- School of Chemistry and Biochemistry, The University of Western Australia, Crawley, WA, Australia
Organic acids are involved in numerous metabolic pathways in all plants. The finding that some plants, known as C4 plants, have four-carbon dicarboxylic acids as the first product of carbon fixation showed these organic acids play essential roles as photosynthetic intermediates. Oxaloacetate (OAA), malate, and aspartate (Asp) are substrates for the C4 acid cycle that underpins the CO2 concentrating mechanism of C4 photosynthesis. In this cycle, OAA is the immediate, short-lived, product of the initial CO2 fixation step in C4 leaf mesophyll cells. The malate and Asp, resulting from the rapid conversion of OAA, are the organic acids delivered to the sites of carbon reduction in the bundle-sheath cells of the leaf, where they are decarboxylated, with the released CO2 used to make carbohydrates. The three-carbon organic acids resulting from the decarboxylation reactions are returned to the mesophyll cells where they are used to regenerate the CO2 acceptor pool. NADP-malic enzyme-type, NAD-malic enzyme-type, and phosphoenolpyruvate carboxykinase-type C4 plants were identified, based on the most abundant decarboxylating enzyme in the leaf tissue. The genes encoding these C4 pathway-associated decarboxylases were co-opted from ancestral C3 plant genes during the evolution of C4 photosynthesis. Malate was recognized as the major organic acid transferred in NADP-malic enzyme-type C4 species, while Asp fills this role in NAD-malic enzyme-type and phosphoenolpyruvate carboxykinase-type plants. However, accumulating evidence indicates that many C4 plants use a combination of organic acids and decarboxylases during CO2 fixation, and the C4-type categories are not rigid. The ability to transfer multiple organic acid species and utilize different decarboxylases has been suggested to give C4 plants advantages in changing and stressful environments, as well as during development, by facilitating the balance of energy between the two cell types involved in the C4 pathway of CO2 assimilation. The results of recent empirical and modeling studies support this suggestion and indicate that a combination of transferred organic acids and decarboxylases is beneficial to C4 plants in different light environments.
Introduction
Organic acids are of fundamental importance in all plant species. They are involved in many and diverse metabolic pathways, including energy production, carbon storage, stomatal conductance, the biosynthesis of amino acids, plant–microbe interactions, and mechanisms allowing plants to deal with excess cations, changing osmotic conditions, and soils low in nutrients as well as those with high metal content (reviewed in López-Bucio et al., 2000). In addition to these varied roles, organic acids play a major part in the C4 photosynthetic pathway as the intermediates connecting CO2 uptake and fixation, and this is the focus of this review.
Work on sugarcane published in the 1960s (Burr, 1962; Kortschak et al., 1965; Hatch and Slack, 1966; Hatch et al., 1967) showed that this plant performed a different type of CO2 fixation biochemistry compared to the photosynthetic carbon reduction (PCR) cycle determined in the Calvin laboratory (Bassham and Calvin, 1957). 14CO2 labeling studies indicated the first major products of CO2 assimilation in sugarcane were the 4-carbon (C) organic acids malate and aspartate (Asp) rather than 3-phosphoglycerate (3-PGA) or other intermediates of the PCR cycle (Burr, 1962; Kortschak et al., 1965; Hatch and Slack, 1966; Hatch et al., 1967). Hatch and Slack (1966) described a model of the pathway, which encompassed and extended the earlier work. The pathway is now known as the C4 pathway because of the initial fixation products, and plants using the pathway are known as C4 plants. This seminal work (Hatch and Slack, 1966) also resolved that the fourth C of oxaloacetate (OAA) was labeled along with those of malate and Asp. However, OAA was short-lived, and immediately converted to malate or Asp. The label in these dicarboxylic acids was transferred to the first C of 3-PGA, which then was used for carbohydrate synthesis via the PCR cycle. Their model also included a 3-C organic acid as the acceptor molecule for atmospheric CO2 (Hatch and Slack, 1966).
A generalized scheme of the reactions making up the C4 photosynthetic pathway is shown in Figure 1. Unlike C3 plants, most C4 species use two cell types in CO2 assimilation: the mesophyll (M) and bundle-sheath (BS) cells. The BS cells surround the vascular tissue, and are in turn surrounded by M cells, giving the characteristic wreath-like or Kranz anatomy that was first describe in the late 19th century (Haberlandt, 1896). In C4 leaves, no M cell is more than two cells away from a BS cell, which facilitates rapid metabolite exchange between the two cell types. Atmospheric CO2 entering a C4 leaf is hydrated to bicarbonate in the M cytosol by carbonic anhydrase (CA). The primary carboxylase of C4 plants, phosphoenolpyruvate carboxylase (PEPC), uses the bicarbonate to fix a CO2 group to the 3-C compound phosphoenolpyruvate (PEP), producing OAA, which is rapidly converted to malate and/or Asp. These dicarboxylic acids move into the neighboring BS where they are decarboxylated, with the released CO2 being used for carbohydrate production by the PCR cycle, located in BS cell chloroplasts. The 3-C organic acids released in the decarboxylation reactions return to a neighboring M cell, where they can be used as the CO2 acceptor by PEPC (Figure 1).
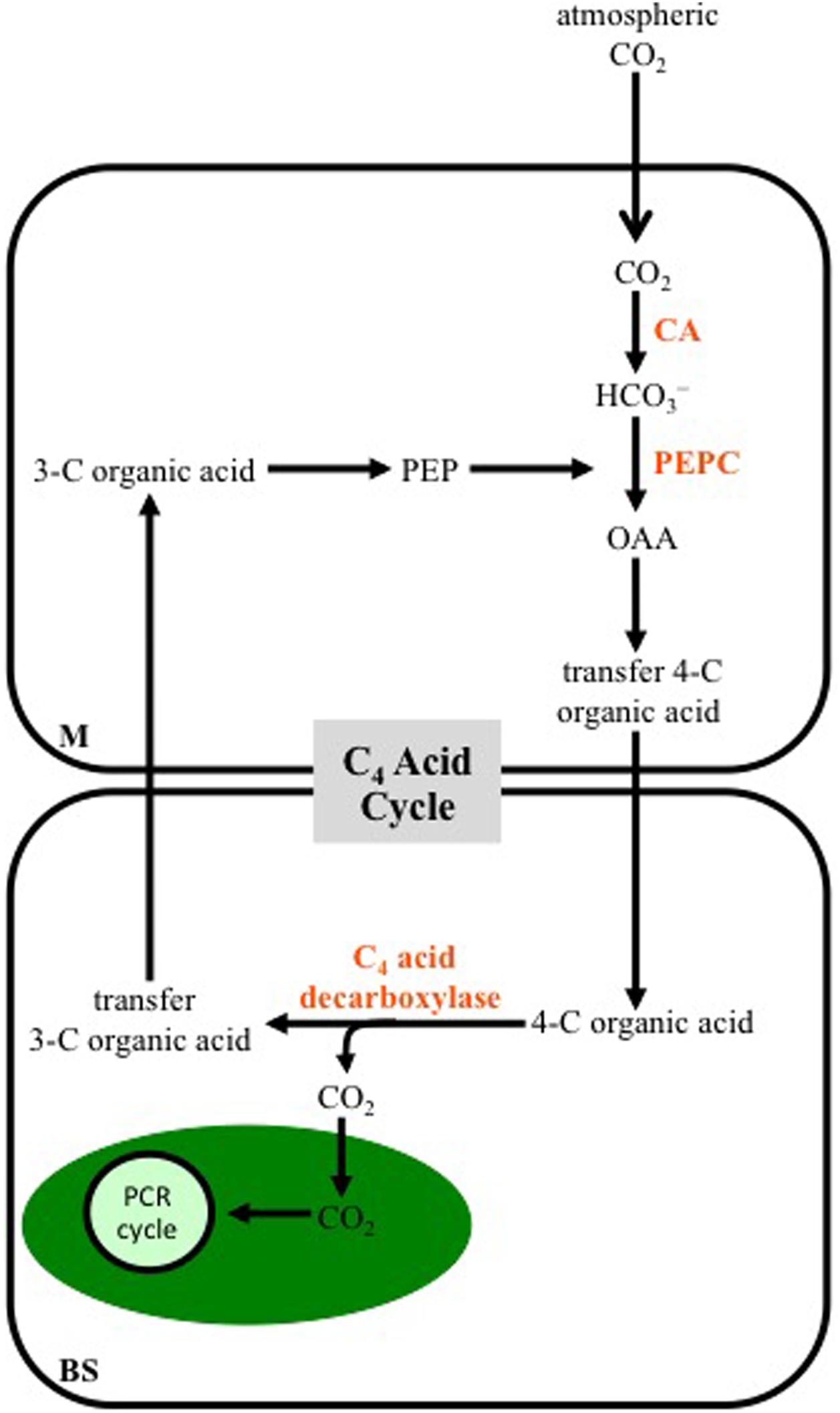
FIGURE 1. A diagram outlining a generalized C4 photosynthetic pathway. Atmospheric CO2 enters a mesophyll (M) cell of a leaf and is converted to bicarbonate (HCO3-) by carbonic anhydrase (CA). The bicarbonate is then used to carboxylate phosphoenolpyruvate (PEP) by PEP carboxylase (PEPC), producing oxaloacetate (OAA), which is immediately converted into another four-carbon (C) organic acid that is transferred to bundle-sheath (BS) cells. Decarboxylation of the 4-C acid occurs in BS cells, releasing CO2, which is fixed into carbohydrates by the photosynthetic carbon reduction (PCR) cycle in the chloroplast (green oval). The 3-C organic acid released at the decarboxylation step, is transferred to M cells, where it contributes to the CO2 acceptor pool. Note the 3-C and 4-C organic acids formed, and the intracellular location of the decarboxylation reactions vary, depending on the C4 subtype (see Figures 2–4).
