- 1National Institute of Plant Genome Research, New Delhi, India
- 2Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi, India
Stress associated proteins (SAPs) are the A20/AN1 zinc-finger containing proteins which can regulate the stress signaling in plants. The rice SAP protein, OsSAP1 has been shown to confer abiotic stress tolerance to plants, when overexpressed, by modulating the expression of endogenous stress-related genes. To further understand the mechanism of OsSAP1-mediated stress signaling, OsSAP1 interacting proteins were identified using yeast two-hybrid analysis. Two novel proteins, aminotransferase (OsAMTR1) and a SCP/TAPS or pathogenesis-related 1 class of protein (OsSCP) were found to interact with OsSAP1. The genes encoding OsAMTR1 and OsSCP were stress-responsive and showed higher expression upon abiotic stress treatments. The role of OsAMTR1 and OsSCP under stress was analyzed by overexpressing them constitutively in Arabidopsis and responses of transgenic plants were assessed under salt and water-deficit stress. The OsAMTR1 and OsSCP overexpressing plants showed higher seed germination, root growth and fresh weight than wild-type plants under stress conditions. Overexpression of OsAMTR1 and OsSCP affected the expression of many known stress-responsive genes which were not affected by the overexpression of OsSAP1. Moreover, the transcript levels of OsSCP and OsAMTR1 were also unaffected by the overexpression of OsSAP1. Hence, it was concluded that OsSAP1 regulates the stress responsive signaling by interacting with these proteins which further regulate the downstream stress responsive gene expression.
Introduction
Plants are constantly challenged by a number of environmental extremities, like water-deficit, temperature, salinity, and pests during their life. To withstand these adversities, they have developed specialized mechanisms which lead to the modulation of their gene expression patterns for adaptive development and growth. Different stress-responsive genes are induced and repressed to play crucial roles in stress signaling. The overall stress signaling is a complex network of different pathways that ultimately confers the appropriate response depending on the type of stress encountered. To understand these signaling mechanisms, both forward and reverse genetics approaches have been employed. Through these approaches, many transcription factors and regulatory proteins have been identified which, if modulated, can affect the whole signal transduction pathway (Bhatnagar-Mathur et al., 2008; Hirayama and Shinozaki, 2010). The regulatory proteins often interlink and control multiple stress signaling pathways and hence can function as the master regulators of stress response (Chinnusamy et al., 2004). Zinc-finger proteins (ZFPs) are a group of regulatory proteins which have diverse functions in a variety of organisms (Takatsuji, 1999). They can function as transcription factors, RNA binding proteins and protein modification enzymes to regulate the growth, development and stress responses of an organism (Eulgem et al., 2000; Englbrecht et al., 2004; Agarwal et al., 2007; Moreno-Risueno et al., 2007).
Stress associated proteins (SAPs) are a class of ZFPs composed of two zinc-finger domains, an N-terminal A20 domain and a C-terminal AN1 domain. These proteins have recently been identified as novel stress regulatory proteins in plants (Giri et al., 2013). These types of proteins have also been identified in animals as regulators of immune signaling and many other responses including biotic/abiotic stresses (Heyninck and Beyaert, 2005). SAP gene family is prevalent in many organisms including plants, animals, protists, and fungi. Majority of SAPs have been found to be stress-inducible and some of the members from different plants have been characterized to confer abiotic stress tolerance in transgenic plants (Giri et al., 2013). In contrast, OsSAP7 has recently been characterized to be a negative regulator of ABA responsive stress signaling (Sharma et al., 2015). Similarly, ZFP185 (OsSAP4) has also been found to be involved in GA and ABA signaling and negatively regulates abiotic stress responses (Zhang et al., 2015). The role of SAPs in regulation of biotic stress responses is also emerging. Banana SAP gene, MusaSAP1 overexpressing transgenic plants showed strong up-regulation of polyphenol oxidase (PPO) encoding transcripts which are well-known to play a role in biotic defense pathway (Sreedharan et al., 2012). A recent study has revealed a role of OsSAP1 in regulating basal defense against pathogen infection via up-regulation of known defense-responsive genes such as PR genes (Tyagi et al., 2014). SAPs are considered to be regulatory proteins and it has been suggested that they can affect the stress signaling by interacting and modulating the activity of target proteins, though their molecular functions are poorly known. The SAPs have been identified as novel E3 ubiquitin ligases in analogy to their animal counterparts. Arabidopsis SAP5, ubiquitinates AtMBP1, a negative regulator of ABA and stress signaling, and targets it for degradation (Kang et al., 2011, 2013). Similarly, OsSAP7 has also been shown to possess E3 ligase activity (Sharma et al., 2015). In addition, SAPs can function as redox sensor as shown for AtSAP12, which can change its oligomeric conformation depending upon the cellular redox potential (Stroher et al., 2009). Besides, SAPs can homo-/hetero-dimerize and interact with other proteins via their zinc-finger domains (Kanneganti and Gupta, 2008; Giri et al., 2011). OsSAP1 and OsSAP11 have been found to interact with a receptor-like cytoplasmic kinase, OsRLCK253, which itself is stress-responsive and its overexpression in Arabidopsis conferred tolerance to abiotic stresses. It was speculated that either the kinase can activate the SAPs through phosphorylation or SAP proteins can regulate the activity of RLCK253 (Giri et al., 2011). In a similar way, it is expected that these proteins can interact with many other proteins and involve in different functions, which needs to be elucidated.
In this study, an attempt has been made to identify the proteins interacting with OsSAP1 using yeast two-hybrid assay and the involvement of interacting proteins in stress response was evaluated by gene overexpression in Arabidopsis.
Materials and Methods
Plant Material, Growth Conditions and Stress Treatments
Rice (Oryza sativa ssp. indica) cv. Pusa Basmati-1 was used for preparation of cDNA library and expression analysis in stress conditions. Surface-sterilized seeds were grown in hydroponics in culture room at 28 ± 2°C with 16 h/8 h light/dark cycle and 100–125 μmol m-2 sec-1 fluence for 7 days. For expression studies, 7-day-old seedlings were given different stress treatments for 3, 8, and 24 h, and unstressed seedlings in RO water were kept as control. For salt and oxidative stress treatments, seedlings were incubated in 100 mM NaCl and 10 μM methyl viologen (MV) solutions, respectively. For water-deficit stress, seedlings were kept on three layers of dry tissue paper in culture room for the indicated time points.
Arabidopsis thaliana (ecotype Col-0) was used for generation of transgenic plants and as wild-type control in transgenic analysis as well as expression analysis of target genes. Arabidopsis plants were grown in culture room maintained at 22 ± 1°C with continuous illumination (100 μmol m-2 sec-1). Ten-day-old seedlings were harvested for gene expression analysis.
Yeast Two-Hybrid Library Screening
The cDNA for yeast two-hybrid screening was prepared from 7-day-old rice seedlings treated with 3 h of water-deficit stress using MatchmakerTM Library Construction & Screening Kit (Clontech, USA) as per the manufacturer’s instructions and was transformed along with pGADT7-Rec vector in yeast AH109 cells to generate the prey library [activation domain (AD) fusion library]. The library was screened with SAP1- binding domain (BD) as bait and the transformants were selected on SD/-His/-Leu/-Trp media supplemented with 2.5 mM 3-AT. The putative positive clones were identified by DNA sequencing. To reconfirm the protein interactions, the positive AD clones (AMTR1 and SCP) were amplified and cloned in pGADT7-Rec vector and co-transformed with the BD clones (SAP1-BD, A20-BD, AN1-BD, SAP1ΔA20ΔAN1-BD) followed by selection on SD/-His/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp. The interactions were further confirmed by the activation of a third reporter gene LacZ by quantitative β-galactosidase assay using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate following the instructions in Yeast Protocol Handbook (Clontech, USA).
Gene Cloning
The cDNA was prepared from total RNA extracted from 7-day-old rice seedlings using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, USA) as per manufacturer’s protocol. The coding sequences of OsAMTR1 and OsSCP were amplified from cDNA using gene-specific primers (Supplementary Table S1) and were cloned in desired vectors. For yeast two-hybrid study, the above sequences were cloned in pGADT7-Rec vector, while OsSAP1 (1–495 bp), A20 domain (10–171 bp), AN1 domain (250–493 bp) and the linker region between A20 and AN1 domain (SAP1ΔA20ΔAN1-BD; 180–305 bp) encoding sequences were cloned in pGBKT7 vectors. To analyze the protein-protein interactions using BiFC (Bimolecular Fluorescence Complementation), the coding sequences of OsSAP1 and the interacting protein genes, i.e., OsAMTR1 and OsSCP, were cloned in pSPYCE(MR) and pSPYNE(R)173 vector, respectively (Waadt et al., 2008) to generate a fusion with the C- and N-terminal fragment of eYFP. For subcellular localization, OsAMTR1 and OsSCP were cloned in frame with the coding sequence of YFP in pSITE3CA vector (Chakrabarty et al., 2007) using Gateway® technology. Similarly, for overexpression studies, OsAMTR1 and OsSCP were cloned in binary vector pMDC32 using Gateway® cloning strategy under the control of dual CaMV35S promoter.
Generation of Arabidopsis Transgenic Plants
The 35S::OsAMTR1 and 35S::OsSCP constructs were transformed in Agrobacterium strain GV3101 and transformed in Arabidopsis plants using floral dip method (Clough and Bent, 1998). The transgene segregation was assessed using Chi-square test to identify the lines with single T-DNA insertion events. The expression of transgene in the homozygous T3 lines was analyzed using qRT-PCR and three lines of each transgenic plant (line # 11, 17, and 23 of 35S::OsAMTR1; line # 4, 22, and 27 of 35S::OsSCP), having single insertion as determined by segregation analysis in T1 generation, were used for further analyses.
Stress Treatment and Assessment of Effects on Plant Growth
To assess the effect of abiotic stress conditions on germination efficiency of 35S::OsAMTR1 and 35S::OsSCP Arabidopsis plants, surface-sterilized seeds of WT and transgenic lines were germinated on MS medium plates supplemented with different concentrations of NaCl (100–150 mM) and mannitol (250–400 mM) for salt stress and water-deficit stress, respectively. As a control, seeds were also germinated on MS basal plates. After stratification at 4°C, plates were incubated in continuous light (100 μmol m-2 sec-1) at 22 ± 1°C and per cent seed germination was scored every day for 4 days. The emergence of radicle was considered as an indication of seed germination.
