- 1Department of Biological Sciences, Korea Advanced Institute of Science and Technology, Daejeon, South Korea
- 2Center for Gas Analysis, Korea Research Institute of Standards and Science, Daejeon, South Korea
- 3School of Life Sciences and Biotechnology, Korea University, Seoul, South Korea
- 4Department of Bioscience and Biotechnology, Chungnam National University, Daejeon, South Korea
- 5Department of Bioenergy Science and Technology, Chonnam National University, Gwangju, South Korea
Plant seedlings germinating under the soil are challenged by rough soil grains that can induce physical damage and sudden exposure to light, which can induce photobleaching. Seedlings overcome these challenges by developing apical hooks and by suppressing chlorophyll precursor biosynthesis. These adaptive responses are, respectively, regulated by the phytochrome and ethylene signaling pathways via the PHYTOCHROME-INTERACTING FACTORs (PIFs) and the ETHYLENE INSENSITIVE 3 (EIN3)/EIN3-LIKE transcription factors. Although many processes downstream of phytochrome and ethylene signaling are similar, it remains unclear if and where these pathways converge. Here, we show PIFs and EIN3 induce similar changes in the transcriptome without robustly regulating each other’s signaling pathways. PIFs and EIN3 target highly overlapped gene promoters and activate subsets of the co-target genes either interdependently or additively to induce plant responses. For chlorophyll biosynthesis, PIFs and EIN3 target and interdependently activate the expression of HOOKLESS1. HOOKLESS1, in turn, represses chlorophyll synthesis genes to prevent photobleaching. Thus, our results indicate an integration of the phytochrome and ethylene signaling pathways at the level of transcriptional gene regulation by two core groups of transcription factors, PIFs and EIN3.
Introduction
Plants use light not only as an energy source but also as a signal that allows them to monitor their environment and neighboring plants. Plants have multiple types of photoreceptors including phytochromes, cryptochromes, phototropins, zeitlupes, and UVR8. This photoreceptor diversity allows plants to detect a broad spectrum of light stimuli and respond with a wide range of developmental and physiological processes. The phytochromes perceive red and far-red light and respond by regulating seed germination, photomorphogenesis, shade avoidance, and senescence (Mathews, 2006; Franklin and Quail, 2010). In the dark, cytosolic phytochromes exist in an inactive Pr form. Light exposure induces a conformational change to the active Pfr form, which then translocates to the nucleus (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999). In the nucleus, active phytochromes interact with various phytochrome-interacting factors to trigger the global gene expression changes that direct appropriate light responses (Castillon et al., 2007; Bae and Choi, 2008; Leivar and Quail, 2011).
The PIFs (i.e., PIF1, PIF3, PIF4, PIF5, and PIF7) are a group of well-characterized bHLH transcription factors that preferentially interact with active Pfr phytochrome (Ni et al., 1998; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Oh et al., 2004). This interaction inhibits the PIFs, either by dissociating them from their target promoters or by inducing their phosphorylation and subsequent degradation by the 26S proteasome (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005; Al-Sady et al., 2006; Lorrain et al., 2008). PIFs inhibit phytochrome-mediated light responses like seed germination (PIF1), seedling photomorphogenesis (PIF1, PIF3, PIF4, and PIF5), shade avoidance (PIF4, PIF5, and PIF7), and senescence (PIF4 and PIF5). The pif quadruple mutant (pif1/pif3/pif4/pif5, pifq) shows constitutive photomorphogenic phenotypes in the dark including short hypocotyls, opened cotyledons without apical hooks, and hypocotyl agravitropism (Leivar et al., 2008b; Shin et al., 2009). In addition, etiolated pifq seedlings accumulate the precursor of chlorophyll protochlorophyllide, which causes photo-oxidation and bleaching upon sudden light exposure (Huq et al., 2004; Shin et al., 2009; Stephenson et al., 2009). In etiolated wild type seedlings, PIF1 and PIF3 inhibit several chlorophyll-biosynthesis genes [e.g., HEMA1 and CHLH/GENOME UNCOUPLED5 (CHELATASE H/GUN5)] to reduce the accumulation of protochlorophyllide and activate the protochlorophyllide oxidoreductases (POR) that convert protochlorophyllide to chlorophylls upon light exposure (Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009). Thus, PIFs prevent seedling photobleaching by inhibiting the over-accumulation of free protochlorophyllide during emergence.
Ethylene, a gaseous plant hormone, induces in etiolated seedlings the so-called “triple response” of a short, thickened hypocotyl and a root with an exaggerated apical hook. Ethylene is synthesized from methionine through S-adenosylmethionine (SAM) and 1-aminocyclopropane-1-carboxylic acid (ACC) intermediates (Adams and Yang, 1979). ACC synthase (ACS) catalyzes the conversion of SAM to ACC, which is the committed step in ethylene biosynthesis. Then, ACC oxidase (ACO) converts ACC to ethylene. In the absence of ethylene, ethylene receptors (e.g., ETHYELENE RESISTENT 1 (ETR1) and ETR2) act with CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) to inhibit ETHYLENE INSENSITIVE 2 (EIN2). This, in turn, inhibits the degradation of two transcription factors, ETHYLENE INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1) via EBF1 and EBF2, or inhibits the translation of EBF1 and 2 (Zhao and Guo, 2011; Merchante et al., 2013, 2015; Li et al., 2015). Ethylene binds and inhibits the ethylene receptors to stabilize EIN3 and EIL1. The stabilized EIN3 and EIL1 then regulate various downstream targets, including the ETHYLENE RESPONSE FACTORs (ERFs) to induce ethylene responses (Chao et al., 1997; Solano et al., 1998). Another downstream signaling gene, HOOKLESS1 (HLS1, an N-acetyltransferase), is important for ethylene-mediated apical hook formation (Lehman et al., 1996). In short, ethylene signaling is essential for the survival of emerging seedlings (Zhong et al., 2014).
Phytochrome-interacting factors regulate several developmental processes via crosstalk with hormone signaling pathways. For example, PIF1 inhibits seed germination in the dark in part by directly activating the expression of gibberellin (GA) and abscisic acid (ABA) signaling genes like GIBBERELLIN INSENSITIVE (GA INSENSITIVE, GAI), REPRESSOR OF GA1 (RGA1), ABSCISIC ACID INSENSITIVE3 (ABA INSENSITIVE3, ABI3), and ABI5 (Oh et al., 2009). It also indirectly regulates GA and ABA metabolic genes to increase ABA levels and decrease GA levels (Oh et al., 2009). BZR1 and ARF6 are key transcription factors in brassinosteroid (BR) and auxin signaling, respectively. In seedlings, PIF4 directly binds BZR1 and ARF6 to cooperatively bind and regulate the promoters of many shared target genes (Oh et al., 2012, 2014). These shared targets include the PACLOBUTRAZOL RESISTANCE (PRE) family of factors that induce hypocotyl elongation in response to hormonal and environmental signals. GA increases the activities of the PIFs by destabilizing DELLA proteins like GAI and RGA. DELLA proteins directly bind and inhibit PIF DNA-binding and indirectly inhibit the formation of BZR1-PIF4 complexes on target promoters via their interaction with BZR1. BR increases PIF activity either by directly stabilizing PIF4 or by indirectly activating BZR1 and inducing the formation of BZR1-PIF4 complexes.
Several lines of evidence suggest significant crosstalk between phytochrome and ethylene signaling in Arabidopsis seedling development. First, PIF5 overexpression increases ethylene levels in etiolated seedlings (Khanna et al., 2007) by directly binding and activating the ACS promoter (Gallego-Bartolome et al., 2011; Oh et al., 2012). Second, PIF1 and EIN3/EIL1 inhibit photobleaching by inhibiting the expression of protochlorophyllide biosynthetic genes and activating the expression of POR genes (Zhong et al., 2009). Although both PIF1 and EIN3/EIL1 inhibit photobleaching, they seem to function independently as exogenous ACC rescues the excessive photobleaching of pif1 mutants. Third, ethylene promotes hypocotyl elongation in light-grown but not dark-grown seedlings by increasing PIF3 expression (Zhong et al., 2012). Fourth, EIN3 and EIL1 inhibit photobleaching in dark-grown seedlings by directly up-regulating PIF3 (Zhong et al., 2014). Contrary to results with pif1 mutants, exogenous ACC treatment does not rescue the excessive bleaching of pif3 mutants. This suggests ethylene inhibits photobleaching by increasing PIF3 mRNA levels.
