- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
- 2College of Horticulture, Shanxi Agricultural University, Taigu, China
Malate dehydrogenases (MDHs) are key metabolic enzymes that play important roles in plant growth and development. In the present study, we isolated the full-length and coding sequences of BraMDH from Chinese cabbage [Brassica campestris L. ssp. pekinensis (Lour) Olsson]. We conducted bioinformatics analysis and a subcellular localization assay, which revealed that the BraMDH gene sequence contained no introns and that BraMDH is localized to the chloroplast. In addition, the expression pattern of BraMDH in Chinese cabbage was investigated, which revealed that BraMDH was heavily expressed in inflorescence apical meristems, as well as the effect of BraMDH overexpression in two homozygous transgenic Arabidopsis lines, which resulted in early bolting and taller inflorescence stems. Furthermore, the fresh and dry weights of aerial tissue from the transgenic Arabidopsis plants were significantly higher than those from the corresponding wild-type plants, as were plant height, the number of rosette leaves, and the number of siliques produced, and the transgenic plants also exhibited stronger aluminum resistance when treated with AlCl3. Therefore, our results suggest that BraMDH has a dramatic effect on plant growth and that the gene is involved in both plant growth and aluminum resistance.
Introduction
Malate plays a key role in plant nutrition and metabolism, as an important intermediate product (Schulze et al., 2002; Scheibe, 2004) and is generated by malate dehydrogenases (MDHs: EC 1.1.1.82 or EC 1.1.1.37), which function as oxidoreductases that catalyze the reversible reactions of malate and oxaloacetic acid, using NAD+ or NADP+ as coenzymes. The occurrence of MDHs is widespread in plants, animals, and microorganisms. In plants, several MDH isoforms possess specific subcellular locations and coenzymes. According to their subcellular location, plant MDHs are classified as mitochondrial, peroxisomal, plastidial, and cytosolic MDHs (Gietl, 1992). In recent years, different MDH isoforms have been reported in different species (Miller et al., 1998; Trípodi and Podestá, 2003; Yao et al., 2011; Abd El-Moneim et al., 2015; Zhang Bao et al., 2015). In the model plant Arabidopsis thaliana, nine MDHs have been identified, including two peroxisomal MDHs (Pracharoenwattana et al., 2007; Eubel et al., 2008), two mitochondrial MDHs (Millar et al., 2001; Tomaz et al., 2010), and two plastidial MDHs (Berkemeyer et al., 1998; Hebbelmann et al., 2012), as well as three cytosolic MDHs (CMDH1, 2, and 3) that were predicted in The Arabidopsis Information Resource1.
The different MDH isoforms play a variety of roles that depend on their subcellular localization and the metabolic activities that occur in their respective cellular compartments. For example, mitochondrial MDHs are key enzymes in tricarboxylic acid cycles and have been reported to be involved in leaf respiration and photorespiration (Tomaz et al., 2010). Peroxisomal MDHs have been proposed to be involved in fatty acid β-oxidation, by supplying the electron acceptor NAD+ via the catalyzation of oxaloacetic acid to malate (Pracharoenwattana et al., 2007), and were also involved in photorespiration, by oxidating malate to generate NADH (Reumann and Weber, 2006). In addition, two MDH isoforms have been reported in plastids, including both NADP-dependent and NAD-dependent MDHs (pdNADP-MDH and pdNAD-MDH) (Berkemeyer et al., 1998; Hebbelmann et al., 2012). Interestingly, pdNADP-MDH is strictly a light-redox enzyme, which is only activated in the light (Berkemeyer et al., 1998; Scheibe, 2004), and in C3 plants, the pdNADP-MDH functions as an important “malate valve”, by regulating the reducing equivalents (malate) to balance the NADPH/ATP ratio (Scheibe, 1987), whereas in C4 plants, pdNADP-MDH is involved in daytime CO2 fixation, by catalyzing the primary CO2 fixation product oxaloacetic acid to the stable CO2 product malate (Scheibe, 1987). However, pdNAD-MDH is active under both light and dark conditions (Berkemeyer et al., 1998), and in Arabidopsis, pdNAD-MDH has been demonstrated to play critical roles in seed development and heterotrophic metabolism (Beeler et al., 2014; Selinski et al., 2014). For example, Arabidopsis mutants that lacking the pdNAD-MDH exhibit defective embryo development, and the disruption of pdNAD-MDH expression results in Arabidopsis plants with abnormal phenotypes, such as reduced chlorophyll and photosynthetic levels, damaged chloroplast ultrastructure, and dwarfed seedlings (Beeler et al., 2014; Selinski et al., 2014). Other research has reported that overexpression of a plastidial MDH gene promotes the growth of transgenic apple plants, especially of roots, which was linked to improved photosynthesis and reduced levels of abscisic acid (Wang et al., 2015). The overexpression of a cytosolic MDH gene in apple was reported to contribute to increased MDH activity and malate accumulation (Yao et al., 2011).
Several reports have indicated that MDHs have a close relationship with stress resistance (Cushman, 1993; Wang et al., 2010); however, direct evidence for this is scarce, and although several studies have reported that overexpression of the Arabidopsis plastidial and cytosolic MDH genes could improve the resistance of transgenic tomato to aluminum and acidic soil (Zhang et al., 2008; Wang et al., 2010), it remains unclear whether other MDH isoforms have the same effect. Furthermore, although the roles of MDH genes have been explored for many years, most of these studies have focused on model plants. However, studies of MDH function should also include non-model plants, such as Chinese cabbage [Brassica campestris L. ssp. pekinensis (Lour) Olsson], which is a popular vegetable in many regions of the world. In addition, further studies are also needed to elucidate the role of MDHs in aluminum tolerance. Because aluminum is the third most abundant element after oxygen and silicon, plant roots are almost always exposed to aluminum in some form, and although most aluminum occurs as oxides and aluminosilicates, which are harmless, the element becomes toxic to plants in acidic soils (Ma et al., 2001). For example, several studies have reported that Al3+ toxicity inhibits root growth and nutrient uptake (Delhaize and Ryan, 1995; Lopez-Bucio et al., 2000).
