- 1Triticease Research Institute, Sichuan Agricultural University, Chengdu, China
- 2National Key Facility of Crop Gene Resources and Genetic Improvement, Institute of Crop science, Chinese Academy of Agricultural Sciences, Beijing, China
Photoperiod response-related genes play a crucial role in duration of the plant growth. In this study, we focused on TaPRR73, a paralog of “Green Revolution” gene Ppd1 (TaPRR37). We found that overexpression of the truncated TaPRR73 form lacking part of the N-terminal PR domain in transgenic rice promoted heading under long day conditions. Association analysis in common wheat verified that TaPRR73 was an important agronomic photoperiod response gene that significantly affected heading date and plant height; expression analysis proved that specific alleles of TaPRR73-A1 had highly expressed levels in earlier heading lines; the distribution of haplotypes indicated that one of these alleles had been selected in breeding programs. Our results demonstrated that TaPRR73 contributed to regulation of heading date in wheat and could be useful in wheat breeding and in broadening adaptation of the crop to new regions.
Introduction
Photoperiod genes in plants affect the timing of transition from the vegetative to reproductive phases. Allelic variation of photoperiod response genes enables common or bread wheat to adapt to different day-lengths characteristic of different latitudes and thus become more widely cultivated. The pseudo-response regulator (PRR) gene family is highly conserved in protein structure. These members have an N-terminal PR (pseudoreceiver) domain and a C-terminal CCT (CONSTANS, CO-like, TOC1) domain (Makino et al., 2000; Wenkel et al., 2006). There are five members in each of Arabidopsis thaliana (TOC1, PRR3, PRR5, PRR7, and PRR9) (Matsushika et al., 2000) and rice (OsPRR1, OsPRR37, OsPRR73, OsPRR59, and OsPRR95) (Murakami et al., 2003). Study of these family members in wheat are less reported, as most attention focused on Ppd (TaPRR37) (Beales et al., 2007; Guo et al., 2010). There has been one expression study of TaPRR73 (Shaw et al., 2012).
PRR73 and PRR37 are paralogous genes that exist in plant genomes (Higgins et al., 2010). Paralogous genes make up a significant proportion of plant genomes, for example 22% of the rice genome (Goff et al., 2002), 50% in modern maize (Schnable et al., 2011), and more than 67% in soybean (Schmutz et al., 2010). Paralogous genes are derived from duplication events that occurred in the ancestors of modern species (Fitch, 1970; Van de Peer et al., 2009), and their functions may duplicate, or be differentiated from, those of their progenitors. Therefore, mining of the functions of paralogous gene series may have significance for both genetic analysis and breeding.
Numerous studies have demonstrated that OsPRR37 plays important roles in increasing photoperiod sensitivity in rice. OsPRR37 delays heading by repressing Hd3a under long day conditions (Koo et al., 2013). TaPRR37 (Ppd), one of the well-known “Green Revolution” genes in wheat, is an important photoperiod gene associated with multiple agronomic traits. Ppd-A1, Ppd-B1, and Ppd-D1, located in the three sub-genomes, are orthologous photoperiod response gene loci, with the a alleles causing early flowering under both short and long photoperiods (Beales et al., 2007; Wilhelm et al., 2009; Bentley et al., 2011) and the b alleles conferring day length sensitivity that delays flowering under SD conditions (Laurie, 1997; Shaw et al., 2012). The Ppd-D1 gene consists of six haplotypes and affects heading time, plant height and 1000-kernel weight (Guo et al., 2010). Multiple copies and/or higher methylation of Ppd-B1 enhance expression levels and promote heading and photoperiod insensitivity (Díaz et al., 2012; Sun et al., 2014). In addition to their effects on flowering Ppd genes regulate inflorescence architecture and paired spikelet behavior (Boden et al., 2015), and may improve grain yield and seed threshability during harvesting (Doebley et al., 2006).
As a paralog of PRR37, PRR73 may also have a potential role in regulation of flowering (Higgins et al., 2010; Shaw et al., 2012). Here, we analyzed the functions of TaPRR73 in wheat by a transgenic approach, expression analysis, linkage mapping, and association analysis. Our results shed light on the potential value of TaPRR73 in genetic improvement of cereals such as wheat, rice and barley.
Materials and Methods
Plant Material
Eleven hexaploid wheat accessions (Chinese Spring, Neixiang 188, Yanzhan 1, Opata M85, W7984, Am3, Am6, Laizhou 953, Fuzhang 30, Hanxuan 10, and Lumai 14) and 6 diploid accessions (UR201, UR203, UR206, ABD104, AB08, AM0102) were used for sequencing. Two hundred and seventy introgression lines (ILs) were derived from crosses of 30 donor varieties to Yanzhan 1, followed by four or five backcrosses to Yanzhan 1, and then selfed without selection for more than three generations. One hundred and fifty-six wild species are listed in Supplementary Tables 1, 2. Three hundred and eighty accessions (including landrace and modern cultivars listed in Supplementary Table 3) from 10 major wheat-growing regions of China were used in determining haplotype distributions. These were planted at Changping in Beijing (116.2°E, 40.2°N), and Luoyang (111.6°E, 33.8°N), Xinxiang (113.8°E, 35.2°N) and Jiaozuo (113.4°E, 35.10°N) in Henan province during years 2011–2014. A recombinant inbred line (RIL) population derived from cross Neixiang 188 × Yanzhan 1 (199 lines) was used for genetic mapping. Transgenic rice lines were planted at Langfang in Heibei province under long day conditions. All materials were provided by the Key Laboratory of Crop Gene Resources and Germplasm Enhancement, Institute of Crop Sciences, CAAS. Genomic DNA was extracted from all materials by a modified CTAB method (Saghai-Maroof et al., 1984).
Phylogenetic Analysis
The sequences of TaPRR1, TaPRR59, and TaPRR95 were obtained from D genome scaffolds, and their protein constructs were predicted by PROSITE (http://www.expasy.ch/prosite/). Mega 5.0 software was used to produce a phylogenetic tree (http://www.megasoftware.net).