Although the PIFs each preferentially regulate specific responses, there is redundancy in their regulation of various aspects of seedling development including seedling morphology, photobleaching, and hypocotyl negative gravitropism. Thus, it is important to determine how PIFs in general interact with other signaling pathways in dark-grown seedlings. Here, we present a systematic study of the relationship between the PIFs and ethylene signaling. The global transcriptional profile of etiolated pifq mutant seedlings is itself suggestive of reduced ethylene signaling. This is consistent with the phenotypic similarities between pifq mutant seedlings and and ethylene-insensitive mutant seedlings. Unexpectedly, however, the lack of PIFs in pifq mutants neither suppresses ethylene biosynthesis during etiolation nor induces significant changes in EIN3 stability. Instead, PIFs and EIN3 bind a highly overlapping set of target genes without affecting one another’s DNA-binding ability. This binding activates their co-targeted genes either interdependently or additively. One of these co-targeted genes, HLS1, prevents photobleaching by regulating the expression of chlorophyll synthesis genes. Together, our results demonstrate the phytochrome and ethylene signaling pathways converge at the promoters of genes simultaneously targeted by PIFs and EIN3.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown at 22–24°C under long days (16 h light/8 h dark) in a growth room with cool-white fluorescent light (90–100 μmol m-2 s-1) for general growth and seed harvest. Mutants and transgenic lines are described in Supplementary Table S1. For phenotypic analyses, surface-sterilized seeds were plated on Murashige and Skoog (MS, Duchefa, M0222) agar plates (half-strength MS, 0.8% phytoagar, and 0.05% MES, pH 5.7), stratified for 3 days at 4°C in darkness, transferred to white light for 3 h to synchronize seed germination, and grown in different experimental conditions. ACC (Sigma, A3903) was dissolved in water, ethephon (Sigma, C-0143) was dissolved in DMSO, and AgNO3 (Sigma, S-0139) was dissolved in either water or DMSO depending on the experiment. For molecular experiments, seedlings were grown under darkness or under red light (13 μmol m-2 s-1) for the indicated period.
Photobleaching and Protochlorophyllide Levels
For photobleaching assays, seedlings grown in the dark for the indicated period were transferred to continuous white light (100 μmol⋅m-2⋅s-1) for 3 days. Then, bleached seedlings were counted. For protochlorophyllide quantification, 10 seedlings grown in the dark for 4 days were gently agitated in 1 mL of ice-cold 80% acetone for 1 h in the dark at 4°C to extract pigments. The protochlorophyllide level in 100 μl of supernatant was determined using a fluorescence spectrophotometer (Tecan, infinite 200 PRO) with an excitation wavelength of 440 nm, a bandwidth of 4 nm, and an emission wavelength of 600–720 nm.
Ethylene Levels
Ethylene levels were measured in 100 seedlings grown in 14 ml vials containing 10 ml growth medium and 4 ml headspace. The vials were refreshed with hydrocarbon-free air before they were sealed gas-tight and further incubated for 24 h in the dark. The headspace air was retrieved and ethylene was quantified by gas chromatography (Hewlett-Packard, 5890 series II).
qPCR
Total RNA from plant tissues was isolated using a plant total RNA extraction kit (Sigma). First-strand cDNAs were prepared with 2 μg of total RNA and M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Gene expression levels were determined by qPCR using SYBR green on a CFX Connect machine (Bio-Rad). Gene expression was normalized to PP2A as an internal control. The gene-specific primers used for qPCR are listed in Supplementary Table S2.
SDS-PAGE and Immunoblot Analysis
Seedlings were harvested and flash-frozen in liquid nitrogen under a dim green light. The seedlings were then ground in liquid nitrogen and homogenized in denaturing buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 8.0) by vigorous vortexing. The debris was removed by centrifugation at 20,000 ×g for 10 m at 4°C. For immunoblot analysis, the supernatants were separated on an 8% SDS-polyacrylamide gel. Then, the proteins were transferred to a nitrocellulose membrane (Hybond ECL, Amersham) using transfer buffer (5.8 g l-1 Tris base, 29 g l-1 glycine, 20% methanol, and 0.01% SDS). A rabbit polyclonal anti-EIN3 antibody for native EIN3 (Kim et al., 2013), a rabbit polyclonal anti-Myc antibody (Santa Cruz, CA, USA) for PIF4-Myc, a mouse monoclonal anti-FLAG antibody (Sigma, USA) for EIN3-FLAG, and a mouse monoclonal anti-tubulin antibody (Sigma, USA) for the loading control were used for protein detection. All antibodies were diluted in PBS buffer containing 0.05% Tween 20. Blots were washed three times with the same buffer and then incubated with the appropriate secondary antibodies. After washing three times, the horseradish peroxidase activity of the secondary antibodies was detected using an ECL detection kit (AbFRONTIER, Korea).
Chromatin Immunoprecipitation (ChIP)
Plants overexpressing GFP-Myc, PIF1-Myc, PIF3-Myc, PIF4-Myc, PIF5-Myc, or EIN3-FLAG were grown for 4 days under the indicated conditions before cross-linking for 20 m with 1% formaldehyde under vacuum. Chromatin complexes were isolated and sonicated as described with slight modifications (Oh et al., 2009). An anti-Myc monoclonal antibody (mouse, Cell Signaling) or an anti-FLAG polyclonal antibody (rabbit, Sigma), and Protein A agarose/salmon sperm DNA (Millipore) were used for immunoprecipitation. After reverse cross-linking and protein digestion, DNA was purified using the QIAquick PCR Purification Kit (Qiagen) before being used for qPCR.
Microarray and ChIP-Chip/Seq Analysis
All microarray analysis was performed with R version 2.15.0. The limma package was used for background correction and intra- and inter-array normalization. Then, lmFit was used to fit a linear model to the data so statistical calculations could be made using ebayes. ChIP-chip/Seq data was mapped to the TAIR10 genome using bowtie, analyzed by CisGenome v2.0, and visualized by IGV v2.3. Gene Set Enrichment Analysis was conducted with the GSEA package v2.08 (Broad Institute, MIT) according to the online user guide. ACC-responsive gene sets were generated based on published microarray data (Nemhauser et al., 2006).
Accession Numbers
PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), ETR1 (AT1G66340), ETR2 (AT3G23150), ERS1 (AT2G40940), ERS2 (AT1G04310), EIN4 (AT3G04580), CTR1 (AT5G03730), EIN2 (AT5G03280), EIN5 (AT1G54490), EBF2 (AT5G25350), EIN3 (AT3G20770), EIL1 (AT2G27050), EBP (AT3G16770), ACO1 (AT2G19590), ERF1 (AT3G23240), EXP9 (AT5G02260), FHL (AT5G02200), HB52 (AT5G53980), AHP1 (AT3G21510), HLS1 (AT4G37580), LOG5 (AT4G35190), GRF2 (AT4G37740), CEL1 (AT1G70710), BRG3 (AT3G12920), SBP1 (AT4G14030), GUN4 (AT3G59400), CHLH/GUN5 (AT5G13630), HEMA1 (AT1G58290), PORA (AT5G54190), PORB (AT4G27440), PORC (AT1G03630), PP2A (AT1G13320), EF1ALPHA (AT5G60390).
Results
The Transcriptome Profile of the pif quadruple Mutant Is Consistent with Reduced Ethylene Responses
The pifq mutant and the ethylene-related mutants etr1, ein2, and ein3 show open cotyledons in the dark and excessive photobleaching when transferred to the light (Supplementary Figure S1). This suggests PIF signaling and ethylene signaling interact. We therefore analyzed four published microarray datasets that compare dark-grown pifq mutants to wild type or ethylene-treated dark-grown wild type to non-treated wild type. These four microarray datasets overlap on 378 genes with significantly altered levels of expression (moderated t-statistic, P < 0.05). Of these 378 genes, 196 show an inverse correlation in the pifq mutants versus wild type ethylene-treated seedlings (Figure 1A). This suggests PIFs positively regulate many ethylene-responsive genes. We next used Gene Set Enrichment Analysis (GSEA) to determine whether ethylene-responsive genes are statistically enriched among PIF-regulated genes. For the analysis, we divided all ethylene-responsive genes into those showing up-regulation of the ethylene precursor ACC (ACC up) and those showing down-regulation of ACC (ACC down). We found ACC up-regulated genes tend to be significantly down-regulated in the pifq mutant, while ACC down-regulated genes tend to be significantly up-regulated in the pifq mutant (Figure 1B). We also observed suppression of several well-known ethylene-responsive markers in both 2- and 4-day-old dark-grown pifq seedlings (Figure 1C). Together, these results indicate PIFs and ethylene regulate large numbers of genes significantly in the same direction. This is consistent with the reduced ethylene signaling phenotypes of the pifq mutants.