Therefore, in the present study, we analyzed the expression of the BraMDH in different tissues of Chinese cabbage. In addition, we also investigated the role of BraMDH in plant growth, development, and aluminum tolerance by overexpression BraMDH in Arabidopsis.
Materials and Methods
Plant Materials and Growing Conditions
Chinese cabbages (line 97C16-3) were planted and maintained in the greenhouse under natural light conditions (Yangling, Shaanxi).
Arabidopsis thaliana ecotype Columbia (Col-0) was used in the production of transgenic plants. Seeds were surface sterilized and sown on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) with 2% sucrose, 0.8% agar, and a pH of 5.7 (adjusted with NaOH). For uniform germination and to break dormancy, the seeds were initially incubated on MS medium for 2–3 days in the dark at 4°C, after which they were transferred to a growth chamber and incubated at 22°C and 50–70% relative humidity, with a photoperiod of 12 h light/12 h dark. The light intensity of the two plant culturing shelves in the growth chamber are slightly different; the light intensity of the first shelf, which held the WT1 and TL3 plants, was 98 μmol m-2s-1, whereas the light intensity of the second shelf, which held WT2 and TL9, was 115 μmol m-2s-1.
Cloning and Bioinformatics Analysis of BraMDH
Genomic DNA was extracted from Chinese cabbage leaves using a modified CTAB protocol (Allen et al., 2006) and total RNA was extracted using a TrizolTM kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA purification and first-strand cDNA synthesis were conducted using the Prime ScriptTM RT Reagent Kit (Takara, Dalian, China). The gene-specific primers BraMDH-F (5′-ATGGCAGCAGCTTCTTCGATTTC-3′) and BraMDH-R (5′- TTAGTTAGCAGGTTTGTTTGCGAATG-3′) were used to amplify the full-length and coding sequences of BraMDH. The PCR products were gel purified and cloned into the pMD18-T vector (Takara, Dalian, China). The positive clones were sequenced by Sangon Biotech Company Limited (Shanghai, China).
Once the sequences were obtained, the DNAMAN software was used to analyze the nucleotide and amino acid sequences of BraMDH and the online software Tmpred2 was used to predict the transmembrane domain.
Testing Tissue-Specific Expression of BraMDH
To investigate the expression pattern of BraMDH in Chinese cabbage, the total RNA were extracted from leaves, flower buds, flowers, inflorescence stems, inflorescence apical meristems, and young siliques using a TrizolTM kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA purification and first-strand cDNA synthesis were conducted using a PrimeScriptTM RT Reagent Kit with gDNA Eraser for Real Time (Takara, Dalian, China), according to the manufacturer’s instructions. The gene specific primer GAPDH-F (5′-TAACTGCCTTGCTCCACTTGC-3′) and GAPDH-R (5′- CGGTGCTGCTGGGAATGAT-3′) were used to amplify the GAPDH (GO0048316), which was used as an internal control. The specific primer qBraMDH-F (5′-GGACCAGATTCCTCTTCTTTCGC-3′) and qBraMDH-F (5′- TGATGTTCAATCCGTAGGGCTTC-3′) were used to amplify the BraMDH. The real-time PCR reaction system was composed of 20 μL mixture that consisted of 10 μL SYBR Premix Ex TaqTM II (Takara, Dalian, China), 0.8 μL forward primer (10 μM), 0.8 μL reverse primer (10 μM), 2 μL diluted cDNA (200 ng), and 6.4 μL ddH2O. All reactions were performed in triplicate. The real-time PCR was conducted in a Bio-Rad IQ5 instrument (Foster City, CA, USA), using a two-step thermal cycling program with the following conditions: 1 cycle of 95°C for 30 s, 40 cycles of 95°C for 5 s, and 62°C for 30 s. Serial dilutions of cDNA were used to calibrate a standard curve for each gene, in order to ensure that the amplification efficiency of actin and target gene primers were almost equal and nearly 100%. The relative expression levels of BraMDH were normalized with that of GAPDH using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Subcellular Localization of BraMDH
In order to determine the subcellular location of BraMDH, we used the online software iPSORT3 and TargetP4 to predict its transit peptide. In addition, we also constructed a BraMDH-green fluorescent protein (GFP) fused expression vector and transiently transformed into isolated Arabidopsis protoplasts. Firstly, The coding region of BraMDH was amplified using the gene specific primers GFP-BraMDH-F (5′-GCTCTAGAATGGCAGCAGCTTCTTCGATTTC-3′ with XbaI site under lined) and GFP-BraMDH -R (5′- AAGGTACCGTTAGCAGGTTTGTTTGCGAATG -3′ with KpnI site underlined). The amplified products were cloned into the PBI221-GFP vector. The constructed vector was then transformed into Arabidopsis protoplasts, which were isolated from callus as described previously (Yoo et al., 2007) with some modification. The transient expression of the BraMDH-GFP fusion protein was observed using a Nikon A1R confoeal laser scanning microscopy (Tokyo, Japan).
Construction of the Overexpression Vector and Its Genetic Transformation into Arabidopsis
The coding region of BraMDH was amplified using the gene specific primers BraMDH-F (5′-GCTCTAGAATGGCAGCAGCTTCTTCGATTTC-3′ with XbaI site underlined) and BraMDH-R (5′- AACTGCAGTTAGTTAGCAGGTTTGTTTGCGAATG-3′ with PstI site underlined), and the amplified product was cloned into the pCAMBIA2301-35S-Nos (Kanr) vector, which was a gift from Professor Yuke He (Shanghai Institutes for Biological Science, Chinese Academy of Sciences). The recombinant vector, p2301-BraMDH was then transformed into Agrobacterium tumefaciens strain GV3101 and was confirmed by PCR, restriction enzyme digestion, and sequence analysis. Subsequently, the p2301-BraMDH vector was transformed into the Arabidopsis ecotype Columbia (Col-0) using the floral dip method (Clough and Bent, 1998). Seeds of the transgenic plants were screened on MS medium (2% sucrose, 0.8% agar) containing 50 μg⋅mL-1 kanamycin, and the T1 positive plants were transferred to a substrate mixture of peat, vermiculite, and pearlite (3:1:0.5, v/v). The surviving transformants were further confirmed by amplifying the target gene BraMDH. The T2 seeds from positive T1 transformants were individually harvested and sown on MS medium (2% sucrose, 0.8% agar) that contained 50 μg⋅mL-1 kanamycin, in order to screen for T2 transgenic lines that corresponded to a 3:1 segregation ratio of survival: lethal. T3 seeds were harvested from the T2 transgenic lines that exhibited 3:1 segregation ratios, and the T3 lines that exhibited 100% kanamycin resistance were considered as homozygous.
