- 1National Research Centre on Plant Biotechnology, Indian Agricultural Research Institute Campus, New Delhi, India
- 2Department of Biotechnology, Periyar University, Salem, India
Aphids, a hemipteran group of insects pose a serious threat to many of the major crop species including Brassica oilseeds. Transgenic strategies for developing aphid-resistant plant types necessitate phloem-bound expression of the insecticidal genes. A few known phloem-specific promoters, in spite of tissue-specific activity fail to confer high level gene-expression. Here, we identified seven orthologues of phloem-specific promoters in B. juncea (Indian mustard), and experimentally validated their strength of expression in phloem exudates. Significant cis-motifs, globally occurring in phloem-specific promoters showed variable distribution frequencies in these putative phloem-specific promoters of B. juncea. In RT-qPCR based gene-expression study promoter of Glutamine synthetase 3A (GS3A) showed multifold higher activity compared to others, across the different growth stages of B. juncea plants. A statistical method employing four softwares was devised for rapidly analysing stability of the promoter-activities across the plant developmental stages. Different statistical softwares ranked these B. juncea promoters differently in terms of their stability in promoter-activity. Nevertheless, the consensus in output empirically suggested consistency in promoter-activity of the six B. juncea phloem- specific promoters including GS3A. The study identified suitable endogenous promoters for high level and consistent gene-expression in B. juncea phloem exudate. The study also demonstrated a rapid method of assessing species-specific strength and stability in expression of the endogenous promoters.
Introduction
Rapeseed-mustard (Brassica spp.) constitute the third most important group of oilseeds in world agriculture. In India, among the three species of rapeseed and mustard Indian-mustard, Brassica juncea (L.) is the chief oil-yielding crop. A major threat to productivity of this crop is posed by aphids, a phloem-feeding hemipteran pest (The International Aphid Genomics Consortium, 2010; Bhatia et al., 2011). Lack of resistance-source within the Brassica spp. limits the conventional breeding for developing aphid-resistant cultivars therefore, the challenging task of aphid-management solely depends on application of systemic insecticides, which inadvertently enter into the food chain. Transgenic approach by utilizing genes from distant sources seems to be a potential avenue for developing aphid resistance in B. juncea. However, the past efforts have led to only moderate success as evident by no commercial release, so far, of any aphid-resistant transgenic plant-type (Dutta et al., 2005; Yu et al., 2014). Whether this bottleneck is due to the lack of effective toxicity of the transgenes or due to its inadequate spatio-temporal expression in the transgenic plants remains enigmatic.
Transgene-expression is governed by the choice of appropriate promoter (Hernandez-Garcia and Finer, 2014). During the early phase of genetic engineering mostly constitutive promoters have been used for a wide range of trait-expression including aphid resistance (Bhatia et al., 2012; Jisha et al., 2015). However, soon it was evident that the constitutive expression of the transgene led to more metabolic pay-offs and often undesirable pleiotropic effects in the transgenic plants (Kasuga et al., 1999; Hsieh et al., 2002; Zhou et al., 2013). Therefore, tailoring transgene-expression with tissue- and temporal-specificity is significant for minimizing such unintended effects of the transgene. In transcriptional control of gene-expression, cis-acting promoter elements play a pivotal role (Hernandez-Garcia and Finer, 2014). In many instances, specific DNA-elements in abiotic stress-inducible promoters conferred specificity of gene-expression and helped in mitigating the undesirable effects of constitutive expression (Lee et al., 2003; Kasuga et al., 2004). Alternatively, for restricting transgene-expression in the desired tissues, tissue-specific promoters have been used (Zheng and Baum, 2008). However, lack of understanding the architecture and DNA-motifs of the tissue-specific promoters necessitates more efforts in this direction.
At molecular level, plants respond to aphid-probing by eliciting phloem-based defense (Louis and Shah, 2013). Therefore, phloem-specific promoters have been considered most suitable for transgenic-expression of defense proteins against aphids (Sadeghi et al., 2007; Chakraborti et al., 2009). To that end, several phloem-specific promoters have been isolated from viruses, bacteria and plants; for example, viral promoters (Medberry et al., 1992; Bhattacharyya-Pakrasi et al., 1993; Rohde et al., 1995; Dinant et al., 2004), agrobacterial promoter rolC (Yokoyama et al., 1994), Arabidopsis promoter AtSUC2, and several SUS promoters from diverse species (Truernit and Sauer, 1995; Singer et al., 2011; Dutt et al., 2012). Notwithstanding apparent redundancy in phloem-specific promoters from viruses and bacteria, endogenous plant promoters are preferred for efficient and consistent transgene-expression in plants (Furtado et al., 2008; Hernandez-Garcia et al., 2009).
Availability of genome sequences, microarray database, and transcriptome data has enabled rapid identification of orthologous promoters across the plant species (Lim et al., 2012; Geng et al., 2014). Full-genome array of more and more number of plants has led to the development of several online bioinformatics tools for classification of related sequences and identification of conserved DNA motifs. Analysis tool, such as MEME suit, allows associating these motifs with gene ontology for deriving their functional significance in gene-function relationship (Bailey et al., 2009). Specific activity of the promoters is commonly identified through expression behavior of their cognate genes across the tissues, developmental stages, and environmental conditions (Kasuga et al., 2004; Ruiz-Medrano et al., 2011). Constitutive promoters, despite high level activity, may suffer inconsistency in certain tissues and at certain developmental stages of the plant (Sunilkumar et al., 2002). For example, CaMV 35S is poor in driving transgene-expression during boll formation in cotton, in germ line tissues of B. juncea, and in dark grown tissues of moss Physcomitrella patens (Arumugam et al., 2007; Saidi et al., 2009; Bakhsh et al., 2012). Therefore, in addition to strength, the spatio-temporal stability of activity is crucial in assessing suitability of a promoter for attaining the desired gene-expression.
We report here on in silico identification of seven orthologues of phloem-specific promoters in a major oilseed-crop, B. juncea and experimental validation of their strength in driving gene expression in the phloem-sap. As structural insight into their architecture, distribution of significant motif-patterns globally associated with phloem-specific promoters were analyzed. In addition, a statistical method based on RT-qPCR data was devised for assessing stability of the promoter-activities over different growth stages of the plant. The study highlights appropriate endogenous promoters for gene-expression in the phloem-sap of B. juncea.
Materials and Methods
Gene Ontology Analysis
The phloem-specific promoters from diverse plant species were identified based on published literature. The nucleotide sequences of the promoters and their cognate genes were retrieved from NCBI GenBank. Arabidopsis homologs of the promoters and their cognate genes were identified in TAIR database using BlastX programme of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Gene ontology analysis of the promoters was performed based on functional classification of their cognate genes. The genes were grouped into functional categories using the GO slim terms from the Arabidopsis information resource annotation (http://www.arabidopsis.org/tools/bulk/go/index.jsp).