An important outcome of the C4 acid cycle is the concentration of CO2 around ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in the BS to levels at least 10-times higher than those of the surrounding atmosphere (Jenkins et al., 1989). This results in C4 plants requiring less Rubisco for carbohydrate production than C3 species, which translates into increased nitrogen-use efficiency (Ghannoum et al., 2011). In contrast to Rubisco, which can fix oxygen in addition to CO2, leading to photorespiration and the loss of fixed CO2 and consumption of ATP, PEPC has only carboxylase activity. Consequently C4 plants have little photorespiratory activity, and through a combination of PEPC kinetics and leaf anatomy, also show increased water-use efficiency compared to C3 plants (Ghannoum et al., 2011).
As more C4 plants were examined, it became clear that while the first two steps in the pathway, those catalyzed by CA and PEPC, are the same in all species (Hatch, 1987; Kanai and Edwards, 1999), differences exist in the 4-C organic acid transferred to the BS; the decarboxylating enzymes and their activities and intracellular locations; the 3-C organic acid returned to the M cells; and regeneration of the CO2 acceptor. As a result, three subtypes of C4 photosynthesis were recognized (Gutierrez et al., 1974; Hatch et al., 1975; Hatch, 1987; Kanai and Edwards, 1999), based on the decarboxylating enzyme with the greatest activity in the leaf tissue, and C4 species were categorized into one of these subtypes: NADP-malic enzyme (NADP-ME), NAD-malic enzyme (NAD-ME), or PEP carboxykinase (PCK).
This review will summarize the three C4 subtypes, focusing on the organic acids utilized in the course of CO2 fixation, and the enzymes responsible for their metabolism. The roles of the 4-C organic acid decarboxylases in C3 plants will be presented along with the current understanding of their co-option into C4 biochemistry. Consideration will then be given to evidence suggesting that C4 plants are more flexible with respect to the types of organic acids and decarboxylases used than previously thought, and the apparent advantages this plasticity gives the plants in fluctuating environments will be discussed.
C4 Photosynthetic Subtypes
NADP-ME-Subtype
Malate is the major 4-C organic acid transferred to the BS in C4 species designated as belonging to the NADP-ME subtype (Figure 2). The OAA synthesized from PEPC activity in the cytoplasm of M cells is transferred to the chloroplast and reduced to malate by NADP-malate dehydrogenase (NADP-MDH). The malate is then transported out of the M chloroplasts, diffuses into the BS, and in the BS chloroplasts is decarboxylated by NADP-ME in a reaction producing NADPH, CO2, and pyruvate. The released CO2 is fixed by Rubisco in the BS chloroplasts, while the pyruvate is transported out of the BS chloroplasts, and into the chloroplasts of M cells, where it is used to regenerate PEP by pyruvate Pi dikinase (PPDK).
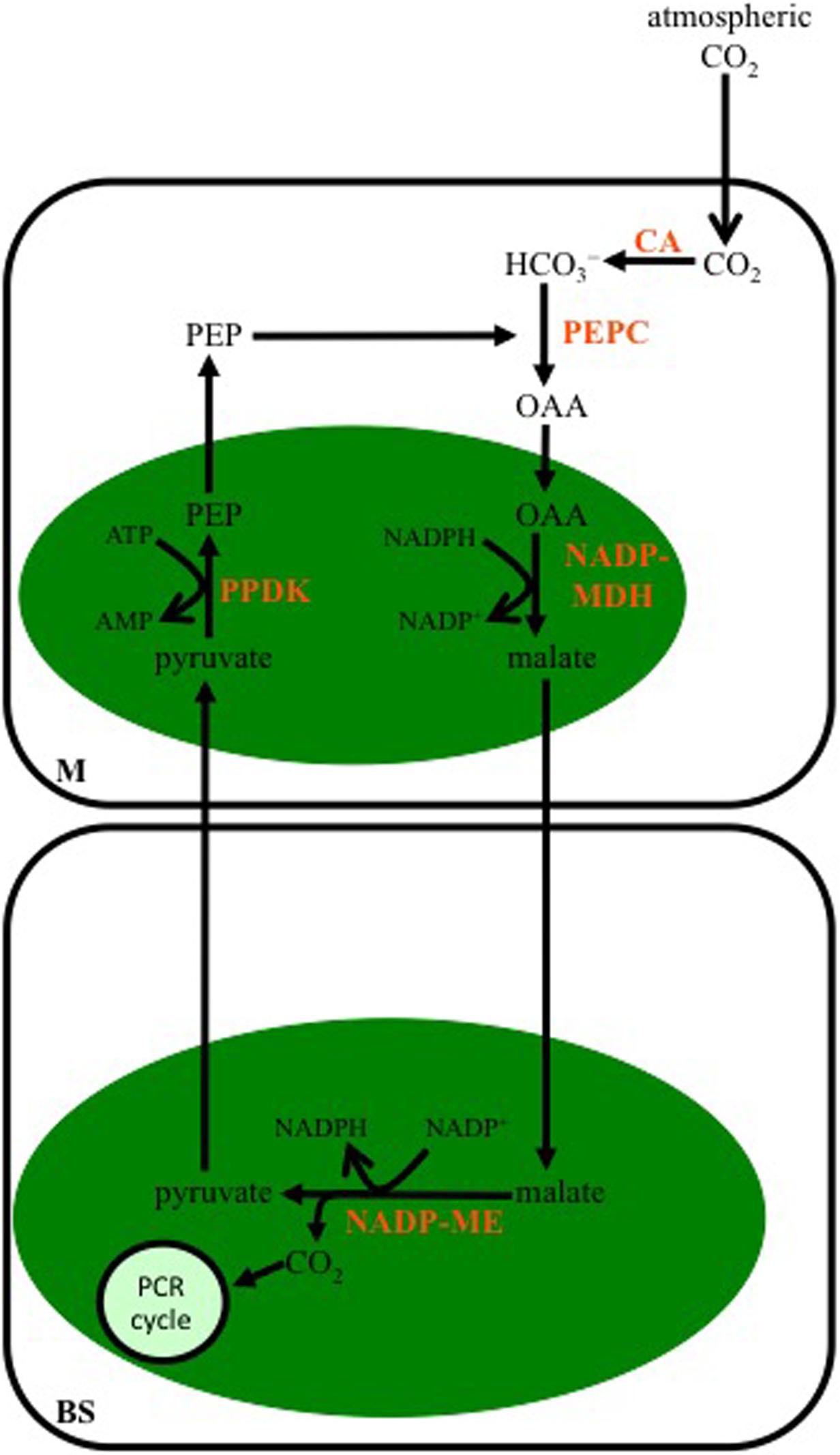
FIGURE 2. Schematic of the C4 NADP-malic enzyme-subtype pathway. In mesophyll (M) cells of this C4 subtype, the bicarbonate (HCO3-) produced from atmospheric CO2 by CA is used to carboxylate phosphoenolpyruvate carboxylase (PEP) by PEPC. The OAA formed is reduced to malate in M chloroplasts (green oval), by NADP-malate dehydrogenase (NADP-MDH), and then diffuses into BS cells, where it is decarboxylated in the chloroplast by NADP-malic enzyme (NADP-ME). The CO2 released is used to make carbohydrates by the PCR cycle, while the pyruvate diffuses to the M where it is converted to PEP by pyruvate Pi dikinase (PPDK).
The NADP-ME subtype is the most prevalent C4 subtype, and is found in both monocotyledonous and dicotyledonous species (Gutierrez et al., 1974; Sage et al., 2011). Agronomically important monocots such as maize, sorghum, and sugarcane are categorized as NADP-ME species. C4 species of Flaveria, a dicotyledonous genus that has been used to examine the evolution of C4 photosynthesis for more than 25 years, also show high levels of malate production during CO2 fixation, as well as high activity of NADP-ME in leaf tissue.
NAD-ME-Subtype
C4 plants using NAD-ME as their primary decarboxylase include Atriplex, and C4 species of Cleome and Amaranthus (Gutierrez et al., 1974; Sage et al., 2011). Some C4 species of Panicum also have high NAD-ME activity in BS mitochondria.
