- 1Key Laboratory of Aquatic Botany and Watershed Ecology, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2Beijing Genome Institute-Shenzhen, Beishan Industrial Zone, Shenzhen, China
- 3Department of Biological Sciences, University of Texas at El Paso, El Paso, TX, USA
Introduction
Ranunculus L. is an early diverging clade in the Angiosperm phylogeny. The split between terrestrial and aquatic/semi-aquatic lineages within the genus occurred within 20 Ma (Emadzade and Horandl, 2011), which is much younger than for some well-known aquatic lineages such as Ceratophyllum and Potamogeton. Ranunculus sceleratus Linn. is a semi-aquatic plant commonly found in paddy fields, streams and lakes in Asia, Europe and North America (eFlora of China, http://www.efloras.org/). The plant has also been studied for its unique toxicological and pharmacological properties (Prieto et al., 2003). Its karyotype is 2n = 32, x = 8 in mainland China (Yang, 2000). It usually roots in inundated soils, with stems and leaves emergent. The semi-aquatic R. sceleratus has been hypothesized to be an inter-mediate type in the transition from a terrestrial to aquatic habitat within the genus (Barrett et al., 1993; Prieto et al., 2003).
In this study, we generated RNA-seq data of R. sceleratus and analyzed this in relation to the RNA-seq data of Ranunculus bungei Steud., Ranunculus cantoniensis DC., and Ranunculus brotherusii Freyn (Chen et al., 2015). Our aim was to generate RNA-seq data of R. sceleratus and identify the orthologous genes among the four species.
Links to Deposited Data
RNA-seq data of R. sceleratus was deposited at NCBI sequence Reads Archive (no. SRP072329). It is in SRA format. ITS sequence of R. sceleratus was deposited in GenBank (no. KT957621). All the unigenes of R. sceleratus, data matrices of ITS and chloroplast DNA, sequences of the 3455 putative orthologous clusters identified using the program OrthoMCL, and 884 clusters after filters were deposited in figshare (https://figshare.com/s/f40d8f9bd8f894f2d630; sequences in fasta format). These data can be used for further transcriptome assembly, selective pressure estimation, phylogenetic analyses, etc.
Materials and Methods
Plant Material, Sequencing, and de novo Assembly
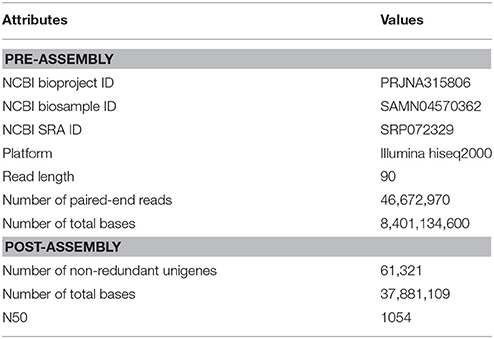
R. sceleratus was collected from East Lake (30°32′44.97″ N, 114°42′10.07″ E), Hubei, China, in Oct. 2014. Living plant material was brought to laboratory within 20 min. Leaves, stems, and roots from one individual were immediately sampled and frozen with liquid nitrogen. RNA extraction using RNAisoTM Plus (Takara, Qingdao, China) and quality checking were performed using the methodology in Chen et al. (2015). The RNA sample was transported to a laboratory of Beijing Genomics Institute (BGI) in Wuhan. mRNA was extracted using oligo (dT) magnetic beads, followed by breaking into small fragments. A cDNA library with inserted size c. 200 bp was constructed. Paired-end sequencing (2 × 90 bp) was performed using an Illumina HiSeq™ 2000 sequencer.
Raw reads were processed using Filter_fq (an internal program of BGI) to remove adaptor sequences, reads with unknown base calls (N) larger than 5%, and reads with low quality bases (quality value ≤ 10) more than 20%. De novo assembly using the clean reads was performed using Trinity v. 20130225 (Grabherr et al., 2011) with parameters—min_contig_length 100,—group_pairs_distance 250,—path_reinforcement_distance 85, and default parameters. Unigenes recovered from Trinity were clustered to get long sequence unigenes and remove redundancies using program TGI Clustering Tool (TGICL) v. 2.1 (Pertea et al., 2003) with parameter -l 40, -c 10, and -v 20.