To analyze the seedling growth under stress, the WT and transgenic seeds were germinated on MS plates supplemented with 100 mM NaCl or 200 mM mannitol. After stratification, the plates were incubated in continuous light (100 μmol m-2 sec-1) at 22 ± 1°C in vertical position to allow root growth. After 10 days, root length and fresh weight of seedlings were recorded.
Subcellular Localization of Fusion Proteins
The 35S::YFP-OsAMTR1 and 35S::YFP-OsSCP constructs were transiently transformed in onion epidermal cells using particle bombardment (Bio-Rad, USA). The cells were analyzed under TCS-SP2 Confocal Laser Scanning Microscope (Leica, Germany) for YFP fluorescence after 18–20 h of incubation in dark at room temperature. To plasmolyse the epidermal cells, the onion peels were incubated in 5% NaCl (w/v) solution for 2–3 min before visualizing under microscope.
Protein Interaction Using BiFC (Bimolecular Fluorescence Complementation) Approach
For BiFC assay, the protocol described by Waadt and Kudla (2008) was used. In brief, the fusion construct 35S::YFPC155-OsSAP1, 35S::YFPN173-OsAMTR1 and 35S::YFPN173-OsSCP were transformed in Agrobacterium strain GV3101. The transformed strains at an OD600 of 0.5 were mixed with Agrobacterium p19 strain (suppressor of silencing) at an OD600 of 0.3. The mixture was infiltrated in 4–5 weeks old Nicotiana benthamiana leaves and observed for YFP fluorescence (488 nm excitation laser) 5–6 days after infiltration under TCS SP5 confocal microscope (Leica, Germany).
Gene Expression Analysis
The expression levels of transgene and target genes in Arabidopsis and/or rice plants were analyzed by qRT-PCR. For expression analysis of transgene, leaf tissues from mature plants were harvested, while for stress marker genes, 10-day-old seedlings of Arabidopsis transgenic and WT (Col-0) plants were harvested. For expression analysis in stress condition, the 7-day-old rice seedlings were used for stress treatments and RNA extraction, as described above. The cDNA was prepared from 1 μg of total RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). The primers for qRT-PCR were designed by Primer Express (version 3.0) software (Supplementary Table S2). Quantification of amplified product was done by Fast SYBR® Green Master Mix (Applied Biosystems, USA) and relative expression levels were analyzed by comparative Ct (2-ΔΔCt) method.
Results
Identification of OsSAP1 Interacting Proteins
OsSAP1 has been shown to be involved in abiotic stress response (Mukhopadhyay et al., 2004; Giri et al., 2013; Dansana et al., 2014). Its interacting proteins under stress were identified using the yeast two-hybrid library screening approach (Clontech, USA). The cDNAs generated from water-deficit stress treated seedlings were fused with GAL4 AD to generate the prey library. This library was screened using OsSAP1, fused with the GAL4 BD, as bait (Figure 1A). After initial screening of the putative positive colonies by sequencing and removal of false positives, nine prey clones were found to have inserts in frame with AD domain (Supplementary Figure S1). For the next round of screening, the prey clones were amplified (full length) and cloned in fusion with AD followed by two-hybrid analysis with OsSAP1-BD. To further determine the region of OsSAP1 that can specifically interact with the AD clones, different deletions of OsSAP1, consisting of A20, AN1 and the linker region between A20 and AN1 domain (SAP1ΔA20ΔAN1), were fused with BD domain (Figure 1A) and co-transformed with the AD clones followed by selection on SD/-Ade/-His/-Leu/-Trp media (AHLT). Out of nine clones, two were found to interact with full length OsSAP1 and its domains. These were identified as novel uncharacterised genes namely, OsAMTR1 (LOC_Os05g39770) and OsSCP (LOC_Os07g03710) (Figure 1B). OsAMTR1 was predicted to be a member of Aminotransferase class-III family of proteins. The 391 amino acid long OsAMTR1 protein carries a pyridoxal phosphate dependent transferase domain lying between 3 and 389 amino acid residues (Supplementary Figure S2). OsSCP was found to be a member of ‘SCP/TAPS (Sperm Coating Protein/Tpx-1/Ag5/PR-1/Sc7)’ protein family (Mizuki and Kasahara, 1992; Cantacessi et al., 2009). The coding sequence of OsSCP was found to be 507 nucleotides long encoding a protein of 168 amino acids (Supplementary Figure S3). The OsSCP protein contains a CAP (Cysteine-rich secretory proteins, Antigen 5, Pathogenesis-related 1 proteins; InterProScan: IPR014044) domain between 11 and 168 amino acid residues. The literature survey identified OsSCP to encode a pathogenesis-related 1a (OsPR-1a) protein (Agrawal et al., 2000). OsSCP was found to interact strongly and with equal efficiency with both A20 and AN1 domains of OsSAP1, whereas, OsAMTR1 interacted more strongly with linker region between A20 and AN1 domains (SAP1ΔA20ΔAN1). The interactions of these two proteins were further confirmed by the activation of LacZ reporter using quantitative β-galactosidase assay (Figure 1C; Supplementary Figure S4).
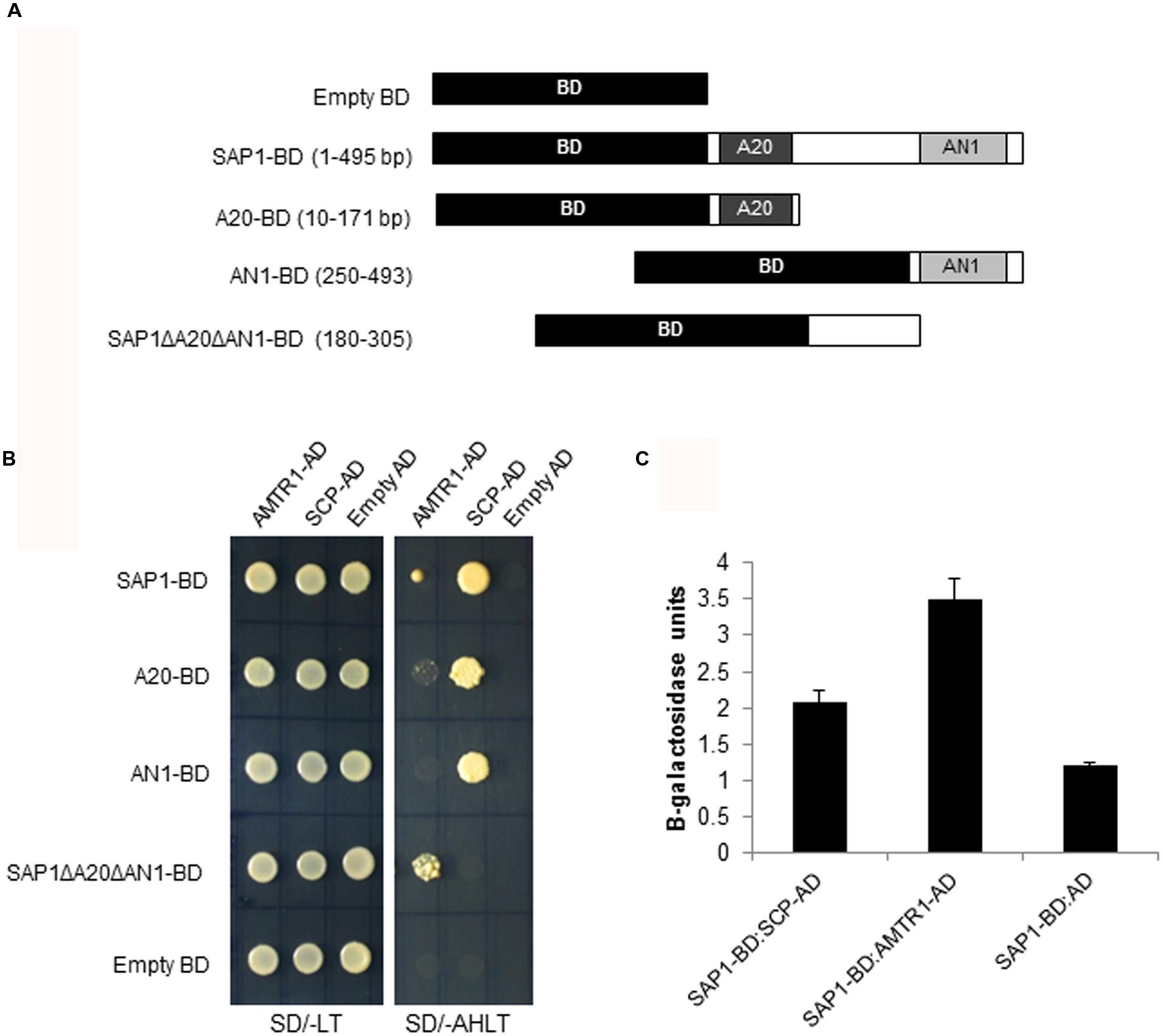
FIGURE 1. Identification of proteins interacting with OsSAP1. (A) The DNA binding domain (BD) was fused to different deletions of OsSAP1 to prepare bait constructs. (B) Yeast cells co-transformed with bait constructs shown in (A) and AD clones (SCP-AD, and AMTR1-AD) were plated on SD/-Leu/-Trp (SD/-LT) and SD/-Ade/-His/-Leu/-Trp (SD/-AHLT) media to test the interaction. Empty BD vector was co-transformed with empty AD clones to serve as negative control. (C) Quantification of β-galactosidase activity in the transformed yeast cells using o-nitrophenyl-β-galactopyranoside (ONPG) as a substrate. Negative controls: Empty AD transformed with SAP1:BD. The histogram represents mean ± SE of three biological replicates.
Subcellular Localization of OsAMTR1 and OsSCP
To delineate their cellular functions, the coding regions of these genes were fused to the C-terminal of YFP fluorescent marker gene and subcellular localization was studied. The YFP-OsAMTR1 fusion protein was found to be distributed in whole cytoplasm including cytoplasmic strands and some dotted structures, although no signal was observed in the nucleus of onion epidermal cells. However, YFP-OsSCP was localized in cytoplasm as well as nucleus, evenly (Figure 2). Further, being a secretory protein, OsSCP was expected to localize in extracellular space, However, YFP-OsSCP remained in cytoplasm as revealed in plasmolysed cells (Supplementary Figure S5). The interaction of OsSAP1 with OsAMTR1 and OsSCP was analyzed in vivo using BiFC approach. OsSAP1 was fused with C-terminal half of YFP protein while OsAMTR1 and OsSCP were fused with N-terminal half of YFP and co-infiltrated in N. benthamiana leaves to analyze the YFP fluorescence using confocal microscope. The complex of YFPC155-OsSAP1 and YFPN173-OsAMTR1 showed YFP signal in whole cell. Similarly, YFPC155-OsSAP1/YFPN173-OsSCP complex also showed signal in tobacco cell (Figure 3). This confirmed the interactions of OsSAP1 with OsAMTR1 and OsSCP.
