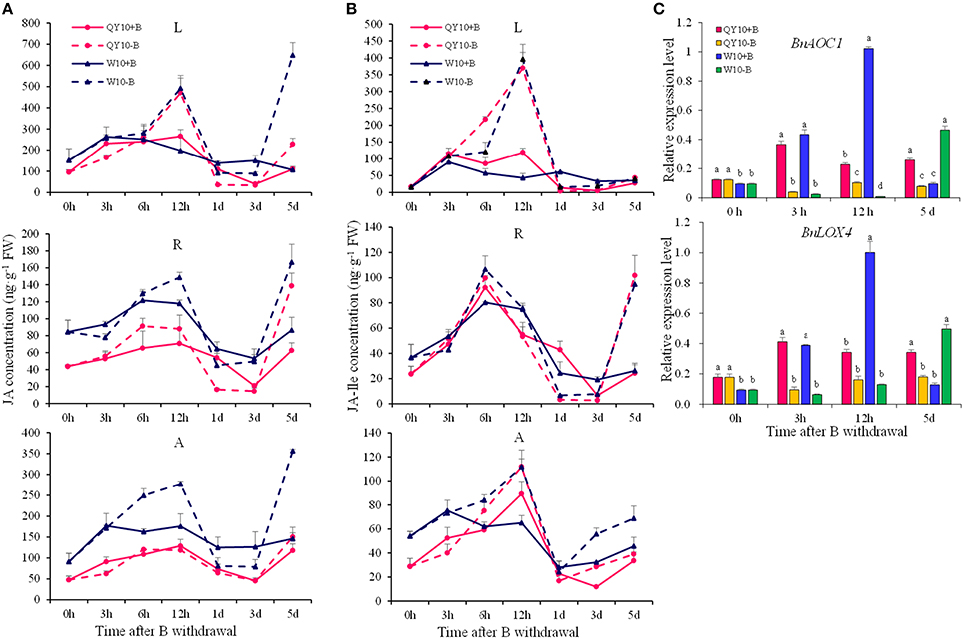
Figure 8. JAs concentrations and related-gene expression of ‘QY10’ and ‘W10’ after been transferred to a B-free solution. (A) JA concentration. (B) JA-Ile concentration. (C) Expression levels of the JAs biosynthesis genes BnAOC1 and BnLOX4 in the leaves of ‘QY10’ and ‘W10.’ After been grown hydroponically under sufficient B (+B, 25 μM B) for 12 days, half of the plants were transplanted to B-free solution (−B) and then sampled at different time points, and the other plants, which served as the control, were grown continuously with sufficient B. L, leaves; R, root; A, shoot apex. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same time among the treatments (ANOVA, LSD, p < 0.05).
Effect of B Deficiency on ABA in ‘QY10’ and ‘W10’
The ABA concentration did not show significant changes under long-term low-B stress (Figure 9). While the expression of the ABA biosynthesis gene BnNCED3 and the ABA sensor gene BnPYL4 exhibited a rather strange alteration trend in ‘QY10’ and ‘W10’ for the whole plant. The expression of BnNCED3 was up-regulated in the root crown in both ‘QY10’ and ‘W10,’ and the expression of BnPYL4 was up-regulated in the leaves, root crown and root of ‘QY10.' However, in most cases, the expression of BnNCED3 and BnPYL4 was down-regulated in ‘W10,’ which may be attributed the disorder caused by long-term B deficiency in this genotype.
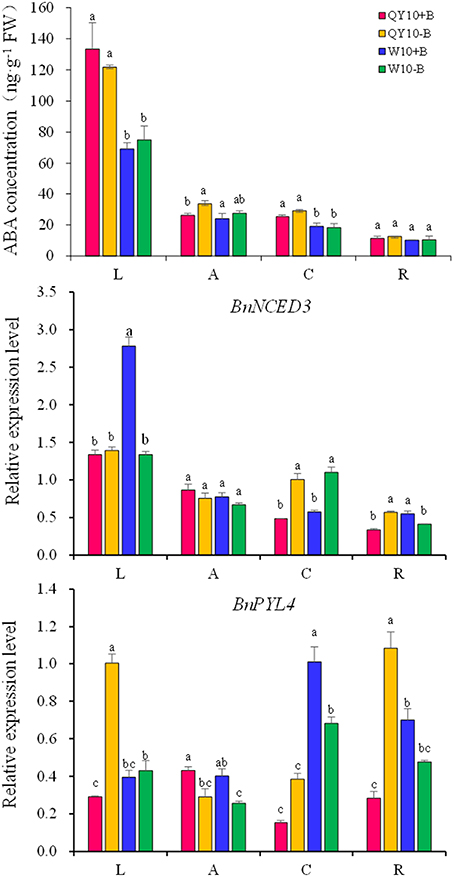
Figure 9. ABA concentration and related-gene expression under long-term low-B stress. BnNCED3, ABA biosynthesis-related gene; BnPYL4, ABA sensor gene. Plants were grown hydroponically under sufficient B (+B, 25 μM B) or low-B (−B, 0.25 μM B) for 20 days. L, leaves; A, shoot apex; C, root crown; R, root. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same organ among the treatments (ANOVA, LSD, p < 0.05).
