- 1Laboratory for Chemistry and Life Science, Institute of Innovative Research, Tokyo Institute of Technology, Yokohama, Japan
- 2Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Saitama, Japan
Abscisic acid (ABA), a pivotal phytohormone that is synthesized in response to abiotic stresses and other environmental changes, induces various physiological responses. Heme, in its unbound form, has a positive signaling role in cell-cycle initiation in Cyanidioschyzon merolae. ABA induces heme accumulation, but also prevents cell-cycle initiation through the titration of the unbound heme by inducing the heme scavenging protein tryptophan-rich sensory protein-related protein O. In this study, we analyzed the accumulation of tetrapyrrole biosynthetic gene transcripts after the addition of ABA to the medium and found that transcripts of a ferrochelatase and a magnesium-chelatase subunit increased, while other examined transcripts decreased. Under the same conditions, the heme and magnesium-protoporphyrin IX contents increased, while the protoporphyrin IX content decreased. Thus, ABA may regulate the intracellular heme and other tetrapyrrole contents through the transcriptional regulation of biosynthetic genes.
Introduction
Abscisic acid (ABA) is a phytohormone of land plants that is involved in many aspects of plant physiology. ABA induces stress tolerance under various stressful conditions, such as drought and high salt (Skriver and Mundy, 1990; Tuteja, 2007). ABA induces the growth of roots, as well as dormancy in buds and seeds, and in leaf organs it induces the stomatal closure, which helps the plant to preserve water during droughts. It is widely believed that these responses are mostly mediated by transcriptional activation, and an underlying mechanism involving the specific receptors pyrabactin resistance 1 (PYR1)/PYR1-like/regulatory components of ABA receptors, SNF1-related kinases 2 and Protein phosphatase 2C has been clarified (Ma et al., 2009; Park et al., 2009; Hartung, 2010; Hauser et al., 2011). In addition, ABA also positively regulates heme biosynthesis in Arabidopsis (Vanhee and Batoko, 2011; Vanhee et al., 2011). Exogenous ABA addition induces the transient increase of intracellular unbound heme, which is required to activate ABA-8′-hydroxylase, an ABA degradation enzyme to prevent continuous signaling events (Vanhee and Batoko, 2011; Vanhee et al., 2011). At the same time, ABA induces a heme-scavenger protein, tryptophan-rich sensory protein-related protein O (TSPO), to quench excess unbound heme that may cause oxidative damage to the cell (Vanhee and Batoko, 2011; Vanhee et al., 2011).
Abscisic acid has also been found in eukaryotic algae and cyanobacteria. Thus, ABA signaling could be very ancient, but the physiological significance has been poorly understood. In a previous study, we analyzed ABA function in the unicellular red alga Cyanidioschyzon merolae, which has available complete nuclear, mitochondrial, and chloroplast genome sequences and molecular genetic tools (Kuroiwa, 1998; Matsuzaki et al., 2004; Nozaki et al., 2007; Kobayashi et al., 2010). In Arabidopsis and other land plants, ABA can be synthesized from the common precursor zeaxanthin by three enzymes, zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED), and short-chain dehydrogenase/reductase (SDR). The C. merolae genome was found to encode orthologous proteins for NCED and SDR, and the presence of zeaxanthin was previously confirmed in C. merolae (Cunningham et al., 2007). Therefore, we checked for endogenous ABA from C. merolae culture, and found that C. merolae accumulates ABA under the salt stressed condition (Kobayashi et al., 2016). As the NCED knock out strain could not accumulate ABA, the common ABA biosynthetic pathway as in land plants was confirmed in C. merolae. While no orthologous gene encoding ZEP was specified from the C. merolae genome, it was reported that an Arabidopsis plant lacking the functional ZEP still accumulates ABA (Barrero et al., 2005). Thus, there is possibly another type enzyme that directs the reaction, which could be common with the C. merolae enzyme.
