- Key Laboratory of Plant Pathology of Hubei Province, College of Plant Science & Technology, Huazhong Agricultural University, Wuhan, China
Secreted effectors in plant root-knot nematodes (RKNs, or Meloidogyne spp.) play key roles in their parasite processes. Currently identified effectors mainly focus on the early stage of the nematode parasitism. There are only a few reports describing effectors that function in the latter stage. In this study, we identified a potential RKN effector gene, Misp12, that functioned during the latter stage of parasitism. Misp12 was unique in the Meloidogyne spp., and highly conserved in Meloidogyne incognita. It encoded a secretory protein that specifically expressed in the dorsal esophageal gland, and highly up-regulated during the female stages. Transient expression of Misp12-GUS-GFP in onion epidermal cell showed that Misp12 was localized in cytoplast. In addition, in planta RNA interference targeting Misp12 suppressed the expression of Misp12 in nematodes and attenuated parasitic ability of M. incognita. Furthermore, up-regulation of jasmonic acid (JA) and salicylic acid (SA) pathway defense-related genes in the virus-induced silencing of Misp12 plants, and down-regulation of SA pathway defense-related genes in Misp12-expressing plants indicated the gene might be associated with the suppression of the plant defense response. These results demonstrated that the novel nematode effector Misp12 played a critical role at latter parasitism of M. incognita.
Introduction
Root-knot nematodes (RKNs, or Meloidogyne spp.) are a large kind of plant-parasite nematodes. It is thought that they could infest more than 5,000 plant species (Blok et al., 2008) and are responsible for substantial economic crop loss (Elling, 2013). As a major specie of RKNs, Meloidogyne incognita has a remarkably wide host range, including some principal crops, and causes more economical loss than other species (Trudgill and Blok, 2001). Elucidating the mechanisms of RKNs parasitism would facilitate the development of new control strategies for nematode diseases.
As sedentary parasites, RKNs usually establish a feeding site composed of giant cells (GCs) to obtain nutrients from plants (Bird et al., 2009). During the process of nematodes infection, proteins secreted through the stylet of RKNs induce vascular cells nuclear division without cytokinesis and ultimately transform four to eight root cells into the GCs (Jaubert et al., 2004; Huang et al., 2006; Caillaud et al., 2008). Although the molecular pathogenic mechanisms of RKNs are still largely uncharacterized, it is believed that proteins synthesized in oesophageal glands and injected through the stylet into plant tissue play key roles in the parasitism of nematodes (Hewezi and Baum, 2013). These secretory proteins function as effectors in GCs formation and maintenance to support the nematode parasitism (Davis et al., 2000; Quentin et al., 2013). Some cell wall-degrading and cell wall-modifying enzymes, such as β-1,4-endoglucanase, β-1,4-endoxylanase, pectate lyase, and cellulose-binding protein, have been characterized as potential effectors involved in the invasion of root tissues by preparasitic juveniles and the migration of nematodes (Ding et al., 1998; Rosso et al., 1999; Popeijus et al., 2000; Dautova et al., 2001; Doyle and Lambert, 2002; Huang et al., 2005a; Ledger et al., 2006). Some effectors including chorismate mutase, venom allergen-like protein, and glutathione S-transferase, are involved in the suppression of defense reactions of the host cell during the infection stages (Lambert et al., 1999; Ding et al., 2000; Huang et al., 2005b; Long et al., 2006; Dubreuil et al., 2007; Wang et al., 2007). Some effectors, such as MiMsp40 and MeTCTP, can suppress programmed cell death (PCD) in host plants to promotes parasitism (Niu et al., 2016; Zhuo et al., 2016). Notably, recently identified effectors function in disturbing the cells metabolism. The M. incognita effector Mi8D05 was proved to play an important role in the regulation of solute and water transport within GCs (Xue et al., 2013), and M. javanica effector MjTTL5 could activate the host reactive oxygen species-scavenging system(Lin et al., 2016). Additionally, some effectors, including MiEFF1, MiCRT, MjNULG1, and 7H08, were found to target the host plant cell nuclei, manipulate the host cell processes and exhibit the transcriptional activation activity (Jaouannet et al., 2012, 2013; Lin et al., 2013; Zhang et al., 2015a).
To identify more novel effectors, transcriptomic approaches were used to analyze the secreted genes from the microaspiration of esophageal gland cells (Gao et al., 2003; Huang et al., 2003). Since the genome of M. incognita and M. hapla were sequenced (Abad et al., 2008; Opperman et al., 2008), the increasing genomic data has provided a convenient way to identify RKNs effector proteins and other essential genes.
The studies focusing on the effectors not only promotes an understanding of nematode-host interaction mechanisms, but also screens new target genes that could be applied for nematocides and breeding of nematode resistant plants to control nematode diseases. The RNA interference of the effector gene Mc16D10L confers resistance against M. chitwood in Arabidopsis and potato plants (Dinh et al., 2014), and the suppression of NGB and NAB/ERabp1 in tomatoes resulted in the reduction in the number of Globodera rostochiensis (Dąbrowska-Bronk et al., 2014).
In this study, combined with the genomic sequences, proteins, an EST library of M. incognita and bioinformatics tools, a potential effector Misp12 (M. incognita putative esophageal gland cell secretory protein 12) was selected. After BLAST against NCBI database, we found that this gene was also pointed out via transcriptomic approaches and was named Msp12 (Huang et al., 2003). We further analyzed the developmental expression profiles and investigated subcellular location in planta of Misp12. In addition, the VIGS (virus induced gene silencing) approaches and the transiently expression of Misp12 in plants were also carried out to examine the function of Misp12 during M. incognita parasitism.
Materials and Methods
Nematodes and Plants
Meloidogyne incognita were collected and identified from 8 different areas of P. R. China and reared on tomato plants in greenhouses at 25°C. Pre-parasitic second-juveniles (J2s) and parasitic stages were collected as described previously (Huang et al., 2005a). Lycopersicon esculentum and Nicotiana benthamiana plants routinely grow in pots at 25°C in the greenhouse.
Nucleic Acid Extraction and RT-PCR Analysis
Genomic DNA was extracted from M. incognita eggs and pre-parasitic J2s by using the cetyltrimethylammonium bromide (CTAB) method, as described by Goetz et al. (2001). Total RNA of nematodes was isolated using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Dalian, China) after grinding nematodes in a 1.5 mL sterile tube with liquid nitrogen and then treated by DNase I (Thermo Scientific, Shanghai, China) at 37°C for 30 min to remove genomic DNA. First-strand cDNA was synthesized using the SuperScript® III Reverse Transcriptase kit (Invitrogen, Shanghai, China). Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed on CFX96TM Real TimeSystem (BIO-RAD, USA) with the following conditions: 95°C for 30 s and 40 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s, using the SYBR Green PCR Master Mix (TaKaRa, Dalian, China). Quantification of the relative changes in gene expression was performed using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The experiments were repeated three times, with three technical replicates for each reaction.
Sequence Analysis and Gene Cloning
The whole genome and protein sequences of M. incognita were downloaded from the M. incognita resources database1. The EST library of M. incognita was obtained from NCBI. SignalP 3.0 (Bendtsen et al., 2004), TMHMM 2.0 (Krogh et al., 2001), and MERCI (Vens et al., 2011) were used to predict potential effector proteins.
The 3′-RACE-Ready cDNA was synthesized from 1 μg of total RNA using the BD SMART RACE cDNA amplification kit (TaKaRa, Dalian, China). Based on the predicted sequence of Misp12 described above, the primers RACE12S1 and RACE12S2 were designed. The 3′-terminal sequence was amplified by PCR using RACE12S1 and the 3′-anchor UPM primes, followed by a second-round PCR with RACE12S2 and NUP primers using the first-round PCR products as template. PCR was performed following the BD SMART RACE cDNA amplification kit user manual.
To confirm the predicted sequence, primers covering the whole sequence, Misp12QS and Misp12QA were designed to perform PCR from cDNA and DNA templates. For PCR amplification, 0.1 μg of cDNA or DNA template was used in a 50 μl reaction mixture consisting of 1 × PCR buffer for Phusion High-Fidelity DNA Polymerase, 0.2 mM of each dNTP, 1.5 mM MgSO4, 0.3 μM primers, and 2 units of Phusion High-Fidelity DNA Polymerasee (Thermo Scientific, Shanghai, China). PCR conditions were as follows: predenaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 45 s, annealing at 57°C for 45 s, and polymerization at 72°C for 90 s; with a final incubation step at 16°C. All primers used in this study were synthesized by Invitrogen Biotechnology Co. Ltd. (Shanghai, China) and are listed in Supplementary Table S1.
Sequence Comparisons and Secondary Structure Predictions
The signal peptide and its cleavage site were predicted by the SignalP 3.0 (Bendtsen et al., 2004) and the TargetP 1.0 (Emanuelsson et al., 2000). Calculation of the predicted Misp12 molecular weight and isoelectric point were performed using the ProtParam program (Gasteiger et al., 2005). Secondary structure prediction of the protein sequence was performed using the PHD program (Rost and Sander, 1993) and the motifs were predicted by Motif Scan (Pagni et al., 2007). The ClustalW program (Thompson et al., 1994) was used to generate an alignment of M. incognita nematodes in eight different areas of China. The HHPred program (Remmert et al., 2011) was used to predict conserved domains.
Developmental Expression Analyses
Total RNA samples were prepared from 200 M. incognita nematodes at different life stages by the method described above. The expression level of Misp12 was analyzed via qRT-PCR with the primes qPCRmi12S/qPCRmi12A. Total cDNA abundance in the samples was normalized using the Actin as controls, which were amplified by primes QactinS/QactinA. These experiments were repeated three times, with three technical replicates for each reaction.
In Situ mRNA Hybridization
In situ hybridization was performed as previously described with a slight modification (de Boer et al., 1998). The hybridization temperature was adjusted to 48°C. Primers Misp12HS and Misp12HA (Supplementary Table S1) were employed to synthesize digoxigenin (DIG)-labeled sense and antisense cDNA probes (Roche, USA) by asymmetric PCR. The sense cDNA probes serve as control (Huang et al., 2003).