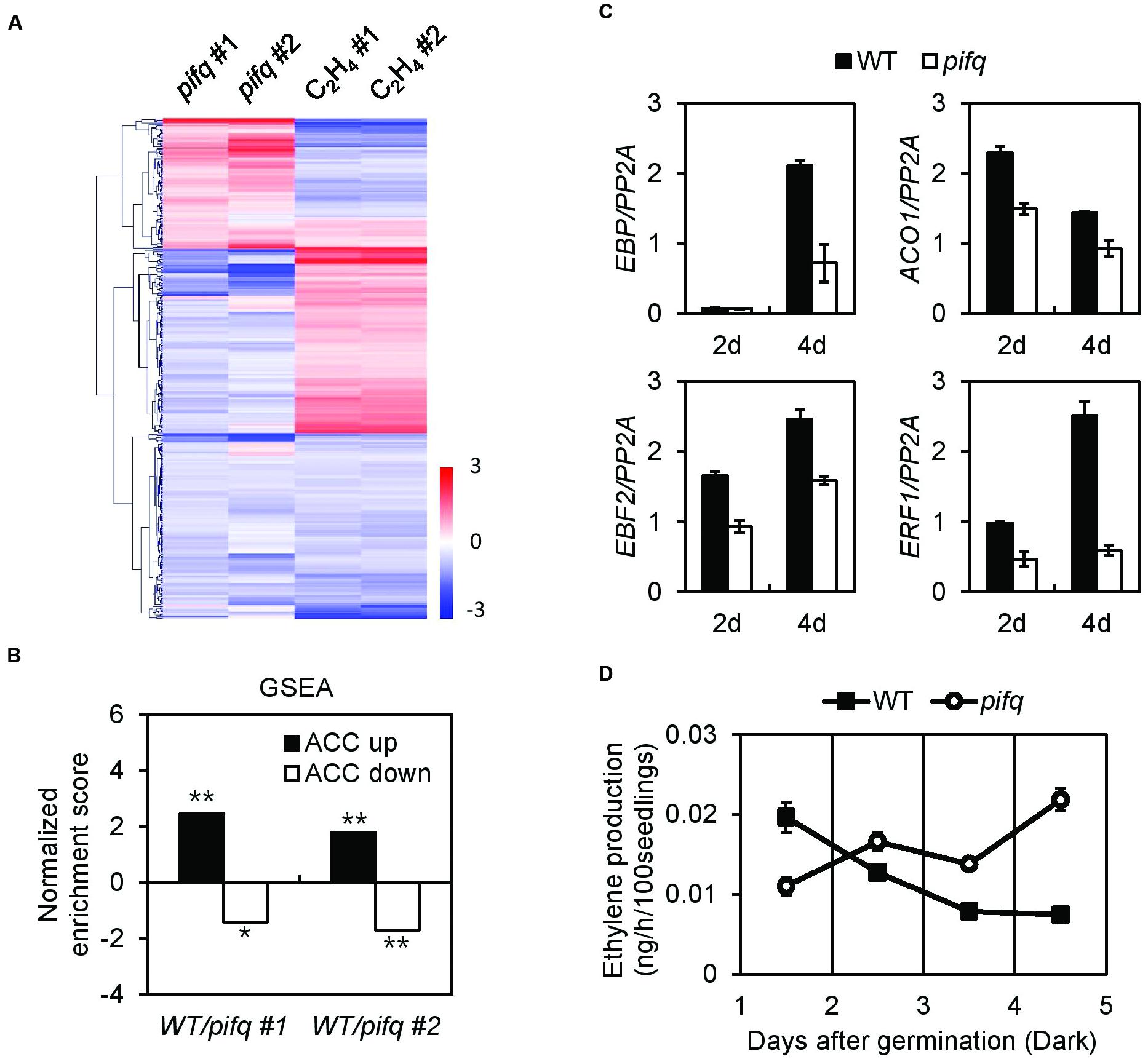
FIGURE 1. Ethylene responses are suppressed in pifq mutants regardless of endogenous ethylene levels. (A) Hierarchical clustering of two independent pifq microarrays and two independent ethylene treatment microarrays from dark-grown seedlings. 378 genes with P < 0.05 from the four microarrays were selected and analyzed. (B) Gene set enrichment analysis (GSEA) of the two independent pifq microarrays shows PIFs significantly up-regulate an ACC-inducible gene set and significantly down-regulate an ACC-repressible gene set (∗FDR = 0.086, ∗∗FDR < 0.005). (C) Expression of four representative ethylene marker genes from 2- and 4-day-old WT and pifq mutant etiolated seedlings. PP2A was used as an internal qPCR control. SD, n = 3 biological replicates. (D) Quantification of endogenous ethylene from WT and pifq mutants (SD, n = 3 biological replicates; ∗P < 0.05, Student’s t-test).
Ethylene-Responsive Gene Suppression in pifq Mutants Is Uncorrelated with Endogenous Ethylene Levels
Since dark-grown PIF5-overexpressing seedlings synthesize far more ethylene than wild type seedlings, we asked whether the suppression of ethylene-responsive genes in pifq mutants is due to low levels of ethylene production. Thus, we measured endogenous ethylene levels in dark-grown pifq mutant seedlings at 24 h intervals (Figure 1D). Surprisingly, we found PIFs do not robustly promote ethylene biosynthesis. From 1 to 2 days after germination, we found the pifq mutants produce roughly half the ethylene wild type seedlings produce. This pattern reverses after day 2 with the pifq mutants producing higher levels of ethylene than wild type (Figure 1D). These results are inconsistent with the hypothesis that reduced ethylene biosynthesis is responsible for the suppression of ethylene-responsive genes in pifq mutants (Figure 1C). We further examined the expression of well-known ethylene-inducible genes (i.e., EBP, ETR2, and ERS2) in the presence of either a saturating level of the ethylene perception inhibitor silver nitrate (AgNO3) or the ethylene-producing compound ethephon. Ethylene induces and AgNO3 represses these ethylene-inducible markers in both wild type and pifq mutants (Figure 2A). Interestingly, the expression of these marker genes is lower in the pifq mutants than in wild type seedlings at all doses of AgNO3 and ethephon (Figure 2A). On the other hand, PIF4- and PIF5-overexpressing lines show increased marker gene expression for the same doses of AgNO3 (Figure 2B). In these experiments, the AgNO3 dose (20 μM) was high enough to fully suppress ethylene responses even with simultaneous treatment of excessive ethylene (Figures 2C,D). These results, thus, suggest PIFs promote the expression of ethylene-responsive genes regardless of endogenous ethylene levels.
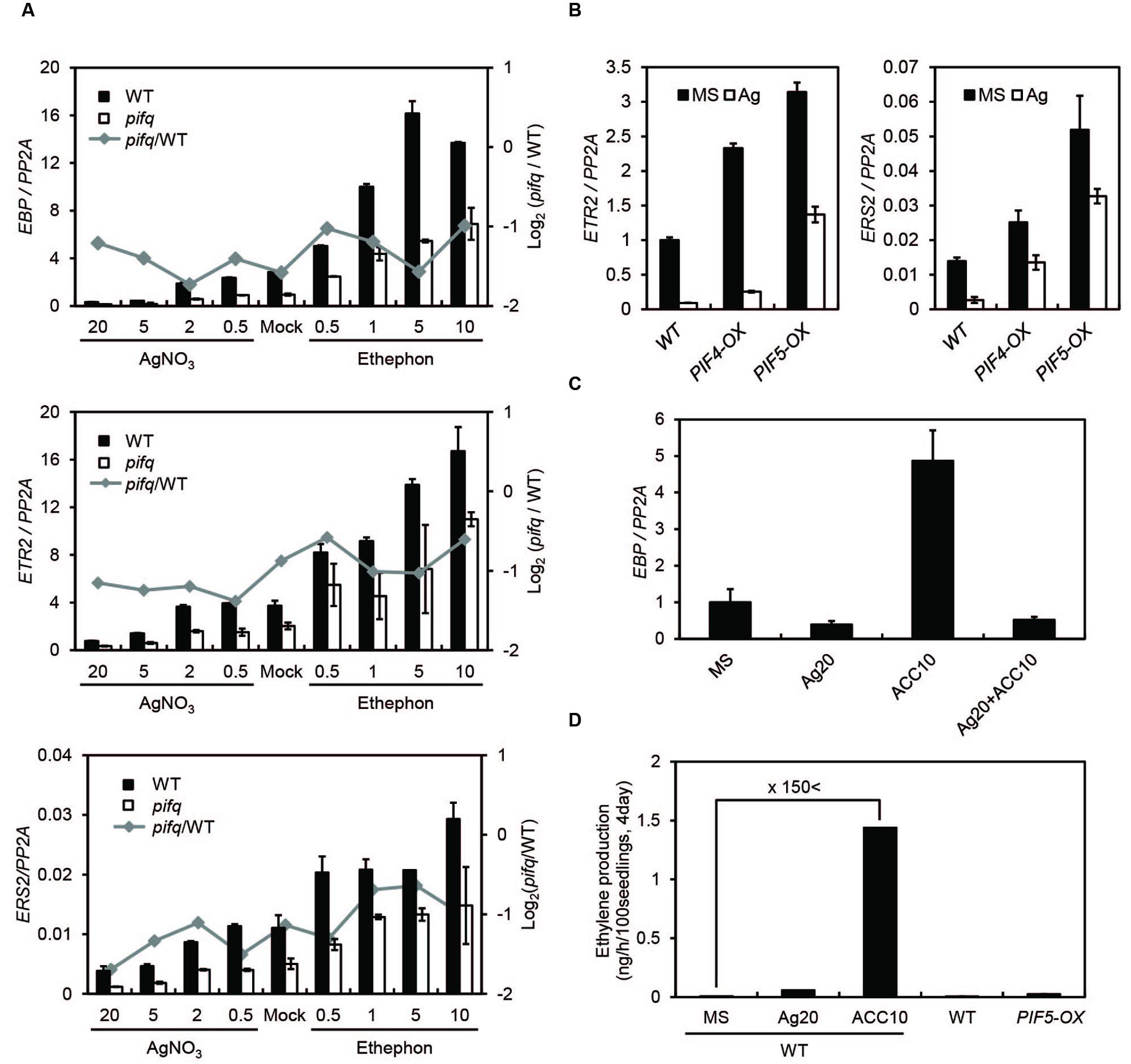
FIGURE 2. Phytochrome-interacting factors promote ethylene marker gene expression independent of ethylene. (A) Expression of the representative ethylene markers EBP, ETR2, and ERS2 in 4-day-old WT and pifq mutant etiolated seedlings. Seedlings were untreated (mock) or treated with AgNO3 or ethephon at the indicated concentrations (μM). The gray line indicates log2 fold changes between pifq mutants and WT. (B) Expression of the representative ethylene markers ETR2 and ERS2 in 4-day-old WT, PIF4-OX, and PIF5-OX etiolated seedlings untreated (mock) or treated with AgNO3 (20 μM). (C) Expression levels of EBP from 4-day-old WT etiolated seedlings untreated (mock) or treated with AgNO3 (20 μM), ACC (10 μM), or AgNO3+ACC. PP2A was used as an internal qPCR control (SD, n = 3 biological replicates). (D) Quantification of endogenous ethylene from 4-day-old WT and PIF5-OX seedlings treated as indicated (SD, n = 2 biological replicates).
