- 1Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Nakhon Pathom, Thailand
Mungbean (Vigna radiata L.) is a cleistogamous plant in which flowers are pollinated before they open, which prevents yield improvements through heterosis. We previously generated a chasmogamous mutant (CM) mungbean in which open flowers are pollinated. In this study, we developed insertion/deletion (indel) markers based on the transcriptome differences between CM and Sulu-1 (i.e., normal flowering) plants. An F2 population derived from a cross between CM and Sulu-1 was used for gene mapping. Segregation analyses revealed that a single recessive gene regulates the production of chasmogamous flowers. Using newly developed indel and simple sequence repeat markers, the cha gene responsible for the chasmogamous flower trait was mapped to a 277.1-kb segment on chromosome 6. Twelve candidate genes were detected in this segment, including Vradi06g12650, which encodes a YUCCA family protein associated with floral development. A single base pair deletion producing a frame-shift mutation and a premature stop codon in Vradi06g12650 was detected only in CM plants. This suggested that Vradi06g12650 is a cha candidate gene. Our results provide important information for the molecular breeding of chasmogamous mungbean lines, which may serve as new genetic resources for hybrid cultivar development.
Introduction
Exploiting heterosis is an effective way to increase crop yield. As an important legume in Asia, several studies have been conducted on mungbean (Vigna radiata L. Wilczek) to investigate the importance of heterosis for seed yield and other yield-related traits (Chen et al., 2003; Soehendi and Srinives, 2005; Sorajjapinun et al., 2012; Yashpal et al., 2015). However, a major obstacle to producing mungbean hybrid seeds is floral architecture. Mungbean plants have papilionaceous flowers with five differently shaped petals, including one standard petal, two wing petals, and two keel petals (Verdcourt, 1970). The anthers and stigmas are enclosed within the pocket-like keel petals, making mungbean a cleistogamous plant whose flowers are pollinated before they open. The natural outcrossing rate of cultivated mungbean is approximately 1.68% (Sangiri et al., 2007), which is too low for the commercial production of hybrid seeds. Some accessions of cleistogamous plants exhibit a specific floral architecture that promotes natural outcrossing. Studies in rice revealed that exerted stigmas and anthers increase the outcrossing rate and considerably help the production of hybrid seeds (Kato and Namai, 1987). Some mutant legume crops with exerted stigmas and anthers have been described (Dahiya et al., 1984; Pundir and Reddy, 1998; Cherian et al., 2006; Srinivasan and Gaur, 2012; Dheer et al., 2014). We previously identified a chasmogamous mutant (CM), which was generated in the mungbean accession V1197 by gamma irradiation (Sorajjapinun and Srinives, 2011). The outcrossing rate of CM plants increased to 9.6%, while outcrossing was undetectable in wild-type controls. Additionally, the yield and agronomic traits of CM plants differed from those of V1197 plants, with fewer pods per plant, seeds per pod, and yield per plant. Genetic analyses using F1, F2, and backcross populations from the cross between CM and V1197 plants revealed that the production of chasmogamous flowers is regulated by a single gene, cha (Sorajjapinun and Srinives, 2011). Thus, additional research focusing on the genetic characterization of this mutant mungbean should be conducted.
In terms of molecular genetics and genomics, mungbean has not been as extensively studied as other legumes, including soybean [Glycine max (L.) Merr.], common bean (Phaseolus vulgaris L.), cowpea [V. unguiculata (L.) Walp.], and chickpea (Cicer arietinum L.). DNA markers represent important tools for genetic analyses and the mapping of genes or quantitative trait loci. Many DNA markers, especially simple sequence repeats (SSRs), have been developed for mungbean (Gwag et al., 2007; Somta et al., 2008, 2009; Tangphatsornruang et al., 2009; Gupta et al., 2014; Chen et al., 2015) or introduced from other closely related species. These markers have been used to develop linkage maps and map quantitative trait loci associated with important mungbean traits (e.g., yield and resistance to biotic and abiotic stresses; Kasettranan et al., 2010; Chankaew et al., 2011, 2013; Isemura et al., 2012; Kajonphol et al., 2012; Prathet et al., 2012; Sompong et al., 2012; Kitsanachandee et al., 2013; Alam A.K.M.M. et al., 2014; Alam A.M. et al., 2014; Chotechung et al., 2016). However, most of these markers are monomorphic or weakly polymorphic. One of the most important advances in mungbean genomics research was the release of the VC1973A whole genome sequence by Kang et al. (2014). This sequence is relevant for research related to marker development, gene mapping, and gene function analyses.
The molecular mechanisms regulating floral development can be summarized using an “ABCE” model (Krizek and Fletcher, 2005). Additionally, some hormones (e.g., auxin) are important for floral development (Okada et al., 1991; Bennett et al., 1995; Przemeck et al., 1996; Galweiler et al., 1998; Aloni et al., 2006; Cheng et al., 2006). Some Arabidopsis thaliana mutants have abnormal flowers, and the genes responsible for this mutation have been identified as pin1, pinoid, mp, and yuc (Cheng and Zhao, 2007). The YUCCA (YUC) family consists of flavin monooxygenases (FMOs) related to the biosynthesis of indole-3-acetic acid. The FMOs are key enzymes that convert indole-3-pyruvic acid to indole-3-acetic acid by catalyzing the hydroxylation of the amino group of tryptophan, which is the rate-limiting step in tryptophan-dependent auxin biosynthesis (Zhao et al., 2001). There are 11 YUC genes in the A. thaliana genome (Zhao et al., 2001; Cheng et al., 2006, 2007). Double, triple, and quadruple mutants of some YUC family genes exhibit severe defects in floral patterns, vascular formations, and other developmental processes (Cheng et al., 2006). Therefore, YUC genes play important roles during floral formation and development, with implications for flower shape.
