- 1State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Taian, China
- 2College of Life Science, Shanghai University, Shanghai, China
- 3Cotton Research Institute – Chinese Academy of Agricultural Sciences, Anyang, China
- 4Cotton Research Center, Shandong Academy of Agricultural Sciences, Jinan, China
A corrigendum on
Overexpression of an Apocynum venetum DEAD-Box Helicase Gene (AvDH1) in Cotton Confers Salinity Tolerance and Increases Yield in a Saline Field
by Chen, J., Wan, S., Liu, H., Fan, S., Zhang, Y., Wang, W., et al. (2015). Front. Plant Sci. 6:1227. doi: 10.3389/fpls.2015.01227
Reason for Corrigendum:
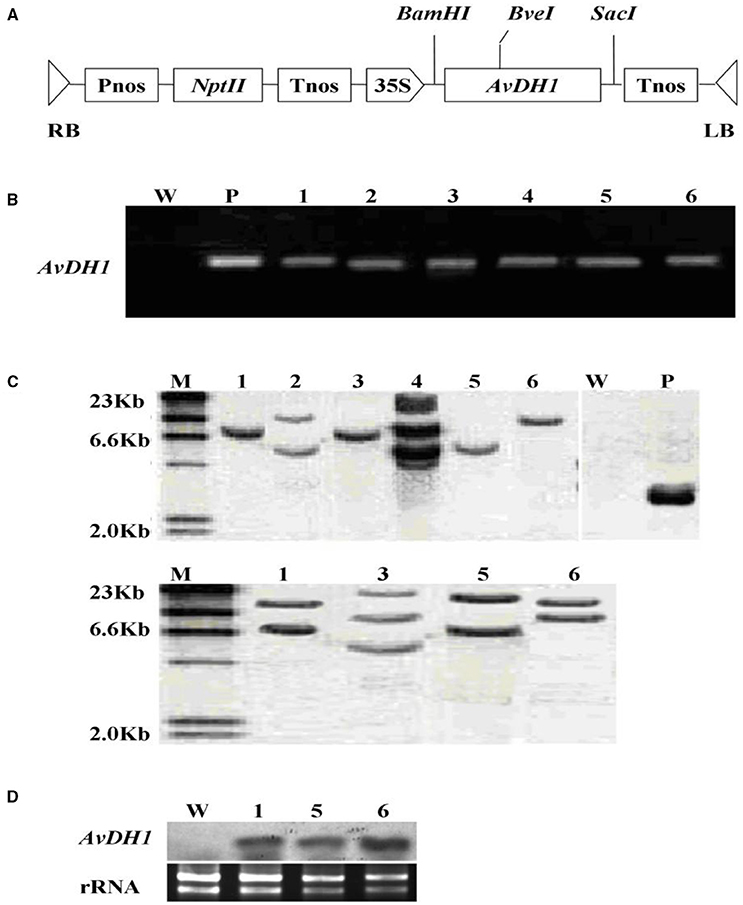
In the Original Research article, the “W” and “P” lanes in the published version of Figure 1C were spliced into the figure rather than run out as part of a contiguous gel/blot, we have regenerated new, unprocessed images via further experimentation. The revised version of Figure has been provided below in the last page and modifications have been made to the corresponding sections in the main text.