PIF Signaling Does Not Significantly Affect Ethylene Signaling Upstream of EIN3
Reduced expression of ethylene signaling genes may explain the reduced responsiveness of ethylene marker genes in pifq mutants in response to exogenous ethylene. We thus examined whether PIFs regulate the expression of ethylene signaling genes (Supplementary Figure S2A). Although we did find via microarray (Supplementary Figure S2B) and qRT-PCR (Supplementary Figure S2C) that some ethylene signaling genes are significantly suppressed in pifq mutants, the suppressed genes include both positive (EIN2) and negative regulators of ethylene signaling (ETR2 and CTR1). This complicates any prediction of their net effect on ethylene responses. Since ethylene signaling pathway ultimately impinges on EIN3 protein stabilization (Supplementary Figure S2A), we examined whether the ethylene-mediated EIN3 protein stability is affected in the pifq mutant. However, the EIN3 proteins were stabilized by both ACC and ethephon in the pifq mutants, just as it is in wild type (Supplementary Figure S3A). Red light treatment has no effect on ethephon-induced stabilization of EIN3 (Supplementary Figure S3B), nor is EIN3 expression significantly altered in pifq mutants or by red light treatment (Supplementary Figure S3C). These results suggest the transcriptional regulation of ethylene signaling genes by PIFs does not significantly affect ethylene signaling upstream of EIN3. Instead, the altered expression of ethylene responsive genes in pifq mutants may relate to the activity of EIN3 itself.
PIFs and EIN3 Directly Regulate Overlapping Target Genes
It is possible EIN3 activity depends on the activity of the PIFs. By comparing the known genome-wide targets of four PIFs (PIF1, PIF3, PIF4, and PIF5) with those of EIN3, we found a significant number of shared targets (584 genes, hypergeometric test, p < 10-90; Figure 3A). These shared targets account for 11% of the PIF targets and 44% of the EIN3 targets. We next examined pifq and ein3/eil1 microarrays (Shin et al., 2009; Zhong et al., 2009) to determine how PIFs and EIN3 regulate the expression of these shared targets (class c from Figure 3A). Although only 331 of the 584 shared targets were included in the microarray analyses (Figure 3B), we found 51 of 331 are significantly regulated by both PIFs and EIN3/EIL1, mostly in the same direction (Figure 3C). We observed enriched binding of both PIF4 and EIN3 in the promoter regions of their shared targets (Supplementary Figure S4B). Furthermore, many of the PIF4 and EIN3 binding peaks precisely overlap (Figures 3D,E) and most of the binding peaks fall within 200 bp of each other (Supplementary Figure S4C, blue). In other words, these two transcription factors bind closely to one another on their shared target promoters. To exclude the possibility that this proximity of the PIF4 and EIN3 binding peaks is attributable to chance, we selected PIF4 and EIN3 binding peaks from random target promoters instead of from the same promoter and then calculated the distances between each peak (Supplementary Figure S4A). Compared to the random peak distances (Supplementary Figure S4C, orange), we found the actual PIF4-EIN3 peaks (Supplementary Figure S4C, blue) are strongly biased toward shorter inter-peak distances. This supports the binding of PIFs and EIN3 to their shared targets in close proximity.
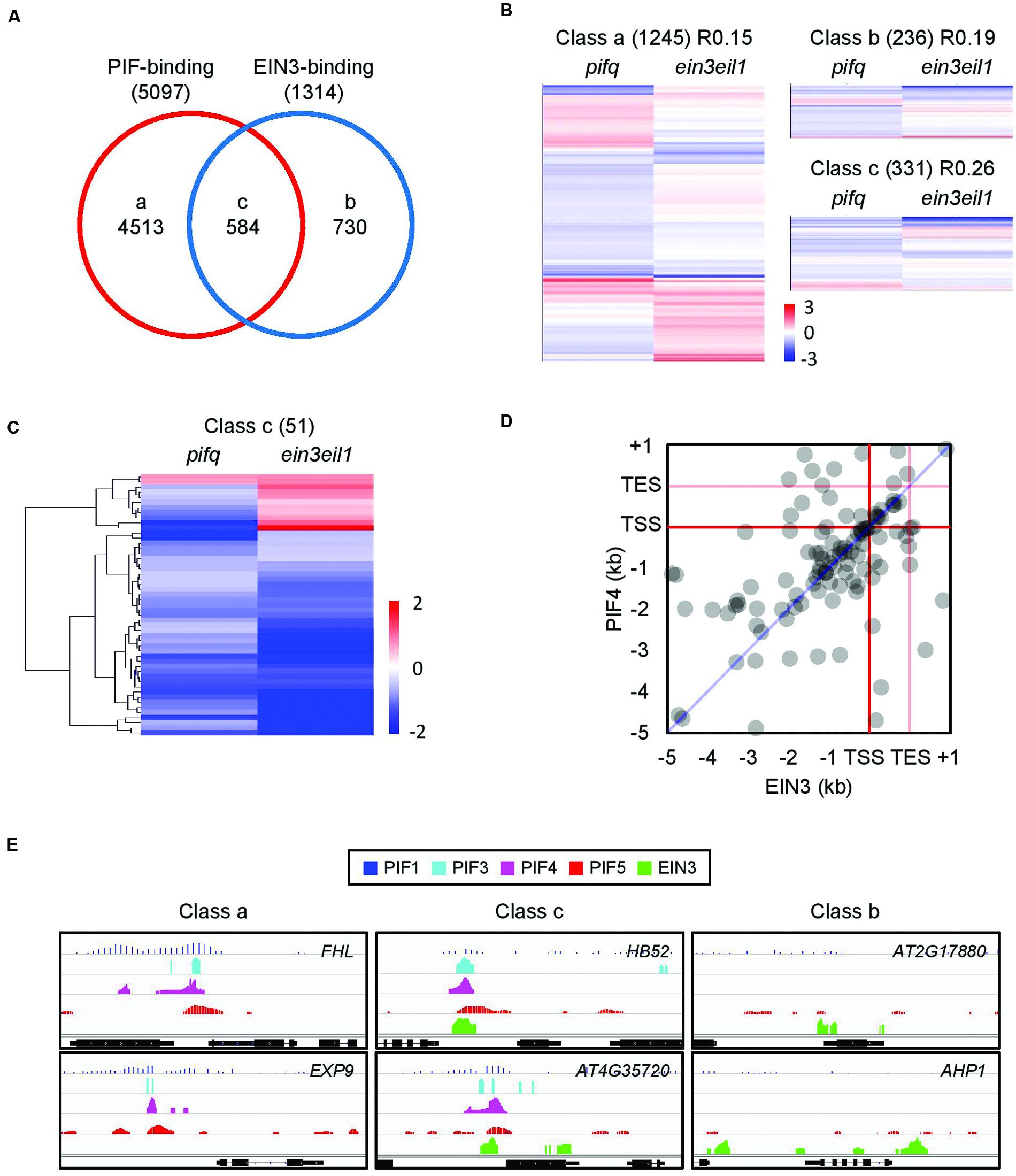
FIGURE 3. Phytochrome-interacting factors and EIN3 bind and regulate overlapping target genes. (A) PIF-binding and EIN3-binding genes significantly overlap. Data were retrieved from relevant ChIP-chip or ChIP-seq databases (Oh et al., 2009, 2012; Hornitschek et al., 2012; Chang et al., 2013; Zhang et al., 2013). Lowercase letters indicate PIF and/or EIN3 binding classes. (B) Hierarchical clustering of the expression patterns for each gene class from (A). Gene expression data were retrieved from previous pifq and ein3/eil1 microarrays (Shin et al., 2009; Zhong et al., 2009). Genes whose expression data correspond to P < 0.05 in at least one of the two microarrays are shown. (C) Hierarchical clustering of the expression patterns for class c genes. Fold changes were rescaled to ± 2 (Log2) and only genes whose expression data correspond to P < 0.05 in both microarrays are shown. Gene expression data were retrieved from the same source as (B). (D) Spatial distribution of PIF4- and EIN3-binding peaks along the promoter (-5 kb to start), coding (TSS, TES) and 3′ (TES to +1 kb) regions of shared target genes. Binding peak data were retrieved from published ChIP-seq experiments performed on PIF4 and EIN3 (Oh et al., 2012; Chang et al., 2013). The analysis was re-done as described in the section “Materials and Methods.”. (E) Representative PIF- and EIN3-binding peaks in the promoters of shared or non-shared target genes from the gene classes defined in (A).