The addition of exogenous ABA induced a block in the cell-cycle G1/S transition and the homologous TSPO gene’s expression. Because TSPO scavenges the intracellular unbound heme in Arabidopsis, we wondered whether this unbound heme was required for the cell-cycle initiation, and found that the inhibitory effect by ABA was canceled by the addition of exogenous heme. Thus, ABA prevents the cell-cycle G1/S transition through the reduction of intracellular unbound heme accumulation in C. merolae (Kobayashi et al., 2016). As the interconnection among ABA, TSPO, and heme was likely conserved between primitive algae and land plants, the regulatory scheme is likely an ancient trait of ABA signaling conserved during plant evolution. However, the underlying mechanism of ABA-induced heme accumulation has not been clearly elucidated. In this study, we examined the expression of tetrapyrrole biosynthetic genes and the cellular contents of tetrapyrrole intermediates, and hypothesize that ABA affects the tetrapyrrole contents through transcriptional control.
Materials and Methods
Materials and Culture Conditions
Cells of C. merolae 10D were cultured and their growth synchronized as described previously (Kobayashi et al., 2010).
Quantitative PCR
Cells were cultured under constant light or synchronizing conditions, with or without ABA (10 μM). Total RNA was extracted from C. merolae cells as described previously (Kobayashi et al., 2011). First-strand synthesis of cDNA was performed using 5 μg RNA and random primers with ReverTra Ace (Toyobo, Osaka, Japan), and the abundance of the each transcript was quantified using real-time PCR. Real-time PCR was performed as described previously (Kobayashi et al., 2016), using the primers shown in Supplementary Table S1.
Measurement of Tetrapyrrole Molecules
Protoporphyrin IX (ProtoIX), magnesium-protoporphyrin IX (Mg-ProtoIX), and chlorophyll-a were measured by high-performance liquid chromatography (HPLC) as described previously (Zapata et al., 2000), with minor modifications. Synchronized cells were homogenized in 80% acetone and centrifuged at 10,000 × g for 5 min. The supernatant was mixed with water to a final concentration of 75% before the HPLC analysis. According to the method of Zapata et al. (2000), pigments were separated on a reversed-phase column, Symmetry C8 (150 mm × 4.6 mm; Waters, Milford, MA, USA) using a Nexera X2 HPLC system (Shimadzu, Kyoto, Japan). Mg-ProtoIX was detected with an excitation wavelength at 417 nm and emission at 600 nm. ProtoIX was detected with an excitation wavelength at 400 nm and emission at 635 nm. Chlorophyll-a was detected by measuring the absorbance at 410 nm. Standard curves were made using authentic standards.
Northern Blot Analysis
Total RNA preparation and northern blot analyses were performed as described previously (Kobayashi et al., 2011). Gene-specific probes for northern blot analyses were generated with specific primers (Supplementary Table S1) and C. merolae genomic DNA as the template.
Results and Discussion
Genes Involved in Heme Homeostasis in C. merolae
Because most known ABA responses are mediated by the transcriptional regulation of nuclear genes, we examined whether this was also the case for the ABA induced-heme increase. In C. merolae, the tetrapyrrole biosynthetic pathway was previously analyzed and localization of each enzyme to specific cell compartments was proposed (Watanabe et al., 2013) (Figure 1). Here, we focused on 13 enzymes, glutamyl-tRNA reductase (HemA; CMJ054C), glutamate-1-semialdehyde 2, 1- aminomutase (HemL; CMP285C), aminolevulinic acid dehydrase (HemB; CMD104C), porphobilinogen deaminase (CME132C), uroporphyrinogen III synthase (HemD; CML040C), uroporphyrinogen decarboxylase (HemE; CME194C, CMP083C), coproporphyrinogen III oxidase (HemF; CMO136C), oxygen-independent coproporphyrinogen III oxidase (HemN; CMR445C), protoporphyrinogen IX oxidase (HemY; CMB025C), ferrochelatase (FeCh; CMS035C), heme oxygenase (HO; CMH209C), and, three Mg-chelatases, ChlH (CMB093C), ChlD (CMM270C), and ChlI (CMV024C), that were considered to be involved in the heme homeostasis. These enzymes are encoded by the nuclear genome, except for one of the three Mg-chelatase subunits, ChlI.