Subcellular Localization of Misp12
The open reading frame (ORF) of Misp12, with and without a signal peptide sequence, was cloned with primes 12S/12BamHI-A and 12nospS/12nospBamHI-A, respectively (Supplementary Table S1). The GUS gene was cloned with primer pairs GusBamHI-S/GusA. After BamHI digestion, the Misp12 and the GUS gene were ligated together. Then, the Misp12:GUS and ΔspMisp12:GUS were cloned with primer pairs12EcoRIS/GusHindIIIA. The sequenced PCR products were digested with EcoRI and HindIII and then ligated into EcoRI/HindIII-digested vector pEGAD (Cutler et al., 2000) to generate the vectors 35S:Misp12:GUS:eGFP and 35S:ΔspMisp12:GUS:eGFP. The recombinant vectors were transferred into the Agrobacterium tumefaciens GV3101 using standard cloning techniques (Sambrook and MacCallum, 2012). Healthy and fresh onion scales (1–1.5 cm × 1 cm) were placed on a 9 cm plate and their inner surfaces were immersed into 6 mL resuspension of A. tumefaciens solution (OD600 = 1–1.5) consisting of 5% (g/v) sucrose, 100 mg L-1 acetosyringone and 0.02% (v/v) Silwet-77 for 6–12 h at 28°C. Then, the onion scales were transferred to a petri dish containing 25 mL 1/2 MS (Murashige and Skoog salts, 30 g L-1 sucrose and 0.7% (g/v) agar, pH 5.7) for 1–2 days. The subcellular localization of the fused proteins was visualized using fluorescence microscopy (Nikon 80i, Nikon, Japan) at an excitation wavelength of 488 nm.
In Planta RNAi
The tobacco rattle virus (TRV)-based vectors pTRV1 and pTRV2 were used for gene expression in N. benthamiana as previously described (Liu et al., 2002). The unique 249 bp fragment of the gene Misp12 was amplified with the primer pairs U12RNAiS/U12RNAiA. The PCR product was digested with XbaI and SacI and then inserted into pTRV2 to generate vector pTRV2::Misp12. The vectors pTRV1, pTRV2::00 (negative control), and pTRV2::Misp12 were transferred into the Agrobacterium tumefaciens GV3101, respectively. A. tumefaciens cultures containing pTRV1 and pTRV2::Misp12 were injected into Nicotiana benthamiana as the previously described (Xiao et al., 2014), while A. tumefaciens cultures containing pTRV1 and pTRV2::00 were used as controls. Five days after inoculation (DAI), primer pairs TRVcpF/TRVcpR (Anand et al., 2007) were used to test the TRV coat protein transcription by qRT-PCR to check whether TRV invasion was successful.
To detect the parasitism ability of Misp12-RNAi nematodes, 150 N. benthamiana plants agroinfiltrated with pTRV::Misp12 or pTRV::00 were inoculated with 300 pre-J2 nematodes, respectively. Additionally, 150 untreated plants were used as blank controls. At 5, 10, 15, and 28 DAI, tomato plant roots were stained with byacid fuchsin to count the numbers of parasitic nematodes (Bybd et al., 1983). At 45 DAI, the galls and eggs in tomato plant roots were counted. Ten N. benthamiana plants were checked at each time point. The same experiments were repeated three times. Statistically significant differences between each treatments and the corresponding control were determined by Student’s t-test using SAS version 9.0.
To evaluate the Misp12 expression level in gene-RNAi nematodes, mRNA was extracted from mix-stages nematodes isolated from Misp12-RNAi and control plants at 15 DAI and 28 DAI. Each treatment was sampled three times. The relative expression level of Misp12 was checked with the primer pair qPCRmi12S/qPCRmi12A by qRT-PCR as described above.
Transiently Expressing Misp12 in Plants
The suppression of PCD in N. benthamiana leaves was assessed as previously described (Bos et al., 2006). The ORF of Misp12, with and without a signal peptide sequence, was cloned into the PVX vector pGR107, respectively (Article et al., 2011). The INF1 gene was amplified from Phytophthora infestans isolate 88069 genomic DNA and cloned into pGR107 (Article et al., 2011).
The confirmed constructs were introduced into the A. tumefaciens strain GV3101 by electroporation. The cultured A. tumefaciens cells (OD600 = 0.4) carrying Misp12 and ΔspMisp12 were initially infiltrated into the leaves of N. benthamiana plants, which were grown in the greenhouse for 6 weeks at 25–28°C under 16 h light/8 h dark. The identical infiltration site was then challenged with A. tumefaciens cells carrying the INF1 gene at 24 h after initial inoculation. Simultaneously, the INF1 gene was expressed alone as controls. The plants were monitored for symptoms, images were acquired 2 days after the last infiltration. The experiment was repeated at least three times, and each assay consisted of at least five plants with three leaves inoculated similarly.
Detection of JA and SA Signaling Pathways in Plants
The jasmonic acid (JA) and salicylic acid (SA) signaling pathways genes expression levels were tested in the VIGS of Misp12 plants at 28 DAI and Misp12-expressing plants at 2 DAI compared to their control plants by qRT-RCR, respectively.
The SA signaling pathways molecular markers were the transcript of pathogenesis-related gene PR-1(Accession no. JN247448) and phenylalanine ammonia lyase gene PAL5 (Accession no. EB684217.1). For the activation of JA signaling pathways, transcripts of proteinase inhibitor gene Pin2 (Accession no. EH368183.1), 12-oxophytodienoate reductase gene OPR3 (Accession no. CN745683) and β-thionin gene (Accession no. EH368982.1) were analyzed. For all qRT-PCR analysis, transcripts were normalized against the geometric mean of the expression levels of two N. benthamiana reference genes Actin and GAPDH that were amplified with the two pairs of primes NbactinS/A and NbGAPDHS/A, respectively.
Results
Identification of the Misp12
The whole protein sequences of M. incognita were used the bioinformatics tools SignalP 3.0 and TMHMM 2.0 to predict the secreted proteins. Then, the MERCI tools were used to predict the potential effectors. Misp12 was found to be a candidate secretory protein. After cloning the cDNA and the genome sequence of Misp12 via 3′RACE and genome walking, it revealed that Misp12 was 1202-bp in length, encompassing a 450 bp ORF (GeneBank: KU737535), which encoded a deduced protein of 149 amino acids.
SignalP and TargetP analysis results revealed that a signal peptide sequence existed at the N-terminal region, where a predicted cleavage site was located between amino acid positions A19 and A20 (Figure 1), generating a small mature protein with a theoretical molecular weight of 15.9 kDa and an isoelectric point of 8.81. Furthermore, no transmembrane region was detected by TMHMM2.0 (Krogh et al., 2001).
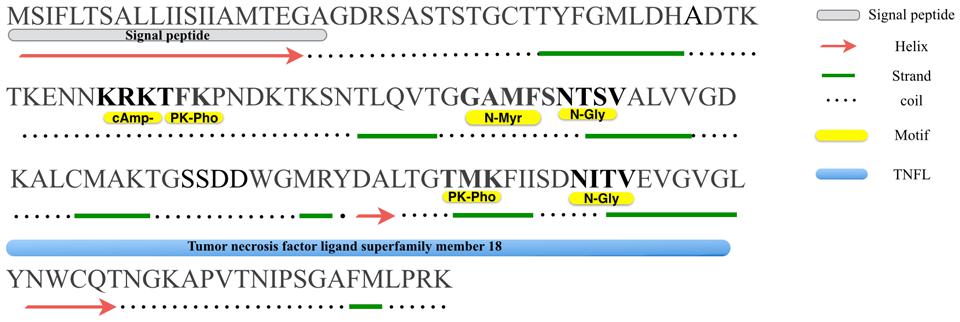
FIGURE 1. Primary and secondary structures analysis of Misp12. A putative hydrophobic leader/signal peptide, as predicted by the SignalP program is shown at the N terminus along with a cleavage site between two alanine residues at position 20–21. Consensus N-linked glycosylation site (N-gly), protein kinase C phophorylation sites (PK-pho), cAMP-and cGMP-dependent glycosylation site (cAmp-) and N-myristoylation site (N-Myr) are indicated.
Based on PHD program analysis, we found that Misp12 consisted of 8 beta-strands and 2 helixes (except the signal peptide) (Figure 1). Motif Scan analysis revealed that two N-linked glycosylation sites presented at residues N74 and N115, two Protein kinase C phosphorylation sites at residues T52 and T107, a cAMP-and cGMP-dependent glycosylation site at residue K49, and an N-myristoylation site at residue G69 (Figure 1). HHPred analysis showed that Misp12 has a region similar to tumor necrosis factor ligand superfamily in the C-terminal between residues K84 and G121 (Figure 1).
In addition, no ortholog of Misp12 was found in other organisms when searching in the NCBI database using BLAST. The ClustalW (Thompson et al., 1994) was used to align 9 Misp12 alignments of M. incognita from eight different areas of China and 1 Misp12 homolog sequence (accession no. AY134431.1) in NCBI, and the result showed that they have 99% identity, with 16 points mutations exist in the gene sequence and 12 of the 16 were in ORF, and only 2 are missense mutations (Supplementary Figure S1).
The Misp12 Gene Was Highly Expressed during the Nematode Mature Stage
To figure out which developmental stage Misp12 mainly takes part in, the expression pattern of the gene was evaluated via qRT-PCR between non-parasitic stages and parasitic stages. The result showed that Misp12 transcript accumulated the most in the mature female stage, which increased 1200-fold when compared to the non-parasitic stages (eggs and pre-parasitic J2s) (Figure 2). In addition, the expression level of Misp12 at parasitic J2 stage and third- and fourth-stage juveniles (J3/J4) stage were also higher than non-parasitic stages (Figure 2). These results suggested that Misp12 was more important at latter parasitism.
