- Department of Nematology, University of California, Riverside, Riverside, CA, USA
Polyphagous aphid pests cause considerable economic damage to crop plants, primarily through the depletion of photoassimilates and transfer of viruses. The potato aphid (Macrosiphum euphorbiae) is a notable pest of solanaceous crops, however, the molecular mechanisms that underpin the ability to colonize these hosts are unknown. It has recently been demonstrated that like other aphid species, M. euphorbiae injects a battery of salivary proteins into host plants during feeding. It is speculated that these proteins function in a manner analagous to secreted effectors from phytopathogenic bacteria, fungi and oomycetes. Here, we describe a novel aphid effector (Me47) which was identified from the potato aphid salivary secretome as a putative glutathione-S-transferase (GST). Expression of Me47 in Nicotiana benthamiana enhanced reproductive performance of green peach aphid (Myzus persicae). Similarly, delivery of Me47 into leaves of tomato (Solanum lycopersicum) by Pseudomonas spp. enhanced potato aphid fecundity. In contrast, delivery of Me47 into Arabidopsis thaliana reduced GPA reproductive performance, indicating that Me47 impacts the outcome of plant–aphid interactions differently depending on the host species. Delivery of Me47 by the non-pathogenic Pseudomonas fluorescens revealed that Me47 protein or activity triggers defense gene transcriptional upregulation in tomato but not Arabidopsis. Recombinant Me47 was purified and demonstrated to have GST activity against two specific isothiocyanates (ITCs), compounds implicated in herbivore defense. Whilst GSTs have previously been associated with development of aphid resistance to synthetic insecticides, the findings described here highlight a novel function as both an elicitor and suppressor of plant defense when delivered into host tissues.
Introduction
Aphids are a large family of hemipteran insects that feed from the vasculature tissue of plants. They feed by inserting their flexible hypodermal needle-like mouthpart or stylets into plant tissue and navigate mostly between cells until they puncture the phloem tissue and feed from the sugar-rich sap (Tjallingii and Esch, 1993; Tjallingii, 2006). Both during initial probing and feeding, aphids secrete watery saliva from their stylets (Tjallingii, 2006). It is known that salivary secretions from aphids play important roles in the establishment and maintenance of successful feeding sites (Will et al., 2007). For example, phloem-plugging in fava bean is dependent on the expansion of forisomes in sieve elements. This process can be inhibited by application of aphid salivary extracts (Will et al., 2007). Saliva from numerous aphid species is known to contain a complex mix of proteins (Harmel et al., 2008; Carolan et al., 2009, 2011; Cooper et al., 2011; Nicholson et al., 2012; Rao et al., 2013; Nicholson and Puterka, 2014; Vandermoten et al., 2014; Chaudhary et al., 2015). It is speculated that salivary proteins act in ways similar to protein effectors from plant microbial pathogens. That is, to inhibit or suppress the activation of host immune processes and enable successful colonization. Whilst the salivary protein complement of several aphid species is now known, functional characterization of individual proteins has extended to just a handful of examples (Jaouannet et al., 2014; Kaloshian and Walling, 2016). The pea aphid Acyrthosiphon pisum protein C002 is injected into fava bean during feeding and is required for effective feeding behavior (Mutti et al., 2008). Two proteins (Mp10, Mp42) were identified from the green peach aphid Myzus persicae that reduced aphid performance when transiently expressed in Nicotiana benthamiana by Agrobacterium tumefaciens (Bos et al., 2010). Mp10 induces chlorosis/cell death in N. benthamiana suggesting direct recognition of this salivary protein through mechanisms that are distinct from Mp42 (Bos et al., 2010; Rodriguez et al., 2014). Further effectors from M. persicae (Mp1, Mp2, Mp55, Mp56, Mp57, and Mp58) have been reported that have various impact on aphid fecundity when either transiently or stably expressed in hosts (Pitino and Hogenhout, 2013; Elzinga et al., 2014). However, the molecular functions of these proteins are unknown. Two effectors (Me10, Me23) have to date been identified from the potato aphid Macrosiphum euphorbiae (Atamian et al., 2013). Me10 and Me23 both increase aphid performance when delivered by the bacterium Pseudomonas syringae type three secretion system (T3SS) into N. benthamiana; however, only Me10 had a similar effect when introduced into tomato (Solanum lycopersicum) leaves using the same delivery method (Atamian et al., 2013). As for other aphid effectors, the specific performance-enhancing activities of Me10 and Me23 are unknown.
The mechanisms by which plants defend themselves against aphid attack are wide-ranging. Preformed physical defenses include barriers such as trichomes, waxy cuticles and oily secretions to discourage aphid settling. There are also inducible changes that occur following the onset of aphid feeding. These include transcriptional modifications, generation of reactive oxygen species (ROS), callose deposition and the production of toxic phytoalexins (Moran et al., 2002; Martinez de Ilarduya et al., 2003; De Vos et al., 2005; Kusnierczyk et al., 2008; Louis et al., 2010; Kettles et al., 2013). The perception of microbial plant attackers, lately shown for aphids as well, has been conceptualized in a multi-layered model of plant defense (Jones and Dangl, 2006; Kaloshian and Walling, 2016). In the first instance, immune recognition of conserved Pathogen-Associated Molecular Patterns (PAMPs) results in PAMP-triggered immunity (PTI) which in most cases is enough to prevent infection or colonization. Only if the pathogen or pest has means to overcome PTI and suppress these inducible changes, typically through the action of proteinaceous effectors or other metabolites, can disease or colonization be achieved.
In order to overcome powerful host defenses, aphids must evolve ways of either suppressing the activation of plant immune processes or detoxifying the resulting chemical assault mounted by the host. Glutathione-S-transferases (GSTs) are a class of detoxification enzyme found throughout the eukaryotic kingdom that catalyzes conjugation of reduced glutathione (GSH) to both natural and synthetic xenobiotics (Li et al., 2007). Specifically for insect pests of plants, they are often grouped with classes of other detoxifying enzymes such as cytochrome P450s and carboxy/cholinesterases (Li et al., 2007; Ramsey et al., 2010) and have been linked to the development of resistance against chemical insecticides (Vontas et al., 2001, 2002). In addition to their role in insecticide resistance, GSTs are assumed to protect insects from xenobiotics encountered in nature. Aphid GSTs are induced when feeding on resistant plants (Bansal et al., 2014) or when fed on toxins in artificial diet (Francis et al., 2005). It has been speculated that diversity of GSTs may contribute to host-range of aphids due to capacity to metabolize a greater variety of host toxins (Ramsey et al., 2010). Study of GSTs in insect pests has largely focussed on those present in gut tissue and their interaction with compounds ingested during feeding. The role of GSTs deployed out on or into plant tissues and their interaction with host immune systems is unexplored.
Recent bioinformatic and proteomic analyses of the M. euphorbiae salivary secretome (Atamian et al., 2013; Chaudhary et al., 2014, 2015) revealed the presence of a single putative GST in aphid saliva. In this investigation, we describe the functional characterization of this candidate effector which we have named Me47. The impact of Me47 expression on performance of two aphid species across three different hosts was examined. Additionally, we find an inverse correlation between Me47-dependent activation of defense responses and aphid performance. Finally, we present evidence of substrate specificity of Me47 which helps explain the role of this GST in plant–aphid interactions.
Materials and Methods
Phylogenetic Analysis and Secretion Signal Prediction
Glutathione sequences from A. pisum (Ramsey et al., 2010) were obtained from AphidBase 2.1 (INRA). M. persicae GST sequences were recovered from Myzus DB (INRA) by low stringency (E < 0.1) Blastp analysis of both M. persciae clone O and clone G006 genomes using Me47 sequence as query. Me47 coding sequence was aligned to GST sequences from A. pisum and M. persicae (both clones G006 and O; Supplementary Table S2) using ClustalW and displayed using a Neighbor-Joining tree with 100 bootstrap replicates using Geneious software (Biomatters). M. persicae protein identifiers are presented as MpG006 or MpO to indicate clonal origin.
The predicted amino acid sequences of the GSTs were subjected to de novo signal peptide prediction analysis using SignalP 4.1 and TargetP 1.1 programs (Emanuelsson et al., 2000; Petersen et al., 2011). For SignalP a Hidden Markov model scores higher than 0.45 was used. For TargetP predictions were determined by predefined set of cutoffs that yielded specificity >0.95 on the test sets.