The RNA-seq data of R. bungei, R. cantoniensis, and R. brotherusii (NCBI Sequence Read Archive, no. SRR1822558, SRR1822529, SRR173752s) were also used.
Nuclear ITS and Chloroplast DNA Sequences
(1) The internal transcribed spacer regions (ITS1, ITS2) and 5.8S ribosomal RNA gene for R. sceleratus was generated with the methodology in Chen et al. (2015); ITS for R. bungei, R. cantoniensis, and R. brotherusii were accessed from GenBank (nos. KP336399, KP336398, KP336400). Close relative of Ranunculus, viz. Laccopetalum giganteum and Krapfia clypeata (GenBank nos. GU552271, GU552272), were used as outgroups following Emadzade and Horandl (2011). (2) cpDNA sequences for the four Ranunculus species were extracted from non- redundant unigenes of the four species; the chloroplast genome of Clematis terniflora was accessed (GenBank no. NC_028000). C. terniflora was used as an outgroup as it is the closest relative of Ranunculus with a complete chloroplast genome available in GenBank. The DNA alignment was conducted using Mauve v. 20150226 (Darling et al., 2004). Conserved sequences for the five species were concatenated and yielded a matrix with 29,654 bp length for each species. Maximum likelihood (ML) analyses were conducted for ITS and cpDNA datasets separately using RAxML v. 8.1.20 (Stamatakis, 2006) with the GTRGAMMA model and fast bootstrap (1000 replicates) analyses. Phylogenetic relationships of the four Ranunculus species recovered from ITS and cpDNA are congruent with each other. ITS phylogeny: [(L. giganteum, K. clypeata), (R. cantoniensis, (R. brotherusii, (R. bungei, R. sceleratus)71)100)85); cpDNA phylogeny: (C. terniflora, (R. cantoniensis, (R. brotherusii, (R. bungei, R. sceleratus)100)100)].
Identification of Orthologous Genes
Step by step strategies were adopted to find orthologs and exclude possible paralogs:
(1) Orthologous clusters among R. bungei, R. sceleratus, R. cantoniensis, R. brotherusii, and Vitis vinifera were identified using OrthoMCL (Li et al., 2003) with default settings according to Wissler et al. (2011). More species may increase efficiency in computational screening for orthologs. Protein sequences of V. vinifera, which show high similarity to Ranunculus species, were downloaded from Genoscope (http://www.genoscope.cns.fr/spip/). Clusters with ≥1 sequence per species were used. If >1 sequence of any species was included in a cluster, only the sequence showing the highest similarity to sequences of other species was kept. (2) After removing the sequences of V. vinifera, orthologous clusters were aligned by MUSCLE (Edgar, 2004) with default parameters. Clusters with alignment length < 200 bps and including unexpected stop codons were discarded. (3) Saturation tests were performed to remove orthologs saturated at synonymous sites. The third codon positions of each ortholog were extracted, then used to estimate branch lengths of the gene tree with the general time reversible model in the PAML package v. 4.8 (Yang, 2007). The cluster was discarded from further analyses if branch length of one or more branches was < 1.
(4) ML analyses were also conducted for all the clusters from the last step using the same methods as for the nuclear ITS and chloroplast DNA sequence data sets, but without the outgroup setting. The clusters, which had different species relationships than the ITS and cpDNA trees, were excluded from further analyses. This method is able to exclude the possible paralogs (Zeng et al., 2014). (5) A local blast database was constructed using protein sequences accessed from NCBI (Aug. 2015) with the software package NCBI BLAST+ v. 2.2.31. Protein sequences of 19 species, which showed high similarity to sequences of Ranunculus in our preliminary analyses, were incorporated, viz. Amborella trichopoda, Arabidopsis thaliana, Brassica napus, Camelina sativa, Citrus sinensis, Cucumis sativus, Elaeis guineensis, Fragaria vesca, Glycine max, Gossypium raimondii, Malus domestica, Medicago truncatula, Nelumbo nucifera, Populus trichocarpa, Prunus mume, Ricinus communis, Sesamum indicum, V. vinifera, and Zea mays. In total, 752,004 sequences were incorporated. All the orthologs from the last step were annotated to the local database using BLASTX with E = 10−3 and default settings. If any of the four orthologs within a cluster were matched to different sequences, the cluster was excluded from further analyses using perl scripts. (6) The aligned clusters were inspected by eye in BioEdit v. 7.2.5 (Hall, 1999). Alignment problems at either end of sequences were deleted, and clusters with problems in the middle were excluded.