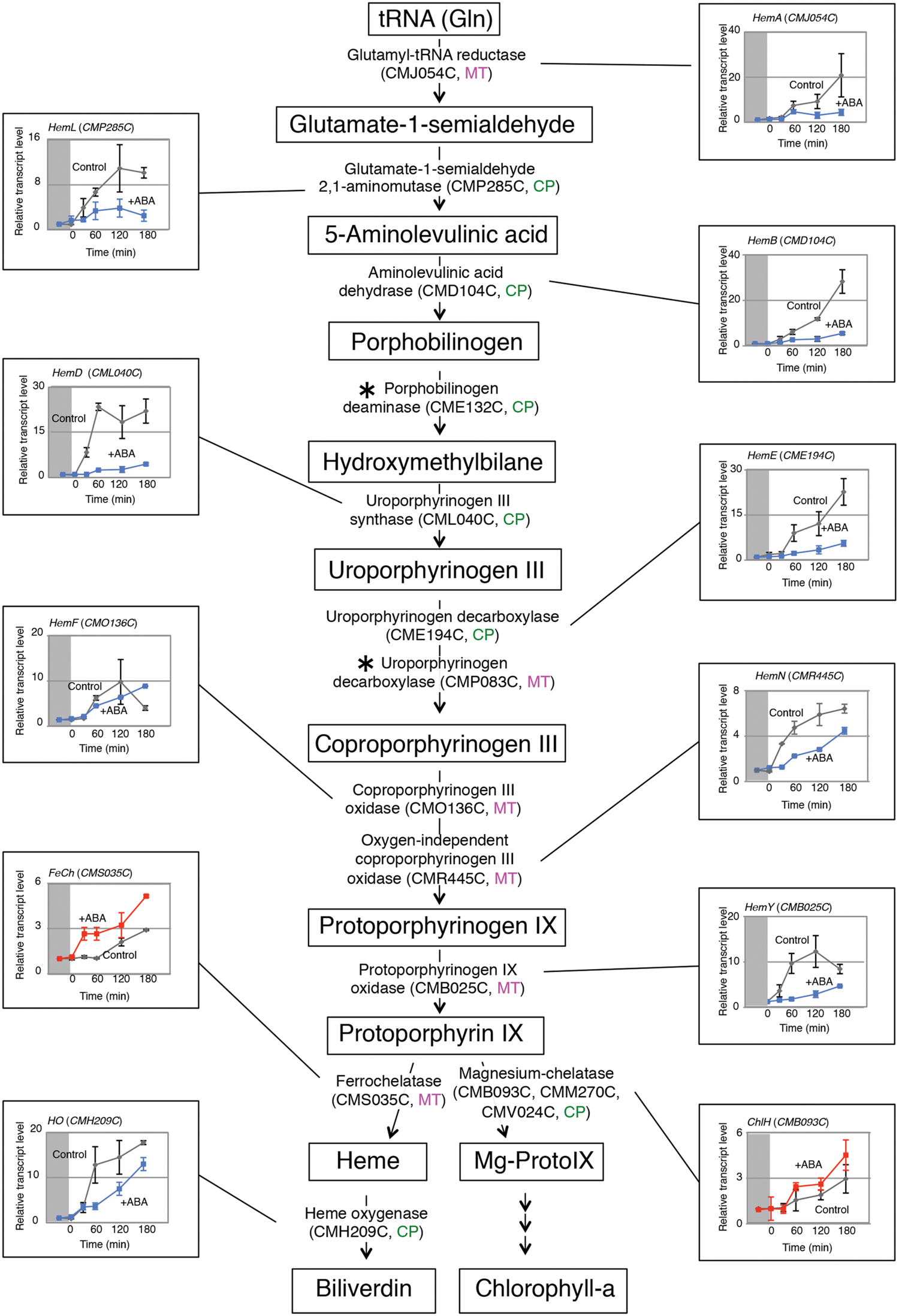
FIGURE 1. ABA affected the accumulation of heme-related gene transcripts. The tetrapyrrole biosynthetic pathway is schematically represented. C. merolae cells were dark adapted for 16 h, and the transcript accumulation for each gene was monitored after illumination by quantitative PCR in the absence or presence of ABA. Cells were sampled at the indicated times, and ABA (10 μM) was added 30 min before the light illumination. Data represent the average of three independent experiments (n = 3 SD). Boxes indicate tetrapyrrole intermediates, and the localization of each enzyme is indicated as in the mitochondrion (MT) or chloroplast (CP) as predicted (Watanabe et al., 2013). The light-dependent increase in the transcript levels were repressed (blue) or activated (red) by ABA addition. Asterisks indicate genes whose transcripts were not detected by the quantitative PCR analysis.