Analysis of BraMDH Expression in Transgenic Arabidopsis
Total RNA was isolated from the rosette leaves of wild-type (WT) and transgenic Arabidopsis lines (T3 and T9) using a TrizolTM kit according to the manufacturer’s instruction (Invitrogen, Carlsbad, CA, USA), and a PrimeScriptTM RT Reagent Kit with gDNA Eraser for Real Time (Takara, Dalian, China) was used for cDNA synthesis. Real-time PCR was performed using SYBR® Premix Ex TaqTM II fluorochrome (Takara, Dalian, China). The specific primers Tub-F (5′-TTTGTGCTCATCTTGCCACGGAAC-3′) and Tub-R (5′-CTCAAGAGGTTCTCAGCAGTACC-3′) were used to amplify Tub (AT5G62690), which was used as an internal reference gene. The specific primers qBraMDH-F and qBraMDH-R were used to amplify BraMDH. The amplification efficiencies of the primers were tested as described above. The real-time PCR was conducted using the following conditions: 1 cycle of 95°C for 30 s, 40 cycles of 95°C for 5 s and 62°C for 30 s; and all reactions were performed in triplicate. The relative expression of BraMDH were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Growth Phenotypes of Transgenic Arabidopsis Plants
Seeds of the homozygous transgenic Arabidopsis lines (TL3 and TL9) and their corresponding wild-type lines (WT1 and WT2) were sown on MS medium (2% sucrose, 0.8% agar), and after 10–12 days, the seedlings were transferred to pots filled with the same weight of substrate mixture. The occurrence of bolting was recorded for each plant at 30 and 35 days after the seedlings were transplanted. In addition, the plant height, fresh and dry weights of aerial tissue, number of rosette leaves, and number of siliques were recorded at 45 days after the seedlings were transplanted.
AlCl3 Treatment of Arabidopsis Plants
Both homozygous transgenic plants (TL3 and TL9) and wild-type plants were used for AlCl3 treatments. For the first treatment, sterilized seeds were sown on MS medium (2% sucrose, 0.8% agar) and grown for 10 days in the growth chamber, after which the WT and homozygous transgenic seedlings (n = 15 each) were transferred to 0, 50, and 100 μM AlCl3 solutions. After 3 days of treatment, all seedlings cultured in 0, 50, and 100 μM AlCl3 solutions were investigated, and the root activity of WT and homozygous transgenic seedlings cultured in 50 μM AlCl3 were determined using triphenyl tetrazolium chloride (TTC) reduction method as described previously (Clemensson-Lindell, 1994; Mugai et al., 2000). TTC reduction method can examine the dehydrogenase activity, and thus it is often used to assess the root activity (Clemensson-Lindell, 1994; Mugai et al., 2000). Firstly, the roots of the pre- and post-treatment WT and homozygous transgenic seedlings were cut and put into centrifugal tubes with 1 mL 1% (m/v) TTC and 1 mL 0.1 mol/L Na2HPO4-NaH2PO4 solution, and after that the tubes were kept at 37°C for 2 h. Then, the roots were observed by a dissecting microscope (Olympus, Tokyo, Japan). For the second treatment, both homozygous transgenic lines (TL3 and TL9) and their corresponding WT plants were cultivated in the pots filled with the same weight of substrate mixture, and watered with equal volumes of water. Then, after reaching the rosette stage, the plants were subjected to a water-deficit condition, after which they were irrigated with a 150 μM AlCl3 solution (Wang et al., 2010; Ding et al., 2013), and the SPAD values and chlorophyll fluorescence of all the plants were measured both before treatment (pre-treatment) and 2 days (post-treatment) after treatment. In addition, the morphological changes in the Arabidopsis plants were observed continuously during the 5-day treatment. The SPAD values and chlorophyll fluorescence parameters were measured using a SPAD-502 chlorophyll meter (Konica Minolta, Japan) and a portable PAM-2500 system (Walz Heinz, Effeltrich, Germany).
Results
BraMDH Nucleotide Sequence and Protein Properties
BLAST analysis of the full-length and coding sequences of BraMDH indicated that the gene contained no introns (Supplementary Figure S1). Bioinformatics analysis indicated that BraMDH encodes a 402-amino acids transmembrane protein that has a single transmembrane domain, molecular mass of 42.3 kDa, and isoelectric point (pI) of 8.347. In addition, multiple sequences alignment analysis revealed that the BraMDH amino acid sequence shared 89, 88, 78, and 78% identity with AlMDH (XP_002875844.1), AtMDH (AT3G47520), RicMDH (XP_002514750.1), and TheMDH (XP_007037368.1), respectively (Figure 1). The BraMDH amino acid sequence shared high homology with the A. lyrata and A. thaliana chloroplast MDH protein (AlMDH and AtMDH), which provides justification for the subcellular localization of BraMDH in Arabidopsis protoplasts.

FIGURE 1. Alignment of the predicted BraMDH protein sequence and other known MDH sequences. The GenBank accession numbers of these proteins are as follows: AlMDH (XP_002875844.1), AtMDH (AT3G47520), RicMDH (XP_002514750.1), TheMDH (XP_007037368.1); Al, Arabidopsis lyrata; At, Arabidopsis thaliana; Ric, Ricinus communis; The, Theobroma cacao; Bra, Brassica campestris.
Localization of BraMDH in Chloroplast
Both iPSORT and TargetP identified a putative chloroplast transit peptide in the BraMDH amino acid sequence, and the co-localization of chlorophyll autofluorescence and BraMDH-GFP fusion protein fluorescence in Arabidopsis protoplasts confirmed that BraMDH was a chloroplast protein (Figure 2).
