- Department of Plant Biology, University of California, Davis, Davis, CA, USA
The phytochrome (phy) family of red and far-red photoreceptors provides plants with critical information about their surrounding environment and can signal downstream developmental and physiological changes. Neighboring plants compete for limited light resources, and their presence is detected by the phytochrome photoreceptors as a reduced ratio of red: far-red light. One common response to shade is increased elongation of petioles and internodes to compete with their neighbors. While the phytochrome family, phyB in particular, has been well studied in Arabidopsis, information about the other phytochrome family members is limited, especially in sympodial crop plants such as tomato, that have a very different architecture from that of the model plant. To study the tomato phytochrome family we took advantage of several existing mutants and generated an artificial miRNA (amiRNA) line to target SlPHYE, the remaining phytochrome B subfamily member with no currently available mutant line. Here, we characterize internode elongation and shade avoidance phenotypes of the SlPHYE amiRNA line (PHYE amiRNA). In addition, higher order phytochrome subfamily B mutants were generated with the PHYE amiRNA line to investigate the role of SlphyE within the phyB subfamily. We find that the PHYE amiRNA line has no detectable phenotype on its own, however in higher order combinations with SlphyB1 and/or SlphyB2 there are notable defects in shade avoidance. Most notably, we find that the triple mutant combination of SlPHYE amiRNA, SlphyB1, and SlphyB2 has a phenotype that is much stronger than the SlphyB1 SlphyB2 double, showing constitutive shade avoidance and little to no response to shade. This indicates that SlphyE is required for the shade avoidance response in the absence of SlphyB1 and SlphyB2.
Introduction
Plant growth and development are dependent not only on internal signals, but environmental factors as well. Light is essential for the growth and development of all plants; not only because light is the essential energy source, but also because light signals provide plants with information about their surrounding environment. Plants monitor changes in the quality and direction of light to optimize germination, growth, and development, and to allow optimal capture of light for photosynthesis (Franklin and Whitelam, 2005). Because light is vital for plants, shade poses a significant challenge. Plants detect foliar shade (which has a low ratio of red to far-red light) through the red and far-red light phytochrome photoreceptors. When shade is detected, plants are able to undergo a developmental response, termed shade avoidance (Smith and Whitelam, 1990). This response includes internode and petiole elongation, early flowering, and an increase in apical dominance to allow the plant to better compete with neighbors for light resources.
The plant species Arabidopsis thaliana provides a powerful genetic and molecular tool for studying shade avoidance, but with its rosette architecture, research on this species is ultimately limited because it does not have substantial vegetative internodes. This is significant because internode elongation is the hallmark of shade avoidance in most plants and little is known about genes important for shade-regulated internode elongation. Studying shade avoidance in tomato provides an excellent opportunity to overcome this limitation as tomato has vegetative internodes that are strongly shade responsive. While the phytochrome family is well-known to play a significant role in shade avoidance, it is unknown which of these phytochromes are important for internode elongation. It is also not fully known how well the Arabidopsis phytochrome family serves as a model for other organisms, especially those with a very different architecture than Arabidopsis.
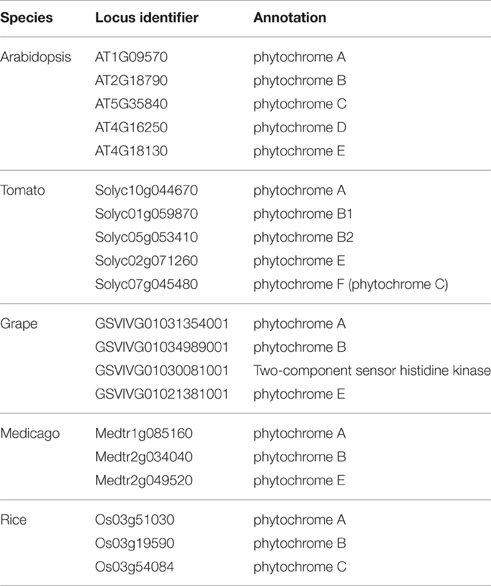
Phytochrome photoreceptors are present in all plants; in flowering plant lineages there are two phytochrome subfamilies, PHYA/PHYC, and PHYB. PHYB has undergone several independent duplications such that not every PHYB subfamily member is present in every lineage (Clack et al., 1994; Devlin et al., 1999; Mathews, 2010). The phytochrome family of both Arabidopsis and tomato consist of five genes, PHYA, PHYC, and three members of the PHYB subfamily, PHYE and two paralogs of PHYB that have arisen through independent duplication (PHYB and PHYD in Arabidopsis, PHYB1 and PHYB2 in tomato; Clack et al., 1994; Alba et al., 2000; Figure 1).