Me47 Cloning and Bacterial Transformation
Me47 coding sequence lacking the secretion signal was amplified from 100 ng of potato aphid cDNA using primers attB1 Me47-F and attB2 Me47-R (Supplementary Table S1) and high-fidelity Phusion polymerase (New England Biolabs) with the following thermocycle (30 s at 95°C, 30 s at 55°C, 30 s at 72°C × 30 cycles). The attB-flanked Me47 PCR product was recombined into pDONRzeo using BP clonase (Invitrogen) following the manufacturer’s instructions. Me47 was sequence verified by Sanger sequencing before subsequent shuttling into the destination vectors pEARLEYGATE100 for in planta Agroexpression (Earley et al., 2006), pVSP_PsSPdes for bacterial delivery in tomato and Arabidopsis (Arabidopsis thaliana; Rentel et al., 2008) and pDEST17 (Invitrogen) for recombinant protein expression using LR clonase (Invitrogen). For initial cloning, electrocompetent DH5α cells were used for all transformations and Agrobacterium strain GV3101 was used for Agroexpression following standard procedures
Plant Materials and Aphid Colonies
Tomato cultivars (cv.) UC82B and Moneymaker, N. benthamiana, tobacco (Nicotiana tabacum) NC-95, mustard India, and Arabidopsis Col-0 were used. Seeds were planted directly into autoclaved soil or transplanted after seeding into soil. Plants were maintained in growth rooms at 22–24°C with 16 h day length and 200 μmol m-2 s-1 light intensity. Solanaceous plants were weekly fertilized with MiracleGro (18-18-21; Stern’s MiracleGro Products).
Colonies of the parthenogenetic M. euphorbiae were reared on tomato cv. UC82B, while M. persicae was reared on tobacco NC-95 or mustard plants. The colonies were maintained in insect cages in a pesticide-free greenhouse at 22–26°C supplemented with light for 16 h day length. One-day old age synchronized M. euphorbiae adults were produced as described by Bhattarai et al. (2007).
Aphid Performance Assays
To assess M. persicae performance on N. benthamiana, Agrobacterium carrying either pEARLEYGATE100-GFP or pEARLEYGATE100-Me47 were grown in LB supplemented with appropriate antibiotics for 36 h at 28°C. Cells were washed thrice and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, 100 μM acetosyringone, pH 5.6) to an OD600 = 0.3. Bacteria were infiltrated into fully expanded leaves using a needleless syringe. After 24 h, four adult M. persicae were applied to infiltrated leaves within clip cages and left to produce a population of age-synchronized nymphs (day 0). After 48 h, all adults and excess nymphs were removed leaving five nymphs on each leaf (day 2). Nymphs were allowed to feed for two further days before being transferred to a second set of plants which had been similarly infiltrated 24 h previously (day 4). Aphids were allowed to feed from the second set of leaves for 4 days, before transfer to a final set of plants infiltrated 24 h previously (day 8). Experiments were terminated on day 12. This method allowed nymphs to mature to adulthood whilst being continuously exposed to high levels of transgene expression. Aphids typically began production of the next generation of nymphs on day 8. Aphid counts were made daily on days 8–12 and nymphs were continuously removed, such that each count represented fecundity over a 24 h period. Counts from all days were pooled for analysis. The experiment was conducted three times with similar results. Comparison of aphid fecundity on GFP-expressing and Me47-expressing leaves was assessed by two-tailed t-test.
To assess M. euphorbiae performance on tomato, GUS or Me47 was delivered by either semi-virulent P. syrinagae pv. tomato (Pst) DC3000 ΔAvrPto ΔAvrPtoB or non-pathogenic P. fluorescens (Pfo) EtHAn engineered with a T3SS (Thomas et al., 2009). In both systems, bacteria were cultured on Kings B plates with appropriate antibiotics for 36 h at 30°C. Cells were washed from plates in 10 mM MgCl2 and resuspended to a density of 1 × 103 CFU/mL in infiltration buffer (1 mM MgCl2, 0.02% Silwet L-77) in 2.5 L total volume. Whole plants were upturned and submerged in infiltration buffer, placed in a vacuum chamber and infiltrated for 2 min at 20 inHg. Plants were immediately transferred to a growth cabinet and allowed to recover overnight. At 24 h post infection (hpi), 10 mature age-synchronized adult M. euphorbiae were applied to the leaves of each plant with a fine paintbrush. Counts of both the surviving adults and newly born nymphs were made daily for 5 days and all nymphs were removed each day such that each count represented fecundity over a 24 h period. The counts from all days were pooled for analysis and each experiment was conducted three times with similar results. Comparison of aphid fecundity on GUS-expressing and Me47-expressing leaves was assessed by two-tailed t-test.
For Arabidopsis performance assays, Pfo EtHAn strain was prepared as for the tomato assay except that leaves were individually syringe-infiltrated rather than whole-plant submersion infiltration. At 24 hpi, single age-synchronized adult M. euphorbiae were applied to the center of each rosette and the whole plant caged. Counts of newly born nymphs were made daily for 5 days as described for the tomato assay. The experiment was conducted three times (Figure 2B; Supplementary Figure S3B) and results were analyzed as for the tomato assay.
Induction of Plant PAMP Responses in Tomato and Arabidopsis
High-dose Pfo inoculum was prepared following the method described above, with the exception that bacteria were infiltrated at OD = 0.01 (∼1 × 106 CFU/mL; Nguyen et al., 2010) compared to the lower dose used for aphid performance assays. Following infiltration, plants were returned to growth conditions until sample harvest at 6 hpi. Experiments were conducted twice with three biological replicates per experiment. Expression data from both experiments were combined and analyzed together. Comparisons of expression levels of defense-related genes in GUS-expressing and Me47-expressing leaves were made using a two-tailed t-test.
qRT-PCR
Leaf tissues from Pfo-infiltrated tomato or Arabidopsis plants were harvested at 6 hpi and snap frozen. Samples were ground in collection tubes using pellet pestles (Sigma) and total RNA extracted using Trizol (Invitrogen) as per the manufacturer’s instructions. The 260/280 ratios of all samples were checked using a Nanodrop spectrophotometer and all were between 1.8 and 2.1 μg of total RNA was treated with DNaseI (NEB) and samples were subsequently tested for gDNA contamination by PCR amplification using either UBI3 (tomato) or PEX4 (Arabidopsis) primer pairs. First strand cDNA synthesis was performed using the SuperScript III kit (Invitrogen). cDNA was diluted 1:10 with dH2O prior to qRT-PCR and 1 μl of this dilution was used per reaction.
Duplicate reactions for each sample/primer-pair combination were conducted using clear 96-well PCR plates (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). Reactions were carried out using an iCycler real-time PCR system (Bio-Rad) using the following thermocycle (5 min at 95°C followed by 30 s at 95°C, 30 s at 58°C, 30 s at 72°C × 40 cycles). Relative expression values for defense-related genes were calculated using the formula 2-ΔCt (Pfaffl, 2001) relative to the TIP41 reference gene (tomato) or PEX4 (Arabidopsis). Expression values were rescaled for presentation such that the buffer treatment is equal to 1.
Protein Purification and Western Blot Analysis
Escherichia coli ArcticExpress cells (Agilent) carrying the pDEST17-Me47 construct were grown in LB media at 37°C to an OD600 of 0.8. Recombinant Me47 production was induced by addition of 1 mM IPTG followed by incubation at 12°C for 16 h. Cells were recovered by centrifugation, resuspended in chilled lysis buffer (300 mM NaCl, 50 mM sodium phosphate, pH7.2) and lysed using sonication (6 × 30 s pulses). The soluble protein fraction was collected and incubated with Ni-NTA agarose beads (Qiagen) for 1 h at 4°C with gentle agitation. Non-specifically bound proteins were removed with four washes of lysis buffer containing 25 mM imidazole. His-tagged Me47 protein was eluted with two washes of lysis buffer containing 250 mM imidazole. Aliquots were taken at all stages of the purification process and protein content assessed by Bradford assay. Twenty micrograms of all samples were separated by SDS-PAGE using a 12% acrylamide gel. To confirm the identity of purified His-tagged Me47, protein was transferred to nitrocellulose membrane and probed with HisProbe-HRP conjugate antibody (Santa Cruz Biotechnology) in PBST with 2% milk powder. Signal was detected by Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) and imaged using X-ray film.
