- 1Department of Bioinformatics and Genomics, University of North Carolina at Charlotte, Charlotte, NC, USA
- 2Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, NC, USA
- 3Key Laboratory of Tropical Forest Ecology, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, China
- 4Miller Lab, Department of Biological Science, Florida State University, Tallahassee, FL, USA
Introduction
Soil salinity is one of the major environmental factors causing crop loss worldwide. Currently, 33% of the global arable land is affected by salinity, hampering crop production in these fields (Flowers and Colmer, 2008). As the global population continues to rise, crop production is facing increasing demands (Flowers and Muscolo, 2015). With the combined pressures to sustain or even increase the world's food supply, salt tolerance is becoming an important agronomic trait to support crop plant growth and production in marginal and high saline soils.
Salt tolerance is a genetically complex trait that has evolved independently by different mechanisms in numerous lineages (Bromham, 2015). Efforts to improve salt tolerance in crops through selective breeding have proven difficult (Ruan et al., 2010) due to a lack of genetic resources and limited salt tolerance associated with known molecular markers (Deinlein et al., 2014).
We conducted a transcriptome analysis of a wild relative of the salt-sensitive sweet potato (Ipomoea batatas): the beach morning glory (Ipomoea imperati). Beach morning glory is a halophyte that thrives in beach ecosystems of high salt content. Our objective was to better understand the genetic basis for salt tolerance in I. imperati, so that future studies might transfer the salt tolerance genes into sweet potatoes.
Value of the Data
• Beach Morning is closely related to sweet potatoes, but commonly grows in high salt conditions. This creates a potential genetic source for adding much-needed salt tolerance into future sweet potato breeding strategies.
• To date, there is no well-characterized transcriptome for either sweet potato or morning glory and no source for gene annotation when exposed to high levels of salt. This dataset of biological triplicates can help in the further understanding of the plant pathways involved under varying salt levels.
• These data will help identify relevant genes that are significant differentially expressed under salt stress as well as identify genes that are detectable under normal growing conditions in both root and leaf tissue. Gene expression can be compared between the 2 tissue types to identify how different tissues respond within the plant to salt exposure.
Data
Experimental Design, Materials, and Methods
Plant Materials
Total RNA Extraction and Quality Control, Library Preparation, and RNA-Seq
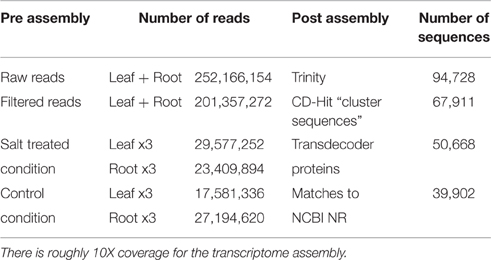
Seeds from I. imperati were collected from St. George Island, Florida as seed and grown in the lab. At 2 weeks of growth 600 mM NaCl solution (treatment) or water (control) was applied to the soil, daily, for 7 days. Three biological replicates for each treatment and species were harvested at 0, 3, 24 h, and 7 days. Total RNA was extracted from the roots and leaves using the Qiagen protocol and treated with DNAse (Qiagen). Quantity and integrity of the extracted total RNA were determined using an Agilent 2100 bioanalyzer (Agilent), respectively, to be RIN >9. A total of 12 RNA-Seq libraries, including three biological replicates, were prepared using Illumina TruSeqRNA sample Preparation Kit (Illumina). Twelve normalized cDNA libraries were constructed and sequenced using the Illumina Hiseq2500 platform (North Carolina State University) to generate 100 bp paired-end raw reads.
Raw reads were deposited into the Short Read Archive (SRA database, http://www.ncbi.nlm.nih.gov/sra) with the following accession information:
• Bioproject ID = PRJNA322032
• Biosample accession
○ Roots = SAMN05007696, SAMN05271550, SAMN05271551
○ Leafs = SAMN05271552, SAMN05271553, SAMN05271554
• SRA Root tissue experiment = SRX1771615, SRX1858743, SRX1858745
• SRA Leaf tissue experiment = SRX1858747, SRX1858786, SRX1858810
Root and leaf tissue experiments contain sequence reads of triplicate runs for both salt treated and control.