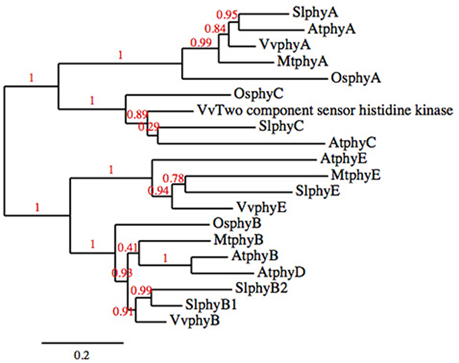
Figure 1. Phylogenetic tree of the phytochrome families of tomato, Arabidopsis, rice, grape, and Medicago truncatula.
Previous research in Arabidopsis showed the phyB family is largely responsible for shade avoidance with phyB as the dominant photoreceptor and phyD and phyE playing minor and/or redundant roles (Franklin et al., 2003). Interestingly, this work and that of others (Weller et al., 2000) shows a different role for the tomato phytochrome family members as compared to Arabidopsis. In order to investigate the role of the tomato PHYB family members, we took advantage of the previously characterized phyA (Lazarova G. I. et al., 1998b), phyB1 (Lazarova G. I. et al., 1998a), and phyB2 (Weller et al., 2000) null mutants and generated an artificial microRNA (amiRNA; Alvarez et al., 2006; Schwab et al., 2006; Warthmann et al., 2008) to knock down expression of PHYE mRNA (PHYE amiRNA). In this manuscript, we characterize the role of phyE in both internode elongation and shade avoidance and demonstrate that phyE is required for shade avoidance in the absence of phyB1 and phyB2.
Materials and Methods
Plant Material
Wild-type cultivar Moneymaker (accession LA2706) and phytochrome mutants in the Moneymaker background; phyA (fri-1, accession LA3809), phyB1 (tri-1, accession LA4357), phyB2 (accession LA4358), phyB1/phyB2 (accession LA4364), and phyA/phyB1/phyB2 (accession LA4366) seed were obtained from the UC Davis/C.M. Rick Tomato Genetics Resource Center, maintained by the Department of Plant Sciences, University of California, Davis, CA.
PHYE amiRNA Generation
The PHYE amiRNA was designed using the WMD-3 Web MicroRNA designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) and generated by modifying the pRS500 plasmid (Addgene, plasmid #22846) through PCR (see Table 1 for PHYE amiRNA generation primers and PCR conditions) to generate the amiRNA precursor fragment of interest following the WMD-3 Web MicroRNA protocol (http://wmd3.weigelworld.org/downloads/Cloning_of_artificial_microRNAs.pdf, Schwab et al., 2006). This PCR product was cloned into expression vector pMDC32 behind a 2x 35S promoter. The pMDC32 vector contains a 35S:HPT hygromycin resistance gene for plant selection (Curtis and Grossniklaus, 2003). The pMDC32 vector containing the PHYE amiRNA was transformed into Agrobacterium strain GV3101 via heat shock transformation.

Table 1. Primer sequences and PCR/ qPCR conditions for genotyping, PHYE amiRNA generation, and expression analysis.
Transgenic Plant Generation
The PHYE amiRNA transgenic lines were generated through Agrobacterium mediated tissue culture transformation by The Ralph M. Parsons Foundation Plant Transformation Facility, University of California, Davis, One Shields Ave, Davis, CA 95616 using a protocol modified from Fillatti et al. (1987) and propagated in the UC Davis College of Biological Sciences Orchard Park Greenhouse Facility.
Higher Order Mutant Generation
To generate the higher order phytochrome mutants with the PHYE amiRNA, a single insertion homozygous PHYE amiRNA was crossed with the phyA/phyB1/phyB2 triple mutant to generate the heterozygous F1. Single insertion was determined through the segregation ratio of the PHYE amiRNA transgene in the T2 generation. Fifty seedlings from each of four independent insertion lines were grown, DNA extracted using a CTAB with chloroform protocol, and genotyped by PCR (see Table 1 for genotyping primers and PCR conditions) for presence or absence of the transgene. Lines with a 3:1 transgene present: transgene absent segregation ratio were determined to be single insertion lines. The F1 was allowed to self to generate the F2 populations segregating for phyA, phyB1, phyB2, and the PHYE amiRNA transgene. Approximately 300 F2 seeds from this population were grown and genotyped by PCR (see Table 1 for genotyping primers and PCR conditions) to identify progeny that represent the different mutant combinations needed to fully characterize the role of PHYE in tomato shade avoidance. These genotypes were phyB1/PHYE amiRNA and phyB2/PHYE amiRNA doubles, the phyB1/phyB2/PHYE amiRNA triple, and the phyA/phyB1/phyB2/PHYE amiRNA quadruple. Plants containing the PHYE amiRNA transgene and homozygous for the phytochrome mutant combinations of choice were transplanted and moved to the greenhouse to bulk seeds for future experiments.