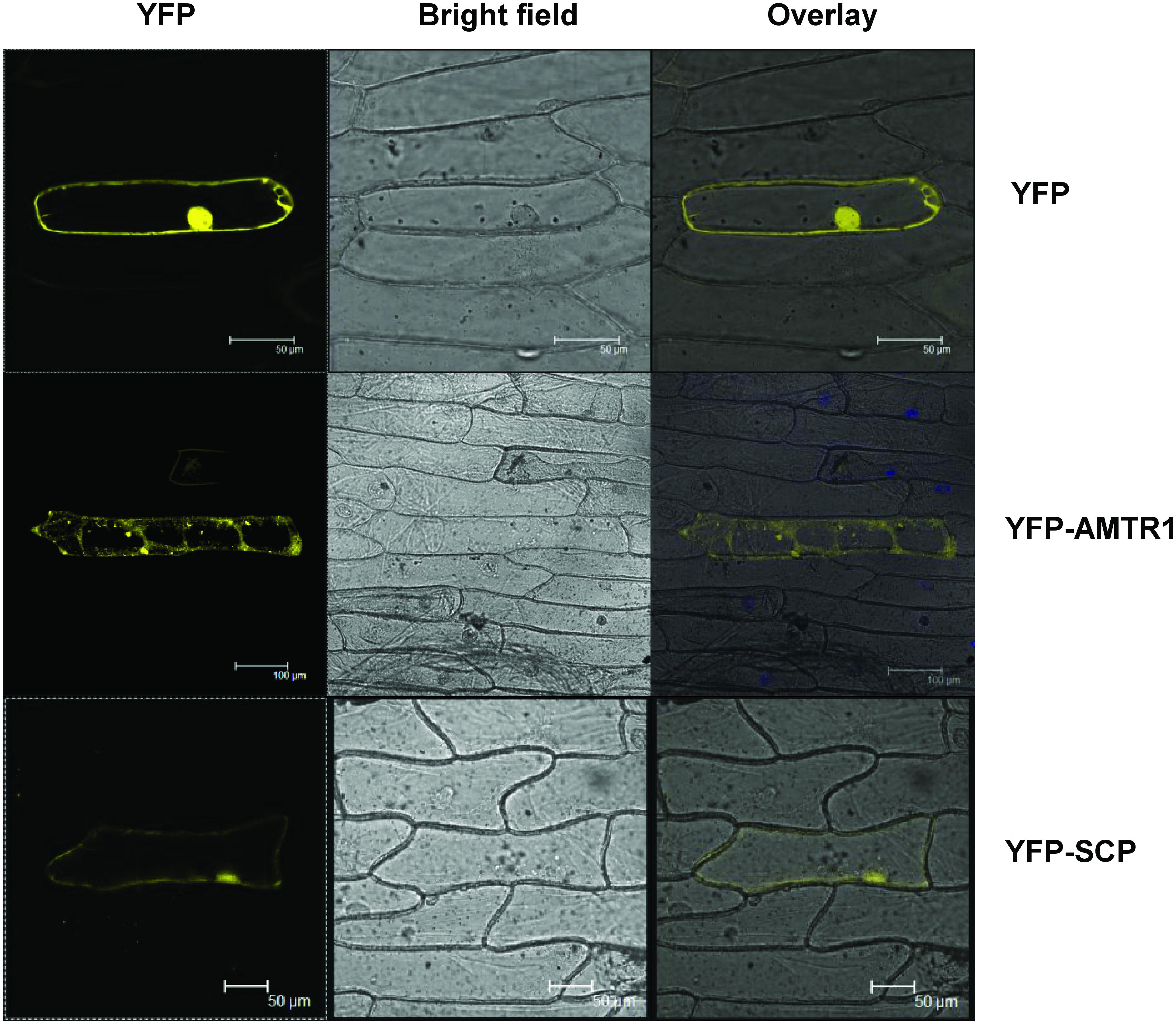
FIGURE 2. Subcellular localization of OsAMTR1 and OsSCP. The YFP fusion constructs (YFP-AMTR1 and YFP-SCP) were transformed transiently in onion epidermal cells and were visualized by confocal microscope. Empty vector (YFP) was used as a negative control.
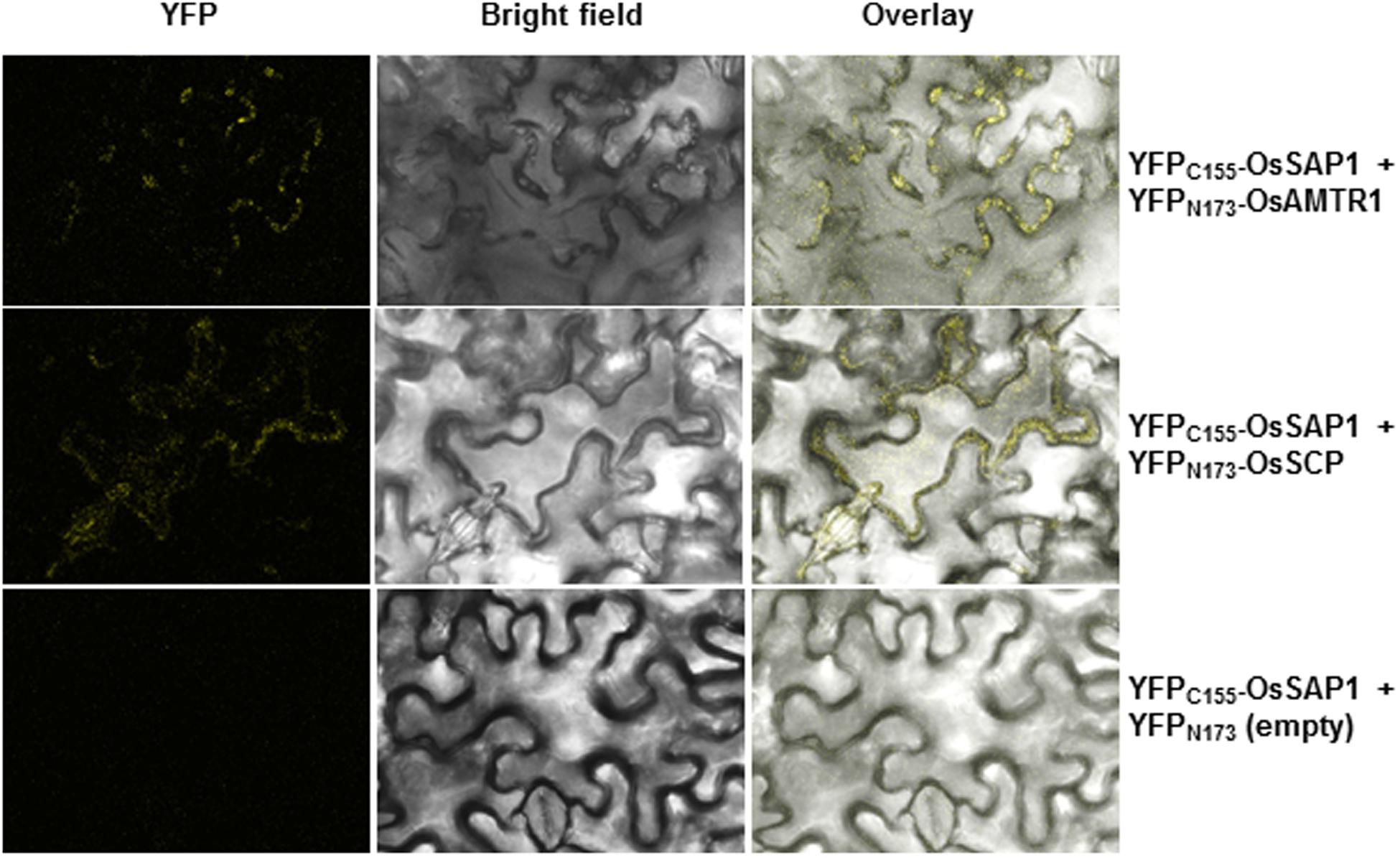
FIGURE 3. Confirmation of interaction of OsSAP1 with OsAMTR1 and OsSCP using BiFC approach. OsSAP1 was cloned in pSPYCE(MR) while OsAMTR1 and OsSCP were cloned in pSPYNE(R)173 vector to generate the fusions with C- and N-terminus of YFP, respectively. 35S::YFPC155-OsSAP1 was co-infiltrated separately with 35S::YFPN173-OsAMTR1 and 35S::YFPN173-OsSCP in Nicotiana benthamiana leaves and visualized under confocal microscope for the YFP fluorescence. As a negative control, 35S::YFPC155-OsSAP1 was co-transformed with empty SPYNE(R)173.
The Genes Encoding OsSAP1 Interacting Proteins Were Stress-Inducible
The expression patterns of OsAMTR1 and OsSCP in different abiotic stress conditions was analyzed by treating the 7-day-old rice seedlings with salt, water-deficit and oxidative stress for different time durations and expression analysis was done using qRT-PCR (Figure 4). Under water-deficit treatment, very high induction (~23-folds) was seen for OsSAP1 at 3 h, which is consistent with its early induction under water-deficit condition as reported earlier (Mukhopadhyay et al., 2004; Dansana et al., 2014). The OsSAP1 expression decreases at later time-points. At 3 h, OsAMTR1 showed about 10-fold induction and the expression further increased with time. Similar pattern of expression was observed for OsSCP, with 2.5–4 folds upregulation. Under salt stress, the highest expression was seen in OsAMTR1, which gets induced to a very high level (~17-folds) at 3 h and the expression decreases, thereafter. The expression of OsSAP1 was about five folds higher than control and was almost constant at all time-points. But, weak induction was observed for OsSCP. All the genes showed up-regulation ≥2 folds under oxidative stress and expression increased with time except in OsSAP1 where after 8 h, expression level decreased slightly. These expression profiles imply that although expression patterns of genes encoding OsSAP1 interacting proteins are not exactly similar to that of OsSAP1, all the genes are responsive to abiotic stress conditions.
