- 1Department of Animal and Plant Sciences, University of Sheffield, Sheffield, UK
- 2Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, UK
Receptor-like kinases (RLKs) are regulators of plant development through allowing cells to sense their extracellular environment. They facilitate detection of local endogenous signals, in addition to external biotic and abiotic stimuli. The Catharanthus roseus RLK1-like (CrRLK1L) protein kinase subfamily, which contains FERONIA, plays a central role in regulating fertilization and in cell expansion mechanisms such as cell elongation and tip growth, as well as having indirect links to plant–pathogen interactions. Several components of CrRLK1L signaling pathways have been identified, including an extracellular ligand, coreceptors, and downstream signaling elements. The presence and abundance of the CrRLK1L proteins in the plant kingdom suggest an origin within the Streptophyta lineage, with a notable increase in prevalence in the seeded land plants. Given the function of the sole CrRLK1L protein in a charophycean alga, the possibility of a conserved role in detection and/or regulation of cell wall integrity throughout the Strephtophytes is discussed. Orthologs of signaling pathway components are also present in extant representatives of non-vascular land plants and early vascular land plants including the liverwort Marchantia polymorpha, the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii. Deciphering the roles in development of the CrRLK1L protein kinases in early diverging land plants will provide insights into their ancestral function, furthering our understanding of this diversified subfamily of receptors in higher plants.
Receptor-Like Kinases (RLKs) in Plants
Plant cells sense their extracellular environment and moderate their developmental programs accordingly. Extracellular signals range from tissue-specific cues to indicators of abiotic and biotic environmental conditions such as drought or disease. A critical component of environment-sensing is plasma membrane-localized receptors. Receptor-like kinases (RLKs) are among the most expanded protein families in plants, comprising ∼600 members in Arabidopsis thaliana (Shiu and Bleecker, 2001). A small proportion of plant RLKs have been characterized, with comprehensive functional descriptions for relatively few members, including BRASSINOSTEROID-INSENSITIVE 1, CLAVATA1 and FLAG-ELLIN-SENSITIVE 2 (Wang et al., 2001; Chinchilla et al., 2006; DeYoung et al., 2006). Understanding RLKs is critical as they regulate many aspects of plant function from development to stress responses.
Receptor-like kinases have a modular organization consisting of an amino-terminal extracellular domain (ECD), a transmembrane (TM) domain, and an intracellular kinase domain (Walker, 1994). Classification by their variable ECDs defines 15 RLK subfamilies including the leucine-rich repeat (LRR) and Catharanthus roseus RLK 1-like (CrRLK1L) subfamilies (Shiu and Bleecker, 2003). The RLKs share a common mechanism for signal perception and transmission. Firstly, the ECD recognizes a specific ligand, the biochemical nature of which depends on the ECD, with ligand binding inducing receptor-coreceptor dimerization. This intermolecular interaction promotes signal transduction through conformational change, leading to kinase domain activation via auto- or trans-phosphorylation (Afzal et al., 2008). Finally, phosphorylation of downstream pathway components results in signal transmission and activation of adaptive responses to the extracellular stimulus.
The Catharanthus roseus RLK1-Like (CrRLK1L) Subfamily
Named after Catharanthus roseus, the species in which its first member (CrRLK1) was identified (Schulze-Muth et al., 1996), the CrRLK1L subfamily has received increasing attention over the past decade. Members of the CrRLK1L subfamily have two ECD regions with similarity to the putative carbohydrate-binding malectin domain (MD). Malectin, first described in Schallus et al. (2008) in Xenopus laevis, acts in the endoplasmic reticulum N-glycosylation surveillance system.
The two tandem MDs are annotated as a malectin-like domain (MLD) (Figure 1A), a conformation specific to algae and plants. Within the RLK family, the MLD comprises the main ECD in the CrRLK1L, LRK10L-2, LRR-1c, and LRR-1a subfamilies. The MLD is classified as a rapidly evolving domain in the CrRLK1L subfamily (Escobar-Restrepo et al., 2007). Although, MLDs are highly divergent in primary sequence, suggesting functional divergence, conservation of predicted secondary structures allows inference of a role in carbohydrate binding (Boisson-Dernier et al., 2011). However, while the structure of malectin is available (Schallus et al., 2008, 2010), no three-dimensional structure of the MLD has been resolved, preventing a more definitive comparison.
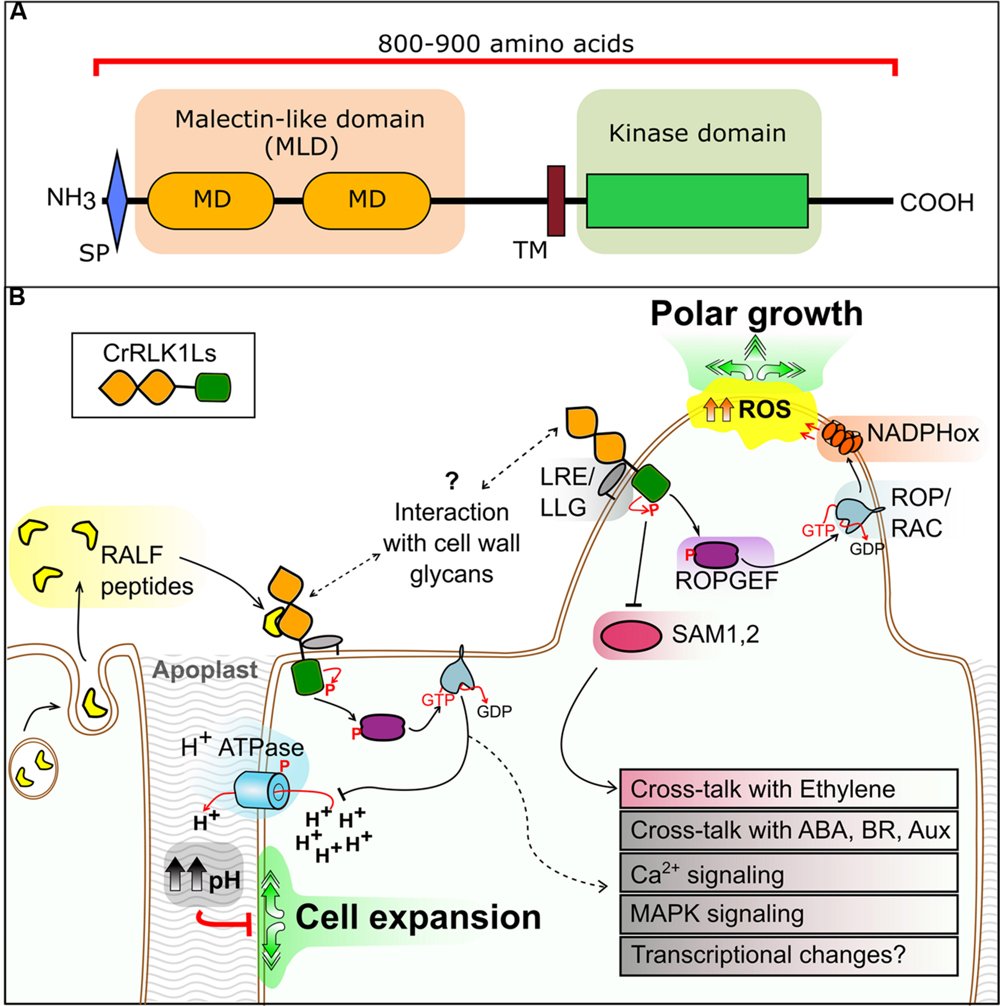
FIGURE 1. (A) Domain organization of the CrRLK1L proteins. Domains are to scale and based on FER as given in the NCBI Conserved Domain Database. SP, signal peptide; MD, malectin domain; TM, transmembrane domain. (B) Overview of CrRLK1L protein kinase signaling pathway components as described in the main text. RALF, rapid alkalinization factor; LRE/LLG, LORELEI/LRE-like GPI-AP; ROPGEF, RAC/ROP guanine exchange factor; ROP/RAC, Rho of plants/RAC GTPase; SAM1, 2, S-adenosylmethionine synthetase 1, 2; NADPHox, NADPH oxidase; ROS, reactive oxygen species. Speculative steps are represented by dashed lines.