Collection of Phloem Exudates
Seeds of Indian mustard (B. juncea) line Bio-YSR were germinated and grown in 12 inch pots in a net house during the mustard growing season (November-February) of Delhi, India. Since purpose of this study was assessing suitability of the promoters in expressing genes in the phloem-sap, all the gene-expression studies were carried out in the phloem exudates. Phloem exudates were collected at three different growth stages of the B. juncea plants viz. vegetative stage (15 days old), bud initiation stage (30 days old) and flowering stage (45 days old) as described in Buhtz et al. (2008). Fully expanded third to sixth leaves from the apical top were excised using a sharp razor blade and immediately transferred into glass tubes containing 20 mM K2-EDTA solution. After incubation for 30 min and removal of the initial exudates, the petiole ends were freshly cut to avoid sieve tube embolism. The cut ends were dipped in 500 μl freshly prepared 20 mM K2-EDTA solution containing 100 U/ml RNase inhibitor (cat# 1B1410, Ameresco, USA) and left for overnight in a pre-humid plexi-glass box. The phloem exudates collected in the EDTA buffer were frozen in liquid N2 and stored at −80°C until used for RNA extraction.
RNA Isolation and cDNA Synthesis
Total RNA was isolated using Trizol reagent (cat# 15596029, Invitrogen, USA) according to the manufacturer's instruction. The residual DNA was removed by RNase-free DNase (cat# 79254, Qiagen, USA) according to manufacturer's specification. Yield and purity of RNA was determined by NanoDrop ND-1000 spectrophotometer (Nanodrop technologies, USA). RNA samples with an absorbance ratio OD 260/280 between 1.9 and 2.2 and OD 260/230 greater than 2.0 were used for experimentations. RNA-integrity was verified through gel electrophoresis, by resolving the samples on 1.8% agarose-gel in 1X TAE. First stand cDNA was synthesized from 2 μg of total RNA using a cDNA synthesis kit (cat# 6110A, Takara Bio Inc., Japan), diluted 20 times with nuclease free water and used for PCR or RT-qPCR.
Primers and RT-PCR
Gene specific primers were designed based on Brassica specific homologous sequences using IDT primer quest software (http://www.idtdna.com) (Tables S1, S2). Primers' specificity was examined by validating the desired amplifications in RT-PCR as well as by sequencing of the amplicons. GeneRuler 100 bp Plus DNA Ladder (Cat# SM0323; Thermo Fisher Scientific, USA) was used as size marker in agarose gel. For determining amplification efficiency (E) of the primers in RT-qPCR, a serial tenfold dilution (1–1000) of the cDNA pool was used. A standard graph based on their Ct values was generated with linear regression and the slope using Microsoft Excel. The amplification efficiency (E) for each gene-specific primer was calculated according to the equation: E (%) = (10−1∕slope−1) × 100%. All the RT-qPCR reactions were performed using SYBR green detection chemistry, in a StepOne plus Real time PCR machine (Applied Biosystems, USA). A reaction cocktail of 20 μl was constituted by mixing 2 μl diluted cDNA, 10 μl 2X SYBR Premix Ex Taq II (cat# RR820A, Takara Bio Inc, Japan), 0.4 μl of ROX reference dye and 0.4 μl each of the forward and reverse primers. PCR cycling was carried out at an initial denaturation of 1 m at 95°C, followed by 40 repeated cycles each consists of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. To check amplification-specificity, dissociation curve analysis was carried out by constant increase of temperature between 60 and 95°C. All RT-qPCR and the RT-PCR experiments were carried out in three and two biological replicates, respectively with three technical replicates each time. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) was used as reference gene (Chandna et al., 2012; Bhogale et al., 2014).
Identification of Significant cis-Elements in Phloem-Specific Promoters
Over represented motifs were identified using MEME (Multiple Expectation Maximization for Motif Elicitation) suite (Bailey et al., 2009), Oligo-analyzer, and info-gibbs programs of RSAT (Regulatory Sequence Analysis Tools) motif discovery tool. The consensus motifs were identified with the following settings: 6–15 bases width, one or more occurrence per sequence, and E < 0.01 for only in the given strand of input sequences. Sequences were also aligned using the AlignACE program (http://atlas.med.harvard.edu/cgi-bin/alignace.pl), and the motifs with the highest MAP scores were selected and their web logo was plotted using WebLogo tool (http://weblogo.berkeley.edu/logo.cgi). The discovered motifs were further analyzed by homology search in PLACE and STAMP (http://www.benoslab.pitt.edu/stamp) programs to identify their putative functions. Statistically significant motifs were searched in individual promoters by using FIMO program of MEME with p < 0.0001.
Statistical Methods and Expression-Stability Analysis
RT-qPCR data was analyzed by Microsoft Excel based software tools geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeepr (version1) (Pfaffl et al., 2004), and the delta Ct methods (Silver et al., 2006) for ranking the expression-stability of the cognate genes. The geNorm algorithm calculates the gene-stability measure (M) among the genes in a given set of samples based on the principle that the logarithmically transformed expression ratios between any two genes should be invariable if both the genes are expressed equally in a given set of samples. It determines rank order of the genes in a set of samples by calculating expression-stability measure (M)-values based on geometric averaging of multiple genes as well as pair-wise comparison and stepwise exclusion of the genes from other samples. The M-value is inversely proportional to expression-stability of the gene; lower the M-value, higher is the expression-stability. M-value <1.5 is recommended to identify stably expressed genes. NormFinder calculates expression-stability of the genes in all the samples in any number of groups based on intra- and inter-group variations and combines these values to provide gene-rank depending on the variation in gene-expression. Gene with lower value signifies more stable expression. The Bestkeeper software uses geometric mean of Ct values with a standard deviation (SD) and PCR efficiency (E) to determine the best suited standards and combines them into an index by the coefficient of determination and the P-value. A lower index score refers to the higher stability in gene expression. The delta Ct method compares relative expression of genes in a pair-wise manner, in which each of the genes is considered as reference gene and compared with the remaining.
Results
Identification of Homologous Phloem-Specific Promoters in Brassica juncea
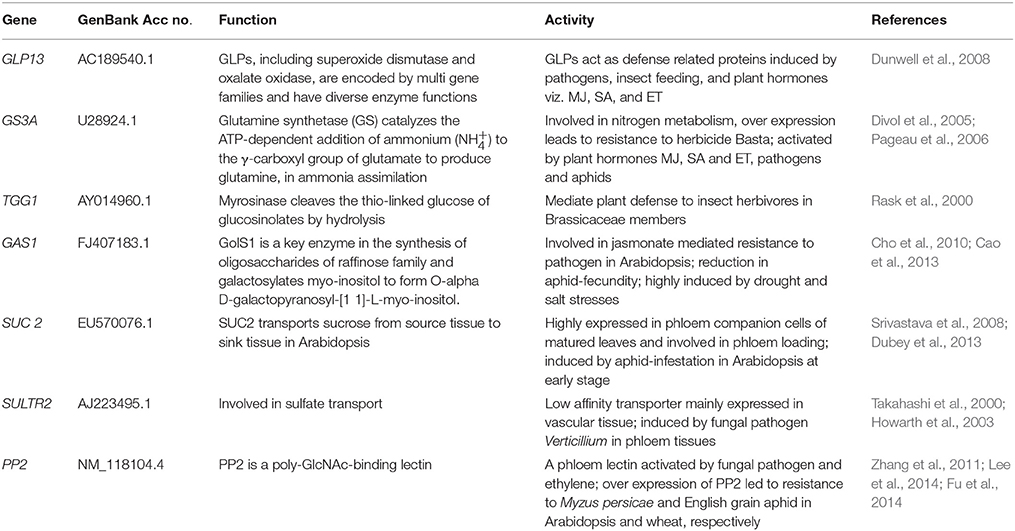
Based on literature, 39 known phloem-specific promoters were chosen and their sequences were downloaded from NCBI database (Table S3). These promoters, from monocot as well as dicot plants, ranged from smallest AtGLP13-promoter of 762 bp to CmGAS1-promoter of 3000 bp. Based on putative function of their cognate genes, the promoters were classified in two GO terms viz. biological process (Figure 1A) and molecular function (Figure 1B). In biological process among the major categories, about 26.6% of the promoters were found to be stress responsive and grouped under response to stresses, followed by 20%, that were associated with developmental processes, and 10% each controlling genes related to transport and transcription processes. Across the ontogenic groups, seven promoters that were associated with host-response against insects and pathogens, either directly or indirectly, were identified (Table 1). Homologues of the seven promoters were predicted in Brassica spp. and identified either in B. rapa or in B. juncea.
