- 1College of Life Sciences, Capital Normal University, Beijing, China
- 2National Research Council Canada, Saskatoon, SK, Canada
- 3Department of Microbiology and Immunology, University of Saskatchewan, Saskatoon, SK, Canada
In this study, we report the identification and functional characterization of three Brachypodium distachyon UEV genes. All three BdUev1s form heterodimers with BdUbc13s, which are capable of catalyzing Lys63-linked polyubiquitination in vitro. The three BdUEV1 genes are also able to functionally complement the budding yeast mms2 mutant defective in DNA-damage tolerance. BdUev1A differs from the other two BdUev1s in that it contains an 18-amino acid tail, which appears to compromise its function in yeast, as deletion of this tail restores full function. BdUev1A is excluded from the nucleus, whereas BdUev1B, BdUev1C and the C-terminally truncated BdUev1A are mainly found in the nucleus. These and the BdUEV1 gene expression analysis allow us to speculate that although all three BdUev1s function by promoting Lys63-linked polyubiquitination, BdUev1B and BdUev1C are involved in DNA-damage response and possibly other nuclear functions, while BdUev1A is required for non-nuclear function(s).
Introduction
Ubiquitin (Ub) is a highly conserved 76-residue protein; its amino acid sequence is conserved in animals, plants, and fungi (Hochstrasser, 1996; Hershko and Ciechanover, 1998). One ubiquitin C terminus can be covalently linked to any one of the seven surface lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the N-terminal methionine residue of a second ubiquitin via isopeptide bond, forming a ubiquitin chain. Substrates can be modified by ubiquitin chains (polyubiquitination), multiple single ubiquitin (multi-monoubiquitination) or a single ubiquitin (monoubiquitination). Ub-activating enzyme (Uba or E1), Ub-conjugating enzyme (Ubc or E2) and Ub ligase (E3) are involved in this process. Lys48-linked chains are the predominant linkage type, and play a role in targeting protein for degradation by the 26S proteasome (Hershko and Ciechanover, 1998). Lys63-linked chains are the second abundant form and their role is to target proteins to primarily serve as a signal (Pickart, 2000; Chen and Sun, 2009). Ubc13 is the only known Ubc dedicated to catalyzing Lys63-linked polyubiquitination reaction in eukaryotes. This reaction requires a Ubc-like or Ubc/E2 variant (Uev), which forms a stable heterodimer with Ubc13 (Hofmann and Pickart, 1999; McKenna et al., 2001).
The first UEV gene, denoted MMS2, was isolated from budding yeast cells and is involved in the error-free DNA-damage tolerance (DDT) pathway (Broomfield et al., 1998; Xiao et al., 1999). Mms2 itself is homologous to an E2 enzyme, but lacks the active site cysteine residue (Broomfield et al., 1998). Several structural studies revealed that Mms2 is required for catalyzing Lys63-linked poly-Ub chains (Moraes et al., 2001; VanDemark et al., 2001). In budding yeast, Mms2, Ubc13 and Rad5 form a E2-E3 complex and are all required for the sequential Lys63-linked polyubiquitination of PCNA after it is monoubiquitinated at the Lys164 residue by another E2–E3 complex, Rad6-Rad18 (Hoege et al., 2002). Yeast has only one UEV (MMS2) gene (Broomfield et al., 1998) while higher eukaryotic genomes appear to contain multiple UEV genes (Xiao et al., 1998; Wen et al., 2008, 2012) whose products form stable complexes with Ubc13 and may participate in different biological processes. For example, mammalian Uev1A-Ubc13 is involved in NF-κB activation, while Mms2-Ubc13 is required for DNA-damage response (Andersen et al., 2005). It has been reported that Arabidopsis UBC13 genes are involved in DNA-damage response (Wen et al., 2006), apical dominance (Yin et al., 2007), iron metabolism (Li and Schmidt, 2010), immunity (Mural et al., 2013) and auxin signaling (Wen et al., 2014). In contrast, little is known about plant UEV genes. Among Arabidopsis UEV1 genes, only AtUEV1D has been shown to be involved in DNA-damage response (Wen et al., 2008). So far there has been no report on the characterization of other plant UEV1 genes.