Cellular Functions of CrRLK1L Receptor-Like Kinases
Since Escobar-Restrepo et al. (2007) discovered a role for FERONIA (FER, AT3G51550) in pollen tube (PT) reception at the female gametophyte, the CrRLK1L subfamily has been linked to an increasing number of biological processes. Generally regarded as cell wall stability sensors, CrRLK1L proteins have been implicated in cell elongation and cell shape, polarized growth, and plant–pathogen interactions (reviewed in Boisson-Dernier et al., 2011; Cheung and Wu, 2011; Lindner et al., 2012; with a summarizing table of CrRLK1L functions in Nissen et al., 2016).
Cell Wall Integrity
Genetics provided the first evidence of CrRLK1L surveillance of cell wall stability (Hematy et al., 2007), with mutations in THESEUS1 (THE1, AT5G54380) partially rescuing the hypocotyl growth defect of cellulose deficient cesA6 (cellulose synthase A, catalytic subunit 6) mutants without influencing the cellulose deficiency (Hematy et al., 2007). THE1 activation may be cell wall perturbation-dependent, leading to reinforcement by ectopic lignification, and preventing excessive loosening and consequent damage to the cell wall.
Cell Elongation
Cell elongation during vegetative growth involves several CrRLK1L subfamily members. HERCULES RECEPTOR KINASES 1 and 2 (HERK1 and HERK2, AT3G46290 and AT1G30570), THE1, FER, CURVY1 (CVY1, AT2G39360) and uncharacterized CrRLK1L AT5G24010 are among the most up-regulated genes upon brassinosteroid (BR) treatment (Guo et al., 2009a,b). BR functions in cell elongation and Guo et al. (2009a,b) described reduced hypocotyl and petiole cell elongation in herk1the1 and fer mutants, and a stronger phenotype in the herk1herk2the1 triple mutant. BR treatment further reduces hypocotyl length in these lines, indicating redundancy between cell elongation pathways (Guo et al., 2009a).
Balancing of cell wall rigidification/loosening has been proposed as the underlying regulatory mechanism for CrRLK1L-mediated cell elongation (Hematy et al., 2007). Cell wall-loosening enzymes, such as expansin and pectin lyase-like genes, are down-regulated in the herk1the1 double mutant, which could promote cell wall stiffening and reduced cell elongation (Guo et al., 2009a; Boisson-Dernier et al., 2011). In contrast, expansin and pectin lyase-like gene expression is up-regulated in the1 mutants in a cesA6 background, suggesting context-dependent function.
Tip Growth in Root Hairs, Pollen Tubes, and Trichomes
Tip growth represents an extreme form of polarized cell expansion that occurs during development of root hairs, PTs, and trichomes (Yang, 1998). For tip growth to occur, various mechanisms must be spatiotemporally coordinated, including polarized exocytosis of vesicles, cytoskeletal reorganization, and generation of second messenger gradients (Lee and Yang, 2008). Many key genetic and exogenous factors have been described (Li et al., 1999; Baluska et al., 2000; Rigas et al., 2001; Foreman et al., 2003; Bloch et al., 2011). Genetic factors include five CrRLK1L proteins: FER, ANXUR1 (ANX1, AT3G04690), ANX2 (AT5G28680), CVY1 and [Ca2+]cyt-associated protein kinase 1/ERULUS (CAP1/ERU, AT5G61350). Although, described in different structures (FER and CVY1 in root hairs; ANX1 and ANX2 in PTs), the former four proteins are localized to the plasma membrane of the growing tip where they increase reactive oxygen species (ROS) production by a common mechanism (Duan et al., 2010; Gachomo et al., 2014). Both fer and cvy1 mutants have defects in other tip growth-requiring structures such as the leaf trichomes (Duan et al., 2010; Gachomo et al., 2014). In contrast, CAP1 is localized to the tonoplast in root hairs where it activates nitrogen permease in response to ammonium/nitrogen levels and facilitates accumulation of Ca2+ (Bai et al., 2014), demonstrating the potential for CrRLK1L proteins to function in a wider range of cellular processes.
Pollen Tube-Female Gametophyte Interactions
During fertilization in angiosperms, the PT grows through the pistil, transporting two sperm cells. At the female gametophyte synergid cells (SCs), the PT bursts to release the sperm cells (Dresselhaus and Franklin-Tong, 2013). Three CrRLK1L subfamily members regulate this process in Arabidopsis: FER, ANX1, and ANX2. Maternal fer mutants fail to induce PT rupture and growth arrest (Escobar-Restrepo et al., 2007). FER is asymmetrically localized to the filiform apparatus in the SCs, and regulates a local increase in ROS (Escobar-Restrepo et al., 2007). Exogenous quenching of ROS in the SC results in a fer-like PT overgrowth phenotype, implying FER-triggered, ROS-mediated, PT growth arrest (Duan et al., 2014). Domain swaps between FER and closely related CrRLK1Ls indicate that, while the FER ECD is needed to complement reproductive PT reception defects in fer mutants, the kinase domain can be replaced with that of related CrRLK1Ls (Kessler et al., 2015). Furthermore, FER kinase activity is not required, suggesting that co-receptors may compensate or provide signal transduction capacity (Kessler et al., 2015).
The two closest homologs to FER, ANX1 and ANX2, act redundantly as its male equivalents. While FER is expressed in all tissues except for mature pollen, ANX1 and ANX2 localization is restricted to the PT tip (Boisson-Dernier et al., 2009). Both ANX proteins are proposed to sense cell wall integrity and maintain its stability at the PT tip (Boisson-Dernier et al., 2009, 2011; Miyazaki et al., 2009). ROS production and a Ca2+ gradient are required downstream of ANX1 and ANX2 in polarized PT growth (Boisson-Dernier et al., 2013).