Growth Measurements in High R:FR and Low R:FR Light Conditions
Hypocotyl
Seeds were surface sterilized with 50% household bleach and plated on 0.5x MSMO (Sigma-Aldrich) and 0.7% phytagar (Sigma-Aldrich) in Phytatrays (tomato, Sigma-Aldrich) or round petri dishes (Arabidopsis, Fisher). Plated seeds were kept in the dark (tomato) or dark and 4°C (Arabidopsis) for 2 days before they were moved to constant light conditions set to 34 μE red and 7 μE blue light in an LED chamber for 1 day. LED chambers are custom made and are equipped with Quantum Devices Snap-Lite LED modules, part # SL1515-470-670-735, with peak emissions at 470 nm (blue), 670 nm (red), and 735 nm (far-red). After 1 day, seeds were scored for germination to ensure only seeds with synchronized germination were used. After scoring, the Phytatrays/petri dishes of each genotype were split into two different LED chambers. Far-red light was added to each chamber to bring the red: far-red (R:FR) ratio to 1.5 for high R:FR (R:FR 1.5, 34 μE red, 7 μE blue) or 0.5 for low R:FR to induce the shade avoidance response (R:FR 0.5, 34 μE red, 7 μE blue) in a randomized design in LED chambers. At 10 days post plating, seedlings with synchronized germination were collected onto transparencies, scanned, and hypocotyl lengths were measured from the scanned images using ImageJ (Schneider et al., 2012). This experiment was repeated for a total of two replicates, switching the R:FR treatments for each chamber. An average of 20 plants per genotype/treatment/replicate were measured.
Internode
Seeds were surfaced sterilized with 50% household bleach and plated on moist paper towels in Phytatrays (Sigma-Aldrich). Plated seeds were kept in the dark for 2 days and then moved into 16 h light:8 h dark under high R:FR conditions [R:FR 1.5, photosynthetically active radiation (PAR) 100 μE] in the growth chamber. One week after plating, uniform seedlings were transplanted to soil (commercial Sunshine Mix No. 1, Sun Gro Horticulture) in four-inch pots in a randomized block design and placed in either high R:FR (R:FR 1.5, PAR 100 μE) or low R:FR (R:FR 0.5, PAR 100 μE) conditions to induce the shade avoidance response in a randomized block design in the growth chamber. In these chambers light was provided by a mixture of far-red fluorescent bulbs with a maximum emission at 750 nm (Interlectric Corporation, Warren, PA) and cool-white fluorescent bulbs (GE, F48T12-CW-1500).
For the PHYE amiRNA characterization, 3, 4, and 5 weeks after planting, the epicotyl and first three internodes were measured with digital calipers (Mitutoyo) to capture organ length over time. Six plants per genotype/treatment were measured.
For the higher order mutant analysis, planting was split into three groups staggered by 1 day to allow proper measurements to be obtained. Five weeks after plating the epicotyl and first three internodes were measured with digital calipers to capture organ length. The higher order mutant growth characterization was repeated for a total of two replicates, switching the high R:FR and low R:FR sides of the growth chamber. An average of eight plants per genotype/treatment/replicate was measured.
Light Measurements
Measurements to determine LED chamber μE red, μE blue, and R:FR ratio and growth chamber PAR and R:FR ratio were taken with a BLACK-Comet CXR-SR-50 (StellarNet Inc.) Light spectra for high R:FR and low R:FR LED and growth chambers are in Supplemental Figure 1.
Statistical Analyses
Wild Type vs. Double Mutants (Figure 2)
For each wild-type or mutant strain a t-test was performed to determine if the different light conditions caused a significant difference in length. P-values were adjusted for multiple comparisons. Scripts for this analysis are available at https://github.com/MaloofLab/Lavelle-phyE-Frontiers-2016.
Wild Type vs. Single Mutants (Figure 4)
A linear mixed effects model was fit using the lme4 (Bates et al, 2013, 2015) and lmerTest (Kuznetsova et al., 2014) packages in R (R Core Team, 2014). Genotype, treatment, and measurement day, along with two and three-way interactions were used as fixed effects and flat was used as a random effect. Scripts for this analysis are available at https://github.com/MaloofLab/Lavelle-phyE-Frontiers-2016.