Software Analysis
Cis-regulatory elements were predicted by PLACE (Higo et al., 1999). Statistical analyses were conducted with SPSS 15.0 (SPSS Inc. Chicago, IL, USA) and Power Marker V3.25 (Liu and Muse, 2005).
Primer Design and PCR
Primers for amplifying the TaPRR73 gene included the A genome-specific primer TaPRR73AF1/TaPRR73AR1 and B and D genome primers TaPRR73BDF1/TaPRR73BDR1, TaPRR73AF1: GCACCACCACTTCTCTCCTC; and TaPRR73AR1: CTACTGGCTTGCTCCTTCTT; TaPRR73BDF1: AAACGAGGACAAGGAATGGAGG; and TaPRR73BDR1: GGGACAATAATCATACGGGTGG.
RT-qPCR primers used for wheat were TaPRR73-A1F/TaPRR73-A1R, TaPRR73-B1F/TaPRR73-B1R, TaPRR73-D1F/TaPRR73-D1 (Shaw et al., 2012) and TaPRR73-F/PRR73-R; and primer sets OsHd1-F/OsHd1-R (Kojima et al., 2002), OsGI-F/OsGI-R and OsMADS51-F2/OsMADS51-R2 were used in transgenic rice (OsGI-F2: CCGAATACTCTCCCAACCGA and OsGI-R2: AAACCATACGCAGCCTCCCA; OsMADS51-F2: GTCTCTCCAAAACAATGC; and OsMADS51-R2: TCTGCTCCTACTCCCTTC). High-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) was used to isolate T-DNA-flanking sequences from transgenic rice plants (Liu and Chen, 2007). All primers were synthesized by Sangon (www.sangon.com). LA-Taq enzyme from TaKaRa (www.takara.com.cn) was used for PCR amplification, and Pfu was included at 1/10th of the total enzyme concentration to ensure amplification accuracy. The PCR mixture comprised 5 μL of 2 × GC buffer, 2.5 μL ddH2O, 1.5 μL DNA (20 ng/μL) or cDNA as template, 0.4 μL of each primer (10 μmol/L), 0.1 μL dNTP (25 mmol/L), and 0.1 μL LA-Taq (5 U/μL) in a total volume 10 μL. The PCR protocol was 95°C for 5 min; 95°C for 40 s, primer annealing at 58°C for 40 s, and extension at 72°C for 1 Kb/min for 32 cycles and a final extension at 72°C for 10 min.
Marker Development
Marker PASF2/PASR2 was developed based on the 9 bp indel difference between Hap 1 and Hap 2 of TaPRR73-A1 (PASF2: TTTGTAGTTATCGCTGCTGAGAA; PASR2: AAC AAGGACCAAAATAAGCGTAT). Marker URSF1/URSR1 was designed according to the 10 bp indel present in the third exon of diploid lines (URSF1: ACGGGTGGGTCTTTATTTGTT; URSR1: GCCTCATCTGCTTGGCTATTT). Marker 73ASF1/73ASR1 was designed according to a 306 bp indel differentiating hexaploid (Hap 1 and Hap 2) and diploid (Hap 3 and Hap IV) haplotypes (73ASF1: GTCGTTTGTCAACCGTCTCT; 73ASR1: CAGGGCATTACCTTCATAGC). Allele-specific markers (CF4/R4; TF4/R4) to distinguish TaPRR73-B1 haplotypes were designed according to SNPs in the two haplotypes (CF4: ATG ACTGTACCCGACATATC; TF4: ATGACTGTACCCGACATGCT; R4: CAGCCAACCATTGCATGCA).
Transgenic Vector Construction
We firstly constructed a binary vector by combing the pCUbi1390 vector and Gateway cassete A including the maize ubiquitin promoter, NOS terminator, and hygromycin resistance. We inserted the truncated cDNA of TaPRR73 partially lacking the N-terminal PR domain into the binding vector by Gateway technology (Life technologies, Invitrogen), and then transformed it into japonica rice cultivar (cv.) Nipponbare.
Expression Analysis
Wheat accession Hussar and rice accession Nipponbare and transgenic lines were planted under long (15 h light, 9 h darkness) and short (9 h light, 15 h darkness) day conditions. Each treatment was sampled every 3 h during a 48 h period; at each time-point samples from three plants were pooled. Plant organs including roots, shoots, leaves, and young ears were taken from one plant of Chinese Spring under natural long day conditions (LDs). Total mRNA was extracted with an RNA extraction kit (Tiangen Biotech, Beijing) and reverse transcribed with Moloney Murine Leukemia Virus (M-MLV) (Invitrogen). Real-time qPCR (RT-qPCR) was performed on an ABI PRISM 7900 (Applied Biosystems, USA) with SYBR Premix ExTaqII (Takara), and data were quantified by the 2−ΔΔCt method (Livak and Schmittgen, 2001). Wheat and transgenic rice expression data were normalized by GADPH and tubulin, respectively. Microarray data for TaPRR73, Ppd, HvPRR73, OsPRR73, and OsPRR37 were obtained from the Genevestigator database (https://www.genevestigator.com/gv/) (Zimmermann et al., 2004). Expression patterns of these genes were compared after all data were standardized by the Z-score method (Benedito et al., 2008). A heatmap was drawn by MeV4.9 (http://www.tm4.org/mev.html).
Results
Overexpression of a Truncated TaPRR73 Promoted Heading in Rice under Long Day Conditions by Suppressing Expression of OsGI
To explore the role of TaPRR73 in heading, we constructed a truncated TaPRR73 expression vector, beginning with the second ATG in the intact gene sequence (Supplementary sequence 1), and transformed it into cv. Nipponbare. The sequence was based on a platform of wheat genes transformed into rice to mine new functional genes. Five lines including the truncated TaPRR73 were obtained, but three of them contained other inserted genes. Two independent T0 transgenic lines were 6 days earlier in heading than the controls under long day conditions (LDs). Next we determined the transgene position in the rice genome by hiTAIL-PCR (Liu and Chen, 2007) and found that it was inserted into a retrotransposon rather than a promoter or coding region. T1 generation transgenic lines planted in natural LDs were significantly earlier heading (113.7 days) than wild type (119.9 days) (p < 0.01) (Table 1). T2 generation plants were even earlier heading (nearly 15 days) than the Nipponbare control (Figure 1).