In this study, we mapped the cha gene regulating the production of chasmogamous flowers in CM plants using new insertion/deletion (indel) and SSR markers. We identified a gene encoding a YUC-like protein as a likely candidate for the cha gene.
Materials and Methods
Mapping Population and DNA Extraction
We previously identified a CM mungbean line in the M2 generation of accession V1197 following gamma irradiation. The CM plants lacked wing and keel petals, which exposed the stigmas and anthers (Sorajjapinun and Srinives, 2011). A stable CM line was selected from the M3 and M4 generations. A CM plant was pollinated using pollen from Sulu-1, which is a mungbean line with normal flowers. Three F1 plants were grown and the resulting flowers were morphologically analyzed. The hybrids were verified using two polymorphic indel markers (i.e., VRID001 and VRID002; Supplementary Table S1). Seeds from one F1 plant were then harvested to produce an F2 population consisting of 127 plants. The F2 plants and their parents were grown in a field at Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand from May to July 2014. Flowers from each plant were individually examined to determine whether they were normal or chasmogamous.
Total genomic DNA was extracted from fresh leaf tissue of individual plants according to a slightly modified version of the method described by Lodhi et al. (1994). All DNA samples were diluted to 5 ng μl-1 according to lambda DNA, and analyzed by 1.0% agarose gel electrophoresis.
Transcriptome Sequencing and Development of Molecular Markers
Total RNA was extracted from young flowers of CM and Sulu-1 mungbean plants using the EasyPure Plant RNA kit (Transgene Biotech, Beijing, China). The RNA samples were used to prepare libraries for sequencing by the Illumina HiSeq 2000 sequencer at the Beijing Genomics Institute (Shenzhen, China). The resulting sequences were assembled using the Trinity program (Grabherr et al., 2011). The Sulu-1 and CM transcriptome sequences were aligned using the NCBI BLAST+ 2.2.31 program with an E-value cutoff of 10.0. Sequences with indels that were 5 bp or larger were randomly chosen for marker development. Primers specific for the selected indels were designed using Primer3 (Untergasser et al., 2012) with the following criteria: primer length: 18–27 nucleotides; melting temperature: 50–65°C; GC content: 40–60%; and polymerase chain reaction (PCR) product size: 100–300 bp. Transcript sequences used for indel marker development were blasted against the mungbean whole genome sequence1 (Kang et al., 2014) to determine the physical locations of markers.
In addition to indel markers, we developed new SSR markers to fine map the cha locus. The mungbean chromosome 6 sequence was downloaded (Kang et al., 2014) and scanned for di-, tri-, and tetra-nucleotide repeats using SSR Hunter 1.3 (Li and Wan, 2005). Based on our initial mapping of the cha locus using indel markers, we focused on a 2.2-Mb genomic region of chromosome 6 (i.e., 29.9–32.1 Mb) carrying cha, and selected the SSRs therein. Primers for the SSRs were designed as described for the indel markers.
Molecular Marker Analysis
The newly developed indel and SSR markers were used to detect polymorphisms between CM and Sulu-1 sequences. The PCR analyses were completed using a 10-μl solution containing 2 ng genomic DNA, 1x Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 1 U Taq DNA polymerase (Thermo Scientific), and 0.5 μM forward and reverse primers. The PCR was conducted in an MJ Research PTC-200 Thermal Cycler (Bio-Rad) using the following program: 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30s; 72°C for 5 min. The amplicons were separated in a 6% denaturing polyacrylamide gel or a 3% agarose gel, and visualized with silver or ethidium bromide staining, respectively. The polymorphic markers were used to analyze the F2 population.
Data Analysis and Gene Mapping
We counted the number of plants with normal and chasmogamous flowers, and analyzed the data using a χ2 test (Mather, 1951) to confirm the monogenic inheritance in CM plants as reported earlier by Sorajjapinun and Srinives (2011). Segregation data for the floral traits and DNA markers were used to construct a linkage map with MapMaker/EXP 3.0 (Lander et al., 1987). A minimum logarithm of odds value of 3.0 and maximum recombination frequency of 4.0 were used to group the markers. The genetic map distance was calculated using the Kosambi mapping function. Linkage groups were anchored to chromosomes by the physical location of markers. The map was drawn using MapChart 2.30 (Voorrips, 2002).
Identifying and Sequencing the Candidate Gene
Based on the locations of the markers flanking cha on the linkage map, the predicted genes on the mungbean reference sequence (Kang et al., 2014) between the flanking markers were downloaded. Deduced protein sequences for these genes were subjected to a BLASTP search against the NCBI database to obtain information regarding their homologs and functions.
The following two pairs of primers were designed to amplify the genomic region containing a candidate gene responsible for the production of chasmogamous flowers: Seq-1-F (5′-AAGAACGAGGTTTGGCTTCA-3′)/Seq-1-R (5′-AAC TTGACCCACATCAAGGA-3′) and Seq-2-F (5′-GGAAAC ACAAATCACTATGGCA-3′)/Seq-2-R (5′-ATTGCATGTACA TGCCAGCTA-3′). The PCR was conducted using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) following the manufacturer’s instructions with some modifications (i.e., annealing temperature of 58°C and elongation time of 90 s). The PCR products were separated by electrophoresis on 1% agarose gels stained with ethidium bromide. The DNA fragments of the expected size were purified using the E.Z.N.A Gel Extraction Kit (Omega Bio-tek), and sequenced using the 3730xl DNA Analyzer (Applied Biosystems). The deduced Vr06g12650 protein and 11 A. thaliana YUC proteins (i.e., AT4G32540.1, AT4G13260.1, AT1G04610.1, AT5G11320.1, AT5G43890.1, AT5G25620.2, AT2G33230.1, AT4G28720.1, AT1G04180.1, and AT1G48910.1) obtained from The Arabidopsis Information Resource2 database were used to construct a phylogenetic tree using the “One Click” mode and LRT statistical test of Phylogeny.fr (Dereeper et al., 2010).