PIFs and EIN3 Bind Independently to Their Target Promoters
The proximity of the PIF and EIN3 binding peaks suggests PIFs and EIN3 may enhance one another’s binding to their shared target promoters. Since red light dissociates PIFs from their target promoters and enhances PIF degradation (Park et al., 2004, 2012; Shen et al., 2005; Al-Sady et al., 2006; Oh et al., 2006; Lorrain et al., 2008), it may also inhibit EIN3 binding to promoters it co-targets with PIFs. We therefore performed a ChIP assay with transgenic plants expressing FLAG-tagged EIN3 grown either in the dark or under red light (Figure 4A). After confirming EIN3 protein stability is unaffected by red light (Figure 4B), we found red light does not significantly affect EIN3 binding to four PIF co-targeted promoters (i.e., those of HLS1, GRF2, LOG5, and SOB3) or to two non-binding control promoters (i.e., those of EF-1alpha and FHL; Figure 4A). This suggests EIN3 binding to the target promoters it shares with the PIFs is independent of PIF binding. We next asked whether ethylene signaling enhances PIF binding to co-targeted promoters by performing a ChIP assay with transgenic plants expressing MYC-tagged PIF4 grown in the presence of either AgNO3 or ACC (Figure 4C). Since ethylene stabilizes EIN3, AgNO3 should inhibit and ACC should enhance the binding of PIF4 to PIF4/EIN3 co-targeted promoters if EIN3 is required for PIF4 binding. After confirming neither AgNO3 nor ACC treatment significantly alters PIF4 protein stability (Figure 4D), we found PIF4 binds equally to four co-targeted promoters regardless of the presence of AgNO3 or ACC (Figure 4C). This suggests the binding of PIF4 to its target promoters is independent of ethylene signaling. Together, these results suggest PIFs and EIN3 independently bind their target promoters.
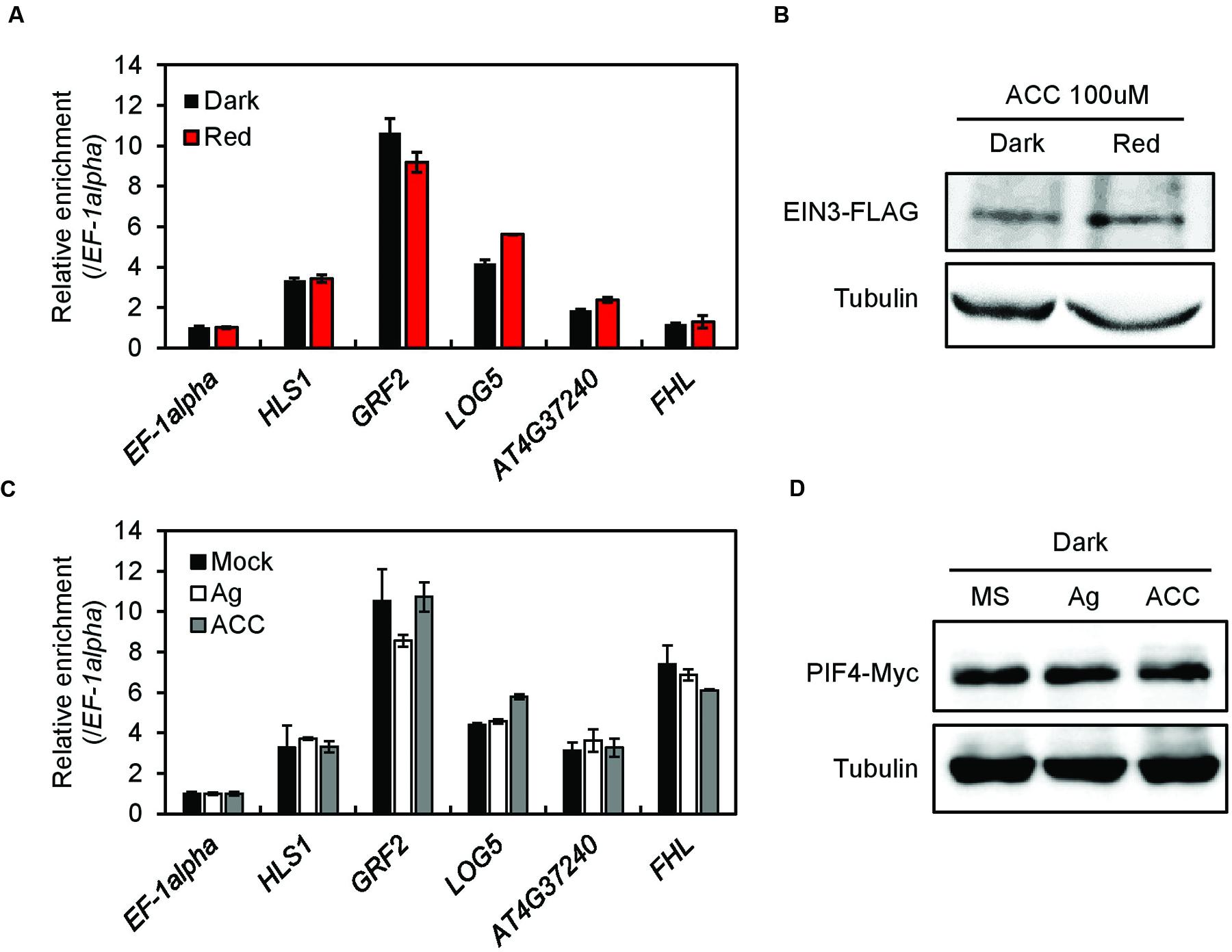
FIGURE 4. Phytochrome-interacting factors and EIN3 bind independently to their shared target promoters. (A,C) ChIP assay for EIN3 and PIF4 binding to shared target promoters (HLS1, GRF2, LOG5, and AT4G37240) under dark and red light conditions (A) or under darkness untreated or treated with 20 μM AgNO3 or 10 μM ACC (C). Fold enrichment was calculated by normalizing the results with respect to an input control and a control region, EF-1-alpha (SD, n = 3 biological replicates). FHL was used as a PIF-specific target gene. (B,D) EIN3-FLAG and PIF4-Myc protein levels were measured under the indicated conditions. Tubulin was used as a loading control.
PIFs and EIN3 Regulate the Expression of Their Shared Targets Either Interdependently or Additively
Phytochrome-interacting factors and EIN3 bound to the same promoters may independently or interdependently regulate the expression of their shared targets. We therefore measured the expression of their shared targets in pifq, ein2, and pifq;ein2 quintuple mutants. EIN3 and EIL protein levels are very low in ein2 mutants because they are constitutively degraded by EBF1 and EBF2 (Guo and Ecker, 2003; Potuschak et al., 2003). Some of the shared target genes (e.g., LOG5 and AT4G37240) are equally repressed in the pifq, ein2, and pifq;ein2 mutants compared to wild type, suggesting PIFs and EIN3 interdependently activate their expression (Figure 5A). The expression levels of other shared target genes (e.g., HLS1 and GRF2) are slightly higher in the pifq mutant than in the ein2 and pifq;ein2 mutants (Figure 5A). Since the rest of the PIFs (e.g., PIF7) remain active in pifq mutants, they may be responsible for the residual HLS1 and GRF2 expression observed in pifq mutants. To remove the residual PIF activities, we treated wild type seedlings with red light because red light robustly suppresses all PIFs (including PIF7) via Pfr phytochrome (Leivar et al., 2008a). The red light treatment of wild type seedlings reduced shared target expression to the levels observed in ein2 mutants and unlike in wild type seedlings, ein2 mutants showed low expression of co-targeted genes regardless of red light treatment (Figure 5B). On the other hand, the expression of shared target genes (HLS1 and LOG5) in ein3;eil1 double mutants was reduced by the red light treatment (Supplementary Figure S5) suggesting that the activity of residual EIN3-like protein (e.g., EIL2) is also dependent on PIF activities. Taken together, our results suggest both PIFs and ethylene signaling are required for high expression of these co-targeted genes in the dark. However, not all the shared target genes are interdependently regulated by PIFs and EIN3 as shown by further decreased expression of other subset of co-targeted genes (BRG3 and SBP1; Figure 5B). These results support the hypothesis that PIFs and EIN3 either interdependently or additively activate the expression of their shared targets.