Table 1. T-test of days to heading of truncated TaPRR73-B1 T1 transgenic and nontransgenic rice plants.
To investigate the mechanism of action of TaPRR73 in rice we compared the expression patterns of heading date-related genes OsGI, OsHd1, and OsMADS51 in transgenic lines under LDs and SDs (Figure 2). Under LDs, OsGI and OsHd1 suppress flowering in rice (Yano et al., 2000; Hayama et al., 2003) whereas expression of the genes in our transgenic lines was suppressed, suggesting that truncated TaPRR73 promoted heading by inhibiting expression of heading date suppressors under LDs. Under SDs, OsGI suppresses flowering (Hayama et al., 2003) whereas OsHd1 and OsMADS51 promote heading in rice (Yano et al., 2000; Kim et al., 2007). Expression levels of OsGI, OsHd1, and OsMADS51 in the transgenic lines increased during darkness.
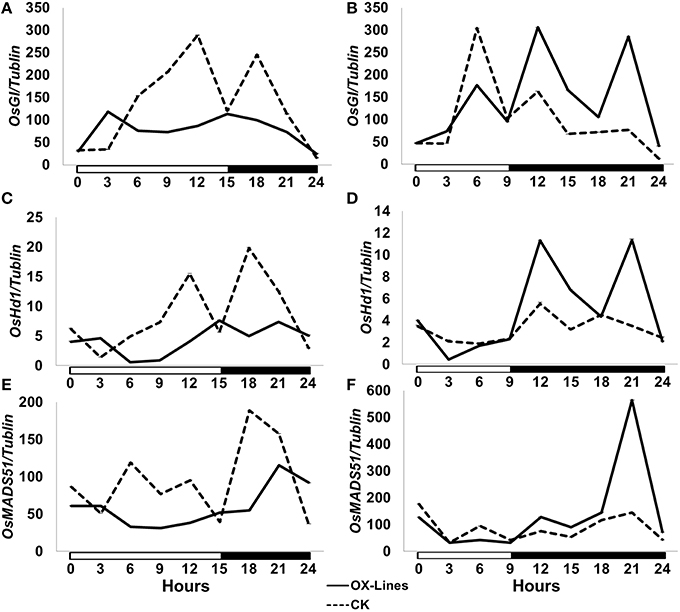
Figure 2. Expression of major photoperiod genes in truncated TaPRR73-OX rice lines and cv. Nipponpare check. (A,C,E) Expression levels of OsHd1, OsGI, and OsMADS51 under LDs (15 h light/ 9 h darkness). (B,D,F) The expression level of OsGI, OsHd1, and OsMADS51 under SDs (9 h light/ 15 h darkness). Four biologically independent replications were performed for each sample. White boxes below the graphs indicate light periods; dark boxes indicate darkness. The data were normalized by tubulin.
The PRR Gene Family Members Have a Conserved Protein Structure and Might be Similar in Function
To study variation and function of PRR73 in common wheat and compare it with other PRR families, we isolated TaPRR73-A1, TaPRR73-B1, and TaPRR73-D1 in Chinese Spring according to the cDNA sequence in transgenic lines and D genome scaffolds (Jia et al., 2013), and also compared the amino acid sequences of family members in Triticum aestivum, Oryza sativa, Arabidopsis thaliana, Hordeum vulgare, Sorghum bicolor, Brachypodium distachyon, and Zea mays by constructing a genetic phylogenic tree. The phylogenic tree divided into three clusters, revealing that the PRRs were very similar in monocots Triticum aestivum, Oryza sativa, Hordeum vulgare, Sorghum bicolor, Brachypodium distachyon, and Zea mays (Figure 3). PRR73 and PRR37 were highly similar in monocots and closest to PRR3 and PRR7 in the dicotyledon A. thaliana. PRR59 and PRR95 were also similar in monocots and orthologous with PRR9 and PRR5 in A. thaliana. The PRR1 cluster had a similar pattern. However, the PRR1 cluster had least similarity with the other clusters, indicating a difference in amino acid sequence and possibly in function. Research on PRR gene function has shown that PRR37 and PRR73 were morning-expressed circadian genes, whereas TOC1 was evening-expressed (Higgins et al., 2010). This suggested that PRR37 and PRR73 might have different ways of regulating photoperiod response compared to TOC1. All the above results demonstrated that PRRs are highly conserved in monocots.
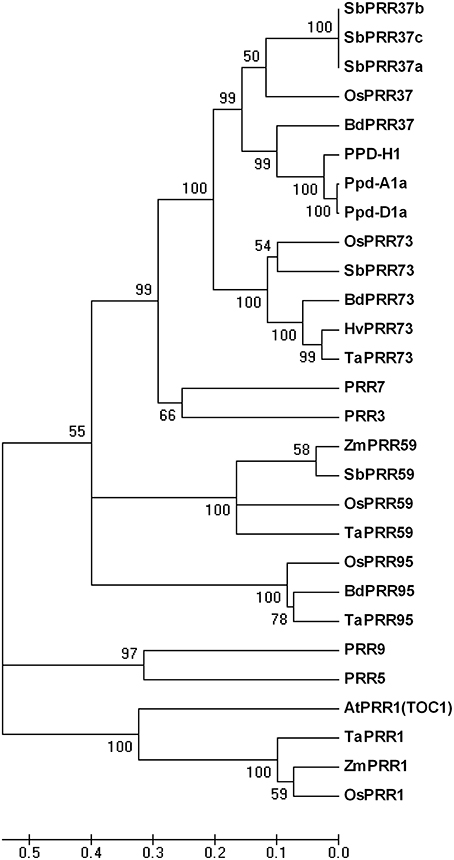
Figure 3. Phylogenetic tree of PRR proteins from Triticum aestivum, Oryza sativa, Hordeum vulgare, Sorghum bicolor, Brachypodium distachyon, Zea mays, and Arabidopsis thaliana. Accession no. of Oryza sativa, Hordeum vulgare, Sorghum bicolor, Brachypodium distachyon, Zea mays, and A. thaliana proteins were obtained from NCBI; Triticum aestivum Ppd-D1a, ABL09464; and Ppd-A1a, ABW93666. The amino acid sequences of TaPRR1, TaPRR59, and TaPRR95 were predicted from the D genome scaffold.