Aspartate is the first stable organic acid of this C4 subtype, and results from the transamination of OAA by an Asp aminotransferase (AST) located in the cytosol of M cells (Figure 3). The Asp enters the mitochondria of BS cells where it is converted back to OAA by a mitochondrial isoform of AST, and through the activity of a mitochondrial NAD-malate dehydrogenase (NAD-MDH), the OAA is reduced to malate. NAD-ME is also active in the BS mitochondria, and catalyzes the release of CO2 from malate, along with the reduction of NAD+. The CO2 diffuses into the chloroplasts of the BS, and is fixed by Rubisco. The pyruvate, resulting from the decarboxylation reaction, is transported out of the mitochondria, and transaminated to Ala by a cytosolic Ala aminotransferase (ALT). The Ala diffuses into a neighboring M cell, and is converted back to pyruvate in the reverse reaction catalyzed by an ALT active in the M cell cytosol. The pyruvate is then used by PPDK to regenerate PEP in M cell chloroplasts.
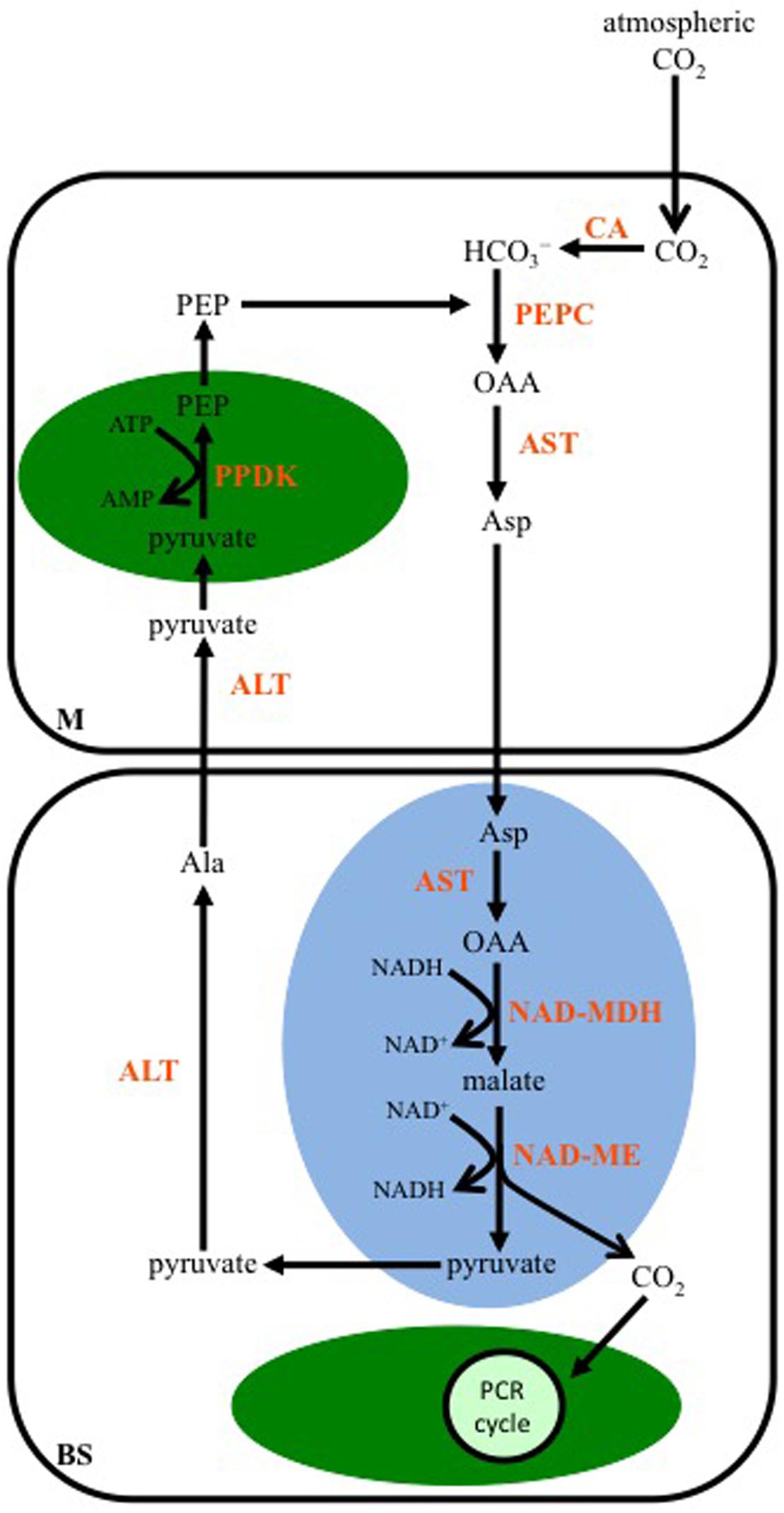
FIGURE 3. A diagram showing the steps in the C4 NAD-malic enzyme-subtype pathway. In mesophyll (M) cells, PEP is carboxylated by PEPC using the CO2 group from bicarbonate (HCO3-), which was produced from the hydration of atmospheric CO2 by CA. The resulting OAA is transaminated to Asp by Asp aminotransferase (AST). In the mitochondria (blue oval) of BS cells, OAA is generated from Asp by a mitochondrial AST, and then reduced to malate by NAD-malate dehydrogenase (NAD-MDH). Malate is decarboxylated by NAD-malic enzyme (NAD-ME), and the released CO2 diffuses into the BS chloroplast (green oval), where it is fixed by the PCR cycle. The pyruvate resulting from the decarboxylation reaction is converted to alanine (Ala) by Ala aminotransferase (ALT) in the BS cytosol. The Ala is transaminated back to pyruvate by ALT activity in the M cytosol. PPDK activity in M chloroplasts converts pyruvate to PEP.
PCK-Subtype
As for the NAD-ME-subtype, multiple transamination reactions characterize the C4-PCK-subtype pathway. In plants using this enzyme as their primary decarboxylase, both malate and Asp are formed from OAA (Figure 4). A cytosolic AST in M cells produces Asp, while OAA is also transported into M cell chloroplasts and converted to malate by NADP-MDH. The Asp diffuses into the BS, and there a cytosolic AST converts it back to OAA, which is then decarboxylated by PCK in a reaction requiring ATP. The released CO2 enters the PCR cycle in the BS chloroplasts. The PEP generated from PCK activity diffuses back into the M to be used by PEPC. The malate formed in M cell chloroplasts is transported out of these organelles, and into the mitochondria of the BS. As for the NAD-ME-subtype, a mitochondrial NAD-ME isoform decarboxylates the malate, the released CO2 enters BS chloroplasts and the PCR cycle. The pyruvate formed is ultimately used to regenerate PEP in the M cell chloroplasts, following the same transamination reactions as those of the NAD-ME-subtype pathway. The NADH produced from NAD-ME activity is used in mitochondrial respiration to make ATP, supporting PCK activity in the BS cytosol.