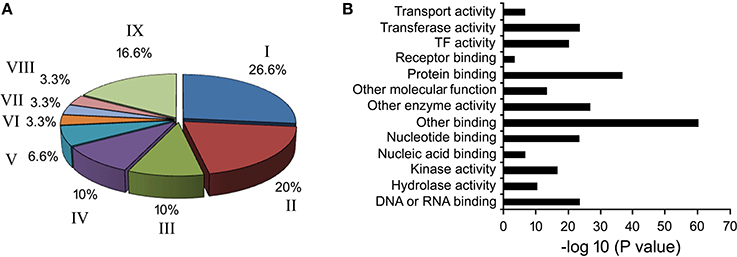
Figure 1. Gene ontology of phloem-specific transcripts. (A) Nine functional categories of the transcripts are represented by different letters in the pie chart. I. Response to biotic and abiotic stimulus, II. Developmental processes, III. Transcription, IV. Transport, V. Signal transduction, VI. Electron transport, VII. Cell organization and biogenesis, VIII. Protein metabolism, and IX. Unknown biological processes. (B) Distribution of Go terms based on their molecular function.
Functional Validation of the Predicted Promoters
RT-PCR based detection of the cognate mRNAs in the phloem exudates of B. juncea and their RT-qPCR based quantitative analysis validated activity of the in silico identified promoters and their relative strength, respectively in the phloem exudates of B. juncea. Limited availability of sequence information on B. juncea genome in public domain database led us to retrieve the cognate gene sequences from other species of Brassica. For each of the target transcripts, PCR and RT-qPCR conditions were optimized so that the gene specific primer-pairs amplified single PCR product of desired size and single peak in melt curve analysis (Figure S1). The amplicons were further validated through sequencing. The standard curve analysis for RT-qPCR efficiency revealed a linear regression R2 of all the primer-pairs ranging between 0.999 and 1.000 and the efficiency ranging from 93 to 105% (Figure S2). Phloem exudates were collected from excised petioles of B. juncea plants. The RNAs isolated from the phloem exudates were assayed for the presence of any non-vascular cellular contamination. For that, the RNAs were analyzed for the presence of RbcS and Lhca2 specific amplification in RT-PCR. RbcS and Lhca2 encode two highly abundant photosynthetic proteins rubisco small subunit and chlorophyll a/b binding protein, respectively. RbcS and Lhca2 gene expression is excluded from vascular cells (Sawchuk et al., 2008) and therefore, their mRNAs, despite showing different dynamics during leaf development, are likely to be completely absent in phloem exudates (Buhtz et al., 2008). No amplification of either RbcS or Lhca2 transcripts in RT-PCR unambiguously indicated purity of the phloem exudates (Figure 2A). On the other hand, specific amplification of the cognate genes in the same cDNA sample indicated phloem-bound activity of the seven orthologous B. juncea promoters (Figure 2B). The relative transcript levels of the cognate genes were estimated in RT-qPCR analysis and expressed as multifold ratio of their normalized level to the least abundant transcript level of GAS1 (Figure 2C). The results empirically showed significant variation among the transcript levels indicating significant differences in strength of the promoters. Amongst the variability, GS3A showed the highest transcript level followed by PP2, GLP13 and SULTR2. However, quantitative expressions of PP2, GLP13 and SULTR2 were statistically similar. Co-amplification of UBC9 in the exudate-cDNA indicated integrity and optimum level of cDNA in the reactions.
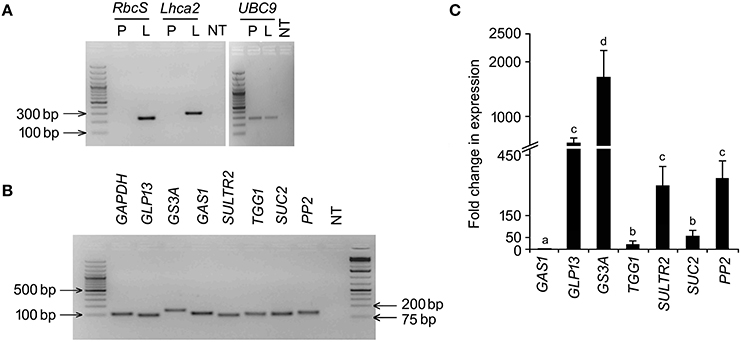
Figure 2. RT-PCR analysis for the cognate-transcripts of the phloem-specific promoters. Total RNA was isolated from phloem exudates collected from 45 days old B. juncea leaves and assayed for the cognate-transcripts of the in silico identified B. juncea phloem-specific promoters by RT-PCR and RT-qPCR. (A) RT-PCR amplification of RbcS, Lhca2 and UBC9 in phloem exudates (P), and leaf tissues (L) along with non-template (NT) control. (B) RT-PCR amplification of the cognate-transcripts in phloem exudates. (C) RT-qPCR based analysis of the cognate-transcript levels in phloem exudates.
Identification of Signature cis-Elements in Phloem-Specific Promoters
DNA-motifs globally associated with phloem-specific promoters were not known. Therefore, discovery of such motifs was mandatory for understanding the architecture of the putative phloem-specific promoters identified in B. juncea. For that, comprehensive set of 39 promoter sequences from diverse origin (described in Table S3) were analyzed, and the over-represented motifs in them were discovered through string based (oligo-analysis of RSAT) and position weight matrix-based (info-gibbs of RSAT, AlignACE, and MEME) motif discovery programmes (Van Helden et al., 1998; Hughes et al., 2000; Defrance and van Helden, 2009). Oligo-analysis revealed the commonly occurring over-represented motifs in all the promoter sequences. Among the identified motifs, the two with lowest expectation (E)-values were scored as significant and their frequency of occurrence per promoter has been shown in Figure 3A. The two motifs showed similarity with known plant cis-regulatory elements which are responsive to plant hormones, such as auxin and salycilic acid. Independent analysis based on info-gibbs identified a CT-rich signature motif (Figure 3B) specifically present in the promoters of phloem-specific transcripts. Info-gibbs predicted another AG-rich motif showing similarity to CTRMCAMV 35S motif which was found in -60 nt downstream of transcription start site of the CaMV 35S RNA and known to enhance gene expression driven by the CaMV 35S (Pauli et al., 2004). Analysis based on MEME predicted three significant motifs with low (E)-values that have been shown in Figure 4. Among the three, two motifs were grouped in CT/GA-rich repeat motifs specific to phloem-specific promoters. The other one showed similarity with 314MOTIFZMSBE1 which is a positive cis-element located between –314 and –295 region of maize Sbe1 promoter and required for high level as well as sugar responsive expression. The AlignACE programme was used to identify two A/T rich degenerate motifs in phloem-specific promoter sequences which are widespread in eukaryotic genomes (Figure S3). Use of multiple motif discovery programmes led to the identification of nine significant signature cis-elements specific to the phloem-specific promoters across the vascular plant species.