Biotic and Abiotic Responses
The CrRLK1L subfamily is linked to plant–pathogen interactions, with multiple CrRLK1L genes up-regulated upon elicitor treatments including bacterial flagellin (epitope flg22) and fungal chitin (Lindner et al., 2012). FER becomes phosphorylated upon treatment with flg22 and is speculated to act as a coreceptor with FLS2 (Keinath et al., 2010). FER triggers an intracellular ROS burst in Arabidopsis suspension cells in response to flg22 (Keinath et al., 2010). Kessler et al. (2010) identified FER as a pathogen resistance determinant in powdery mildew infections. They described the similarity between PT reception at the SCs and the first steps in infection, identifying shared molecular components (Kessler et al., 2010). In addition, a member of the mildew resistance locus O (MLO) family, NORTIA (NTA, AT2G17430), re-locates to the filiform apparatus upon PT reception at the SCs in a FER-dependent fashion and is necessary for correct PT response (Kessler et al., 2010). Together these results shed new light on plant–pathogen interactions.
Additionally, CrRLK1L genes respond to a variety of stress and hormone treatments. From genome-wide expression data, Lindner et al. (2012) summarized the general down-regulation of CrRLK1L subfamily expression in response to abiotic stresses such as heat, drought, high osmolarity, cold and hypoxia. Since CrRLK1Ls are linked to ROS production, down-regulation could indicate a strategy to minimize the deleterious oxidative burst that is common to abiotic stress responses. Hormone treatments such as BRs also increase transcript levels of a subset of CrRLK1Ls (Guo et al., 2009a,b).
CrRLK1L Signaling Pathways
A number of proteins have been identified as either direct interaction partners of CrRLK1L kinases, or as phosphorylated/activated upon CrRLK1L signaling. Although our understanding is incomplete, conserved signaling mechanisms are emerging. CrRLK1L signaling generally involves activation of downstream elements through phosphorylation of guanine exchange factors, activation of plasma membrane NADPH oxidase-dependent ROS production and calcium ion fluctuations (Figure 1B).
Upstream Effectors
Perhaps the most striking recent discovery is identification of a ligand for FER. A phosphoproteomic screen of seedlings exposed to the peptide rapid alkalinization factor 1 (RALF1, AT1G02900) identified FER as its receptor (Haruta et al., 2014). Additionally, putative downstream elements of RALF1-FER intracellular signaling were identified, including a plasma membrane H+-ATPase that increases apoplastic pH and cell wall rigidity upon activation (Figure 1B).
Surprisingly, as MDs were anticipated to bind carbohydrate ligands, RALF1 is a small secreted peptide that lacks N-glycosylation (Haruta et al., 2014). The RALF family comprises 34 members in Arabidopsis. The ubiquitous, pollen-specific or SC-specific expression patterns of some RALF-encoding genes correlate with those of CrRLK1L genes, leading to the hypothesis that additional RALF peptides may be ligands for CrRLK1Ls in different processes or developmental stages (Murphy and De Smet, 2014; Wolf and Hofte, 2014). For example, two RALF peptides, SlRALF in tomato and AtRALF4 in Arabidopsis, regulate PT elongation (Covey et al., 2010; Morato do Canto et al., 2014). Further evidence supporting a link between the CrRLK1L and RALF families was provided when Srivastava et al. (2009) described down-regulation of AtRALF23 upon BR treatment. AtRALF23 appears to counteract BR effects on cell growth and elongation (Srivastava et al., 2009), a process in which HERK1, HERK2, FER and THE1 participate (Guo et al., 2009a,b). Additionally, RALF1 suppresses BR effects on root cell elongation by inducing several BR-down-regulated genes involved in BR biosynthesis (Bergonci et al., 2014).
Although unexpected, the discovery of a peptide ligand for FER does not exclude carbohydrate binding by CrRLK1L proteins. As suggested by Wolf and Hofte (2014), the RALF1-CrRLK1L interaction may not involve the MLD, and each CrRLK1L kinase may have multiple biochemically diverse ligands, a property described for animal receptors such as the epidermal growth factor receptor and beta-adrenergic receptor kinase (Touhara, 1997; Moghal and Sternberg, 1999).
Coreceptors and Chaperones
Other RLKs such as FLS2 and CrRLK1L subfamily member HERK1 may act as coreceptors with FER (Guo et al., 2009a; Keinath et al., 2010), however, only two glycosylphosphatidylinositol-anchored proteins (GPI-APs) have been biochemically confirmed to interact with FER and mediate downstream signaling (Li et al., 2015; Liu et al., 2016). LORELEI (LRE) and LRE-like GPI-AP1 (LLG1) bind the extracellular juxtamembrane domain of FER and, in different developmental contexts, are essential for deposition and stability of FER as well as effective RALF1-FER signaling (Li et al., 2015). Considering that there are two further LRE homologs in Arabidopsis (LLG2 and LLG3) with different expression patterns (Capron et al., 2008; Tsukamoto et al., 2010), we speculate that different LLGs could act as chaperones and co-receptors for different CrRLK1L proteins in diverse developmental contexts.
Downstream Signaling Pathways
Downstream components of CrRLK1L signaling have been identified, including RAC/ROP guanine exchange factors (ROPGEFs), a plant-specific subfamily of RHO-GTPases that convert GDP into GTP and activate RAC/ROPs (Carol et al., 2005). Two studies have identified five ROPGEFs that interact with the kinase domain of FER, and two RAC/ROPs as potential downstream components (Duan et al., 2010; Yu et al., 2012). One RAC/ROP (ROP11; AT5G62880) phosphorylates and inactivates PP2C phosphatase ABI2, an integral element in abscisic acid (ABA) signaling (Nishimura et al., 2010; Yu et al., 2012). Adding further complexity, the stunted phenotype of fer-4 mutants has been linked to the ethylene biosynthetic pathway through characterization of S-adenosylmethionine synthetase 1 and 2 (SAM1 and SAM2) as direct interactors of the FER kinase domain (Mao et al., 2015). Mao et al. (2015) propose a scenario in which FER inhibits SAM1 and SAM2 through phosphorylation, suppressing S-adenosylmethionine production and down-regulating ethylene biosynthesis. Together, these findings depict the first steps in a branched CrRLK1L pathway, where a single receptor can interact with multiple partners to activate different downstream elements (Figure 1B). Crosstalk of CrRLK1L signaling with the hormones ABA, ethylene, and BR is also likely.