Results
De novo Assembly
We generated 46.7 million of clean paired reads totaling 8.4 × 109 bp of RNA-seq data for R. sceleratus. De novo assembly yielded 111,101 contigs, with total length 37,271,711 bp, mean length of 335 bp and N50 at 635 bp. TGICL yielded 61,321 non-redundant unigenes, with total length of 37,881,109 bp, mean length of 618 bp, and N50 at 1054 bp (Table 1).
Orthologous Clusters
A total of 3455 putative orthologs were recovered for R. sceleratus, R. bungei, R. cantoniensis, and R. brotherusii using OrthoMCL. After removing the clusters with aligned length < 200 bp, including unexpected stop codons, and saturation at synonymous sites, 2637 orthologous clusters were retained. After removing the clusters that were not consistent with phylogeny, blast or alignment, 884 clusters were retained.
Author Contributions
SZ and QW conceived this study; SZ and LC carried out experimental works and with MM drafted the manuscript; SZ, LC, YW and ZH performed data analyses. All authors gave approval of this manuscript to be published.
Funding
This research was supported by grants from the National Natural Science Foundation of China (no. 31500192), the Chinese Academy of Sciences (no. XDAO5090305), and the Special Foundation for State Basic Working Program of China (no. 2013FY112300).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AC and handling Editor AM declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Wei Li for discussion on this study; Kuo Liao for assistance in plant collection.
References
Barrett, S. C. H., Eckert, C. G., and Husband, B. C. (1993). Evolutionary processes in aquatic plant populations. Aquat. Bot. 44, 105–145. doi: 10.1016/0304-3770(93)90068-8
Chen, L. Y., Zhao, S. Y., Wang, Q. F., and Moody, M. L. (2015). Transcriptome sequencing of three Ranunculus species (Ranunculaceae) reveals candidate genes in adaptation from terrestrial to aquatic habitats. Sci. Rep. 5:e10098. doi: 10.1038/srep10098
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emadzade, K., and Horandl, E. (2011). Northern Hemisphere origin, transoceanic dispersal, and diversification of Ranunculeae DC. (Ranunculaceae) in the Cenozoic. J. Biogeogr. 38, 517–530. doi: 10.1111/j.1365-2699.2010.02404.x
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid 41, 95–98.
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Prieto, J. M., Recio, M. C., Giner, R. M., Máñez, S., and Ríos, J. L. (2003). Pharmacological approach to the pro-and anti-inflammatory effects of Ranunculus sceleratus L. J. Ethnopharmacol. 89, 131–137. doi: 10.1016/S0378-8741(03)00271-X
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Wissler, L., Codoner, F. M., Gu, J., Reusch, T. B. H., Olsen, J. L., Procaccini, G., et al. (2011). Back to the sea twice: identifying candidate plant genes for molecular evolution to marine life. BMC Evol. Biol. 1:e8. doi: 10.1186/1471-2148-11-8
Yang, Q. E. (2000). Cytology of eleven species in the genus Ranunculus L. and five in its four related genera from China (in Chinese). Acta Phytotaxon. Sin. 39, 405–422.
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Keywords: transcriptome, Ranunculus, orthologous genes
Citation: Zhao SY, Chen LY, Wei YL and Wang QF and Moody ML (2016) RNA-seq of Ranunculus sceleratus and Identification of Orthologous Genes among Four Ranunculus Species. Front. Plant Sci. 7:732. doi: 10.3389/fpls.2016.00732
Received: 13 April 2016; Accepted: 12 May 2016;
Published: 31 May 2016.
Edited by:
Alessio Mengoni, Università degli Studi di Firenze, ItalyReviewed by:
Gabriel Rocha Fernandes, Centro de Pesquisa René Rachou-Fiocruz, BrazilAndrea Coppi, Università degli Studi di Firenze, Italy
Copyright © 2016 Zhao, Chen, Wei, Wang and Moody. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Feng Wang, cWZ3YW5nQHdiZ2Nhcy5jbg==
 Shu-Ying Zhao1
Shu-Ying Zhao1 Ling-Yun Chen
Ling-Yun Chen Michael L. Moody
Michael L. Moody