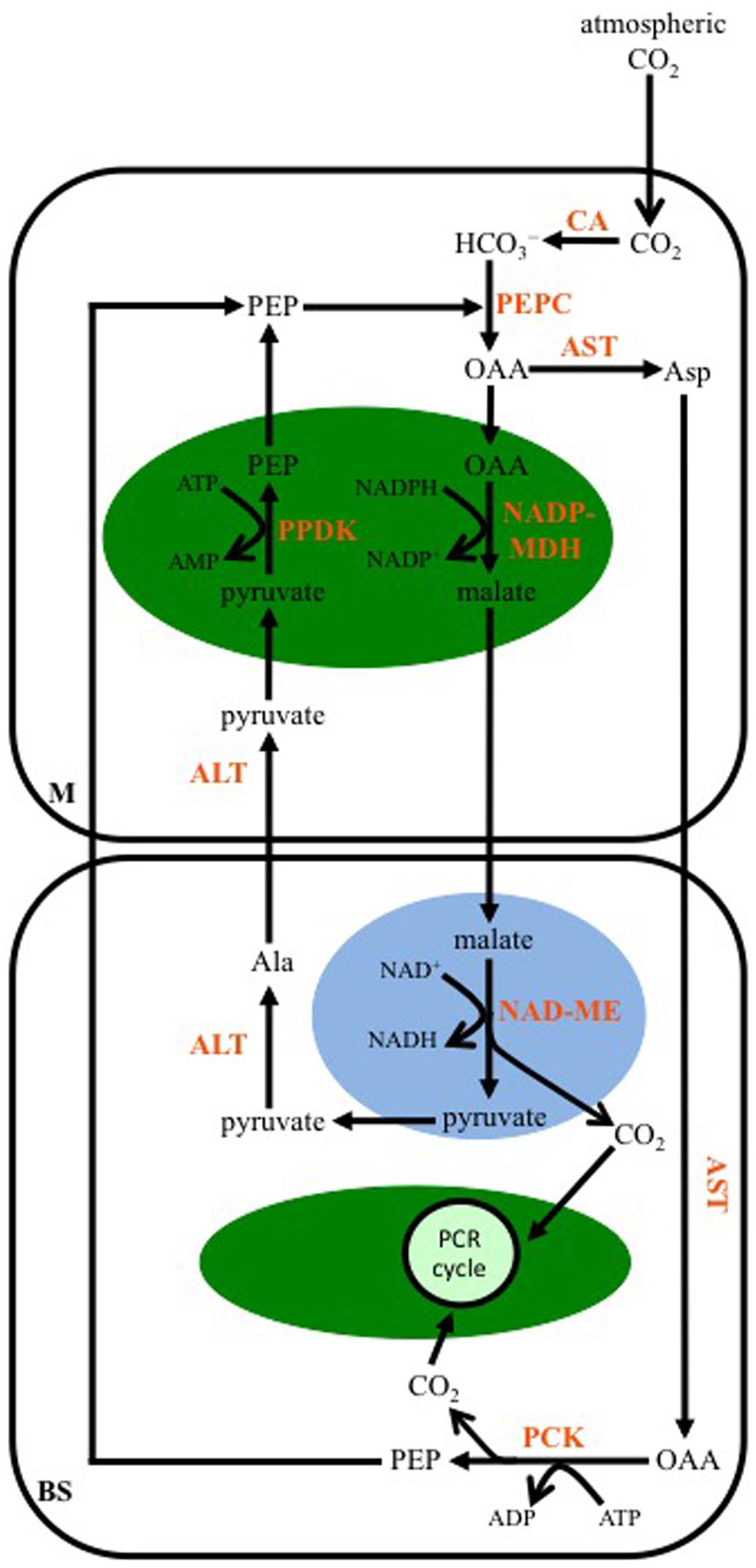
FIGURE 4. A representation of the C4 phosphoenolpyruvate carboxykinase-subtype pathway. Atmospheric CO2 is converted to bicarbonate (HCO3-) by CA in the mesophyll (M) cytosol. PEP is carboxylated, producing OAA, by PEPC. The OAA is either transaminated to Asp by AST in the cytosol, or is reduced to malate in the chloroplast (green oval) by NADP-MDH. The Asp diffuses into the BS cell cytosol, where it is transaminated back to OAA, which is then decarboxylated by PEP carboxykinase (PCK). The resulting PEP diffuses into the M, while the CO2 is used to make carbohydrates by the PCR cycle. The malate formed in M chloroplasts is transferred to BS mitochondria (blue oval), where it is decarboxylated by NAD-ME, with the released CO2 diffusing into BS chloroplasts for fixation, and the pyruvate being transaminated to Ala in the BS cytosol by ALT. The Ala is transferred to the M, where another ALT converts it back to pyruvate, which is then converted to PEP in the chloroplast by PPDK.
PCK-type grasses include Urochloa panicoides, Chloris gayana, and some C4 species of Panicum (Gutierrez et al., 1974; Sage et al., 2011). It was thought PCK did not play a role in CO2 assimilation in C4 dicots; however, significant PCK activity has been reported in several C4 dicot lineages, including members of the Sesuvioideae and Cleome (Meister et al., 1996; Muhaidat et al., 2007; Sommer et al., 2012; Muhaidat and McKown, 2013). In contrast, a more recent study examining C4 species in the Cleomaceae, Aizoaceae, and Chenopodiaceae detected only low levels of PCK in these dicot groups (Koteyeva et al., 2015).
Evolution of C4 Acid Decarboxylases
All the C4 cycle enzymes involved in the production and utilization of malate, OAA, Asp, Ala, PEP, and pyruvate have counterparts in C3 species, as these organic acids are involved in a myriad of roles in all plants, as noted above (López-Bucio et al., 2000). Many of the genes encoding the C4 proteins appear to have resulted from duplication events, which allowed ancestral function to be maintained, while also permitting neofunctionalization of the other copy, leading to the C4-specific roles and expression patterns (Ludwig, 2013). Interestingly, a number of the genes encoding the enzymes involved in the metabolism of the organic acids appear to have been co-opted from the same gene lineage in numerous independent C4 lineages (Christin et al., 2013, 2015). Changes in regulatory mechanisms of the ancestral genes, which may involve sequences in the promoter, untranslated and/or coding regions, and trans-acting factors led to the distinctive C4 expression levels and cell-specific patterns, while modifications to the coding regions were responsible for differences in the kinetic properties seen between the C3 and C4 enzymes (Williams et al., 2012; Ludwig, 2013). The evolution of the decarboxylases catalyzing the release of CO2 from either malate or OAA in the three C4 subtypes has been studied to varying levels, and our current knowledge is summarized below.
NADP-Malic Enzyme
Cytosolic and chloroplastic forms of NADP-ME exist in both C3 and C4 plants. The cytosolic proteins play roles in defense, development, and stress responses by coordinating the levels of malate, pyruvate, and reducing power needed by a plant during these processes (Table 1) (Drincovich et al., 2011; Maier et al., 2011; Badia et al., 2015). These cytosolic enzymes are thought to represent the ancestral form of the protein. It has been suggested for maize that the gene encoding a cytosolic NADP-ME was duplicated, with one of the resulting copies acquiring a sequence encoding a chloroplast transit peptide (Tausta et al., 2002). A duplication of this gene led to the C4 isoform with its ability to decarboxylate malate in BS chloroplasts, and its characteristic properties of tetramerization and regulation, including inhibition by high malate concentrations at pH 7 and redox modulation (Detarsio et al., 2007; Alvarez et al., 2012; Saigo et al., 2013).
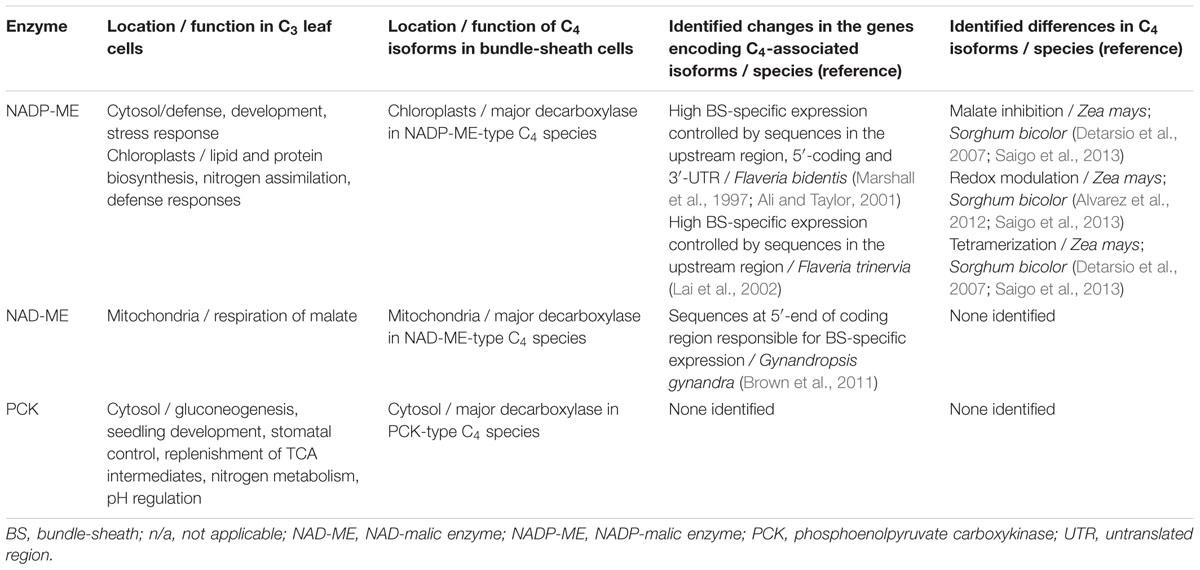
TABLE 1. Characteristics of four-carbon organic acid decarboxylating enzymes in the leaves of C4 plants and the orthologous gene products from C3 species.
A similar duplication of the gene encoding a chloroplastic, non-C4-associated enzyme has been proposed for the evolution of the C4 NADP-ME in Flaveria (Marshall et al., 1996). Potentially interacting regulatory elements found in the 5′- and 3′-ends of the F. bidentis gene encoding the C4-associated NADP-ME control the level of gene activity, while other sequences in the 5′-end were found to determine BS specificity (Table 1) (Marshall et al., 1997; Ali and Taylor, 2001). In the closely related C4 species F. trinervia, the high levels of BS-specific expression of the C4-associated NADP-ME are apparently controlled only by elements in the 5′-region of the gene (Table 1) (Lai et al., 2002).
Non-C4-associated NADP-ME isoforms targeted to the chloroplast in C3 and C4 plants also appear to be involved in defense responses, as well as providing pyruvate and NADPH for lipid and amino acid biosynthesis, and nitrogen assimilation (Table 1) (Maurino et al., 2001; Drincovich et al., 2011; Maier et al., 2011; Saigo et al., 2013). Phylogenetic and genomic analyses have shown that in independent C4 grass lineages, the same ancestral ortholog was used as the template for evolution of the C4-associated NADP-ME isoform, and that positive selection acted on particular codons, which resulted in adaptive parallel changes in the cognate proteins (Christin et al., 2009). These observations support the idea that the number of paths leading to the evolution of a C4-associated enzyme is constrained by the subcellular compartment and milieu in which it must function, and that, more generally, if suitable evolutionary enablers were not present in the C3 ancestor, then the C4 syndrome could not evolve (Christin and Osborne, 2013; Christin et al., 2013).