Accumulation of the Heme-Related Gene Transcripts Was Affected by ABA
We first incubated C. merolae cells under dark conditions, and the accumulations of heme-related gene transcripts were examined after illumination by quantitative PCR in the absence or the presence of ABA. We could not detect transcripts for two genes, CME132C and CMP083C, probably because of the limited transcript abundance. For the other genes examined, the detected transcripts increased in response to light while the relative increase and the time course profiles differed for each gene (Figure 1). However, the effect of ABA was divided into two categories: ABA repressively affected the light-dependent increase of most gene transcripts, while conversely activating the accumulation of FeCh and ChlH gene transcripts. As FeCh is the enzyme directly responsible for heme biosynthesis, the activation by ABA may result in the increased heme accumulation. HO is an enzyme involved in heme degradation (Shan et al., 2004; Shekhawat and Verma, 2010), and thus the decrease of HO transcripts in the presence of ABA is also consistent with heme accumulation. However, the relevance of the ChlH transcript increase, which encodes for a subunit of Mg-chelatase for the chlorophyll biosynthesis branch, is not clear. As we initially only examined the ChlH transcripts, we further checked for the expression of other Mg-chelatase subunit genes, ChlD and ChlI, by northern blot analysis, and confirmed that their transcripts’ accumulation was also activated by ABA (Supplementary Figure S1).
ProtoIX and Mg-ProtoIX Pools Were also Affected by ABA
If the transcripts levels explain the change in the heme content, then not only would the increase in heme be expected in the presence of ABA, but also the decrease in ProtoIX and the increase in Mg-ProtoIX. To test this hypothesis, we determined the cellular contents of heme, ProtoIX and Mg-ProtoIX, as well as chlorophyll-a as a control, under the same conditions as in Figure 1. As shown in Figure 2, the ABA-induced decrease in ProtoIX and increases in heme and Mg-ProtoIX were observed, which was consistent with the gene expression changes. The depletion of ProtoIX, the direct precursor of these compounds, explains the transient nature of the heme and Mg-ProtoIX increases. The chlorophyll-a content was not affected by ABA probably because the cellular content was rather abundant as compared with other tetrapyrrole intermediates and not easily affected by the change in the biosynthesis rate.
