- Institute of Crop Science, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
Eukaryotic translation initiation factor 3 (eIF3) is a large protein complex that participates in most translation initiation processes. While eIF3 has been well characterized, less is known about the roles of individual eIF3 subunits, particularly in plants. Here, we identified and characterized OseIF3e in rice (Oryza sativa L.). OseIF3e was constitutively expressed in various tissues, but most strongly in vigorously growing organs. Transgenic OseIF3e-silenced rice plants showed inhibited growth in seedling and vegetative stages. Repression of OseIF3e led to defects in pollen maturation but did not affect pollen mitosis. In rice, eIF3e interacted with eIF3 subunits b, d, e, f, h, and k, and with eIF6, forming homo- and heterodimers to initiate translation. Furthermore, OseIF3e was shown by yeast two-hybrid assay to specifically bind to inhibitors of cyclin-dependent kinases 1, 5, and 6. This interaction was mediated by the sequence of amino acid residues at positions 118–138, which included a conserved motif (IGPEQIETLYQFAKF). These results suggested although OseIF3e is not a “functional core” subunit of eIF3, it still plays crucial roles in rice growth and development, in combination with other factors. We proposed a pathway by which OseIF3e influence organ size and pollen maturation in rice, providing an opportunity to optimize plant architecture for crop breeding.
Introduction
In the process of translation initiation, eukaryotic initiation factors (eIFs) participate in the recruitment of initiator tRNA (Met-tRNAiMet) and mRNA to the 40S ribosomal subunit, as well in scanning for the AUG start codon (Browning et al., 2001; Kapp and Lorsch, 2004; Hinnebusch, 2006). Of the 12 known eIFs, eukaryotic translation initiation factor 3 (eIF3) is the largest and most complex. It is involved in assembling the eIF2-GTP-Met-tRNAiMet ternary complex and recruiting it to the 40S subunit, recruiting mRNA to the 43S pre-initiation complex, and scanning for and recognizing AUG start codons (Burks et al., 2001; Kawaguchi and Bailey-Serres, 2002; Siridechadilok et al., 2005; Hinnebusch, 2006).
Mammalian eIF3 contains 13 non-identical subunits designated eIF3a–m (Asano et al., 1997; Browning et al., 2001). In contrast, eIF in Saccharomyces cerevisiae comprises only six subunits (eIF3a, eIF3b, eIF3c, eIF3g, eIF3i, and eIF3j). Five of these (eIF3a, eIF3b, eIF3c, eIF3g, and eIF3i) are conserved in all eukaryotes (Phan et al., 1998; Browning et al., 2001). The non-conserved nature of subunit e indicates that it may not be essential for translation initiation (Asano et al., 1998; Burks et al., 2001; Zhou et al., 2005; Xia et al., 2010).
The eIF3e subunit, also known as Int6, is a common integration site for the mouse mammary tumor virus (MMTV) genome (Marchetti et al., 1995), which plays multiple roles in translation, as indicated by its association with the COP9 signalosome (CSN). The CSN is known to be involved in the regulation of proteolysis (Yahalom et al., 2001), control of 26S proteasome activity (Yen et al., 2003), and spindle organization (Yen and Chang, 2000; Morris and Jalinot, 2005). These findings suggest its potential as a regulatory subunit for gene translation (von Arnim and Chamovitz, 2003).
Few studies have examined the functions of the various eIF3 subunits in plants, some of those have been conducted in Arabidopsis (Kim et al., 2004; Yahalom et al., 2008; Xia et al., 2010). Two Arabidopsis thaliana eIF3e mutants are known (Yahalom et al., 2008). AteIF3e-Tp, which carries an insertion (T) 150 bp upstream of the first exon, leads to reduced fertility and reproductive defects (Yahalom et al., 2008). The mutant eIF3e-Tnull, containing an insertion (T) in the middle of the third exon, results in lethality of the male gametophyte. These results suggest that AteIF3e is necessary for male gametogenesis. Mutations in subunits eIF3f and eIF3h have also been characterized in Arabidopsis. A Ds (transposon element) insertion mutation in AteIF3f has been found to disrupt pollen germination and embryonic development (Xia et al., 2010). Plants homozygous for AteIF3h mutation exhibit pleiotropic growth defects throughout development, including low fertility, reduced stamen number, partial seed abortion, and inhibition of root hair formation (Kim et al., 2004, 2007). Subsequently, Zhou et al. (2014) described a mutation in AteIF3h led to expansion of shoot apical meristem (SAM) size accompanied by a failure to initiate new organs. Recently, the biological function of OseIF3f has been studied by Li et al. (2016). The OseIF3f-RNAi plants showed a higher percentage of arrested unicellular pollen at bicellular stage and aborted pollen at the tricellular stage, it is suggest that OseIF3f plays a vital role in microgametogenesis. Overall, even eIF3 subunits are not part of the functional core, it’s also play important roles in the growth and development of Arabidopsis and rice (Li et al., 2016).
Organ size is controlled by two fundamental processes: cell proliferation and cell expansion, which are strictly regulated by cyclin-dependent kinases (CDKs) together with their specific cyclin partners (Mizukami and Fischer, 2000; Sugimoto-Shirasu and Roberts, 2003). Other factors act as inhibitors of CDK (ICK) during plant development and in response to environmental changes (Sherr and Roberts, 1999). Studies in plants, particularly Arabidopsis and rice, have shown that overexpression of various ICK genes results in phenotypic effects similar to those produced by mutations of eIF3 subunits, including small organ sizes, reduced cell numbers, pollen sterility, and low seed setting (Wang et al., 2000; De Veylder et al., 2001; Zhou et al., 2002; Barroco et al., 2006; Bemis and Torii, 2007; Kang et al., 2007). For example, overexpression of either AtICK1 or AtICK2 in Arabidopsis induces cells to initiate endoreduplication earlier than normal, resulting in a higher ploidy numbers (Verkest et al., 2005; Weinl et al., 2005). Similarly, overexpression of rice OsiICK6 results in multiple phenotypic effects on plant growth, pollen viability, and seed setting (Yang et al., 2011).
A previous study revealed that inhibition of Osj10gBTF3 (Oryza sativa BTF3) results in dramatic plant miniaturization. Furthermore, pollen is completely sterile, an effect correlated with the altered expression of two Rf (fertility restorer)-like genes encoding pentatricopeptide repeat-containing proteins (OsPPRs); two translation initiation factors, OseIF3e and OseIF3h; and the heat shock protein OsHSP82 (Wang et al., 2012). The present study sought to confirm the functions of OseIF3e in plant growth and development. Specifically, protein–protein interactions demonstrated that OseIF3e plays important roles in rice growth and pollen development and interacts with eIF3 subunits OseIF3b, OseIF3d, OseIF3e, OseIF3f, OseIF3h, OseIF3k, as well as eIF6 and ICKs. Taken together, these results help to unravel a possible pathway for OseIF3e involvement in organ growth and pollen development in rice.
Materials and Methods
Plant Materials, Growth Conditions, and Stress Treatments
Rice (O. sativa L ssp japonica cv Nipponbare) was used in this study for various experiments. All plants were grown on the experimental field of Zhejiang University in Hangzhou (30°16′N, temperate climate, China) or Sanya (19°2′N, tropical climate, China) during the natural growing season. Rice seedling plant were grown at 28°C with 16 h light/8 h dark cycle, 75% relative humidity in greenhouse. For expression studies of OseIF3e in response to various treatments, 2-week-old seedlings were transferred to Yoshida solution (Yoshida et al., 1976) supplemented with 200 mM NaCl, 10 μM ABA, 100 μM GA. Seedlings grown in the same liquid medium without any supplementary component were used as controls. For cold stress, 4-week-old seed-derived seedlings were transferred from semi-solid 1/2MS medium (Murashige and Skoog, 1962) to Yoshida solution, were exposed in 4°C for 24 h.
Vector Construction and Rice Transformation
To generate OseIF3e knock-down transgenic lines, the OseIF3e cDNA fragments of 325 bp (from 174 to 499 bp of OseIF3e, Supplementary Figure S3) was inserted into pTCK303 vector (Wang et al., 2004) to produce RNAi repression vectors. The resultant vector was introduced into Agrobacterium tumefaciens strain EHA105, which was used to infect rice embryogenic calli from Nipponbare. Transgenic plants were screened by PCR amplification with hygromycin B phosphotransferase gene (Hpt). All primers used in this study are listed in Supplementary Table S2.
Phenotypic Analysis of Transgenic Plants
The evaluation of phenotypic traits of three independent transformants OseIF3eRi-2, OseIF3eRi-4, OseIF3eRi-7 were performed in the T1–T3 generation. Seeds of OseIF3eRi and wild-type (WT) plants were collected and germinated by soaking in water for 2 days at 37°C. Germinating seeds were sown in experimental field as described above during the natural growing season at five-leaf and maturity stage, the phenotypic characteristics were measured and photographed, including plant height, tiller number, the internode length, panicle length, the spikelet number, the grain length and width. The data were analyzed by ANOVA, and mean values were separated by least significant difference at the 5 and 1% probability level using Statistical software (Sigmaplot10.0.).