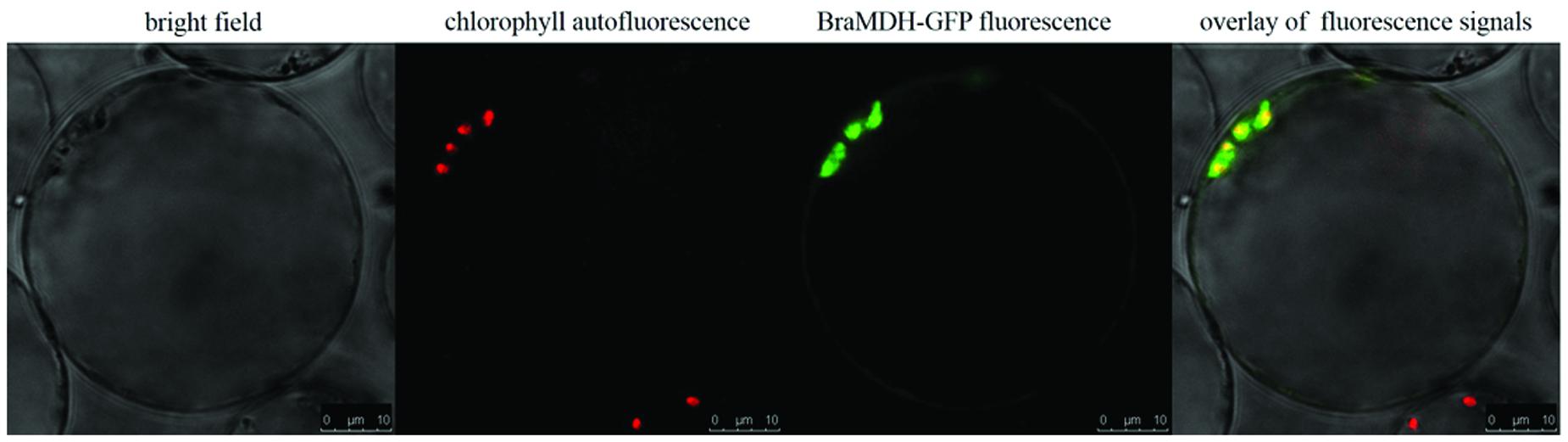
FIGURE 2. Subcellular localization of BraMDH in Arabidopsis protoplasts. From left to right: bright field, chlorophyll autofluorescence, BraMDH-GFP fused fluorescence, and the overlay of fluorescence signals. Bars = 10 μm.
The Expression of BraMDH in Chinese Cabbage
The expression level of BraMDH was highest in the inflorescence apical meristem, followed by the levels observed in flower buds, flower, young siliques, leaves, and inflorescence stems, respectively (Figure 3), and the expression level in inflorescence apical meristem was more than twice that in leaves. The expression level of BraMDH in flower buds and flowers were almost identical, and the expression level in young siliques was higher than that in leaves. It has been reported that the pdNAD-MDH gene in Arabidopsis is crucial for seed development (Beeler et al., 2014; Selinski et al., 2014). Indeed, our results indicate that the BraMDH gene may play an important role in the reproductive organs of Chinese cabbage as well, especially during flower development and growth.
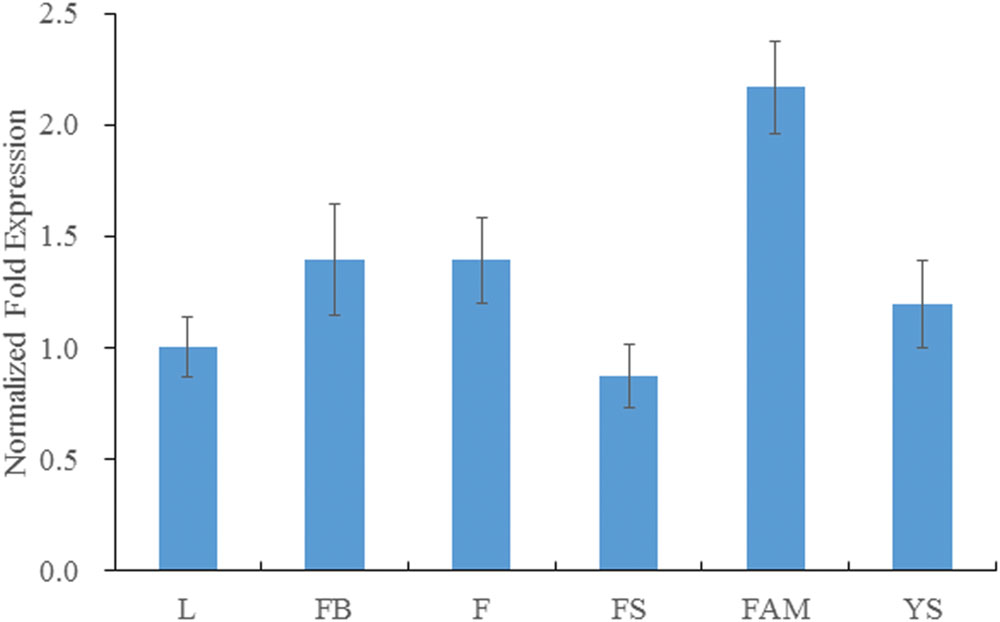
FIGURE 3. Expression of BraMDH in Chinese cabbage. L, leaves; FB, flower buds; F, flowers; FS, inflorescence stems; FAM, inflorescence apical meristems; YS, young siliques. Values indicate mean ± SD (n = 3).
BraMDH Expression in Transgenic Arabidopsis Plants
The full-length BraMDH fragment was amplified from all surviving T1 transformants (Figure 4A), whereas no gene fragments were detected in the WT plants, which together indicated that the BraMDH gene was successfully transmitted into the Arabidopsis genome. Then, after screening T2 lines which corresponded to a 3:1 segregation ratio of survival: lethal, we eventually identified two T3 homozygous transgenic lines (TL3 and TL9), as indicated by their 100% kanamycin resistance. In addition, we also confirmed that BraMDH was expressed in the TL3 and TL9 lines and found that the expression of BraMDH was higher in the TL9 line than in the TL3 line (Figure 4B). Thus, the transgenic lines TL3 and TL9 were considered homozygous and were used in subsequent studies.