NAD-Malic Enzyme
Although malate is the substrate of NAD-ME, in C4 species using this enzyme as the primary decarboxylase, Asp is actually the first stable organic acid formed. The C4-associated NAD-ME is a heterodimer made up of α and β subunits, and as described above, is highly active in the mitochondria of BS cells. However, NAD-ME isoforms are found in all plant mitochondria, where they play a key role in the respiration of malate in the tricarboxylic acid (TCA) cycle (Table 1) (Drincovich et al., 2011; Maier et al., 2011). Given the ubiquitous nature of this decarboxylase, it is interesting that not more is known of the evolutionary history of the C4-associated enzyme in any C4 species. The number of active NAD-ME isoforms is also not clear for many C3 and C4 species that have been examined.
Regulatory sequences directing BS-specific expression have been identified in the 5′-end of the coding regions of the NAD-ME α and β subunit genes from the C4 species Gynandropsis gynandra (previously Cleome gynandra; Table 1) (Brown et al., 2011). As corresponding sequences from the orthologous genes of Arabidopsis thaliana also drive expression in the BS of G. gynandra, it appears that the specific NAD-ME expression pattern of C4 species likely resulted from changes affecting the activity of a trans-acting factor (Brown et al., 2011).
PEP Carboxykinase
ATP-dependent PCK isoforms are found in all plants. They are cytosolic enzymes and in C3 species, have been found to play significant roles in numerous metabolic processes that require or utilize PEP or OAA (Table 1). These include the mobilization of carbon from lipids and amino acids in seeds during gluconeogenesis and seedling development (Leegood and ap Rees, 1978), the control of stomatal aperture (Penfield et al., 2012), replenishment of TCA intermediates (Walker et al., 1999), pH regulation (Walker et al., 1999, 2001; Chen et al., 2004), synthesis of amino acids, and metabolism of nitrogenous assimilates during transport to storage tissues (Walker et al., 1999; Delgado-Alvarado et al., 2007).
In some PCK-type species, gene families have been identified (Finnegan et al., 1999; Christin et al., 2008) although overall, there is limited information regarding the size of the PCK gene family in both C3 and C4 plants, the evolutionary origin of the C4-associated PCK isoforms, and the regulatory mechanism(s) responsible for BS expression. Some of this lack of knowledge can be explained by the few species that have been identified as utilizing PCK as their primary decarboxylase, and the lack of information on the closest C3 relative of the C4 species that have been classified as PCK-subtype. A phylogenetic and genomic study has indicated that in the monocots, gene duplication events gave rise to different PCK gene lineages, and as for NADP-ME, convergence is seen, with parallel adaptive amino acid changes in the C4 isoforms in distinct C4 lineages, suggesting a predisposition of these orthologs for “C4-ness” (Christin et al., 2009).
Flexibility in Organic Acid Production and Utilization in C4 Plants
The observation that in each C4 species, the activity of one 4-C organic acid decarboxylating enzyme predominates in leaf tissue offered a simple criterion by which to categorize C4 plants, and stimulated comparative analyses between and within C4 subtypes at the leaf anatomical, biochemical, physiological, and molecular biological levels. For example, correlations have been made between leaf anatomy and primary C4 decarboxylases (Gutierrez et al., 1974; Hatch et al., 1975; Kanai and Edwards, 1999; Edwards and Voznesenskaya, 2011). However, work with a number of C4 species has also shown that in addition to the primary decarboxylase, there is increased activity of another of the C4 decarboxylases (Gutierrez et al., 1974; Meister et al., 1996; Kanai and Edwards, 1999; Wingler et al., 1999; Muhaidat et al., 2007; Pick et al., 2011; Sommer et al., 2012; Muhaidat and McKown, 2013; Bräutigam et al., 2014; Koteyeva et al., 2015), and increased levels of the 4-C organic acid substrate (Kortschak et al., 1965; Hatch et al., 1967; Hatch, 1971; Meister et al., 1996; Pick et al., 2011; Sommer et al., 2012). This was recognized early on for C4 species that use Asp as the transferred organic acid and PCK as the primary decarboxylase, where malate levels and NAD-ME activity are also appreciable (Edwards et al., 1971). However, even in species such as sugarcane and maize, clear NADP-ME subtypes based on the above criteria, significant Asp formation and PCK activity were detected in the early 14CO2 labeling studies (Kortschak et al., 1965; Hatch and Slack, 1966). Both empirical and modeling studies now argue that nearly all C4 plants transfer more than one 4-C and 3-C organic acid during CO2 assimilation, and alternative decarboxylation pathways function in most C4 species (Pick et al., 2011; Sommer et al., 2012; Bellasio and Griffiths, 2014; Kromdijk et al., 2014; Wang et al., 2014). Insights into the consequences of C flux through the alternative pathways on plant metabolism suggest they extend beyond the obvious provision of CO2 for carbohydrate production, and likely impact on the ability of C4 plants to cope with diverse and fluctuating environments (Furbank, 2011; Bellasio and Griffiths, 2014; Kromdijk et al., 2014; Sharwood et al., 2014; Stitt and Zhu, 2014; Wang et al., 2014), and potentially play a role in development (Sommer et al., 2012).
Maize – A Case Study in Flexibility of C4 Acid Production and Utilization
Early studies using 14CO2 to label actively photosynthesizing maize leaves showed that about 75% the label was quickly incorporated into malate, and 25% into Asp at a slower rate (Kortschak et al., 1965; Hatch and Slack, 1966; Hatch et al., 1967; Hatch, 1971). Significant PCK activity was subsequently found in maize leaves (Walker et al., 1997) and isolated BS cells (Wingler et al., 1999). The scheme of decarboxylase activities in maize leaves proposed by Wingler et al. (1999) included the “classical” NADP-ME pathway with the decarboxylation of malate in BS chloroplasts by NADP-ME, but also the transamination of Asp to OAA in BS mitochondria, followed by the release of CO2 in the BS cytosol by PCK, using ATP, the origin of which was unresolved. Later proposals have suggested that both malate and Asp are produced from OAA either in the M chloroplasts via NADP-MDH or through cytosolic AST activity (Pick et al., 2011), and OAA resulting from Asp transamination in the BS may be either decarboxylated directly by PCK in the BS cytosol, using ATP generated from chloroplast activities (Furbank, 2011), or transported into BS chloroplasts, reduced to malate by NADP-MDH, with the malate then decarboxylated by chloroplastic NADP-ME (Figure 5) (Furbank, 2011; Pick et al., 2011; Wang et al., 2014).