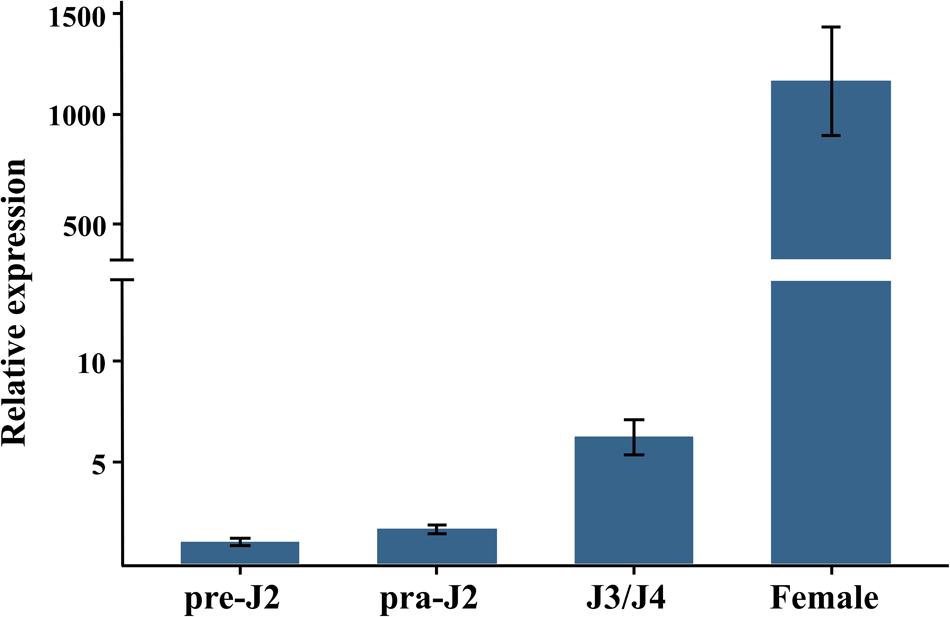
FIGURE 2. Developmental expression pattern of Misp12. The relative expression of Misp12 was quantified using quantitative RT-PCR in four M. incognita life stages, pre-parasitic second-stage juvenile (pre-J2), parasitic second-juveniles (par-J2), third- and fourth-stage juveniles (J3/J4), and mature females. Fold change values were calculated using the 2-ΔΔCT method and normalized to the Meloidogyne incognita Actin gene and relative to expression in pre-J2s. Each column represents the mean of three independent experiments with standard deviation.
Misp12 Was Specifically Expressed in the Dorsal Esophageal Gland
In situ mRNA hybridization was used to determine the tissue localization of Misp12 in the nematode. No hybridization signals were detected in the control treatment when using the DIG-labeled sense cDNA probe (Figure 3A). And the antisense Misp12 cDNA probe hybridized with mRNA within the dorsal esophageal gland cell of parasitic J2 and female (Figures 3B,C). The results indicated that Misp12 was synthesized at dorsal esophageal gland.
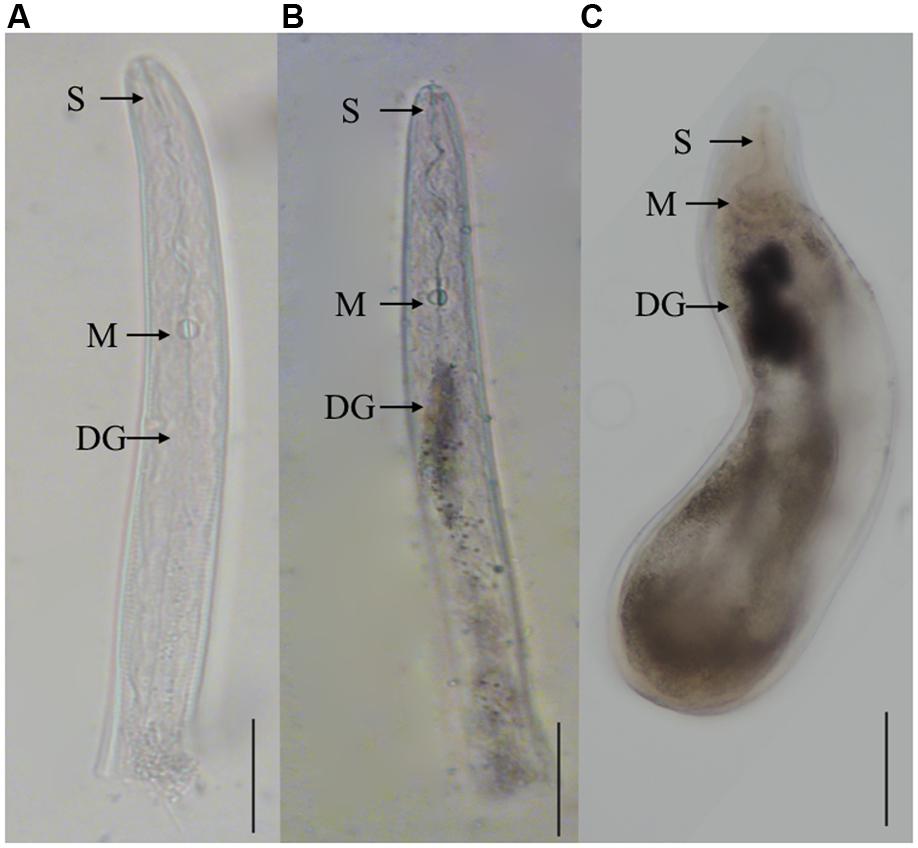
FIGURE 3. In situ hybridization of the Misp12 transcripts in parasitic second-stage juveniles and females. (A) The sense Misp12 DIG-labeled cDNA probes as a negative control in parasitic second-stage juveniles. (B,C) Signal of antisense probes localized within the dorsal esophageal gland (DG) in parasitic second-stage juveniles and females. The DG, metacorpus (M), and stylet (S) are indicated with arrows. Scale bar = 50 μm.
Misp12 was Secreted into the Cytoplasm of Plant Cell
To evaluate the subcellular localization of Misp12 in planta, Misp12 with or without signal peptide gene fragment fused with eGFP and GUS genes was transiently expressed in onion epidermal cells. The result showed that Misp12 without signal peptide was located in the cytoplasm (Figure 4A). However, full-length Misp12 was observed in the apoplast (Figure 4B). Free eGFP was present in the cytoplasm of the cell (Supplementary Figure S2). The results showed that Misp12 was located at the cytoplasm of the plant cell during infection.
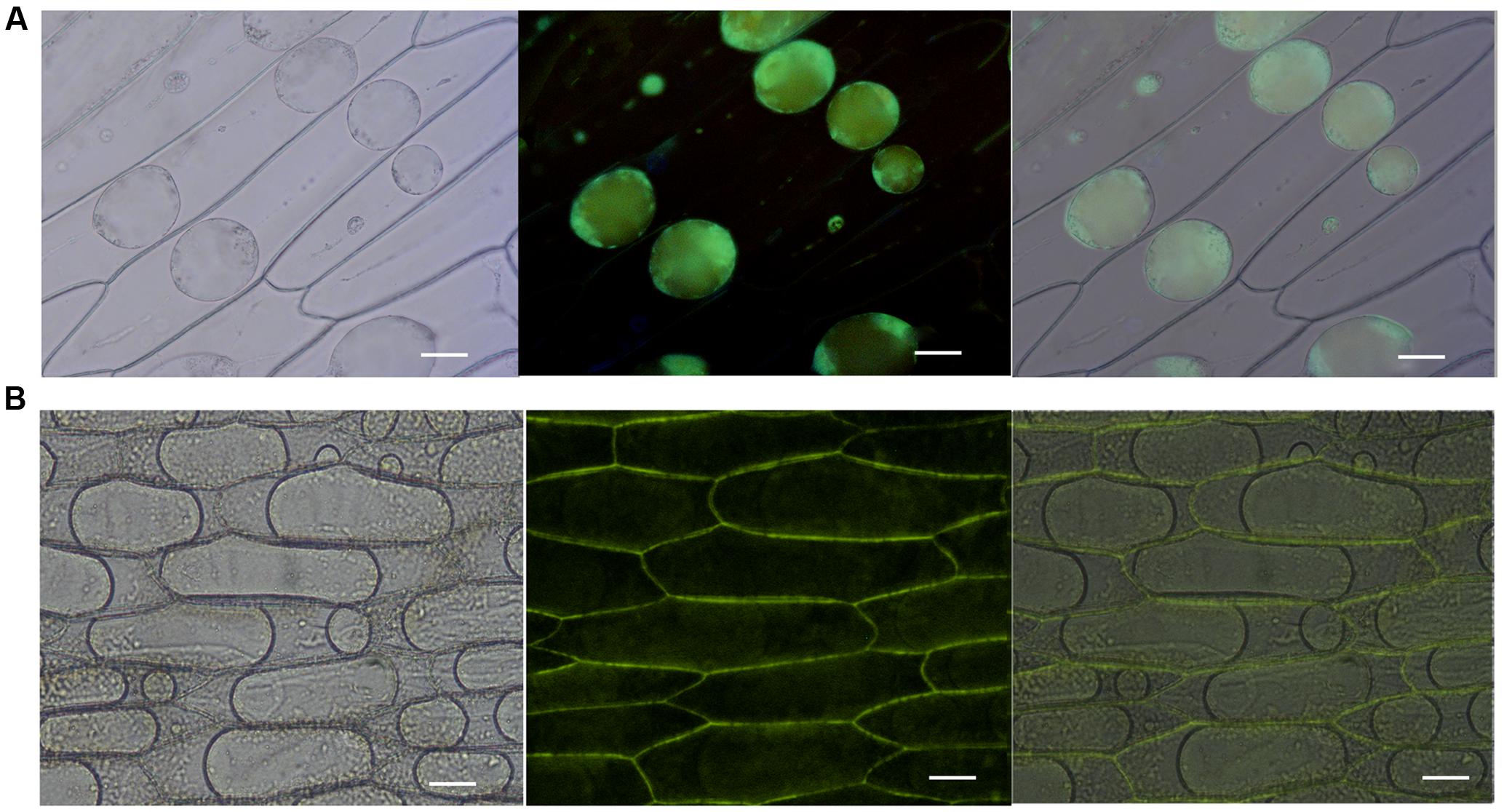
FIGURE 4. Subcellular localization of Misp12 in the plant cell. (A) Agrobacterium tumefaciens cells carrying 35S:ΔspMisp12:GUS:GFP fusion were transiently expressed in onion epidermal cells. The images present sequentially: bright-field, dark-field, and composite image. (B) Agrobacterium tumefaciens cells carrying 35S:Misp12:GUS:GFP fusion. The images present sequentially: bright-field, dark-field, and composite image. Scale bar = 100 μm.