Figure 3. Significant cis-motifs identified by RSAT oligo-analyzer and info-gibbs. The motifs were identified by RSAT oligo-analyzer (A) and info-gibbs (B) motif discovery tool. Frequency distribution of significant motifs were searched by using FIMO program of MEME with p < 0.0001. Two motifs showing lowest expectation (E)-values and high log likelihood ratio (Avg.llr) in each case and their frequencies in the phloem-specific promoters have been shown. The X- and Y-axis show the position of nucleotides and the bits score, respectively.
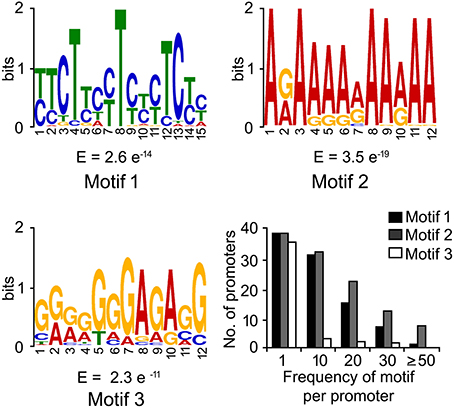
Figure 4. Motifs with lowest expectation (E)-value, identified by MEME suit. The signature motifs across the phloem-specific promoters were identified by MEME suit. Three motifs showing lowest expectation (E)-values and their frequencies are shown. The X- and Y-axis show the position of nucleotides and the bits score, respectively.
Signature cis-Elements in B. juncea Phloem-Specific Promoters
The global frequency of the nine identified signature cis-elements (Figure 5A) and their distribution in the seven putative phloem-specific promoters of B. juncea were analyzed (Figure 5B). The B. juncea promoters showed occurrence of all the signature cis-elements, albeit their distribution and frequency on each promoter varied (Figure 5B). Promoters of GS3A and PP2 contained eight of the nine signature motifs followed by promoter of SUC2 with six motifs. Promoters of GLP13, TGG1, and SULTR2 each contained four of the nine motifs. However, no apparent correlation between the frequency of the cis-elements and the relative strength of the promoters could be established. For example, PP2 promoter in spite of containing equal number of the motif elements as GS3A generated much lesser transcript level compared to GS3A. Interestingly, a notable difference between the GS3A and PP2 promoter was exclusive presence of a degenerate motif (GRRRGGGAGASG; R = A/G, S=G/C), identified in MEME based analysis, only in case of GS3A promoter. However, further experimentation is warranted before attributing this degenerate motif to high expression of GS3A. Nevertheless, the results of RT-qPCR analysis validated the applicability of the identified signature elements in identifying the phloem-specific promoters.
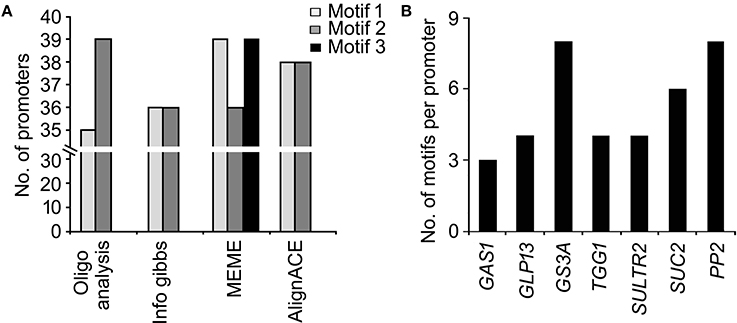
Figure 5. Significant cis-elements globally associated with phloem-specific promoters and their frequency distribution in B. juncea promoters. (A) Global occurrence of nine signature cis-elemnts in phloem-specific promoters of diverse vascular plants. (B) Frequency distribution of signature cis- elements in the seven phloem-specific promoters of B. juncea. Frequency distribution of significant motifs were searched by using FIMO program of MEME with p < 0.0001.
Stability Analysis of Promoter Activity
Promoter-activity of the in silico identified B. juncea promoters at different growth stages of the plant were analyzed by assessing their cognate-transcript levels at the vegetative, bud initiation and flowering stage. RT-qPCR analysis of the cognate-transcripts captured at different growth stages empirically showed likely influence of plant developmental stages on the activity of these promoters (Figure 6A). Based on mean Ct values, GS3A showed highest average transcript levels, followed by GLP13 and PP2, across the developmental stages of B. juncea plants. Independent analysis of the cognate-transcript levels within the vegetative and reproductive stage also showed the highest relative expression of GS3A at both the growth-stages. However, in general the transcript levels of all the cognate genes were consistently lower during the reproductive stage compared to their levels at the vegetative stage, and also the expressions were further reduced as the plants grew toward maturity.
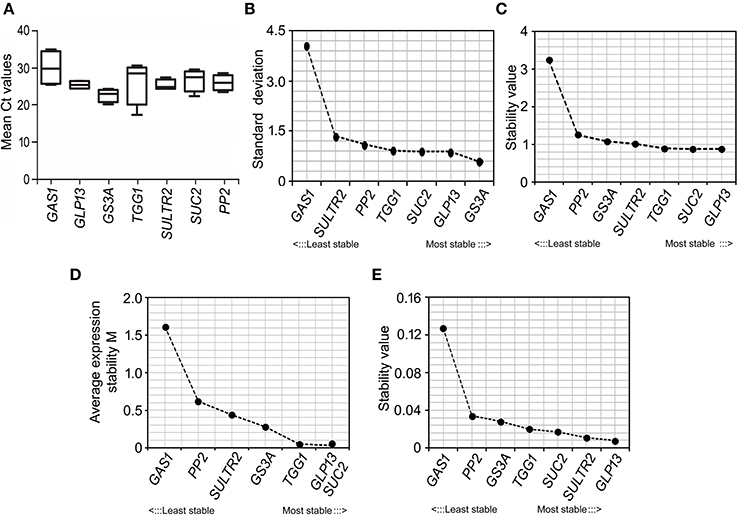
Figure 6. RT-qPCR based analysis of expression-stability of the B. juncea phloem-specific promoters. (A) Mean Ct values of the cognate-transcripts analyzed by RT-qPCR in phloem-cDNA samples collected at different growth stages of B. juncea plants. (B–E) Ranking of the B. juncea phloem-specific promoters in terms of expression-stability measured by four Excel based statistical methods, BestKeeper (B), deltaCt method (C), geNorm (D), and NormFinder (E) and plotted in increasing manner from left to right.
Irrespective of strength, the consistency in promoter-activity across different growth stages was compared in mathematical terms. For that the gene-expression data were analyzed by four Excel based statistical methods, BestKeeper, deltaCt method, geNorm, and NormFinder. In these methods the transcript stability was measured as a function of standard deviation in transcript abundance across the samples. Based on stability of their cognate-transcripts, the promoters were ranked by stepwise exclusion of one at a time with lowest stability. The output of the individual statistical method indicated different ranking pattern of the promoters in terms of consistent activity (Figures 6B–E). The analysis by BestKeeper empirically showed GS3A as the most stably expressed whereas, the other three methods ranked GLP13 to be the most stable across the growth stages. In contrary, irrespective of statistical methods GAS1 was ranked as the least stable in expression. In geNorm based analysis, the M-values of the six promoters except GAS1 promoter were <1.5 and therefore they were categorized as stable promoters (Vandesompele et al., 2002). The apparent variation in ranking within the six promoters, depending on the statistical method used, was insignificant. In spite of differential ranking pattern, all the methods unambiguously indicated higher stability in promoter activity in case of all the B. juncea promoters except GAS1.