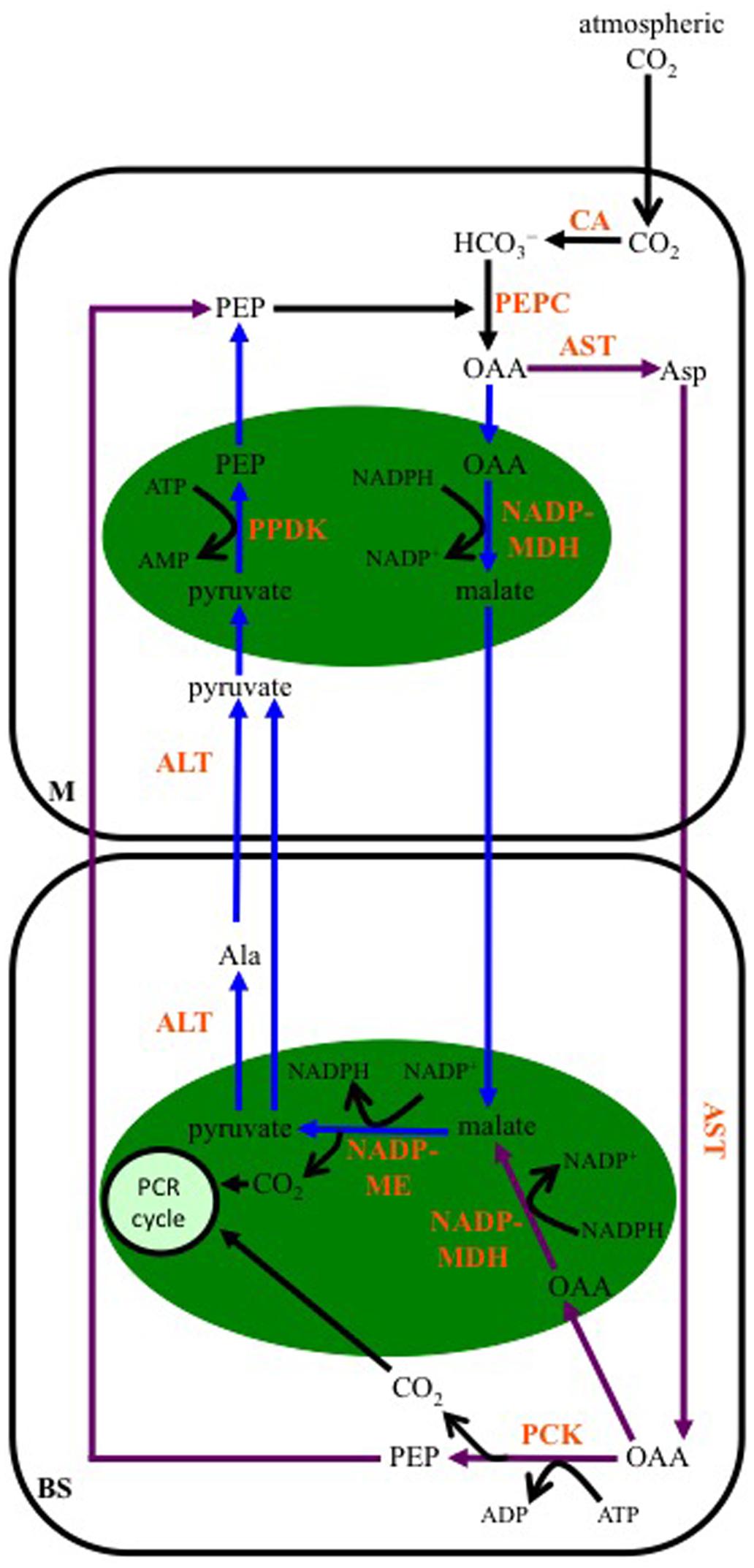
FIGURE 5. Schematic of the proposed carbon flow in maize leaves when both malate and Asp are the transferred 4-carbon organic acids. The initial steps of carbon flow are identical, with CA activity converting atmospheric CO2 to bicarbonate (HCO3-), and PEP being carboxylated by PEPC to produce OAA in M cells. Purple arrows: When Asp is transferred to BS cells, there is no net movement of reducing equivalents to the BS. ATP is required for PCK activity in the BS cytosol; however, there is reduced ATP demand in the M for PEP regeneration. This is the scenario predicted when the absorbed irradiance is relatively high in BS cells (Kromdijk et al., 2014; Wang et al., 2014). Blue arrows: When light striking a leaf is preferentially absorbed by the M, the flux through PCK decreases. Malate and reducing equivalents are transferred to the BS. Demand for ATP is lowered in BS cells due to reduced PCK activity. However, ATP requirements increase in M cells for PEP regeneration from pyruvate. ALT, Ala aminotransferase; AST, Asp aminotransferase; NADP-MDH, NADP-malate dehydrogenase; NADP-ME, NADP-malic enzyme; PCR cycle, photosynthetic carbon reduction cycle; green ovals, chloroplasts. Pathways modified from Furbank (2011), Pick et al. (2011) and Wang et al. (2014).
The transfer of malate and Asp result in different energy scenarios in the M and BS cells of maize. NADP-ME-type species, like maize, are typically described as having fewer grana in BS chloroplasts than in chloroplasts of M cells (Gutierrez et al., 1974; Hatch et al., 1975; Kanai and Edwards, 1999; Edwards and Voznesenskaya, 2011), indicating limited capacity for PSII activity and NADPH production via linear electron transport in the BS (Leegood et al., 1983; Hatch, 1987; Meierhoff and Westhoff, 1993). As a result, about half the 3-PGA produced by Rubisco in BS chloroplasts is transported to the M for phosphorylation and reduction to triose phosphate (triose-P; Hatch, 1987). Some of the triose-P is then returned to the BS for regeneration of ribulose-1,5-bisphosphate and starch production, while the rest is used in carbohydrate synthesis in M chloroplasts. When malate is decarboxylated by NADP-ME, reducing equivalents are moved from the M to the BS, contributing to the NADPH requirements of triose-P generation in the BS (Figure 5). The pyruvate returned to the M following NADP-ME decarboxylation is used by PPDK to generate PEP in a reaction that uses ATP. This adds to the ATP demand in the M over that needed for triose-P production. In the case of Asp being transferred to the BS, no reducing equivalents are moved, and ATP is required by PCK in the BS for the decarboxylation of OAA; however, the need for ATP is lessened in the M as PEP is returned to these cells following PCK activity (Figure 5).
Clearly, for efficient functioning of the maize C4 pathway overall, coordination of ATP and reducing equivalent supply and use must occur between the M and BS. In this regard, it has been proposed that the ability to move carbon through both malate and Asp decarboxylation pathways plays a role in adjusting M and BS energy balance to facilitate efficient functioning of maize C4 photosynthesis in diverse and changing environments, and during development (Furbank, 2011; Pick et al., 2011; Bellasio and Griffiths, 2014; Kromdijk et al., 2014; Stitt and Zhu, 2014; Wang et al., 2014). It has also been postulated that the large pools of 3-C and 4-C organic acids, which support their diffusion between M and BS cells in C4 species (Leegood, 1985; Stitt and Heldt, 1985), act as reserves of ATP and NADPH to buffer against rapid changes in light availability over longer time scales than is possible for C3 plants (Stitt and Zhu, 2014). Being able to switch between the species of 4-C and 3-C organic acids that are transferred, with their differing contributions to M and BS energy balance, strengthens this buffering capacity, and the ability to maintain efficient CO2 assimilation. The sharing of C flux between multiple metabolites is also seen as a benefit as it allows the concentration and diffusion gradient of individual 3-C and 4-C organic acids to be lower without compromising overall C4 cycle activity (Pick et al., 2011; Kromdijk et al., 2014; Wang et al., 2014).
So are the suggested advantages of operating a combination system of transferred organic acids and decarboxylating enzymes in maize realized? Experimental and modeling studies have begun to address this question with respect to differing light environments. Sharwood et al. (2014) looked at the effects of shading on maize NADP-ME and PCK activities along with other photosynthetic characteristics. As expected, relative to control plants, overall biomass and photosynthetic rates of the shade plants were significantly lower, and in addition, both NADP-ME and PCK amount and activities were reduced in the shade-grown individuals. Interestingly however, PCK activity was decreased to a greater extent, with a 75% reduction versus a 60% drop for NADP-ME (Sharwood et al., 2014). Although energy partitioning between M and BS cells, BS PSII activity, and/or grana stacking in the chloroplasts of the two cell types were not investigated, the results do indicate that light availability differentially affects the 4-C organic acid produced, and C flux through the two decarboxylation pathways utilized by maize leaves, and consequently the balance of energy in M and BS cells.
Two recent modeling studies examined the effects of differing light regimes on M and BS energy partitioning while considering combination transfer organic acids and decarboxylating pathways (Figure 5) (Bellasio and Griffiths, 2014; Wang et al., 2014). A novel metabolic model along with experimentally measured inputs (Bellasio and Griffiths, 2014) showed that ATP and NADPH production were complementary in maize M and BS cells such that changes in light quality leading to increases in ATP or reducing power production in one cell type were offset by decreases in the other. This work supported the idea that the presence of alternative decarboxylation pathways, with their differing contributions to M and BS cell energy demands, are important in maintaining cell-type energy balance and high leaf CO2 assimilation rates in different light conditions (Figure 5) (Bellasio and Griffiths, 2014). A systems modeling approach taken by Wang et al. (2014) looked at CO2 assimilation rate for combination systems of transfer acids and decarboxylases relative to the standard NADP-ME pathway when differing amounts of light were allocated to the M and BS. This analysis also showed that flexibility in the organic acids transferred between M and BS cells, and the associated decarboxylase activities, as now recognized as operational in maize, would ensure high photosynthetic efficiency in differing light environments.
The proposal that a combination of transfer acids and decarboxylase pathways might contribute to the ability of C4 leaf organic acid pools to act as capacitors of ATP and reducing power equivalents in fluctuating light environments (Stitt and Zhu, 2014) has not been directly tested. However, the systems modeling approach described above (Wang et al., 2014) did support the suggestion that the concurrent transfer of multiple organic acids reduced the concentration gradient of individual species needed for C4 pathway function. The concentration gradients between M and BS cells for the standard NADP-ME pathway, and the combination pathway now recognized in maize leaves were simulated. A slight decrease in the malate gradient was predicted for the combination pathway compared to the standard NADP-ME pathway, while the pyruvate gradient was reduced by about 20%. Concentration gradients for Asp, PEP and Ala were predicted to compensate for these reductions and ensure efficient C4 photosynthesis (Wang et al., 2014).
Conclusion and Perspectives
The identification of dicarboxylic acids as the initial stable products of photosynthesis in some plant species was the first recognition of photosynthetic diversity in the terrestrial plant world. This discovery opened up not only the field of C4 biochemistry, but also all aspects of C4 plant biology, including a multitude of comparative studies in the areas of anatomy, physiology, ecology, evolution, biogeography, and recently, omics. Much of present day C4 photosynthesis research is focused on understanding the steps in the evolution of the pathway with an aim of transferring it into C3 crop plants to increase yield and/or mitigate effects of climate change.