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time RT-PCR
Total RNAs were extracted from different tissues of the WT and OseIF3eRi plant using TRIzol reagent (Invitrogen). Reverse transcription (RT) was performed using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time RT-PCR (qRT-PCR) analysis was conducted with the Lightcycler 480 machine using SYBR Green I (TAKARA). UBIQUITIN (Os03g0234200) mRNA was used as an internal control. The specific primers for qRT-PCR are listed in Supplementary Table S2.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the Matchmaker Two-Hybrid System (Clontech1). The full-length CDS and different truncations of OseIF3e, OsICKs, and other subunits of OseIF3, OseIF1, OseIF2, OseIF4, OseIF5, and OseIF6 were amplified by PCR using the primers listed in Supplementary Table S2. The fragments were cloned into the pGBKT7 or pGADT7 vector. Then co-transformed into yeast strain AH109 first selected on SD/-Leu/-Trp (DDO) plates at 30°C for 3 days, signal colony from yeast transformants including different pair of constructs were diluted in 0.9% NaCl, and a 1/10th dilution was spotted on SD/-Ade/-His/-Leu/-Trp (QDO) plates and incubate at 30°C for 3 days. Yeast cells co-transformed with pGBKT7-53 and pGADT7-T were used as the positive control, pGBKT7-Lam and pGADT7T were used as the negative control.
I2-KI and DAPI Staining
To analyze pollen viability, mature anthers were incubated with 1% (w/v) I2-KI staining, with three biological repetitions. The stained pollen grains were observed and recorded using a Leica DMIRB fluorescence microscope. For 4′,6-diamidino-2-phenylindole (DAPI) staining, pollen grains were fixed in DAPI staining solution (0.1 M sodium phosphate, pH 7.0, 1 mM EDTA, 0.1% Triton X-100 and 0.25 mg/ml DAPI) for 1 h at room temperature. Photography was performed using Leica DMIRB fluorescence microscope under UV light.
Bioinformatics Analysis
To investigate gene’s structure, the exon/intron boundary were predicted with RGAP2, and protein domains were predicted by PROSITE3, PLACE4 was used for analysis cis-elements of OseIF3e promoter region. The primers used in this study were designed by primer primer5.0 and the BLAST5 was used for sequence alignment. Alignment was performed using CLUSTALX1.8 (Thompson et al., 1994) with default settings. All amino acid sequences were obtained from the NCBI database6. Phylogenetic analysis was conducted using MEGA5 via the neighbor-joining method (Kolaczkowski and Thornton, 2004). Motif 1,2 in OsICK1,-5,-6 and consensus sequence of the conserved motif in eIF3e from different species using were identified by the MEME/MAST program7 (Bailey and Elkan, 1994; Bailey and Gribskov, 1998).
Results
Characteristics and Expression Patterns of OseIF3e
In rice, OseIF3e was originally identified via its interaction with the basal transcription factor Osj10gBTF3, inhibition of which results in plant miniaturization and pollen abortion (Wang et al., 2012). Previous studies have characterized a multitude of eIF3e homologs from other species. We constructed a phylogenetic tree of OseIF3e according to sequence homology. This revealed that OseIF3e is most closely related to ZmeIF3e, while homologs in animals and fungi form separate clades (Figure 1a).
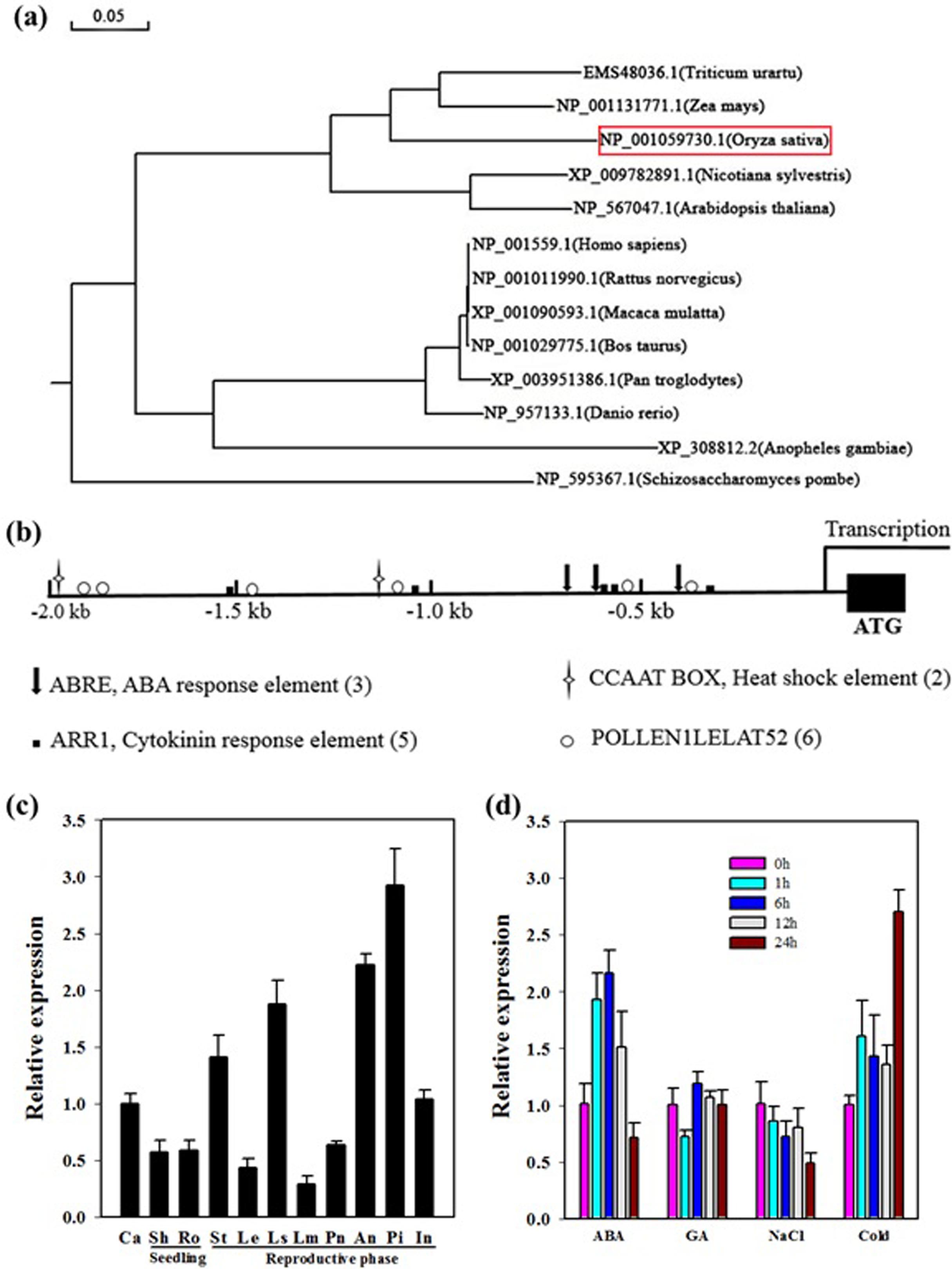
FIGURE 1. Characteristics and expression profile of OseIF3e. (a) Phylogenetic tree constructed by MEGA5 software using the neighbor-joining method. (b) Cis-element analysis of the OseIF3e promoter region. (c) Expression of OseIF3e in various organs. Ca, callus; Sh, shoot; Ro, root; St, stem; Le, leaf; Ls, sheath; Lm, lemma; Pa, palea; An, anther; Pi, pistil; In, internode. (d) Expression of OseIF3e under hormone and stress treatments, including ABA, GA, salt, and cold (0, 1, 6, 12, 24 h) treatments in seedlings.
To investigate the expression profile of OseIF3e, we searched the CREP (Collection of Rice Expression Profiles) database8, which collects genome-wide expression data over the life cycles of two rice varieties (Wang et al., 2010). This revealed OseIF3e to be constitutively expressed in all tissues and organs, with particularly high expression levels in young and developing tissues (Supplementary Table S1 and Figure S1). We then performed qRT-PCR to confirm OseIF3e expression patterns in the following tissues: callus (Ca), shoot (Sh), root (Ro), stem (St), leaf (Le), sheath (Ls), lemma (Lm), palea (Pa), anther (An), pistil (Pi), and internode (In). The results were consistent with the CREP data, with higher OseIF3e expression occurring in vigorously growing tissues (Figure 1c). These results implicate OseIF3e in both vegetative growth and reproductive development in rice.
Next, we analyzed the 2.0-kb promoter region of OseIF3e and found several types of cis-acting elements, including several hormone response elements. These included three ABREs, five ARR1s (cytokinin response elements), and two heat shock elements (Figure 1b). Accordingly, we performed qRT-PCR to determine OseIF3e expression levels under different hormonal and abiotic stress treatments in seedlings (Figure 1d). The results showed OseIF3e to be induced by cold, but repressed by salt treatment. For the ABA treatment, OseIF3e transcripts increased within the first 6 h, but then decreased. GA treatment only slightly affected OseIF3e expression.
Transgenic OseIF3e-Silenced Rice Plants Show Inhibited Growth in Seedling and Vegetative Stages
To determine the function of OseIF3e in rice, we obtained nine OseIF3eRi knockdown lines, in which RNAi reduced the expression of OseIF3e (Figures 2a,c). Eight transgenic plants had significant decreases in OseIF3e compared with WT plants. Then three independent transformants OseIF3eRi-2, OseIF3eRi-4, OseIF3eRi-7 were used for further experiments. Within the first 10 days after germination, these lines did not differ significantly from the WT in terms of seed germination and phenotypic expression (Figure 2c). OseIF3eRi lines gradually became slower than that of the WT (Figure 2e), leading to shorter shoots (Figure 2b) and slightly shorter roots (Figure 2d) in OseIF3e plants. When the plants entered the vegetative period, other organs in the OseIF3eRi plants were reduced, e.g., the length and width of the first flag leaf were shorter than in the WT (Supplementary Figure S2). No differences were observed in tiller number. Overall, before maturity, OseIF3eRi transgenic plants differed most markedly from the WT in seedling and flag leaf phenotypes.