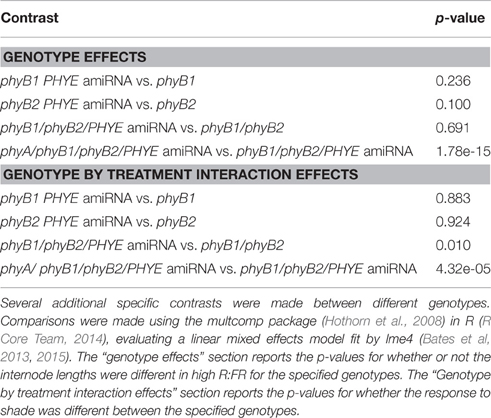
Multiple Mutant Combinations (Figure 6 and Table 2)
To obtain line means and compare the shade avoidance response between Moneymaker and the various mutant combinations a linear mixed effects model was fit using the lme4 (Bates et al, 2013, 2015) and lmerTest (Kuznetsova et al., 2014) packages in R (R Core Team, 2014). Genotype, treatment, and their interaction were used as fixed effects, and stagger date and replicate experiment number were used as random effects. Additional contrasts were tested using the multcomp package (Hothorn et al., 2008). Scripts for this analysis are available at https://github.com/MaloofLab/Lavelle-phyE-Frontiers-2016.
Gene Expression of PHYB Family Genes in the PHYE amiRNA Background
Plants were grown as outlined for internode growth experiments. Young leaf issue from Moneymaker and the PHYE amiRNA line was harvested and flash frozen in liquid nitrogen approximately 3 weeks after planting. Tissue from three plants per genotype was pooled for each of four biological replicates. The Plant RNeasy kit (Qiagen) was used for RNA extraction. qPCR (see Table 1 for qPCR primers and qPCR conditions) was done using a Bio-Rad iCycler to evaluate the affect the PHYE amiRNA has on the transcript levels of PHYB family members PHYB1 Solyc01g059870, PHYB2 Solyc05g053410, and PHYE Solyc02g071260.
Phylogenetic Tree
The phylogenetic analysis of the phytochrome family was done using phylogeny.fr “One-Click” without the use of Gblocks (http://www.phylogeny.fr/index.cgi; Dereeper et al., 2008, 2010). Rice (Ouyang et al., 2007), Medicago (Young et al., 2011), and grape (Jaillon et al., 2007) sequences were obtained from Phytozome (phytozome.org; Goodstein et al., 2012), Arabidopsis sequences were obtained from TAIR (arabidopsis.org; Lamesch et al., 2012), and tomato sequences were obtained from Solgenomics (solgemomics.net; Fernandez-Pozo et al., 2015). Locus identifier and annotation for the sequences used in the analysis are in Table 3.
Results and Discussion
The phyB1/phyB2 Double Mutant Retains a Strong Shade Avoidance Response
Consistent with previous work characterizing the phyB family in Arabidopsis (Aukerman et al., 1997; Franklin et al., 2003) and tomato (Weller et al., 2000) we also documented differences between the Arabidopsis and tomato phytochrome families. Most strikingly, the Arabidopsis phyB/phyD double mutant does not show a hypocotyl elongation shade avoidance response (Figure 2A) while the analogous tomato phyB1/phyB2 double mutant does retain a hypocotyl shade avoidance response (Figure 2B) and has internodes that are strongly shade responsive (Figure 2C). Based on the strong shade avoidance response in the tomato phyB1/phyB2 double mutant we hypothesized that an additional phytochrome, likely the third member of the tomato PHYB family, PHYE, likely plays a role in the tomato shade avoidance response.
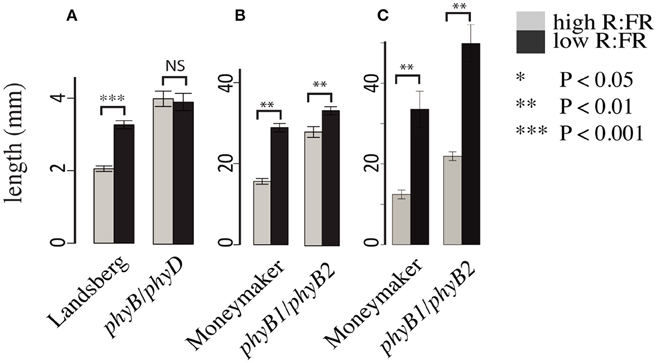
Figure 2. Growth phenotypes of Arabidopsis and tomato phyB paralog double mutants. Hypocotyl lengths of (A) Arabidopsis phyB/phyD double mutants and wild-type Landsberg and (B) tomato phyB1/phyB2 double mutants and wild-type Moneymaker. Measurements were made 10 days post plating on seedlings grown on 0.5x MSMO and 0.7% agar in Phytatrays under constant light in high R:FR and low R:FR conditions. n = an average of 20 for each genotype/treatment. Internode length (C) of 5 week old phyB1/phyB2 and Moneymaker plants grown on soil in four-inch pots under long day (16:8) high R:FR and low R:FR conditions in a growth chamber. n = 5 plants for each genotype/treatment. The error bars show ±SE. *P < 0.05, **P < 0.01, ***P < 0.001, NS = not significant by Student's t-test.