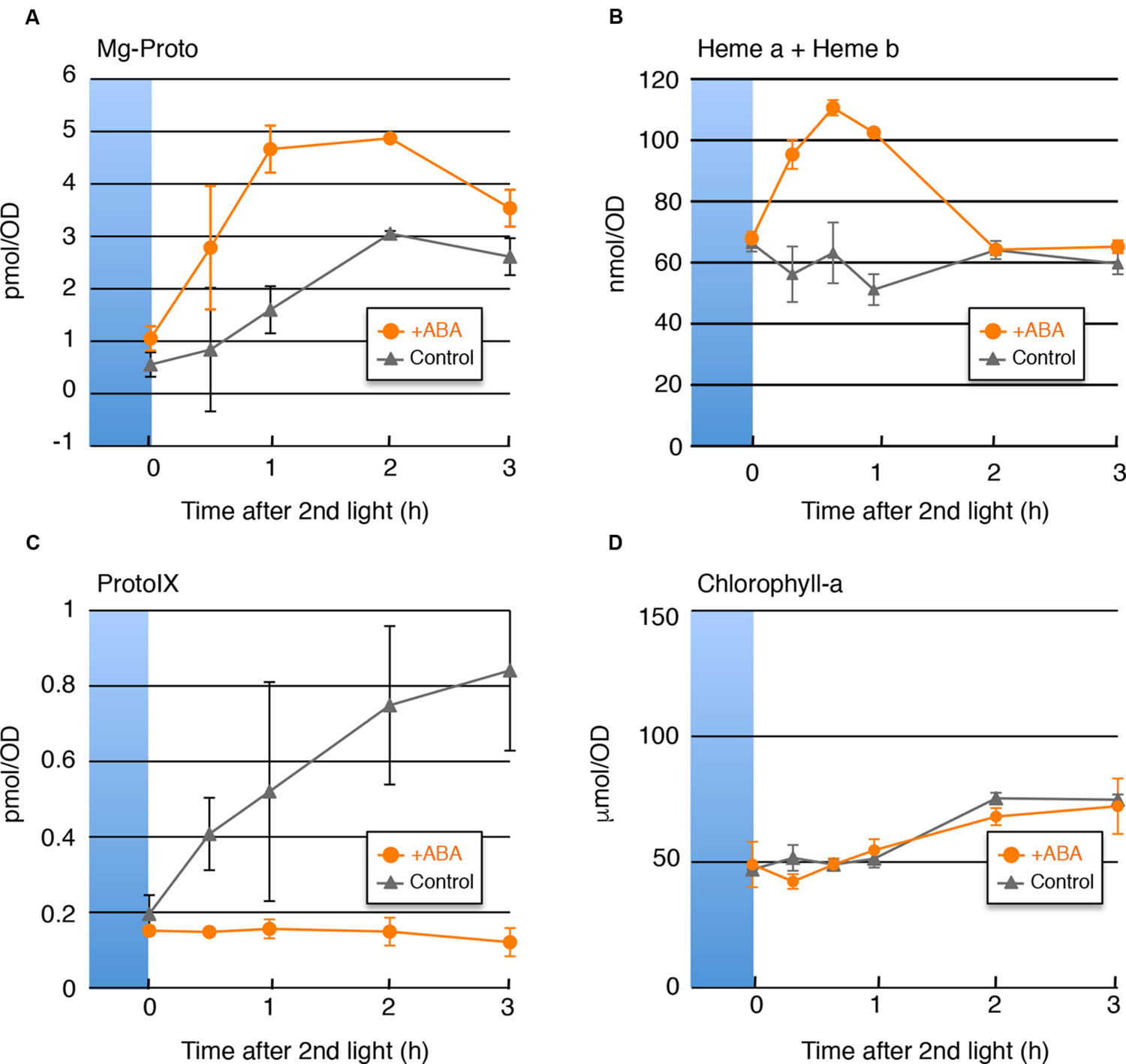
FIGURE 2. Cellular Contents of Mg-ProtoIX, Heme, ProtoIX, and Chlorophyll-a. Changes in accumulation of (A) Mg-ProtoIX, (C) ProtoIX, and, (D) Chlorophyll-a in the absence (gray) or presence (orange) of ABA are shown. The sampled conditions were as in Figure 1. (B) Changes in heme (heme a and heme b) accumulation (from Kobayashi et al., 2016) are also shown compared with previously obtained results. Each data point represents the average of three independent experiments (n = 3 SD). The x-axis indicates the amounts in 1 mL of culture medium (OD750 = 1).