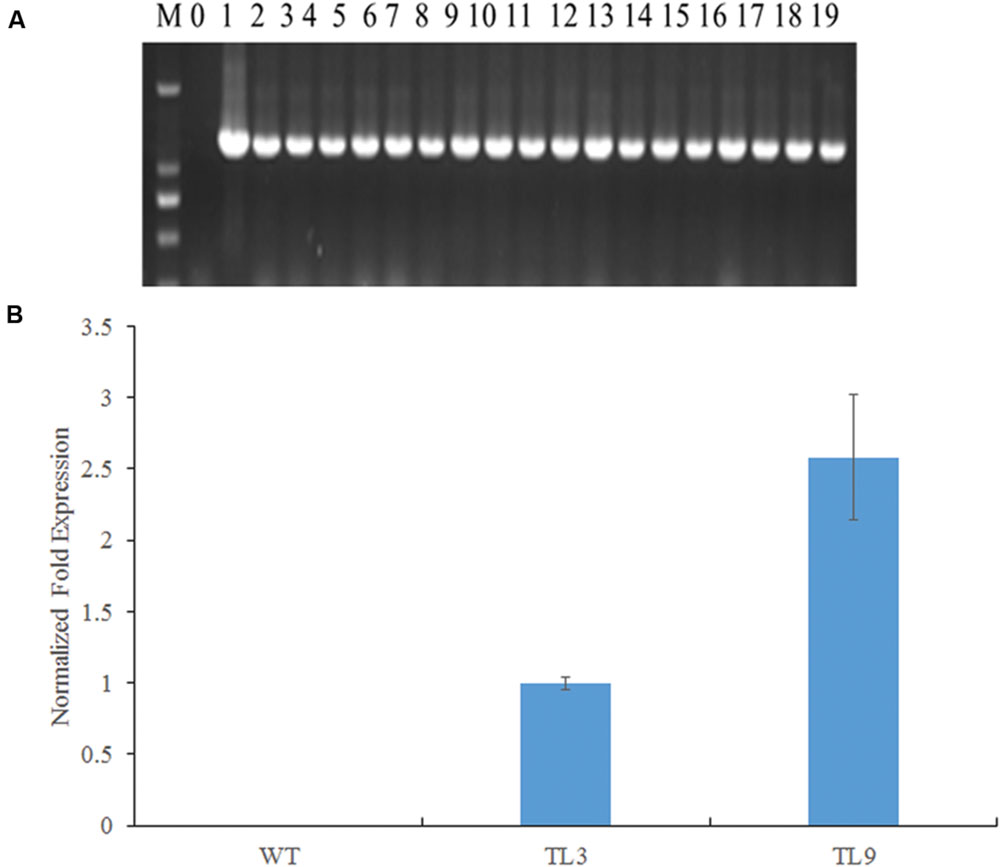
FIGURE 4. Identification of BraMDH-expressing transgenic Arabidopsis lines. (A) Electrophoresis gels of PCR products amplified from independent transgenic Arabidopsis lines (M: DL2000, 0: wild-type, 1: plasmid positive control, 2–19: transgenic plants). (B) Normalized fold expression of BraMDH in rosette leaves of wild-type (WT) and homozygous transgenic lines (TL3 and TL9). Values indicate mean ± SD (n = 3).
Overexpression of BraMDH Induces Early Bolting in Transgenic Arabidopsis Plants
After 30 days of growth, we found that none of the WT1 or WT2 plants had bolted, whereas 66 and 59% of the TL3 and TL9 plants had bolted, respectively; and after 35 days of growth, only to 57 and 60% of the WT1 and WT2 plants had bolted, respectively, whereas all of the transgenic plants had bolted. In addition, we also observed that the inflorescence stems of the WT1, WT2, TL3, and TL9 lines were 4.2 ± 3.0 cm, 7.6 ± 2.4 cm, 10.1 ± 3.7 cm, and 13.5 ± 1.2 cm, respectively, which clearly indicated that the inflorescence stems of the transgenic lines were significantly taller than those of the corresponding WT plants (Figure 5; Supplementary Figure S2). These results indicate that the overexpression of BraMDH contributes to early bolting Arabidopsis.

FIGURE 5. Bolting in wild-type (WT1) and homozygous transgenic lines (TL3) plants. Bolting of WT1 and TL3 plants after 35 days of growth in a substrate mixture of peat, vermiculite, and pearlite (3:1:0.5, v/v). Bars = 5 cm. The bolting of WT2 and TL9 plants is shown in Supplementary Figure S2.
Overexpression of BraMDH Enhanced Growth of Transgenic Arabidopsis Plants
At 45 days after the experimental seedlings were transferred to the substrate mixture, we found that plant height, rosette leaf number, siliques number, and aerial tissue weights of both transgenic lines were greater than those of their corresponding WT controls (Table 1). In particular, the fresh weights of the aerial tissue from TL3 and TL9 plants were 22 and 19% greater than those of their corresponding WT plants, respectively, and the difference both reached significant level (p < 0.05). In addition, the dry weights of the aerial tissue from TL3 and TL9 were 29 and 25% greater than those of their corresponding WT plants, respectively, and the difference both reached extremely significant level (p < 0.01) (Table 1). These data indicate that the overexpression of BraMDH enhanced the vegetative growth of the transgenic Arabidopsis plants, which likely contributed to the earlier bolting phenotype via enhanced nutritional resources. It is also interesting to note that no difference was observed in the dry weight/fresh weight ratio of aerial tissue from the WT and transgenic plants, which suggested that the overexpression of BraMDH did not affected the balance between dry material accumulation and water absorption.