As previously discussed, increased ROS production occurs in various CrRLK1L-mediated processes. NADPH oxidases are required for the CrRLK1L-mediated ROS burst during tip growth in root hairs or PTs, and PT reception at the SCs (Lee and Yang, 2008; Boisson-Dernier et al., 2013). Interestingly, studies in rice identified a NADPH oxidase as a direct interactor of a RAC/ROP protein (Wong et al., 2007). This connection has recently been confirmed in Arabidopsis by characterizing ROP11 as an interaction partner and activator of the NADPH oxidase Rboh F in root hairs (Yan et al., 2016). Finally, NTA and the receptor-like cytoplasmic kinase MARIS also act downstream in this signaling cascade (Kessler et al., 2010; Boisson-Dernier et al., 2015), although the exact mechanisms are yet to be elucidated. The question of whether CrRLK1L pathways indirectly modify gene expression by post-translational modification of transcription factors remains to be addressed.
An Evolutionary Perspective of the CrRLK1L Subfamily
Despite recent advances, only four studies focus on CrRLK1L protein function in species other than Arabidopsis. Beyond the initial identification of CrRLK1 (Schulze-Muth et al., 1996), phylogenomics identified 16 CrRLK1L homologs with Gigantea-mediated circadian regulation of their expression in Oryza sativa (Nguyen et al., 2014). Secondly, Niu et al. (2016) described ∼40 CrRLK1L family members in diploid cotton species, six of which are linked to fiber development. Finally, a closely related protein, CpRLK1, was functionally characterized in a charophycean unicellular alga (Hirano et al., 2015). CpRLK1 is expressed during mating in this heterothallic alga and regulates gamete formation. Hirano’s study is of particular evolutionary interest since comparison of CrRLK1L function between algae and angiosperms demonstrates potentially analogous involvement in reproduction, cell growth, and cell wall stability sensing. CpRLK1 functions in the release of +-type gametes that lack a cell wall, with +-type cells of CpRLK1 knockdown lines generating an abnormally enlarged conjugation papilla but not releasing the gametal protoplast (Hirano et al., 2015). CpRLK1 is suggested to detect cell wall integrity during conjugation to regulate the release of +-type cells, evoking similarities with regulation of PT function by CrRLK1L proteins in angiosperms (Wolf and Hofte, 2014). Similarly, partial protoplast protrusion in Closterium CpRLK1 knockdown lines resembles cellular material discharge in impaired root hair development phenotypes of fer-4 mutants, supporting a possible ancestral role in cell wall integrity surveillance (Li et al., 2015).
Plant RLKs may have originated in the Streptophyta lineage, which is ancestral to both land plants and charophycean algae (Sasaki et al., 2007). Although, limited by a paucity of algal genomes or transcriptomes, CpRLK1 identification in a charophycean alga substantiates divergence of RLK subfamilies and emergence of the CrRLK1L subfamily early in the evolution of Streptophytes (Shiu and Bleecker, 2001; Sasaki et al., 2007). Conservation of cell wall synthesizing enzymes (rosette cellulose, arabinogalactan, and hemicellulose synthases) across the Streptophyta lineage supports the hypothesis of evolutionary conservation of CrRLK1L cell wall surveillance functions (Sorensen et al., 2010; Popper et al., 2011; Fangel et al., 2012).
There has been an expansion of the CrRLK1L subfamily from early diverging to recent Streptophyta lineages, with the greatest increase in prevalence evident between seedless vascular plants and seed plants (Figure 2A). Expansion of the subfamily may have allowed acquisition of new roles in reproductive processes such as PT growth and its recognition at the female gametophyte (see Pollen Tube-Female Gametophyte Interactions), developmental innovations characteristic of the seed plants (Doyle, 2006; Linkies et al., 2010). An extant member of the earliest divergent angiosperm lineage, Amborella trichopoda (Albert et al., 2013) has orthologs of most characterized A. thaliana CrRLK1L proteins (Figure 2B). The conifer Picea abies (Nystedt et al., 2013) has orthologs of the reproductive clade (FER, ANX1, and ANX2) and some non-reproductive members (THE1 and HERK2; Figure 2B). FER and ANX orthologs in A. trichopoda and P. abies suggest a potentially conserved role of these CrRLK1Ls in mediating PT growth and fertilization (see Pollen Tube-Female Gametophyte Interactions). Fertilization in ferns and some gymnosperms (e.g., cycads and ginkgo) depends on multiflagellated sperm rather than on siphonogamous PT transport of non-motile sperm cells, which arose independently in angiosperms and gymnosperms (Doyle, 2006). Given available data, it is impossible to speculate whether the reproductive clade formed independently when siphonogamy established in conifers and angiosperms or was already present before divergence of these lineages of land plants.