Results
Morphological Features of the Chasmogamous Mutant Mungbean and Inheritance of the Floral Trait
The floral architecture of CM plants differed from that of Sulu-1 plants. The CM flowers were missing the wing and keel petals (Figure 1). When the CM and Sulu-1 plants were crossed, the F1 hybrids had normal flowers (Figure 1), which suggested a recessive gene (or genes) regulated the production of chasmogamous flowers in CM plants.
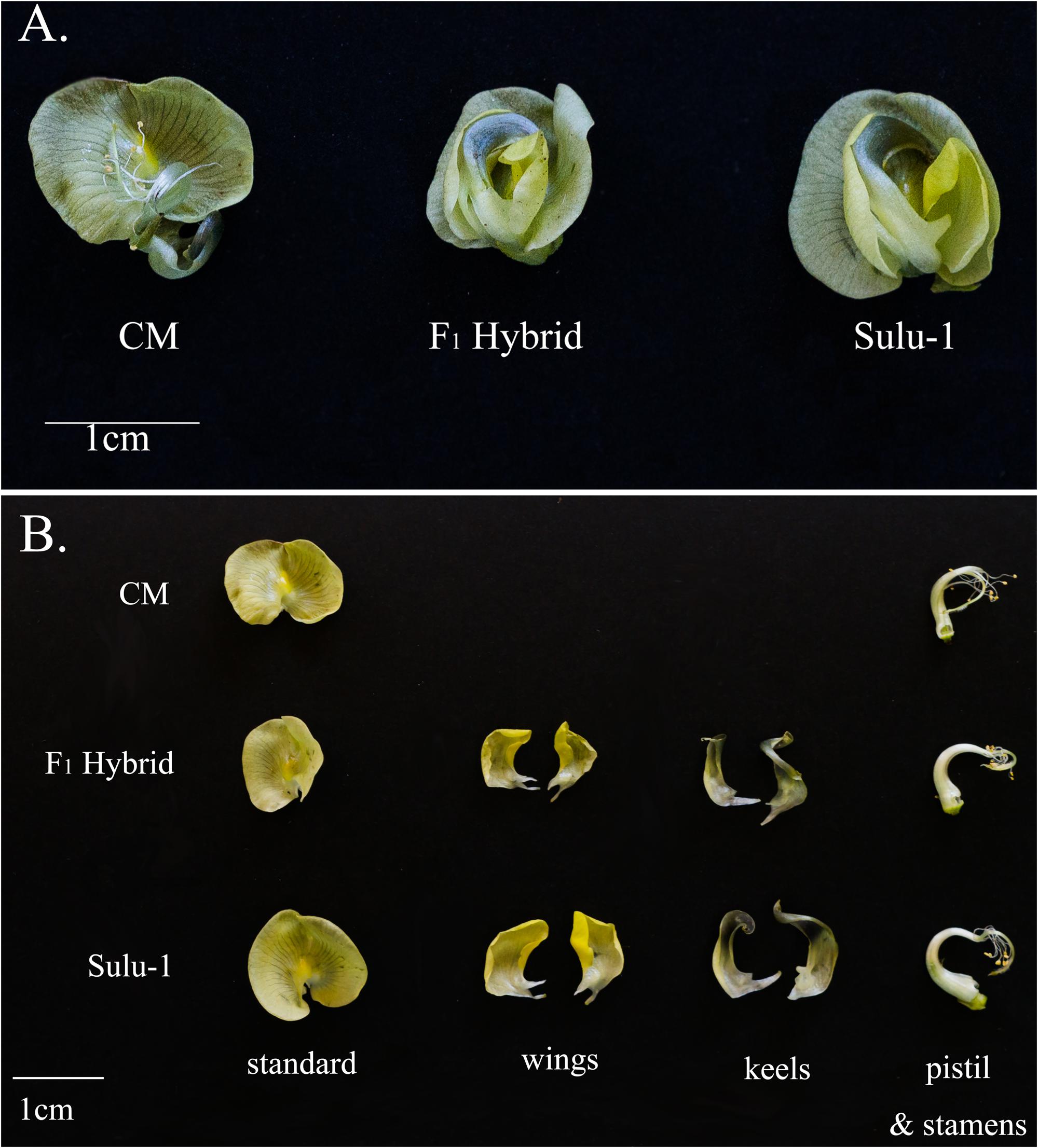
FIGURE 1. Floral morphology of CM and Sulu-1 plants as well as their F1 hybrids. Whole flower (A) and individual flower parts (B).
We determined the segregation ratio of the chasmogamous flower trait in F2 plants. Out of 127 plants, 96 had normal flowers, while 31 had chasmogamous flowers. This segregation corresponded with a 3:1 ratio (χ2 = 0.02, P = 0.88), indicating that a single recessive gene mediated the chasmogamous flower trait in CM plants. This confirmed the results of the study by Sorajjapinun and Srinives (2011).
Development of Indel Markers
To map the gene responsible for the production of chasmogamous flowers, indel markers were developed using the transcriptomes of the Sulu-1 and CM parents. We randomly selected 140 transcript sequences carrying indels to develop markers, and determined that 84 of them (i.e., 57.1%) were polymorphic between the parents (Supplementary Table S1). All of the markers were co-dominant, and able to distinguish between the parents and hybrid progenies. Seventy-four markers were localized to 11 chromosomes by aligning their related transcripts with the whole mungbean genome sequence (Figure 2). Another 10 markers were located on scaffolds that could not be assembled on chromosomes.

FIGURE 2. Physical positions on the mungbean chromosome of the indel markers polymorphic between CM and Sulu-1 plants. Unit: mega base pair (Mbp). Ten markers located on scaffolds are not shown.