Brachypodium distachyon is a model species for monocots, temperate cereals and biofuel plants (Draper et al., 2001; Catalan et al., 2014). It has many advantages including: small genome (300 Mbp), short generation time (8–12 weeks), relative convenience of getting mutants and simpler growth condition. We previously reported that the B. distachyon genome contains two UBC13 genes; their products were able to promote Lys63-linked Ub chain assembly with AtUev1s and functionally rescue yeast ubc13 mutant phenotypes (Guo et al., 2016). Here we report molecular cloning and functional characterization of three UEV1 genes from B. distachyon. All three products can interact and catalyze Lys63-linked Ub chain assembly with BdUbc13; however, BdUev1B and BdUev1C appear to be involved in DDT while BdUev1A is not.
Materials and Methods
Plant Materials and Yeast Cell Culture
B. distachyon 21 (Bd21) seeds were surface sterilized with 20% sodium hypochlorite twice for 10 min, rinsed five times with sterile H2O, incubated in H2O for 12 h at room temperature, and then transferred to a wet filter paper to germinate in darkness for 24 h at 22–25°C. Uniformly germinated seeds were spread on plastic pots containing 1/2 MS (Murashige and Skoog), and the medium was changed every 2 days. After 2 weeks, the seedlings were transferred to soil and grown in a growth chamber with a daily photo cycle of 16/8 h light/dark, 22°C/18°C and 65–75% air humidity.
Yeast strains used in this study are listed in Supplementary Table S1 and the cell culture conditions were as previously described (Guo et al., 2016). Yeast cells were transformed by a lithium acetate method (Ito et al., 1983). The mms2Δ::HIS3 (Xiao et al., 1999) and ubc13Δ::HUH (Wen et al., 2008) disruption cassettes were as previously described.
Molecular Cloning and Plasmid Construction
To clone B. distachyon UEV1 genes, total RNA was extracted from B. distachyon seedlings using a Trizol reagent (Invitrogen), the first-strand of cDNA was synthesized by a RevertAid First Strand cDNA Synthesis Kit (Thermo). The BdUEV1 ORFs were amplified by PCR from the above cDNAs using gene-specific primers (Supplementary Table S2). The yeast two-hybrid vectors pGAD424Bg and pGBT9Bg were derived from pGAD424 and pGBT9 (Bartel and Fields, 1995).
Yeast Two-Hybrid and GST Pull-Down Assays
Constructed Gal4AD and Gal4BD vectors were transformed into the yeast two-hybrid strain PJ69-4a (James et al., 1996) in pairs and allowed to grow at 30°C for 2–3 days. Transformants were selected on SD-Leu-Trp plates. Protein interaction was determined on synthetic complete medium lacking Trp, Leu and His, supplemented with 3-amino-1,2,4-triazole (3-AT, Sigma-Aldrich), or on plates lacking Trp, Leu, and Ade. SD-Leu-Trp plates were used as a control.
Full-length BdUEV1 ORFs were cloned in plasmid pGEX-6p and BdUBC13s were cloned in plasmid pET30a as previously described (Guo et al., 2016). The His6 and GST fusion constructs were transformed into E. coli strain BL21 (DE3) and the recombinant proteins were purified with Ni Sepharose and glutathione (Amersham Pharmacia), respectively. For the pull-down assay, crude cell extracts were loaded on Glutathione SepharoseTM 4B beads and then 10 μg of purified His6-BdUbc13 was later added. After incubation and washing, the GST beads were boiled with SDS-PAGE loading buffer for 10 min prior to Western blotting.
Ub Conjugation Reaction
Ub conjugation reactions were performed by using purified fusion proteins and Ub thioester conjugation reagents (Boston Biochem). The reaction mixture contained 225 nM E1 enzyme, 200 μM Ub (or recombinant Ub-K63R), 1 mM MgATP, 1 mM Ubc13 and 1 mM Uev1 in 20 μl of reaction buffer. The conjugation reactions were performed at 37°C for 2 h, samples were subjected to 12% SDS-PAGE and Ub-containing molecules were detected by Western blotting using the mouse anti-Ub monoclonal antibody P4D1 (Cell Signaling).