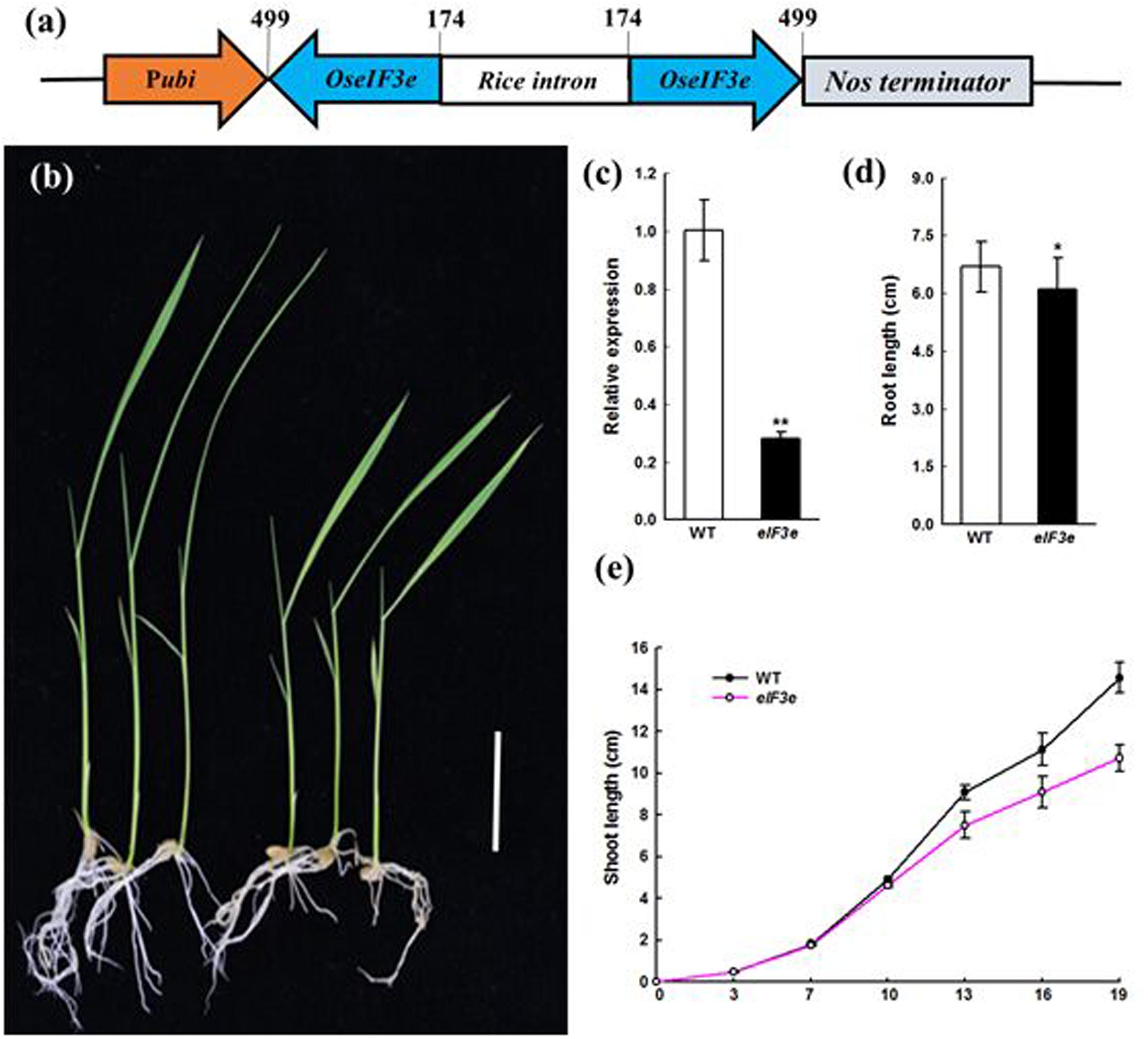
FIGURE 2. Phenotype of the eIF3e knockdown seedling. (a) Schematic representation of the OseIF3e RNAi vector construction. The numbers indicate the nucleotide position in the OseIF3e CDS sequence. Pubi, maize ubiquitin promoter. (b) Wild-type (WT, left) and OseIF3eRi plants (right) at 15 days after sowing. Scale bars: 3 cm. (c) Relative expression level of OseIF3e in WT and OseIF3eRi plants. Two-week-old seedlings were used for the analysis. Rice ubiquitin (OsUBQ) was used as the reference. (d) Two-week-old seedlings root length of WT and OseIF3eRi plants. Values are expressed as means ± SD. ∗P < 0.05; ∗∗P < 0.01 compared with the WT plant using Student’s t-test. (e) Height of WT and OseIF3eRi plants after sowing.
These phenotypic differences between WT and OseIF3eRi plants remained stable in generations T0–T3, confirming that they were indeed due to the suppression of OseIF3e (Figure 2c). The observation that OseIF3e suppression led to stunted rice suggests that OseIF3e is critical to the growth of seedling and vegetative-stage plants.
Aberrant Panicle Phenotype and Low Plant Biomass in OseIF3eRi Lines
OseIF3eRi plants remained notably shorter than WT plants due to reduced internode lengths (Figures 3a,b,m). At the mature stage, the panicle axis of OseIF3eRi was notably shorter than in WT (Figure 3d). We measured the lengths of panicle and primary branches and numbers of primary branches and spikelets. The OseIF3eRi plants displayed shorter panicles and reduced spikelet numbers (Figures 3e–i). Moreover, the grains of OseIF3eRi lines appeared thinner and shorter than WT grains, resulting in lower 100-grain weights (Figures 3c,j–l). These results demonstrated that OseIF3e influences not only panicle size and shape, but also overall plant biomass.
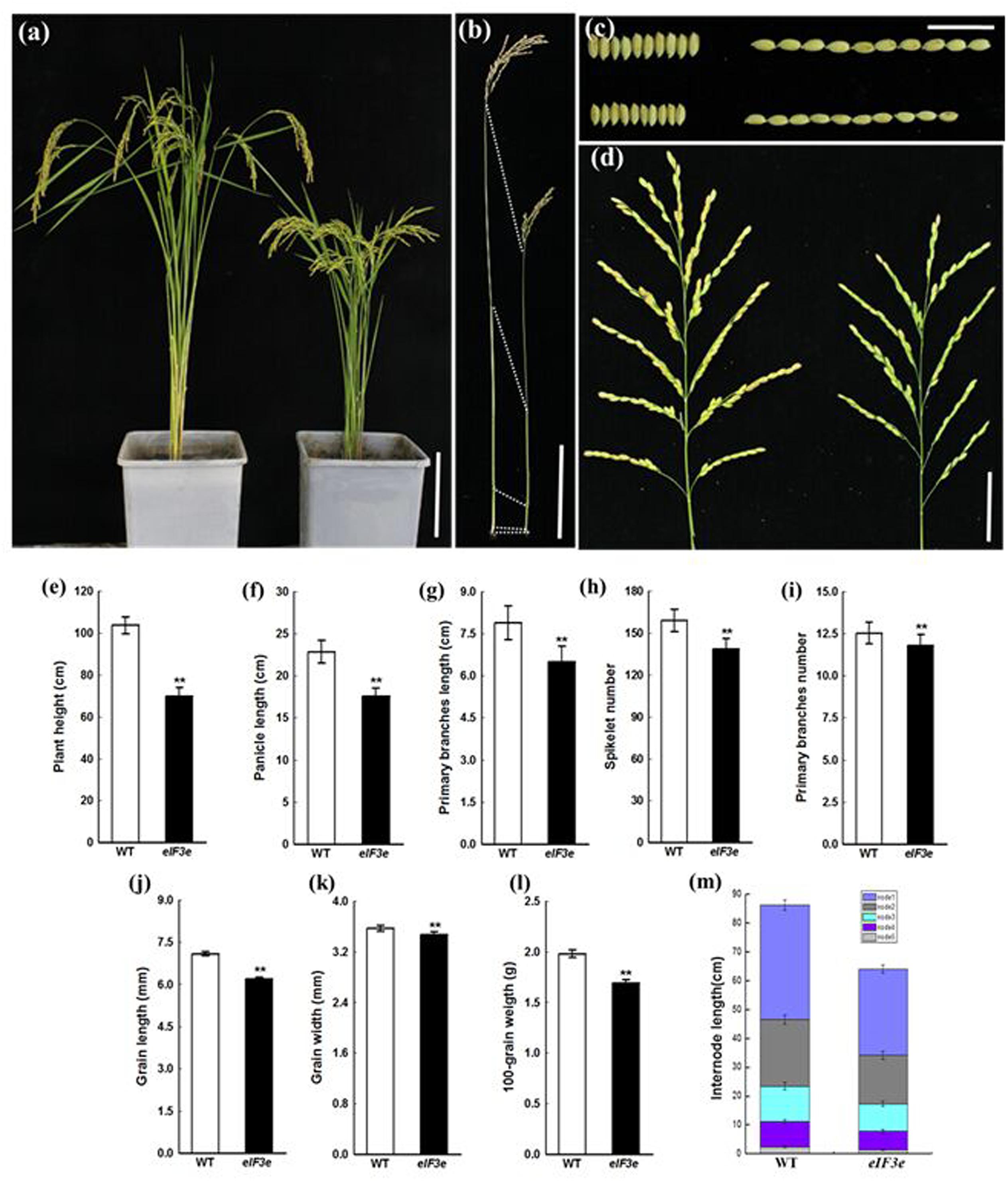
FIGURE 3. Phenotype and statistical analysis of panicle and seed of the OseIF3eRi plant at the maturity stage. (a) Five-month-old wild-type (WT, left) and OseIF3eRi (right) plants. Scale bars: 20 cm. (b) Comparison of the internode in WT (left) and OseIF3eRi (right) plants. Scale bars: 4 cm. (c) Seed width and seed length in WT (upper) and OseIF3eRi (lower) plants. Scale bars: 2 cm. (d) Panicle branching in WT (left) and OseIF3eRi (right) plants. Scale bars: 5 cm. (e) Statistical analysis of plant height in WT and OseIF3eRi plants. (f–i) Statistical analysis of panicle types in WT and OseIF3eRi plants. (j–l) Statistical analysis of seed size in WT and OseIF3eRi plants. (m) Comparison of internode length of the main culm in WT and OseIF3eRi plants. Values are expressed as means ± SD. ∗∗P < 0.01 compared with the WT using Student’s t-test.