Genetic Mapping of the cha Gene
Linkage analysis of the polymorphic indel markers and phenotyping of the flowers were completed using 127 plants from the F2 population. The target cha gene was mapped to chromosome 6 between markers VRID155 and VRID120 at a distance of 1.6 and 3.4 cM, respectively (Figure 3A). To locate cha more precisely, seven polymorphic SSR markers were developed in the target interval by screening SSR motifs in the reference genome sequence (Supplementary Table S2). Eight recombinants were identified in the interval between VRID115 and VRID120. By associating the marker genotypes with floral phenotypes, the 60 and 38 recombinants restricted cha a segment between markers SSR09 and SSR12. These two markers were 277.1 kb apart, and were located at 30.40 and 30.68 Mb of chromosome 6, respectively (Figure 3B). Based on the mungbean whole genome sequence, 12 candidate genes (i.e., Vradi06g12620 to Vradi06g12730; Supplementary Table S3) were detected in this region.
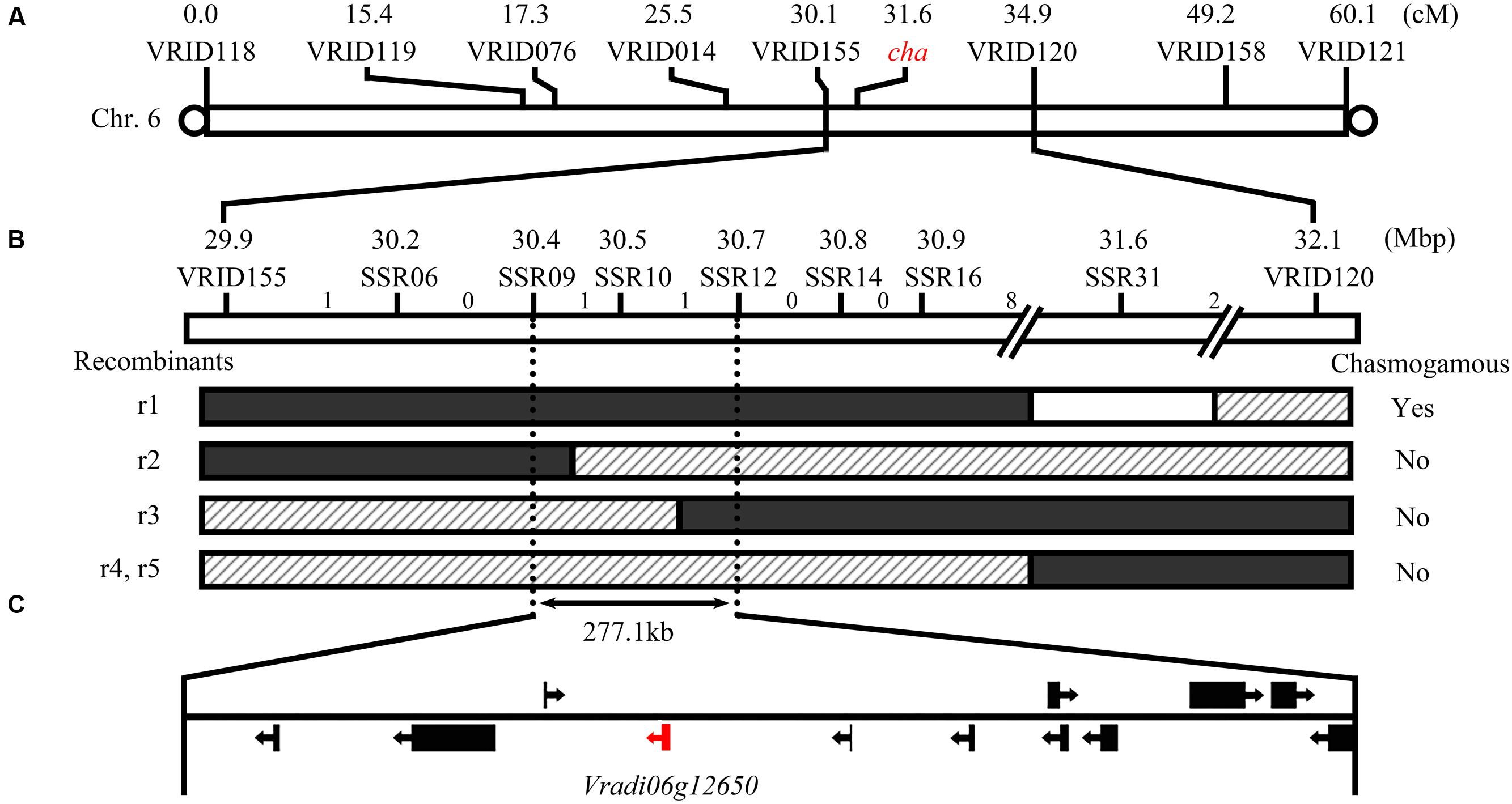
FIGURE 3. Mapping of the cha gene regulating the production of chasmogamous flowers in CM plants. Location of cha (A) and recombinant events on chromosome 6 (B). White, shadow, and black regions in the recombinants indicate homozygous regions for the Sulu-1 allele, and heterozygous and homozygous regions for the CM allele, respectively. The number of recombinant events between adjacent markers are indicated above the bar. Another five recombinants between the homozygous Sulu-1 genotype and heterozygous genotype are not presented because all of the plants had normal flowers. The 12 predicted genes in the candidate gene region are indicated (C).