PHYE mRNA is Reduced 50% in the PHYE amiRNA Transgenic Line While Expression of PHYB Family Members PHYB1 and PHYB2 Are Unaffected
In order to investigate the role of PHYE in internode elongation and shade avoidance, an amiRNA was designed to target PHYE using the WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Schwab et al., 2006) and cloned into a binary vector under the control of the constitutive 35S promoter. The construct was transformed into Moneymaker and transformants were screened for single insertion homozygous lines in the T3 generation. In order to demonstrate that the PHYE amiRNA is functional and specific to the target of interest, gene expression of the tomato phyB subfamily members (PHYB1, PHYB2, and PHYE) were assayed by qPCR in the PHYE amiRNA line. As expected, the PHYE amiRNA does reduce expression of PHYE mRNA (Figure 3A) and does not target related family members PHYB1 (Figure 3B) and PHYB2 (Figure 3C). The results show that PHYE mRNA is reduced about 50% in the transgenic line while expression levels of the other family members are unchanged in the amiRNA line compared to their expression in wild type Moneymaker.
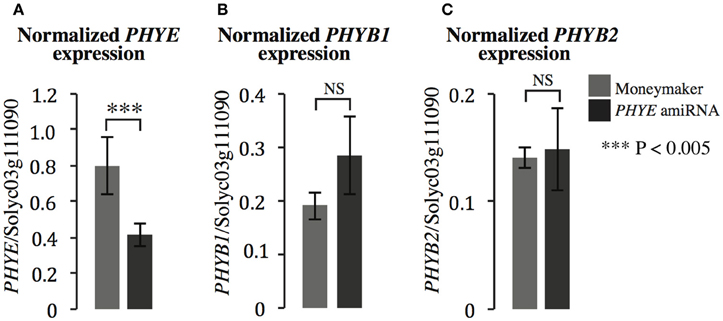
Figure 3. Gene expression of PHYB subfamily members (A) PHYE, (B) PHYB1, and (C) PHYB2 in Moneymaker and the PHYE amiRNA transgenic line. Leaf tissue was collected from 3-week old transgenic PHYE amiRNA and wild type Moneymaker plants grown on soil in four-inch pots under long day (16:8) high R:FR conditions in a growth chamber. n = 4 biological replicates, each consisting of three individuals for each genotype. The error bars show ±SE. ***P < 0.005, NS = not significant by Student's t-test.
PHYE amiRNA Transgenic Plants Have a Wild Type Phenotype
The PHYE amiRNA line, previously characterized phyB1 (Lazarova G. I. et al., 1998a) and phyB2 (Weller et al., 2000) null mutants, and wild-type Moneymaker were characterized for the internode elongation shade avoidance response over time under long day high R:FR and low R:FR to induce the shade avoidance response in a growth chamber. Like phyB2 mutants, PHYE amiRNA plants have wild-type internode lengths in both high R:FR and low R:FR (Figure 4). In contrast the phyB1 mutant plants have elongated internodes in high R:FR but a wild-type response to shade (Figure 4). These similarities are especially apparent at 5 weeks of age as the phyB2, PHYE amiRNA, and wild-type Moneymaker are all indistinguishable from each other for internode length in high R:FR and response to shade. These results suggest that like phyB2, phyE is not required for wild-type shade avoidance in the presence of other phyB family members; it is possible, however, that since some PHYE transcript remains in the PHYE amiRNA line, a true phyE null monogenic mutant would show an elongated internode length phenotype or reduced shade avoidance response. This finding is consistent with Arabidopsis phytochrome family, as the monogenic Arabidopsis phyB mutant is the only phyB subfamily member to display a defective elongation phenotype. The Arabidopsis phyD (Devlin et al., 1999) and phyE (Devlin et al., 1998) monogenic mutants both have a wild-type growth phenotype.