Discussion
Crop losses due to aphids are estimated to be hundreds of millions of dollars annually (Blackman and Eastop, 1984; Morrison and Peairs, 1998). In Indian mustard (B. juncea), aphid-resistant germplasm has been largely unavailable (Bhatia et al., 2011). Therefore, transgenic development for aphid resistance has been given significant priority (The International Aphid Genomics Consortium, 2010). Past efforts on transgenic development utilized mostly the constitutive promoter CaMV 35S in transgene-expression (Sadeghi et al., 2007; Nakasu et al., 2014). Use of CaMV 35S promoter raised several concerns (Ho et al., 1999; Oraby et al., 2014). Moreover, since aphids feed exclusively the phloem-sap use of phloem-specific promoters is more appropriate for the expression of aphid-deterrent genes.
The phloem-expressing promoters play significant roles in many plant processes including organ development, sink-source flow of photoassimilates etc. Gene ontology of the phloem-expressing genes classified them in nine groups depending on their cellular functions. Primarily based on defense associated function we narrowed down to seven of them and identified their homologs in B. juncea (Table 1). Validation of the promoter-function and strength traditionally requires development of large number of transgenics with single copy insertion of the promoter-reporter cassette. Being tedious, such approach limits the number of recruited promoters in such studies (Dutt et al., 2012). Therefore, for verifying the phloem-bound activity and assessing the strength of the in silico identified B. juncea promoters, we resorted into RT-qPCR based analysis of their cognate-transcripts in the phloem exudates. Phloem exudate instead of phloem and companion cells per se were more relevant so as to analyse the promoters in the context of expressing aphid deterrent genes in the phloem-saps.
Collection of phloem exudates is complicated in most species because of the normal occlusion response of the sieve tube upon wounding (Guelette et al., 2012). To prevent this, B. juncea petioles were washed with EDTA immediately upon excision and prior to the collection of exudates. EDTA chelates Ca2+ ions and inhibits reactions leading to blockage of sieve pores (Knoblauch et al., 2001). In experimental validation, all the seven promoters showed activity in the phloem-sap, though at variable strength, as evident from their cognate-transcripts in the phloem exudates. Relative abundance of the cognate-transcripts, which is by far governed by the strength of their promoters, showed three distinct levels of gene-expression. Interestingly, GS3A promoter conferred significantly higher gene-expression compared to the second level of expression shown by PP2, GLP13 and SULTR2, and the least gene-expression by TGG1, SUC2, and GAS1. Strength of the promoters depends on the presence of specific DNA elements in and around it (Hernandez-Garcia and Finer, 2014). Therefore, further studies on significant cis-motifs in these promoters were warranted.
Though, a large number of promoters with activity in the phloem-have been isolated previously, only in case of few the specific cis-elements were proposed (Schneidereit et al., 2008). Therefore, discovery of the major cis-motifs globally associated with the phloem-specific promoters was a prerequisite for structural insight into the B. juncea promoters. Further it was necessary to perform the bioinformatic analysis preferably on a larger and more comprehensive set of promoter sequences retrieved from across the vascular species. Analysis of 39 diverse phloem-specific promoters identified the core promoter elements in addition to other potentially important cis-regulatory elements responsive to hormones, light and stress related cues (data not shown). Frequent occurrence of such elements clearly corroborated influence of most of the biotic and environmental stresses on the vascular gene-expression. For example, phloem-specific expression of calcium sensor gene AmCBL1 in Ammopiptanthus mongolicus was found to be induced by multiple stresses, and various plant hormones at different time points (Guo et al., 2010).
Use of multiple motif-finding algorithms in parallel provided robust result in terms of discovering globally over-represented signature motifs in the phloem-specific promoters. The A/N-rich motifs, identified by AlignACE (Figure S3), were conspicuous and non-specific. Similar kind of A/T rich motif was over-represented in pumpkin promoters of phloem-specific transctipts (Ruiz-Medrano et al., 2011). MEME and RSAT info-gibbs programs identified GA/CT repeat motifs, over-represented in the phloem-specific promoter sequences. Similar GA/CT and CT/GA repeats were also identified in promoter sets of phloem transcripts in pumpkin, cucumber, poplar, and rice (Ruiz-Medrano et al., 2011). It was proved that the motifs with CT/GA-repeat signature could drive vascular-specific gene expression when they were fused with the minimal promoter sequences in Arabidopsis (Ruiz-Medrano et al., 2011). In PLACE database GA/CT-rich motifs showed best match with GAGA8HVBKN3 motif, which is a GA octa-dinucleotide repeat found in intron IV of barley (H.v.) Bkn3 gene and bound by a nuclear protein, Barley B recombinant (BBR) (Santi et al., 2003). In spite of variable number of cis- elements across the phloem-specific promoters any correlation between the dosage of cis-elements and the promoter-strength was not apparent (Figure 5B). Therefore, the likely cis-regulatory logic governing the expression pattern and strength of the promoters lied in the combinatorial interaction among the set of DNA-elements in and around the promoter (Zou et al., 2011). GS3A promoter which showed strikingly higher activity compared to the other promoters, uniquely contained the degenerate motif–GRRRGGGAGASG-(R = A/G, S = G/C), which showed functional homology to GA octadinucleotide repeat of barley Bkn3 gene as evident by lowest E-value in pair-wise comparison.
In most of the studies comparing different phloem-specific promoters for transgene expression (Dutt et al., 2012; Benyon et al., 2013; Miyata et al., 2013) the assay for expression has been carried out at a particular developmental stage of the transgenic plants. However, for expressing transgenic resistance against insects like aphids, trait-expression across the plant developmental stages is pivotal. Therefore, a RT-qPCR based statistical method employing four algorithms has been used for analysing temporal stability of the seven B. juncea promoters. geNorm and NormFinder are mostly used for analysing expression-consistency of the reference genes in RT-qPCR analysis (Chandna et al., 2012; Ling et al., 2014). The analyses empirically demonstrated differential influence of plant growth-stage on the promoter-activities and relatively higher promoter-stability of the six phloem-bound promoters, except GAS1.
Conclusion
Our study identified the conserved DNA-motifs that presumably define the phloem-specificity of gene-expression. However, the complex task of fully understanding architecture of the phloem-specific promoters will essentially require complementation based functional validation of the identified signature-motifs, and particularly their interaction in combinatorial control of gene expression. Nevertheless, the signature-motifs identified in this study can be used for quick identification of potential phloem-specific promoters across the species based on the available sequence data. Also, for the first time MS Excel based statistical algorithms have been used for measuring expression-stability of the promoters based on gene-expression data of their cognate genes. In future perspective, it will be intriguing to examine whether complementation of the phloem-specific motifs to a promoter can endow phloem-specific activity to it.
Author Contributions
RB, MK: Conceived the idea, designed the experiments and wrote the manuscript; MK, DB: Performed the experiments and analyzed the data; MN, PJ, and PV: contributed reagents/analysis tools/logistics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Agricultural Science Fund, Indian Council of Agricultural Research, and Department of Science and Technology, Govt. of India.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00457
Figure S1. RNA quality and validation of amplification-specificity in RT-qPCR. (A) RNA gel visualized under UV. (B) Desired PCR amplification viewed in 2% agarose gel along with 100 bp DNA ladder M. (C) Melt-curves showing single peak generated in RT-qPCR. (TIF).