Function Prediction and Sequencing of cha Candidate Genes
To predict the functions of the 12 detected cha candidate genes, the putative protein sequences encoded by these genes were used as queries to search the NCBI database. Their predicted functions are listed in Table S3. Vradi06g12650, which encoded a YUC homolog (Figure 3) related to the auxin biosynthesis pathway and floral development, was considered the most likely cha gene. A BLASTP search revealed that the YUC protein encoded by Vradi06g12650 was most similar to the A. thaliana proteins AtYUC4 (identity: 68.13%) and AtYUC1 (identity: 62.32%). A protein sequence-based phylogenetic analysis involving Vradi06g12650 and 11 A. thaliana YUC proteins also revealed a close relationship between Vradi06g12650 and AtYUC1/AtYUC4 (Figure 4). We sequenced the Vradi06g12650 coding domain in Sulu-1, CM, and wild-type V1197 mungbean plants. Comparisons among the resulting sequences and the corresponding reference sequence (i.e., from VC1973A) revealed six single nucleotide polymorphisms (SNPs) between the CM and Sulu-1 genes (Figure 5). The SNP at the 382-bp position led to an amino acid substitution from glutamine to glutamic acid, while the other SNPs resulted in non-sense mutations (Figures 5 and 6). The six SNPs were not detected between the CM and V1197 sequences. However, a 1-bp deletion at the 894-bp position was observed in the CM Vradi06g12650 sequence (Figure 5). This deletion produced a frame-shift mutation in the 3′-terminus, resulting in a shorter CM Vradi06g12650 protein with a different C-terminus compared with the corresponding wild-type V1197, Sulu-1, and VC1973A proteins (Figure 6). Additionally, this deletion was detected in a sequence (i.e., Unigene0038420) from the CM transcriptome (Supplementary Data 1). These results suggested that the 1-bp deletion in Vradi06g12650 is responsible for the production of chasmogamous flowers in CM plants.
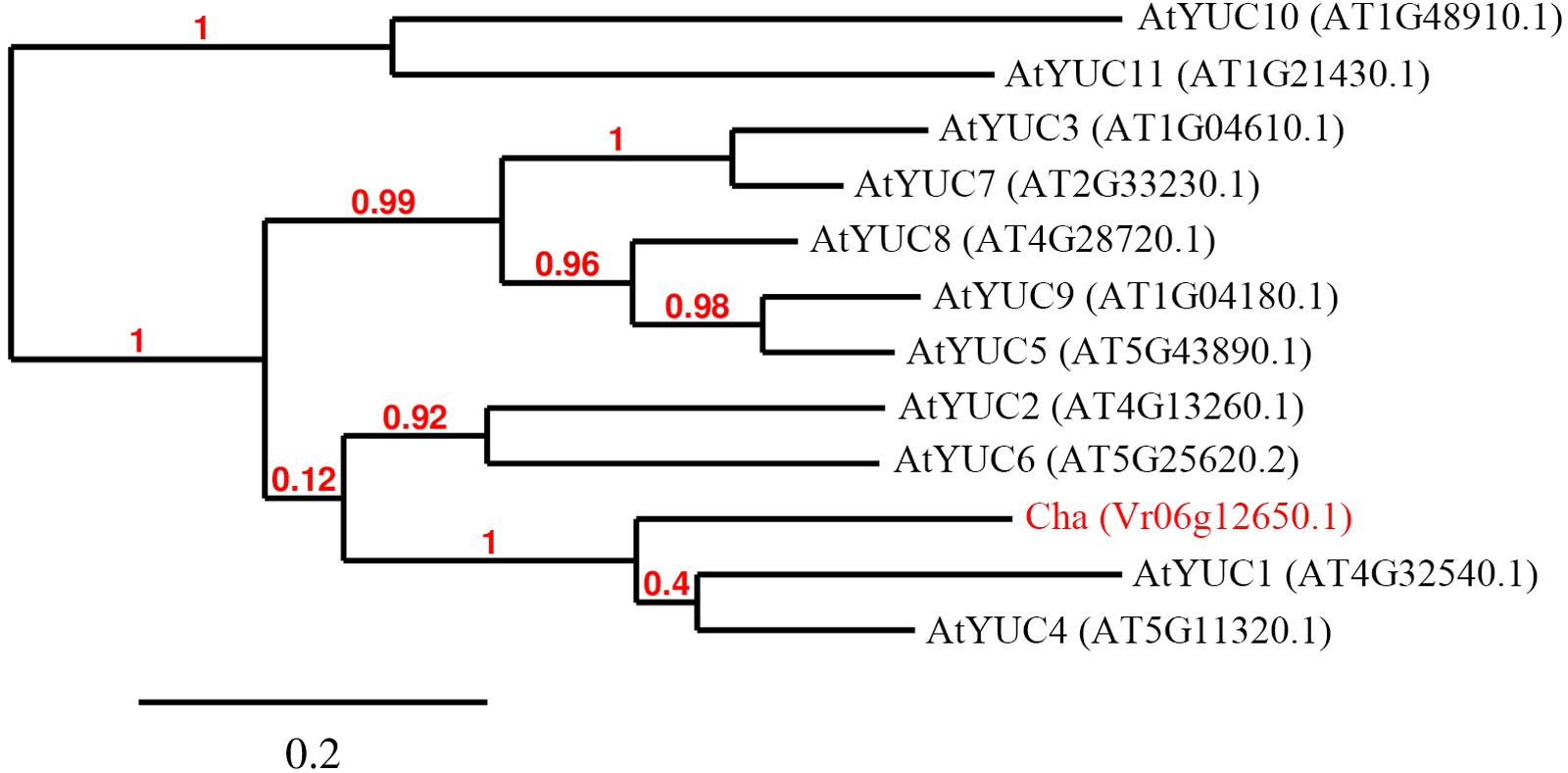
FIGURE 4. Phylogenetic tree of Arabidopsis thaliana YUCCA and Cha proteins. Sequences of 11 A. thaliana YUCCA proteins and the deduced mungbean Vr06g12650.1 protein were used to generate the phylogenetic tree.