Under short-term B-free starvation, the ABA concentration remained in a dynamic balance except that it increased drastically from the 3rd day to the 5th day in the shoot of ‘W10’ (Figure 10A). BnNCED3 and BnPYL4 were up-regulated from 12 h to 5 days except for BnNCED3 in ‘QY10’ on the 5th day, which confirmed the ABA concentration result (Figure 10B). These analyses suggested that the ABA concentration was more likely to be induced in the shoot of ‘W10’ under low-B conditions.
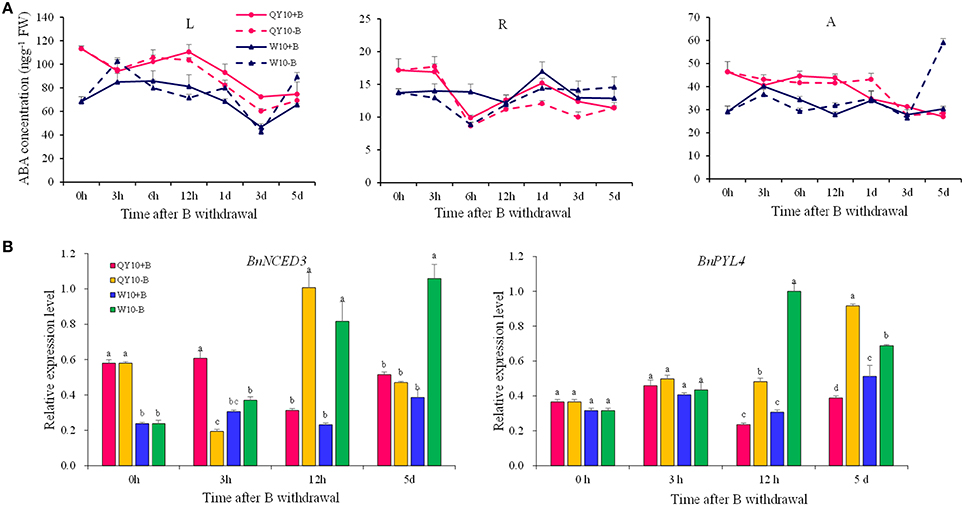
Figure 10. ABA concentration and its biosynthesis gene expression in ‘QY10’ and ‘W10’ after been transferred to a B-free solution. (A) ABA concentration. (B) ABA biosynthesis gene expression in the leaves of ‘QY10’ and ‘W10.’After been grown hydroponically under sufficient B (+B, 25 μM B) for 12 days, half of the plants were transplanted to B-free solution (−B) and then sampled at different time points, and the other plants, which served as the control, were grown continuously with sufficient B. L, leaves; R, root; A, shoot apex. Values represent the mean ± SD of four independent replicates, and bars with different letters show significant differences for the same time among the treatments (ANOVA, LSD, p < 0.05).
Discussion
In the present study, analyses of phytohormone concentrations and related gene expression in different tissues of B. napus revealed for the first time that B deficiency exerted a considerable impact on IAA, Jas, and ABA homeostasis. In addition, the more disordered phytohormone status in the B-inefficient genotype ‘W10’ was attributed to its greater sensitivity to stress caused by B deficiency.