Repression of OseIF3e Affects Pollen Maturation
OseIF3eRi plants exhibited a high rate of sterility in generations T0–T3, which were grown in different locations (Figures 4a,l). Seed setting rate of OseIF3eRi plants ranged from 20.2 to 42.8%, compared to from 88.9 to 94.6% in WT plants (Figure 4l). In addition, OseIF3eRi plants exhibited abnormal anthers (Figures 4c,h). We examined the pollen viability of WT and OseIF3eRi plants with I2-KI staining. Stained WT pollen presented full and black, while OseIF3eRi pollen appeared light brown (Figures 4b,d,e,g,i,j). To visualize possible mitotic defects, pollen grains were stained with DAPI. DAPI staining revealed two brightly stained sperm nuclei and a large, diffusely stained vegetative cell nucleus in both OseIF3eRi and WT pollen grains (Figures 4f,k, arrowhead). Therefore, while repression of OseIF3e led to defects in pollen maturation, it did not appear to affect pollen mitosis.
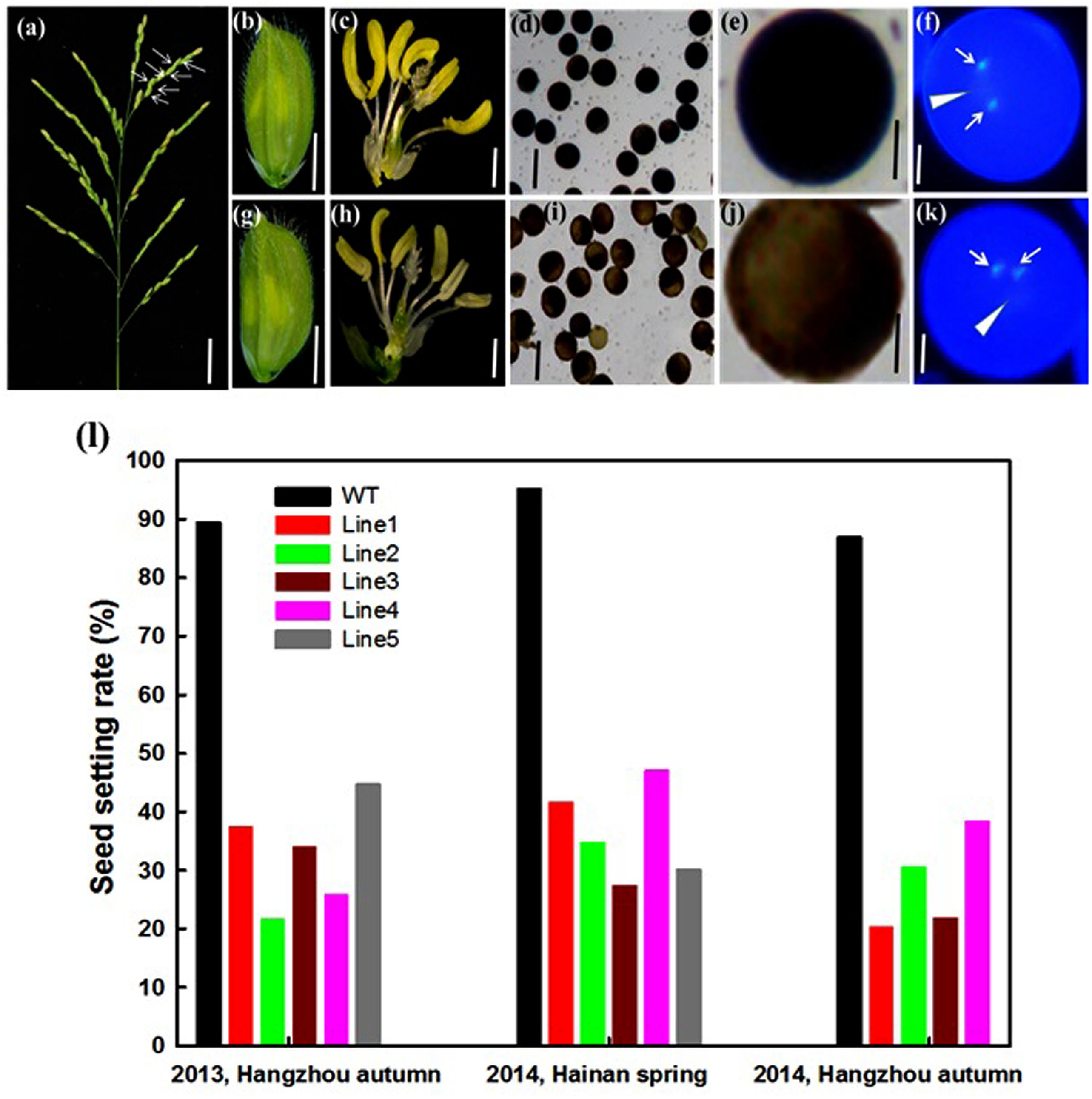
FIGURE 4. Analysis of the sterile phenotype in the OseIF3e RNAi transgenic plants. (a) Panicle of OseIF3eRi plants. The white arrow represents empty seeds. Scale bars: 3 cm. (b,c,g,h) Wild-type spikelet (b,c) and OseIF3eRi spikelet (g,h). The lemma and palea are removed in (c,h). (d,i) I2-KI staining showing pollen viability in the control plant (d) and OseIF3eRi plant (i). (e,j) The higher magnification of I2-KI staining showing pollen viability in the control plant (e) and OseIF3eRi plant (j). (f,k) DAPI staining showing three nuclei of pollen grain in the control plant (f) and OseIF3eRi plant (k). Arrowheads indicate the vegetative nucleus, and arrows indicate sperm-cell nuclei in the pollen of the control plant (f) and abnormal pollen of OseIF3eRi plant (k). (l) Seed setting analysis of OseIF3eRi transgenic plants generated in 2013 autumn, and 2014 spring and autumn, respectively. Scale bars: 2 mm in (b,g), 1 mm in (c,h), 100 μm in (d,i), 20 μm in (e,j), and 10 μm in (f,k).
OseIF3eRi Seedlings Exhibited a Sugar-Sensitive Phenotype
In Arabidopsis, mutation of either of two eIF3 components, eIF3f and eIF3h, produced a biphasic response to exogenous sugars (Kim et al., 2004; Xia et al., 2010). The present study examined the role of the OseIF3e subunit in response to sugar, using the OseIF3eRi knockdown line. WT and OseIF3eRi seeds were germinated on 1/2MS agar plates containing either no sugar (control) or one of the following: 2% (w/v) sucrose, 2% (w/v) mannitol, 2% (w/v) maltose, 1% (w/v) glucose. The results showed nearly no differences in responses of WT seedlings to sugar treatments. However, OseIF3eRi seedlings exhibited stunted growth in 2% (w/v) mannitol, compared with the other sugar treatments (Figures 5a,b). Subsequently, WT and OseIF3eRi seedlings were grown on 1/2MS agar plates containing 0, 1, 2, 3, or 5% mannitol (w/v). As mannitol concentration increased, the growth of OseIF3eRi seedlings appeared more notably stunted, compared to WT (Figures 5c,d). In summary, repression of OseIF3e caused rice seedlings to become sensitive to exogenous mannitol, resulting in stunted growth of the transgenic plants.