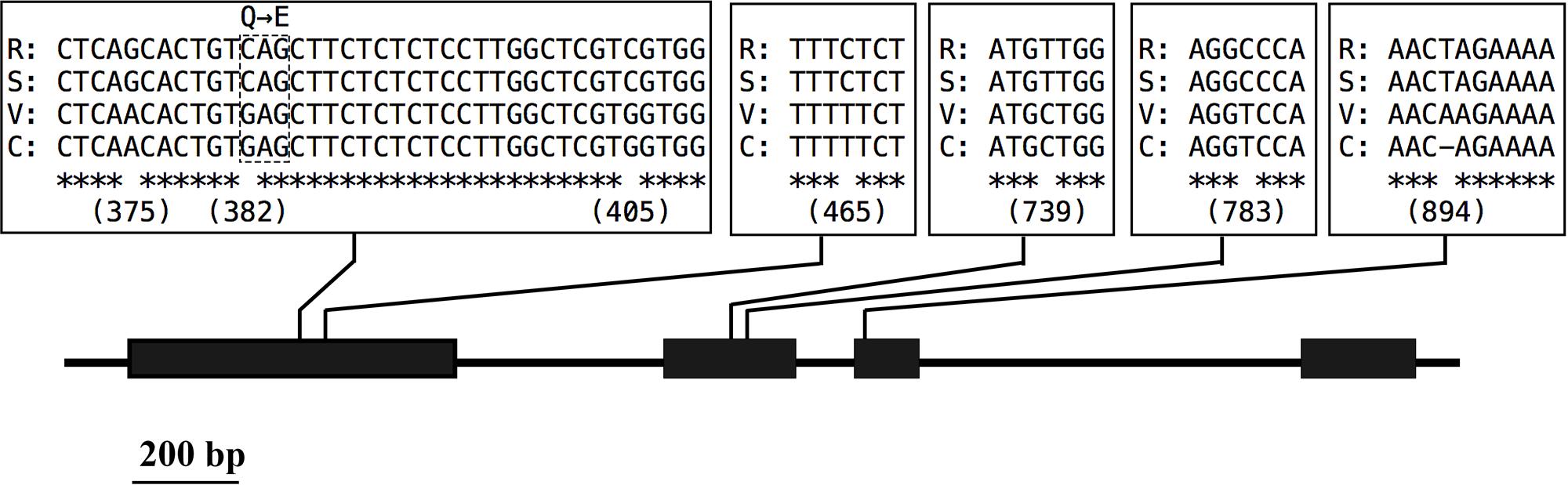
FIGURE 5. Variability in the Vradi06g12650 open reading frame. Consensus sequences are marked by asterisks. Numbers under variant loci indicate the position (in bp) on the open reading frame. The R, S, V, and C correspond to V1973A (reference sequence), Sulu-1, V1197, and CM, respectively.

FIGURE 6. Alignment of the deduced Cha protein sequences from Sulu-1, V1197, and CM plants by ClustalX. Conserved flavin monooxygenase motifs are indicated (on top). Asterisk indicates conserved amino acid.
Discussion
Advantages of Developing Markers from Transcriptomes and the Utility of cha for Mungbean Improvement
Mungbean is not a model plant. Therefore, its molecular genetics and genome have not been as comprehensively studied as in many other crops. The available molecular markers are insufficient for genetics research and mungbean breeding programs. Additionally, some reports have indicated that the polymorphic information content of mungbean SSR markers is low (Tangphatsornruang et al., 2009; Chen et al., 2015). Thus, it is important to develop more mungbean genomic resources. In this study, we developed indel markers by comparing the transcriptomes of two mungbean accessions, which resulted in the detection of several polymorphic markers between the parents of the mapping population. The differences in indel sizes between Sulu-1 and CM plants enabled the indels to be distinguished by PCR followed by 3% agarose gel electrophoresis analysis (Supplementary Figure S1), which is easier to complete than polyacrylamide gel electrophoresis. This resulted in a more efficient genetic mapping procedure.
With the newly developed markers, the cha gene responsible for the production of chasmogamous flowers in mungbean was mapped to chromosome 6. Several newly developed markers were identified closely linked to the target gene. Our data revealed that the marker SSR10 was completely linked to cha. However, this finding is based on a relatively small population of 127 F2 plants. Because the CM floral phenotype was observed to be regulated by a single recessive gene, these linked markers may be useful for the marker-assisted selection of mungbean plants producing chasmogamous flowers.
Functions of the cha Candidate Gene
Vradi06g12650 is the most likely cha gene. This gene putatively encodes a YUC homolog involved in auxin biosynthesis and floral development (Zhao et al., 2001; Cheng et al., 2006). The YUC proteins constitute a family of FMOs containing several conserved sequence motifs, including the FAD-binding motif, FMO-identifying motif, NADPH-binding motif, and F/LATGY motif (Schlaich, 2007). The protein encoded by Vradi06g12650 is highly similar to A. thaliana YUC4 (Supplementary Figure S2). The CM Vradi06g12650 coding sequence differed from that of the wild-type V1197 by a 1-bp deletion at the 894-bp position. This deletion was only detected in CM plants, and results in a frame-shift of the coding sequence leading to the absence of the LATGY motif in the C-terminus of the predicted YUC protein (Figure 6). There are 11 A. thaliana genes encoding YUC proteins, suggesting there may be some functional redundancy among these proteins. Mutational inactivation of a single YUC family gene in A. thaliana caused no obvious developmental defects, while a double yuc1 yuc4 mutant and a quadruple yuc1 yuc4 yuc10 yuc11 mutant exhibited severe defects in the formation of floral organs (Zhao et al., 2001; Cheng et al., 2006). These findings differed from our observation that a defect in a single YUC4-like gene causes a dramatic morphological abnormality in mungbean floral organs. However, it is worth noting that the mutation detected in our study produced an immature YUC4-like protein lacking the F/LATGY motif, which is highly conserved among YUC proteins (Supplementary Figure S2). Therefore, this defective YUC4-like protein may be responsible for the abnormal floral development in CM plants. Additional studies are required to characterize how cha affects the production of chasmogamous flowers in mungbean.