Auxin-Mediated Axillary Bud and Root Growth in Response to B Deficiency
In plants, the awe-inspiring number of processes is regulated by auxin and its directional cell-to-cell transport, e.g., the regulation of apical dominance (Phillips, 1975), promotion of the development of lateral roots (Casimiro et al., 2001), and the promotion of phototropism and gravitropism (Iino, 1995). Auxin homeostasis in plants is regulated by synthesis, catabolism, transport, and conjugation (Woodward and Bartel, 2005). Among the phytohormones, auxins are the most obviously involved with B nutrition of plants. Many B-defective symptoms exhibit considerable similarity to the symptoms of abnormal auxin concentrations, including the inhibition of root elongation, a change in the direction of cell expansion from longitudinal to radial, browning of root tips and proliferation of lateral roots (Pilbeam and Kirkby, 1983). However, auxin concentration changes in response to low-B stress have remained a dilemma for many years (Coke and Whittington, 1968; Crisp et al., 1976; Li et al., 2001; Wang et al., 2006). For the initiation of axillary buds, a minimum auxin accumulation in the leaf axil is necessary, and this auxin distribution is mediated by PIN1 (Wang et al., 2014). Auxin accumulation in the root apex is mediated by both auxin biosynthesis and transport (Grieneisen et al., 2007; Mano and Nemoto, 2012). In this research, it can be concluded that auxin concentration decreased under low-B (Figures 3, 4A), both in the shoot apex and root, which was consistent with previous studies (Li et al., 2001; Wang et al., 2006), and this may be attributed to the down-regulation of the auxin synthesis gene BnNIT1 (Figures 3, 4B). Auxin concentration did not decrease until the 3rd day, indicating auxin concentration decrease was an indirect response, which was also consistent with previous studies (Li et al., 2001; Wang et al., 2006; González-Fontes et al., 2015). As auxin efflux genes, PIN1 and PIN2 play different physiological roles and represent different expression patterns in Arabidopsis. In the shoot, PIN1 mainly regulates the auxin apical-to-basal flow (Tanaka et al., 2006). In the root, PIN1 is expressed in the vascular bundle, charging auxin flow to the root tip to maintain root growth (Tanaka et al., 2006; Krecek et al., 2009). PIN2 is mainly expressed in the epidermis and cortex in the root meristem, regulating meristem enlargement in the root (Tanaka et al., 2006; Kleine-Vehn and Friml, 2008). In the present study, the expression of BnPIN1 in the shoot apex and root paralleled the IAA concentration under long-term B deficiency (Figure 3), which is in agreement with previous studies (Peng et al., 2012; Li et al., 2015). Meanwhile, the expression of BnPIN2 declined in the root of ‘W10’ under long-term B deficiency, which is in agreement with Camacho-Cristobal et al. (2015) (Figure 3). The attenuated auxin transport may function synergistically with the reduced auxin biosynthesis to block auxin accumulation in low-B-treated roots. However, under short-term B-free starvation, the expression of BnPIN1 was induced from 0 to 12 h, followed by a decrease in the root on the 5th day; BnPIN2 exhibited a similar expression pattern (Figure 4B). Therefore, more attention should be paid to the different expression patterns of the PIN family in B. napus under long- and short-term B deficiency.
Combining the phenotypes of ‘QY10’ and ‘W10’ and the results of the IAA concentration, DGE analysis, quantitative gene expression analysis, Arabidopsis root DR5:GFP and PIN1::PIN1-GFP fluorescence observations and the IAA exogenous application experiment, we propose a model accounting for the specific effect of IAA on axillary bud (AB) and root growth under a relatively long-term B deficiency (Figure 11). Two situations were shown to illustrate the effect of auxin on AB and root growth, corresponding to plants supplied with sufficient or low-B. In many plant species, the outgrowth of the AB is inhibited by an active apical bud; this is, generally referred to as apical dominance (Wang et al., 2014). Under B-sufficient conditions, plants are in a suitable auxin level, which slows down the outgrowth and elongation of the AB because of apical dominance. In the root, auxin was distributed mainly in the QC, which guarantees the growth of the primary root (Figure 11A). However, in the absence of B, the expression of BnNIT1 and BnPIN1 was down-regulated, which reduced the synthesis and basipetal transport of auxin in the shoot apex, thus preventing auxin accumulation in the AB, and initiating the outgrowth and elongation of AB. Meanwhile, low-B also reduced the synthesis and acropetal transport of auxin in the root, preventing auxin accumulation in the QC and inhibiting the growth of the primary root (Figure 11B). We speculated a low-B tolerance mechanism based on phenotypic and phytohormonal evidences for ‘QY10’ and ‘W10,’ although the auxin changes in the two genotypes showed the same trends, there were significant differences when they were exposed to low-B stress. The magnitude of the auxin concentration reduction was much greater in ‘W10’ than in ‘QY10,’ and the decrease rate was more rapid (Figures 3, 4A). Meanwhile, auxin biosynthetic gene BnNIT1 declined much more in each organ of ‘QY10’ than ‘W10’ (Figure 3). Transcription profiling proved that the expression of the DEGs involved in the auxin-mediated signaling pathway was almost higher in the leaves and root of ‘QY10’ compared with ‘W10’ under B deficiency (Figures 2C,D).