Glutathione Depletion Assay
Fractions of N-terminal His-tagged Me47 protein were pooled and imidazole removed by buffer exchange using PBS (pH 6.5) and PD10 buffer exchange columns (GE Healthcare). N-terminal His-tagged GroEL was prepared using the same method. To assess activity against ITC substrates, 2 μg of each protein treatment (equine liver GST, His-Me47, His-GroEL) were incubated in the presence of 50 μM glutathione and 200 μM of three ITCs (AITC, BITC, and PEITC) at pH 7.0 for 20 min at room temperature. Buffer-only control reactions with no protein treatment were also included. The concentration of free glutathione remaining in each reaction was assessed using the Glutathione Assay Kit (Sigma) following the manufacturer’s instructions.
ROS Burst Assay
GFP, Me47, and Mp10 (Bos et al., 2010) were expressed in N. benthamiana leaf tissue following Agroinfiltration with GV3101 containing pEARLEYGATE100-GFP, pEARLEYGATE100-Me47, or pEARLEYGATE100-Mp10 as described above. At 2 dpi, 2 mm × 2 mm leaf squares from the Agroinfiltrated leaves were cut using a razor blade and soaked overnight in dH2O. Leaf squares were subsequently exposed to flg22 (100 nM) in a luminol-based assay (Chaudhary et al., 2014) and luminosity was recorded using a Mithras LB 940 Multimode Reader luminometer (Berthold Technologies) for 25 min. For assays to test elicitor activity of Me47, naive N. benthamiana leaf disks were prepared as described above before exposure to reaction cocktail containing flg22 (100 nM), Me47 (1.5 μM) or PBS as negative control.
Results
Identification and Phylogenetic Analysis of Me47
The identification of the proteinaceous components of M. euphorbiae saliva and the correlation with salivary gland EST data has been described previously (Atamian et al., 2013; Chaudhary et al., 2014, 2015). This analysis revealed the presence of a single protein (contig Me_WB05003; Me47), encoding 261 amino acids, with predicted GST activity based on homology to known enzymes of this type. To characterize Me47 and to perform phylogenetic analysis, using BLASTp at low stringency (E < 0.1), we identified the GST homologs from the two aphid species with publically available genome sequences. These are the legume specialist A. pisum and the generalist M. persicae with genome sequences for two distinct clones (The International Aphid Genomics Consortium [TIAGC], 2010; Myzus DB). These analyses identified 17 GSTs (AphidBase 2.1; Ramsey et al., 2010) from A. psium and nine GSTs from each of the M. persicae clones O and G006 (Myzus DB). We identified several alternate spliced forms of some of these GSTs and only one representative of these was included in further analysis. Phylogenetic analysis of Me47 coding sequence relative to the GST predicted proteins identified from A. pisum and M. persicae revealed that Me47 is more similar to GSTs from A. pisum (Figure 1A). A. pisum has three different classes of GST and the closest homolog to Me47 is the delta-class GST ACYPI006899 which encodes a 241 amino acid protein (Chelvanayagam et al., 2001; Ramsey et al., 2010). Direct comparison between Me47 and ACYPI006899 showed 62% identity at the amino acid level (Figure 1B) with conservative or semi-conservative substitutions at 2/6 positions of the GSH-binding site (G site; Figure 1B black asterisks) and 2/9 positions of the substrate-binding pocket (H site; Figure 1B red asterisks).
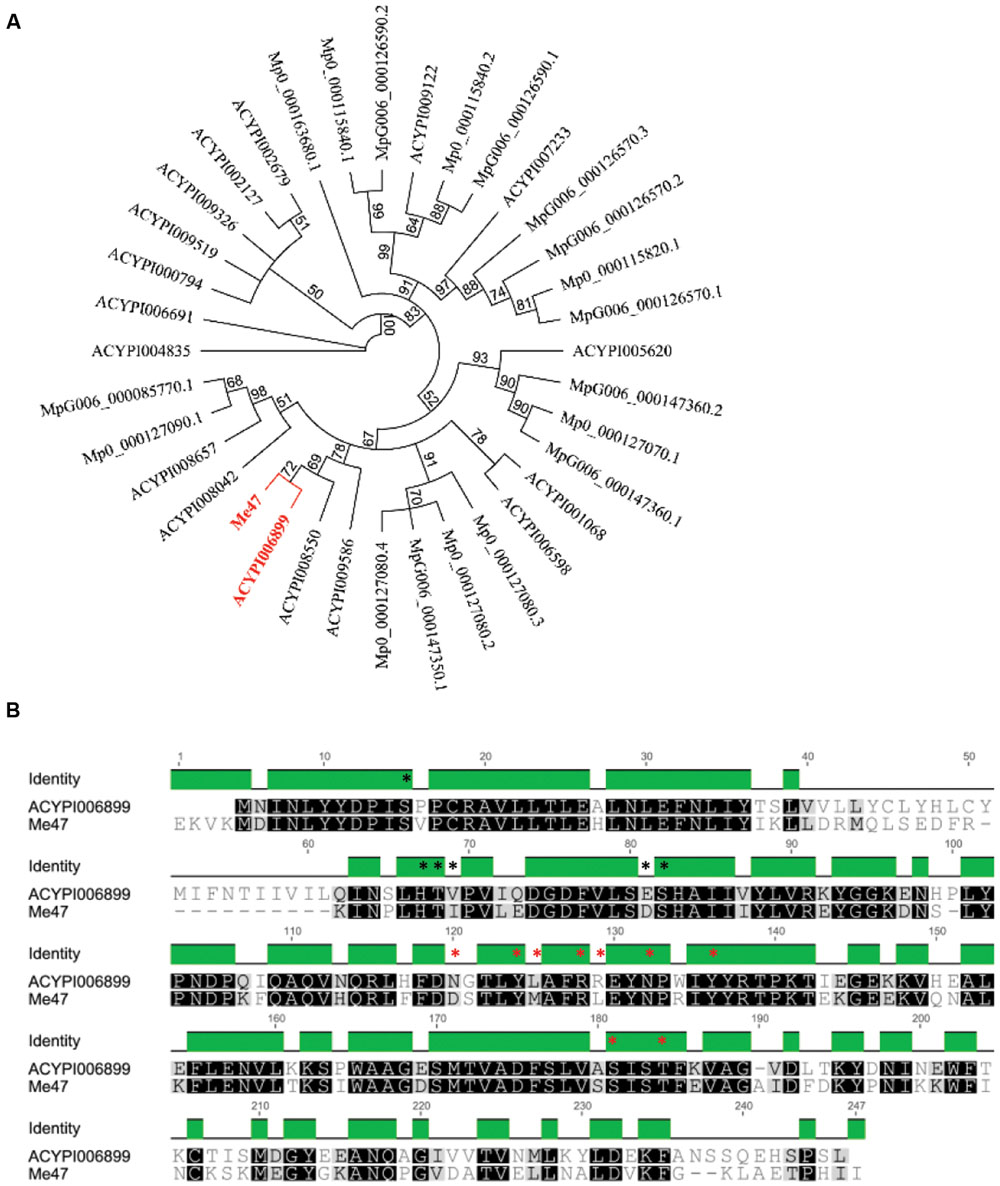
FIGURE 1. Phylogenetic analysis of Me47 relative to A. pisum and M. persicae GST sequences. (A) Predicted Me47 amino acid sequences were aligned with M. persicae (both clones G006 and O) and A. pisum GSTs using ClustalW and presented as a phylogenetic tree using the Neighbor-joining method (Geneious, 100 bootstrap replicates). Me47 and the closest A. pisum ortholog (ACYPI006899) are highlighted in red. (B) Alignment of Me47 with ACYPI006899. Residues forming the GSH binding site (G site; black) and substrate binding pocket (H site; red) are marked with asterisks.
Since Me47 peptides were detected in the M. euphorbiae saliva (Chaudhary et al., 2014, 2015), we investigated the presence of a secretion signal peptide cleavage site in the predicted Me47 protein. Using SignalP, the presence of a 28 amino acid secretion signal was identified in Me47 confirming secretion of this GST in aphid saliva (Petersen et al., 2011). Curiously, ACYPI006899 does not contain a secretion signal cleavage site predicted by SignalP. Indeed, of the 36 aphid GSTs in this analysis, only three (Me47, ACYPI009586 and MpO_000127080.4) contain putative canonical secretion signal cleavage sites. Secretion of proteins could also be predicted by TargetP in the absence of a secretion signal peptide cleavage site (Emanuelsson et al., 2000). Using TargetP with the remaining M. persicae and A. pisum GSTs, predicted secretion for two additional A. pisum GSTs (ACYPI006899 and ACYPI006691) including the Me47 homolog (ACYPI006899) was identified. Taken together this information indicates that aphid GSTs have evolved different mechanisms for secretion and that their secretion into either extracellular spaces or saliva is relatively uncommon.