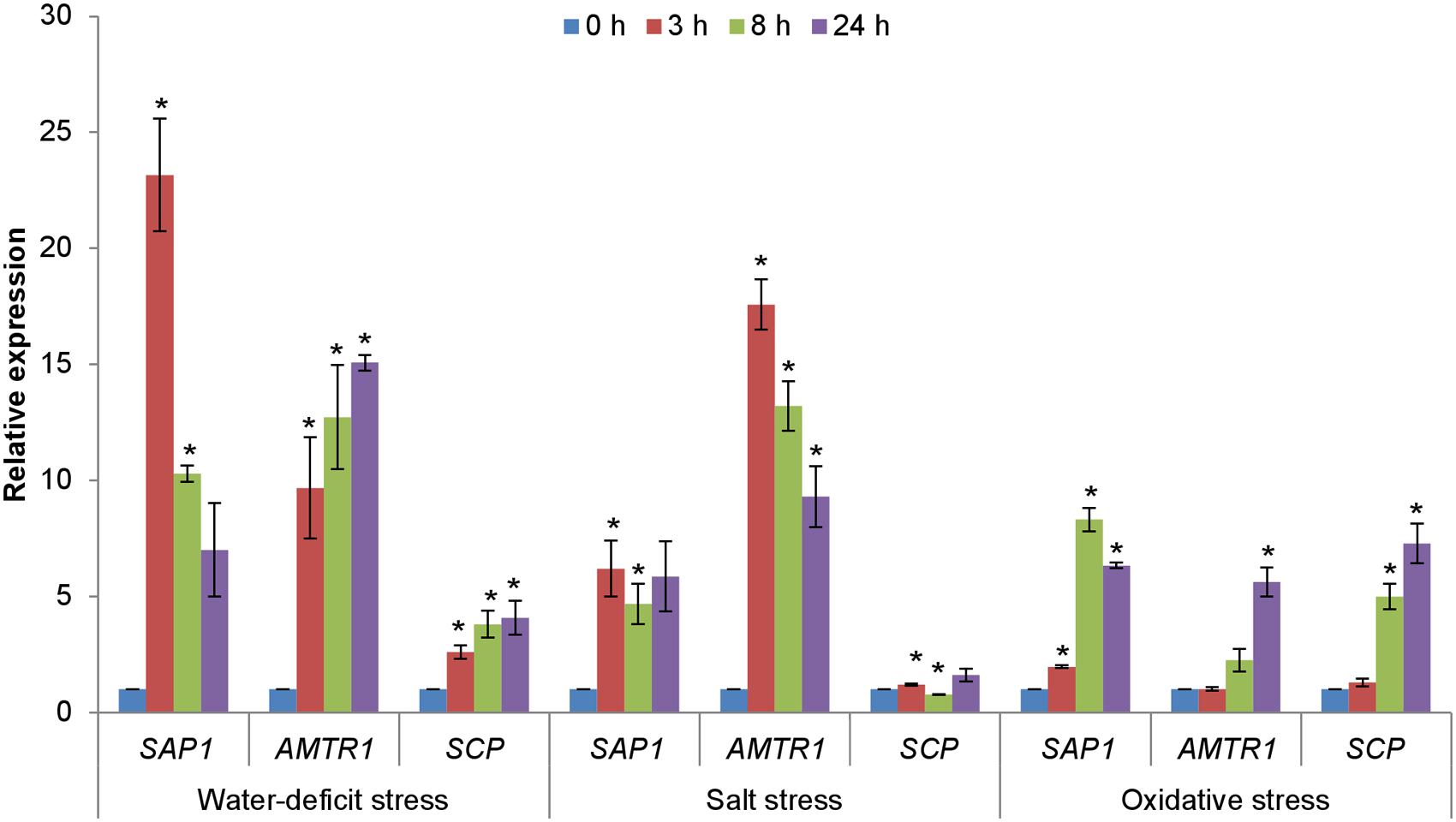
FIGURE 4. Expression analysis of genes encoding interacting proteins of OsSAP1 under different abiotic stress treatments. Seven-day-old seedlings were treated with NaCl (200 mM), water-deficit (air drying), and methyl viologen (MV; 10 μM) induced oxidative stress for different time durations (0, 3, 8, and 24 h) and expression levels were quantified by qRT-PCR. The histogram represents the mean ± SE of three biological replicates. Asterisks represent significant difference with respect to unstressed (0 h) condition as determined by t-test at p-value <0.05.
Evaluation of Response of Arabidopsis Transgenic Plants Overexpressing OsAMTR1 and OsSCP during Stress Condition
To functionally characterize the OsAMTR1 and OsSCP under abiotic stress, these genes were overexpressed in Arabidopsis under the control of CaMV35S promoter. After confirming the transgenic nature by PCR, the expression of transgene was analyzed using qRT-PCR. The transgenic plants showed no visible phenotypic differences with the wild-type plants under normal growth conditions (Supplementary Figures S6 and S7). These lines also showed similar seed germination rate as compared to wild-type under normal growth conditions (Supplementary Figure S8). Three independent lines from each transgenic (AT-11, AT-17, and AT-23 of 35S::OsAMTR1; SCP-4, SCP-22, and SCP-27 of 35S::OsSCP) were selected for further stress analysis.
Functional Characterization of OsAMTR1 under Salt and Water-Deficit Stress
The effect of overexpression of OsAMTR1 on plant during abiotic stress conditions was analyzed by treating the plants with different stresses and evaluation of response of transgenics in comparison to wild-type. To assess the effect of salt and water-deficit stress on the seed germination efficiency, wild-type and 35S::OsAMTR1 transgenic seeds were germinated on MS medium supplemented with different concentrations of NaCl or mannitol and per cent seed germination were scored for 4 days. The transgenic seeds were found to have higher germination efficiency as compared to wild-type seeds under both high salt and mannitol conditions (Figures 5A,C). Even at lower concentrations of NaCl and mannitol, the 35S::OsAMTR1 plants showed higher germination than wild-type (Figures 5B,D). Similarly, to study the effect of salt and water-deficit stress during seedling growth stage, the root growth and biomass accumulation in 10-day-old seedlings of wild-type and 35S::OsAMTR1 transgenic plants were analyzed. The seeds of wild-type and transgenic lines were plated on salt and mannitol containing media and after 10 days of growth, transgenic plants were found to have longer roots than WT plants (Figures 6A–C). Further, the fresh weight of transgenic plants was higher than the wild-type plants (Figures 6D,E). The improvement in root growth and biomass accumulation was more prominent under water-deficit as compared to salt stress.
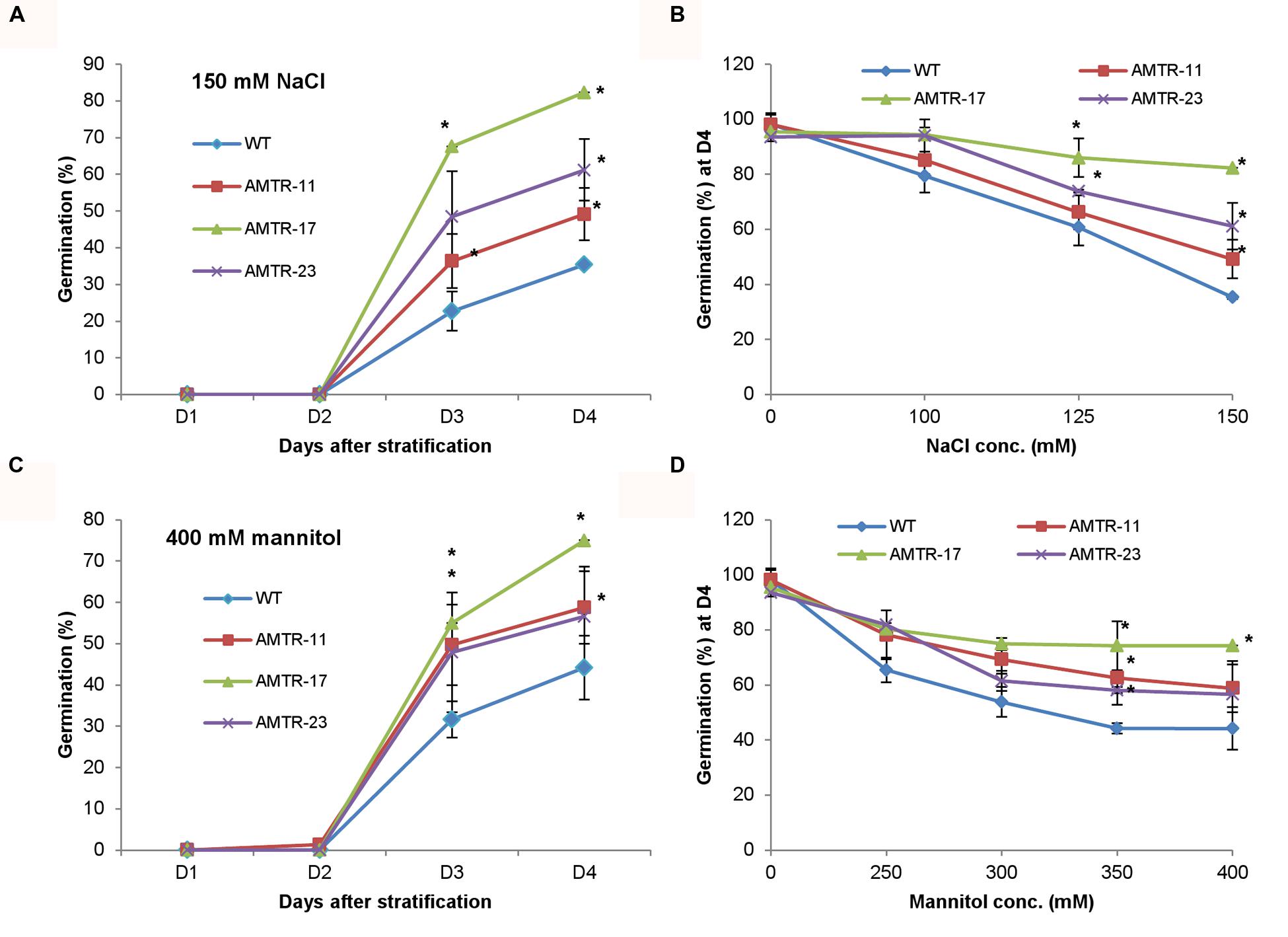
FIGURE 5. Effect of abiotic stress on seed germination efficiency of 35S::OsAMTR1 Arabidopsis plants. Wild-type (WT) and transgenic (AMTR-11, AMTR-17, and AMTR-23) seeds were plated on MS medium supplemented with (A) 150 mM NaCl and (C) 400 mM mannitol and per cent seed germination was scored for 4 days after stratification. (B,D) The per cent seed germination at day 4 after stratification on variable concentrations of NaCl and mannitol, respectively. The graphs represent the mean ± SE of three biological replicates (n = 50). Asterisks represent significant difference with respect to wild-type as determined by t-test at p-value <0.05.