In Planta RNAi of Misp12 Attenuates Nematode Parasitism
A virus-induced gene silencing (VIGS) technique was utilized to silence Misp12 in nematodes and the consequential effects on parasitism were evaluated. TRV-based expression vectors were employed to drive the expression of dsRNA complementary to Misp12 in N. benthamiana. A unique 249 bp fragment amplified from Misp12 cDNA named Umisp12 was ligated into vector pTRV2 to generate the expression constructs TRV2::Misp12. The leaves of N. benthamiana were co-infiltrated with cultures of recombinant Agrobacterium strains containing expression vectors of pTRV1 with TRV2::Misp12 or with empty vector pTRV2::00. At 5 DAI, the 447 bp TRV coat gene was detected by qRT-PCR in plant roots (Figure 5A). qRT-PCR analysis demonstrated that the TRV::Misp12-infiltrated plants showed a 75–80% reduction in Misp12 transcript level at 15 and 28 DAI when compared to the empty vector-infiltrated plants or non-infiltrated plants (Figure 5B).
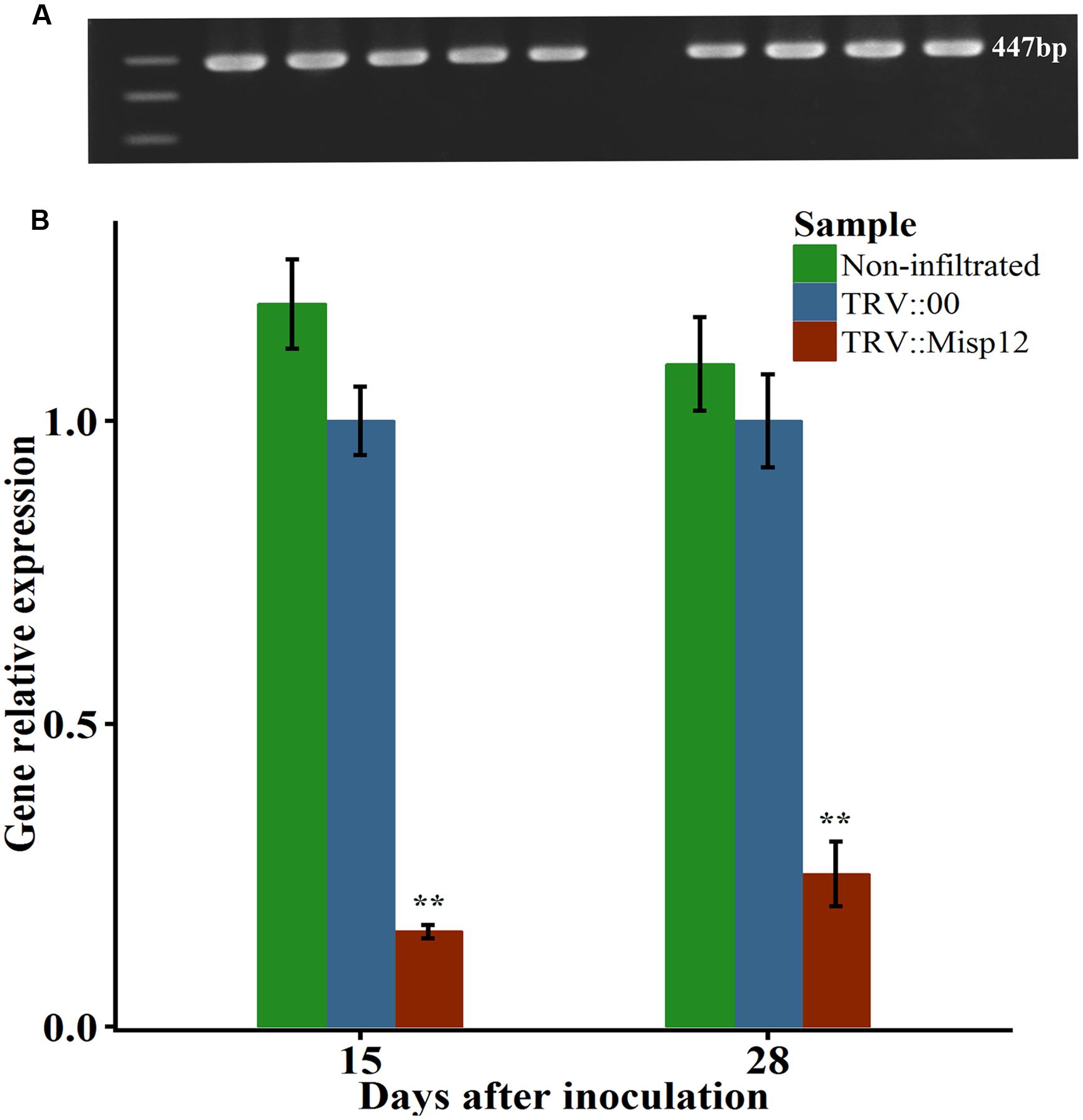
FIGURE 5. The Misp12 silence efficiency. (A) The figure of TRV coat gene sequence by agarose gel electrophoresis. (B) qRT-PCR assays for relative expression levels of Misp12 in M. incognita collected from non-infiltrated plants, TRV-vector transformed control roots (TRV::00) and Misp12-silencing root (TRV::Misp12); Actin and 18S was amplified as control. qRT-PCR experiments were repeated thrice with the similar results. Each bar value represents the mean ± SD of n > 10. Columns for the same time point or treatment marked with ∗∗ are significantly different (P < 0.01) from each other based on Student’s t-test.
Comparing the parasitism of nematodes in transgenic N. benthamiana and non-transgenic plants, we found that the Misp12-RNAi plants grow better than the non-transgenic plants (Figure 6), and the RNAi of Misp12 could significantly affected the galls formation, the numbers of eggs and nematodes (Figure 7).
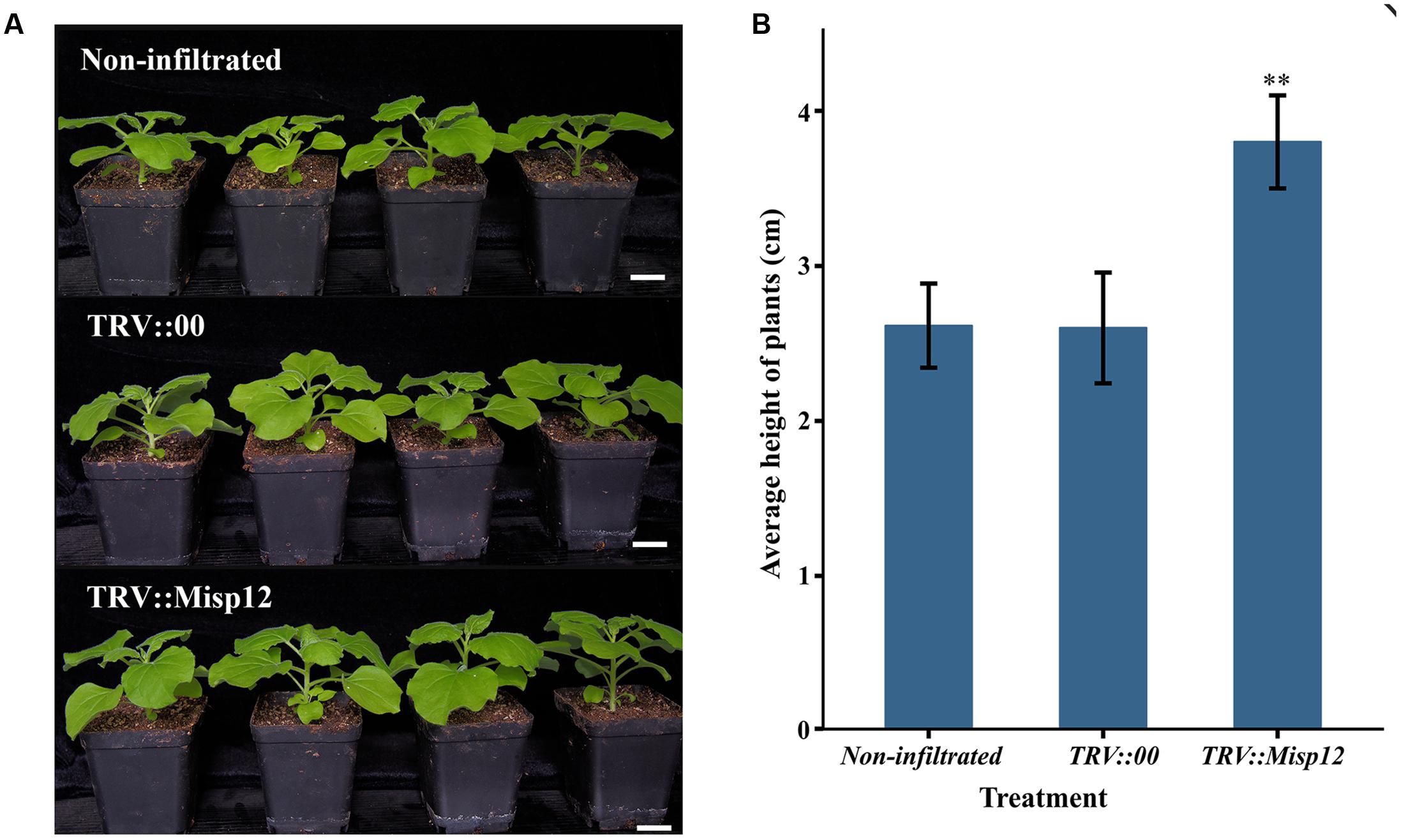
FIGURE 6. The plants comparison of Misp12-silencing plants and the control plant. (A) Photograph of non-infiltrated plants, TRV-vector transformed control plants (TRV::00) and Misp12-silencing plants (TRV::Misp12) at 20 DAI. Scale bar = 2 cm. (B) The average height of non-infiltrated plants, TRV::00 control plants and Misp12-silencing plants at 20 DAI. Columns for the treatment marked with the “∗∗” are significantly different (P < 0.01) from others based on Student’s t-test.