Me47 Modifies Aphid Performance in Multiple Fecundity Systems
To examine the role of Me47 during aphid colonization, we used Agrobacterium-mediated transient expression to express Me47 in leaf tissue of N. benthamiana. As M. euphorbiae does not reproduce successfully on N. benthamiana, we assessed the fecundity of M. persicae, which is able to feed on this host, over a 12-day period in an assay similar to experiments conducted previously (Bos et al., 2010; Atamian et al., 2013). In these experiments, we found that M. persicae fecundity was significantly increased on Me47-expressing leaves compared to GFP-expressing control leaves (Figure 2A, P < 0.001, Supplementary Figure S3A). This indicates that Me47 may function as a suppressor of plant immunity to enhance aphid colonization of tobacco.
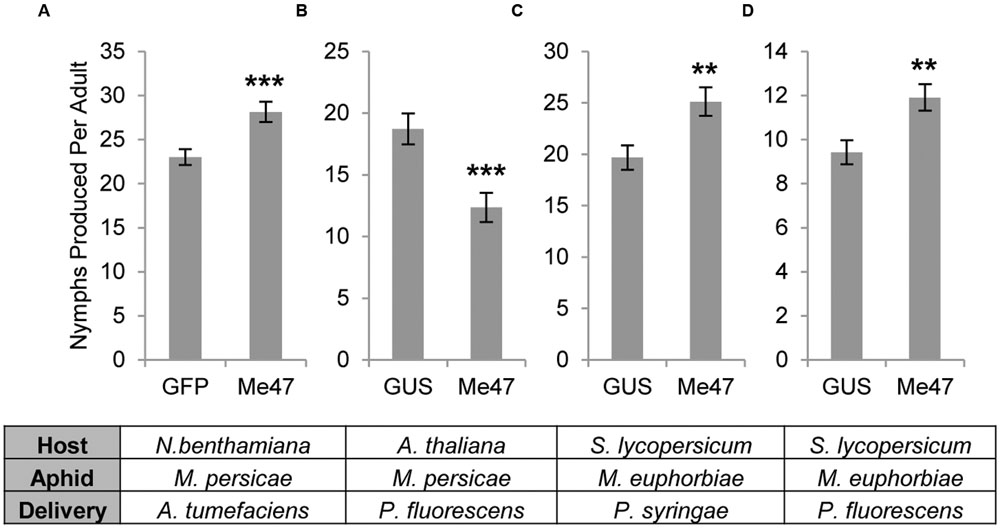
FIGURE 2. Me47 changes aphid reproductive performance. (A) M. persicae fecundity assessed on N. benthamiana transiently expressing either GFP or Me47 by Agroinfiltration. (B) M. persicae fecundity assessed on A. thaliana infected with either Pfo EtHAn (pVSP-GUS) or Pfo EtHAn (pVSP-Me47). (C) M. euphorbiae fecundity assessed on tomato cv. Moneymaker infected with either Pst DC3000 ΔAvrPto/ΔAvrPtoB (pVSP-GUS) or Pst DC3000 ΔAvrPto/ΔAvrPtoB (pVSP-Me47). (D) M. euphorbiae fecundity assessed on tomato cv. Moneymaker infected with either Pfo EtHAn (pVSP-GUS) or Pfo EtHAn (pVSP-Me47). ∗∗P < 0.01 and ∗∗∗P < 0.001 as determined by two-tailed t-test. Data from single experiments presented and data from additional experiments are shown in Supplementary Figure S3.
To assess the role of Me47 in wider plant–aphid interactions, we transformed the semi-virulent bacterial strain Pst DC3000 ΔAvrPto/ΔAvrPtoB with the construct pVSP_PsSPdes Me47 (pVSP-Me47). Whole tomato plants were vacuum-infiltrated with this semi-virulent inoculum. Using this method, the aphid protein of interest is delivered into tomato leaf cells via the bacterial T3SS, allowing performance of M. euphorbiae to be assessed on its natural host (Atamian et al., 2013). In these trials, M. euphorbiae fecundity was significantly increased on plants infected with Pst DC3000 ΔAvrPto/ΔAvrPtoB (pVSP-Me47) relative to those infected with the Pst DC3000 ΔAvrPto/ΔAvrPtoB (pVSP-GUS) control (Figure 2C, p < 0.01, Supplementary Figure S3C). This indicates that Me47 can function as a pathogenicity determinant in at least two host plant species with impact on two distinct aphid pests. However, as Pst DC3000 ΔAvrPto/ΔAvrPtoB possesses its own effector complement and is semi-virulent to tomato, we chose to assess the effect of Me47 delivery in the absence of other pathogen effectors. For this experiment, we used the non-pathogenic Pfo EtHAn strain (Thomas et al., 2009), which has been engineered to express the T3SS. This strain was transformed with the same constructs used in experiments described for Pst DC3000 ΔAvrPto/ΔAvrPtoB. In fecundity assays, M. euphorbiae performed significantly better on tomato infected with Pfo EtHAn (pVSP-Me47) compared to a Pfo EtHAn (pVSP-GUS) control (Figure 2D, p < 0.01, Supplementary Figure S3D). This confirmed our previous result in tomato, and indicates that the choice of Pseudomonas species for delivery of Me47 has minimal impact on the role of this protein in modifying M. euphorbiae fecundity on tomato. Finally, we used Pfo EtHAn with the same constructs to assess M. persicae fecundity on Arabidopsis. Interestingly in these experiments, M. persicae fecundity was significantly reduced on plants infected with Pfo EtHAn (pVSP-Me47) relative to the Pfo EtHAn (pVSP-GUS) control (Figure 2B, p < 0.001, Supplementary Figure S3B). This indicates that in specific host–aphid interactions, Me47 can have a host-dependent deleterious impact on aphid fecundity.
Me47 Induces PAMP-Responsive Genes in Tomato But Not in Arabidopsis
Our data indicated that in some experimental systems, Me47 made a positive contribution to aphid fecundity (Figures 2A,C,D) but in others the impact was negative (Figure 2B). As a non-pathogen, Pfo has been shown to induce PTI-related defense genes following infiltration into leaves of several plant species (Nguyen et al., 2010). We therefore made use of the Pfo EtHAn strain to assess ability of Me47 to suppress the PTI responses induced by this non-pathogenic bacterium. In these experiments, tomato plants were challenged with Pfo EtHAn delivering either GUS or Me47. The tomato defense genes Lrr22 and Pti5 have previously been shown to be inducible at 6 hpi following Pfo treatment (Nguyen et al., 2010). In our experiments, Lrr22 and Pti5 were only slightly induced by Pfo EtHAn (pVSP-GUS) but this increase was not statistically significant relative to the buffer control (Figures 3A,B). However, as the initial study used tomato cultivar Rio Grande-prf3, and the cultivar used in experiments described here is Moneymaker, it is possible that there is temporal variation in defense gene induction between tomato cultivars.