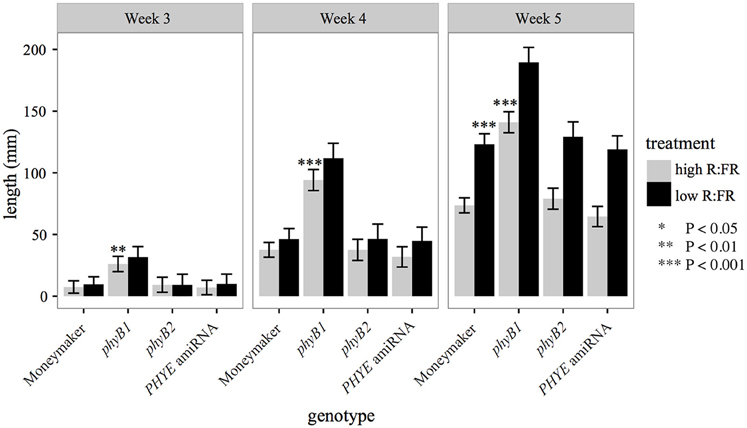
Figure 4. Growth characteristics of phyB family single mutants and amiRNA in high R:FR and in low R:FR. Combined epicotyl, internode 1, and internode 2 length of 3, 4, and 5 week-old phyB1, phyB2, PHYE amiRNA, and Moneymaker plants grown on soil in four-inch pots under long day (16:8) high R:FR and low R:FR conditions. n = 6 for each genotype/treatment. The error bars show ±SE. phyE was not significantly different than wild type in high R:FR or low R:FR at any time point (P > 0.25). Asterisks for “high R:FR” bars indicate that a mutant was significantly different than Moneymaker in high R:FR at that time. Asterisks for “low R:FR” bars indicated that a mutant had a significantly different response to shade as compared to wild type at that time. Asterisks for Moneymaker “low R:FR” are for the high R:FR vs. low R:FR comparison Moneymaker for that time point. *P < 0.05, **P < 0.01, ***P < 0.001 by a linear mixed-effects model.
PHYE Is Redundant for Epicotyl and Internode Elongation and Shade Avoidance
To fully investigate the role of PHYE in the control of epicotyl and internode length and internode elongation response to shade, higher order mutants were generated by crossing the PHYE amiRNA transgenic line with the phyA/phyB1/phyB2 triple mutant. The heterozygous F1 progeny of this cross was then grown and allowed to self-pollinate to generate an F2 population segregating for phyA, phyB1, phyB2, and the PHYE amiRNA transgene. Individuals from this population were genotyped to identify the lines that represent the different double, triple, and quadruple mutant combinations needed to fully characterize the role of PHYE in elongation and shade avoidance. Once the homozygous higher order mutant lines were obtained, epicotyl length, internode length, and increased elongation in response to shade were characterized in phyB subfamily double mutants phyB1/phyB2, phyB1/PHYE amiRNA, and phyB2/PHYE amiRNA, the phyB subfamily triple mutant phyB1/phyB2/PHYE amiRNA, and quadruple mutant phyA/phyB1/phyB2/PHYE amiRNA under long day high R:FR and shade conditions in a growth chamber (Figure 5).

Figure 5. Growth phenotypes of 5-week old high R:FR grown Moneymaker, single mutants phyA, phyB1, phyB2, PHYE amiRNA, double mutants phyB1/phyB2, phyB1/PHYE amiRNA, phyB2/PHYE amiRNA, triple mutant phyB1/phyB2/PHYE amiRNA, and quadruple mutant phyA/phyB1/phyB2/PHYE amiRNA.
Double mutant analysis of the phyB1/phyB2, phyB1/PHYE amiRNA, and phyB2/PHYE amiRNA lines revealed overlapping roles in the tomato PHYB subfamily. Interestingly, like their respective individual single mutants, both internode length and shade avoidance in the phyB2/PHYE amiRNA double mutant are not significantly different from wild type (Figure 6), further suggesting that phyB1 is sufficient for wild-type internode growth and shade avoidance under these conditions. The phyB1 single mutant and phyB1/PHYE amiRNA double mutant are not significantly different from each other with respect to internode length in high R:FR but phyB1/PHYE amiRNA is less responsive than the phyB1 single for internode elongation in response to shade (Figure 6 and Table 2) indicating that phyB2 and/or residual phyE provide some shade responsiveness in these lines. Finally, the phyB1/phyB2 double mutant was significantly taller than wild type in high R:FR but did not show a significantly different response to shade than wild type (Figure 6). Although the double mutant analysis does not provide direct information about the role of PHYE in internode elongation or shade avoidance, the shade responsiveness of the phyB1/phyB2 double mutant shows that an additional gene or genes are involved in the control of shade avoidance. PHYE is the likely candidate as the third member of the PHYB subfamily that has been shown to play a central role in shade avoidance.