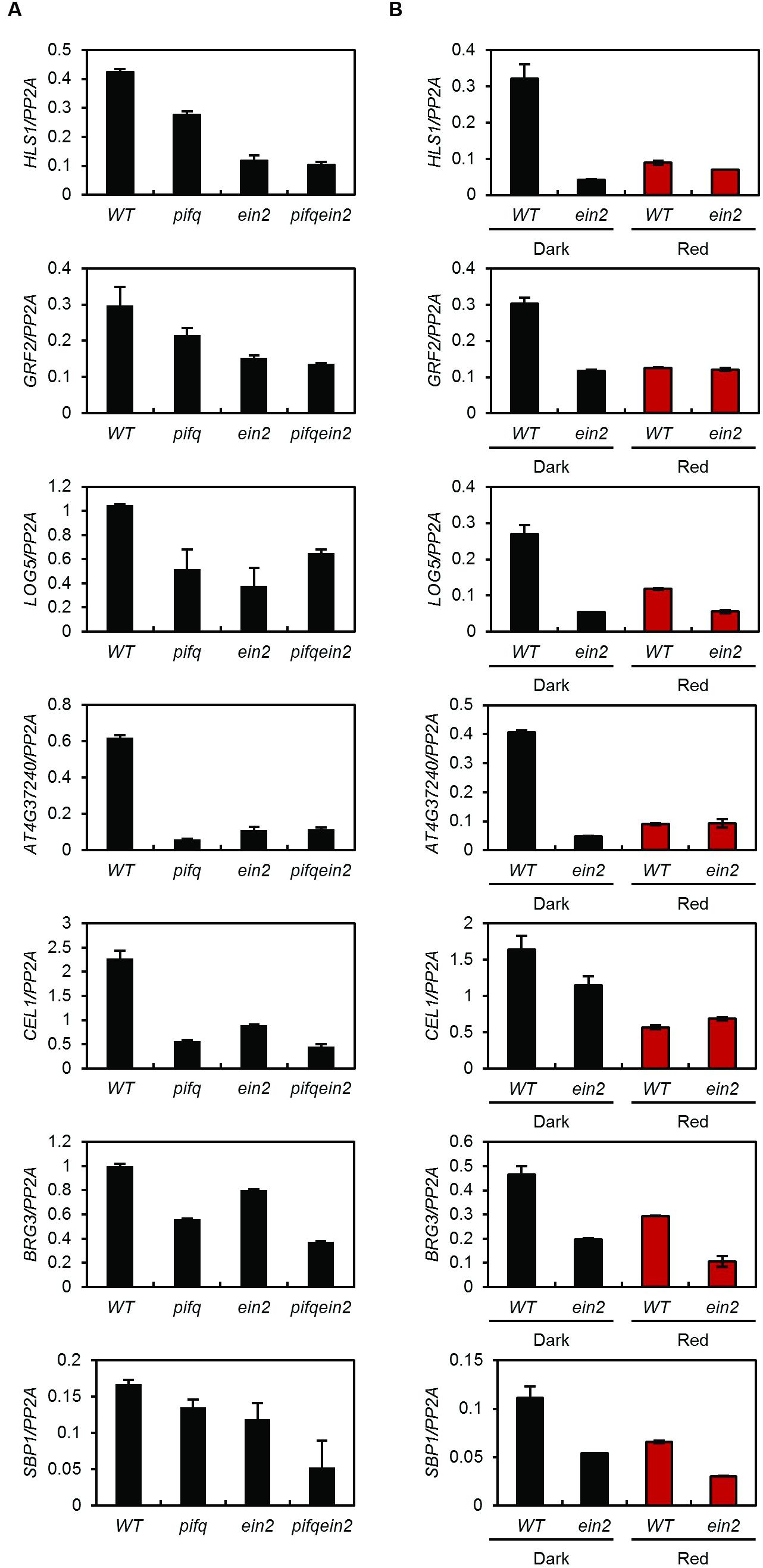
FIGURE 5. Phytochrome-interacting factors and EIN3 interdependently activate a subset of their shared target genes. Expression of shared target genes in 4-day-old WT, pifq, ein2, and pifq ein2 etiolated seedlings treated with 250 nM ACC (A) or in 4-day-old WT and ein2 MS-medium-grown seedlings under dark or red light conditions (B). PP2A was used as an internal control (SD, n = 3 biological replicates).
HLS1 Regulates the Expression of Chlorophyll Biosynthesis Genes
Hookless1 mediates the ethylene-induced formation of the apical hook. Since HLS1 is one of the genes co-targeted by PIFs and EIN3 and since PIFs and EIN3 are required for the prevention of photobleaching in etiolated seedlings, we asked whether HLS1 is also involved in the prevention of photobleaching. When 4-day-old etiolated wild type seedlings are transferred to white light, their cotyledons turn green. However, the cotyledons of hls1 mutant seedlings fail to turn green and are instead photobleached (Figure 6A). Photobleaching occurs when protochlorophyllide over-accumulates, and etiolated hls1 seedlings consistently accumulate more protochlorophyllide than wild type seedlings (Figure 6B). We therefore measured the expression of a series of chlorophyll biosynthesis genes in the hls1 mutants. Consistent with their photobleaching phenotype, hls1 mutant seedlings show high expression of HEMA1 and CHLH and low expression of PORA and PORB. Together, these results suggest PIFs and EIN3 prevent photo-oxidative damages of etiolated seedlings in the dark to light transition by activating HLS1, which represses HEMA1 and CHLH and activates PORA and PORB.
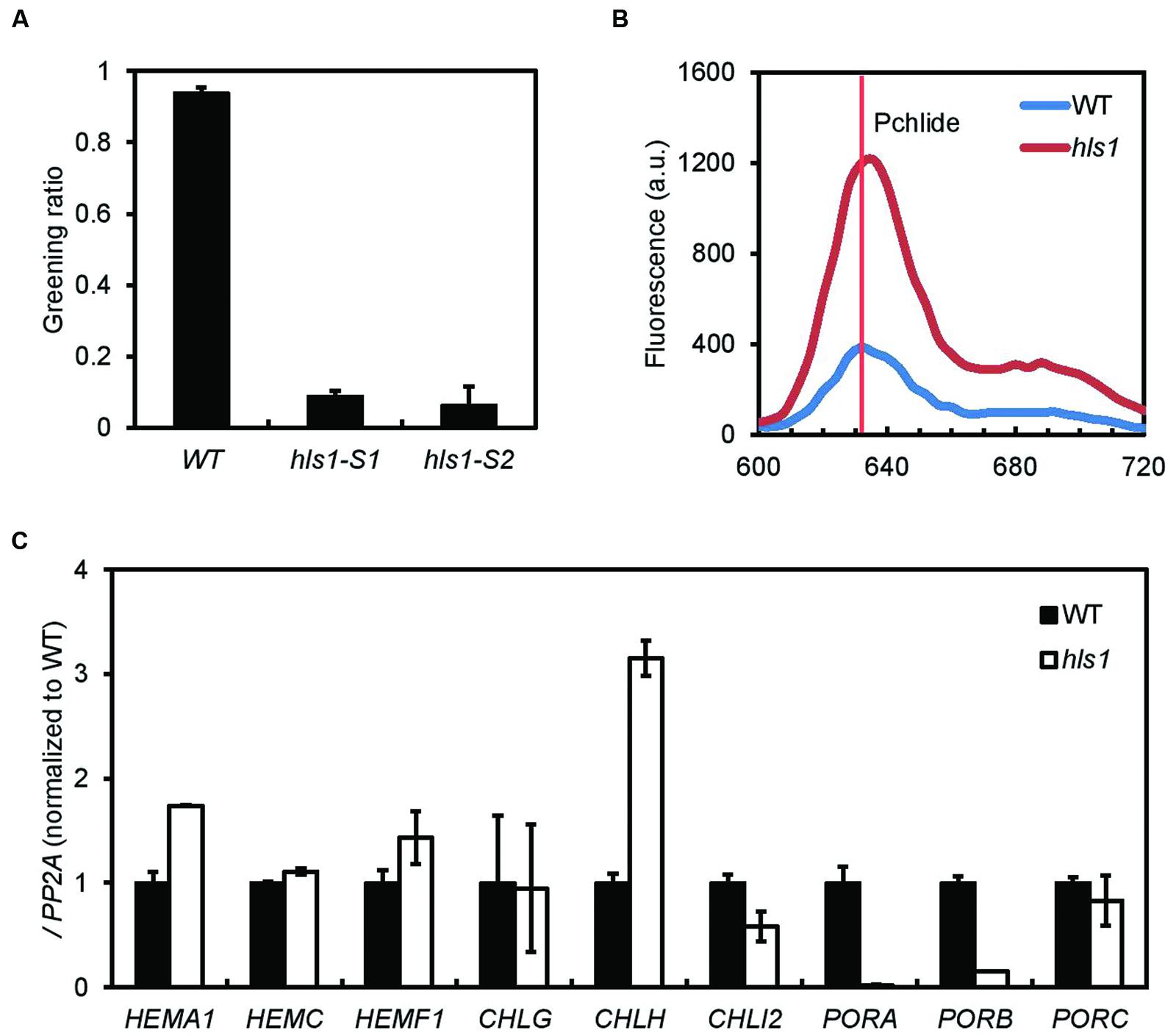
FIGURE 6. Hookless1 inhibits photobleaching by regulating chlorophyll synthesis gene expression. (A) Excessive photobleaching of hls1 mutants during the dark to light transition (SD, n = 2 independent experiments; n > 40 for each experiment). (B) Increased protochlorophyllide levels in dark-grown hls1 mutants as measured by fluorescence. (C) Expression levels of chlorophyll synthesis genes in 4-day-old WT and hls1 etiolated seedlings. PP2A was used as an internal control (SD, n = 3 biological replicates).