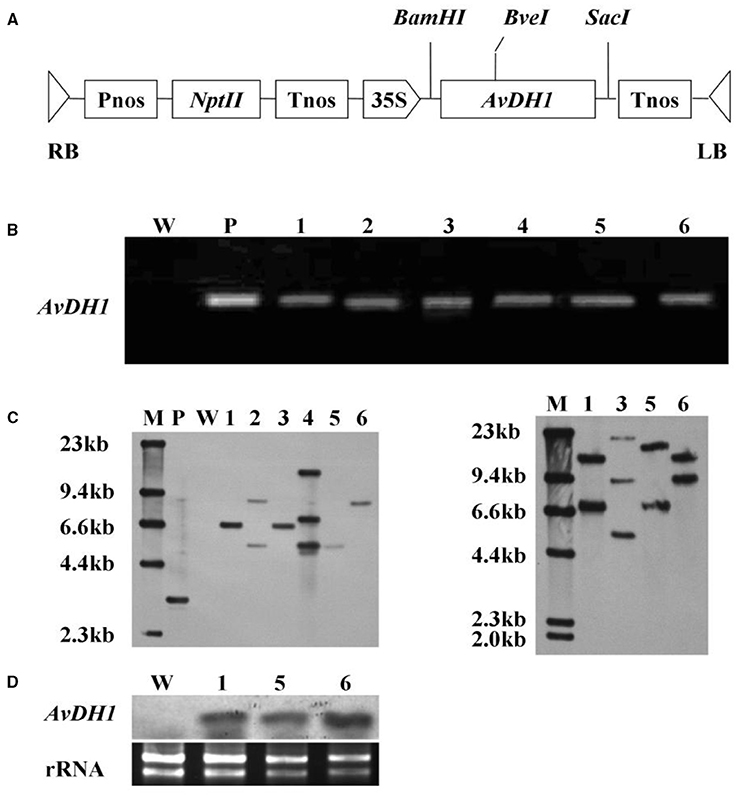
FIGURE 1. Schematic structure of the T-DNA and molecular analysis of AvDH1-expressing cotton. (A) T-DNA region of the vector pBI121-AvDH1. RB, right T-DNA border; LB, left T-DNA border; Pnos, nopaline synthase gene promoter; nptII, neomycin phosphotransferase gene; Tnos, nopaline synthase gene terminator; 35 S, cauliflower mosaic virus 35 s promoter; AvDH1, Apocynum venetum DEAD-box helicase gene. (B) PCR analysis of genomic DNA from an untransformed control (W) and six independent T0 transgenic lines (1–6). P, pBI121-AvDH1 as positive control. (C) Southern blot analysis of AvDH1-transformed cotton lines. Genomic DNA was digested with StuI (left) and BveI (right). The membrane was hybridized with a DIG-labeled AvDH1 probe. M, molecular marker; P, pBI121-AvDH1 as positive control; W, untransformed control; 1–6, transgenic lines 08–66, 08–89, 08–90, 08–92, 08–26, and 08–87, respectively. (D) Three independent T3 transgenic lines were confirmed by Northern blotting. Lower panel shows rRNA to confirm equal loading. 1, line 08–66; 5, line 08–26; 6, line 08–87.
In the “Materials & Methods” section, sub section “PCR and Southern Blot”, the last two sentences of the paragraph has been modified from
A [32P]-labeled AvDH1 gene was used as the probe. Southern blots were hybridized by following the standard procedure provided by the manufacturer. After hybridization and stringent washing, the radioactive membranes were exposed to an imaging plate (Fuji Photo Film, Japan) for 5 h or overnight to record the images.
to
A DIG-labeled marker (molecular weight marker II, Roche Diagnostics, Mannheim, Germany) was used for size estimation, and a digoxigenin (DIG)-labeled AvDH1 gene sequence was used as the probe. Incorporation of digoxigenin-11-dUTP into the AvDH1 probe was done by Taq DNA polymerase during a PCR reaction using forward primer AvDH1-F 5′-TTGGCGGCAATAGCGT-3′ and reverse primer AvDH1-R 5′-CCTTAGTAGCACCACCCT-3′, following the supplier's instruction (Roche Diagnostics). Southern blots were hybridized using a DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Mannheim, Germany) following the standard procedure provided by the manufacturer. After hybridization and stringent washing, X-ray films (Biomax MS, Kodak) exposure was done for 0.5, 1, and 3 h to achieve the desired signal strength.
In the “Results” section, sub section “Regeneration and Analysis of Transgenic Cotton Plants”, the word “Upper” has been changed to “Left” and “lower” has been changed to “right”.
The authors apologize for the ambiguity. This error does not change the scientific conclusions of the article in any way.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was mainly supported by the China Major Projects for Transgenic Breeding (Grant No 2011ZX08005-004 and 2011ZX08005-002) and the China Key Development Project for Basic Research (973; Grant No 2010CB12606).
Keywords: DEAD-box helicase, AvDH1, cotton, salinity, yield, field trial
Citation: Chen J, Wan S, Liu H, Fan S, Zhang Y, Wang W, Xia M, Yuan R, Deng F and Shen F (2016) Corrigendum: Overexpression of an Apocynum venetum DEAD-Box Helicase Gene (AvDH1) in Cotton Confers Salinity Tolerance and Increases Yield in a Saline Field. Front. Plant Sci. 7:1041. doi: 10.3389/fpls.2016.01041
Received: 15 June 2016; Accepted: 01 July 2016;
Published: 13 July 2016.
Edited and reviewed by: Keqiang Wu, National Taiwan University, Taiwan
Copyright © 2016 Chen, Wan, Liu, Fan, Zhang, Wang, Xia, Yuan, Deng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fafu Shen, ZmZzaGVuQHNkYXUuZWR1LmNu
 Jie Chen
Jie Chen Sibao Wan2
Sibao Wan2 Wei Wang
Wei Wang