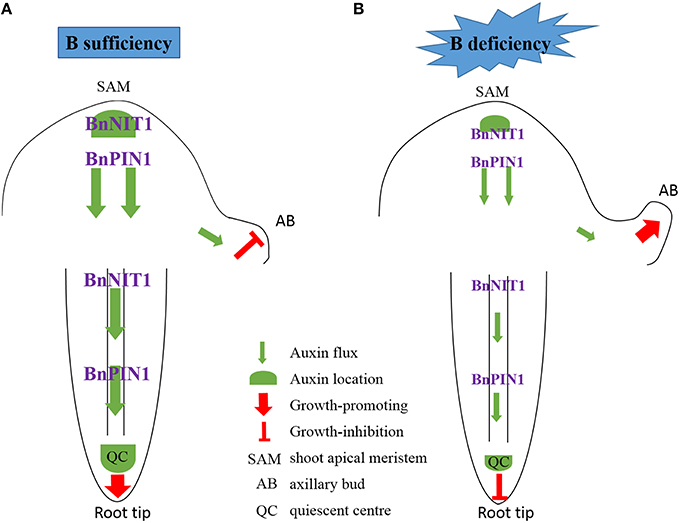
Figure 11. Schematic model for auxin control of axillary bud and root tip growth in response to B deficiency. Two situations were shown to illustrate the effect of auxin on AB and root growth, corresponding to plants supplied with sufficient or low-B. (A) Under B-sufficient conditions, plants are in a suitable auxin level, which slows down the outgrowth and elongation of the AB because of apical dominance. In the root, auxin was distributed mainly in the QC, which guarantees the growth of the primary root. (B) However, in the absence of B, the expression of BnNIT1 and BnPIN1 was down-regulated, which reduced the synthesis and basipetal transport of auxin in the shoot apex, thus preventing auxin accumulation in the AB, and initiating the outgrowth and elongation of AB. Meanwhile, low-B also reduced the synthesis and acropetal transport of auxin in the root, preventing auxin accumulation in the QC and inhibiting the growth of the primary root.
Response of JAs to B Deficiency
JAs are widely distributed in the plant kingdom and play multiple roles in physiological processes, such as inhibiting the growth of Arabidopsis seedlings (Staswick et al., 1992; Feys et al., 1994), regulating the resistance to insects and pathogens (Browse, 2009a) and the development of reproductive organs (Browse, 2009b; Wasternack et al., 2013). Whereas, the role of JAs in the resistance to mineral element deficiencies has been rarely reported. In the present study, the concentration changes of JA and its precursor OPDA or the conjugate JA-Ile were investigated in B. napus under low-B stress, together with the expression of JAs biosynthesis genes. Based on the above-mentioned analysis, JAs were found to be the most abundant phytohormone among the three phytohormones determined in B. napus. In addition, there was an obvious pattern in the concentration of JAs, which increased from morning (9:00 a.m.) to night (9:00 p.m.) and then declined by the next morning, i.e., the circadian rhythm, together with JAs biosynthesis gene expression (Figure 8). A similar phenomenon has been reported in rice (Liu et al., 2012). This may be caused by the self-activation of the JA biosynthesis mechanism (Sasaki et al., 2001), and the positive feedback mechanism of the JA biosynthetic enzymes (Creelman and Mullet, 1997). Under the long-term low-B condition, the alteration trend differed between the two genotypes: the JA concentration increased significantly in ‘QY10’ but decrease slightly in ‘W10.’ Interestingly, the JA concentration and its synthesis genes were up-regulated under low-B stress, especially in the leaves of ‘W10’ (Figure 7). It was likely that ‘W10’ plants were about to die after the 20-day B deficiency, the physiological function of these plants was seriously damaged and the cell biology activity was irreversibly inactivated (Oiwa et al., 2013), some responses measured were those which characterized plants that were severely damaged (Brown et al., 2002), so the JA concentration decreased. Under short-term B-free starvation, the JAs concentration increased on the 5th day and was more erratic in ‘W10’ (Figures 8A,B). However, the expression of JAs related genes was suppressed compared with the B-sufficient condition, except in ‘W10’ on the 5th day (Figure 8C). Therefore, the instant response of JAs to B deficiency may not be an increase, but rather a decrease, which requires further investigation.
The distinct responses of the JAs concentrations between the B-efficient and B-inefficient B. napus genotypes prompted us to determine whether JA interacted with B and if so, whether this interaction contributes to plant tolerance to B deficiency. In the DGE profiling analysis, the expression of all DEGs involved in the JAs biosynthetic process was lower in the leaves of ‘QY10’ than ‘W10’ under B deficiency (Figure 2F), further confirming the results of the measured JAs concentrations (Figures 7, 8A,B). Transcriptional regulation is one of the crucial mechanisms involved in protecting plants from various environmental stresses, and the key roles of many individual transcription regulators and their family classes in plant responses to environmental stresses have been well-demonstrated (Todaka et al., 2012). MYB21, MYB24, MYB34, MYB47, and MYB75 have been reported to be involved in the JA-mediated signaling pathway (Chen Y. H. et al., 2006; Qi et al., 2011). In the present study, the expression of MYB34 and MYB47 were significantly higher in both the root and leaves of ‘QY10’ compared with ‘W10’ (Figures 2A,B), indicating they may mediate the crosstalk between JA and B in B. napus genotypes and may open new avenues for future studies in this field.