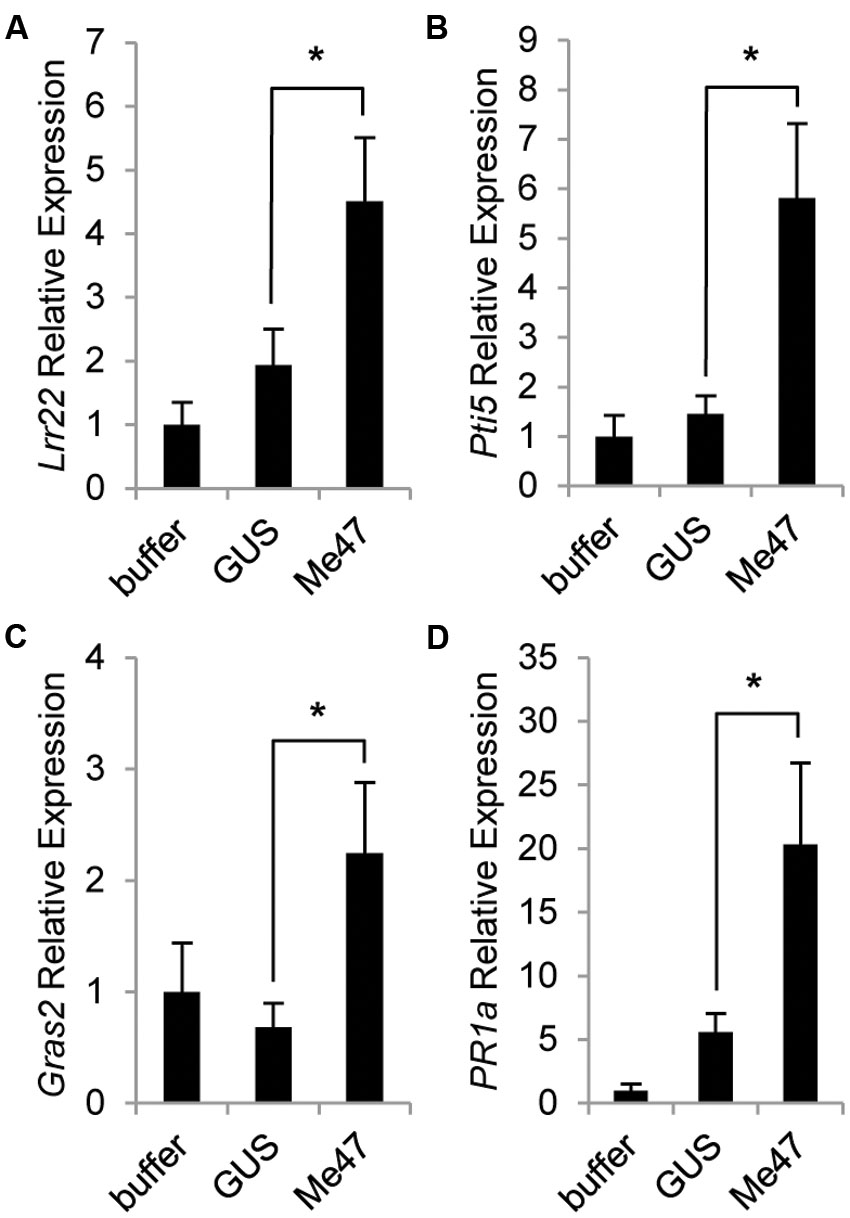
FIGURE 3. Me47 induces defense genes in tomato. Tomato cv. Moneymaker was infiltrated with buffer-only, Pfo EtHAn (pVSP-GUS) or Pfo EtHAn (pVSP-Me47) and leaves harvested at 6 hpi. Expression analysis of genes involved in PTI (Lrr22, Pti5, Gras2) (A–C) and salicylic acid (SA)-dependent defense (PR1a) (D) conducted by qRT-PCR. ∗P < 0.05 as determined by two-tailed t-test. Bars represent means and standard error across six biological replicates from two independent experiments. Buffer treatment rescaled to 1 for presentation.
As we previously showed that Me47 can enhance aphid performance on tomato (Figure 2D), we suspected that Me47 might further suppress expression of these two defense genes. To our surprise, we found that delivery of Me47 by Pfo EtHAn enhanced the induction of both Lrr22 and Pti5 when transcript abundance was assessed at 6 hpi (Figures 3A,B, p < 0.05). For the PAMP-inducible gene Gras2, Nguyen et al. (2010) reported no induction at 6 hpi following Pfo infection. Similarly, we found no change in expression between the buffer and Pfo EtHAn (pVSP-GUS) treatments (Figure 3C). However, Gras2 was significantly induced following Pfo EtHAn (pVSP-Me47) treatment (Figure 3C, p < 0.05). In this experiment, we also analyzed the expression of the PR1a reporter gene as it is frequently observed to be inducible both during pathogen infection and by PAMP treatment. Similar to the other genes tested, PR1a was highly induced following Pfo EtHAn (pVSP-Me47) treatment relative to both Pfo EtHAn (pVSP-GUS) and the buffer control (Figure 3D, p < 0.05). Together, this defense gene expression dataset illustrates the surprising observation that delivery of Me47 into tomato leaves enhances the expression of PAMP-responsive genes during bacterial challenge.
To assess whether a similar phenomenon is present in another host used in our aphid bioassay, we conducted a parallel experiment to monitor defense gene induction in Arabidopsis following delivery of Me47 by Pfo EtHAn (Figures 4A–D). Unlike tomato, a specific defense marker assay for Pfo infection has not been developed for Arabidopsis. However, numerous studies have used Arabidopsis for dissection of aphid-relevant defense pathways (De Vos et al., 2005; Couldridge et al., 2007; Kusnierczyk et al., 2008; Kettles et al., 2013). The camalexin biosynthetic gene PAD3 is known to be involved in resistance to numerous pathogens in addition to aphids. We therefore hypothesized it would be a good choice for assessing defense activation in plants challenged with Pfo. Whilst Pfo infection indeed caused a significant increase in PAD3 expression (Figure 4A), there was no difference in expression levels between the Pfo EtHAn (pVSP-GUS) and Pfo EtHAn (pVSP-Me47) treatments at 6 hpi (Figure 4A). CYP81F2 has been reported to be involved in the production of indolic glucosinolates that have activity against some pathogens and also aphids (Bednarek et al., 2009; Pfalz et al., 2009). Again, we found that expression of this gene was highly responsive to Pfo treatment at 6 hpi irrespective of the expressed construct (Figure 4B). PDF1.2 is routinely used as a defense marker of specific relevance to the jasmonic acid (JA)/ethylene signaling pathways, whilst PR1 has long been known to be highly responsive to many pathogens/pests and as a marker for salicylic acid (SA)-related defense signaling. Neither of these genes showed statistically significant responses either to Pfo treatment or delivery of Me47 relative to GUS at 6 hpi (Figures 4C,D). Together, we found no evidence of enhanced defense marker gene expression in Arabidopsis leaves treated with Pfo EtHAn (pVSP-Me47) relative to Pfo EtHAn (pVSP-GUS) at 6 hpi, thus illustrating differential responses of tomato and Arabidopsis to the Me47 effector protein.
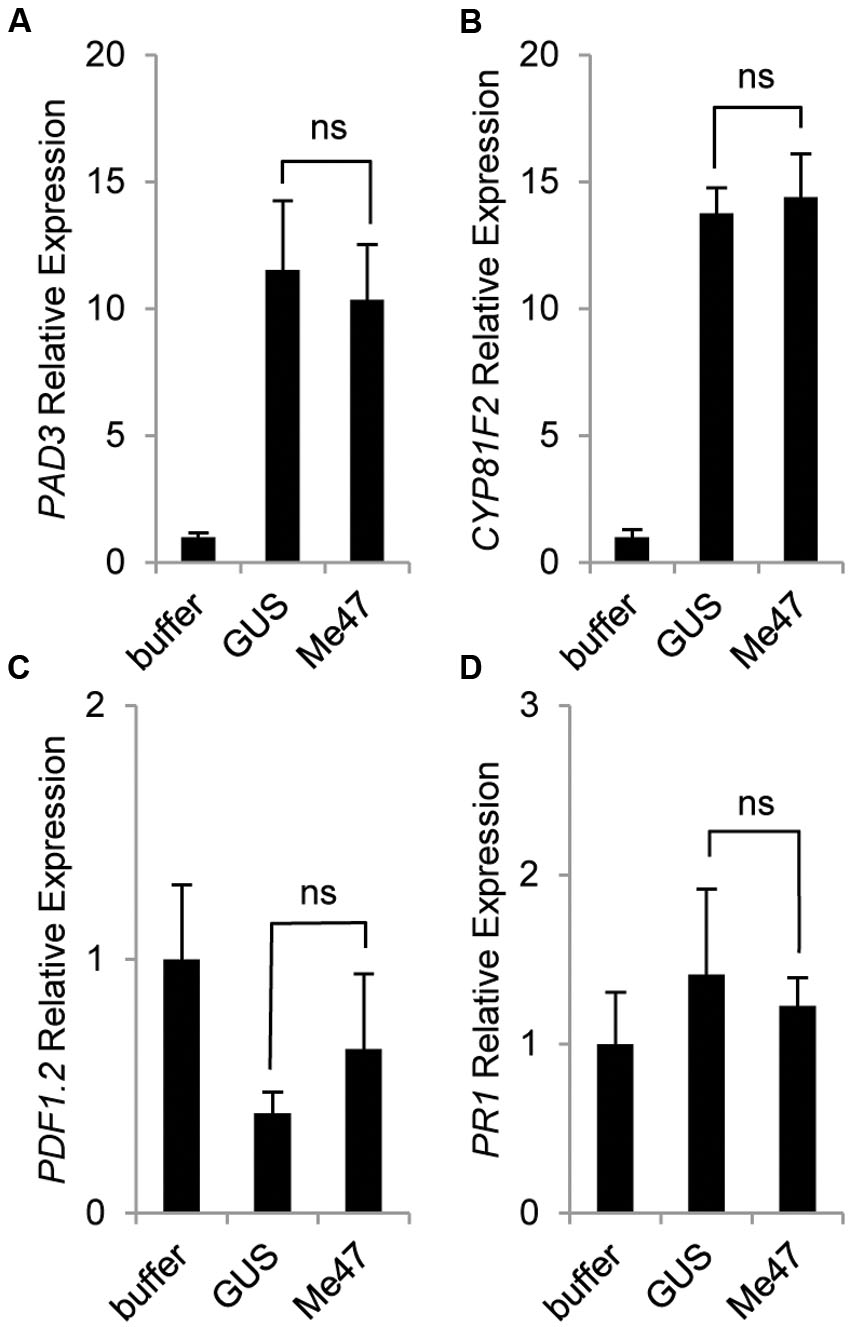
FIGURE 4. Me47 has no effect on defense gene induction in Arabidopsis. Arabidopsis Col-0 was treated as described in Figure 3 and leaves harvested at 6 hpi. Expression analysis of genes involved in (A) camalexin (PAD3), (B) indole glucosinolate (CYP81F2), (C) Jasmonic acid/ethylene (JA/ET; PDF1.2) and (D) SA pathway (PR1) done by qRT-PCR. Bars represent means and standard error across six biological replicates from two independent experiments. Buffer treatment rescaled to 1 for presentation. Differences between GUS and Me47 treatments were not significant (ns).