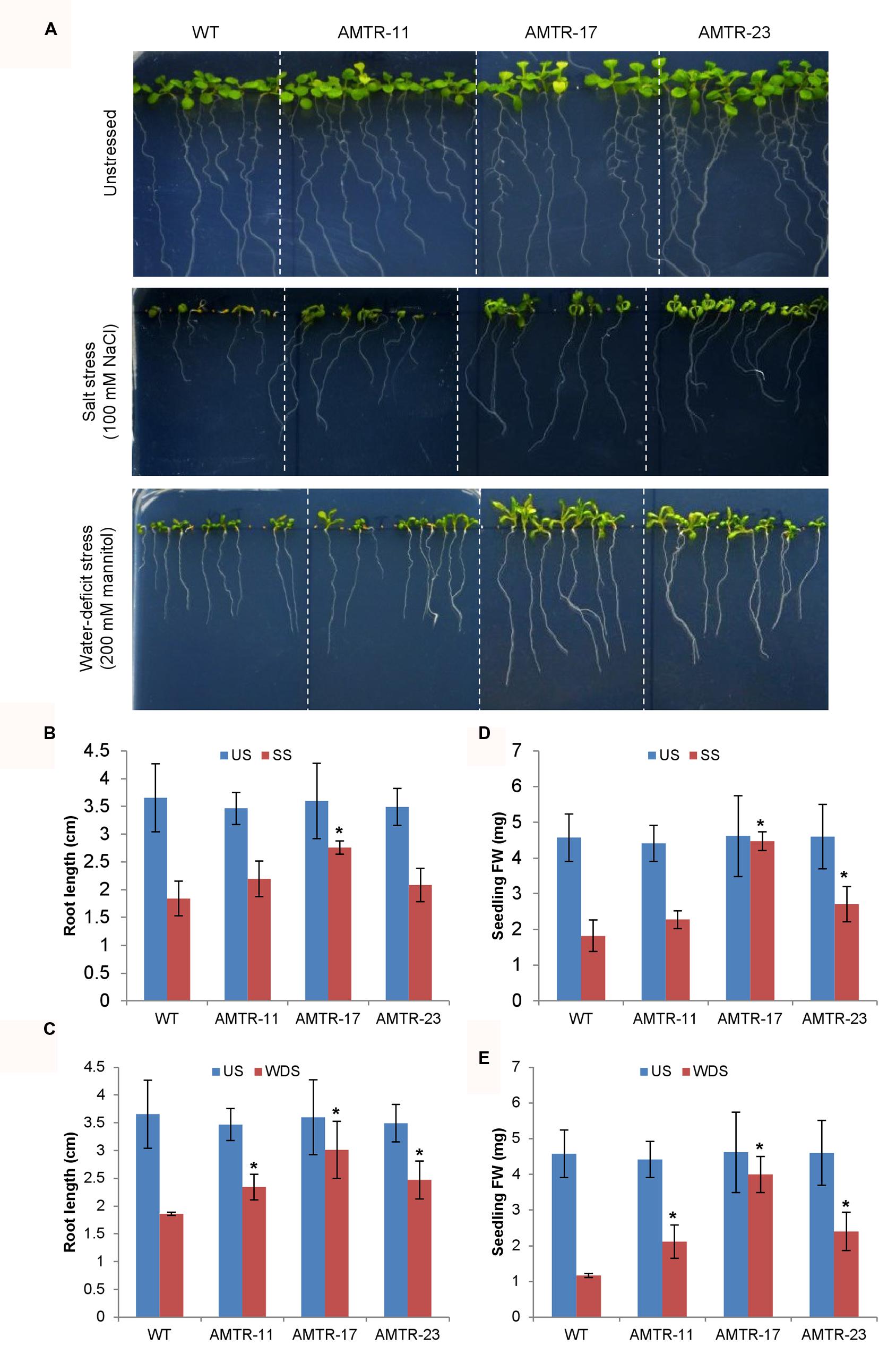
FIGURE 6. Effect of abiotic stress on root growth and biomass accumulation of 35S::OsAMTR1 Arabidopsis plants. Seeds of WT and transgenic lines (AMTR11, AMTR17, and AMTR23) were plated on MS medium supplemented with 100 mM NaCl and 200 mM mannitol and plates were incubated in vertical position. (A) Morphology of WT and transgenic lines, (B,D) root length and (C,E) fresh weight (FW) of individual plant after 10 days of growth. The histograms represent mean ± SE from three biological replicates (n = 10). Asterisks represent significant difference with respect to wild-type as determined by t-test at p-value <0.05. SS, salt stress, US, unstressed; WDS, water-deficit stress.
Functional Characterization of OsSCP under Salt and Water-Deficit Stress
As stated earlier, OsSCP was overexpressed in Arabidopsis under the control of CaMV35S promoter and 35S::OsSCP transgenic plants were assessed for their response under abiotic stress conditions. Transgenic seeds showed higher germination efficiency than wild-type only at high concentration of NaCl (150 mM), while, at lower concentrations, transgenic seeds had slightly higher or almost similar germination efficiency as that of wild-type (Figures 7A,B). Similarly, at different concentrations of mannitol, only marginally higher germination was observed in transgenic seeds than wild-type, except for line SCP-4, which showed significantly high germination (Figures 7C,D). To further analyze the effect of stress on 35S::OsSCP transgenic plants, the root growth and fresh weight accumulation of 10-day-old wild-type and transgenic seedlings growing on 100 mM NaCl and 200 mM mannitol supplemented media were evaluated. The transgenic plants exhibited longer roots and accumulated significantly more fresh weight than wild-type plants at both salt and mannitol induced water-deficit stress condition (Figure 8).
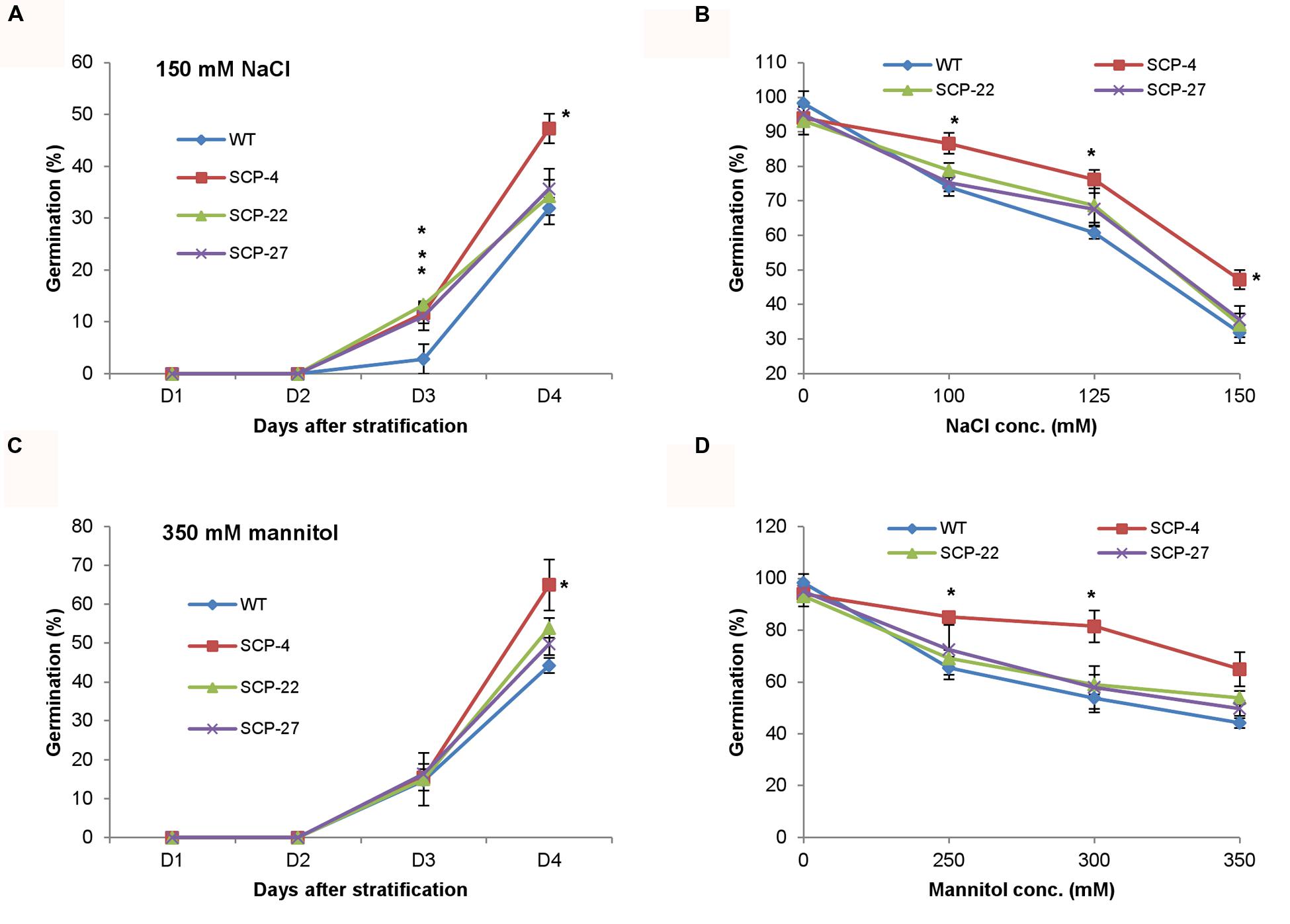
FIGURE 7. Effect of abiotic stress on seed germination efficiency of 35S::OsSCP Arabidopsis plants. Wild-type and transgenic (SCP-4, SCP-22, and SCP-27) seeds were plated on MS medium supplemented with (A) 150 mM NaCl and (C) 350 mM mannitol and per cent seed germination was scored for 4 days after stratification. (B,D) The per cent seed germination at day 4 of stratification on variable concentrations of NaCl and mannitol, respectively. The graphs represent mean ± SE from three biological replicates (n = 50). Asterisks represent significant difference with respect to wild-type as determined by t-test at p-value <0.05.