Author Contributions
JC designed the InDel and SSR markers and prepared the manuscript. PS performed gene mapping and reviewed the manuscript. XChen involved in bioinformatics analysis and reviewed the manuscript. XCui sequenced the candidate gene. XY conducted the hybrids and developed the populations. PS designed the study and refined the manuscript.
Funding
This study was financially supported by grants from the National Natural Science Foundation of P. R. China (Grant no. 31271786) and Jiangsu Planned Projects for Postdoctoral Research Funds (Grant no. 1302034B).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the Center for Agricultural Biotechnology, Kasetsart University, Joint Legume Research Center Kasetsart University and Jiangsu Academy of Agricultural Sciences, and Institute of Vegetable Crops, Jiangsu Academy of Agricultural Sciences/Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, for molecular lab facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00830
FIGURE S1 | Polymorphic InDel markers on 3% agarose gel. Polymorphisms of 48 markers with large variance can be detected by 3% agarose gel. For each marker, bands from left to right are the PCR product from Sulu-1, CM and an F1 hybrid.
FIGURE S2 | Protein sequence alignment of arabidopsis YUC1, YUC4 and Cha candidate. Sequences of AtYUC1 (AT4G32540.1) and AtYUC4 (AT5G11320.1) were downloaded from NCBI. Cha is the deduced protein of mungbean Vr06g12650.1. Alignment was performed by ClustalX, consensus sequence was marked by asterisks. Conserved motifs of FMOs were mark on the top of sequences.
Footnotes
References
Alam, A. K. M. M., Somta, P., and Srinives, P. (2014). Identification and confirmation of quantitative trait loci controlling resistance to mungbean yellow mosaic disease in mungbean [Vigna radiata (L.) Wilczek]. Mol. Breed. 34, 1497–1506. doi: 10.1007/s11032-014-0133-0
Alam, A. M., Somta, P., Muktadir, M., and Srinives, P. (2014). Quantitative trait loci associated with seed weight in mungbean (Vigna radiata (L.) Wilczek). Kasetsart J. (Nat. Sci.) 48, 197–204.
Aloni, R., Aloni, E., Langhans, M., and Ullrich, C. I. (2006). Role of auxin in regulating Arabidopsis flower development. Planta 223, 315–328. doi: 10.1007/s00425-005-0088-9
Bennett, S. R., Alvarez, J., Bossinger, G., and Smyth, D. R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. doi: 10.1046/j.1365-313X.1995.8040505.x
Chankaew, A., Somta, P., Isemura, T., Tomooka, N., Kaga, A., Vaughan, D. A., et al. (2013). QTL mapping reveals conservation of major and minor loci for powdery mildew resistance in four sources of resistance in mungbean [Vigna radiata (L.) Wilczek]. Mol. Breed. 32, 121–130. doi: 10.1007/s11032-013-9856-6
Chankaew, S., Somta, P., Sorajjapinun, W., and Srinives, P. (2011). Quantitative trait loci mapping of Cercospora leaf spot resistance in mungbean, Vigna radiata (L.) Wilczek. Mol. Breed. 28, 255–264.
Chen, H., Qiao, L., Wang, L., Wang, S., Blair, M. W., and Cheng, X. (2015). Assessment of genetic diversity and population structure of mung bean (Vigna radiata) germplasm using EST-based and genomic SSR markers. Gene 566, 175–183. doi: 10.1016/j.gene.2015.04.043
Chen, X., Sorajjapinun, W., Reiwthongchum, S., and Srinives, P. (2003). Identification of parental mungbean lines for production of hybrid varieties. CMU J. 2, 97–106.
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Cheng, Y., and Zhao, Y. (2007). A role for auxin in flower development. J. Integr. Plant Biol. 49, 99–104. doi: 10.1111/j.1744-7909.2006.00412.x
Cherian, C. A., Mallikarjuna, N., Jadhav, D., and Saxena, K. (2006). Open flower segregants selected from Cajanus platycarpus crosses. J. SAT Agric. Res. 2, 1–2.
Chotechung, S., Somta, P., Chen, J., Yimram, T., Chen, X., and Srinives, P. (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. doi: 10.1007/s00122-016-2731-1 [Epub ahead of print].
Dahiya, B., Lather, V., Solanki, I., and Kumar, R. (1984). Genetic resources: useful spontaneous mutants in chickpea. Int. Chick. News. 11, 5–7.
Dereeper, A., Audic, S., Claverie, J. M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dheer, M., Punia, S. S., Ram, B., Jain, N. K., Koli, N. R., Bhatnagar, V., et al. (2014). Morphological features of an open flower mutant plant and characterization of their progenies in lentil (Lens culinaris Medik.). Genet. Resour. Crop Evol. 61, 879–886. doi: 10.1007/s10722-014-0100-y
Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., et al. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. doi: 10.1126/science.282.5397.2226
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gupta, S. K., Bansal, R., and Gopalakrishna, T. (2014). Development and characterizatiom of genic SSR markers for mungbean (Vigna radiata (L.) Wilczek). Euphytica 195, 245–258.
Gwag, J. G., Chung, J. W., Chung, H. K., Lee, J. H., Ma, K. H., Dixit, A., et al. (2007). Characterization of new microsatellite markers in mung bean, Vigna radiata (L.). Mol. Ecol. Resour. 6, 1132–1134. doi: 10.1007/s11033-014-3436-7
Isemura, I., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7:e41304. doi: 10.1371/journal.pone.0041304
Kajonphol, T., Sangsiri, C., Somta, P., Toojinda, T., and Srinives, P. (2012). SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). SABRAO J. Breed. Genet. 44, 71–86.