Transcriptome De novo Assembly
Sequence reads were filtered using the Fastx-toolkit (Gordon and Hannon, 2010) for quality and adapter removal using the fastq_quality_trimmer tool with the following parameters: -Q33 -v -t 20. Paired ends were corrected and repaired using Perl script, PE_FIX_POSTQC.pl (all scripts described herein are available at https://github.com/bioinformagical/SweetPotatoRNA-Seq). Paired reads were validated using validateHiseqPairs.pl.
Reads were combined across all conditions and de novo assembled via Trinity (Grabherr et al., 2011; version r2013-02-25) using default settings in order to build a suitable set of reference contigs (column 4 of Table 1). These contigs are used for the purposes of determining differential gene expression and pathway level analysis (paper in preparation). Assembly is publicly available on Figshare at: https://figshare.com/articles/Morning_Glory_Transcriptome_assembly/3498239.
Trinity contigs of high similarity were clustered into groups with CD-HIT-EST (version v4.6.1-2012-08-27) and a single representative from each cluster was used as a reference sequence for read alignment. Clustered contigs are publicly available on Figshare at: https://figshare.com/articles/CDHIT_Cluster_of_assemblies/3498263.
Predicted Proteins and Initial Annotation
Sequences from each CD-HIT cluster were transdecoded into predicted proteins using Transdecoder (http://transdecoder.github.io), a software tool that identifies the most likely protein sequence by finding the longest open reading frame and comparing the translated protein sequence to known proteins in the PFAM domain (Haas et al., 2013). Only proteins greater than 100 amino acids in length were retained for further annotating. After protein translation, 50,688 predicted proteins were found (column 4 of Table 1). Sequences are available on Figshare at: https://figshare.com/articles/Morning_Glory_Predicted_proteins/3498308.
Sequences were initially annotated by blasting nucleotide sequences against the NCBI NR database (BLASTX, -evalue 1e-10 -soft_masking true -max_target_seqs 1) where 39,902 sequences had a match. The most commonly occurring matches were to genes from Solanum lycopersium. We mapped the CD-HIT clusters onto the records from the GO database and retrieved 21,418 GO annotations. BLAST2GO assigned 18,034 with terms of “Biological process,” 13,322 with terms of “cellular component,” and 17,134 with terms of “molecular functions.”
Author Contributions
BS: conceived the idea and acquired funding; YL, BS, SY, TM: collected seeds and conducted the experiment; RR: performed analysis on the data; RR, YL, BS, SY, TM: wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the University of North Carolina at Charlotte and is part of the Plant Pathways Elucidation Project's (p2ep.org) ongoing effort to improve nutrition through plant genomics based strategies.
References
Bromham, L. (2015). Macroevolutionary patterns of salt tolerance in Angiosperms. Ann. Bot. 115, 333–341. doi: 10.1093/aob/mcu229
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Flowers, T. J., and Muscolo, A. (2015). Introduction to the special issue: halophytes in a changing world. AoB Plants 7:plv020. doi: 10.1093/aobpla/plv020
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Gordon, A., and Hannon, G. J. (2010). Fastx-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools (unpublished). Available online at: http://hannonlab.Cshl.Edu/fastx_toolkit/
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D A., Amit, I., et al. (2011). Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Keywords: sweet potato, morning glory, salt tolerance and sensitivity, RNA-sequencing, halophyte
Citation: Reid RW, Luo Y, Yan S, Miller TE and Song B-H (2016) Transcriptome Dataset of Halophyte Beach Morning Glory, a Close Wild Relative of Sweet Potato. Front. Plant Sci. 7:1267. doi: 10.3389/fpls.2016.01267
Received: 31 May 2016; Accepted: 10 August 2016;
Published: 24 August 2016.
Edited by:
Richard A. Jorgensen, University of Arizona, USAReviewed by:
Michael Deyholos, University of British Columbia, CanadaCaiguo Zhang, University of Colorado Denver, USA
Copyright © 2016 Reid, Luo, Yan, Miller and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Hua Song, YnNvbmc1QHVuY2MuZWR1
 Robert W. Reid
Robert W. Reid Yan Luo2,3
Yan Luo2,3 Thomas E. Miller
Thomas E. Miller Bao-Hua Song
Bao-Hua Song