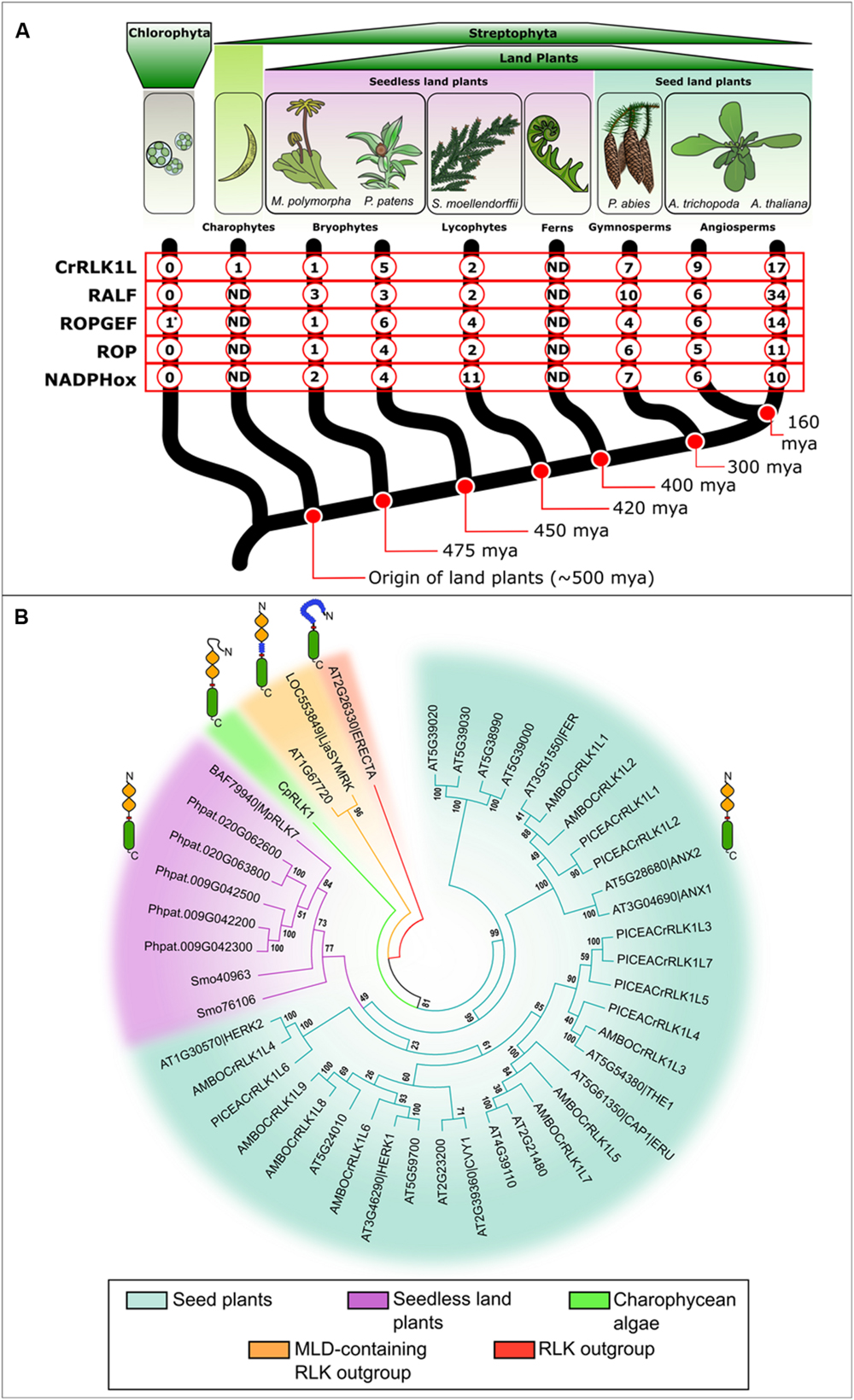
FIGURE 2. (A) Occurrence of CrRLK1L signaling elements across plant lineages. Data is shown for Chlorophyta species Ostreococcus lucimarinus, Chlamydomonas reinhardtii, Volvox carteri, Coccomyxa subellipsoidea, Micromonas pusilla, and Micromonas sp. (Merchant et al., 2007; Palenik et al., 2007; Worden et al., 2009; Prochnik et al., 2010; Blanc et al., 2012); Charophyta Closterium peracerosum-strigosum-littorale complex (Hirano et al., 2015); the liverwort Marchantia polymorpha (Sasaki et al., 2007; These sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community); the model bryophyte Physcomitrella patens (Rensing et al., 2008; Lehti-Shiu and Shiu, 2012); lycophyte Selaginella moellendorffii (Banks et al., 2011; Lehti-Shiu and Shiu, 2012); conifer Picea abies (Nystedt et al., 2013); single living sister species to all other angiosperms Amborella trichopoda (Albert et al., 2013) and Arabidopsis thaliana (Hematy and Hofte, 2008). According to the GenBank, Phytozome and ConGenIE databases, the CrRLK1L subfamily is only present in Streptophyta, increasing in number with developmental complexity. Other signaling elements present similar trends in numbers: RALF peptides, ROPs and membrane bound NADPH oxidases are absent in Chlorophyta and present in land plants (Berken et al., 2005; Torres et al., 2006; Eklund et al., 2010; Cao and Shi, 2012). ROPGEFs are present in land plants and Chlorophyta, although their presence in the latter is restricted to Ostreococcus lucimarinus (∗). Full genomic or transcriptomic records are not available for charophyte and fern lineages included in this figure (ND). (B) Comparative phylogenetic analysis of the CrRLK1L proteins in plants. Sequences were aligned using ClustalX (Larkin et al., 2007). The evolutionary history was inferred using a Neighbor-Joining phylogenetic tree generated with the software MEGA5.2 (Saitou and Nei, 1987; Tamura et al., 2011). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to each branch. Putative CrRLK1L members in A. thaliana (AT4G39110, AT2G21480, AT5G61350|CAP1|ERU, AT5G54380|THE1, AT2G23200, AT5G24010, AT2G39360|CVY1, AT3g46290|HERK1, AT5g59700, AT1g30570|HERK2, AT3G51550|FER, AT3G04690|ANX1, AT5G28680|ANX2, AT5G39000, AT5G38990, AT5G39020, AT5G39030), A. trichopoda (AMBOCrRLK1L1-9; Phyotzome v11 IDs: evm_27.TU.AmTr_v1.0_scaffold – 00077.136, 00016.362, 00003.355, 00092.136, 00045.60, 00080.56, 00024.19, 00001.334, 00001.335), P. abies (PICEACrRLK1L1-7; ConGenIE IDs: MA_71280g0010, MA_21765g0010, MA_10432359g0010, MA_5246g0010, MA_44655g0010, MA_45223g0010, MA_10432359g0020), S. moellendorffii (Smo40963, Smo76106), P. patens (Phpat.020G063800, Phpat.020G062600, Phpat.009G042500, Phpat.009G042200, Phpat.009G042300), M. polymorpha (BAF79940|MpRLK7), and Closterium peracerosum-strigosum-littorale Complex (AB920609|CpRLK1) are shown. Lotus japonicus MLD-LRR-containing RLK SYMRK (LOC553849|LjaSYMRK) and its closest MLD-LRR-RLK homolog in A. thaliana (AT1G67720) were used as an outgroup together with ERECTA from A. thaliana (AT2G26330|ERECTA).
Although Selaginella moellendorffii is a vascular plant, the composition of its CrRLK1L family closely resembles that of non-vascular plants: CrRLK1L subfamily members from the liverwort Marchantia polymorpha, the moss Physcomitrella patens, and the lycophyte S. moellendorffii form a sister clade to non-reproductive CrRLK1L proteins (THE1, CVY1, HERK1, and CAP1; see Cell Wall Integrity, Cell Elongation, Tip Growth in Root Hairs, Pollen Tubes, and Trichomes). Although further research is required to confirm protein function, the phylogeny suggests a potentially conserved function in cell wall stability and regulation of cell elongation across land plants that was recruited in seed plant specific processes as the family expanded. Algal CpRLK1 does not cluster with the early divergent Streptophyta sequences, but is positioned between the CrRLK1L subfamily and the closely related MLD-LRR-RLK subfamily, of which Lotus japonicus SYMRK is best characterized (Antolin-Llovera et al., 2014; Hirano et al., 2015). This may be due to additional extracellular sequences in both CpRLK1 and MLD-LRR-RLKs, which are not present in other CrRLK1L proteins. Ongoing genomic and transcriptomic projects will facilitate more comprehensive phylogenetic studies of RLKs in early diverging Streptophyte lineages and ferns. Phylogenies along with functional studies from diverse species will help us decipher (i) the importance of divergence between CpRLK1 and other CrRLK1Ls, (ii) the evolutionary history of the expansion of this subfamily of RLKs, and (iii) whether the reproductive clade of CrRLK1Ls is specific to seed plants or appeared earlier in the evolution of plants, i.e. in ferns.