TaPRR73 Exhibited a Circadian Rhythm and Higher Expression Level in Leaf Tissue
RT–qPCR was used to investigate the expression patterns of TaPRR73 during a 48 h period in common wheat cv. Hussar grown under SD and LD conditions. TaPRR73 was expressed mainly during the light period under both LD and SD conditions, and its transcript levels peaked 3 h after dawn (Figure 4) as reported in previous studies (Wilhelm et al., 2009; Shaw et al., 2012). OsPRR73 and OsPRR37 in rice also had the same expression peak, and five PRRs expressed in a sequential manner of OsPRR73 (OsPRR37) → OsPRR95 (OsPRR59) → OsPRR1 (Murakami et al., 2005). In Arabidopsis, five members expressed in the order AtPRR9, AtPRR7, AtPRR5, AtPRR3, and TOC1 over a 24 h period (Matsushika et al., 2000). From our results and published data we concluded that the paralogous gene pairs TaPRR73 and TaPRR37, and OsPRR73 and OsPRR37 are orthologs of AtPRR7 and behave in a similar circadian manner.
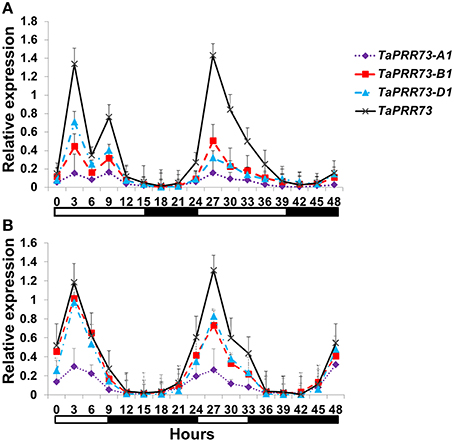
Figure 4. Relative circadian expression of TaPRR73 in cv. Hussar under LD and SD conditions. (A) TaPRR73, TaPRR73-A1, TaPRR73-B1, and TaPRR73-D1 in LDs. (B) TaPRR73, TaPRR73-A1, TaPRR73-B1, and TaPRR73-D1 in SDs. The expression level of TaPRR73 reached a peak 3 h after dawn. The contrasting environments were 15 h light/9 h darkness and 9 h light/15 h darkness. Four biologically independent replications were performed for each sample. White boxes below the graphs indicate light periods; dark boxes indicate darkness. The data were normalized by GADPH.
We investigated expression levels of the three orthologous TaPRR73 genes in six different organs of Chinese Spring (Figure 5), collected from 9 to 10 am during the day. Expression levels of the three homoeologs ranked TaPRR73-B1 > TaPRR73-D1 > TaPRR73-A1 (Figure 5). All three were more highly expressed in leaves than in other organs, with expression levels from highest to lowest being leaves > leaf sheaths > adult roots > young ears > nodes > shoots, in accordance with in-silico expression data (the Genevestigator database) (Supplementary Figure 1). Both TaPRR73 and Ppd had the highest expression levels in leaves and lowest expression levels in endosperm as determined from microarray data for OsPRR73, OsPRR37, and Ppd1 (TaPRR37) (Genevestigator database). However, TaPRR73 also had high expression levels in roots and pistils, whereas the corresponding expression levels of Ppd in roots and pistils were lower than in other organs, suggesting partial differentiation in function of the two genes.
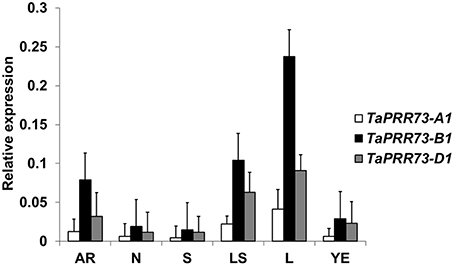
Figure 5. Expression levels of the three TaPRR73 homologs in different organs. AR, adult roots; N, nodes; S, shoots; LS, leaf sheaths; L, leaves; YE, young ears. Ranking of expression levels from highest to lowest was L > LS > AR > YE > N > S.
To investigate expression differences between TaPRR73 and Ppd1 (TaPRR37), we compared their coding and promoter regions. They shared high similarity in amino acid sequence (68.2%), especially in the CCT domain region (95.2%), but had large differences in the promoter regions. We analyzed the cis-regulatory elements in 2000 bp segments upstream of the ATG start codons of TaPRR73-B1 and Ppd1 (TaPRR37-B1). Although, both paralogs had a light-responsive element, a hormone-responsive element, and abiotic-responsive elements, the element sequences were different and bound different proteins. Moreover, there were a root hair-specific cis-element and a dehydration-responsive element in TaPRR73-B1, suggesting specific effects on roots (Supplementary Figure 2). The different cis-regulatory elements in the promoter may cause functional differentiation of these paralogous genes.
Haplotype Variation and Linkage Analysis of TaPRR73
In order to detect variation in TaPRR73-A1, TaPRR73-B1, and TaPRR73-D1, we sequenced the coding and promoter regions in four diploid accessions and 11 hexaploid accessions. There were four haplotypes of TaPRR73-A1 and two haplotypes of TaPRR73-B1, but no variation in TaPRR73-D1 in the tested hexaploid wheat accessions.