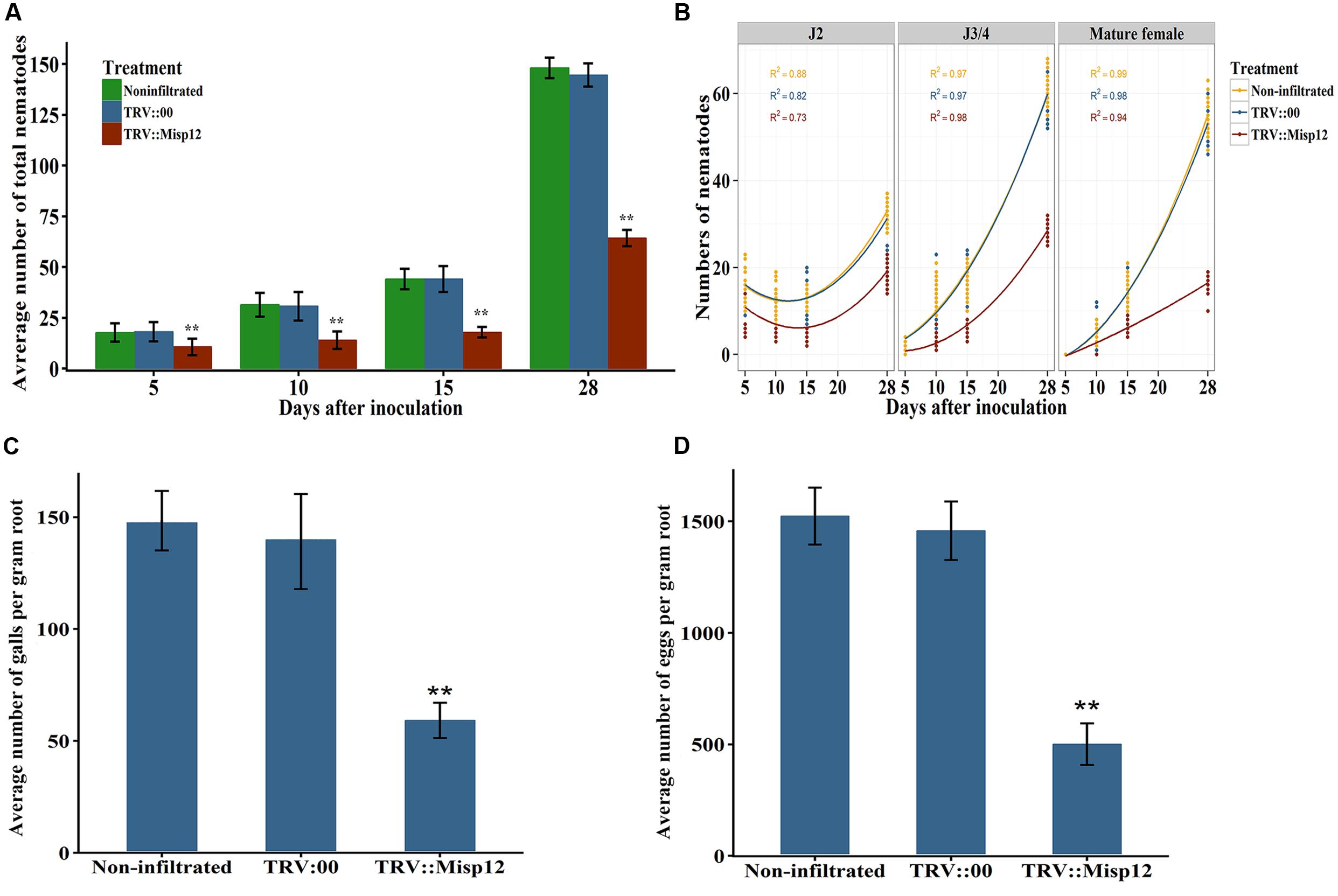
FIGURE 7. Effect of in planta RNAi of Misp12 on M. incognita. (A) Numbers of total nematodes in the N. benthamiana roots at various DAI, as indicated. (B) Numbers of nematodes at different stages in the N. benthamiana roots at various DAI. (C,D), Numbers of galls and eggs, respectively, in the N. benthamiana roots at 45 DAI. Each column represents the mean of three independent experiments with standard deviations. Columns for the same time point or treatment marked with ∗∗ are significantly different (P < 0.01) from each other based on Student’s t-test.
In TRV::Misp12-infiltrated plants roots, the number of parasitic nematodes exhibited much fewer mature females than non-infiltrated plants or plants infiltrated with TRV::00. At 5, 10, 15, and 28 DAI, the numbers of nematodes in the Misp12-RNAi transgenic lines reduced in a range from 41 to 59% when compared to the empty vector-transformed lines (Figure 7A). Moreover, the distribution of nematodes in different developmental stages in the root trended differently between the Misp12-RNAi and the control plants. For migratory parasitic J2s, the trend of nematode numbers in the Misp12-RNAi plants was similar to the control plants. While the J3/J4 and females in the Misp12-RNAi plants were sharply decreased at 28 DAI when compared to the control plants. Especially, the number of the mature females decreased by 69% in the Misp12-RNAi plants (Figure 7B). Additionally, at 45 DAI, both the size and amount of root galls in the Misp12-RNAi plants showed a significant reduction when compared to the empty vector-transformed lines and non-transgenic plants (Figure 7C). Likewise, the number of eggs showed a similar situation with 63% reduction rate (Figure 7D). These results suggested that Misp12 could promote the abilities of the nematode, especially at latter parasitism.
Higher Expression Level of Defense Genes in the Misp12-RNAi Transgenic Plant Roots
At 5 and 28 days after nematodes infection, the expression level of JA and SA signaling pathway defense genes were evaluated in the Misp12-RNAi transgenic plant roots and the control plant roots. At 28 DAI, three genes of OPR3, Pin2 and β-thionin genes in JA signaling pathway has 190-, 280-, and 280-fold up-regulation, respectively, when compared to the empty-vector transgenic plant or non-infiltrated plant roots. Consistently, the expression levels of SA marker genes PAL5 and PR1 were increased by 62- and 37-fold, respectively, at 28 DAI (Figure 8A). At 5 DAI, the JA and SA signaling pathway defense genes had no significant change compared with empty-vector transgenic plant or non-infiltrated plant roots (Figure 8B). These results indicated that Misp12 could suppress the plant defense response to nematodes at the latter stages of nematode infection.
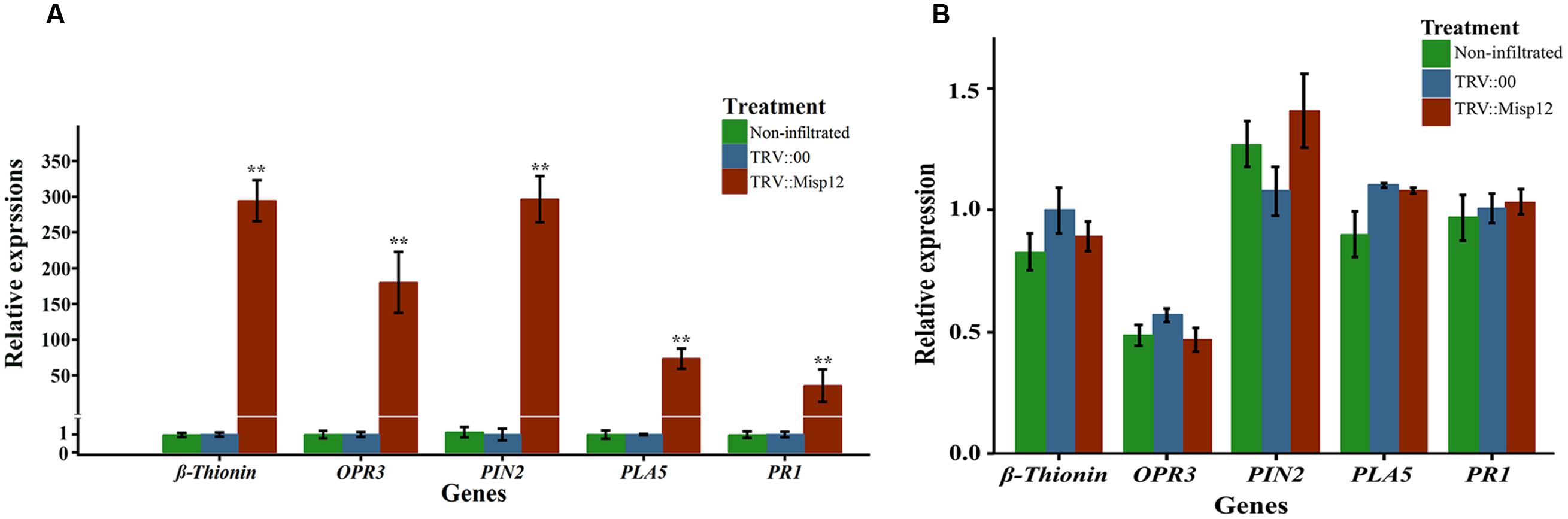
FIGURE 8. Relative expression levels of genes involved in JA and SA signaling defensive pathways. (A) Expression level of five JA and SA signal pathway mark genes in Misp12-silencing root at 28 DAI with M. incognita. (B) Expression level of the same mark genes in Misp12-silencing root at 5 DAI. The graph showed the mean and standard error of the relative amount of transcripts of these genes in Misp12-silencing root (TRV::Misp12) in comparison with TRV-vector transformed control roots (TRV::00) growing under the same conditions. Each reaction was performed in triplicate and the results represented the mean of three independent biological replicates. Columns for the same time point or treatment marked with ∗∗ are significantly different (P < 0.01) from each other based on Student’s t-test.
Down-Regulation of Defensive Genes in Misp12-exprssing Plant
At 2 days after PVX::INF1 infiltrated into the leaves of N. benthamiana, the Misp12 without signal peptide could obviously suppress programmed cell death triggered by INF1 (PT-PCD), and the Misp12 with signal peptide may partially suppressed PT-PCD. (Figure 9A and Supplementary Figure S3). Additionally, the expression levels of SA marker genes PAL5 and PR1 were strongly induced in control plants and were repressed in plants overproducing Misp12 (Figure 9B). However, the JA-related defense genes OPR3, Pin2 and β-thionin had no significant changes (Figure 9B).