Response of ABA to B Deficiency
Ever since its discovery, ABA has been intensively studied due to its versatile functions in plant development and stress physiology (Ye et al., 2012). The physiological roles of ABA were previously elucidated, including the establishment of seed dormancy, root development, stomatal closure, and so on (Cutler et al., 2010; Hirayama and Shinozaki, 2010). ABA is regarded as a phytohormone that can respond to nearly every type of stress (Chen J. et al., 2006; Ye et al., 2012). Wang and Zhou (1994) also reported that considerable amounts of ABA were generated enormously during every development period of cotton under B deficiency. In the present study, the expression of BnNCED3 and BnPYL4 was up-regulated over time (Figure 10B); on the 5th day, ‘W10’ underwent relatively severe B deficiency, and the ABA concentration increased drastically in the shoot of this genotype (Figure 10A). However, under long-term low-B stress, this difference was not apparent, and the ABA concentrations were similar to those measured in plants with sufficient B. A similar result was found in the root of wheat, which showed ABA only increased in the apical of root, not in the whole root under nutrient starvation (Vysotskaya et al., 2008). The expression of the DEGs involved in the ABA-mediated signaling pathway was almost higher in the leaves of ‘W10’ compared with ‘QY10’ (Figure 2G), and this is highly consistent with the result that the ABA concentration increased in the shoot of ‘W10’ but did not change in ‘QY10’ (Figure 10A). A similar alteration pattern was exhibited by the JAs and ABA, i.e., their concentrations increased in ‘QY10’ and decreased in ‘W10’ under long-term B deficiency, while their concentrations increased, especially in ‘W10,’ under short-term B-free starvation (Figures 7–10). The possible explanation for this is the same as that for the JAs, i.e., the leaf physiological function was seriously damaged in ‘W10,’ and the cell biology activity was irreversibly inactivated, leading to a decline in the phytohormone concentration in this genotype.
Although, our knowledge of the mechanism underlying the balance of various phytohormones in B. napus genotypes under B deficiency remains limited, these results indicate a dynamic and complex regulatory network that includes different phytohormonal synthesis/signaling responses to B deficiency in the molecular aspect. In contrast to previously published phytohormonal analyses in other plant species, we used two genotypes with contrasting tolerance to B deficiency, and formulated a comprehensive characterization of different transcriptional behaviors of phytohormones-related genes under B deficiency, so the finding is also potentially valuable for B efficiency identification and rapid nutrient diagnosis in the crop species susceptible to B deficiency.
Author Contributions
TZ and FX designed the experiments; TZ conceived the project, analyzed the data, and wrote the article with contributions of all the authors; YH, YPH, GD, and LS provided technical assistance to TZ; FX supervised and complemented the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 31372129). We are grateful to Professor Hongwei Xue in Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences and Professor Yingtang Lu in Wuhan University for kindly providing Arabidopsis DR5:GFP and PIN1::PIN1-GFP seeds, respectively. We also thank Dr. Hongbo Liu in National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University for his assistance with the phytohormone analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00221
Abbreviations
AB, axillary bud; AOC1, allene oxide cyclase1; B, boron; BEC, B efficiency coefficient; BRs, brassinosteroids; B. napus, Brassica napus; CTK, cytokinin; DEGs, differentially expressed genes; DGE, digital gene expression; DM: dry matter; ETH, ethylene; FW: fresh weight; GA, gibberellin; GFP, green fluorescent protein; JAs, jasmonates; LOX4, lipoxygenase 4; NCED3, nine-cis-epoxycarotenoid dioxygenase 3; NIT1, nitrilase1; OPDA, JA biosynthetic precursor 12-oxophytodienoic; Pi, inorganic phosphate; PI, propidium iodide; PIN1, pin-formed 1; PIN2, pin-formed 2; PYL4, pyr1-like 4; QC, quiescent center; qRT-PCR, quantitative real time PCR; SA, salicylic acid; UFLC-ESI-MS, ultra-fast liquid chromatography-electrospray ionization tandem mass spectrometry.