We compared TaPRR73-A1 haplotypes in common wheat (HapI and HapII) and wild species (Hap3 and Hap4) (Supplementary Figures 3A–C). A 306 bp insertion in common wheat led to an additional exon in TaPRR73-A1 compared TaPRR73-B1 (Figure 6A). Three SNPs and a 276 bp indel in the promoter region, and 12 SNPs and a 9 bp indel in coding region in TaPRR73-A1 formed two haplotypes. The twelve SNPs in the coding region caused no amino acid substitutions, but the 9 bp indel in the third exon resulted in a 3 amino acid indel (GIG) that potentially could lead to functional polymorphism. In TaPRR73-B1 13 SNPs and 2 Indels resulted in two haplotypes (Figure 6B, Supplementary Figure 3E). Differences included two indels (206 and 11 bp) and five SNPs in the promoter region, and eight SNPs in the coding region. The SNPs in the first and sixth exons caused no amino acid substitutions; the SNP in the seventh exon led to an amino acid substitution: asparagine (N) at position 681 in Hap 1 to threonine (T) in Hap II.
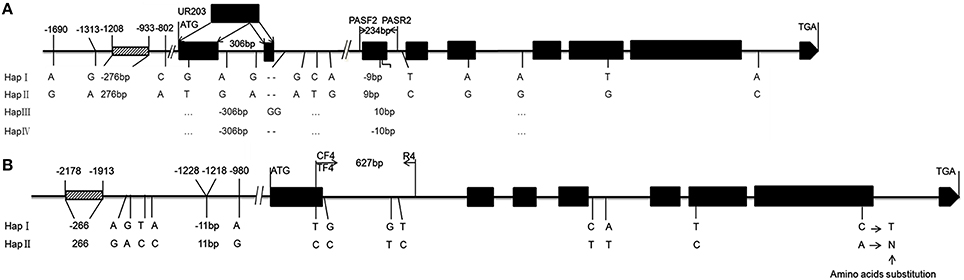
Figure 6. Gene structures of TaPRR73-A1 and TaPRR73-B1. Structure and SNPs and indels in the four TaPRR73-A1 (A) and two TaPRR73-B1 (B) haplotypes are shown below the diagrams.
To map TaPRR73, marker PASF2/PASR2 based on the 9 bp indel was developed to distinguish TaPRR73-A1 HapI and HapII. This marker was then utilized to map TaPRR73-A1 in chromosome 4A in the recombinant inbred line (RIL) population developed from the cross Neixiang 188 × Yanzhan 1. TaPRRR73 was located between markers WMC516 and W7001, with genetic distances of 8.6 and 6.2 cM, respectively (Supplementary Figure 3D).
TaPRR73 is Associated with Heading Date and Plant Height
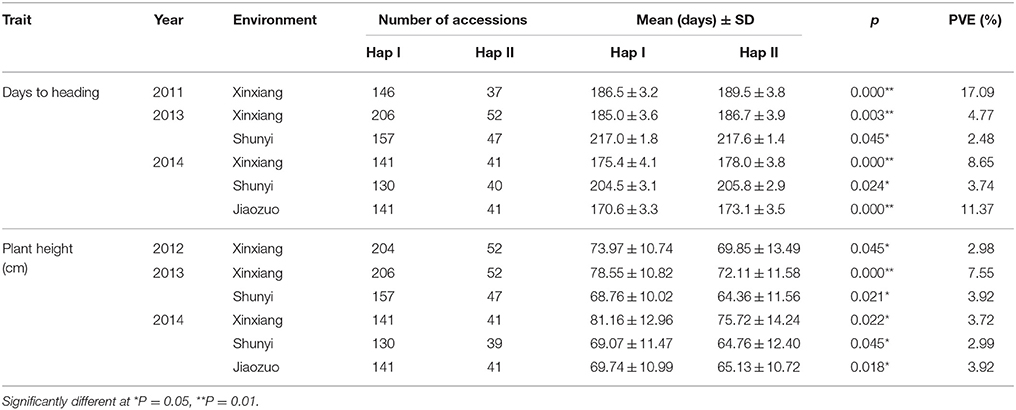
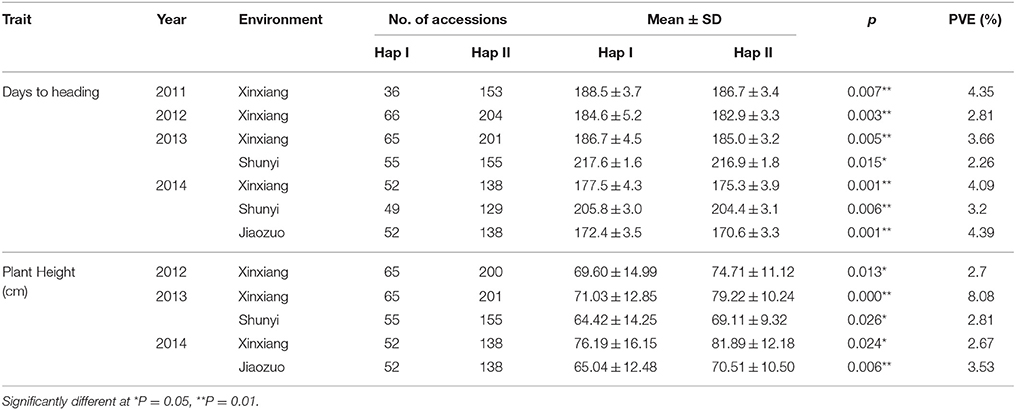
The Yanzhan 1 introgression lines were used to further examine the relationship between TaPRR73 and agronomic traits. TaPRR73-A1 was significantly associated with heading date (phenotypic variation explained (PVE) ranging from 2.48 to 17.09%) and plant height (PVE, 2.98–7.55%). HapI accessions were earlier heading (0.6–3 days) and taller (4.31–6.44 cm) than HapII accessions under long day conditions (Table 2). TaPRR73-B1 was also significantly associated with heading date (PVE ranging from 2.26 to 4.39%), and plant height (PVE, 2.7–8.08%). Hap II was earlier heading (0.7–2.2 days), and taller (5.11–8.19 cm) than Hap I (Table 3).