Me47 Is a Glutathione-S-Transferase with Activity against Isothiocyanates
To confirm that Me47 is a functional GST, recombinant N-terminal His-tagged Me47 (His-Me47) was expressed and purified from bacterial cell lysates (Supplementary Figure S1) for use in a GST activity assay utilizing the broad-spectrum GST substrate 1-Chloro-2,4-dinitrobenzene (CDNB). Surprisingly, purified His-Me47 showed no ability to conjugate glutathione to CDNB when compared to commercially available GST preparations (data not shown). Nonetheless, we developed a glutathione depletion assay based on the method of Wadleigh and Yu (1988) to assess activity of Me47 against a selection of isothiocyanates (ITCs). These volatile defense compounds are specific to members of Brassicaceae, have toxic activity against insects and are known substrates for both insect and human GSTs (Wadleigh and Yu, 1988; Kolm et al., 1995). The bacterial chaperonin GroEL, also expressed and purified with an N-terminal His-tag (His-GroEL), was used as a negative control. In this assay, His-Me47 depleted the free glutathione in the presence of benzyl isothiocyanate (BITC; Figure 5A) and phenylethyl isothiocyanate (PEITC; Figure 5B) to a level comparable to the commercially prepared equine GST (eqGST) positive control. As expected, His-GroEL did not have any glutathione-depleting activity in the presence of either BITC or PEITC similar to the buffer-only control. In contrast, His-Me47 was unable to utilize allyl isothiocyanate (AITC) as substrate and the free glutathione level remained consistent with the buffer and His-GroEL protein controls (Figure 5C). These data illustrate that Me47 is a functional GST with ability to utilize known plant defense compounds as substrates.
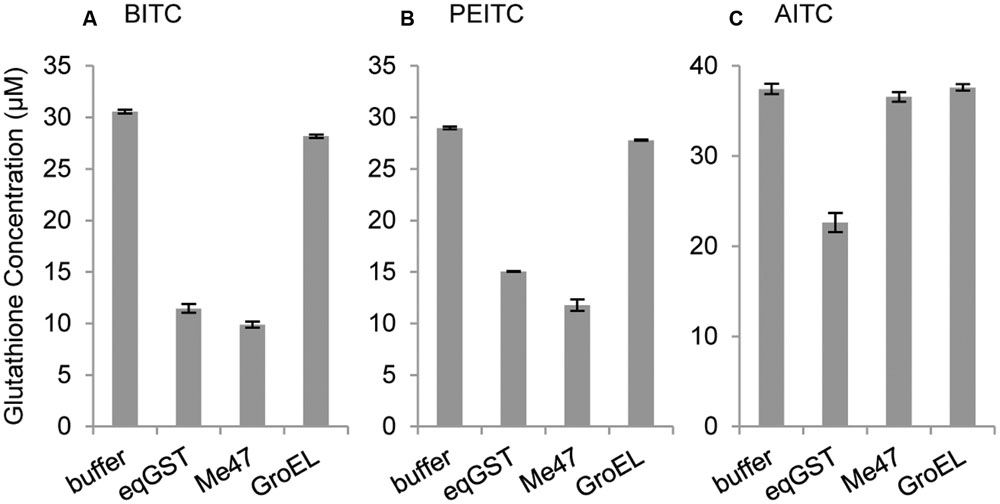
FIGURE 5. Me47 can utilize selected isothiocyanates as substrates. Glutathione depletion assay analysis of substrates (A) benzyl isothiocyanate (BITC), (B) phenylethyl isothiocyanate (PEITC), and (C) allyl isothiocyanate (AITC), incubated with buffer control, commercial equine GST (eqGST; Sigma), recombinant N-terminal His-tagged Me47 (Me47) and non-enzymatic recombinant N-terminal His-tagged GroEL (GroEL).
Me47 Does Not Interfere with the PAMP-Induced ROS Burst in N. benthamiana
The M. persicae effector Mp10 was previously demonstrated to suppress the flg22-induced ROS burst when transiently expressed in N. benthamiana (Bos et al., 2010). We conducted a similar experiment to test whether Me47 might have similar properties. In contrast to Mp10, Me47 had no suppressive effect on the flg22-induced ROS burst relative to leaf tissue expressing a GFP control transgene (Supplementary Figure S2A). Given that Me47 was found to induce PAMP-responsive defense genes when delivered into tomato leaves (Figure 3), we then tested whether Me47 might trigger a ROS burst in N. benthamiana using the same recombinant Me47 protein as used for the glutathione depletion assay (Figure 5, Supplementary Figure S2). Me47 protein was unable to induce ROS production above the PBS control levels (Supplementary Figure S2B) indicating that it is not an elicitor of ROS burst in N. benthamiana.
Discussion
To date, only a handful of aphid effectors have been reported and for all the specific function or activity is unknown. For M. euphorbiae, effectors Me10 and Me23 are the only examples known to have impact on aphid fecundity when expressed in planta (Atamian et al., 2013). Agroexpression of both Me10 and Me23 increased M. persicae fecundity on N. benthamiana, whilst only Me10 increased potato aphid fecundity on tomato when delivered through bacterial T3SS (Atamian et al., 2013). In studies on M. persicae effectors, Mp10 and Mp42 were found to reduce aphid fecundity on N. benthamiana (Bos et al., 2010). Additional M. persicae effectors, Mp55-58, were described by Elzinga et al. (2014) that have either beneficial or deleterious impact on aphid success across N. benthamiana, N. tabacum or Arabidopsis. In each case, the change in aphid performance was consistent across the host species assayed. This indicates that well-conserved defense mechanisms may be subject to manipulation by these effectors.
Previously it has been also demonstrated that some aphid effectors have host-specific activity (Bos et al., 2010; Pitino and Hogenhout, 2013). For example, M. persicae effectors Mp1 and Mp2 enhanced M. persicae performance when stably expressed in Arabidopsis. In contrast, transient expression of the A. pisum orthologs of these genes had no effect on M. persicae performance. Me47 is the first aphid effector observed to have both beneficial and detrimental impact on aphid colonies that is host-dependent. In tomato, Me47 improved M. euphorbiae reproductive success (Figures 2C,D) yet Me47 protein or activity was recognized and induced the expression of multiple defense genes (Figure 3). Remarkably, Me47 decreased M. persicae fecundity on Arabidopsis (Figure 2B), yet there was no immune recognition of Me47 protein or activity, at an early effector delivery time point (6 hpi), as indicated by expression levels of several genes previously linked to aphid defense (Figure 4). These observations are surprising, as it is expected that an increase in aphid fecundity (as on tomato) would align with some degree of immune suppression and not immune activation. Furthermore, a decrease in aphid fecundity (as on Arabidopsis) might be expected to accompany a degree of immune stimulation. Together, these observations suggest that the direct recognition of either Me47 or its activity does not underpin the likelihood of successful aphid colonization. Additionally, the immunogenicity of Me47 may be suppressed in natural aphid infestations by the action of other, as yet unidentified, effector proteins present in the salivary milieu.