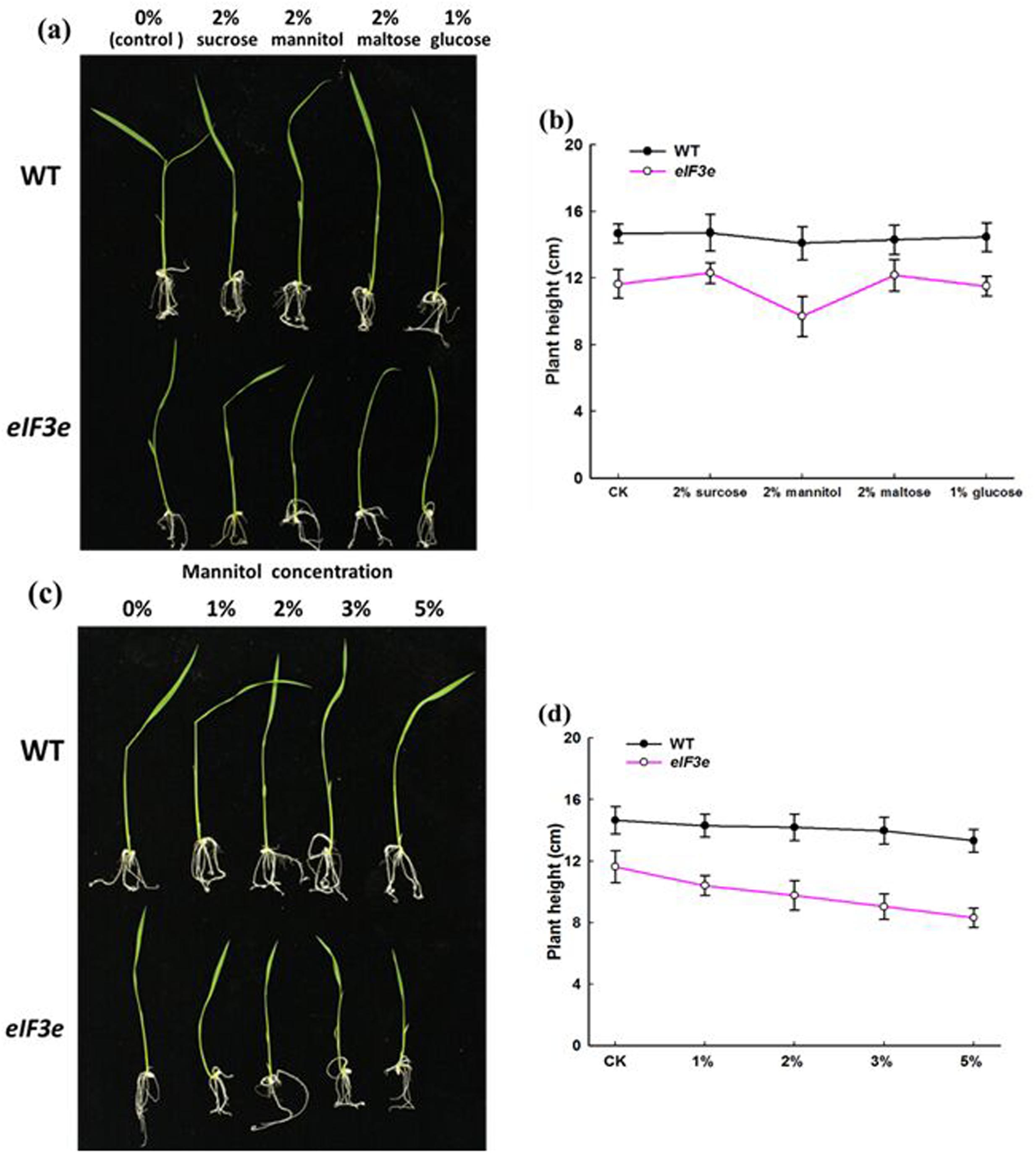
FIGURE 5. OseIF3eRi seedlings are sensitive to exogenous sugars. (a) Wild-type (WT) and OseIF3eRi seedlings cultured on 1/2 MS medium containing various sugars. (b) Comparison of plant height in WT and OseIF3eRi seedlings cultured on 1/2 MS medium containing various sugars. (c) WT and OseIF3eRi seedlings cultured on 1/2MS medium containing various concentrations of mannitol. (d) Comparison of plant height in WT and OseIF3eRi seedlings cultured on 1/2 MS medium containing various concentrations of mannitol.
Yeast Two-Hybrid Assays Reveal that eIF3e Interacts with itself, Other Subunits of eIF3, and eIF6
The components of eIF3 have been identified in many species. Previous studies show that the different subunits of eIF3 form complexes, which allows them to participate in gene regulation (Kim et al., 2004; Xia et al., 2010). In Arabidopsis, eIF3h interacts directly with the eIF3a, eIF3b, eIF3c, and eIF3e subunits (Kim et al., 2004). In addition, the eIF3f subunit has been confirmed to interact with eIF3e and eIF3h (Xia et al., 2010). We performed yeast two-hybrid assays, demonstrating that in rice, eIF3e is able to interact with itself, with other subunits of eIF3 (b, d, f, h, and k), and with eIF6, but does not interact with eIF1, eIF2;1, eIF4, or eIF5 (Figure 6). These protein–protein interactions suggest that the subunits of eIF3 and eIF6 form homo- and heterodimers, in different combinations, to initiate translation and regulate target gene expression in rice.
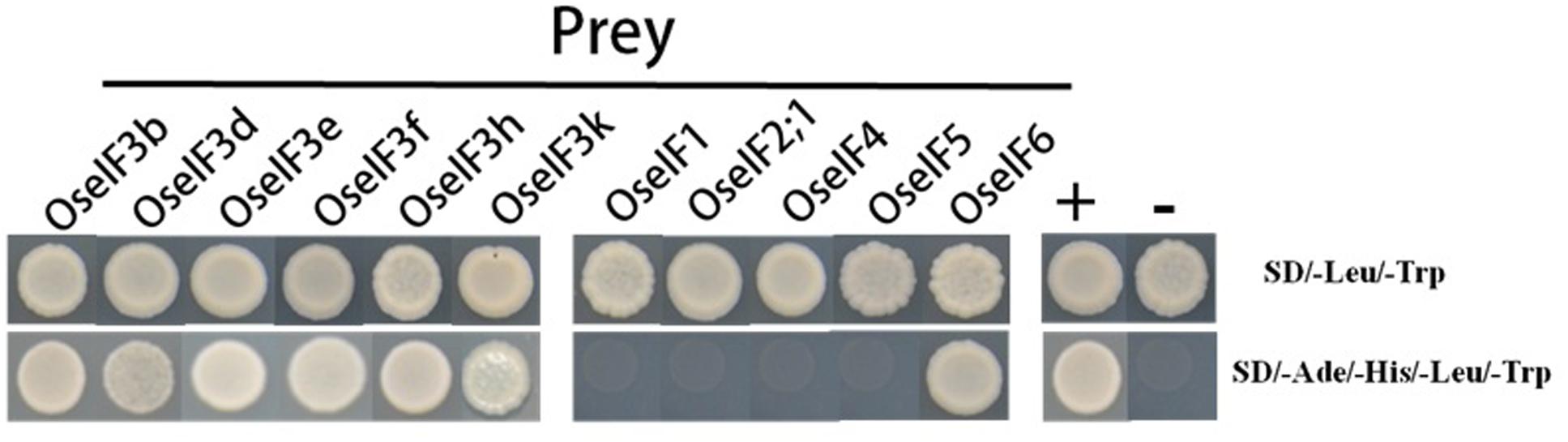
FIGURE 6. OseIF3e interacts with itself, other subunits of OseIF3, and OseIF6 protein in Y2H assays. The bait (BD) vector contained full-length OseIF3e, the prey (AD) vector contained other subunits of OseIF3 and OseIF1,-2,-4,-5,-6, yeast strains were cultured on the QDO (-Ade/-His/-Leu/-Trp) selection medium. p53:T7 (Clontech) and pLam:T7 (Clontech) are positive and negative controls.
Targeting of the OsICK Family by the eIF3 Complex is Mediated by Amino Acids 118–138 of eIF3e
To determine whether the OsICK gene family is regulated by the eIF3 complex, the interaction between OseIF3e and OsICKs was investigates by yeast two-hybrid assay. Moreover, considering that the OseIF3e protein possesses relevant domains in its N- and C-terminal regions, we used fragments of OseIF3e encoding the eIF3_N domain (OseIF3eΔPCI), the PCI domain (OseIF3eΔeIF3_N), and the full-length cDNA as baits. The assay revealed that OsICK1, OsICK5, and OsICK6 interacted with OseIF3e and OseIF3eΔPCI, both of which included the eIF3_N domain, while no interaction between OseIF3eΔeIF3_N and any OsICK was observed (Figure 7a). These results suggest that the OsICK family is a direct target of the eIF3 complex and that this interaction is mediated by the eIF3_N domain of OseIF3e.