Figure S2. Amplification efficiency of the gene-specific primer sets used in qRT-PCR.
Figure S3. Consensus nucleotide sequences identified in plant phloem-specific promoters using the AlignACE program. The X- and Y-axis show the position of nucleotides and the bits score, respectively. Two motifs with highest score are shown.
Table S1. Primer sequences and related information of the genes used in RT-qPCR.
Table S2. List of primer sequences and related information of the genes used in RT-PCR.
Table S3. Phloem-specific promoters from different plant species.
Abbreviations
RT-PCR, Reverse transcription polymerase chain reaction; RT-qPCR, Reverse transcription quantitative real time PCR; MJ, Methyl jasmonate; SA, Salicylic acid; ET, Ethylene; ABA, Abscisic acid; GA, Gibberellic acid.
References
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Arumugam, N., Gupta, V., Jagannath, A., Mukhopadhyay, A., Pradhan, A. K., Burma, P. K., et al. (2007). A passage through in vitro culture leads to efficient production of marker-free transgenic plants in Brassica juncea using the Cre–loxP system. Transgenic Res. 16, 703–712. doi: 10.1007/s11248-006-9058-7
Asnao, T., Kusano, H., Okuda, T., Kubo, N., Shimada, H., and Kadowaki, K. (2002). Rpp16 and Rpp17, from a common origin, have different protein characteristics but both genes are predominantly expressed in rice phloem tissue. Plant Cell Physiol. 43, 668–674. doi: 10.1093/pcp/pcf083
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Bakhsh, A., Siddique, S., and Husnain, T. (2012). A molecular approach to combat spatio-temporal variation in insecticidal gene (Cry1Ac) expression in cotton. Euphytica 183, 65–74. doi: 10.1007/s10681-011-0497-8
Benyon, L. S., Stover, E., Bowman, K. D., Niedz, R., Shatters, R. G., Zale, J., et al. (2013). GUS expression driven by constitutive and phloem-specific promoters in citrus hybrid US-802. In Vitro Cell Dev. Biol. Plant 49, 255–265. doi: 10.1007/s11627-013-9509-9
Bhatia, V., Bhattacharya, R., Uniyal, P. L., Singh, R., and Niranjan, R. S. (2012). Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS ONE 7:e46343. doi: 10.1371/journal.pone.0046343
Bhatia, V., Uniyal, P. L., and Bhattacharya, R. (2011). Aphid resistance in Brassica crops: challenges, biotechnological progress and emerging possibilities. Biotechnol. Adv. 29, 879–888. doi: 10.1016/j.biotechadv.2011.07.005
Bhattacharyya-Pakrasi, M., Peng, J., Elmer, J. S., Laco, G., Shen, P., Kaniewska, M. B., et al. (1993). Specificity of a promoter from the rice tungro bacilliform virus for expression in phloem tissues. Plant J. 4, 71–79. doi: 10.1046/j.1365-313X.1993.04010071.x
Bhogale, S., Mahajan, A. S., Natarajan, B., Rajabhoj, M., Thulasiram, H. V., and Banerjee, A. K. (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164, 1011–1027. doi: 10.1104/pp.113.230714
Blackman, R. L., and Eastop, V. F. (1984). Aphids on the World's Crops: An Identification and Information Guide. Chichester: John Wiley & Sons.
Brears, T., Walker, E. L., and Coruzzi, G. M. (1991). A promoter sequence involved in cell-specific expression of the pea glutamine synthetase GS3A gene in organs of transgenic tobacco and alfaalfa. Plant J. 1, 235–244. doi: 10.1111/j.1365-313X.1991.00235.x
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C., and Kehr, J. (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749. doi: 10.1111/j.1365-313X.2007.03368.x
Cao, T., Lahiri, I., Singh, V., Louis, J., Shah, J., and Ayre, B. G. (2013). Metabolic engineering of raffinose-family oligosaccharides in the phloem reveals alterations in carbon partitioning and enhances resistance to green peach aphid. Front. Plant Sci. 4:263. doi: 10.3389/fpls.2013.00263
Chakraborti, D., Sarkar, A., Mondal, H. A., and Das, S. (2009). Tissue specific expression of potent insecticidal, Allium sativum leaf agglutinin (ASAL) in important pulse crop, chickpea (Cicer arietinum L.) to resist the phloem feeding Aphis craccivora. Transgenic Res. 8, 529–544. doi: 10.1007/s11248-009-9242-7
Chandna, R., Augustine, R., and Bisht, N. C. (2012). Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE 7:e36918. doi: 10.1371/journal.pone.0036918
Cho, S. M., Kang, E. Y., Kim, M. S., Yoo, S. J., Im, Y. J., et al. (2010). Jasmonate-dependent expression of a galactinol synthase gene is involved in priming of systemic fungal resistance in Arabidopsis thaliana. Botany 88, 452–461. doi: 10.1139/B10-009
Defrance, M., and van Helden, J. (2009). Info-gibbs: a motif discovery algorithm that directly optimizes information content during sampling. Bioinformatics 25, 2715–2722. doi: 10.1093/bioinformatics/btp490
Dinant, S., Ripoll, C., Pieper, M., and David, C. (2004). Phloem specific expression driven by wheat dwarf geminivirus V-sense promoter in transgenic dicotyledonous species. Physiol Plant 121, 108–116. doi: 10.1111/j.0031-9317.2004.00296.x
Divol, F., Vilaine, F., Thibivilliers, S., Amselem, J., Palauqui, J. C., Kusiak, C., et al. (2005). Systemic response to aphid infestation by Myzus persicae in the phloem of Apium graveolens. Plant Mol. Biol. 57, 517–540. doi: 10.1007/s11103-005-0338-z
Dubey, N., Idris, A., Verma, A., Chandrashekar, K., and Pandey, K. (2013). Expression pattern of sucrose transporters in Arabidopsis thaliana during aphid (Myzus persicae) infestation. Am. J. Plant Sci. 4, 47–51. doi: 10.4236/ajps.2013.412A3006
Dunwell, J. M., Gibbings, J. G., Mahmood, T., and Naqvi, S. M. S. (2008). Germin and germin-like proteins: evolution, structure, and function. CRC Crit. Rev. Plant Sci. 27, 342–375. doi: 10.1080/07352680802333938
Dutt, M., Ananthakrishnan, G., Jaromin, M. K., Brlansky, R. H., and Grosser, J. W. (2012). Evaluation of four phloem-specific promoters in vegetative tissues of transgenic citrus plants. Tree Physiol. 32, 83–93. doi: 10.1093/treephys/tpr130