Interestingly, CrRLK1L signaling may be conserved from bryophytes to angiosperms (Figure 2A). Downstream signaling components for polarized cell expansion and tip growth mechanisms that are CrRLK1L-mediated are present in liverworts, bryophytes, lycophytes, gymnosperms and angiosperms, including ROPGEFs, RAC/ROPs and NADPH oxidases. Homologs of putative ligands in the form of RALF peptides are also present in these species (Cao and Shi, 2012; Albert et al., 2013; Nystedt et al., 2013). Furthermore, ROPGEFs and RAC/ROPs have conserved functions in cell polarity and tip growth regulation in the filamentous gametophytic (protonemal) stage of P. patens (Ito et al., 2014). In this light, it is plausible that CrRLK1L proteins mediate tip growth from bryophytes to angiosperms via conserved CrRLK1L-ROPGEF-RAC/ROP signaling pathways. A more detailed analysis of when signaling pathway components arose relative to CrRLK1L kinases is precluded by a lack of genomic data from early diverging Streptophyta lineages (excepting the identification of ROPGEF proteins in some Chlorophyta species), however, all signaling components were apparently present in early non-vascular land plants (Figure 2A).
As a final example of how CrRLK1L function and signaling may be conserved across plant evolution, let us consider CAP1 function in root development. Rhizoids are tip-growing root-like structures providing anchorage and water and nutrient uptake for some charophytes and land plants with a free-living gametophytic generation (liverworts, mosses, hornworts, lycophytes, and ferns; Jones and Dolan, 2012). Rhizoids and root hair development share a regulatory transcription network in which transcription factor ROOTHAIR DEFECTIVE 6 (RHD6) family has a central function. RHD6 transcription factors drive expression of root hair-specific genes, including genes required for cell expansion, and are necessary and sufficient for root hair and rhizoid development in angiosperms, liverworts and mosses (Masucci and Schiefelbein, 1994; Menand et al., 2007; Yi et al., 2010). Interestingly, expression of CAP1 (see Tip Growth in Root Hairs, Pollen Tubes, and Trichomes) is positively regulated by RHD6 and RHD6-LIKE 4 (RSL4) in Arabidopsis, suggesting that CAP1 acts downstream of RHD6 and RSL4 in root hair growth (Menand et al., 2007; Bruex et al., 2012). Given functional conservation of RHD6 across distant land plant lineages, we can hypothesize that a similar regulatory network may occur in early diverging land plants, in which RHD6/RSL4 orthologs transcriptionally regulate CrRLK1Ls during tip growth of rhizoids.
Future Directions
CrRLK1L kinases are receiving increasing attention for their roles in regulating developmental and stress responses. A number of questions remain to be addressed: (i) cross-talk with hormonal pathways, (ii) putative carbohydrate-binding capacity via the MLD, and (iii) the ligand-receptor relationships with different RALF peptides. Functional conservation across plant lineages remains speculative, with further research in early diverging lineages key to inferring primordial function, and understanding conserved mechanisms shared by functionally diverse CrRLK1L proteins in higher plants.
Author Contributions
SG-T, JG, and LS conceived and designed the review. SG-T wrote the manuscript draft and all authors edited, read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
LS and SG-T are supported by the Department of Animal and Plant Sciences, University of Sheffield. Research in JG’s lab is supported by RCUK grant BB/N004167/1. Thanks are due to Andrew Fleming for discussion of the manuscript. We apologize to colleagues whose work we did not cite or discuss in depth due to the format length restriction. We also thank John Bowman (Monash) for granting us permission to use unpublished data from the Marchantia genome.
Abbreviations
ABA, abscisic acid; ANX1, ANXUR1; ANX2, ANXUR2; BR, brassinosteroid; CAP1/ERU, [Ca2+]cyt-associated protein kinase 1/ERULUS; CpRLK1, Closterium peracerosum-strigosum-littorale Complex RLK 1; CrRLK1L, Catharanthus roseus RLK 1-like; CVY1, CURVY1; ECD, extracellular domain; FER, FERONIA; flg22, flagellin 22 (epitope of FLAGELLIN); GPI-AP, glycosylphosphatidylinositol-anchored protein; HERK1, HERCULES Receptor Kinase 1; HERK2, HERCULES Receptor Kinase 2; LLG, LRE-like GPI-AP; LRE, LORELEI; LRR, leucine-rich repeat; MD, malectin domain; MLD, malectin-like domain; MLO, mildew resistance locus O; NTA, NORTIA; PAMP, pathogen-associated molecular pattern; PM, powdery mildew; PT, pollen tube; RALF, rapid alkalinization factor; RLK, receptor-like kinase; ROPGEF, RAC/ROP guanine exchange factor; ROS, reactive oxygen species; SAM1, S-adenosylmethionine synthetase 1; SAM2, S-adenosylmethionine synthetase 2; SC, synergid cells; THE1, THESEUS1; TM, transmembrane domain.
References
Afzal, A. J., Wood, A. J., and Lightfoot, D. A. (2008). Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 21, 507–517. doi: 10.1094/MPMI-21-5-0507
Albert, V. A., Barbazuk, W. B., Depamphilis, C. W., Der, J. P., Leebens-Mack, J., Ma, H., et al. (2013). The Amborella genome and the evolution of flowering plants. Science 342, 1241089. doi: 10.1126/science.1241089
Antolin-Llovera, M., Ried, M. K., and Parniske, M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24, 422–427. doi: 10.1016/j.cub.2013.12.053
Bai, L., Zhou, Y., Ma, X., Gao, L., and Song, C. P. (2014). Arabidopsis CAP1-mediated ammonium sensing required reactive oxygen species in plant cell growth. Plant Signal. Behav. 9, e29582.