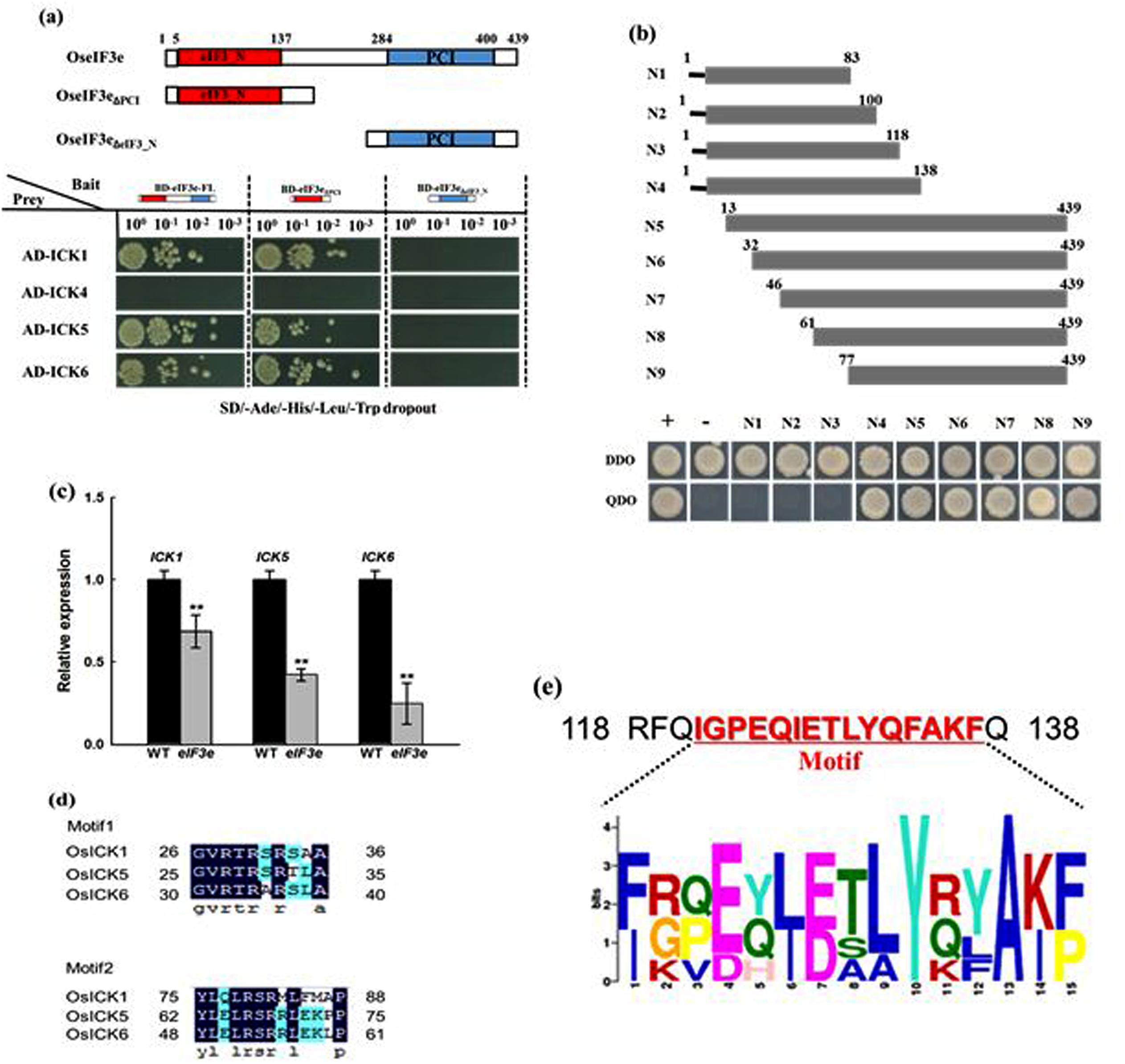
FIGURE 7. Interaction of OseIF3e with OsICKs and identification of the binding region responsible for the interaction. (a) Interaction of OseIF3e with OsICKs in a yeast two-hybrid assay. A schematic diagram of OseIF3e and the truncations. The bait (BD) vector contained full-length OseIF3e, OseIF3eΔPCI, or OseIF3eΔeIF3e_N, the prey (AD) vector contained ICKs (ICK1,-4,-5,-6), yeast strains were cultured on the QDO (-Ade/-His/-Leu/-Trp) selection medium. p53:T7 (Clontech) and pLam:T7 (Clontech) are positive and negative controls. (b) The interaction between OsICK5 and a series of truncated version of eIF3e_N domain (N1–N9) in yeast. The bottom panels show the growth of the transformed yeast cells on DDO (-Trp/-Leu) or QDO (-Ade/-His/-Leu/-Trp) selection medium. (c) Quantitative RT-PCR analysis of gene expression of OsICK1, OsICK5, and OsICK6. (d) Sequences of conserved motifs in OsICK1, OsICK5, and OsICK6 proteins. (e) Consensus sequence of the conserved motif (amino acids 120–137 in rice) in eIF3e from different species. ∗∗indicates P values generated by student’s t-test < 0.01.
We then identified conserved sequence motifs in OsICK1, OsICK5, and OsICK6. In these three OsICKs, two consensus sequence motifs were identified by the MEME/MAST program (Bailey and Elkan, 1994; Bailey and Gribskov, 1998; Torres Acosta et al., 2011; Figure 7d). Examination of OsICK1, OsICK5, and OsICK6 gene expression in WT and OseIF3eRi lines showed that all three genes experienced various degrees of reduction in OseIF3eRi plants, compared with WT plants (Figure 7c).
In order to further characterize the OseIF3e N-terminal motif responsible for its interaction with OsICKs, we cloned fragments encoding different truncations of the OseIF3e N terminus as baits and determined their interaction with OsICK5. As shown in Figure 7b, no interaction was detected if the cloned fragment lacked amino acids 118–138 (N4), suggesting that these 20 amino acids which included a conserved motif (IGPEQIETLYQFAKF, Figure 7e) are necessary for the interaction to occur.
Discussion
OseIF3e Is Involved in the Regulation of Organ Size and Pollen Maturation
Plant organ size is controlled by two successive, overlapping types of cell growth: cell proliferation and cell expansion (Mizukami and Fischer, 2000; Busov et al., 2008). To date, several positive and negative factors affecting organ size have been identified in Arabidopsis and rice. Positive factors include AINTEGUMENTA (ANT; Krizek, 1999; Mizukami and Fischer, 2000), ARGOS (Hu et al., 2003), KLUH/CYP78A5 (Anastasiou et al., 2007), ORGAN SIZE RELATED1 (Feng et al., 2011), and XIAO (Jiang et al., 2012). Negative regulators include BIG BROTHER (Disch et al., 2006), PEAPOD1/2 (White, 2006), DA1 (Li et al., 2008), and MED25 (Xu and Li, 2011). However, the pathways involved in organ size regulation are not yet well understood.
The present study identified a translation initiation factor in rice, OseIF3e, which we found to influence organ size and pollen maturation. During both the vegetative and reproductive stages, all organs of OseIF3eRi plants exhibited significant reductions in size, compared with WT plants. In addition, repression of OseIF3e led to defects in pollen maturation but did not affect pollen mitosis. These results implicate eIF3e as an essential gene in rice growth and development.
The eIF3e gene was first described as Int-6, a common integration site for the MMTV genome (Marchetti et al., 1995). In plants, eIF3e was originally identified as co-purifying with the CSN (Karniol et al., 1998), and its function was verified in Arabidopsis. Targeted expression of AteIF3e results in pleiotropic effects on development, including defects in seedling, vegetative, and floral development (Yahalom et al., 2008). In this respect, our results are consistent with those reported for Arabidopsis. AteIF3f and AteIF3h mutants also exhibit severe defects in plant growth and development (Kim et al., 2004; Yahalom et al., 2008; Xia et al., 2010). These phenotypes are similar to those of the OseIF3hRi plants examined in our study. Besides, repression of OseIF3e led to rice seedlings to become sensitive to mannitol, resulting in stunted growth of OseIF3eRi knockdown lines. These results imply that subunits of eIF3, even though not part of the functional core, are crucial for not only normal plant growth and development, but also abiotic stress response.
The Activity of the OseIF3 Complex may be Regulated by OseIF3e in Combination with OseIF3 Subunits b, d, e, f, h, and k, as well as eIF6
Research in plants has shown that different subunits of eIF3 initiate translation and regulate gene expression through the formation of homo- and heterodimers (Karniol et al., 1998; Yahalom et al., 2001; Kim et al., 2004; Huang et al., 2005). For example, in Arabidopsis, the eIF3h subunit interacts directly with subunits a, b, c, and e (Kim et al., 2004). Similarly, AteIF3f interacts with both the e and h subunits (Xia et al., 2010).
The present study revealed in vivo protein–protein interactions between the OseIF3e subunit and subunits b, d, e, f, h, and k, as well as eIF6. Although it is not part of the highly conserved functional of eIF3, OseIF3e also plays a role in translation processes in combination with other subunits or eIFs. Karniol et al. (1998) first associated subunit eIF3e with the CSN in plants. Further examination by Kim et al. (2004) revealed that AteIF3e interacts directly with AteIF3h. Yahalom et al. (2008) and Xia et al. (2010) subsequently demonstrated that AteIF3e exhibits subcellular co-localization with CSN and is negatively regulated by it. Binding between multiple subunits of eIF3 in Arabidopsis suggests the possibility that its activity is regulated by these interactions. However, interactions between eIF3e and other proteins in plants are still largely unknown.
eIF6 was initially identified as a wheat protein capable of interaction with the 60S ribosome (Russell and Spremulli, 1980). In yeast, disruption of eIF6 results in the abnormal processing of ribosomal RNA precursors and a reduction in abundance of the 60S subunit (Wood et al., 1999; Basu et al., 2001). In Arabidopsis, loss of the AteIF6;1 gene results in embryonic lethality (Kato et al., 2010), suggesting that eIF6 is an essential component of ribosome biogenesis (Si and Maitra, 1999).
Targeting of OsICKs by the OseIF3 Complex, Mediated by Amino Acids 118–138, Is Responsible for Plant Growth and Development in Rice
We used the proteins OseIF3e, OseIF3eΔPCI (which included the eIF3_N domain), and OseIF3eΔeIF3_N (which included the PCI domain) as baits in yeast two-hybrid assays. We thereby determined that three members of the OsICK family (OsICK1, OsICK5, and OsICK6) interacted with OseIF3e and OseIF3eΔPCI, but not OseIF3eΔeIF3_N. This demonstrated that the interactions were mediated by the eIF3_N domain, as the deletion of this region resulted in the lack of interaction. Interestingly, the interaction between eIF3 and CDK was confirmed during apoptosis (Shi et al., 2003), while ICK as inhibitor of CDK which also interact with eIF3, suggesting that eIF3 play a vital role in processes which CDK and ICK participate in, such as cell cycle and cell proliferation.