It has been speculated that aphids might actively trigger host defenses that have little efficacy against this class of plant attacker (Walling, 2008). For example, it is known that the JA and SA signaling pathways can act antagonistically where induction of one leads to suppression of the other. Aphid infestations primarily elicit SA-dependent defenses (De Vos et al., 2005; Kettles et al., 2013), yet other studies have reported JA-mediated defense to be more effective (Ellis et al., 2002; Zhu-Salzman et al., 2004). It is possible that the defense pathways triggered by Me47 delivery in tomato have little impact on aphid colonization, but might supress more effective defense responses not included as part of this investigation. Indeed, PR1a is frequently used as a marker gene for SA-dependent defense responses and was highly induced by Me47 in tomato (Figure 3D) but not in Arabidopsis (Figure 4D) consistent with this hypothesis.
Me47 delivery in tomato induces defense-related genes as determined by qRT-PCR (Figure 3). However, the elicitor activity of Me47 remains to be determined. From limited available data, it appears many endogenous Arabidopsis GSTs have non-specific subcellular localization and are present in the cytosol (Dixon et al., 2009), although a limited number are nuclear- or peroxisome-localized. We were unable to precisely localize Me47 in plant cells as transient expression of yellow fluorescent protein (YFP)-tagged Me47 in N. benthamiana revealed localization to both the cytosol and nucleus similar to YFP control (Data not shown). Since the Me47-YFP size is 57 kDa the protein could defuse through the nuclear pore. Nevertheless, it is unlikely that Me47 is present in plant organelles in the absence of endogenous GSTs. One possibility is that it is not Me47 that is recognized but the metabolomic products of its activity. Me47 substrate specificity may be different from endogenous GSTs, such that Me47-catalyzed reaction products are hallmarks of a foreign GST. Aberrant GST activity might therefore be open to recognition and stimulate immunity in a manner analogous to the perception of PAMPs during the PTI phase of host–microbe interactions.
A plethora of secondary plant metabolites exists and is speculated the primary function of many is for defense against herbivory. To overcome these defenses, insects have evolved large and diverse classes of detoxification enzymes, including GSTs, to negate the potentially lethal effects of toxic phytochemicals (Li et al., 2007; Ramsey et al., 2010). In aphids, GSTs have been linked to detoxification of glucosinolates (Francis et al., 2005) and nicotine (Ramsey et al., 2014) and the cereal hydroxamic acid 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA; Mukanganyama et al., 2003). A single GST from the cotton bollworm (Helicoverpa armigera) was also found to detoxify the plant JA pathway precursor 12-oxophytodienoic acid (cis-OPDA; Dabrowska et al., 2009). However, the focus of insect GSTs has been almost exclusively on their role in the gut (for phytochemical detoxification) or in the cuticle/body (for insecticide detoxification; Vontas et al., 2001, 2002). To our knowledge, no prior study has described the role of a single GST, from any insect, out of the producing organism and in direct mediation of host–pest interactions. It is perhaps not surprising that such a mechanism has evolved in insects, as longer exposure time of toxic phytochemicals to detoxification enzymes likely reduces the concentration of toxin ingested and exposed to cells in the gut. Whilst this function for Me47 is therefore novel, it is unlikely to be the only example of such a phenomenon. Indeed, a catalytically active GST, expressed in salivary glands of wheat-infesting Hessian fly (Mayetiola destructor; Yoshiyama and Shukle, 2004) suggest that additional examples will exist in other groups of plant pests.
In our characterization of Me47, we found that Me47 substrate specificity did not include CDNB, a model substrate found to be metabolized by total GST preparations from M. persicae (Francis et al., 2005). The activity spectrum of total GST extracts from M. euphorbiae have not been described, but it is possible that other GSTs aside from Me47 show activity against this model substrate. In this study, we identified two ITCs (BITC, PEITC) as Me47 substrates (Figure 5). Me47 did not metabolize AITC, however, suggesting some degree of enzymatic specificity within this class of defensive metabolite. ITCs are defense compounds associated with insect resistance and are specific for cruciferous plants. During aphid infestations of plants belonging to this family, such as Arabidopsis, the function of Me47 is therefore clear. However, natural substrates of Me47 from the other hosts used in this study, tobacco and tomato, remain to be identified. It is therefore not yet possible to assess whether Me47 is a highly promiscuous, broad-spectrum GST or moderately promiscuous in its activity against plant defense compounds of relevance to natural M. euphorbiae infestations. Our initial data regarding metabolism of ITCs, coupled with the inability of Me47 to metabolize the model substrate CDNB would suggest the latter, but this requires further biochemical investigation.
Author Contributions
GK and IK conceived and planned the experiments. GK performed the experiments. GK and IK analyzed the data. GK wrote the manuscript with help from IK.
Funding
This work was funded by grant (2010-65106-20675) to IK from the United States Department of Agriculture, National Institute of Food and Agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors would like to thank Hagop Atamian and Ritu Chaudhary for helpful discussions during the course of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01142
References
Atamian, H. S., Chaudhary, R., Cin, V. D., Bao, E., Girke, T., and Kaloshian, I. (2013). In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol. Plant Microbe Interact. 26, 67–74. doi: 10.1094/MPMI-06-12-0144-FI
Bansal, R., Mian, M. A. R., Mittapalli, O., and Michel, A. P. (2014). RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean. BMC Genomics 15:972. doi: 10.1186/1471-2164-15-972
Bednarek, P., Piślewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Bhattarai, K. K., Xie, Q., Pourshalimi, D., Younglove, T., and Kaloshian, I. (2007). Coi1-dependent signaling pathway is not required for Mi-1—Mediated potato aphid resistance. Mol. Plant Microbe Interact. 20, 276–282. doi: 10.1094/MPMI-20-3-0276
Bos, J. I., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216. doi: 10.1371/journal.pgen.1001216
Carolan, J. C., Caragea, D., Reardon, K. T., Mutti, N. S., Dittmer, N., Pappan, K., et al. (2011). Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. J. Proteome Res. 10, 1505–1518. doi: 10.1021/pr100881q
Carolan, J. C., Fitzroy, C. I., Ashton, P. D., Douglas, A. E., and Wilkinson, T. L. (2009). The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9, 2457–2467. doi: 10.1002/pmic.200800692
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., and Kaloshian, I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. U.S.A. 111, 8919–8924. doi: 10.1073/pnas.1407687111
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., and Kaloshian, I. (2015). Potato aphid salivary proteome: enhanced salivation using resorcinol and identification of aphid phosphoproteins. J. Proteome Res. 14, 1762–1778. doi: 10.1021/pr501128k
Chelvanayagam, G., Parker, M. W., and Board, P. G. (2001). Fly fishing for GSTs: a unified nomenclature for mammalian and insect glutathione transferases. Chem. Biol. Interact. 133, 256–260.