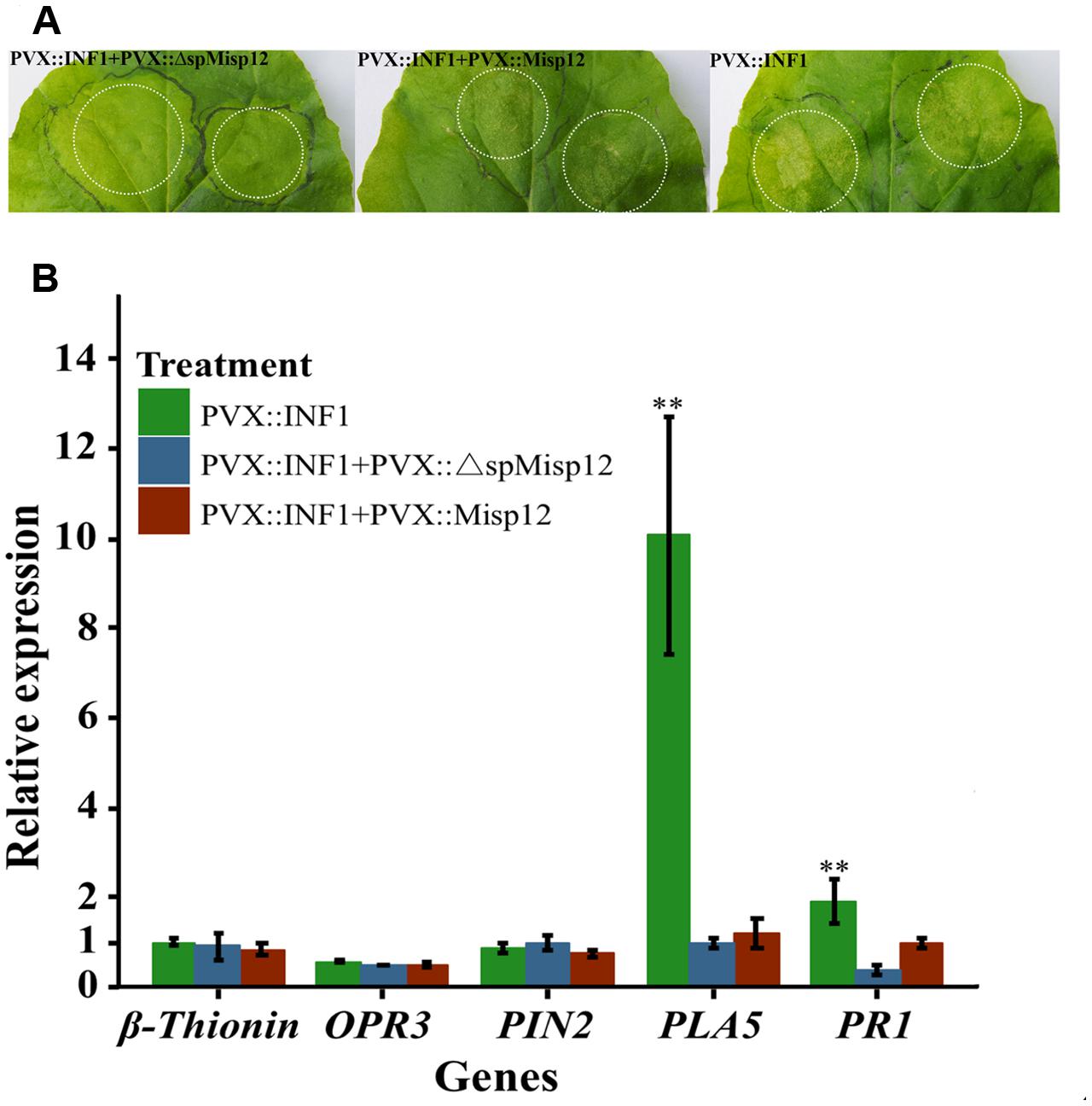
FIGURE 9. Misp12 suppresses INF1-triggered cell death (A) Graph showing the necrosis triggered by co-infiltration. N. benthamiana leaves were infiltrated Agrobacterium cells carrying the Misp12 gene with or without signal peptide 24 h before infiltration with Agrobacterium cells carrying the INF1 gene (PVX::Misp12+PVX::INF1 or PVX:: ΔspMisp12+PVX::INF1), the INF1 genes (PVX::INF1) were infiltrated alone as control. Representative images were acquired 2 days after the last infiltration. (B) Expression level of five JA and SA signal pathway mark genes mark genes in Misp12-expressing plant at 2 DAI with PVX::INF1. The plants growing under the same conditions. Each reaction was performed in triplicate and the results represented the mean of three independent biological replicates. Columns for the same time point or treatment marked with ∗∗ are significantly different (P < 0.01) from each other based on Student’s t-test.
Discussion
Since the completion of genome sequencing of different kinds of nematodes, tremendous progress has been made toward the building of effectors repertoire. Current studies mainly focused on the early parasitism of plant-nematode interactions. Most of the identified effectors were highly expressed in the migratory parasitic J2s, which suggested that these effectors are involved in the root invasion, larvae migration and the formation of the GCs (Rehman et al., 2016). The GCs need to be maintained throughout of the whole life of the nematodes, so some effectors must exist at the later parasitism. Here, we reported a novel potential effector, Misp12, that might play a key role at the later parasitic stage by using VIGS and transcription analysis.
It is reported that an abundance existence of unique genes contributes to M. incognita’s wide host range, a range that exceeds other parasitic nematodes (Bird et al., 2009). The BLAST results indicated that no other similar sequences of Misp12 exists in the other organisms, and the further gene amplification of M. incognita collected from eight different areas of China showed that Misp12 had only a few nucleotide mutations. These results suggested that Misp12 was highly conserved and specific in M. incognita and might be involved in the improvement of host range.
In parasitic nematodes, mature effector proteins need the signal peptide to be imported into the endoplasmic reticulum of the cells and then to be secreted into the host via the nematode stylet (Elling et al., 2007; Sacco et al., 2009). Based on the bioinformatics tools analysis, the Misp12 protein was predicted to be a potential effector. Consistent with this prediction, the subcellular localization of Misp12 in planta indicated that the Misp12 protein was delivered to the cell cytoplasm and targeted cell cytoplasmic proteins. Additionally, in situ mRNA hybridization suggested that the Misp12 was specifically expressed in the dorsal esophageal gland, which confirmed the results of a previously report (Huang et al., 2003). Esophageal is the common origin of nematode secretory proteins localization (Davis et al., 2004). Thus, consistent with previously report (Huang et al., 2003), we deduced that Misp12 was a secretory protein and a novel effector of M. incognita. It is believed that the subventral glands show activity at the stage of nematode penetration and migration, whereas the dorsal glands tend to respond during the formation and maintenance of the nematode feeding cells in the sedentary nematode life stages (Davis et al., 2008). The expression level of Misp12 was significantly greater at the mature female stages. Hence, as a secretory protein derived from the dorsal esophageal gland, Misp12 might be involved in the maintenance of GCs during the latter parasitism.
VIGS is an alternate useful tool to study target genes as the knock-out of specific nematode genes is not feasible for plant-parasitic nematodes at present (Becker and Lange, 2010; Senthil-Kumar and Mysore, 2011; Lee et al., 2012). To observe the consequential effects of Misp12 on nematode parasitism, VIGS was employed to silence the Misp12 gene. It resulted in fewer galls, eggs, and parasitic nematodes in the gene silenced-plant roots than that in the negative control plants group. Notably, gene silencing induced numbers of J3/J4 and mature female nematodes a more significant reduction. The numbers of J3/J4 showed a 65% reduction and the mature female showed a 69% reduction at 28 DAI. These results were consistent with the dynamics of Misp12 expression levels. Thereby, we concluded that Misp12 played a critical role during latter parasitism, especially at the mature stage of M. incognita.
Root-knot nematodes are biotrophic pathogens and the success of infection depends on the ability of the pathogen to overcome the plant defense system. SA and JA hormones can activate the plant defense system (Grant and Jones, 2009; Pieterse et al., 2009; Yan and Dong, 2014; Yang et al., 2015; Zhang et al., 2015b) and play crucial roles in plant defense responses against RKNs and other parasitic nematodes (Molinari and Loffredo, 2006; Bhattarai et al., 2008; Molinari et al., 2014). Several effectors that influence the plant defense responses have been characterized (Quentin et al., 2013; Rehman et al., 2016). Chorismate mutases, which are secreted by the RKNs, can affect the plant shikimate pathway, thereby decreasing the synthesis of SA and phytoalexin through competition with chorismate, and can prevent the triggering of host defense (Huang et al., 2005b; Long et al., 2006). The effectors Hs10A06 from Heterodera schachtii targets Arabidopsis spermidine synthase and suppresses the SA signaling pathway in the syncytia (Hewezi et al., 2010). And Mi-CRT from M. incognita also acts on the SA signaling pathway to promote the nematodes parasitism (Jaouannet et al., 2013). In this study, the results showed that PR1 and PAL5 were up-regulated in the virus-induced silencing of Misp12 plants and down-regulated in the Misp12-expressing plants. It is reported that PR-1 was highly induced in Arabidopsis roots after M. incognita infection (Hamamouch et al., 2011). These may imply that SA signaling pathways could be manipulated by Misp12 in the root cells to support nematode parasitism at the latter stages.