The ICK family of CDK inhibitors have been identified as key genes in plant growth and development. Seven ICK genes, along with a pseudogene, have been reported in rice and several studies have reported notable effects on plant growth and development due to their over-expression (Wang et al., 2000; Barroco et al., 2006; Yang et al., 2011). For example, over-expression of OsICK6 results in multiple phenotypic effects on plant growth, morphology, pollen viability, and seed setting (Yang et al., 2011). Transgenic overexpression of OsiICK1 affects endosperm development and greatly reduces seed filling (Barroco et al., 2006). In the present study, we analyzed the phenotypes of OseIF3e RNAi-mediated knockdown transgenic rice plants, which revealed pleiotropic growth inhibition throughout development. The phenotypes of the OseIF3eRi plants were similar to those reported in transgenic plants that overexpress OsICK1 and OsICK6.
Consistent with this finding, in the present study, knockdown of OseIF3e dramatically reduced the expression of OsICK1, OsICK5, and OsICK6, suggesting that OseIF3e may influence the cell division cycle via interaction with ICKs. In addition, we found that the OseIF3e amino acids 118–138 are necessary for its interaction with OsICKs. This interaction may be mediated by either motif1 or motif2 in OsICKs and by amino acids residues 118–138 in OseIF3e (Figure 7d). The eIF3e subunit may interact with various proteins that possess different binding specificities to initiate translation and regulate the expression genes involved in the development of plants.
In summary, this study points to OseIF3e as a crucial regulator of rice seedling development and reproductive processes, including pollen maturation. Based on previous results and the present findings, we propose a possible regulatory model (Figure 8) for the role of OseIF3e in these processes: (i) Osj10gBTF3 regulates transcription of OseIF3e and OseIF3h in the nucleus (Wang et al., 2012); (ii) in the cytosol, OseIF3e interacts with OseIF3 subunits b, d, e, f, h, and k, and with eIF6, to form homo- and heterodimers; (iii) the eIF complex interacts with OsICKs to regulate cell division, affecting plant growth and development. These findings may lead to a better understanding of the factors influencing plant growth and pollen development. Moreover, OseIF3e-mediated regulation of OsICKs genes pleiotropically modulates several plant characteristics (e.g., plant height, spikelet number, and seed size), providing an opportunity to optimize crop architecture for crop breeding.
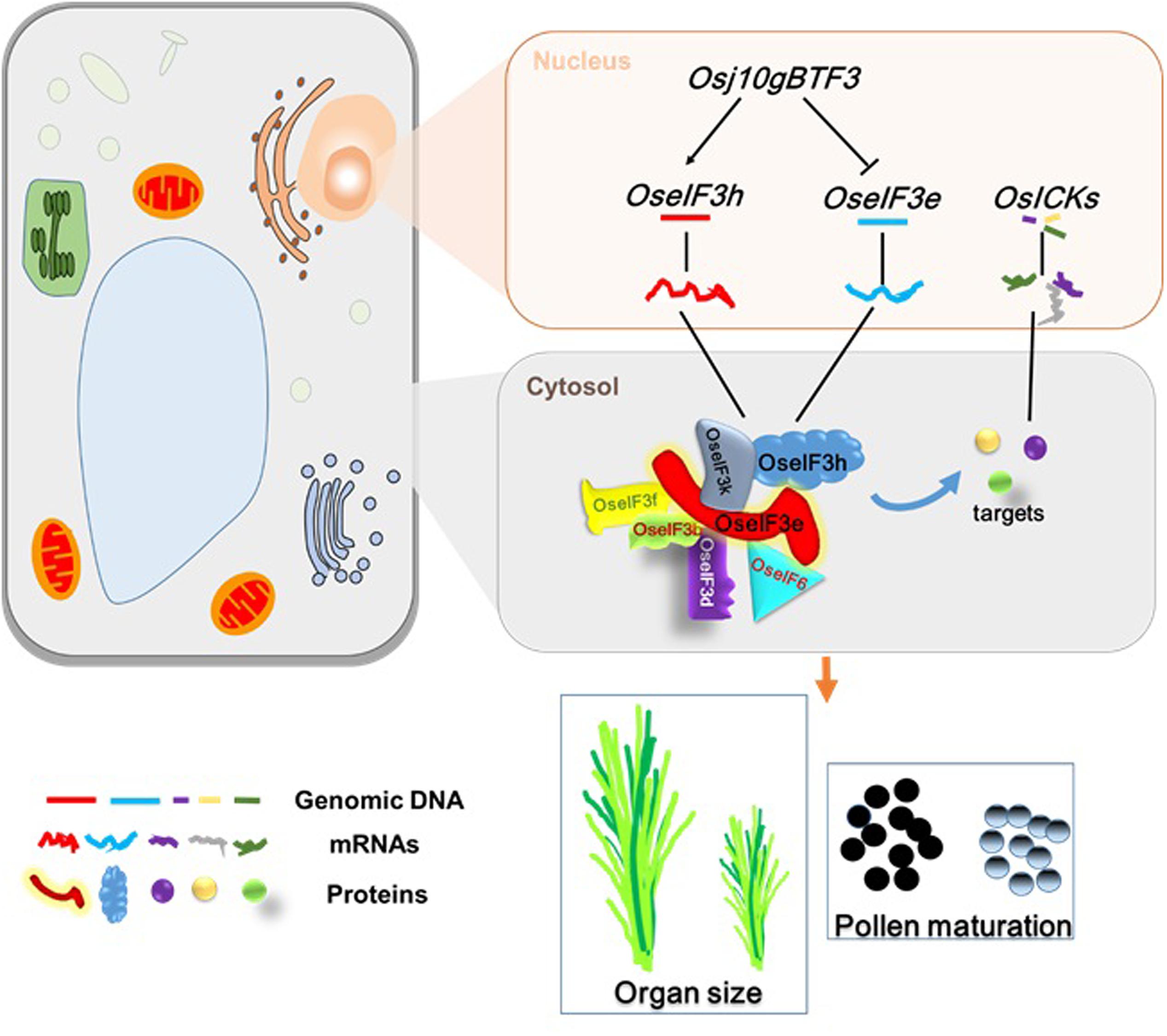
FIGURE 8. A general model of OseIF3e involved in rice growth and pollen development. The expression of OseIF3e and OseIF3h were regulated by Osj10gBTF3 in the nucleus. In the cytosol, OseIF3e interacts with OseIF3 subunits b, d, e, f, h, k and eIF6 to form homo- and heterodimers. Then, eIF complex binds to OsICKs, thereby affecting their functions and leading to wide-ranging defects.
Author Contributions
WW and JT conceived and designed the project, analyzed the data, and wrote the manuscript. MX and XL helped with data analysis. The manuscript was approved by all other authors.
Funding
This work was supported by National Nature Science Foundation of China (grant No. 30871502).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01399
Abbreviations
ABA, abscisic acid; ABREs, ABA response elements; BTF3, basal transcription factor 3; CDS, coding sequence; eIF3e, eukaryotic translation initiation factor 3 subunit E; eIF3h, eukaryotic translation initiation factor 3 subunit H; GA, gibberellin; ICK, inhibitors of cyclin-dependent kinases; Os, Oryza sativa; qRT-PCR, quantitative reverse transcription-PCR; RNAi, RNA interference.
Footnotes
- ^ http://www.clontech.com/
- ^ http://rice.plantbiology.msu.edu
- ^ http://prosite.expasy.org/
- ^ http://www.dna.affrc.go.jp/htdocs/PLACE/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://meme-suite.org/tools/tomtom
- ^ http://crep.ncpgr.cn
References
Anastasiou, E., Kenz, S., Gerstung, M., MacLean, D., Timmer, J., Fleck, C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856. doi: 10.1016/j.devcel.2007.10.001
Asano, K., Phan, L., Anderson, J., and Hinnebusch, A. G. (1998). Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 18573–18585.
Asano, K., Vornlocher, H. P., RichterCook, N. J., Merrick, W. C., Hinnebusch, A. G., and Hershey, J. (1997). Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J. Biol. Chem. 272, 27042–27052. doi: 10.1074/jbc.272.43.27042
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. .
Bailey, T. L., and Gribskov, M. (1998). Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14, 48–54. doi: 10.1093/bioinformatics/14.1.48
Barroco, R. M., Peres, A., Droual, A., De Veylder, L., Nguyen, L. S. L., De Wolf, J., et al. (2006). The cyclin-dependent kinase inhibitor orysa; KRP1 plays an important role in seed development of rice. Plant Physiol. 142, 1053–1064. doi: 10.1104/pp.106.087056
Basu, U., Si, K., Warner, J. R., and Maitra, U. (2001). The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21, 1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001
Bemis, S. M., and Torii, K. U. (2007). Autonomy of cell proliferation and developmental programs during Arabidopsis aboveground organ morphogenesis. Dev. Biol. 304, 367–381. doi: 10.1016/j.ydbio.2006.12.049
Browning, K. S., Gallie, D. R., Hershey, J., Hinnebusch, A. G., Maitra, U., Merrick, W. C., et al. (2001). Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem. Sci. 26:284. doi: 10.1016/S0968-0004(01)01825-4
Burks, E. A., Bezerra, P. P., Le, H., Gallie, D. R., and Browning, K. S. (2001). Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J. BiolChem. 276, 2122–2131.
Busov, V. B., Brunner, A. M., and Strauss, S. H. (2008). Genes for control of plant stature and form. New Phytol. 177, 589–607. doi: 10.1111/j.1469-8137.2007.02324.x
De Veylder, L., Beeckman, T., Beemster, G., Krols, L., Terras, P., Landrieu, I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. doi: 10.2307/3871392
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279.