Discussion
The phytochrome and ethylene signaling pathways antagonistically regulate apical hook formation and chlorophyll biosynthesis in etiolated seedlings. The precise molecular integrations between these two signaling pathways in seedling development, however, are unknown. In this study, we present evidence the phytochrome and ethylene signaling pathways are integrated at the level of the transcriptional control of shared targets by the PIFs and EIN3. We found via microarray analysis a down-regulation of ethylene-responsive genes in pifq mutants. Since the pifq mutants show neither reduced ethylene nor reduced EIN3 protein levels, the down-regulated ethylene response suggests a reduction in EIN3 activity in the absence of PIFs. We found via ChIP analysis that PIFs and EIN3 share a significant number of direct target genes, which they either interdependently or additively activate (Figure 7). HLS1, one of these shared PIF/EIN3 targets, is required in etiolated seedlings to prevent photobleaching and to form the apical hook. This demonstrates the phytochrome and ethylene signaling pathways converge at the promoters of genes co-regulated by PIFs and EIN3.
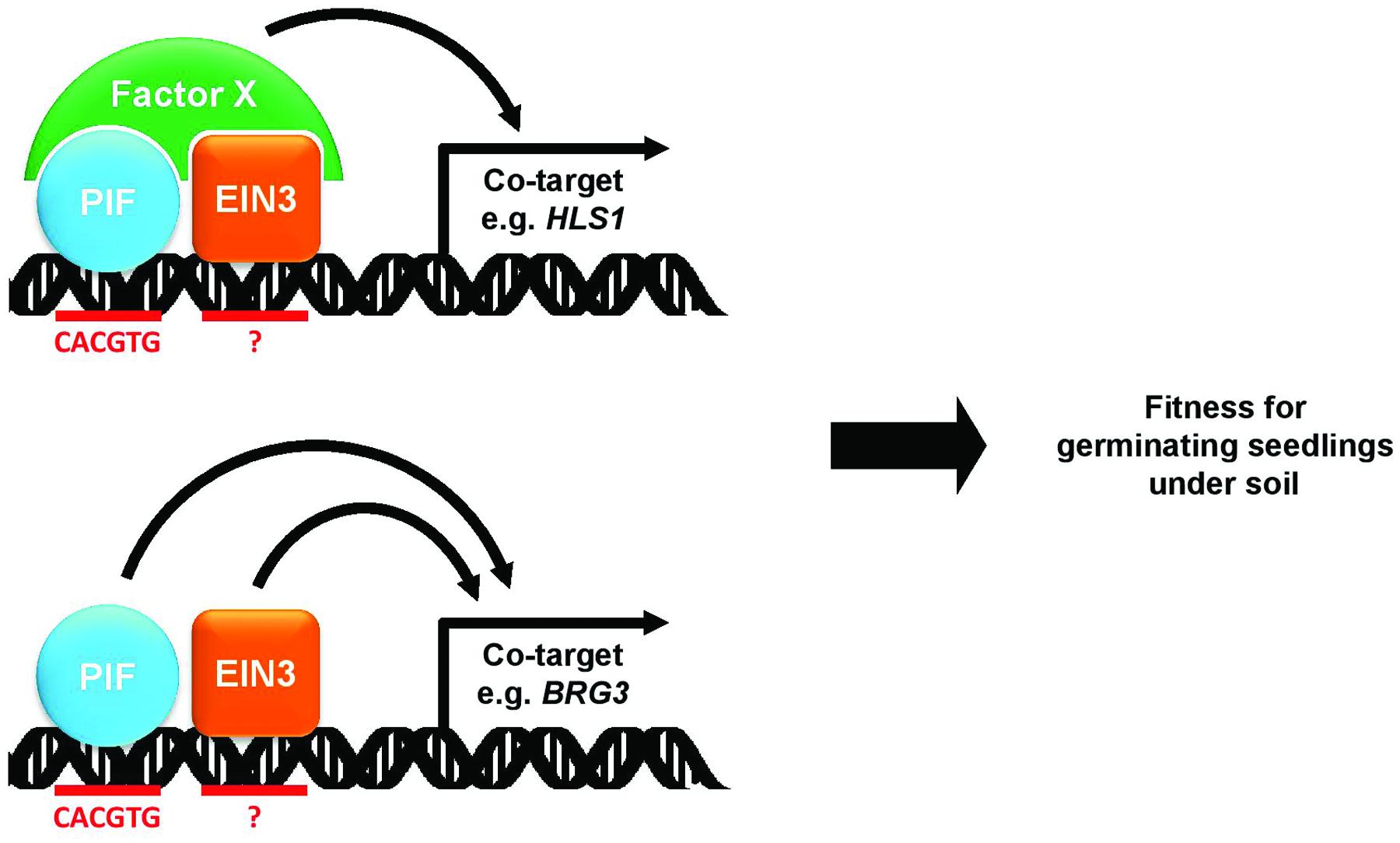
FIGURE 7. A model for the interdependent and additive activation of shared PIF and EIN3 target genes. PIF and EIN3 transcription factor bind to shared targets in a close proximity. In the interdependent mode (upper), two transcription factors recruit factor X which in turn activates the target gene expression. In the additive mode (bottom), two transcription factors independently activate the target gene expression. Through both modes of target gene regulation, PIF and EIN3 increase fitness of seedlings that germinate under soil.
In a microarray analysis, we found a significant overlap between PIF- and ethylene-regulated genes in etiolated seedlings. Consistent with the similar roles these signaling pathways play in etiolated seedling development, most of these overlapping target genes are regulated by PIFs and EIN3/EIL1 in the same direction (Figure 3C). According to published reports, PIFs activate the expression of some ethylene biosynthesis genes including ACSs, and PIF5-OX seedlings produce more ethylene than wild type seedlings (Khanna et al., 2007). We found, however, pifq mutants only produce less ethylene than wild type for the first 2 days post-germination. After that, pifq mutants produce more ethylene than wild type (Figure 1D). Thus, PIF-induced ethylene biosynthesis cannot be responsible for the reduced expression of ethylene-responsive genes in 4-day-old pifq mutant seedlings (Figure 1C). Moreover, PIF4/5-OX increases the expression of ethylene-responsive genes even in the presence of saturating concentrations of the ethylene perception inhibitor AgNO3 (Figure 2B). These results suggest PIFs directly regulate ethylene signaling independent of ethylene biosynthesis and ethylene perception.
The pifq mutants also show repression of some ethylene signaling components compared to wild type. These repressed genes, however, include both positive and negative regulators of ethylene responses (Supplementary Figure S2). In addition, neither the pifq mutation nor red light treatment alter EIN3 protein levels (Supplementary Figures S3A,B; Figure 4B). Since EIN3 protein levels are tightly regulated by ethylene signaling (Guo and Ecker, 2003; Potuschak et al., 2003), PIFs likely affect ethylene signaling downstream rather than upstream of EIN3. Indeed, we found PIFs cooperate with EIN3 to regulate the expression of ethylene-responsive genes. Our ChIP-Seq analyses showed PIFs and EIN3 share many target genes and bind to their co-regulated promoters in close proximity (Figures 3A,D,E). This suggests the large overlap we observed between PIF- and ethylene-regulated genes is due to extensive overlap of the DNA binding loci of the PIFs and EIN3. Furthermore, PIFs and EIN3 interdependently activate the expression of a subset of their shared targets (Figure 5). This ensures the transcriptional activation of these shared target genes only when light signaling is inactive and ethylene signaling is active.
We expected these two transcription factors interdependently regulate gene expression by enhancing one another’s DNA-binding ability, presumably via a direct protein–protein interaction. We were unable to observe, however, any change in PIF4 binding to PIF/EIN3 shared target promoters in response to ethylene or AgNO3 treatment (Figure 4C). We were also unable to observe any change in EIN3 binding in response to red light treatment (Figure 4A) even though red light dramatically reduces PIF levels. It thus seems the PIFs and EIN3 bind their shared targets independent of one another. It is also possible, though, PIFs and EIN3 cooperatively recruit transcriptional co-activators or chromatin modifying enzymes to activate target gene expression (Figure 7). PIF3 is known to interact with the chromatin remodeling factor PICKLE to regulate gene expression, and PICKLE is required for hypocotyl elongation and apical hook formation in seedling etiolation (Zhang et al., 2014). In a future study, we will determine whether PICKLE is recruited cooperatively by PIFs and EIN3 to their shared target promoters.