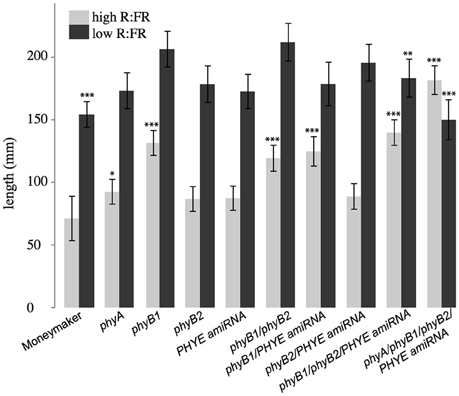
Figure 6. Growth phenotypes of higher order phytochrome mutants. Total length of epicotyl, internode 1, and internode 2 of 5-week old Moneymaker and phytochrome single, double triple, and quadruple mutant plants grown on soil in four-inch pots under long day (16:8) high R:FR and low R:FR conditions in a growth chamber. n = an average of eight plants for each genotype/treatment/replicate. The error bars show ±SE. Asterisks for mutant “high R:FR” bars indicate that a mutant was significantly different than Moneymaker in high R:FR. Asterisks for mutant “low R:FR” bars indicated that a mutant had a significantly different response to shade as compared to wild type at that time. Asterisks for Moneymaker “low R:FR” are for the high R:FR vs. low R:FR comparison in Moneymaker. P-values for additional contrasts are provided in Table 3. *P < 0.05, **P < 0.01, ***P < 0.001 by a linear mixed-effects model.
Triple and Quadruple Mutants Reveal a Role for phyE in Internode Elongation and Shade Avoidance
Although neither single nor double mutant analysis uncovered a specific role for PHYE in either internode elongation or shade avoidance, characterization of the phyB1/phyB2//PHYE amiRNA triple mutant revealed a role for PHYE in shade avoidance. While the phyB1/phyB2/PHYE amiRNA triple mutant had a similar internode length to the phyB1 single and phyB1 double mutants, shade avoidance is significantly reduced in the phyB1/phyB2/PHYE amiRNA triple mutant as compared to wild type or the phyB1/phyB2 double mutant (p = 0.009, 0.01, respectively; Figure 6 and Table 2). These results show that PHYE is required for shade avoidance in the absence of PHYB1 and PHYB2, indicating a distinct role for PHYE in the internode elongation response to shade. Beyond the tomato PHYB subfamily, the phyA/phyB1/phyB2/PHYE amiRNA quadruple mutant shows both significant internode elongation and loss of a shade avoidance response.
Conclusions
The internode length and internode shade avoidance response of higher order tomato PHYB subfamily mutants highlight both similarities and differences between the tomato and Arabidopsis phytochrome families. Tomato PHYB1, like Arabidopsis PHYB, is the dominant phytochrome in both internode elongation and shade avoidance while tomato PHYB2 and PHYE and Arabidopsis PHYD and PHYE play largely redundant roles as monogenic mutants in any of the genes display a wild-type phenotype. Although it is possible that PHYE plays a larger role in both internode elongation and shade avoidance that cannot be seen due to the incomplete knockdown of PHYE mRNA in the PHYE amiRNA line, like Arabidopsis PHYD and PHYE, the role of tomato PHYB2 and PHYE are more readily seen in the tomato phyB1 background. There is a clear role for PHYE in tomato shade avoidance. The most interesting difference between the tomato and Arabidopsis phytochrome families comes with the comparison between the shade responsive tomato phyB1/phyB2 double mutant and non-shade responsive phyB1/phyB2/ PHYE amiRNA triple mutant. This result clearly outlines a role for PHYE in the internode elongation response for shade.
Author Contributions
AS and JM conceived and designed the experiments; AS and JM wrote the article; AS, LH, and JM conducted the experiments, analyzed data, and contributed to the study design.