Cooper, W. R., Dillwith, J. W., and Puterka, G. J. (2011). Comparisons of salivary proteins from five aphid (Hemiptera: Aphididae) species. Environ. Entomol. 40, 151–156. doi: 10.1603/EN10153
Couldridge, C., Newbury, H. J., Ford-Lloyd, B., Bale, J., and Pritchard, J. (2007). Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull. Entomol. Res. 97, 523–532. doi: 10.1017/S0007485307005160
Dabrowska, P., Freitak, D., Vogel, H., Heckel, D. G., and Boland, W. (2009). The phytohormone precursor OPDA is isomerized in the insect gut by a single, specific glutathione transferase. Proc. Natl. Acad. Sci. U.S.A. 106, 16304–16309. doi: 10.1073/pnas.0906942106
De Vos, M., Van Oosten, V. R., Van Poecke, R. M. P., Van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact 18, 923–937. doi: 10.1094/MPMI-18-0923
Dixon, D. P., Hawkins, T., Hussey, P. J., and Edwards, R. (2009). Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60, 1207–1218. doi: 10.1093/jxb/ern365
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Ellis, C., Karafyllidis, I., and Turner, J. G. (2002). Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol. Plant Microbe Interact. 15, 1025–1030. doi: 10.1094/MPMI.2002.15.10.1025
Elzinga, D. A., De Vos, M., and Jander, G. (2014). Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 27, 747–756. doi: 10.1094/MPMI-01-14-0018-R
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Francis, F., Vanhaelen, N., and Haubruge, E. (2005). Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Arch. Insect. Biochem. Physiol. 58, 166–174. doi: 10.1002/arch.20049
Harmel, N., Letocart, E., Cherqui, A., Giordanengo, P., Mazzucchelli, G., Guillonneau, F., et al. (2008). Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Mol. Biol. 17, 165–174. doi: 10.1111/j.1365-2583.2008.00790.x
Jaouannet, M., Rodriguez, P. A., Thorpe, P., Lenoir, C. J., MacLeod, R., Escudero-Martinez, C., et al. (2014). Plant immunity in plant-aphid interactions. Front. Plant Sci. 5:663. doi: 10.3389/fpls.2014.00663
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaloshian, I., and Walling, L. L. (2016). Hemipteran and dipteran pests: effectors and plant host immune regulators. J. Integr. Plant Biol. 58, 350–361. doi: 10.1111/jipb.12438
Kettles, G. J., Drurey, C., Schoonbeek, H. J., Maule, A. J., and Hogenhout, S. A. (2013). Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 198, 1178–1190. doi: 10.1111/nph.12218
Kolm, R. H., Danielson, U. H., Zhang, Y., Talalay, P., and Mannervik, B. (1995). Isothiocyanates as substrates for human glutathione transferases: structure-activity studies. Biochem. J. 311, 453–459. doi: 10.1042/bj3110453
Kusnierczyk, A., Winge, P., Jorstad, T. S., Troczynska, J., Rossiter, J. T., and Bones, A. M. (2008). Towards global understanding of plant defence against aphids–timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115. doi: 10.1111/j.1365-3040.2008.01823.x
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Louis, J., Lorenc-Kukula, K., Singh, V., Reese, J., Jander, G., and Shah, J. (2010). Antibiosis against the green peach aphid requires the Arabidopsis thaliana MYZUS PERSICAE-INDUCED LIPASE1 gene. Plant J. 64, 800–811. doi: 10.1111/j.1365-313X.2010.04378.x
Martinez de Ilarduya, O., Xie, Q., and Kaloshian, I. (2003). Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant Microbe Interact. 16, 699–708. doi: 10.1094/MPMI.2003.16.8.699
Moran, P. J., Cheng, Y., Cassell, J. L., and Thompson, G. A. (2002). Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch. Insect. Biochem. Physiol. 51, 182–203. doi: 10.1002/arch.10064
Mukanganyama, S., Figueroa, C. C., Hasler, J. A., and Niemeyer, H. M. (2003). Effects of DIMBOA on detoxification enzymes of the aphid Rhopalosiphum padi (Homoptera: aphididae). J. Insect Physiol. 49, 223–229. doi: 10.1016/s0022-1910(02)00269-x
Mutti, N. S., Louis, J., Pappan, L. K., Pappan, K., Begum, K., Chen, M. S., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U.S.A. 105, 9965–9969. doi: 10.1073/pnas.0708958105
Nguyen, H. P., Chakravarthy, S., Velásquez, A. C., McLane, H. L., Zeng, L., Nakayashiki, H., et al. (2010). Methods to Study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 23, 991–999. doi: 10.1094/MPMI-23-8-0991
Nicholson, S. J., Hartson, S. D., and Puterka, G. J. (2012). Proteomic analysis of secreted saliva from Russian wheat aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J. Proteomics 75, 2252–2268. doi: 10.1016/j.jprot.2012.01.031
Nicholson, S. J., and Puterka, G. J. (2014). Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum Rondani) biotypes. J. Proteomics 105, 186–203. doi: 10.1016/j.jprot.2013.12.005
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pfalz, M., Vogel, H., and Kroymann, J. (2009). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21, 985–999. doi: 10.1105/tpc.108.063115
Pitino, M., and Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26, 130–139. doi: 10.1094/MPMI-07-12-0172-FI
Ramsey, J. S., Elzinga, D. A., Sarkar, P., Xin, Y. R., Ghanim, M., and Jander, G. (2014). Adaptation to nicotine feeding in Myzus persicae. J. Chem. Ecol. 40, 869–877. doi: 10.1007/s10886-014-0482-5
Ramsey, J. S., Rider, D. S., Walsh, T. K., De Vos, M., Gordon, K. H., Ponnala, L., et al. (2010). Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect. Mol. Biol. 19(Suppl. 2), 155–164. doi: 10.1111/j.1365-2583.2009.00973.x
Rao, S. A., Carolan, J. C., and Wilkinson, T. L. (2013). Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS ONE 8:e57413. doi: 10.1371/journal.pone.0057413
Rentel, M. C., Leonelli, L., Dahlbeck, D., Zhao, B., and Staskawicz, B. J. (2008). Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc. Natl. Acad. Sci. U.S.A. 105, 1091–1096. doi: 10.1073/pnas.0711215105
Rodriguez, P. A., Stam, R., Warbroek, T., and Bos, J. I. (2014). Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant Microbe Interact. 27, 30–39. doi: 10.1094/MPMI-05-13-0156-R
The International Aphid Genomics Consortium [TIAGC] (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
Thomas, W. J., Thireault, C. A., Kimbrel, J. A., and Chang, J. H. (2009). Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 60, 919–928. doi: 10.1111/j.1365-313X.2009.03998.x
Tjallingii, W. F. (2006). Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 57, 739–745. doi: 10.1093/jxb/erj088
Tjallingii, W. F., and Esch, T. H. (1993). Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol. Entomol. 18, 317–328. doi: 10.1111/j.1365-3032.1993.tb00604.x
Vandermoten, S., Harmel, N., Mazzucchelli, G., De Pauw, E., Haubruge, E., and Francis, F. (2014). Comparative analyses of salivary proteins from three aphid species. Insect Mol. Biol. 23, 67–77. doi: 10.1111/imb.12061
Vontas, J. G., Small, G. J., and Hemingway, J. (2001). Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 357, 65–72. doi: 10.1042/0264-6021:3570065
Vontas, J. G., Small, G. J., Nikou, D. C., Ranson, H., and Hemingway, J. (2002). Purification, molecular cloning and heterologous expression of a glutathione S-transferase involved in insecticide resistance from the rice brown planthopper, Nilaparvata lugens. Biochem. J. 362, 329–337. doi: 10.1042/0264-6021:3620329
Wadleigh, R. W., and Yu, S. J. (1988). Detoxification of isothiocyanate allelochemicals by glutathione transferase in three lepidopterous species. J. Chem. Ecol. 14, 1279–1288. doi: 10.1007/BF01019352
Walling, L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Will, T., Tjallingii, W. F., Thonnessen, A., and van Bel, A. J. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U.S.A. 104, 10536–10541. doi: 10.1073/pnas.0703535104
Yoshiyama, M., and Shukle, R. H. (2004). Molecular cloning and characterization of a glutathione s-transferase gene from hessian fly (Diptera: Cecidomyiidae). Annal. Entomol. Soc. Am. 97, 1285–1293. doi: 10.1603/0013-8746(2004)097[1285:MCACOA]2.0.CO;2
Keywords: effector, potato aphid, glutathione-S-transferase, GST, secretome
Citation: Kettles GJ and Kaloshian I (2016) The Potato Aphid Salivary Effector Me47 Is a Glutathione-S-Transferase Involved in Modifying Plant Responses to Aphid Infestation. Front. Plant Sci. 7:1142. doi: 10.3389/fpls.2016.01142
Received: 25 April 2016; Accepted: 18 July 2016;
Published: 03 August 2016.
Edited by:
Jyoti Shah, University of North Texas, USACopyright © 2016 Kettles and Kaloshian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isgouhi Kaloshian, aXNnb3VoaS5rYWxvc2hpYW5AdWNyLmVkdQ==
†Present address: Graeme J. Kettles, Plant Biology and Crop Science, Rothamsted Research, Harpenden, Hertfordshire, AL5 2JQ, UK
 Graeme J. Kettles
Graeme J. Kettles Isgouhi Kaloshian
Isgouhi Kaloshian