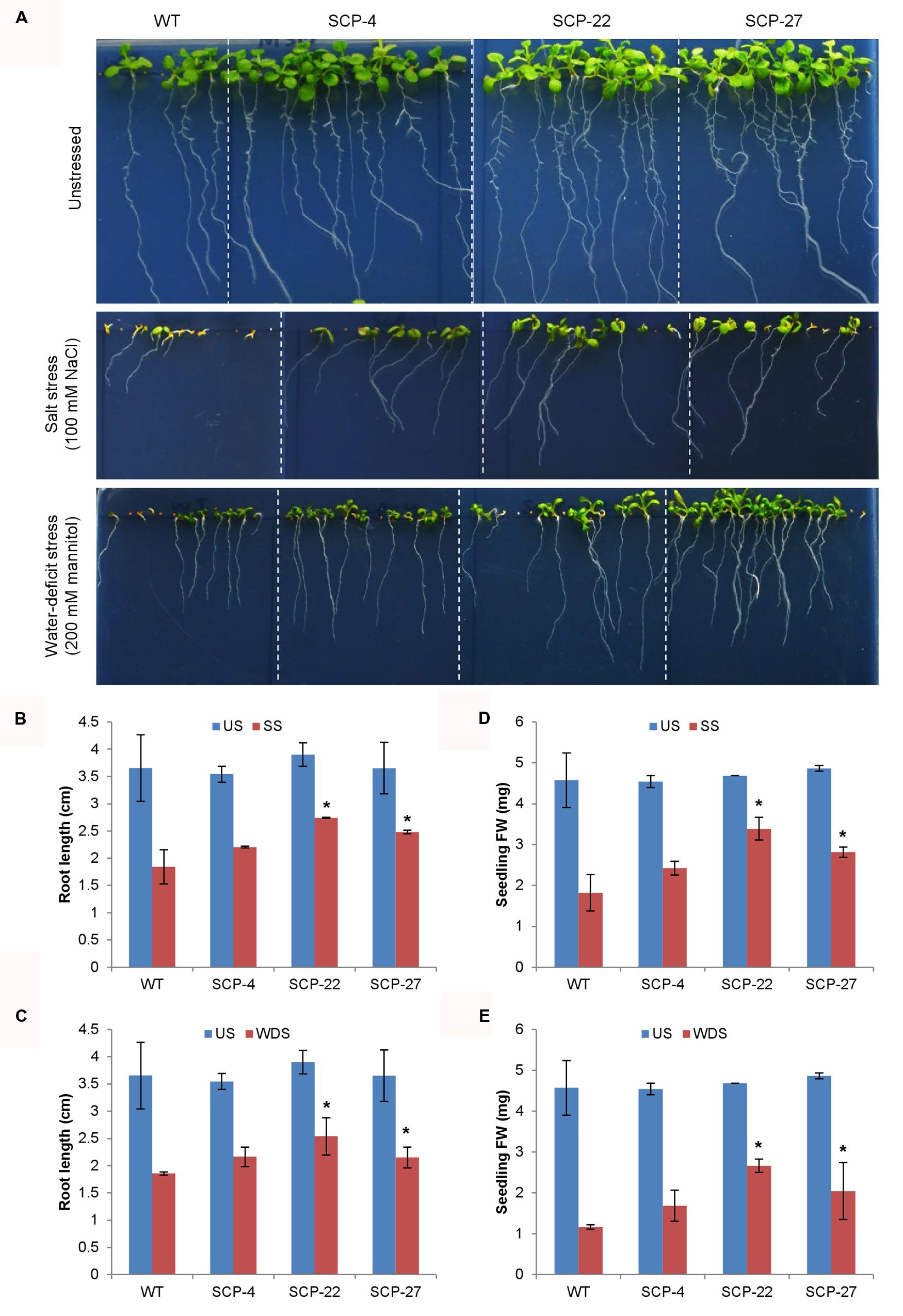
FIGURE 8. Effect of abiotic stress on root growth and biomass accumulation of 35S::OsSCP Arabidopsis plants. Seeds of WT and transgenic lines (SCP-4, SCP-22, and SCP-27) were plated on MS medium supplemented with 100 mM NaCl and 200 mM mannitol and plates were incubated in vertical position. (A) Morphology of WT and transgenic lines, (B,D) root length and (C,E) FW of individual plant after 10 days of growth. The histograms represent mean ± SE from three biological replicates (n = 10). Asterisks represent significant difference with respect to wild-type as determined by t-test at p-value<0.05. SS, salt stress, US, unstressed; WDS, water-deficit stress.
Effect of Overexpression of OsAMTR1 and OsSCP on Stress Responsive Genes
To analyze the effect of overexpression of OsAMTR1 and OsSCP on stress signaling, the expression patterns of Arabidopsis known stress-responsive genes were analyzed in the 35S::OsAMTR1 and 35S::OsSCP transgenic plants under normal growth conditions. A group of genes including RD22, COR47, ADH1, P5CS1, and RAB18 were found to have increased expression in both the transgenics as compared to wild-type, which probably imparts higher stress tolerance to the transgenic plants (Figure 9A). In contrast, some genes like RD29A, RD29B, and KIN1 are upregulated in OsSCP overexpressing plants but show similar expression levels as wild-type in OsAMTR1 overexpressing plants, implying gene-specific effect on expression of Arabidopsis endogenous genes (Figure 9B). Likewise, some other genes, ABF3, ABF4, and COR15a, might not be involved in the stress responsive signaling in 35S::OsAMTR1 and 35S::OsSCP transgenic plants as no change in expression of these genes was observed between wild-type and transgenic plants (Figure 9C).
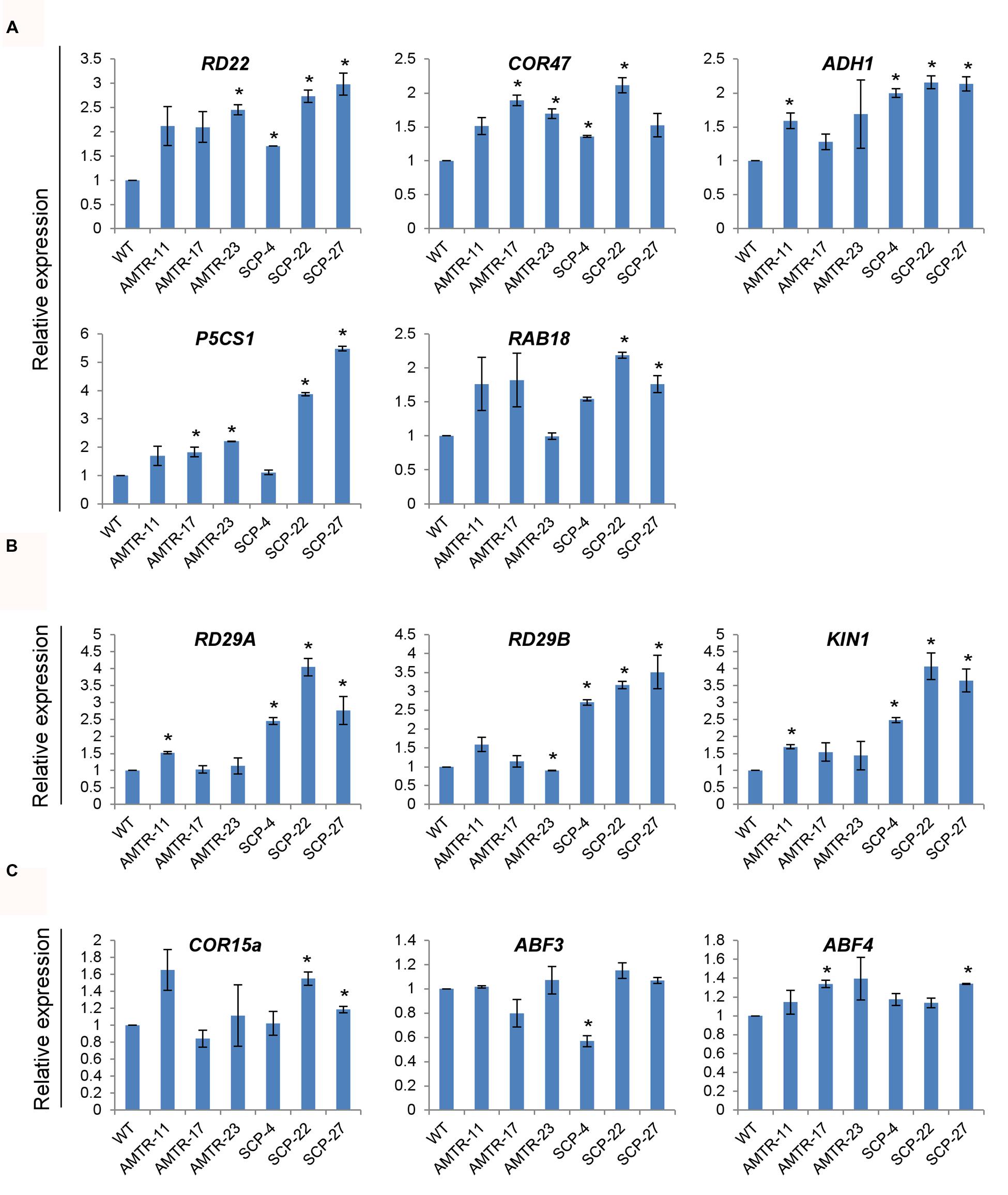
FIGURE 9. Effect of overexpression of OsAMTR1 and OsSCP in Arabidopsis on the expression of stress-responsive genes. qRT-PCR analysis of stress-responsive genes in 10-day-old seedlings of 35S::OsAMTR1 and 35S::OsSCP transgenic plants and WT. (A) Genes upregulated in both 35S::OsAMTR1 and 35S::OsSCP plants, (B) genes upregulated in 35S::OsSCP plants, (C) genes which have no change in expression in WT and either of transgenics. The graphs represent mean ± SE from three biological replicates. Asterisks represent significant difference with respect to wild-type as determined by t-test at p-value <0.05.