Funding
This work was supported in part by the Elsie Taylor Stocking Memorial Fellowship awarded to AS in 2013, by NSF grant IOS-0820854, by United States Department of Agriculture NIFA project (http://www.csrees.usda.gov/; CA-D-PLB-7226-H) to JM, and by internal UC Davis funds.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Tomato Genetics Resource Center for providing seed of the Moneymaker cultivar and phytochrome mutant lines. The MATERIAL was developed by and/or obtained from the UC Davis/C.M. Rick Tomato Genetics Resource Center, maintained by the Department of Plant Sciences, University of California, Davis, CA 95616. We thank undergraduate interns Natalie Gath and Deyu (Judy) Wang for their technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01275
References
Alba, R., Kelmenson, P. M., Cordonnier-Pratt, M.-M., and Pratt, L. H. (2000). The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 17, 362–373. doi: 10.1093/oxfordjournals.molbev.a026316
Alvarez, J. P., Pekker, I., Goldshmidt, A., Blum, E., Amsellem, Z., and Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18, 1134–1151. doi: 10.1105/tpc.105.040725
Aukerman, M. J., Hirschfeld, M., Wester, L., Weaver, M., Clack, T., Amasino, R. M., et al. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9, 1317–1326. doi: 10.1105/tpc.9.8.1317
Bates, D., Maechler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Soft. 67, 1–48. doi: 10.18637/jss.v067.i01
Bates, D., Maechler, M., Bolker, B., and Walker, S. (2013). lme4: Linear Mixed-Effects Models Using Eigen and S4. R Package Version 1.
Clack, T., Mathews, S., and Sharrock, R. A. (1994). The phytochrome apoprotein family inArabidopsis is encoded by five genes: the sequences and expression ofPHYD andPHYE. Plant Mol. Biol. 25, 413–427. doi: 10.1007/BF00043870
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Dereeper, A., Audic, S., Claverie, J.-M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Devlin, P. F., Patel, S. R., and Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1487. doi: 10.1105/tpc.10.9.1479
Devlin, P. F., Robson, P. R. H., Patel, S. R., Goosey, L., Sharrock, R. A., and Whitelam, G. C. (1999). Phytochrome D acts in the shade-avoidance syndrome in arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119, 909–916. doi: 10.1104/pp.119.3.909
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Fillatti, J. J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. doi: 10.1038/nbt0787-726
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., and Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. doi: 10.1104/pp.102.015487
Franklin, K. A., and Whitelam, G. C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96, 169–175. doi: 10.1093/aob/mci165
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biom. J. 50, 346–363. doi: 10.1002/bimj.200810425
Jaillon, O., Aury, J.-M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Kuznetsova, A., Brockhoff, P., and Christensen, R. (2014). lmerTest: Tests in Linear Mixed Effects Models. R Package Version 2.0–20.
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lazarova, G. I., Kerckhoffs, L. H. J., Brandstädter, J., Matsui, M., Kendrick, R. E., Cordonnier-Pratt, M.-M., et al. (1998b). Molecular analysis of PHYA in wild-type and phytochrome A-deficient mutants of tomato. Plant J. 14, 653–662. doi: 10.1046/j.1365-313x.1998.00164.x
Lazarova, G. I., Kubota, T., Frances, S., Peters, J., Hughes, M. G., Brandstädter, J., et al. (1998a). Characterization of tomato PHYB1 and identification of molecular defects in four mutant alleles. Plant Mol. Biol. 38, 1137–1146. doi: 10.1023/A:1006068305454
Mathews, S. (2010). Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22, 4–16. doi: 10.1105/tpc.109.072280
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl.976
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial MicroRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Smith, H., and Whitelam, G. (1990). Phytochrome, a family of photoreceptors with multiple physiological roles. Plant Cell Environ. 13, 695–707. doi: 10.1111/j.1365-3040.1990.tb01084.x
Warthmann, N., Chen, H., Ossowski, S., Weigel, D., and Hervé, P. (2008). Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3:e1829. doi: 10.1371/journal.pone.0001829
Weller, J. L., Schreuder, M. E. L., Smith, H., Koornneef, M., and Kendrick, R. E. (2000). Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant J. 24, 345–356. doi: 10.1046/j.1365-313x.2000.00879.x
Keywords: elongation, internode, phytochrome, shade avoidance, Solanum lycopersicum, tomato
Citation: Schrager-Lavelle A, Herrera LA and Maloof JN (2016) Tomato phyE Is Required for Shade Avoidance in the Absence of phyB1 and phyB2. Front. Plant Sci. 7:1275. doi: 10.3389/fpls.2016.01275
Received: 30 March 2016; Accepted: 10 August 2016;
Published: 16 September 2016.
Edited by:
Tohru Hashimoto, Uozaki Life Science Laboratory, JapanReviewed by:
Aaron I. Velez Ramirez, Ghent University, BelgiumGolam Jalal Ahammed, Zhejiang University, China
Copyright © 2016 Schrager-Lavelle, Herrera and Maloof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julin N. Maloof, am5tYWxvb2ZAdWNkYXZpcy5lZHU=
 Amanda Schrager-Lavelle
Amanda Schrager-Lavelle Leslie A. Herrera
Leslie A. Herrera Julin N. Maloof
Julin N. Maloof