TSPO May Scavenge the ABA-Induced Unbound Mg-ProtoIX
In a previous study, we showed that a transient increase in ProtoIX or Mg-ProtoIX induces nuclear DNA replication (Kobayashi et al., 2009). This could have indicated that ABA induces nuclear DNA replication, but this was not the case, and ABA inhibited cell-cycle initiation (Kobayashi et al., 2016). The total heme content was increased by ABA, but the unbound heme that is required for cell-cycle initiation was decreased through the induction of the heme scavenger protein TSPO (Kobayashi et al., 2016). TSPO has an affinity not only to heme but also for dicarboxylic porphyrins, including Mg-ProtoIX (Papadopoulos et al., 2006; Vanhee and Batoko, 2011). Thus, even when the total Mg-ProtoIX content increased, its unbound form, required for the signaling events, was likely quenched by TSPO and not available for cell-cycle activation.
Physiological Significance of the Heme Accumulation
It was previously shown that salt stress induced ABA accumulation in C. merolae (Kobayashi et al., 2016). While ABA is the phytohormone that transmits the stress signal to other cells in land plants, ABA in C. merolae is likely an intracellular signaling molecule since ABA rapidly loses its activity in the sulfur acidic culture medium (Kobayashi et al., 2016). As the signaling molecule, it is important to prevent the continuous ABA signaling, and thus the degradation enzyme ABA-8′-hydroxylase should be activated subsequently. In addition, salt stress induces generation of oxygen radicals, which needs to be scavenged by enzymes such as catalase and peroxidase. These enzymes require heme as the prosthetic group, and therefore it is reasonable that salt stress-induced ABA activates the heme accumulation. The excess unbound heme itself may cause the radical formation, which requires quenching by the ABA-induced TSPO.
Evolutionary Implications
The interrelationship among ABA, heme, and TSPO was first shown in Arabidopsis (Vanhee and Batoko, 2011; Vanhee et al., 2011), and a similar scheme was subsequently revealed in C. merolae (Kobayashi et al., 2016). Thus, this regulatory scheme was likely conserved during plant evolution. Presently, two issues remain to be addressed from mechanical and evolutionary points of view. First, although heme activates the cell-cycle G1/S progression in C. merolae, the presence of a similar mechanism has not been found in land plants. Future studies on the underlying molecular mechanism in C. merolae would clarify the significance for plant cell-cycle control. Second, an increase in the heme content was observed in C. merolae, as well as in Arabidopsis, but almost no information on the underlying mechanism in Arabidopsis has been provided. ABA-induced changes in the Arabidopsis transcriptome have been reported previously (Matsui et al., 2008), but it is difficult to determine the relevance to the heme content because the experimental time scale was rather different. Whether the expression of tetrapyrrole biosynthetic genes is regulated as in C. merolae needs to be clarified. The elucidation and comparison of these whole schemes in C. merolae and Arabidopsis would provide insights on the evolution of plant cell physiology.
Author Contributions
YK performed the experiments and contributed to the writing of the manuscript. KT designed and had overall responsibility for the study, and wrote the manuscript.
Funding
This study was supported by the Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI (Grant Nos. 21370015, 23120505, 2424806, and 15K14539 to KT, and 13274350 and 15621958 to YK).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01300
References
Barrero, J. M., Piqueras, P., González-Guzmán, M., Serrano, R., Rodríguez, P. L., Ponce, M. R., et al. (2005). A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 56, 2071–2083. doi: 10.1093/jxb/eri206
Cunningham, F. X. Jr., Lee, H., and Gantt, E. (2007). Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae. Eukaryot. Cell 6, 533–545. doi: 10.1128/EC.00265-06
Hartung, W. (2010). The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 37, 806–812. doi: 10.1071/FP10058
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Kobayashi, Y., Ando, H., Hanaoka, M., and Tanaka, K. (2016). Abscisic acid participates in the control of cell-cycle initiation through heme homeostasis in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol. 57, 953–960. doi: 10.1093/pcp/pcw054
Kobayashi, Y., Imamura, S., Hanaoka, S., and Tanaka, K. (2011). A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat. Cell Biol. 13, 483–487. doi: 10.1038/ncb2203
Kobayashi, Y., Kanesaki, Y., Tanaka, A., Kuroiwa, H., Kuroiwa, T., and Tanaka, K. (2009). Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. U.S.A. 106, 803–807. doi: 10.1073/pnas.0804270105
Kobayashi, Y., Ohnuma, M., Kuroiwa, T., Tanaka, K., and Hanaoka, M. (2010). The basics of cultivation and molecular genetic analysis of the unicellular red alga Cyanidioschyzon merolae. Endocytobiosis Cell Res. 20, 53–61.