Because of the significant associations between haplotypes and traits we examined gene expression levels by RT-qPCR. Relative expression of TaPRR73-A1 HapI (0.250) was higher than HapII (0.191) (p = 0.01), whereas relative expression of TaPRR73-B1 HapII (0.258) was higher than HapI (0.207) (Figure 7A). There was a significant negative correlation between relative expression and days to heading in TaPRR73-A1 (R2 = 0.4574, r = −0.676, p < 0.01; Figure 7B), but not in TaPRR73-B1 (R2 = 0.5501, r = −0.575, p > 0.05; Figure 7C). Higher expression levels of TaPRR73-A1 HapI led to earlier heading.
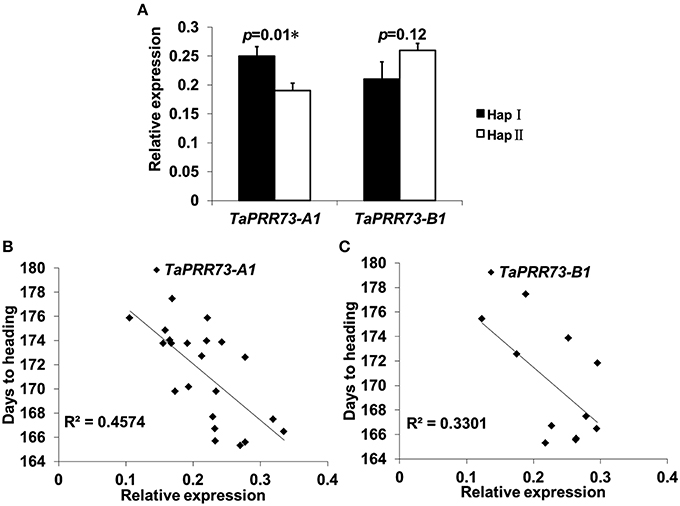
Figure 7. Relative expression levels of different haplotypes and correlations between relative expression levels and days to heading. (A) Relative peak time expression of TaPRR73-A1 and TaPRR73-B1 for different haplotypes grown under long day conditions. Three biologically independent replications were performed for each sample, and five or six accessions were tested for each haplotype. Standard deviations of means are indicated by error bars. Correlation analyses of relative expression levels and days to heading for TaPRR73-A1 (B) and TaPRR73-B1 (C).
Haplotype Analyses of TaPRR73-A1 and TaPRR73-B1
Allele frequency is an indicator of past selection in breeding. TaPRR73-A1 Hap I (7.69%), Hap II (75%), Hap III (9.62%), and Hap IV (7.69%) were detected in wild species, but Hap III and Hap IV were absent in common wheat landraces and modern cultivars. Hap II was the dominant haplotype in wild species (46.34%) and landraces (86.52%), but its frequency was lower (33.06%) in modern cultivars. Hap I was present in 6.10 and 13.48% of wild species accessions and landraces, and 66.94% in modern cultivars (Figure 8A). We analyzed haplotype frequencies of TaPRR73-A1 by PowerMarker V3.25 (Liu and Muse, 2005), and found that the frequencies for TaPRR73-A1 differed significantly between landraces and modern cultivars (u = 7.99 > 3.29; p < 0.001), suggesting that Hap I was a favored haplotype selected in modern breeding programs. When the frequencies of the two TaPRR73-B1 haplotypes were tested in the same way (u = 0.49, p > 0.05) there was no difference between landraces and modern cultivars and hence no evidence of previous selection for either haplotype (Figure 8B).
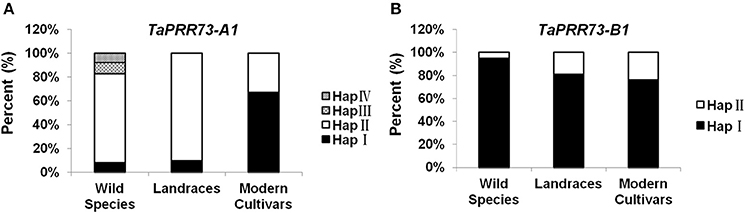
Figure 8. Distributions of haplotypes of TaPRR73-A1 (A) and TaPRR73-B1 (B) in wild species, landraces, and modern cultivars.
Discussion
Overexpression of Truncated TaPRR73 Accelerates Heading by Regulating Expression of OsGI in Transgenic Rice under LDs
Gene expression analyses showed that overexpression of truncated TaPRR73 promotes heading in transgenic rice by reducing expression of OsGI, OsHd1, and OsMADS51 under LDs. OsHd1 and OsMADS51 are downstream of OsGI in the rice flowering pathway (Hayama et al., 2003; Kim et al., 2007). OsHd1 activates OsHd3a to promote flowering in SDs, and down-regulates OsHd3a to inhibit heading under LDs (Yano et al., 2000; Kim et al., 2007). OsGI is an ortholog of GI (Fowler et al., 1999) and suppresses flowering when overexpressed in transgenic rice, leading to late flowering under both SDs and LDs (Hayama et al., 2003). OsGI regulates expression of OsHd1 and OsMADS51, and both OsHd1 and OsMADS51 promote heading under short-day conditions (Yano et al., 2000; Kim et al., 2007). In truncated TaPRR73-OX lines, expression of OsGI was repressed and transgenic plants flowered earlier under LDs. We therefore concluded that overexpression of truncated TaPRR73 advances heading date by reducing the expression level of OsGI in transgenic rice under LDs.
Transgenic rice lines with the truncated TaPRR73 exhibited earlier heading indicating that TaPRR73 may function as a regulator of heading in common wheat. However, we also raise the question of whether overexpression of truncated TaPRR73 affects heading in transgenic rice plants by interfering with the function of the rice orthologs of TaPRR73 or acts independently. This will be addressed in future studies.