Four- and 3-C organic acids are the substrates and products of the C4 acid transfer cycle that links CO2 uptake with CO2 fixation into carbohydrates. In most C4 plants, these reactions take place over two cell types and function to concentrate CO2 in internal cells of a C4 leaf. The genes encoding the decarboxylases that catalyze the release of CO2 from the 4-C organic acids near the sites of fixation and carbohydrate production have been co-opted for the C4 pathway from ancestral C3 genes. Historically, three subtypes of C4 plants have been recognized, based on the transferred 4-C acid and the decarboxylase with the highest activity in leaf tissue; however, recent work suggests that the majority of C4 plants transfer more than one type of 4-C acid, as well as multiple 3-C acids, during CO2 fixation, and have significant activity of the required secondary decarboxylase. Modeling studies indicate that the evolutionary routes to the C4 syndrome favored combination pathways.
The realization that multiple organic acids are transferred and combination decarboxylation pathways exist in C4 species has expanded our conception of C4 plant metabolism. However, we have only begun to comprehend the consequences of this more complex biochemistry on the overall biology of a C4 plant. Increasing evidence indicates that combination pathways allow flexibility in differing light regimes to meet the energy demands of the M and BS cells for CO2 fixation, thereby ensuring efficient functioning of the C4 CO2 concentrating mechanism. However, little to no information is available on how combination pathways may enable C4 plants to mitigate the effects of other fluctuating environmental factors or stresses, or how they may play a role during development.
Future work with C4 plants should consider the effects of differing light environments, nutrient availabilities, salinity, and leaf development on the levels of organic acids and other metabolites, enzyme and photosystem activities, CO2 assimilation rates, leaf anatomy, chloroplast ultrastructure, and M and BS energy status. The inter- and intracellular location of AST and ALT isoforms, and the identification of additional transporters on organelle membranes would contribute to the clarification of actual paths of C flux. Future studies should also consider the evolutionary history of C4-associated NAD-ME and PCK isoforms and identify the molecular changes responsible for their expression, location and activity. With C4 species, along with groups containing closely related C3, C3–C4 intermediate, and C4-like species, increasingly being examined in genomic, transcriptomic, proteomic, metabolomic, and flux studies, the resolution of the components and mechanisms of combination pathways will be a focus for future research. All of the above multifaceted approaches will allow a more comprehensive understanding of the costs and/or benefits of operating combination pathways on C4 plant metabolism, growth and productivity. In turn, the knowledge will give insights into the significance of these systems on C4 plant physiology and ecology, and contribute to attempts to increase C4 crop yield and ensure global food security, predict the effects of different climate change scenarios on natural and agricultural C4 species-rich environments, and inform future strategies in plant biotechnology.
Author Contributions
ML wrote the entire review and contributed all of the intellectual content.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GG, and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgment
Funding from the Australian Research Council is gratefully acknowledged (award number DP130102243).
References
Ali, S., and Taylor, W. C. (2001). Quantitative regulation of the Flaveria Me1 gene is controlled by the 3′-untranslated region and sequences near the amino terminus. Plant Mol. Biol. 46, 251–261. doi: 10.1023/A:1010669204137
Alvarez, C. E., Detarsio, E., Moreno, E., Andreo, C. S., and Drincovich, M. F. (2012). Functional characterization of residues involved in redox modulation in maize photosynthetic NADP-malic enzyme activity. Plant Cell Physiol. 53, 1144–1153. doi: 10.1093/pcp/pcs059
Badia, M. B., Arias, C. L., Tronconi, M. A., Maurino, V. G., Andreo, C. S., Drincovich, M. F., et al. (2015). Enhanced cytosolic NADP-ME2 activity in A. thaliana affects plant development, stress tolerance and specific diurnal and nocturnal processes. Plant Sci. 240, 193–203. doi: 10.1016/j.plantsci.2015.09.015
Bassham, J. A., and Calvin, M. (1957). The Path of Carbon in Photosynthesis. New Jersey, NJ: Prentice-Hall.
Bellasio, C., and Griffiths, H. (2014). The operation of two decarboxylases, transamination, and partitioning of C4 metabolic processes between mesophyll and bundle sheath cells allows light capture to be balanced for the maize C4 pathway. Plant Physiol. 164, 466–480. doi: 10.1104/pp.113.228221
Bräutigam, A., Schliesky, S., Külahoglu, C., Osborne, C. P., and Weber, A. P. M. (2014). Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 65, 3579–3593. doi: 10.1093/jxb/eru100
Brown, N. J., Newell, C. A., Stanley, S., Chen, J. E., Perrin, A. J., Kajala, K., et al. (2011). Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 331, 1436–1439. doi: 10.1126/science.1201248
Burr, G. O. (1962). The use of radioisotopes by the Hawaiian Sugar Plantations. Int. J. Appl. Radiat. 13, 365–374. doi: 10.1016/0020-708X(62)90008-X
Chen, Z. H., Walker, R. P., Tecsi, L. I., Lea, P. J., and Leegood, R. C. (2004). Phosphoenolpyruvate carboxykinase in cucumber plants is increased both by ammonium and acidification, and is present in the phloem. Plant 219, 48–58. doi: 10.1007/s00425-004-1220-y
Christin, P. A., Arakaki, M., Osborne, C. P., and Edwards, E. J. (2015). Genetic enablers underlying the clustered evolutionary origins of C4 photosynthesis in angiosperms. Mol. Biol. Evol. 32, 846–858. doi: 10.1093/molbev/msu410
Christin, P. A., Boxall, S. F., Gregory, R., Edwards, E. J., Hartwell, J., and Osborne, C. P. (2013). Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. 5, 2174–2187. doi: 10.1093/gbe/evt168
Christin, P. A., and Osborne, C. P. (2013). The recurrent assembly of C4 photosynthesis. Photosyn. Res. 117, 163–175. doi: 10.1007/s11120-013-9852-z
Christin, P. A., Petitpierre, B., Salamin, N., Buchi, L., and Besnard, G. (2008). Evolution of C4 phosphoenolpyruvate carboxykinase in grasses, from genotype to phenotype. Mol. Biol. Evol. 26, 357–365. doi: 10.1093/molbev/msn255
Christin, P. A., Samaritani, E., Petitpierre, B., Salamin, N., and Besnard, G. (2009). Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol. Evol. 1, 221–230. doi: 10.1093/gbe/evp020
Delgado-Alvarado, A., Walker, R. P., and Leegood, R. C. (2007). Phosphoenolpyruvate carboxykinase in developing pea seeds is associated with tissues involved in solute transport and is nitrogen-responsive. Plant Cell Environ. 30, 225–235. doi: 10.1111/j.1365-3040.2006.01622.x
Detarsio, E., Alvarez, C. E., Saigo, M., Andreo, C. S., and Drincovich, M. F. (2007). Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme. J. Biol. Chem. 282, 6053–6060. doi: 10.1074/jbc.M609436200
Drincovich, M. F., Lara, M. V., Andreo, C. S., and Maurino, V. G. (2011). “C4 decarboxylases: different solutions for the same biochemical problem, the provision of CO2 to Rubisco in the bundle sheath cells,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage (Dordrecht: Springer), 277–300.
Edwards, G. E., Kanai, R., and Black, C. C. (1971). Phosphoenolpyruvate carboxykinase in leaves of certain plants which fix CO2 by the C4-dicarboxylic acid cycle of photosynthesis. Biochem. Biophs. Res. Commun. 45, 278–285. doi: 10.1016/0006-291X(71)90814-X
Edwards, G. E., and Voznesenskaya, E. V. (2011). “C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage (Dordrecht: Springer), 29–61.
Finnegan, P. M., Suzuki, S., Ludwig, M., and Burnell, J. B. (1999). Phosphoenolpyruvate carboxykinase in the C4 monocot Urochloa panicoides is encoded by four differentially expressed genes. Plant Physiol. 120, 1033–1041. doi: 10.1104/pp.120.4.1033
Furbank, R. T. (2011). Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. doi: 10.1093/jxb/err080
Ghannoum, O., Evans, J. R., and von Caemmerer, S. (2011). “Nitrogen and water use efficiency of C4 plants,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage (Dordrecht: Springer), 129–146.
Gutierrez, M., Gracen, V. E., and Edwards, G. E. (1974). Biochemical and cytological relationships in C4 plants. Planta 119, 279–300. doi: 10.1007/BF00388331
Hatch, M. D. (1971). The C4-pathway of photosynthesis. Evidence for an intermediate pool of carbon dioxide and the identity of the donor C4-dicarboxylic acid. Biochem. J. 125, 425–432. doi: 10.1042/bj1250425
Hatch, M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106. doi: 10.1016/S0304-4173(87)80009-5
Hatch, M. D., Kagawa, T., and Craig, S. (1975). Subdivision of C4-pathway species based on differing C4 decarboxylating systems and ultrastructural features. Aust. J. Plant Physiol. 2, 111–128. doi: 10.1071/PP9750111
Hatch, M. D., and Slack, C. R. (1966). Photosynthesis by sugar-cane leaves: a new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101, 103–111. doi: 10.1042/bj1010103
Hatch, M. D., Slack, C. R., and Johnson, H. S. (1967). Further studies on a new pathway of photosynthetic carbon dioxide fixation in sugar-cane and its occurrence in other plant species. Biochem. J. 102, 417–422. doi: 10.1042/bj1020417
Jenkins, C. L. D., Furbank, R. T., and Hatch, M. D. (1989). Mechanism of C4 photosynthesis. A model describing the inorganic carbon pool in bundle sheath cells. Plant Physiol. 91, 1372–1381. doi: 10.1104/pp.91.4.1372
Kanai, R., and Edwards, G. E. (1999). “The biochemistry of C4 photosynthesis,” in C4 Plant Biology, eds R. F. Sage and R. K. Monson (London: Academic Press), 49–87.