Yeast Survival Assays
Yeast strain HK578-10D, its isogenic mms2Δ single and ubc13Δ mms2Δ double mutants were transformed with pGAD-BdUEV1s, or co-transformed with pGBT-BdUEV1 and pGAD-BdUBC13. Transformants were selected on SD-Leu or SD-Leu-Trp plates. The gradient plate and serial dilution assays were as described (Xu et al., 2014).
Spontaneous Mutagenesis Assay
Spontaneous mutagenesis was measured by the reverse mutation rate of the trp1-289 allele in the DBY747 strain (Xiao and Samson, 1993). DBY747 cells with the mms2Δ mutation were transformed with pGAD-BdUEV1s and the transformants were selected on SD-Leu plates. This modified Luria–Delbruck fluctuation test was adapted as described (Blackwell et al., 2014) and each treatment contained five independent cultures.
Subcellular Localization of BdUev1s
GFP-BdUEV1 fusion genes were constructed by cloning BdUEV1 ORFs into the modified binary vector pCAMBIA1300-GFP downstream of GFP at the SacI and KpnI sites so that the fusion gene was driven by a CaMV 35S promoter. The constructed plasmids were introduced into A. tumefaciens PMP90, which was then co-infiltrated with A. tumefaciens P19 into N. benthamiana leaves (Waadt and Kudla, 2008). GFP and DAPI fluorescence was observed 2 days after transformation using a Zeiss 780 confocal microscope.
Expression Analysis by Droplet Digital RT-PCR (ddRT-PCR)
To determine the expression level of BdUEV1 genes in different development stages and in response to abiotic stress, ddRT-PCR was performed by using a Bio-Rad QX200TM ddPCR System. All reagents and consumables for the experiments including droplet generator oil, DG8TM cartridges and gaskets, droplet reader oil, and ddPCR supermix for EvaGreen were purchased from Bio-Rad. Briefly, the Droplet Digital PCR began by partitioning the reaction mix containing EvaGreen Supermix, primers and sample cDNA into aqueous droplets in oil via the QX200 Droplet Generator; after transfer of droplets to a 96-well PCR plate, a thermocycling protocol including 95°C for 5 min; 95°C for 30 s, 40 cycles; 60°C for 60 s, 40 cycles (ramp rate set to 2°C/s); 4°C for 5 min; 90°C for 5 min and 4°C infinite was carried out in a conventional thermal cycler. The PCR plate was then transferred to the QX200 Droplet Reader for automatic reading of samples. QuantaSoftTM Software was used for data analysis. The primers used to amplify the BdUEV1s are listed in Supplementary Table S3.
Results
Identification and Sequence Analysis of the BdUEV1 Genes
To identify B. distachyon UEV1 genes, Arabidopsis UEV1 genes were used to blast the B. distachyon genomic database. Three genes, BRADI1G11410.1, BRADI4G02400.1 and BRADI4G27530.1, were obtained and named BdUEV1A, BdUEV1B and BdUEV1C, respectively. Protein sequence alignment of BdUev1s reveals 81% identity between BdUev1A and BdUev1B, 77% identity between BdUev1A and BdUev1C, and 82% identity between BdUev1B and BdUev1C. The predicted BdUev1A, BdUev1B and BdUev1C proteins contain 160, 147 and 148 amino acids, respectively. It is noted that BdUev1A contains a C-terminal tail but the other two BdUev1s do not (Figure 1A). The sequences of BdUev1 proteins were also aligned with those of Uev1 proteins from six other eukaryotic organisms. As shown in Figure 1A, several critical residues deemed to be necessary for the Uev activity are also conserved in BdUev1s, including Phe13 of hMms2 (red asterisk) required for the physical interaction with Ubc13 (Pastushok et al., 2005), and Ser27 and Ile57 of ScMms2 (blue asterisks) required for the non-covalent interaction with Ub and poly-Ub chain assembly (Tsui et al., 2005; Eddins et al., 2006; Pastushok et al., 2007).