Baluska, F., Salaj, J., Mathur, J., Braun, M., Jasper, F., Samaj, J., et al. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227, 618–632. doi: 10.1006/dbio.2000.9908
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., Depamphilis, C., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. doi: 10.1126/science.1203810
Bergonci, T., Ribeiro, B., Ceciliato, P. H. O., Guerrero-Abad, J. C., Silva-Filho, M. C., and Moura, D. S. (2014). Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 65, 2219–2230. doi: 10.1093/jxb/eru099
Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436, 1176–1180. doi: 10.1038/nature03883
Blanc, G., Agarkova, I., Grimwood, J., Kuo, A., Brueggeman, A., Dunigan, D. D., et al. (2012). The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13, R39. doi: 10.1186/gb-2012-13-5-r39
Bloch, D., Monshausen, G., Singer, M., Gilroy, S., and Yalovsky, S. (2011). Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant Cell Environ. 34, 76–88. doi: 10.1111/j.1365-3040.2010.02227.x
Boisson-Dernier, A., Franck, C. M., Lituiev, D. S., and Grossniklaus, U. (2015). Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc. Natl. Acad. Sci. U.S.A. 112, 12211–12216. doi: 10.1073/pnas.1512375112
Boisson-Dernier, A., Kessler, S. A., and Grossniklaus, U. (2011). The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 62, 1581–1591. doi: 10.1093/jxb/erq445
Boisson-Dernier, A., Lituiev, D. S., Nestorova, A., Franck, C. M., Thirugnanarajah, S., and Grossniklaus, U. (2013). ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tipviaNADPHoxidases. PLoS Biol. 11:e1001719. doi: 10.1371/journal.pbio.1001719
Boisson-Dernier, A., Roy, S., Kritsas, K., Grobei, M. A., Jaciubek, M., Schroeder, J. I., et al. (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288. doi: 10.1242/dev.040071
Bruex, A., Kainkaryam, R. M., Wieckowski, Y., Kang, Y. H., Bernhardt, C., Xia, Y., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8:e1002446. doi: 10.1371/journal.pgen.1002446
Cao, J., and Shi, F. (2012). Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol. Bioinform. Online 8, 271–292. doi: 10.4137/EBO.S9652
Capron, A., Gourgues, M., Neiva, L. S., Faure, J.-E., Berger, F., Pagnussat, G., et al. (2008). Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20, 3038–3049. doi: 10.1105/tpc.108.061713
Carol, R. J., Takeda, S., Linstead, P., Durrant, M. C., Kakesova, H., Derbyshire, P., et al. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438, 1013–1016. doi: 10.1038/nature04198
Cheung, A. Y., and Wu, H. M. (2011). THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 14, 632–641. doi: 10.1016/j.pbi.2011.09.001
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Covey, P. A., Subbaiah, C. C., Parsons, R. L., Pearce, G., Lay, F. T., Anderson, M. A., et al. (2010). A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153, 703–715. doi: 10.1104/pp.110.155457
DeYoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., and Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem functionin in Arabidopsis. Plant J. 45, 1–16. doi: 10.1111/j.1365-313x.2005.02592.x
Doyle, J. A. (2006). Seed ferns and the origin of angiosperms. J. Torrey Bot. Soc. 133, 169–209. doi: 10.3159/1095-5674(2006)133[169:SFATOO]2.0.CO;2
Dresselhaus, T., and Franklin-Tong, N. (2013). Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. doi: 10.1093/mp/sst061
Duan, Q., Kita, D., Johnson, E. A., Aggarwal, M., Gates, L., Wu, H. M., et al. (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 5, 3129. doi: 10.1038/ncomms4129
Duan, Q., Kita, D., Li, C., Cheung, A. Y., and Wu, H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107, 17821–17826. doi: 10.1073/pnas.1005366107
Eklund, D. M., Svensson, E. M., and Kost, B. (2010). Physcomitrella patens: a model to investigate the role of RAC/ROP GTPase signalling in tip growth. J. Exp. Bot. 61, 1917–1937. doi: 10.1093/jxb/erq080
Escobar-Restrepo, J. M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W. C., et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660. doi: 10.1126/science.1143562
Fangel, J. U., Ulvskov, P., Knox, J. P., Mikkelsen, M. D., Harholt, J., Popper, Z. A., et al. (2012). Cell wall evolution and diversity. Front. Plant Sci. 3:152. doi: 10.3389/fpls.2012.00152
Foreman, J., Demidchik, V., Bothwell, J. H., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Gachomo, E. W., Jno Baptiste, L., Kefela, T., Saidel, W. M., and Kotchoni, S. O. (2014). The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biol. 14:221. doi: 10.1186/s12870-014-0221-7
Guo, H., Li, L., Ye, H., Yu, X., Algreen, A., and Yin, Y. (2009a). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106, 7648–7653. doi: 10.1073/pnas.0812346106
Guo, H., Ye, H., Li, L., and Yin, Y. (2009b). A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal. Behav. 4, 784–786. doi: 10.4161/psb.4.8.9231
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., and Sussman, M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. doi: 10.1126/science.1244454
Hematy, K., and Hofte, H. (2008). Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 11, 321–328. doi: 10.1016/j.pbi.2008.02.008
Hematy, K., Sado, P. E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., et al. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17, 922–931. doi: 10.1016/j.cub.2007.05.018
Hirano, N., Marukawa, Y., Abe, J., Hashiba, S., Ichikawa, M., Tanabe, Y., et al. (2015). A receptor-like kinase, related with cell wall sensor of higher plants, is required for sexual reproduction in the unicellular charophycean alga, Closterium peracerosum-strigosum-littorale complex. Plant Cell Physiol. 56, 1456–1462. doi: 10.1093/pcp/pcv065
Ito, K., Ren, J., and Fujita, T. (2014). Conserved function of Rho-related Rop/RAC GTPase signaling in regulation of cell polarity in Physcomitrella patens. Gene 544, 241–247. doi: 10.1016/j.gene.2014.04.057
Jones, V. A., and Dolan, L. (2012). The evolution of root hairs and rhizoids. Ann. Bot. 110, 205–212. doi: 10.1093/aob/mcs136
Keinath, N. F., Kierszniowska, S., Lorek, J., Bourdais, G., Kessler, S. A., Shimosato-Asano, H., et al. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39140–39149. doi: 10.1074/M110.160531
Kessler, S. A., Lindner, H., Jones, D. S., and Grossniklaus, U. (2015). Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 16, 107–115. doi: 10.15252/embr.201438801
Kessler, S. A., Shimosato-Asano, H., Keinath, N. F., Wuest, S. E., Ingram, G., Panstruga, R., et al. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971. doi: 10.1126/science.1195211
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, Y. J., and Yang, Z. (2008). Tip growth: signaling in the apical dome. Curr. Opin. Plant Biol. 11, 662–671. doi: 10.1016/j.pbi.2008.10.002
Lehti-Shiu, M. D., and Shiu, S. H. (2012). Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2619–2639. doi: 10.1098/rstb.2012.0003
Li, C., Yeh, F.-L., Cheung, A. Y., Duan, Q., Kita, D., Liu, M.-C., et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife 4, e06587. doi: 10.7554/eLife.06587
Li, H., Lin, Y., Heath, R. M., Zhu, M. X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11, 1731–1742. doi: 10.1105/tpc.11.9.1731
Lindner, H., Muller, L. M., Boisson-Dernier, A., and Grossniklaus, U. (2012). CrRLK1L receptor-like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15, 659–669. doi: 10.1016/j.pbi.2012.07.003
Linkies, A., Graeber, K., Knight, C., and Leubner-Metzger, G. (2010). The evolution of seeds. New Phytol. 186, 817–831. doi: 10.1111/j.1469-8137.2010.03249.x
Liu, X., Castro, C., Wang, Y., Noble, J., Ponvert, N., Bundy, M., et al. (2016). The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28, 1035–1052. doi: 10.1105/tpc.15.00703
Mao, D., Yu, F., Li, J., Van De Poel, B., Tan, D. A. N., Li, J., et al. (2015). FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ. 38, 2566–2574. doi: 10.1111/pce.12570
Masucci, J. D., and Schiefelbein, J. W. (1994). The rhd6 Mutation of Arabidopsis thaliana alters root-hair initiation through an Auxin- and Ethylene-associated process. Plant Physiol. 106, 1335–1346.