Kortschak, H. P., Hartt, C. E., and Burr, G. O. (1965). Carbon dioxide fixation in sugarcane leaves. Plant Physiol. 40, 209–213. doi: 10.1104/pp.40.2.209
Koteyeva, N. K., Voznesenskaya, E. V., and Edwards, G. E. (2015). An assessment of the capacity for phosphoenolpyruvate carboxykinase to contribute to C4 photosynthesis. Plant Sci. 235, 70–80. doi: 10.1016/j.plantsci.2015.03.004
Kromdijk, J., Ubierna, N., Cousins, A. B., and Griffiths, H. (2014). Bundle-sheath leakiness in C4 photosynthesis: a careful balancing act between CO2 concentration and assimilation. J. Exp. Bot. 65, 3443–3457. doi: 10.1093/jxb/eru157
Lai, L. B., Wang, L., and Nelson, T. M. (2002). Distinct but conserved functions of two chloroplastic NADP-malic enzyme isoforms in C3 and C4 Flaveria species. Plant Physiol. 128, 125–139. doi: 10.1104/pp.010448
Leegood, R. C. (1985). The intercellular compartmentation of metabolites in leaves of Zea mays L. Planta 164, 163–171. doi: 10.1007/BF00396078
Leegood, R. C., and ap Rees, T. (1978). Identification of the regulatory steps in gluconeogenesis in cotyledons of Cucurbita pepo. Biochim. Biophys. Acta 542, 1–11. doi: 10.1016/0304-4165(78)90226-X
Leegood, R. C., Crowther, D., Walker, D. A., and Hind, G. (1983). Energetics of photosynthesis in Zea mays. I. Studies of the flash-induced electrochromic shift and fluorescence induction in bundle sheath cells. Biochim. Biophys. Acta 722, 116–126. doi: 10.1016/0005-2728(83)90164-0
López-Bucio, J., Nieto-Jacobo, M. F., Ramírez-Rodríguez, V., and Herrera-Estrella, L. (2000). Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 160, 1–13. doi: 10.1016/S0168-9452(00)00347-2
Ludwig, M. (2013). Evolution of the C4 pathway: events at the cellular and molecular levels. Photosyn. Res. 117, 147–161. doi: 10.1007/s11120-013-9853-y
Maier, A., Zell, M. B., and Maurino, V. G. (2011). Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 62, 3061–3069. doi: 10.1093/jxb/err024
Marshall, J. S., Stubbs, J. D., Chitty, J. A., Surin, B., and Taylor, W. C. (1997). Expression of the C4 Me1 gene from Flaveria bidentis requires an interaction between 5′ and 3′ sequences. Plant Cell 9, 1515–1525. doi: 10.2307/3870440
Marshall, J. S., Stubbs, J. D., and Taylor, W. C. (1996). Two genes encode highly similar chloroplastic NADP-malic enzymes in Flaveria. Implications for the evolution of C4 photosynthesis. Plant Physiol. 111, 1251–1261. doi: 10.1104/pp.111.4.1251
Maurino, V. G., Saigo, M., Andreo, C. S., and Drincovich, M. F. (2001). Non-photosynthetic malic enzyme from maize: a constitutively expressed enzyme that responds to plant defense inducers. Plant Mol. Biol. 45, 409–420. doi: 10.1023/A:1010665910095
Meierhoff, K., and Westhoff, P. (1993). Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: the non-stoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191, 23–33.
Meister, M., Agostino, A., and Hatch, M. D. (1996). The roles of malate and aspartate in C4 photosynthetic metabolism of Flaveria bidentis (L.). Planta 199, 262–269. doi: 10.1007/BF00196567
Muhaidat, R., and McKown, A. D. (2013). Significant involvement of PEP-CK in carbon assimilation of C4 eudicots. Ann. Bot. 111, 577–589. doi: 10.1093/aob/mct017
Muhaidat, R., Sage, R. F., and Dengler, N. G. (2007). Diversity of Kranz anatomy and biochemistry in C4 eudicots. Am. J. Bot. 94, 362–381. doi: 10.3732/ajb.94.3.362
Penfield, S., Clements, S., Bailey, K. J., Gilday, A. D., Leegood, R. C., Gray, J. E., et al. (2012). Expression and manipulation of phosphoenolpyruvate carboxykinase 1 identifies a role for malate metabolism in stomatal closure. Plant J. 69, 679–688. doi: 10.1111/j.1365-313X.2011.04822.x
Pick, T. R., Bräutigam, A., Schluter, U., Denton, A. K., Colmsee, C., Scholz, U., et al. (2011). Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23, 4208–4220. doi: 10.1105/tpc.111.090324
Sage, R. F., Christin, P. A., and Edwards, E. J. (2011). The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169. doi: 10.1093/jxb/err048
Saigo, M., Alvarez, C. E., Andreo, C. S., and Drincovich, M. F. (2013). Plastidial NADP-malic enzymes from grasses: unraveling the way to the C4 specific isoforms. Plant Physiol. Biochem. 63, 39–48. doi: 10.1016/j.plaphy.2012.11.009
Sharwood, R. E., Sonawane, B. V., and Ghannoum, O. (2014). Photosynthetic flexibility in maize exposed to salinity and shade. J. Exp. Bot. 65, 3715–3724. doi: 10.1093/jxb/eru130
Sommer, M., Bräutigam, A., and Weber, A. P. M. (2012). The dicotyledonous NAD malic enzyme C4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biol. 14, 621–629. doi: 10.1111/j.1438-8677.2011.00539.x
Stitt, M., and Heldt, H. W. (1985). Generation and maintenance of concentration gradients between mesophyll cell and bundle sheath in maize leaves. Biochim. Biophys. Acta 808, 400–414. doi: 10.1016/0005-2728(85)90148-3
Stitt, M., and Zhu, X. G. (2014). The large pools of metabolites involved in intercellular metabolite shuttles in C4 photosynthesis provide enormous flexibility and robustness in a fluctuating light environment. Plant Cell Environ. 37, 1985–1988. doi: 10.1111/pce.12290
Tausta, S. L., Coyle, H. M., Rothermel, B., Stiefel, V., and Nelson, T. (2002). Maize C4 and non-C4-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol. Biol. 50, 635–652. doi: 10.1023/A:1019998905615
Walker, R. P., Acheson, R. M., Tésci, L. I., and Leegood, R. C. (1997). Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust. J. Plant Physiol. 24, 459–468. doi: 10.1071/PP97007
Walker, R. P., Chen, Z. H., Johnson, K. E., Famiani, F., Tecsi, L., and Leegood, R. C. (2001). Using immunocytochemistry to study plant metabolism: the examples of its use in the localization of amino acids in plant tissue, and of phosphoenolpyruvate carboxykinase and its possible role in pH regulation. J. Exp. Bot. 52, 565–576. doi: 10.1093/jexbot/52.356.565
Walker, R. P., Chen, Z. H., Tésci, L. I., Famiani, F., Lea, P. J., and Leegood, R. C. (1999). Phosphoenolpyruvate carboxykinase plays a role in interactions of carbon and nitrogen metabolism during grape seed development. Planta 210, 9–18. doi: 10.1007/s004250050648
Wang, Y., Bräutigam, A., Weber, A. P. M., and Zhu, X. G. (2014). Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65, 3567–3578. doi: 10.1093/jxb/eru058
Williams, B. P., Aubry, S., and Hibberd, J. M. (2012). Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 17, 213–220. doi: 10.1016/j.tplants.2012.01.008
Keywords: C4 photosynthesis, C4 acid, malate, aspartate, NAD-malic enzyme, NADP-malic enzyme, oxaloacetate, phosphoenolpyruvate carboxykinase
Citation: Ludwig M (2016) The Roles of Organic Acids in C4 Photosynthesis. Front. Plant Sci. 7:647. doi: 10.3389/fpls.2016.00647
Received: 23 March 2016; Accepted: 27 April 2016;
Published: 17 May 2016.
Edited by:
Veronica Graciela Maurino, Heinrich-Heine-Universität Düsseldorf, GermanyReviewed by:
Xinguang Zhu, Chinese Academy of Sciences, ChinaGeorg Groth, University of Duesseldorf, Germany
Copyright © 2016 Ludwig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martha Ludwig, bWFydGhhLmx1ZHdpZ0B1d2EuZWR1LmF1
 Martha Ludwig
Martha Ludwig