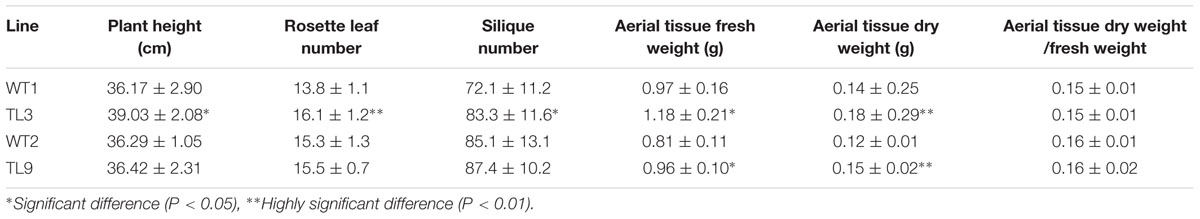
TABLE 1. Growth traits of wild-type (WT1 and WT2) and homozygous transgenic (TL3 and TL9) Arabidopsis plants.
Overexpression of BraMDH Enhanced Al Tolerance of Arabidopsis
After 3-day AlCl3 exposure of the first treatment, we found that all the seedlings cultured in 0 μM AlCl3 (distilled water) grew normally, that all the seedlings cultured in 100 μM AlCl3 solution were nearly dead, and that the WT and transgenic seedlings cultured in 50 μM AlCl3 solution exhibited significantly different phenotypes (Figures 6A,B). More specifically, we found that 63% of the WT seedlings had purple petioles, whereas only 40% of the transgenic lines had purple petioles. In addition, no significant difference of the root activity, which was assessed by the dehydrogenase activity according to the reduction level of TTC, had been found between the pre-treatment WT and transgenic seedlings; whereas the red color of transgenic seedlings was darker than that of WT (Supplementary Figure S4), and which indicated that the root of transgenic lines have greater TTC reduction level and dehydrogenase activity, after they had been exposed in 50 μM AlCl3 solution for 3 days (Steponkus, 1971).

FIGURE 6. Effect of aluminum stress on the growth of wild-type (WT1) and homozygous transgenic line (TL3) plants. (A,B) Whole plants and leaf stalk of seedlings treated with 50 μM AlCl3 (Treatment 1), bars = 3 mm. (C) Rosette-stage WT1 and TL3 plants treated with 150 μM AlCl3 (Treatment 2) for 2 days (left) and 5 days (right), bars = 5 cm. Col: untreated wild type plants, WT: treated wild-type plants, TL: treated transgenic plants. The results of WT2 and TL9 plants are shown in Supplementary Figure S3.
Meanwhile, in the second treatment, we found that the rosette leaves of WT1 and WT2 plants became wilted after 2 days of treatment and then turned yellow and dry after 5 days. However, the wilting of the transgenic plants was less pronounced, and after 5 days of treatment, the color of the TL3 and TL9 leaves remained green and did not appear to dry out (Figure 6C; Supplementary Figure S3). In order to define the reason for wilting and chlorosis phenotypes, the chlorophyll content (SPAD value) and chlorophyll fluorescence of WT1, WT2, TL3, and TL9 treated with 150 μM AlCl3 solution were measured (Table 2). The mean SPAD values of WT1, WT2, TL3, and TL9 leaves were 25.7, 28.8, 28.0, and 30.4, respectively, before treatment. Obviously, the SPAD values of transgenic lines were higher than those of the corresponding WT lines. However, after being exposed to AlCl3 for 2 days, the SPAD values of WT1, WT2, TL3, and TL9 lines dropped to 16.3, 16.8, 23.1, and 23.6, respectively. The decreased ratio of SPAD values of WT1 and WT2 were 36.6 and 41.7%, respectively, which were higher than 17.5 and 22.4% in TL3 and TL9. These indicated that, under AlCl3 treatment, the chlorophyll content of transgenic plant leaves was more stable than that of WT leaves. Furthermore, in regards to chlorophyll fluorescence, we observed no significant differences, in pre- and post-treatment measurements of non-photochemical quenching (NPQ) and variable fluorescence/maximum fluorescence yield (Fv/Fm) of either the WT or transgenic plants (Table 2). However, the pre-treatment values of ground state fluorescence (Fo), maximum fluorescence (Fm), and photochemical quenching (qP) were slightly higher in transgenic plants than that in WT plants; and the Fo, Fm, and qP values of WT plants decreased significantly in response to the AlCl3 treatment, along with the chlorophyll content, whereas the parameters of the transgenic plants did not change significantly (Table 2). As a result, the post-treatment Fo, Fm, and qP values of TL3 and TL9 plants were significantly higher than those of WT1 and WT2 (Table 2), which indicated that the greater aluminum resistance of transgenic lines was associated with the higher light absorption, but not photosynthetic efficiency.
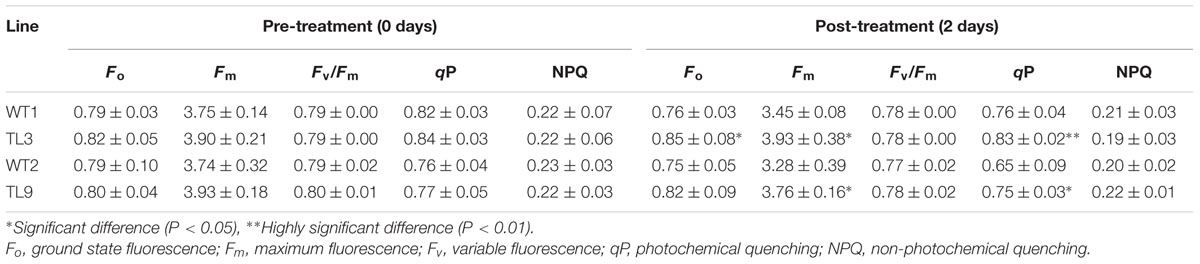
TABLE 2. Effect of aluminum (AlCl3) stress on the chlorophyll fluorescence parameters of wild-type (WT1 and WT2) and homozygous transgenic (TL3 and TL9) Arabidopsis plants.