The intermediate region (IR) and CCT domains were present and involved no frame-shifts. The repression motif and CCT domain are important parts of PRRs in regulating downstream genes in Arabidopsis (Nakamichi et al., 2010; Gendron et al., 2012). TOC1 is a critical circadian component of a feedback loop acting as a DNA-binding transcription repressor. It binds directly to the promoters of CCA1/LHY by its CCT domain to repress their expression (Gendron et al., 2012). The repression motif is in the pseudoreceiver (PR) domain of TOC1, and it alone cannot repress CCA1 expression in the absence of the CCT and IR domains (Gendron et al., 2012). However, the repression motif is present in the IR between the PR and CCT domains of PRR5, PRR7, and PRR9 (Nakamichi et al., 2010). Moreover, the CCT motif mutation (toc1-2) reduces the expression level of CCA1/LHY in Arabidopsis under LDs, whereas the PR domain mutation (toc1-1) continues to have some TOC1 function (Millar et al., 1995; Strayer et al., 2000; Alabadí et al., 2001).
TaPRR73 is an Agronomically Important Heading Date Gene in Wheat Breeding
PRR37 (Ppd1) is an agronomically important photoperiod response gene that made a significant contribution in wheat, barley and rice breeding in the “Green Revolution”. In the present study, we found that TaPRR73, a paralog of TaPRR37, is also an important heading date-related gene lso affects plant height. The expression of TaPRR73 in roots is relatively high, whereas there is negligible expression of TaPRR37. A comparison of TaPRR37 and TaPRR73 revealed differences in the promoter regions that could be the underlying reason for differences in expression. Zawaski et al. (2012) reported two putative PHOTOPERIOD RESPONSE 1 (PHOR1) orthologs, PtPHOR1_1 and PtPHOR1_2, in Populus. PtPHOR1_1 was most highly expressed in, and restricted to, the roots, whereas PtPHOR1_2 was more uniformly expressed throughout all plant tissues with similar effects in aerial and below-ground tissues. We therefore speculate that TaPRR37 mainly affects aerial tissues and that TaPRR73 has effects on both aerial parts and roots. Many important agronomic traits, such as drought and salinity tolerances, are related to root development, therefore justifying further investigation of the functions of TaPRR73 in roots. Functional divergence of paralogs might result from differences in the promotor regions. The functions of TaPRR37 and TaPRR73 are either similar or diverged in functional complementarity, but work together in plant growth and development.
PRR37 has been clearly recognized and widely employed in wheat and rice breeding and has made enormous contributions worldwide. Here, we identified functions of PRR73 that could be applied in crop improvement. For example, favorable haplotypes of TaPRR73-A1 were selected in past breeding programs. Although, no favorable allele was found in the D genome of hexapolid wheat, perhaps due to the domestication bottleneck associated with hexaploidisation, more diversity may be present the diploid progenitor Ae. tauschii, which is known to be rich in genetic diversity (Jia et al., 2013). Any favorable allele in the species can easily be transferred to common wheat by development of synthetic wheat followed by introgression to agronomically adapted cultivars.
Development of an Efficient Platform to Mine Paralogous Gene Function
Plant genome sequencing has revealed that many plant genomes have paralogous gene sets. However, their individual functions are rarely reported (Xu et al., 2015). In the present study, we investigated TaPRR73, a paralog of the well-known Ppd1 gene series, as a target gene, and employed a series of approaches (including transformation experiments, expression analysis, haplotype analysis, and association analysis) to mine its function. We transferred about 4000 wheat transcription factors to rice to observe their functions by over-expression. We constructed a core collection and a series of introgression lines for association analysis. With rapid advances in plant genomics techniques, increasing numbers of platforms and databases are available for paralogous gene analysis. RNA-seq databases (Wang et al., 2009; Bansal et al., 2014) also provide a platform to detect paralogous gene expression patterns. Techniques for achieving high transformation rates in rice (Duan et al., 2012), wheat (Ishida et al., 2015), and other crops provide effective platforms to confirm gene function. Genotypes of core collections and ILs generated from re-sequencing and high density SNP arrays together with their phenotypes provide a platform for GWAS (Topol and Frazer, 2007). All of these platforms and methods will accelerate mining of paralogous gene function.
Author Contributions
WZ, JJ, and BW designed the study. WZ, GZ, LG, XK, and ZG collected data and performed the research. WZ, JJ Wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Robert A McIntosh (University of Sydney, Australia) for English editing, Dr. Haiyang Wang (Institute of Biotechnology, CAAS, China) and Dr. Jiaqiang Sun (Institute of Crop Science, CAAS, China) for critical reading of our manuscript. This work was supported by the National Natural Science Foundation of China (grant nos. 39893352, 31271292, and 31571668) and National Basic Research Program of China (grant no. 2014CB138103).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00772
Abbreviations
PRR, pseudo-response regulator; PR, pseudoreceiver; IR, intermediate region; CCT, CONSTANS, CO-like, TOC1; RIL, recombinant inbred line; ILs, introgression lines; LDs, long day conditions; SDs, short day conditions; PVE, phenotypic variation explained.