References
Abreu, I., Poza, L., Bonilla, I., and Bolanos, L. (2014). Boron deficiency results in early repression of a cytokinin receptor gene and abnormal cell differentiation in the apical root meristem of Arabidopsis thaliana. Plant Physiol. Biochem. 77, 117–121. doi: 10.1016/j.plaphy.2014.02.008
Armengaud, P., Breitling, R., and Amtmann, A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136, 2556–2576. doi: 10.1104/pp.104.046482
Asad, A., Bell, R. W., Dell, B., and Huang, L. (1997). External boron requirements for Canola (Brassica napus L.) in boron buffered solution culture. Ann. Bot. 80, 65–73. doi: 10.1006/anbo.1997.0408
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Brown, P. H., Bellaloui, N., Wimmer, M. A., Bassil, E. S., Ruiz, J., Hu, H., et al. (2002). Boron in plant biology. Plant Biol. 4, 205–223. doi: 10.1055/s-2002-25740
Browse, J. (2009a). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Browse, J. (2009b). The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70, 1539–1546. doi: 10.1016/j.phytochem.2009.08.004
Camacho-Cristobal, J. J., Martin-Rejano, E. M., Herrera-Rodriguez, M. B., Navarro Gochicoa, M. T., Rexach, J., and Gonzalez-Fontes, A. (2015). Boron deficiency inhibits root cell elongation via an ethylene/auxin/ROS-dependent pathway in Arabidopsis seedlings. J. Exp. Bot. 66, 3831–3840. doi: 10.1093/jxb/erv186
Casimiro, I., Marchant, A., Bhalerao, R. P., Beeckman, T., Dhooge, S., Swarup, R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. doi: 10.1105/tpc.13.4.843
Chen, J., Pan, K. W., and Gu, B. (2006). Physiological function and mechanism of abscisic acid in plants under stress. Plant Physiol. Commun. 42, 1176–1182.
Chen, Y. H., Yang, X. Y., He, K., Liu, M. H., Li, J. G., Gao, Z. F., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Ciereszko, I., and Kleczkowski, L. A. (2002). Effects of phosphate deficiency and sugars on expression of rab18 in Arabidopsis: hexokinase-dependent and okadaic acid-sensitive transduction of the sugar signal. Biochim. Biophys. Acta 1579, 43–49. doi: 10.1016/S0167-4781(02)00502-X
Coke, L., and Whittington, W. J. (1968). The role of boron in plant growth IV. Interralationships between boron and indol-3yl-acetic acid in the metabolism of bean radicals. J. Exp. Bot. 19, 295–308. doi: 10.1093/jxb/19.2.295
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Crisp, P., Collier, G. F., and Thomas, T. H. (1976). The effect of boron on tipburn and auxin activity in lettuce. Sci. Hortic. 5, 215–226. doi: 10.1016/0304-4238(76)90085-6
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dan, H., Yang, G., and Zheng, Z. L. (2007). A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 63, 221–235. doi: 10.1007/s11103-006-9084-0
Feys, B., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. doi: 10.1105/tpc.6.5.751
Goldbach, H. E., Wimmer, M. A., and Findeklee, P. (2000). Discussion paper: Boron-how can the critical level be defined? J. Plant Nutr. Soil Sci. 163, 115–121. doi: 10.1002/(SICI)1522-2624(200002)163:1<115::AID-JPLN115>3.0.CO;2-#
González-Fontes, A., Herrera-Rodríguez, M., Martín-Rejano, E., Navarro-Gochicoa, M., Rexach, J., and Camacho-Cristóbal, J. (2015). Root responses to boron deficiency mediated by ethylene. Front. Plant Sci. 6:1103. doi: 10.3389/fpls.2015.01103
Grieneisen, V. A., Xu, J., Marée, A. F. M., Hogeweg, P., and Scheres, B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013. doi: 10.1038/nature06215
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hoagland, D. R., and Arnon, D. I. (1950). “The water-culture method for growing plants without soil,” in California Agricultural Experiment Station Circular, Vol. 347, ed D. I. Arnon (Berkeley, CA: The College of Agriculture; University of California), 1–39.
Iino, M. (1995). Gravitropism and phototropism of maize coleoptiles: evaluation of the cholodny-went theory through effects of auxin application and decapitation. Plant Cell Physiol. 36, 361–367.