Discussion
Malate is an important intermediate product, and plays a key role in plant nutrition, metabolism, and cellular energy supply (Schulze et al., 2002). Because MDHs are crucial for generating malate, they have drawn increasing attentions from the scientific community (Schulze et al., 2002; Scheibe, 2004), and many researchers have studied the function of MDH genes; however, most of these studies have been in the model plants (Gietl, 1992; Pracharoenwattana et al., 2007; Tomaz et al., 2010; Selinski et al., 2014), and the role of MDH genes in economic crops remains unclear. In the present study, we isolated BraMDH which located in chloroplast from Chinese cabbage and discovered that ectopic expression of BraMDH augmented the growth and aluminum resistance of transgenic Arabidopsis plants. More specifically, the transgenic Arabidopsis lines exhibited greater fresh and dry weights of aerial tissue, plant height, number of rosette leaves, and number of siliques than the WT plants, and in addition, the transgenic plants also bolted earlier and exhibited more stable chlorophyll content and fluorescence (Fo, Fm, and qP).
Previous studies have reported that malate valves act as a powerful system for balancing ATP/NAD(P)H ratio required in energy-consuming reactions within plant cell (Scheibe, 2004). Therefore, the changes observed in the transgenic plants could indicated that the overexpression of BraMDH contributes to the production of malate, which can be shuttled from organelles and be used as a base material in ATP production within chloroplast and mitochondria (Scheibe, 2004; Tomaz et al., 2010; Selinski et al., 2014) and, subsequently, accelerate plant growth and development. Rosette leaves are the main photosynthetic organ of Arabidopsis plants, therefore, enhanced plant growth (i.e., more rosette leaves) results in more photosynthetic products, which increased the yield of biomass, so that the transgenic plants exhibit augmented plant height, siliques number, and bolting phenomenon. Indeed, the transgenic Arabidopsis plants in present study exhibited increased chlorophyll content and fluorescence, as well as increased growth, which supports earlier finding that Arabidopsis plants lacking the plastid NAD-MDH gene showed reduced levels of chlorophyll and photosynthesis, damaged chloroplast ultrastructure, and the inability to develop seeds (Beeler et al., 2014; Selinski et al., 2014) and also indicates that the function of BraMDH is similar to that of pdNAD-MDH in Arabidopsis.
Interestingly, our study also demonstrated that BraMDH contributes to stress tolerance, which supports previous findings regarding plastidial and cytosolic MDH genes in transgenic tomato plants (Zhang et al., 2008; Wang et al., 2010). In the present study, we found that the overexpression of BraMDH enhanced the aluminum tolerance of transgenic Arabidopsis (Figure 6; Supplementary Figure S3) and that also increased the stability of the plants’ chlorophyll content and fluorescence over that of WT plants (Table 2). It follows, then, that the stability of chlorophyll content in transgenic plants could be correlated with aluminum resistance. Indeed, Fo, which is the minimal fluorescence in PSII reaction centers, has a close relationship to the chlorophyll content (van Kooten and Snel, 1990), and both Fm and qP, which represent electron transfer and photosynthetic activity in PSII reaction centers (van Kooten and Snel, 1990), were reduced slightly than in treated WT plants. This indicates that the electron transfer and photosynthetic activity were reduced in PSII reaction centers of WT plants, but those of transgenic plants were not impacted. We further speculate that the wilting of leaves in the treated WT plants reduced the plants’ light absorption abilities, which then contributed to reduced Fo and Fm levels, whereas the more stable chlorophyll content, Fo, and Fm of the transgenic lines allowed the transgenic plants to outperform the WT plants when under the AlCl3 stress. Overall, the reason for increase of the aluminum resistance in the transgenic Arabidopsis is speculated that overexpression of BraMDH could have catalyzed conversions of the oxaloacetic acid to malate and other organic acids. This statement could be supported indirectly by the greater dehydrogenases activity that reflected by the higher TTC reduction level of transgenic Arabidopsis under the AlCl3 stress (Supplementary Figure S4). It is well established that, when faced with aluminum stress, some plants are capable of releasing organic acids (e.g., citric acid, malate, and oxalate) into the rhizosphere and that the organic acids form non-toxic complexes with Al3+ to preventing the toxic element from entering the roots (Pellet et al., 1996; Ma et al., 2001; Kochian et al., 2004), and moreover, overexpression of neMDH in transgenic alfalfa was reported to enhance both organic acid synthesis and aluminum tolerance (Tesfaye et al., 2001).
Conclusion
The present study demonstrates that overexpression of BraMDH promotes growth and enhances aluminum resistance in transgenic Arabidopsis plants. The results suggest that BraMDH possesses potential value for genetic breeding of high-yield and aluminum-resistance Chinese cabbage cultivars.
Author Contributions
QL and LZ conceived the experiments; QL and JZo performed the experiments; JZg participated in gene cloning and sequence analysis; ZD participated in seedling culture; LZ contributed materials, reagents, and analysis tools; QL and LZ wrote the paper.
Funding
This research was supported by the National Science and Technology Support Program (2012BAD02B01 and 2014BAD01B0802).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01180
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.arabidopsis.org/index.jsp
- ^ http://www.ch.embnet.org/software/TMPRED_form.html
- ^ http://ipsort.hgc.jp/
- ^ http://www.cbs.dtu.dk/services/TargetP/
References
Abd El-Moneim, D., Contreras, R., Silva-Navas, J., Gallego, F. J., Figueiras, A. M., and Benito, C. (2015). On the consequences of aluminium stress in rye: repression of two mitochondrial malate dehydrogenase mRNAs. Plant Biol. (Stuttg) 17, 123–133. doi: 10.1111/plb.12219
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissue using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Beeler, S., Liu, H. C., Stadler, M., Schreier, T., Eicke, S., Lue, W. L., et al. (2014). Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 164, 1175–1190. doi: 10.1104/pp.113.233866
Berkemeyer, M., Scheibe, R., and Ocheretina, O. (1998). A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 273, 27927–27933. doi: 10.1074/jbc.273.43.27927
Clemensson-Lindell, A. (1994). Triphenyltetrazolium chloride as an indicator of fine-root vitality and environmental stress in coniferous forest stands: applications and limitations. Plant Soil 159, 297–300.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cushman, J. C. (1993). Molecular cloning and expression of chloroplast NADP-malate dehydrogenase during Crassulacean acid metabolism induction by salt stress. Photosynth. Res. 35, 15–27.
Delhaize, E., and Ryan, P. R. (1995). Aluminum toxicity and tolerance in plants. Plant Physiol. 107, 315–321.