Menand, B., Yi, K., Jouannic, S., Hoffmann, L., Ryan, E., Linstead, P., et al. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480. doi: 10.1126/science.1142618
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Miyazaki, S., Murata, T., Sakurai-Ozato, N., Kubo, M., Demura, T., Fukuda, H., et al. (2009). ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19, 1327–1331. doi: 10.1016/j.cub.2009.06.064
Moghal, N., and Sternberg, P. W. (1999). Multiple positive and negative regulators of signaling by the EGF-receptor. Curr. Opin. Cell Biol. 11, 190–196. doi: 10.1016/S0955-0674(99)80025-8
Morato do Canto, A., Ceciliato, P. H. O., Ribeiro, B., Ortiz Morea, F. A., Franco Garcia, A. A., Silva-Filho, M. C., et al. (2014). Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiol. Biochem. 75, 45–54. doi: 10.1016/j.plaphy.2013.12.005
Murphy, E., and De Smet, I. (2014). Understanding the RALF family: a tale of many species. Trends Plant Sci. 19, 664–671. doi: 10.1016/j.tplants.2014.06.005
Nguyen, Q. N., Lee, Y. S., Cho, L. H., Jeong, H. J., An, G., and Jung, K. H. (2014). Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 241, 603–613. doi: 10.1007/s00425-014-2203-2
Nishimura, N., Sarkeshik, A., Nito, K., Park, S. Y., Wang, A., Carvalho, P. C., et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299. doi: 10.1111/j.1365-313X.2009.04054.x
Nissen, K. S., Willats, W. G., and Malinovsky, F. G. (2016). Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci. 21, 516–527. doi: 10.1016/j.tplants.2015.12.004
Niu, E., Cai, C., Zheng, Y., Shang, X., Fang, L., and Guo, W. (2016). Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol. Genet. Genomics 291, 1137–1154. doi: 10.1007/s00438-016-1169-0
Nystedt, B., Street, N. R., Wetterbom, A., Zuccolo, A., Lin, Y. C., Scofield, D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. doi: 10.1038/nature12211
Palenik, B., Grimwood, J., Aerts, A., Rouze, P., Salamov, A., Putnam, N., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710. doi: 10.1073/pnas.0611046104
Popper, Z. A., Michel, G., Herve, C., Domozych, D. S., Willats, W. G., Tuohy, M. G., et al. (2011). Evolution and diversity of plant cell walls: from algae to flowering plants. Annu. Rev. Plant Biol. 62, 567–590. doi: 10.1146/annurev-arplant-042110-103809
Prochnik, S. E., Umen, J., Nedelcu, A. M., Hallmann, A., Miller, S. M., Nishii, I., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223–226. doi: 10.1126/science.1188800
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Rigas, S., Debrosses, G., Haralampidis, K., Vicente-Agullo, F., Feldmann, K. A., Grabov, A., et al. (2001). TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13, 139–151. doi: 10.1105/tpc.13.1.139
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasaki, G., Katoh, K., Hirose, N., Suga, H., Kuma, K.-I., Miyata, T., et al. (2007). Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene 401, 135–144.
Schallus, T., Feher, K., Sternberg, U., Rybin, V., and Muhle-Goll, C. (2010). Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology 20, 1010–1020. doi: 10.1093/glycob/cwq059
Schallus, T., Jaeckh, C., Feher, K., Palma, A. S., Liu, Y., Simpson, J. C., et al. (2008). Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 19, 3404–3414. doi: 10.1091/mbc.E08-04-0354
Schulze-Muth, P., Irmler, S., Schroder, G., and Schroder, J. (1996). Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus). cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. J. Biol. Chem. 271, 26684–26689. doi: 10.1074/jbc.271.43.26684
Shiu, S. H., and Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Sorensen, I., Domozych, D., and Willats, W. G. (2010). How have plant cell walls evolved? Plant Physiol. 153, 366–372. doi: 10.1104/pp.110.154427
Srivastava, R., Liu, J.-X., Guo, H., Yin, Y., and Howell, S. H. (2009). Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59, 930–939. doi: 10.1111/j.1365-313X.2009.03926.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Touhara, K. (1997). Binding of multiple ligands to pleckstrin homology domain regulates membrane translocation and enzyme activity of beta-adrenergic receptor kinase. FEBS Lett. 417, 243–248. doi: 10.1016/S0014-5793(97)01294-5
Tsukamoto, T., Qin, Y., Huang, Y., Dunatunga, D., and Palanivelu, R. (2010). A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J. 62, 571–588. doi: 10.1111/j.1365-313X.2010.04177.x
Walker, J. C. (1994). Structure and function of the receptor-like protein kinases of higher plants. Plant Mol. Biol. 26, 1599–1609. doi: 10.1007/BF00016492
Wang, Z.-Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wolf, S., and Hofte, H. (2014). Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell 26, 1848–1856.
Wong, H. L., Pinontoan, R., Hayashi, K., Tabata, R., Yaeno, T., Hasegawa, K., et al. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034. doi: 10.1105/tpc.107.055624
Worden, A. Z., Lee, J. H., Mock, T., Rouze, P., Simmons, M. P., Aerts, A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. doi: 10.1126/science.1167222
Yan, M., Jing, W., Xu, N., Shen, L., Zhang, Q., and Zhang, W. (2016). Arabidopsis thaliana constitutively active ROP11 interacts with the NADPH oxidase respiratory burst oxidase homologue F to regulate reactive oxygen species production in root hairs. Funct. Plant Biol. 43, 221–231. doi: 10.1071/FP15090
Yang, Z. (1998). Signaling tip growth in plants. Curr. Opin. Plant Biol. 1, 525–530. doi: 10.1016/S1369-5266(98)80046-0
Yi, K., Menand, B., Bell, E., and Dolan, L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42, 264–267. doi: 10.1038/ng.529
Keywords: CrRLK1L, signaling pathway, cell expansion, functional conservation, kinase, Streptophyta
Citation: Galindo-Trigo S, Gray JE and Smith LM (2016) Conserved Roles of CrRLK1L Receptor-Like Kinases in Cell Expansion and Reproduction from Algae to Angiosperms. Front. Plant Sci. 7:1269. doi: 10.3389/fpls.2016.01269
Received: 13 June 2016; Accepted: 10 August 2016;
Published: 29 August 2016.
Edited by:
Elena M. Kramer, Harvard University, USAReviewed by:
Verónica S. Di Stilio, University of Washington, USAIve De Smet, Vlaams Instituut voor Biotechnologie and UGent, Belgium
Martin Parniske, Ludwig Maximilian University of Munich, Germany
Copyright © 2016 Galindo-Trigo, Gray and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa M. Smith, TGlzYS5NLlNtaXRoQHNoZWZmaWVsZC5hYy51aw==
 Sergio Galindo-Trigo
Sergio Galindo-Trigo Julie E. Gray
Julie E. Gray Lisa M. Smith
Lisa M. Smith