Kuroiwa, T. (1998). The primitive red algae: Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20, 344–354. doi: 10.1002/(SICI)1521-1878(199804)20:4<344::AID-BIES11>3.0.CO;2-2
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 8, 1135–1149. doi: 10.1093/pcp/pcn101
Matsuzaki, M., Misumi, O., Shin-I, T., Maruyama, S., Takahara, M., Miyagishima, S., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
Nozaki, H., Takano, H., Misumi, O., Terasawa, K., Matsuzaki, M., Maruyama, S., et al. (2007). A 100%-complete sequence reveals unusually simple genomic features in the hot spring red alga Cyanidioschyzon merolae. BMC Biol. 5:28. doi: 10.1186/1741-7007-5-28
Papadopoulos, V., Baraldi, M., Guilarte, T. R., Knudsen, T. B., Lacapère, J. J., Lindemann, P., et al. (2006). Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 27, 402–409.
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Shan, Y., Lambrecht, R. W., Ghaziani, T., Donohue, S. E., and Bonkovsky, H. L. (2004). Role of Bach-1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAS. J. Biol. Chem. 279, 51769–51774. doi: 10.1074/jbc.M409463200
Shekhawat, G. S., and Verma, K. (2010). Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. J. Exp. Bot. 61, 2255–2270. doi: 10.1093/jxb/erq074
Skriver, K., and Mundy, J. (1990). Gene expression in response to abiscisic acid and osmotic stress. Plant Cell 2, 503–512. doi: 10.2307/3869112
Tuteja, N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428, 419–438. doi: 10.1016/S0076-6879(07)28024-3
Vanhee, C., and Batoko, H. (2011). Arabidopsis TSPO and porphyrins metabolism. Plant Signal. Behav. 6, 1383–1385. doi: 10.4161/psb.6.9.16477
Vanhee, C., Zapotoczny, G., Masquelier, D., Ghislain, M., and Batoko, H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805. doi: 10.1105/tpc.110.081570
Watanabe, S., Hanaoka, M., Ohba, Y., Ono, Y., Ohnuma, M., Yoshikawa, H., et al. (2013). Mitochondrial localization of ferrochelatase in a red alga Cyanidioschyzon merolae. Plant Cell Physiol. 54, 1289–1295. doi: 10.1093/pcp/pct077
Keywords: abscisic acid, Cyanidioschyzon merolae, heme, tetrapyrrole, transcriptional regulation, TSPO
Citation: Kobayashi Y and Tanaka K (2016) Transcriptional Regulation of Tetrapyrrole Biosynthetic Genes Explains Abscisic Acid-Induced Heme Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae. Front. Plant Sci. 7:1300. doi: 10.3389/fpls.2016.01300
Received: 28 May 2016; Accepted: 15 August 2016;
Published: 29 August 2016.
Edited by:
Henri Batoko, Université catholique de Louvain, BelgiumReviewed by:
Shan Lu, Nanjing University, ChinaMartha Gledhill, GEOMAR Helmholtz-Zentrum für Ozeanforschung, Germany
Copyright © 2016 Kobayashi and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kan Tanaka, a250YW5ha2FAcmVzLnRpdGVjaC5hYy5qcA==
 Yuki Kobayashi
Yuki Kobayashi Kan Tanaka
Kan Tanaka