Dutta, I., Majumder, P., Saha, P., Ray, K., and Das, S. (2005). Constitutive and phloem specific expression of Allium sativum leaf agglutinin (ASAL) to engineer aphid (Lipaphis erysimi) resistance in transgenic Indian mustard (Brassica juncea). Plant Sci. 169, 996–1007. doi: 10.1016/j.plantsci.2005.05.016
Fu, M., Xu, M., Zhou, T., Wang, D., Tian, S., Han, L., et al. (2014). Transgenic expression of a functional fragment of harpin protein Hpa1 in wheat induces the phloem-based defence against English grain aphid. J. Exp. Bot. 65, 1439–1453. doi: 10.1093/jxb/ert488
Furtado, A., Henry, R. J., and Takaiwa, F. (2008). Comparison of promoters in transgenic rice. Plant Biotechnol. J. 6, 679–693. doi: 10.1111/j.1467-7652.2008.00352.x
Geng, L, Duan, X., Liang, C., Shu, C., Song, F., and Zhang, J. (2014). Mining tissue-specific contigs from peanut (Arachis hypogaea L.) for promoter cloning by deep transcriptome sequencing. Plant Cell Physiol. 55, 1793–1801. doi: 10.1093/pcp/pcu111
Guelette, B. S., Benning, U. F., and Hoffmann-Benning, S. (2012). Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 63, 3603–3616. doi: 10.1093/jxb/ers028
Guo, H., Chen, X., Zhang, H., Fang, R., Yuan, Z., Zhang, Z., et al. (2004). Characterization and activity enhancement of the phloem-specific pumpkin PP2 gene promoter. Transgenic Res. 13, 559–566. doi: 10.1007/s11248-004-2738-2
Guo, L., Yu, Y., Xia, X., and Yin, W. (2010). Identification and functional characterization of the promoter of the calcium sensor gene CBL1 from the xerophyte Ammopiptanthus mongolicus. BMC Plant Biol. 10:18. doi: 10.1186/1471-2229-10-18
Haritatos, E., Ayre, B. G., and Turgeon, R. (2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 3, 929–937. doi: 10.1104/pp.123.3.929
Hedley, P. E., Maddison, A. L., Davidson, D., and Machray, G. C. (2000). Differential expression of invertase genes in internal and external phloem tissues of potato (Solanum tuberosum L.). J. Exp. Bot. 51, 817–821. doi: 10.1093/jexbot/51.345.817
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217-218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hernandez-Garcia, C. M., Martinelli, A. P., Bouchard, R. A., and Finer, J. J. (2009). A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 28, 837–849. doi: 10.1007/s00299-009-0681-7
Ho, M. W., Ryan, A., and Cummins, J. (1999). Cauliflower mosaic viral promoter – recipe for disaster? Microb. Ecol. Health Dis. 11, 194–197. doi: 10.1080/08910609908540827
Howarth, J. R., Fourcroy, P., Davidian, J.-C., Smith, F. W., and Hawkesford, M. J. (2003). Cloning of two contrasting high-affinity sulfate transporters from tomato induced by low sulfate and infection by the vascular pathogen Verticillium dahliae. Planta 218, 58–64. doi: 10.1007/s00425-003-1085-5
Hsieh, T. H., Lee, J. T., Charng, Y. Y., and Chan, M. T. (2002). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 130, 618–626. doi: 10.1104/pp.006783
Hughes, J. D., Estep, P. W., Tavazoie, S., and Church, G. M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296, 1205–1214. doi: 10.1006/jmbi.2000.3519
Husebye, H., Chadchawan, S., Winge, P., Thangstad, O. P., and Bones, A. M. (2002). Guard cell- and phloem idioblast-specific expression of Thioglucoside Glucohydrolase 1 (Myrosinase) in Arabidopsis. Plant Physiol. 128, 1180–1188. doi: 10.1104/pp.010925
Jisha, V., Dampanaboina, L., Vadassery, J., Mithöfer, A., Kappara, S., and Ramanan, R. (2015). Overexpression of an AP2/ERF type Transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 10:e0127831. doi: 10.1371/journal.pone.0127831
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Knoblauch, M., Peters, W. S., Ehlers, K., and van Bel, A. J. (2001). Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell 13, 1221–1230. doi: 10.1105/tpc.13.5.1221
Lee, J. R., Boltz, K. A., and Lee, S. Y. (2014). Molecular chaperone function of Arabidopsis thaliana phloem protein 2-A1, encodes a protein similar to phloem lectin. Biochem. Biophys. Res. Commun. 443, 18–21. doi: 10.1016/j.bbrc.2013.11.034
Lee, J. T., Prasad, V., Yang, P. T., Wu, J. F., Ho, T. H. D., Charng, Y. Y., et al. (2003). Expression of Arabidopsis CBF1 regulated by an ABA/stress promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ. 26, 1181–1190. doi: 10.1046/j.1365-3040.2003.01048.x
Lim, C. J., Lee, H. Y., Kim, W. B., Lee, B. S., Kim, J., Ahmad, R., et al. (2012). Screening of tissue-specific genes and promoters in tomato by comparing genome wide expression profiles of Arabidopsis orthologs. Mol. Cells 34, 53–59. doi: 10.1007/s10059-012-0068-4
Ling, H., Wu, Q., Guo, J., Xu, L., and Que, Y. (2014). Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE. 9:e97469. doi: 10.1371/journal.pone.0097469
Louis, J., and Shah, J. (2013). Arabidopsis thaliana-Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front Plant Sci. 4:213. doi: 10.3389/fpls.2013.00213
Martin, T., Frommer, W. B., Salanoubat, M., and Willmitzer, L. (1993). Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J. 4, 367–377. doi: 10.1046/j.1365-313X.1993.04020367.x
Medberry, S. L., Lockhart, B. E., and Olszewski, N. E. (1992). The Commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 4, 185–192. doi: 10.1105/tpc.4.2.185
Miyata, L. Y., Harakava, R., Stipp, L. C., Mendes, B. M., Appezzato-da-Glória, B., and de Assis Alves MourãoFilho, F. (2013). GUS expression in sweet oranges (Citrus sinensis L. Osbeck) driven by three different phloem-specific promoters. Plant Cell Rep. 31, 2005–2013. doi: 10.1007/s00299-012-1312-2
Morrison, W. P., and Peairs, F. B. (1998). “Response model concept and economic impact,” in Response Model for an Introduced Pest-the Russian Wheat Aphid, eds S. S. Quisenberry and F. B. Peairs (Lanham, MD: Thomas Say Publications in Entomology), 1–11.