Both PIFs and EIN3 protect etiolated seedlings from photo-oxidative damage upon sudden exposure to light, enhancing survival. Several mechanisms have been proposed to account for this protective effect. PIF1 and PIF3 inhibit the accumulation of protochlorophyllide in the dark by repressing chlorophyll biosynthesis genes like HEMA1 and CHLH (Shin et al., 2009; Stephenson et al., 2009). In addition, PIF1 directly activates PORC expression and indirectly activates PORA and PORB expression (Moon et al., 2008). Ethylene-activated EIN3 directly binds the promoters of PORA and PORB to activate their expression (Zhong et al., 2009). EIN3 also directly increases PIF3 expression (Zhong et al., 2012), which, in turn, inhibits the accumulation of protochlorophyllide (Zhong et al., 2014). In addition to these mechanisms, we propose PIFs and EIN3 indirectly regulate chlorophyll biosynthesis through HLS1, which is known to regulate ethylene-induced apical hook formation (Lehman et al., 1996). We found PIFs and EIN3 cooperatively regulate HLS1 expression (Figures 4A,C and 5; Supplementary Figure S4), and HLS1 is required for preventing photo-oxidative damage (Figures 6A,B). HLS1 also represses HEMA1 and CHLH expression and activates PORA and PORB expression (Figure 6C). Thus, PIFs and EIN3 directly and indirectly regulate chlorophyll biosynthesis gene expression via their shared target HLS1. This ensures etiolated seedlings complete the greening process without photo-oxidative damage when they are exposed to light.
Phytochrome signaling is interconnected with various hormone signaling pathways. PIF4 directly interacts with the BR-regulated transcription factor BZR1 and the auxin-regulated transcription factor ARF6 (Oh et al., 2012, 2014). The interactions interdependently regulate the expression of thousands of target genes to achieve proper hypocotyl elongation by integrating phytochrome, brassinosteroid, and auxin signalings. Our study demonstrates a similar integration of phytochrome signaling with the ethylene signaling pathway via the transcriptional co-regulation of targets shared by PIFs and EIN3. The co-regulation of shared targets by key signaling transcription factors seems to be a common mechanism integrating phytochrome signaling with hormonal signaling. Since PIF4 directly interacts with other hormone signaling transcription factors, it is possible that PIFs directly interact with EIN3 to co-regulate target genes, which should be determined by a future study.
Author Contributions
JJ, EO, and GC designed the study. JJ, KK, and EO performed the overall experiments. JJ performed bioinformatics analysis. MK and GH consulted and performed the gas chromatography. HK and OP consulted and performed the Western blots of native EIN3 protein. Y-IP and GC supervised the work. JJ, EO, and GC wrote the manuscript. All authors discussed the results and made substantial contributions to the manuscript.
Funding
This work was supported in part by grants from the National Research Foundation of Korea (2015R1A2A1A05001091 and 2011-0031955) and the Rural Development Administration (SSAC-PJ011073) to GC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HQ and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgment
We thank TAIR and NASC for providing information and mutant seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01055
References
Adams, D. O., and Yang, S. F. (1979). Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U.S.A. 76, 170–174. doi: 10.1073/pnas.76.1.170
Al-Sady, B., Ni, W., Kircher, S., Schäfer, E., and Quail, P. H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 23, 439–446. doi: 10.1016/j.molcel.2006.06.011
Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311. doi: 10.1146/annurev.arplant.59.032607.092859
Bauer, D., Viczián, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., et al. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. doi: 10.1105/tpc.021568
Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521. doi: 10.1016/j.tplants.2007.10.001
Chang, K. N., Zhong, S., Weirauch, M. T., Hon, G., Pelizzola, M., Li, H., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2:e00675. doi: 10.7554/eLife.00675
Chao, Q. M., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
Franklin, K. A., and Quail, P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. doi: 10.1093/jxb/erp304
Gallego-Bartolome, J., Arana, M. V., Vandenbussche, F., Zadnikova, P., Minguet, E. G., Guardiola, V., et al. (2011). Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 67, 622–634. doi: 10.1111/j.1365-313X.2011.04621.x
Guo, H., and Ecker, J. R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. doi: 10.1016/S0092-8674(03)00969-3
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., López-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P. H. (2004). PHYTOCHROME-INTERACTING FACTOR 1 Is a critical bHLH regulator of chlorophyll biosynthesis. Science 305, 1937–1941. doi: 10.1126/science.1099728
Huq, E., and Quail, P. H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. doi: 10.1093/emboj/21.10.2441
Khanna, R., Huq, E., Kikis, E. A., Al-Sady, B., Lanzatella, C., and Quail, P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16, 3033–3044. doi: 10.1105/tpc.104.025643
Khanna, R., Shen, Y., Marion, C. M., Tsuchisaka, A., Theologis, A., Schafer, E., et al. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929. doi: 10.1105/tpc.107.051508
Kim, H. G., Kwon, S. J., Jang, Y. J., Nam, M. H., Chung, J. H., Na, Y.-C., et al. (2013). GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 163, 1776–1791. doi: 10.1104/pp.113.225649
Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., et al. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. doi: 10.2307/3870974
Lehman, A., Black, R., and Ecker, J. R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85, 183–194. doi: 10.1016/S0092-8674(00)81095-8
Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J. M., et al. (2008a). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. doi: 10.1105/tpc.107.052142
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008b). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, W., Ma, M., Feng, Y., Li, H., Wang, Y., Ma, Y., et al. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163, 670–683. doi: 10.1016/j.cell.2015.09.037
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Mathews, S. (2006). Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15, 3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x
Merchante, C., Alonso, J. M., and Stepanova, A. N. (2013). Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. doi: 10.1016/j.pbi.2013.08.001
Merchante, C., Brumos, J., Yun, J., Hu, Q., Spencer, K. R., Enríquez, P., et al. (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule. Cell 163, 684–697. doi: 10.1016/j.cell.2015.09.036
Moon, J., Zhu, L., Shen, H., and Huq, E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 9433–9438. doi: 10.1073/pnas.0803611105
Nemhauser, J. L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475. doi: 10.1016/j.cell.2006.05.050
Ni, M., Tepperman, J. M., and Quail, P. H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667.
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., et al. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21, 403–419. doi: 10.1105/tpc.108.064691
Oh, E., Kim, J., Park, E., Kim, J. I., Kang, C., and Choi, G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16, 3045–3058. doi: 10.1105/tpc.104.025163
Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W. I., and Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. doi: 10.1111/j.1365-313X.2006.02773.x
Oh, E., Zhu, J.-Y., Bai, M.-Y., Arenhart, R. A., Sun, Y., and Wang, Z.-Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. doi: 10.7554/eLife.03031
Oh, E., Zhu, J.-Y., and Wang, Z.-Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W. I., et al. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975. doi: 10.1093/pcp/pch125
Park, E., Park, J., Kim, J., Nagatani, A., Lagarias, J. C., and Choi, G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72, 537–546. doi: 10.1111/j.1365-313X.2012.05114.x
Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., et al. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. doi: 10.1016/S0092-8674(03)00968-1
Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10, 859–868. doi: 10.1046/j.1365-313X.1996.10050859.x
Shen, H., Moon, J., and Huq, E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44, 1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Solano, R., Stepanova, A., Chao, Q. M., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Stephenson, P. G., Fankhauser, C., and Terry, M. J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 106, 7654–7659. doi: 10.1073/pnas.0811684106
Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S. A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. doi: 10.1083/jcb.145.3.437
Zhang, D., Jing, Y., Jiang, Z., and Lin, R. (2014). The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26, 2472–2485. doi: 10.1105/tpc.113.121848
Zhang, Y., Mayba, O., Pfeiffer, A., Shi, H., Tepperman, J. M., Speed, T. P., et al. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9:e1003244. doi: 10.1371/journal.pgen.1003244
Zhao, Q., and Guo, H.-W. (2011). Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4, 626–634. doi: 10.1093/mp/ssr042
Zhong, S., Shi, H., Xue, C., Wang, L., Xi, Y., Li, J., et al. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22, 1530–1535. doi: 10.1016/j.cub.2012.06.039
Zhong, S., Shi, H., Xue, C., Wei, N., Guo, H., and Deng, X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. U.S.A. 111, 3913–3920. doi: 10.1073/pnas.1402491111
Keywords: phytochrome, ethylene signaling, phytochrome-interacting factors PIFs, EIN3, transcription factors, signaling crosstalk, Photobleaching
Citation: Jeong J, Kim K, Kim ME, Kim HG, Heo GS, Park OK, Park Y -I, Choi G and Oh E (2016) Phytochrome and Ethylene Signaling Integration in Arabidopsis Occurs via the Transcriptional Regulation of Genes Co-targeted by PIFs and EIN3. Front. Plant Sci. 7:1055. doi: 10.3389/fpls.2016.01055
Received: 14 May 2016; Accepted: 05 July 2016;
Published: 19 July 2016.
Edited by:
Enamul Huq, University of Texas at Austin, USAReviewed by:
Caiji Gao, The Chinese University of Hong Kong, ChinaHong Qiao, University of Texas at Austin, USA
Copyright © 2016 Jeong, Kim, Kim, Kim, Heo, Park, Park, Choi and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giltsu Choi, Z2Nob2lAa2Fpc3QuZWR1 Eunkyoo Oh, ZW9oQGpudS5hYy5rcg==
†Present address: Jinkil Jeong, Molecular and Cell Biology Laboratory, Salk Institute for Biological Studies, La Jolla, CA, USA
 Jinkil Jeong
Jinkil Jeong Keunhwa Kim1
Keunhwa Kim1 Hye G. Kim
Hye G. Kim Youn-Il Park
Youn-Il Park