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5, 5543. doi: 10.1038/ncomms6443
Kasettranan, W., Somta, P., and Srinives, P. (2010). Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean (Vigna radiata (L.) Wilczek). J. Crop Sci. Biotechnol. 13, 155–161. doi: 10.1007/s12892-010-0052-z
Kato, H., and Namai, H. (1987). Floral characteristics and environmental factors for increasing natural outcrossing rate for F1 hybrid seed production of rice Oryza sativa L. Jpn. J. Breed. 37, 318–330. doi: 10.1270/jsbbs1951.37.318
Kitsanachandee, R., Somta, P., Chatchawankanphanich, O., Akhtar, K. P., Shah, T. M., Nair, R. M., et al. (2013). Detection of quantitative trait loci for mungbean yellow mosaic India virus (MYMIV) resistance in mungbean (Vigna radiata (L.) Wilczek) in India and Pakistan. Breed. Sci. 63, 367–373. doi: 10.1270/jsbbs.63.367
Krizek, B. A., and Fletcher, J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Li, Q., and Wan, J.-M. (2005). SSRHunter: development of a local searching software for SSR sites. Hereditas 27, 808–810.
Lodhi, M. A., Ye, G.-N., Weeden, N. F., and Reisch, B. I. (1994). A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 12, 6–13. doi: 10.1007/BF02668658
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684. doi: 10.1105/tpc.3.7.677
Prathet, P., Somta, P., and Srinives, P. (2012). Mapping QTL conferring resistance to iron deficiency chlorosis in mungbean [Vigna radiata (L.) Wilczek]. Field Crops Res. 137, 230–236. doi: 10.1016/j.fcr.2012.08.002
Przemeck, G. K., Mattsson, J., Hardtke, C. S., Sung, Z. R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. doi: 10.1007/BF00208313
Pundir, R., and Reddy, G. (1998). Two new traits-open flower and small leaf in chickpea (Cicer arietinum L.). Euphytica 102, 357–361. doi: 10.1023/A:1018307709788
Sangiri, C., Kaga, A., Tomooka, N., Vaughan, D., and Srinives, P. (2007). Genetic diversity of the mungbean (Vigna radiata, Leguminosae) genepool on the basis of microsatellite analysis. Aust. J. Bot. 55, 837–847. doi: 10.1071/Bt07105
Schlaich, N. L. (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12, 412–418. doi: 10.1016/j.tplants.2007.08.009
Soehendi, R., and Srinives, P. (2005). Significance of heterosis and heterobeltiosis in F1 hybrid of mungbean (Vigna radiata (L.) Wilczek) for hybrid seed production. SABRAO J. Breed. Genet. 37, 97–105.
Sompong, U., Somta, P., Raboy, V., and Srinives, P. (2012). Mapping of quantitative trait loci for phytic acid and phosphorus contents in seed and seedling of mungbean (Vigna radiata (L.) Wilczek). Breed. Sci. 62, 87–92. doi: 10.1270/jsbbs.62.87
Somta, P., Musch, W., Kongsamai, B., Chanprame, S., Nakasathien, S., Toojinda, T., et al. (2008). New microsatellite markers isolated from mungbean (Vigna radiata (L.) Wilczek). Mol. Ecol. Resour. 8, 1155–1157. doi: 10.1111/j.1755-0998.2008.02219.x
Somta, P., Seehalak, W., and Srinives, P. (2009). Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv. Genet. 10, 1939–1943. doi: 10.1007/s10592-009-9860-x
Sorajjapinun, W., Chankaew, S., and Srinives, P. (2012). Comparison of hybrid vigor based on parental distance in SSR markers and agronomic traits in mungbean (Vigna radiata (L.) Wilczek). Sonklanakarin J. Sci. Technol. 34, 143–151.
Sorajjapinun, W., and Srinives, P. (2011). Chasmogamous mutant, a novel character enabling commercial hybrid seed production in mungbean. Euphytica 181, 217–222. doi: 10.1007/s10681-011-0408-z
Srinivasan, S., and Gaur, P. M. (2012). Genetics and characterization of an open flower mutant in chickpea. J. Hered. 103, 297–302. doi: 10.1093/jhered/esr125
Tangphatsornruang, S., Somta, P., Uthaipaisanwong, P., Chanprasert, J., Sangsrakru, D., Seehalak, W., et al. (2009). Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Biol. 9:137. doi: 10.1186/1471-2229-9-137
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Verdcourt, B. (1970). Studies in the Leguminosae-Papilionoïdeae for the ‘Flora of Tropical East Africa’: IV. Kew Bull. 24, 507–569. doi: 10.2307/4103051
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Yashpal, Singh, M. N., Pathak, N., and Saroj, S. K. (2015). Combining ability, heterosis and inbreeding depression in inter specific hybrids involving greengram [Vigna radiata (L.) Wilczek] and blackgram [Vigna mungo (L.) Hepper]. Electron. J. Plant Breed. 6, 87–92.
Keywords: mungbean, Vigna radiata, chasmogamy, outcrossing, gene mapping, hybrid development
Citation: Chen J, Somta P, Chen X, Cui X, Yuan X and Srinives P (2016) Gene Mapping of a Mutant Mungbean (Vigna radiata L.) Using New Molecular Markers Suggests a Gene Encoding a YUC4-like Protein Regulates the Chasmogamous Flower Trait. Front. Plant Sci. 7:830. doi: 10.3389/fpls.2016.00830
Received: 12 March 2016; Accepted: 26 May 2016;
Published: 10 June 2016.
Edited by:
Nicolas Rispail, Institute for Sustainable Agriculture-CSIC, SpainReviewed by:
Vikas Kumar Singh, International Crops Research Institute for the Semi Arid Tropics, IndiaRupesh Kailasrao Deshmukh, Laval University, Canada
Copyright © 2016 Chen, Somta, Chen, Cui, Yuan and Srinives. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peerasak Srinives, YWdycHNzQHlhaG9vLmNvbQ==; Xin Chen, Y3hAamFhcy5hYy5jbg==
†These authors have contributed equally to this work.
 Jingbin Chen
Jingbin Chen Prakit Somta
Prakit Somta Xin Chen1*
Xin Chen1* Peerasak Srinives
Peerasak Srinives