Feng, G., Qin, Z., Yan, J., Zhang, X., and Hu, Y. (2011). Arabidopsis organ size related1 regulates organ growth and final organ size in orchestration with argos and arl. New Phytol. 191, 635–646. doi: 10.1111/j.1469-8137.2011.03710.x
Hinnebusch, A. G. (2006). eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562. doi: 10.1016/j.tibs.2006.08.005
Hu, Y. X., Xie, O., and Chua, N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. doi: 10.1105/tpc.013557
Huang, X. H., Hetfeld, B., Seifert, U., Kahne, T., Kloetzel, P. M., Naumann, M., et al. (2005). Consequences of COP9 signalosome and 26S proteasome interaction. FEBS J. 272, 3909–3917. doi: 10.1111/j.1742-4658.2005.04807.x
Jiang, Y., Bao, L., Jeong, S., Kim, S., Xu, C., Li, X., et al. (2012). XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 70, 398–408. doi: 10.1111/j.1365-313X.2011.04877.x
Kang, J., Mizukami, Y., Wang, H., Fowke, L., and Dengler, N. G. (2007). Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta 226, 1207–1218. doi: 10.1007/s00425-007-0567-2
Kapp, L. D., and Lorsch, J. R. (2004). The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657–704. doi: 10.1146/annurev.biochem.73.030403.080419
Karniol, B., Yahalom, A., Kwok, S., Tsuge, T., Matsui, M., Deng, X. W., et al. (1998). The Arabidopsis homologue of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett. 439, 173–179. doi: 10.1016/S0014-5793(98)01367-2
Kato, Y., Konishi, M., Shigyo, M., Yoneyama, T., and Yanagisawa, S. (2010). Characterization of plant eukaryotic translation initiation factor 6 (eIF6) genes: the essential role in embryogenesis and their differential expression in Arabidopsis and rice. Biochem. Biophys. Res. Commun. 397, 673–678. doi: 10.1016/j.bbrc.2010.06.001
Kawaguchi, R., and Bailey-Serres, J. (2002). Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 5, 460–465. doi: 10.1016/S1369-5266(02)00290-X
Kim, B., Cai, X., Vaughn, J. N., and von Arnim, A. G. (2007). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8:R60. doi: 10.1186/gb-2007-8-4-r60
Kim, T. H., Kim, B. H., Yahalom, A., Chamovitz, D. A., and von Arnim, A. G. (2004). Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis, translation initiation factor subunit elF3h. Plant Cell 16, 3341–3356. doi: 10.1105/tpc.104.026880
Kolaczkowski, B., and Thornton, J. W. (2004). Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 431, 980–984. doi: 10.1038/nature02917
Krizek, B. A. (1999). Ectopic expression AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y
Li, Q., Deng, Z., Gong, C., and Wang, T. (2016). The rice eukaryotic translation initiation factor 3 subunit f (OseIF3f) is involved in Microgametogenesis. Front. Plant Sci. 7:532. doi: 10.3389/fpls.2016.00532
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Marchetti, A., Buttitta, F., Miyazaki, S., Gallahan, D., Smith, G. H., and Callahan, R. (1995). Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J. Virol. 69, 1932–1938.
Mizukami, Y., and Fischer, R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947. doi: 10.1073/pnas.97.2.942
Morris, C., and Jalinot, P. (2005). Silencing of human Int-6 impairs mitosis progression and inhibits cyclin B-Cdk1 activation. Oncogene 24, 1203–1211. doi: 10.1038/sj.onc.1208268
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15, 473–493.
Phan, L., Zhang, X. L., Asano, K., Anderson, J., Vornlocher, H. P., Greenberg, J. R., et al. (1998). Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 18, 4935–4946. doi: 10.1128/MCB.18.8.4935
Russell, D. W., and Spremulli, L. (1980). Mechanism of action of the wheat-germ ribosome dissociation factor - interaction with the 60-s subunit. Arch. Biochem. Biophys. 201, 518–526. doi: 10.1016/0003-9861(80)90540-8
Sherr, C. J., and Roberts, J. M. (1999). CDK inhibitors: positive and negative regulators of G(1)-phase progression. Genes Dev. 13, 1501–1512. doi: 10.1101/gad.13.12.1501
Shi, J., Feng, Y., Goulet, A. C., Vaillancourt, R. R., Sachs, N. A., Hershey, J. W., et al. (2003). The p34 cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of Eukaryotic initiation factor 3 during apoptosis. J. Biol. Chem. 278, 5062–5071. doi: 10.1074/jbc.M206427200
Si, K., and Maitra, U. (1999). The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 19, 1416–1426. doi: 10.1128/MCB.19.2.1416
Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A., and Nogales, E. (2005). Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515. doi: 10.1126/science.1118977
Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Torres Acosta, J. A., Fowke, L. C., and Wang, H. (2011). Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann. Bot. 107, 1141–1157. doi: 10.1093/aob/mcr034
Verkest, A., Manes, C., Vercruysse, S., Maes, S., Van der Schueren, E., Beeckman, T., et al. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736. doi: 10.1105/tpc.105.032383
von Arnim, A. G., and Chamovitz, D. A. (2003). Protein homeostasis: a degrading role for Int6/eIF3e. Curr. Biol. 13, R323–R325.
Wang, H., Zhou, Y. M., Gilmer, S., Whitwill, S., and Fowke, L. C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. doi: 10.1046/j.1365-313x.2000.00899.x
Wang, L., Xie, W., Chen, Y., Tang, W., Yang, J., Ye, R., et al. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61, 752–766. doi: 10.1111/j.1365-313X.2009.04100.x
Wang, M., Chen, C., Xu, Y. Y., Jiang, R. X., Han, Y., Xu, Z. H., et al. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22, 409–417. doi: 10.1007/BF02772683
Wang, Y., Zhang, X., Lu, S., Wang, M., Wang, L., Wang, W., et al. (2012). Inhibition of a basal transcription factor 3-like gene Osj10gBTF3 in rice results in significant plant miniaturization and typical pollen abortion. Plant Cell Physiol. 53, 2073–2089. doi: 10.1093/pcp/pcs146
Weinl, C., Marquardt, S., Kuijt, S., Nowack, M. K., Jakoby, M. J., Hulskamp, M., et al. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17, 1704–1722. doi: 10.1105/tpc.104.030486
White, D. W. R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wood, L. C., Ashby, M. N., Grunfeld, C., and Feingold, K. R. (1999). Cloning of murine translation initiation factor 6 and functional analysis of the homologous sequence YPR016c in Saccharomyces cerevisiae. J. Biol. Chem. 274, 11653–11659.
Xia, C., Wang, Y., Li, W., Chen, Y., Deng, Y., Zhang, X., et al. (2010). The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 63, 189–202. doi: 10.1111/j.1365-313X.2010.04237.x
Xu, R., and Li, Y. (2011). Control of final organ size by mediator complex subunit 25 in Arabidopsis thaliana. Development 138, 4545–4554. doi: 10.1242/dev.071423
Yahalom, A., Kim, T., Roy, B., Singer, R., Von Arnim, A. G., and Chamovitz, D. A. (2008). Arabidopsis eIF3e is regulated by the COP9 signalosome and has an impact on development and protein translation. Plant J. 53, 300–311. doi: 10.1111/j.1365-313X.2007.03347.x
Yahalom, A., Kim, T. H., Winter, E., Karniol, B., von Arnim, A. G., and Chamovitz, D. A. (2001). Arabidopsis eIF3e (INT-6) associates with both eIF3c and the COP9 signalosome subunit CSN7. J. Biol. Chem. 276, 334–340.
Yang, R., Tang, Q., Wang, H., Zhang, X., Pan, G., Wang, H., et al. (2011). Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann. Bot. 107, 1087–1101. doi: 10.1093/aob/mcr057
Yen, H., and Chang, E. C. (2000). Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. Proc. Natl Acad. Sci. U.S.A. 97, 14370–14375. doi: 10.1073/pnas.97.26.14370
Yen, H., Gordon, C., and Chang, E. C. (2003). Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell 112, 207–217. doi: 10.1016/S0092-8674(03)00043-6
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). “Routine procedures for growing rice plants in culture solution,” in Laboratory Manual for Physiological Studies of Rice, eds S. Yoshida, D. A. Forno, J. H. Cook, and K. A. Gomez (Los Banos, CA: International Rice Research Institute), 61–66.
Zhou, C. S., Arslan, F., Wee, S., Krishnan, S., Ivanov, A. R., Oliva, A., et al. (2005). PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 3:14. doi: 10.1186/1741-7007-3-14
Zhou, F., Roy, B., Dunlap, J. R., Enganti, R., and von Arnim, A. G. (2014). Translational control of Arabidopsis meristem stability and organogenesis by the eukaryotic translation factor eIF3h. PLoS ONE 9:e95396. doi: 10.1371/journal.pone.0095396
Keywords: OseIF3e, translation initiation, OsICKs, Oryza sativa L., reproductive development, pollen maturation
Citation: Wang W, Xu M, Liu X and Tu J (2016) The Rice Eukaryotic Translation Initiation Factor 3 Subunit e (OseIF3e) Influences Organ Size and Pollen Maturation. Front. Plant Sci. 7:1399. doi: 10.3389/fpls.2016.01399
Received: 12 July 2016; Accepted: 02 September 2016;
Published: 20 September 2016.
Edited by:
Johannes Hanson, Umeå University, SwedenReviewed by:
Alberto A. Iglesias, National University of the Littoral, ArgentinaChristian Meyer, Institut National de la Recherche Agronomique, France
Copyright © 2016 Wang, Xu, Liu and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jumin Tu, anR1QHpqdS5lZHUuY24=
 Wenyi Wang
Wenyi Wang Mengyun Xu
Mengyun Xu Xuejiao Liu
Xuejiao Liu Jumin Tu
Jumin Tu