References
Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Bansal, R., Mian, M. A. R., Mittapalli, O., and Michel, A. P. (2014). RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean. BMC Genomics. 15:972. doi: 10.1186/1471-2164-15-972
Beales, J., Turner, A., Griffths, S., Snape, J. W., and Laurie, D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. doi: 10.1007/s00122-007-0603-4
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Bentley, A. R., Turner, A. S., Gosman, N., Leigh, F. J., Maccaferri, M., Dreisigacker, S., et al. (2011). Frequency of photoperiod-insensitive Ppd-A1a alleles in tetraploid, hexaploid and synthetic hexaploid wheat germplasm. Plant Breed. 130, 10–15. doi: 10.1111/j.1439-0523.2010.01802.x
Boden, S. A., Cavanagh, C., Cullis, B. R., Ramm, K., Greenwood, J., Finnegan, E. J., et al. (2015). Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plants. 14016, 1–6. doi: 10.1038/nplants.2014.16
Díaz, A., Zikhali, M., Turner, A. S., Isaac, P., and Laurie, D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 7:e33234. doi: 10.1371/journal.pone.0033234
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Duan, Y. B., Zhai, C. G., Li, H., Li, J., Mei, W. Q., Gui, H. P., et al. (2012). An efficient and high-throughput protocol for Agrobacterium-mediated transformation based on phosphomannose isomerase positive selection in Japonica rice (Oryza sativa L.). Plant Cell Rep. 31, 1611–1624. doi: 10.1007/s00299-012-1275-3
Fitch, W. M. (1970). Distinguishing homologous from analogous proteins. Syst. Zool. 19, 99–106. doi: 10.2307/2412448
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Coupland, G., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. Embo J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R. L., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Guo, Z., Song, Y., Zhou, R., Ren, Z., and Jia, J. (2010). Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 185, 841–851. doi: 10.1111/j.1469-8137.2009.03099.x
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Higgins, J. A., Bailey, P. C., and Laurie, D. A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5:e10065. doi: 10.1371/journal.pone.0010065
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Ishida, Y., Tsunashima, M., Hiei, Y., and Komari, T. (2015). Wheat (Triticum aestivum. L.) transformation using immature embryos. Agrobacterium Protocols. 1223, 189–198. doi: 10.1007/978-1-4939-1695-5_15
Jia, J. Z., Zhao, S. C., Kong, X. Y., Li, Y. R., Zhao, G. Y., He, W. M., et al. (2013). Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496, 91–95. doi: 10.1038/nature12028
Kim, S. L., Lee, S. Y., Kim, H. J., Nam, H. G., and An, G. H. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a(1[W][OA]). Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant. 6, 1877–1888. doi: 10.1093/mp/sst088
Laurie, D. A. (1997). Comparative genetics of flowering time. Plant Mol. Biol. 35, 167–177. doi: 10.1023/A:1005726329248
Liu, K. J., and Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. doi: 10.1093/bioinformatics/bti282
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–656. doi: 10.2144/000112601
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., et al. (2000). Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. doi: 10.1093/pcp/41.6.791
Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. doi: 10.1093/pcp/pcd043
Millar, A. J., Carré, I. A., Strayer, C. A., Chua, N. H., and Kay, S. A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. doi: 10.1126/science.7855595
Murakami, M., Ashikari, M., Miura, K., Yamashino, T., and Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44, 1229–1236. doi: 10.1093/pcp/pcg135
Murakami, M., Matsushika, A., Ashikari, M., Yamashino, T., and Mizuno, T. (2005). Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci. Biotech. Bioch. 69, 410–414. doi: 10.1271/bbb.69.410
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). Pseudo-response regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J. X., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Schnable, J. C., Springer, N. M., and Freeling, M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. U.S.A. 108, 4069–4074. doi: 10.1073/pnas.1101368108
Shaw, L. M., Turner, A. S., and Laurie, D. A. (2012). The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 71, 71–84. doi: 10.1111/j.1365-313X.2012.04971.x
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., et al. (2000). Cloning of the Arabidopsis clock cone TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Sun, H., Guo, Z., Gao, L., Zhao, G., Zhang, W., Zhou, R., et al. (2014). DNA methylation pattern of Photoperiod-B1 is associated with photoperiod insensitivity in wheat (Triticum aestivum). New Phytol. 204, 682–692. doi: 10.1111/nph.12948
Topol, E. J., and Frazer, K. A. (2007). The resequencing imperative. Nat. Genet. 39, 439–440. doi: 10.1038/ng0407-439
Van de Peer, Y., Fawcett, J. A., Proost, S., Sterck, L., and Vandepoele, K. (2009). The flowering world: a tale of duplications. Trends Plant Sci. 14, 680–688. doi: 10.1016/j.tplants.2009.09.001
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wenkel, S., Turck, F., Singer, K., Gissot, L., Gourrierec, J. L., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Wilhelm, E. P., Turner, A. S., and Laurie, D. A. (2009). Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor. Appl. Genet. 118, 285–294. doi: 10.1007/s00122-008-0898-9
Xu, C. Z., Tai, H. H., Saleem, M., Ludwig, Y., Majer, C., Berendzen, K. W., et al. (2015). Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol. 207, 1123–1133. doi: 10.1111/nph.13420
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Zawaski, C., Ma, C., Strauss, S. H., French, D., Meilan, R., and Busov, V. B. (2012). Photoperiod response 1 (PHOR1)-like genes regulate shoot/root growth, starch accumulation, and wood formation in Populus. J. Exp. Bot. 63, 5623–5634. doi: 10.1093/jxb/ers217
Keywords: TaPRR73, association analysis, heading date, transgenic gene, Triticum aestivum
Citation: Zhang W, Zhao G, Gao L, Kong X, Guo Z, Wu B and Jia J (2016) Functional Studies of Heading Date-Related Gene TaPRR73, a Paralog of Ppd1 in Common Wheat. Front. Plant Sci. 7:772. doi: 10.3389/fpls.2016.00772
Received: 07 March 2016; Accepted: 17 May 2016;
Published: 01 June 2016.
Edited by:
Paula Casati, Centro de Estudios Fotosintéticos y Bioquímicos-CONICET, ArgentinaReviewed by:
Mingsheng Chen, Institute of Genetics and Developmental Biology-Chinese Academy of Sciences, ChinaTao Sun, Stanford University, USA
Copyright © 2016 Zhang, Zhao, Gao, Kong, Guo, Wu and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bihua Wu, d3ViaWh1YTIwMDVAMTI2LmNvbQ==;
Jizeng Jia, anpqaWFAbWFpbC5jYWFzLm5ldC5jbg==
 Wenping Zhang
Wenping Zhang Guangyao Zhao2
Guangyao Zhao2