Jasmonic acid pathways also play a major role in the defense against the RKNs (Nahar et al., 2011). The effector Mi-CRT mentioned above can also suppressed the defense genes from the JA pathway (Jaouannet et al., 2013). Mj-FAR-1, a secreted fatty acid and retinol binding proteins from M. javanica, could manipulate the lipid based signaling to suppress the JA related defense gene (Iberkleid et al., 2013). In this study, the proteinase inhibitors(Pin2), 12-oxophyto-dienoate reductase (OPR3) and β-thionin genes, associating with the JA metabolic pathway (Wasternack, 2007; Fujimoto et al., 2011) were significantly up-regulated in the virus-induced silencing of Misp12 plants, indicating the Misp12 gene could manipulate these defense genes. Interestingly, it was reported that β-thionin is involved in inhibiting mammalian cell growth by membrane permeabilization (Li et al., 2002) and able to inhibit insect amylases and proteinases activity (Li et al., 2002; Melo et al., 2002). Moreover, previous researches has shown that proteinase inhibitors (Pin2) respond to wounding when attacked by tobacco hornworm larvae (Howe et al., 1996), and predicated that protease inhibitor could enhance plant resistance to nematodes (McPherson and Harrison, 2001). Thus, up-regulation of β-thionin and Pin2 found in the virus-induced silencing of Misp12 plant roots might also contribute to the suppression of nematode parasitism. However, in the Misp12-expressing plants, in which the INF1 gene was inoculated, the JA-related defense genes had no significant changes; this may be due to JA defense genes not being active in the progression of PT-PCD.
Interestingly, Misp12 with and without signal peptide have different localizations in the plant cell, but both of them suppressed programed cell death (PCD) triggered by INF1 through transient expression in N. benthamiana. Recently, two secreted nematode effectors, MeTCTP from M. enterolobii and MiMsp40 from M. incognita, were demonstrated to suppress the PCD triggered by BAX (Niu et al., 2016; Zhuo et al., 2016), but both effectors were up-regulated during the early parasitic stages. The Misp12 up-regulated at the J3/4 and female stages, which suggested that the Misp12 might suppress the PCD at the nematode mature stages to maintain the GCs for nutrients.
Taken together, although the mechanism of host defense suppression by Misp12 and the plant target for Misp12 remains unclear, our data indicated that Misp12 protein has a potential through down-regulation of SA and JA-dependent defense responses genes to promote the latter parasitism of M. incognita during the mature stages. This study revealed a novel effector Misp12, which functions at the latter nematode parasitic stage, and it would be a new complement to the effectors of the parasitic nematode. This gene plays a key role in manipulating plant defense signal responsive genes to maintain the GCs, thus promoting successful parasitism in the host plant. Furthermore, Misp12 could be a new target gene to control nematode disease.
Author Contributions
YX and XX initiated and designed the research. JX and SL wrote the manuscript. JX, SL, and CM performed experiments and analyzed data. YX, XX, and GW reviewed the paper. All authors read and approved the final manuscript.
Funding
This study was carried out at the Key Laboratory of Plant Pathology of Hubei Province at Huazhong Agricultural University in Wuhan. This project was supported by the Special Fund for Agro-scientific Research in the Public Interest 201503114 and 201103018, the Major State Basic Research Development Program (973) 2013CB127504, and Fundamental Research Funds for the Central Universities 2662015QC037.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors appreciate Prof. Daohong Jiang of Key Laboratory of Plant Pathology of Hubei Province in Huazhong Agricultural University for offering the TRV vector and Dr. Weixiao Yin of Key Laboratory of Plant Pathology of Hubei Province in Huazhong Agricultural University for providing the pGR107 vector.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00964
FIGURE S1 | The alignment of Misp12 from 8 different area of China.
FIGURE S2 | The free eGFP location in the onion cells.
FIGURE S3 | Misp12 suppresses INF1-triggered cell death using A. tumefaciens cells with OD600 = 1.0 at 2 DAI.
Footnotes
References
Abad, P., Gouzy, J., Aury, J.-M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi: 10.1038/nbt.1482
Anand, A., Krichevsky, A., Schornack, S., Lahaye, T., Tzfira, T., Tang, Y., et al. (2007). Arabidopsis VIRE2 INTERACTING PROTEIN2 Is Required for Agrobacterium T-DNA Integration in Plants. Plant Cell. 19, 1695–1708. doi: 10.1105/tpc.106.042903
Article, L. B., Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 23, 2064–2086.
Becker, A., and Lange, M. (2010). VIGS–genomics goes functional. Trends Plant Sci. 15, 1–4. doi: 10.1016/j.tplants.2009.09.002
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bhattarai, K. K., Xie, Q.-G., Mantelin, S., Bishnoi, U., Girke, T., Navarre, D. A., et al. (2008). Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant Microbe Interact. 21, 1205–1214. doi: 10.1094/MPMI-21-9-1205
Bird, D. M., Williamson, V. M., Abad, P., McCarter, J., Danchin, E. G. J., Castagnone-Sereno, P., et al. (2009). The genomes of root-knot nematodes. Annu. Rev. Phytopathol. 47, 333–351. doi: 10.1146/annurev-phyto-080508-081839
Blok, V. C., Jones, J. T., Phillips, M. S., and Trudgill, D. L. (2008). Parasitism genes and host range disparities in biotrophic nematodes: the conundrum of polyphagy versus specialisation. Bioessays 30, 249–259. doi: 10.1002/bies.20717
Bos, J. I. B., Kanneganti, T.-D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Bybd, D. W., Kirkpatrick, T., and Barker, K. R. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143.
Caillaud, M.-C., Lecomte, P., Jammes, F., Quentin, M., Pagnotta, S., Andrio, E., et al. (2008). MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20, 423–437. doi: 10.1105/tpc.107.057422
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723. doi: 10.1073/pnas.97.7.3718
Dąbrowska-Bronk, J., Czarny, M., Wiśniewska, A., Fudali, S., Baranowski, L., Sobczak, M., et al. (2014). Suppression of NGB and NAB/ERabp1 in tomato modifies root responses to potato cyst nematode infestation. Mol. Plant Pathol. 16, 1–15. doi: 10.1111/mpp.12183
Dautova, M., Gommers, F., Abad, P., Smant, G., Bakker, J., and Rosso, M.-N. (2001). Single pass cDNA sequencing – a powerful tool to analyse gene expression in preparasitic juveniles of the southern root-knot nematode Meloidogyne incognita. Nematology 3, 129–139. doi: 10.1163/156854101750236259
Davis, E. L., Hussey, R. S., and Baum, T. J. (2004). Getting to the roots of parasitism by nematodes. 20, 134–141.
Davis, E. L., Hussey, R. S., Baum, T. J., Bakker, J., Schots, A., Rosso, M.-N., et al. (2000). NEMATODE PARASITISM GENES. Annu. Rev. Phytopathol. 38, 365–396. doi: 10.1146/annurev.phyto.38.1.365
Davis, E. L., Hussey, R. S., Mitchum, M. G., and Baum, T. J. (2008). Parasitism proteins in nematode-plant interactions. Curr. Opin. Plant Biol. 11, 360–366. doi: 10.1016/j.pbi.2008.04.003
de Boer, J. M., Yan, Y., Smant, G., Davis, E. L., and Baum, T. J. (1998). In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 30, 309–312.
Ding, X., Shields, J., Allen, R., and Hussey, R. (2000). Molecular cloning and characterisation of a venom allergen AG5-like cDNA from Meloidogyne incognita1Note: Nucleotide sequence data reported in this paper are available in the EMBL. GenBank and DDJB databases under the accession number AF013289.1. Int. J. Parasitol. 30, 77–81.
Ding, X., Shields, J., Allen, R., and Hussey, R. S. (1998). A secretory cellulose-binding protein cDNA cloned from the root-knot nematode (Meloidogyne incognita). Mol. plant microbe interact. 11, 952–959. doi: 10.1094/MPMI.1998.11.10.952
Dinh, P., Brown, C., and Elling, A. (2014). RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in Arabidopsis and Potato. Phytopathology 104, 1–41. doi: 10.1094/PHYTO-03-14-0063-R
Doyle, E. A., and Lambert, K. N. (2002). Cloning and characterization of an esophageal-gland-specific pectate lyase from the root-knot nematode Meloidogyne javanica. Mol. Plant Microbe Interact. 15, 549–556. doi: 10.1094/MPMI.2002.15.6.549
Dubreuil, G., Magliano, M., Deleury, E., Abad, P., and Rosso, M. N. (2007). Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New Phytol. 176, 426–436. doi: 10.1111/j.1469-8137.2007.02181.x
Elling, A. A. (2013). Major emerging problems with minor meloidogyne species. Phytopathology 103, 1092–1102. doi: 10.1094/PHYTO-01-13-0019-RVW
Elling, A. A., Davis, E. L., Hussey, R. S., and Baum, T. J. (2007). Active uptake of cyst nematode parasitism proteins into the plant cell nucleus. Int. J. Parasitol. 37, 1269–1279. doi: 10.1016/j.ijpara.2007.03.012
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Fujimoto, T., Tomitaka, Y., Abe, H., Tsuda, S., Futai, K., and Mizukubo, T. (2011). Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J. Plant Physiol. 168, 1084–1097. doi: 10.1016/j.jplph.2010.12.002
Gao, B., Allen, R., Maier, T., Davis, E. L., Baum, T. J., and Hussey, R. S. (2003). The parasitome of the phytonematode Heterodera glycines. Mol. plant microbe interact. 16, 720–726. doi: 10.1094/MPMI.2003.16.8.720
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M., Appel, R., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. Walker (New York, NY: Humana Press), 571–607.