Nakasu, E. Y., Edwards, M. G., Fitches, E., Gatehouse, J. A., and Gatehouse, A. M. (2014). Transgenic plants expressing ω-ACTX-Hv1a and snowdrop lectin (GNA) fusion protein show enhanced resistance to aphids. Front. Plant Sci. 5:673. doi: 10.3389/fpls.2014.00673
Nontachaiyapoom, S., Scott, P. T., Men, A. E., Kinkema, M., Schenk, P. M., and Gresshoff, P. M. (2007). Promoters of orthologous Glycine max and Lotus japonicus nodulation auto regulation genes interchangeably drive phloem-specific expression in transgenic plants. Mol. Plant Microbe Interact. 20, 769–780. doi: 10.1094/MPMI-20-7-0769
Oraby, H. A. S., Kandil, M. M. H., Hassan, A. A. M., and Sharawi, H. A. (2014). Addressing the issue of horizontal gene transfer from a diet containing genetically modified components into rat tissues. Afr. J. Biotechnol. 13, 4410–4418. doi: 10.5897/AJB2014.14088
Pageau, K., Reisdorf-Cren, M., Morot-Gaudry, J. F., and Masclaux-Daubresse, C. (2006). The two senescence-related markers GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilisation are differentially regulated during pathogen attack, by stress hormones and reactive oxygen species in Nicotiana tabacum L. J. Exp. Bot. 57, 547–557. doi: 10.1093/jxb/erj035
Pauli, S., Rothnie, H. M., Chen, G., He, X., and Hohn, T. (2004). The cauliflower mosaic virus 35S promoter extends into the transcribed region. J. Virol. 78, 12120–12128. doi: 10.1128/JVI.78.22.12120-12128.2004
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–Excel-based tool using pairwise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Pommerrenig, B., Feussner, K., Zierer, W., Rabinovych, V., Klebl, F., Feussner, I., et al. (2011). Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23, 1904–1919. doi: 10.1105/tpc.110.079657
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. doi: 10.1023/A:1006380021658
Rohde, W., Becker, D., and Randles, J. W. (1995). The promoter of coconut foliar decay-associated circular single-stranded DNA directs phloem-specific reporter gene expression in transgenic tobacco. Plant Mol. Biol. 27, 623–628. doi: 10.1007/BF00019328
Ruiz-Medrano, R., Xoconostle-Cázares, B., Ham, B. K., Li, G., and Lucas, W. J. (2011). Vascular expression in Arabidopsis is predicted by the frequency of CT/GA-rich repeats in gene promoters. Plant J. 67, 130–144. doi: 10.1111/j.1365-313X.2011.04581.x
Sadeghi, A., Broeders, S., De Greve, H., Hernalsteens, J. P., Peumans, W. J., Van Damme, E. J., et al. (2007). Expression of garlic leaf lectin under the control of the phloem-specific promoter Asus1 from Arabidopsis thaliana protects tobacco plants against the tobacco aphid (Myzus nicotianae). Pest Manag. Sci. 63, 1215–1223. doi: 10.1002/ps.1455
Saidi, Y., Schaefer, D. G., Goloubinoff, P., Zrÿd, J. P., and Finka, A. (2009). The CaMV 35S promoter has a weak expression activity in dark grown tissues of moss Physcomitrella patens. Plant Signal. Behav. 4, 457–459. doi: 10.4161/psb.4.5.8541
Santi, L., Wang, Y., Stile, M. R., Berendzen, K., Wanke, D., Roig, C., et al. (2003). The GA octodinucleotide repeat binding factor BBR participates to the transcriptional regulation of selected plant homeobox genes. Plant J. 34, 813–826. doi: 10.1046/j.1365-313X.2003.01767.x
Sawchuk, M. G., Donner, T. J., Head, P., and Scarpella, E. (2008). Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol. 148, 1908–1924. doi: 10.1104/pp.108.126946
Schnabel, E., Karve, A., Kassaw, T., Mukherjee, A., Zhou, X., Hall, T., et al. (2012). The M. truncatula SUNN gene is expressed in vascular tissue, similarly to RDN1, consistent with the role of these nodulation regulation genes in long distance signaling. Plant Signal. Behav. 7, 4–6. doi: 10.4161/psb.7.1.18491
Schneidereit, A., Imlau, A., and Sauer, N. (2008). Conserved cis-regulatory elements for DNA-binding-with-one-finger and homeo-domain-leucine-zipper transcription factors regulate companion cell-specific expression of the Arabidopsis thaliana SUCROSE TRANSPORTER 2 gene. Planta 228, 651–662. doi: 10.1007/s00425-008-0767-4
Shi, Y., Wang, M. B., Powell, K. S., Van Damme, E., Hilder, V. A., Gatehouse, A. M. R., et al. (1994). Use of the rice sucrose synthase-1 promoter to direct phloem-specific expression of β-glucuronidase and snowdrop lectin genes in transgenic tobacco plants. J. Exp. Bot. 45, 623–631. doi: 10.1093/jxb/45.5.623
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using realtime PCR. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-33
Singer, S. D., Hily, J. M., and Cox, K. D. (2011). The sucrose synthase-1 promoter from Citrus sinensis directs expression of the β-glucuronidase reporter gene in phloem tissue and in response to wounding in transgenic plants. Planta 234, 623–637. doi: 10.1007/s00425-011-1432-x
Srivastava, A. C., Ganesan, S., Ismail, I. O., and Ayre, B. G. (2008). Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ Symporter by tissue- specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148, 200–211. doi: 10.1104/pp.108.124776
Sunilkumar, G., Mohr, L., Lopata-Finch, E., Emani, C., and Rathore, K. S. (2002). Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol. 50, 463–474. doi: 10.1023/A:1019832123444
Takahashi, H., Watanabe-Takahashi, A., Smith, F. W., Blake-Kalff, M., Hawkesford, M. J., and Saito, K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23, 171–182. doi: 10.1046/j.1365-313x.2000.00768.x
The International Aphid Genomics Consortium (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis Thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. doi: 10.1007/BF00203657
Van Helden, J., André, B., and Collado-Vides, J. (1998). Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281, 827–842. doi: 10.1006/jmbi.1998.1947
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:7. doi: 10.1186/gb-2002-3-7-research0034
Yang, L., Li, T., Zhang, S. C., Gao, G. L., and Yang, C. W. (2013). Characterization of the GLP13 gene promoter in Arabidopsis thaliana. Biol. Plant. 57, 231–237. doi: 10.1007/s10535-012-0273-1
Yang, N. S., and Russell, D. (1990). Maize sucrose synthase-1 promoter directs phloem specific expression of Gus gene in transgenic tobacco plants. Proc. Natl. Acad. Sci. U.S.A. 87, 4144–4148. doi: 10.1073/pnas.87.11.4144
Yokoyama, R., Hirose, T., Fujii, N., Aspuria, E. T., Kato, A., and Uchimiya, H. (1994). The rolC promoter of Agrobacterium rhizogenes Ri plasmid is activated by sucrose in transgenic tobacco plants. Mol. Gen. Genet. 244, 15–22. doi: 10.1007/BF00280182
Yu, X., Wang, G., Huang, S., Ma, Y., and Xia, L. (2014). Engineering plants for aphid resistance: current status and future perspectives. Theor. Appl. Genet. 127, 2065–2083. doi: 10.1007/s00122-014-2371-2
Zhang, C., Shi, H., Chen, L., Wang, X., Lü, B., Zhang, S., et al. (2011). Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 11:11. doi: 10.1186/1471-2229-11-11
Zheng, C., and Baum, B. J. (2008). Evaluation of promoters for use in tissue-specific gene delivery. Methods Mol. Biol. 434, 205–219. doi: 10.1007/978-1-60327-248-3_13
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of amiR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bent grass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
Keywords: promoter analysis, aphid-resistance, plant-promoters, phloem promoters, transgenic resistance, promoter elements, promoter stability
Citation: Koramutla MK, Bhatt D, Negi M, Venkatachalam P, Jain PK and Bhattacharya R (2016) Strength, Stability, and cis-Motifs of In silico Identified Phloem-Specific Promoters in Brassica juncea (L.). Front. Plant Sci. 7:457. doi: 10.3389/fpls.2016.00457
Received: 21 November 2015; Accepted: 24 March 2016;
Published: 18 April 2016.
Edited by:
Tiegang Lu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Swarup Kumar Parida, National Institute of Plant Genome Research, IndiaYongqing Li, Chinese Academy of Sciences, China
Copyright © 2016 Koramutla, Bhatt, Negi, Venkatachalam, Jain and Bhattacharya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramcharan Bhattacharya, cmNiQG5yY3BiLm9yZw==; cmNiaGF0dGFjaGFyeWExQGdtYWlsLmNvbQ==
 Murali Krishna Koramutla
Murali Krishna Koramutla Deepa Bhatt1
Deepa Bhatt1 Pradeep K. Jain
Pradeep K. Jain Ramcharan Bhattacharya
Ramcharan Bhattacharya