Discussion
OsSAP1 Could Regulate Stress Responses by Interacting with Other Stress-Responsive Proteins
To understand the signaling mechanism of any protein, it is necessary to identify interacting proteins which are required for relay of signal. It is well-studied that homo- and hetero-complex formation of proteins can serve as a mechanism for their responses under stress (Nakashima et al., 2012; Nie et al., 2014; Zhu et al., 2014). For example, the homo- and hetero-dimerization between WRKY18, WRKY40, and WRKY60 has been shown to significantly affect their DNA binding activities (Xu et al., 2006). The self-association of SAPs can also have an effect on their stress responses. As SAPs are regulatory proteins, they can associate with many other proteins in the signaling pathway to regulate their activity under stress. With this idea in mind, the proteins which interact with OsSAP1 were identified using yeast two-hybrid library screening in rice. Two proteins, an aminotransferase (OsAMTR1) and a pathogenesis-related protein (OsSCP), were found to interact with OsSAP1 (Figure 1). Further, OsAMTR1 localizes in the form of irregular subcellular bodies in cytoplasm. These could be high molecular weight aggregates formed due to homo-heterodimers of OsAMTR1 or OsSAP1.
It is known that under salt and water-deficit stress, OsSAP1 transcript starts accumulating within 15 min of stress (Mukhopadhyay et al., 2004), whereas OsSAP1 protein starts accumulating as early as 30 min of water-deficit stress and 3 h of salt treatment and keeps on increasing thereafter (Dansana et al., 2014). Similar to OsSAP1, OsAMTR1, and OsSCP were found to be responsive to salt and water-deficit stress condition at different levels. The expression profiles of all the genes were more or less similar under oxidative stress treatment and expression increased with time. This indicates high level of overlap in expression patterns between OsSAP1 and its interacting proteins. The temporal expression patterns can give an idea about the possible role of a gene during stress (Hasegawa et al., 2000; Singh et al., 2002). For instance, early induction of any gene under stress might be related to its function in early stress response. While, late expressing genes under stress can have a function under prolonged stress. Similarly, it has been suggested that co-expressing genes can be functionally related as it can be assumed that genes regulated by similar set of transcription factors can have similar expression patterns under a set of condition (van Noort et al., 2003; Allocco et al., 2004). Based on these hypotheses, OsSAP1 interacting proteins, OsAMTR1 and OsSCP, having almost similar expression patterns as OsSAP1, were selected for further analyses.
OsAMTR1 Positively Regulates Salt and Water-Deficit Stress Response
Aminotransferases are a group of pyridoxal-5’-phosphate dependent enzymes that catalyze the reversible transfer of amino group from amino acids to oxo-acids and are involved in a number of metabolic activities (Liepman and Olsen, 2001). In plants, many aminotransferases have been found to play important roles in stress regulation, both biotic and abiotic. For instance, AlaATs from different plants have been suggested to play role in hypoxic stress (Ricoult et al., 2006; Rocha et al., 2010a,b). The homologs of AlaAT, alanine:glyoxylate aminotransferase (AGT) and glutamate:glyoxylate aminotransferase (GGAT) are involved in photorespiration and overexpression of these aminotransferases has shown higher resistance to many plant pathogens (Liepman and Olsen, 2001; Taler et al., 2004). Another aminotransferase ALD1 has been found to be a positive regulator of salicylic acid (SA)-mediated plant defense responses (Song et al., 2004; Nie et al., 2011). Besides this, a GABA transaminase (GABA-T) involved in GABA catabolism has been shown to play a role in plant salt stress tolerance (Renault et al., 2010).
Further, the YFP-OsAMTR1 fusion protein was found to be cytosolic. The plant alanine aminotransferases have been found to have different isoforms expressing in peroxisomes, mitochondria and cytosol and are involved in different cellular functions (Liepman and Olsen, 2003; Ricoult et al., 2006). In general, the peroxisomal forms have been identified to function in photorespiration while the mitochondrial forms are involved in hypoxic stress responses, but no specific function for cytosolic form has been described till date. All these evidences suggest that aminotransferases can have significant effect in the stress response of plants. OsAMTR1 overexpressing plants showed enhanced tolerance under salt and water-deficit stress as evident from higher seed germination efficiency, root growth and biomass accumulation in transgenics as compared to wild-type. The observed positive regulation of known stress-responsive genes namely, RD22, COR47, ADH1, P5CS1, and RAB18 in the transgenic plants might be responsible for imparting the stress tolerance to transgenic plants. In contrast, no deviation in the expression of RD29A, RD29B, KIN1, ABF3, ABF4, and COR15a was found in the transgenics than the wild-type plants suggesting these genes are not involved in the stress signaling pathway affected by OsAMTR1.
Pathogenesis Related 1 (PR1) Protein Encoding Gene (OsSCP) Can Play a Role in Abiotic Stress Responses by Regulating Stress Responsive Genes
Pathogenesis-related (PR) proteins are group of proteins induced in plants in response to pathogen infection and are divided into 17 families (PR1 to PR17) based on the specific molecular function played by the family members (van Loon et al., 2006). In rice, PR1 gene family comprises of 12 members and all of them have been found to be inducible by pathogen infection (Mitsuhara et al., 2008). Transcripts of two of the PR1 genes, OsPR1a and OsPR1b, have been found to be inducible by JA, ethylene, SA, ABA, H2O2, protein phosphatase inhibitors and fungal infection (Agrawal et al., 2000). These inductions were dose-dependent and require de novo synthesis of some protein factor(s). In the present study, OsPR1a, named as OsSCP, has been identified to interact with OsSAP1 in yeast. The OsSCP transcript was found to be inducible by abiotic stress treatments such as water-deficit and MV induced oxidative stress. Earlier studies have also shown induction of PR genes in response to abiotic stress conditions such as salt stress, low temperature, and light (Snider et al., 2000; Zeier et al., 2004; Seo et al., 2008, 2010) suggesting important roles played by PR proteins in abiotic stress adaptation in addition to plant defense response against pathogens. Overexpression of OsSCP in Arabidopsis could enhance the tolerance of transgenic plants to salt and water-deficit stress as compared to wild-type plants. The OsSCP transgenic plants were also found to upregulate the expressions of RD22, COR47, ADH1, P5CS1, and RAB18 but unlike in 35S::OsAMTR1 plants, expressions of RD29A, RD29B, and KIN1 were also increased in 35S::SCP plants, implying that in addition to some overlap, OsAMTR1 and OsSCP also have independent effects on stress related genes.
Plant PR1 proteins have been reported to exist in acidic and basic forms based on the isoelectric point (pI) of the protein (Van Loon and Van Strien, 1999; Mitsuhara et al., 2008). The acidic isoforms have been found to be secreted in extracellular space, whereas, the basic isoforms are localized in vacuoles (Dixon et al., 1991; Sessa et al., 1995). As expected, the subcellular localization of OsSCP, the acidic OsPR1a, has been predicted to be extracellular. However, OsSCP fused with YFP was found to be localized in the whole cell of onion epidermis and not in the extracellular space (Supplementary Figure S5). There could be a possibility that translocation of OsSCP to the extracellular space is condition dependent. So, the subcellular localization of OsSCP needs to be analyzed in detail by considering these possibilities.
Possible Regulatory Mechanism of OsSAP1 under Abiotic Stress
The A20/AN1 containing proteins in animals possess E3 ubiquitin ligase activity and are involved in the regulation of inflammatory pathway via 26S proteasomal degradation pathway (Hishiya et al., 2006; Chang et al., 2011; Shembade and Harhaj, 2012). The plants SAPs have also been shown to possess E3 ligase activity (Kang et al., 2013; Sharma et al., 2015). Based on the recognition of different type of polyubiquitin chains, Lys48- or Lys63-linked, the ubiquitin binding protein can function in either degradation or regulating different cellular processes such as protein trafficking, DNA repair, protein activation or signal transduction (Pickart, 2001; Mural et al., 2013). The polyubiquitin recognition via A20 domain has been shown for Arabidopsis SAPs along with different affinities of AtSAP5 for Lys63- and Lys48-linked polyubiquitin chains (Choi et al., 2012). A similar activity has also been presumed for other SAPs, owing to high degree of homology in their A20 domain. This allows us to speculate that if OsSAP1 can add Lys63-linked polyubiquitin chains to its interacting proteins, then this interaction might affect activity of OsAMTR1 and OsSCP, which in turn influence the abiotic stress signaling. Also, OsSAP1 can regulate the biotic stress signaling pathway by regulating the expression of defense related genes including PR genes (Tyagi et al., 2014). Although, expression of SCP, also a PR gene, is not directly regulated by OsSAP1 as its expression did not change in OsSAP1 overexpression rice transgenics (Dansana et al., 2014), it could be speculated that OsSAP1-mediated alteration of OsSCP protein can converge the biotic and abiotic stress responses.
The signaling mechanisms involved in the stress response are complex and often interconnected. However, some elements are responsible for invoking specific stress responses that provide the specificity in the pathway (e.g., SOS pathway in salt stress response or ICE1 mediated cold stress response; Knight and Knight, 2001; Chinnusamy et al., 2004). It is evident that, except for RD29A and RD29B, all the stress responsive genes which were differentially expressed by the overexpression of OsAMTR1 and OsSCP were not affected by the overexpression of OsSAP1 (Giri, 2009). So, it could be speculated that OsSAP1 can regulate the stress responses by either modulating the expression of stress responsive genes or by affecting activity of its interacting proteins, which in turn can regulate the downstream stress responsive genes.
Conclusion
The present study suggests that the OsAMTR1 and OsSCP interact with OsSAP1 and positively regulate the abiotic stress response in transgenic Arabidopsis. It may be stated that OsSAP1 overexpression does not directly affect the expression of OsAMTR1 and OsSCP in transgenic rice (Dansana et al., 2014). However, these proteins interact with OsSAP1 and SAPs have been shown to act as ubiquitin ligase, thereby affecting the interacting proteins which may be considered as possible mechanism of action for investigation in future. It may also be noted that the mechanism of action of AMTR1 and SCP may be variable or overlapping as discussed earlier.