Goetz, M., Godt, D. E., Guivarc’h, A., Kahmann, U., Chriqui, D., and Roitsch, T. (2001). Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. U.S.A. 98, 6522–6527. doi: 10.1073/pnas.091097998
Grant, M. R., and Jones, J. D. G. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
Hamamouch, N., Li, C., Seo, P. J., Park, C.-M., and Davis, E. L. (2011). Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 12, 355–364. doi: 10.1111/j.1364-3703.2010.00675.x
Hewezi, T., and Baum, T. J. (2013). Manipulation of plant cells by cyst and root-knot nematode effectors. Mol. plant microbe interact. 26, 9–16. doi: 10.1094/MPMI-05-12-0106-FI
Hewezi, T., Howe, P. J., Maier, T. R., Hussey, R. S., Mitchum, M. G., Davis, E. L., et al. (2010). Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol. 152, 968–984. doi: 10.1104/pp.109.150557
Howe, G. A., Lightner, J., Browse, J., and Ryan, C. A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. doi: 10.2307/3870413
Huang, G., Dong, R., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2005a). Developmental expression and molecular analysis of two Meloidogyne incognita pectate lyase genes. Int. J. Parasitol. 35, 685–692. doi: 10.1016/j.ijpara.2005.01.006
Huang, G., Dong, R., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2005b). Two chorismate mutase genes from the root-knot nematode Meloidogyne incognita. Mol. Plant Pathol. 6, 23–30. doi: 10.1111/j.1364-3703.2004.00257.x
Huang, G., Dong, R., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. plant microbe interact. 19, 463–470. doi: 10.1094/MPMI-19-0463
Huang, G., Gao, B., Maier, T., Allen, R., Davis, E. L., Baum, T. J., et al. (2003). A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Mol. plant microbe interact. 16, 376–381. doi: 10.1094/MPMI.2003.16.5.376
Iberkleid, I., Vieira, P., de Almeida Engler, J., Firester, K., Spiegel, Y., and Horowitz, S. B. (2013). Fatty acid-and retinol-binding protein. Mj-FAR-1 induces tomato host susceptibility to root-knot nematodes. PLoS ONE 8:e64586. doi: 10.1371/journal.pone.0064586
Jaouannet, M., Magliano, M., Arguel, M. J., Gourgues, M., Evangelisti, E., Abad, P., et al. (2013). The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol. plant microbe interact. 26, 97–105. doi: 10.1094/MPMI-05-12-0130-R
Jaouannet, M., Perfus-Barbeoch, L., Deleury, E., Magliano, M., Engler, G., Vieira, P., et al. (2012). A root-knot nematode-secreted protein is injected into giant cells and targeted to the nuclei. New Phytol. 194, 924–931. doi: 10.1111/j.1469-8137.2012.04164.x
Jaubert, S., Laffaire, J.-B., Ledger, T. N., Escoubas, P., Amri, E.-Z., Abad, P., et al. (2004). Comparative analysis of two 14-3-3 homologues and their expression pattern in the root-knot nematode Meloidogyne incognita. Int. J. Parasitol. 34, 873–880. doi: 10.1016/j.ijpara.2004.02.008
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lambert, K. N., Allen, K. D., and Sussex, I. M. (1999). Cloning and characterization of an esophageal-gland-specific chorismate mutase from the phytoparasitic nematode Meloidogyne javanica. Mol. plant microbe interact. 12, 328–336. doi: 10.1094/MPMI.1999.12.4.328
Ledger, T. N., Jaubert, S., Bosselut, N., Abad, P., and Rosso, M.-N. (2006). Characterization of a new beta-1,4-endoglucanase gene from the root-knot nematode Meloidogyne incognita and evolutionary scheme for phytonematode family 5 glycosyl hydrolases. Gene 382, 121–128. doi: 10.1016/j.gene.2006.06.023
Lee, W.-S., Hammond-Kosack, K. E., and Kanyuka, K. (2012). Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590.
Li, S. S., Gullbo, J., Lindholm, P., Larsson, R., Thunberg, E., Samuelsson, G., et al. (2002). Ligatoxin B, a new cytotoxic protein with a novel helix-turn-helix DNA-binding domain from the mistletoe Phoradendron liga. Biochem. J. 366, 405–413. doi: 10.1042/bj20020221
Lin, B., Zhuo, K., Chen, S., Hu, L., Sun, L., Wang, X., et al. (2016). A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 209, 1159–1173. doi: 10.1111/nph.13701
Lin, B., Zhuo, K., Wu, P., Cui, R., Zhang, L.-H., and Liao, J. (2013). A novel effector protein, MJ-NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. plant microbe interact. 26, 55–66.
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ÄÄCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, H., Wang, X., and Xu, J. (2006). Molecular cloning and life-stage expression pattern of a new chorismate mutase gene from the root-knot nematode Meloidogyne arenaria. Plant Pathol. 55, 559–563. doi: 10.1111/j.1365-3059.2006.01362.x
McPherson, M. J., and Harrison, D. J. (2001). Protease inhibitors and directed evolution: enhancing plant resistance to nematodes. Biochem. Soc. Symp. 125–142.
Melo, F. R., Rigden, D. J., Franco, O. L., Mello, L. V., Ary, M. B., Grossi de Sá, M. F., et al. (2002). Inhibition of trypsin by cowpea thionin: characterization, molecular modeling, and docking. Proteins 48, 311–319. doi: 10.1002/prot.10142
Molinari, S., Fanelli, E., and Leonetti, P. (2014). Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 15, 255–264. doi: 10.1111/mpp.12085
Molinari, S., and Loffredo, E. (2006). The role of salicylic acid in defense response of tomato to root-knot nematodes. Physiol. Mol. Plant Pathol. 68, 69–78. doi: 10.1007/s00425-014-2138-7
Nahar, K., Kyndt, T., De Vleesschauwer, D., Hofte, M., and Gheysen, G. (2011). The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in Rice. Plant Physiol. 157, 305–316. doi: 10.1104/pp.111.177576
Niu, J., Liu, P., Liu, Q., Chen, C., Guo, Q., Yin, J., et al. (2016). Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 6, 19443. doi: 10.1038/srep19443
Opperman, C. H., Bird, D. M., Williamson, V. M., Rokhsar, D. S., Burke, M., Cohn, J., et al. (2008). Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. U.S.A. 105, 14802–14807. doi: 10.1073/pnas.0805946105
Pagni, M., Ioannidis, V., Cerutti, L., Zahn-Zabal, M., Jongeneel, C. V., Hau, J., et al. (2007). MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35, 433–437. doi: 10.1093/nar/gkm352
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Popeijus, H., Overmars, H., Jones, J., Blok, V., Goverse, A., Helder, J., et al. (2000). Degradation of plant cell walls by a nematode. Nature 406, 36–37. doi: 10.1038/35017641
Quentin, M., Abad, P., and Favery, B. (2013). Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 4:53. doi: 10.3389/fpls.2013.00053
Rehman, S., Gupta, V. K., and Goyal, A. K. (2016). Identification and functional analysis of secreted effectors from phytoparasitic nematodes. BMC Microbiol. 16:48. doi: 10.1186/s12866-016-0632-8
Remmert, M., Biegert, A., and Hauser, A. (2011). HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175.
Rosso, M. N., Favery, B., Piotte, C., Arthaud, L., De Boer, J. M., Hussey, R. S., et al. (1999). Isolation of a cDNA encoding a beta-1,4-endoglucanase in the root-knot nematode Meloidogyne incognita and expression analysis during plant parasitism. Mol. plant microbe interact. 12, 585–591. doi: 10.1094/MPMI.1999.12.7.585
Rost, B., and Sander, C. (1993). Secondary structure prediction of all-helical proteins in two states. Protein Eng. 6, 831–836. doi: 10.1093/protein/6.8.831
Sacco, M. A., Koropacka, K., Grenier, E., Jaubert, M. J., Blanchard, A., Goverse, A., et al. (2009). The Cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog. 5:e1000564. doi: 10.1371/journal.ppat.1000564
Sambrook, J., and MacCallum, P. (2012). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Senthil-Kumar, M., and Mysore, K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665. doi: 10.1016/j.tplants.2011.08.006
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trudgill, D. L., and Blok, V. C. (2001). Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39, 53–77. doi: 10.1146/annurev.phyto.39.1.53
Vens, C., Rosso, M.-N., and Danchin, E. G. J. (2011). Identifying discriminative classification-based motifs in biological sequences. Bioinformatics 27, 1231–1238. doi: 10.1093/bioinformatics/btr110
Wang, X., Li, H., Hu, Y., Fu, P., and Xu, J. (2007). Molecular cloning and analysis of a new venom allergen-like protein gene from the root-knot nematode Meloidogyne incognita. Exp. Parasitol. 117, 133–140. doi: 10.1016/j.exppara.2007.03.017
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. doi: 10.1093/aob/mcm079
Xiao, X., Xie, J., Cheng, J., Li, G., Yi, X., Jiang, D., et al. (2014). Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. plant microbe interact. 27, 40–55. doi: 10.1094/MPMI-05-13-0145-R
Xue, B., Hamamouch, N., Li, C., Huang, G., Hussey, R. S., Baum, T. J., et al. (2013). The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 103, 175–181. doi: 10.1094/PHYTO-07-12-0173-R
Yan, S., and Dong, X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68. doi: 10.1016/j.pbi.2014.04.006
Yang, L., Li, B., Zheng, X., Li, J., Yang, M., Dong, X., et al. (2015). Corrigendum: salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. commun. 6, 8145. doi: 10.1038/ncomms8309
Zhang, L., Davies, L. J., and Elling, A. A. (2015a). A Meloidogyne incognita effector is imported into the nucleus and exhibits transcriptional activation activity in planta. Mol. Plant Pathol. 16, 48–60. doi: 10.1111/mpp.12160
Zhang, L., Yao, J., Withers, J., Xin, X.-F., Banerjee, R., Fariduddin, Q., et al. (2015b). Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proc. Natl. Acad. Sci. U.S.A. 112, 14354–14359. doi: 10.1073/pnas.1510745112
Keywords: Misp12, Meloidogyne incognita, effector protein, defense suppression, latter parasit
Citation: Xie J, Li S, Mo C, Wang G, Xiao X and Xiao Y (2016)A Novel Meloidogyne incognita Effector Misp12 Suppresses Plant Defense Response at Latter Stages of Nematode Parasitism. Front. Plant Sci. 7:964. doi: 10.3389/fpls.2016.00964
Received: 15 March 2016; Accepted: 16 June 2016;
Published: 30 June 2016.
Edited by:
Dirk Balmer, Syngenta Crop Protection, SwitzerlandReviewed by:
Lei Zhang, Washington State University, USADaolong Dou, Nanjing Agricultural University, China
Copyright © 2016 Xie, Li, Mo, Wang, Xiao and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yannong Xiao, eGlhb3lhbm5vbmdAbWFpbC5oemF1LmVkdS5jbg== Xueqiong Xiao, eHVlcWlvbmd4aWFvQG1haWwuaHphdS5lZHUuY24=
 Jialian Xie
Jialian Xie Shaojun Li
Shaojun Li Chenmi Mo
Chenmi Mo Xueqiong Xiao
Xueqiong Xiao