Iqbal, N., Trivellini, A., Masood, A., Ferrante, A., and Khan, N. A. (2013). Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol. Biochem. 73, 128–138. doi: 10.1016/j.plaphy.2013.09.011
Jung, J. Y., Shin, R., and Schachtman, D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21, 607–621. doi: 10.1105/tpc.108.063099
Kapulnik, Y., Resnick, N., Mayzlish-Gati, E., Kaplan, Y., Wininger, S., Hershenhorn, J., et al. (2011). Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 62, 2915–2924. doi: 10.1093/jxb/erq464
Kiba, T., Kudo, T., Kojima, M., and Sakakibara, H. (2011). Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409. doi: 10.1093/jxb/erq410
Kleine-Vehn, J., and Friml, J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24, 447–473. doi: 10.1146/annurev.cellbio.24.110707.175254
Krecek, P., Skupa, P., Libus, J., Naramoto, S., Tejos, R., Friml, J., et al. (2009). The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 10:249. doi: 10.1186/gb-2009-10-12-249
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Lee, H. G., Lee, K., and Seo, P. J. (2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 87, 371–381. doi: 10.1007/s11103-015-0283-4
Li, C. J., Pfeffer, H., Dannel, F., Römheld, V., and Bangerth, F. (2001). Effects of boron starvation on boron compartmentation, and possibly hormone-mediated elongation growth and apical dominance of pea (Pisum sativum) plants. Physiol. Plant. 111, 212–219. doi: 10.1034/j.1399-3054.2001.1110212.x
Li, K., Kamiya, T., and Fujiwara, T. (2015). Differential roles of PIN1 and PIN2 in root meristem maintenance under low-B Conditions in Arabidopsis thaliana. Plant Cell Physiol. 56, 1205–1214. doi: 10.1093/pcp/pcv047
Liu, H. B., Li, X. H., Xiao, J. H., and Wang, S. P. (2012). A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8:2. doi: 10.1186/1746-4811-8-2
Liu, T. Y., Chang, C. Y., and Chiou, T. J. (2009). The long-distance signaling of mineral macronutrients. Curr. Opin. Plant Biol. 12, 312–319. doi: 10.1016/j.pbi.2009.04.004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mano, Y., and Nemoto, K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63, 2853–2872. doi: 10.1093/jxb/ers091
Martin-Rejano, E. M., Camacho-Cristobal, J. J., Herrera-Rodriguez, M. B., Rexach, J., Navarro-Gochicoa, M. T., and Gonzalez-Fontes, A. (2011). Auxin and ethylene are involved in the responses of root system architecture to low boron supply in Arabidopsis seedlings. Physiol. Plant. 142, 170–178. doi: 10.1111/j.1399-3054.2011.01459.x
Meyer, M. (2009). Rapeseed oil fuel - the crisis-proof home-made eco-fuel. Agrarforschung 16, 262–267.
Oiwa, Y., Kitayama, K., Kobayashi, M., and Matoh, T. (2013). Boron deprivation immediately causes cell death in growing roots of Arabidopsis thaliana (L.) Heynh. Soil Sci. Plant Nutr. 59, 621–627. doi: 10.1080/00380768.2013.813382
Peng, L., Zeng, C., Shi, L., Cai, H., and Xu, F. (2012). Transcriptional profiling reveals adaptive responses to boron deficiency stress in Arabidopsis. Z. Naturforsch. C. 67, 510–524. doi: 10.5560/ZNC.2012.67c0510
Philippar, K., Fuchs, I., Lüthen, H., Hoth, S., Bauer, C. S., Haga, K., et al. (1999). Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc. Natl. Acad. Sci. U.S.A. 96, 12186–12191. doi: 10.1073/pnas.96.21.12186
Phillips, I. D. J. (1975). Apical dominance. Annu. Rev. Plant Physiol. 26, 341–367. doi: 10.1146/annurev.pp.26.060175.002013
Pilbeam, D. J., and Kirkby, E. A. (1983). The physiological role of boron in plants. J. Plant Nutr. 6, 563–582. doi: 10.1080/01904168309363126
Potters, G., Pasternak, T. P., Guisez, Y., Palme, K. J., and Jansen, M. A. (2007). Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci. 12, 98–105. doi: 10.1016/j.tplants.2007.01.004
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Romera, F. J., García, M. J., Alcántara, E., and Pérez-Vicente, R. (2011). Latest findings about the interplay or auxin, ethylene and nitric oxide in the regulation of Fe deficiency responses by Strategy I plants. Plant Signal. Behav. 6, 167–170. doi: 10.4161/psb.6.1.14111
Rubio, V., Bustos, R., Irigoyen, M. L., Cardona-Lopez, X., Rojas-Triana, M., and Paz-Ares, J. (2009). Plant hormones and nutrient signaling. Plant Mol. Biol. 69, 361–373. doi: 10.1007/s11103-008-9380-y
Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., et al. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153–161. doi: 10.1093/dnares/8.4.153
Scheible, W. R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., et al. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. doi: 10.1104/pp.104.047019