Author Contributions
KK performed all the experiments and wrote the manuscript. PD did BiFC experiment. AT conceived the idea. AT and JG designed the experiments, analyzed data and wrote manuscript. All authors approved final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
KK and PD acknowledge the Council for Scientific and Industrial Research [Award No. 09/803(0064)/2009-EMR-I] and University Grants Commission, India, respectively, for research fellowship. JG acknowledges the core grant from National Institute of Plant Genome Research, India. Our research is supported by Department of Biotechnology, Government of India.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01057
References
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485. doi: 10.1007/s11103-007-9199-y
Agrawal, G. K., Jwa, N. S., and Rakwal, R. (2000). A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem. Biophys. Res. Commun. 274, 157–165. doi: 10.1006/bbrc.2000.3114
Allocco, D. J., Kohane, I. S., and Butte, A. J. (2004). Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 5:18. doi: 10.1186/1471-2105-5-18
Bhatnagar-Mathur, P., Vadez, V., and Sharma, K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 27, 411–424. doi: 10.1007/s00299-007-0474-9
Cantacessi, C., Campbell, B. E., Visser, A., Geldhof, P., Nolan, M. J., Nisbet, A. J., et al. (2009). A portrait of the “SCP/TAPS” proteins of eukaryotes–developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27, 376–388. doi: 10.1016/j.biotechadv.2009.02.005
Chakrabarty, R., Banerjee, R., Chung, S. M., Farman, M., Citovsky, V., Hogenhout, S. A., et al. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20, 740–750. doi: 10.1094/MPMI-20-7-0740
Chang, E. J., Ha, J., Kang, S. S., Lee, Z. H., and Kim, H. H. (2011). AWP1 binds to tumor necrosis factor receptor-associated factor 2 (TRAF2) and is involved in TRAF2-mediated nuclear factor-kappaB signaling. Int. J. Biochem. Cell Biol. 43, 1612–1620. doi: 10.1016/j.biocel.2011.07.010
Chinnusamy, V., Schumaker, K., and Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Choi, H., Han, S., Shin, D., and Lee, S. (2012). Polyubiquitin recognition by AtSAP5, an A20-type zinc finger containing protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 419, 436–440. doi: 10.1016/j.bbrc.2012.02.044
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dansana, P. K., Kothari, K. S., Vij, S., and Tyagi, A. K. (2014). OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 33, 1425–1440. doi: 10.1007/s00299-014-1626-3
Dixon, D. C., Cutt, J. R., and Klessig, D. F. (1991). Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 10, 1317–1324.
Englbrecht, C. C., Schoof, H., and Bohm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39. doi: 10.1186/1471-2164-5-39
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Giri, J. (2009). Investigations on Function of Abiotic Stress-Related Genes (OsSAPs, OsRLCKs and codA) and their Influence on Transcript Profiles of Native Genes in Transgenics. Ph.D. thesis, University of Delhi, New Delhi.
Giri, J., Dansana, P. K., Kothari, K. S., Sharma, G., Vij, S., and Tyagi, A. K. (2013). SAPs as novel regulators of abiotic stress response in plants. Bioessays 35, 639–648. doi: 10.1002/bies.201200181
Giri, J., Vij, S., Dansana, P. K., and Tyagi, A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191, 721–732. doi: 10.1111/j.1469-8137.2011.03740.x
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Heyninck, K., and Beyaert, R. (2005). A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem. Sci. 30, 1–4. doi: 10.1016/j.tibs.2004.11.001
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Hishiya, A., Iemura, S., Natsume, T., Takayama, S., Ikeda, K., and Watanabe, K. (2006). A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 25, 554–564. doi: 10.1038/sj.emboj.7600945
Kang, M., Abdelmageed, H., Lee, S., Reichert, A., Mysore, K. S., and Allen, R. D. (2013). AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 76, 481–493. doi: 10.1111/tpj.12312
Kang, M., Fokar, M., Abdelmageed, H., and Allen, R. D. (2011). Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 75, 451–466. doi: 10.1007/s11103-011-9748-2
Kanneganti, V., and Gupta, A. K. (2008). Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 66, 445–462. doi: 10.1007/s11103-007-9284-2
Knight, H., and Knight, M. R. (2001). Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6, 262–267. doi: 10.1016/S1360-1385(01)01946-X
Liepman, A. H., and Olsen, L. J. (2001). Peroxisomal alanine : glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 25, 487–498. doi: 10.1046/j.1365-313x.2001.00961.x
Liepman, A. H., and Olsen, L. J. (2003). Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 131, 215–227. doi: 10.1104/pp.011460
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Mizuki, N., and Kasahara, M. (1992). Mouse submandibular glands express an androgen-regulated transcript encoding an acidic epididymal glycoprotein-like molecule. Mol. Cell. Endocrinol. 89, 25–32. doi: 10.1016/0303-7207(92)90207-M
Moreno-Risueno, M. A., Martinez, M., Vicente-Carbajosa, J., and Carbonero, P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390. doi: 10.1007/s00438-006-0186-9
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Mural, R. V., Liu, Y., Rosebrock, T. R., Brady, J. J., Hamera, S., Connor, R. A., et al. (2013). The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25, 3615–3631. doi: 10.1105/tpc.113.117093
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nie, H., Wu, Y., Yao, C., and Tang, D. (2011). Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J. Genet. Genomics 38, 137–148. doi: 10.1016/j.jgg.2011.03.001
Nie, X., Ji, X., Liu, Y., Zheng, L., and Wang, Y. (2014). Elucidation of the specific formation of homo- and heterodimeric forms of ThbZIP1 and its role in stress. Int. J. Mol. Sci. 15, 10005–10017. doi: 10.3390/ijms150610005
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Renault, H., Roussel, V., El Amrani, A., Arzel, M., Renault, D., Bouchereau, A., et al. (2010). The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 10:20. doi: 10.1186/1471-2229-10-20
Ricoult, C., Echeverria, L. O., Cliquet, J. B., and Limami, A. M. (2006). Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J. Exp. Bot. 57, 3079–3089. doi: 10.1093/jxb/erl069
Rocha, M., Licausi, F., Araujo, W. L., Nunes-Nesi, A., Sodek, L., Fernie, A. R., et al. (2010a). Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 152, 1501–1513. doi: 10.1104/pp.109.150045
Rocha, M., Sodek, L., Licausi, F., Hameed, M. W., Dornelas, M. C., and van Dongen, J. T. (2010b). Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 39, 1043–1053. doi: 10.1007/s00726-010-0596-1
Seo, P. J., Kim, M. J., Park, J. Y., Kim, S. Y., Jeon, J., Lee, Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61, 661–671. doi: 10.1111/j.1365-313X.2009.04091.x
Seo, P. J., Lee, A. K., Xiang, F., and Park, C. M. (2008). Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol. 49, 334–344. doi: 10.1093/pcp/pcn011
Sessa, G., Yang, X. Q., Raz, V., Eyal, Y., and Fluhr, R. (1995). Dark induction and subcellular localization of the pathogenesis-related PRB-1b protein. Plant Mol. Biol. 28, 537–547. doi: 10.1007/BF00020400
Sharma, G., Giri, J., and Tyagi, A. K. (2015). Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 237, 80–92. doi: 10.1016/j.plantsci.2015.05.011
Shembade, N., and Harhaj, E. W. (2012). Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cell. Mol. Immunol. 9, 123–130. doi: 10.1038/cmi.2011.59
Singh, K. B., Foley, R. C., and Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Snider, C. S., Hsiang, T., Zhao, G., and Griffith, M. (2000). Role of ice nucleation and antifreeze activities in pathogenesis and growth of snow molds. Phytopathology 90, 354–361. doi: 10.1094/PHYTO.2000.90.4.354
Song, J. T., Lu, H., McDowell, J. M., and Greenberg, J. T. (2004). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40, 200–212. doi: 10.1111/j.1365-313X.2004.02200.x
Sreedharan, S., Shekhawat, U. K., and Ganapathi, T. R. (2012). MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 80, 503–517. doi: 10.1007/s11103-012-9964-4
Stroher, E., Wang, X. J., Roloff, N., Klein, P., Husemann, A., and Dietz, K. J. (2009). Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2, 357–367. doi: 10.1093/mp/ssn084
Takatsuji, H. (1999). Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. doi: 10.1023/A:1006184519697
Taler, D., Galperin, M., Benjamin, I., Cohen, Y., and Kenigsbuch, D. (2004). Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16, 172–184. doi: 10.1105/tpc.016352
Tyagi, H., Jha, S., Sharma, M., Giri, J., and Tyagi, A. K. (2014). Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 225, 68–76. doi: 10.1016/j.plantsci.2014.05.016
Van Loon, L., and Van Strien, E. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
van Noort, V., Snel, B., and Huynen, M. A. (2003). Predicting gene function by conserved co-expression. Trends Genet. 19, 238–242. doi: 10.1016/S0168-9525(03)00056-8
Waadt, R., and Kudla, J. (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. 2008:pdb.prot4995. doi: 10.1101/pdb.prot4995
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Zeier, J., Pink, B., Mueller, M. J., and Berger, S. (2004). Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219, 673–683. doi: 10.1007/s00425-004-1272-z
Zhang, Y., Lan, H., Shao, Q., Wang, R., Chen, H., Tang, H., et al. (2015). An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa L.). J. Exp. Bot. 67, 315–326. doi: 10.1093/jxb/erv464
Keywords: abiotic stress, Arabidopsis, A20/AN1, protein interactions, zinc-finger
Citation: Kothari KS, Dansana PK, Giri J and Tyagi AK (2016) Rice Stress Associated Protein 1 (OsSAP1) Interacts with Aminotransferase (OsAMTR1) and Pathogenesis-Related 1a Protein (OsSCP) and Regulates Abiotic Stress Responses. Front. Plant Sci. 7:1057. doi: 10.3389/fpls.2016.01057
Received: 22 March 2016; Accepted: 06 July 2016;
Published: 19 July 2016.
Edited by:
Niranjan Baisakh, Louisiana State University, USAReviewed by:
Ing-Feng Chang, National Taiwan University, TaiwanSitakanta Pattanaik, University of Kentucky, USA
Copyright © 2016 Kothari, Dansana, Giri and Tyagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akhilesh K. Tyagi, YWtoaWxlc2hAZ2Vub21laW5kaWEub3Jn
 Kamakshi S. Kothari
Kamakshi S. Kothari Prasant K. Dansana
Prasant K. Dansana Jitender Giri
Jitender Giri Akhilesh K. Tyagi
Akhilesh K. Tyagi