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Eubel, H., Meyer, E. H., Taylor, N. L., Bussell, J. D., O’Toole, N., Heazlewood, J. L., et al. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148, 1809–1829. doi: 10.1104/pp.108.129999
Gietl, C. (1992). Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta 1100, 217–234.
Hebbelmann, I., Selinski, J., Wehmeyer, C., Goss, T., Voss, I., Mulo, P., et al. (2012). Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 63, 1445–1459. doi: 10.1093/jxb/err386
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Bucio, J., de la Vega, O. M., Guevara-Garcia, A., and Herrera-Estrella, L. (2000). Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat. Biotechnol. 18, 450–453.
Ma, J. F., Ryan, P. R., and Delhaize, E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/S1360-1385(01)01961-6
Millar, A. H., Sweetlove, L. J., Giege, P., and Leaver, C. J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. doi: 10.1104/pp.010387
Miller, S. S., Driscoll, B. T., Gregerson, R. G., Gantt, J. S., and Vance, C. P. (1998). Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J. 15, 173–184. doi: 10.1046/j.1365-313X.1998.00192.x
Mugai, E. N., Agong, S. G., and Matsumoto, H. (2000). Aluminium tolerance mechanisms in Phaseolus vulgaris L.: citrate synthase activity and TTC reduction are well correlated with citrate secretion. Soil Sci. Plant Nutr. 46, 939–950. doi: 10.1080/00380768.2000.10409159
Murashige, T., and Skoog, F. (1962). A Revised Medium for Rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
Pellet, D. M., Papernik, L. A., and Kochian, L. V. (1996). Multiple aluminum-resistance mechanisms in wheat – Roles of root apical phosphate and malate exudation. Plant Physiol. 112, 591–597.
Pracharoenwattana, I., Cornah, J. E., and Smith, S. M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J. 50, 381–390. doi: 10.1111/j.1365-313X.2007.03055.x
Reumann, S., and Weber, A. P. (2006). Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim. Biophys. Acta 1763, 1496–1510. doi: 10.1016/j.bbamcr.2006.09.008
Scheibe, R. (1987). NADP+ -malate dehydrogenase in C3 -plants: regulation and role of a light-activated enzyme. Physiol. Plant. 71, 393–400.
Scheibe, R. (2004). Malate valves to balance cellular energy supply. Plant Physiol. 120, 21–26. doi: 10.1111/j.0031-9317.2004.0222.x
Schulze, J., Tesfaye, M., Litjens, R. H. M. G., Bucciarelli, B., Trepp, G., Miller, S., et al. (2002). Malate plays a central role in plant nutrition. Plant Soil 247, 133–139. doi: 10.1023/A:1021171417525
Selinski, J., Konig, N., Wellmeyer, B., Hanke, G. T., Linke, V., Neuhaus, H. E., et al. (2014). The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol. Plant 7, 170–186. doi: 10.1093/mp/sst151
Steponkus, P. L. (1971). Effect of freezing on dehydrogenase activity and reduction of triphenyl tetrazolium chloride. Cryobiology 8, 570–573.
Tesfaye, M., Temple, S. J., Allan, D. L., Vance, C. P., and Samac, D. A. (2001). Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol. 127, 1836–1844. doi: 10.1104/Pp.010376
Tomaz, T., Bagard, M., Pracharoenwattana, I., Linden, P., Lee, C. P., Carroll, A. J., et al. (2010). Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154, 1143–1157. doi: 10.1104/pp.110.161612
Trípodi, K. J., and Podestá, F. (2003). Purification and characterization of an NAD-dependent malate dehydrogenase from leaves of the crassulacean acid metabolism plant Aptenia cordifolia. Plant Physiol. Biochem. 41, 97–105.
van Kooten, O., and Snel, J. F. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25, 147–150.
Wang, Q. F., Zhao, Y., Yi, Q., Li, K. Z., Yu, Y. X., and Chen, L. M. (2010). Overexpression of malate dehydrogenase in transgenic tobacco leaves: enhanced malate synthesis and augmented Al-resistance. Acta Physiol. Plant 32, 1209–1220. doi: 10.1007/s11738-010-0522-x
Wang, Q. J., Jin, Z. X., Zhou, L. J., Hao, Y. J., and Yao, Y. X. (2015). Impacts of MdcyMDH overexpression on photosynthesis, hormone and growth in apple. Sci. Agric. Sin. 48, 2868–2875.
Yao, Y. X., Li, M., Zhai, H., You, C. X., and Hao, Y. J. (2011). Isolation and characterization of an apple cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J. Plant Physiol. 168, 474–480. doi: 10.1016/j.jplph.2010.08.008
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, M., Luo, X. Y., Bai, W. Q., Li, Y. R., Hou, L., Pei, Y., et al. (2008). Characterization of malate dehydrogenase gene from citrus junos and its transgenic tobacco’s tolerance to aluminium toxicity. Acta Horticul. Sin. 35, 1751–1758.
Keywords: malate dehydrogenase, aluminum stress, growth potential, Chinese cabbage, overexpression, Arabidopsis
Citation: Li Q-F, Zhao J, Zhang J, Dai Z-H and Zhang L-G (2016) Ectopic Expression of the Chinese Cabbage Malate Dehydrogenase Gene Promotes Growth and Aluminum Resistance in Arabidopsis. Front. Plant Sci. 7:1180. doi: 10.3389/fpls.2016.01180
Received: 27 May 2016; Accepted: 22 July 2016;
Published: 03 August 2016.
Edited by:
Nokwanda Makunga, Stellenbosch University, South AfricaReviewed by:
Khurram Bashir, Riken Brain Science Institute, JapanTaras P. Pasternak, Institute of Biology II, Germany
Copyright © 2016 Li, Zhao, Zhang, Dai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu-Gang Zhang, bHVnYW5nemhAMTYzLmNvbQ==
 Qing-Fei Li
Qing-Fei Li Jing Zhao
Jing Zhao Jing Zhang
Jing Zhang Zi-Hui Dai
Zi-Hui Dai Lu-Gang Zhang
Lu-Gang Zhang