Shen, C., Yue, R., Sun, T., Zhang, L., Yang, Y., and Wang, H. (2015). OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 231, 148–158. doi: 10.1016/j.plantsci.2014.12.003
Shi, W. G., Li, H., Liu, T. X., Polle, A., Peng, C. H., and Luo, Z. B. (2015). Exogenous abscisic acid alleviates zinc uptake and accumulation in Populus x canescens exposed to excess zinc. Plant Cell Environ. 38, 207–223. doi: 10.1111/pce.12434
Shin, R., and Schachtman, D. P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl Acad. Sci. U.S.A. 101, 8827–8832. doi: 10.1073/pnas.0401707101
Staswick, P. E., Su, W., and Howell, S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89, 6837–6840. doi: 10.1073/pnas.89.15.6837
Tanaka, H., Dhonukshe, P., Brewer, P. B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol. Life Sci. 63, 2738–2754. doi: 10.1007/s00018-006-6116-5
Tanaka, M., and Fujiwara, T. (2008). Physiological roles and transport mechanisms of boron: perspectives from plants. Pflugers. Archiv. 456, 671–677. doi: 10.1007/s00424-007-0370-8
Todaka, D., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice 5:6. doi: 10.1186/1939-8433-5-6
Trull, M. C., Guiltinan, M. J., Lynch, J. P., and Deikman, J. (1997). The responses of wild-type and ABA mutant Arabidopsis thaiiana plants to phosphorus starvation. Plant Cell Environ. 20, 85–92. doi: 10.1046/j.1365-3040.1997.d01-4.x
Vysotskaya, L. B., Korobova, A. V., and Kudoyarova, G. R. (2008). Abscisic acid accumulation in the roots of nutrient-limited plants: its impact on the differential growth of roots and shoots. J. Plant Physiol. 165, 1274–1279. doi: 10.1016/j.jplph.2007.08.014
Wang, G., Romheld, V., Li, C., and Bangerth, F. (2006). Involvement of auxin and CKs in boron deficiency induced changes in apical dominance of pea plants (Pisum sativum L.). J. Plant Physiol. 163, 591–600. doi: 10.1016/j.jplph.2005.09.014
Wang, Q., Kohlen, W., Rossmann, S., Vernoux, T., and Theres, K. (2014). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26, 2068–2079. doi: 10.1105/tpc.114.123059
Wang, Y. H., and Zhou, X. F. (1994). Effect of boron on some element, phenol, enzyme activity and hormone in cotton Petiole. Plant Nutr. Fertil. Sci. 1, 61–66.
Wang, Z., Wang, Z., Chen, S., Shi, L., and Xu, F. (2011). Proteomics reveals the adaptability mechanism of Brassica napus to short-term boron deprivation. Plant Soil 347, 195–210. doi: 10.1007/s11104-011-0838-3
Wasternack, C., Forner, S., Strnad, M., and Hause, B. (2013). Jasmonates in flower and seed development. Biochimie 95, 79–85. doi: 10.1016/j.biochi.2012.06.005
Woodward, A. W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Xiong, S. L., Wu, L. S., and Wang, Y. H. (2001). Study on relationship between B-deficiency symptoms of cucumber and changes of endogenous hormone. Plant Nutr. Fertil. Sci. 7, 194–198.
Xu, F. S., Yang, Y. H., Wang, Y. H., and Wu, L. S. (2002). “Boron uptake and retranslocation in cultivars of Brassica napus differing in boron efficiency,” in Boron in Plant and Animal Nutrition, eds H. E. Goldbach, B. Rerkasem, M. A. Wimmer, P. H. Brown, M. Thellier, and R. W. Bell (New York, NY: Academic Press), 127–135.
Ye, N., Jia, L., and Zhang, J. (2012). ABA signal in rice under stress conditions. Rice 5:1. doi: 10.1186/1939-8433-5-1
Yu, L. J., Luo, Y. F., Liao, B., Xie, L. J., Chen, L., Xiao, S., et al. (2012). Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 195, 97–112. doi: 10.1111/j.1469-8137.2012.04154.x
Zhang, Z., Liao, H., and Lucas, W. J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56, 192–220. doi: 10.1111/jipb.12163
Zhao, H., Shi, L., Duan, X., Xu, F., Wang, Y., and Meng, J. (2008). Mapping and validation of chromosome regions conferring a new boron-efficient locus in Brassica napus. Mol. Breed. 22, 495–506. doi: 10.1007/s11032-008-9193-3
Keywords: ABA, boron deficiency response, Brassica napus, gene expression profiling, IAA, JAs
Citation: Zhou T, Hua Y, Huang Y, Ding G, Shi L and Xu F (2016) Physiological and Transcriptional Analyses Reveal Differential Phytohormone Responses to Boron Deficiency in Brassica napus Genotypes. Front. Plant Sci. 7:221. doi: 10.3389/fpls.2016.00221
Received: 02 December 2015; Accepted: 09 February 2016;
Published: 26 February 2016.
Edited by:
Raul Antonio Sperotto, Centro Universitário Univates, BrazilReviewed by:
Nacer Bellaloui, U.S. Department of Agriculture-Agricultural Research Service, USAYuan Mei Zuo, China Agricultural University, China
Copyright © 2016 Zhou, Hua, Huang, Ding, Shi and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangsen Xu, ZmFuZ3Nlbnh1QG1haWwuaHphdS5lZHUuY24=
 Ting Zhou
Ting Zhou Yingpeng Hua
Yingpeng Hua Yupu Huang
Yupu Huang Guangda Ding
Guangda Ding Lei Shi
Lei Shi Fangsen Xu
Fangsen Xu