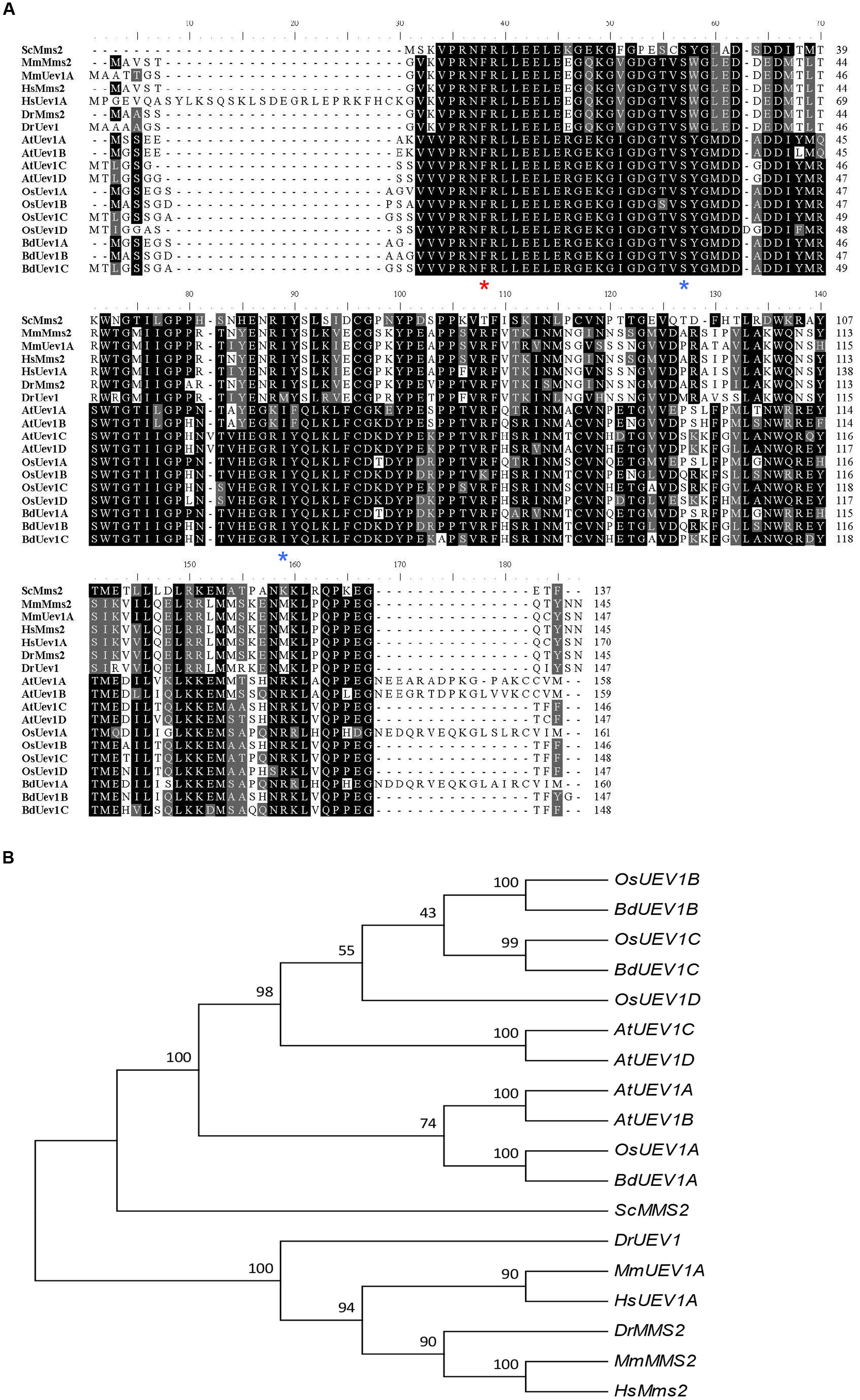
FIGURE 1. Analysis of UEV1s from different organisms. (A) Amino acid sequence alignment of BdUev1s and Uevs from other organisms. The sequences were aligned and edited using BioEdit 7.0.9. Critical residues for UEV functions are indicated with asterisks underneath the residue. Sc, S. cerevisiae (NP_011428); Mm, Mus musculus (MmMMS2 = NP_076074, MmUEV1A = NP_075719.1); Hs, Homo sapiens (HsMMS2 = NP_003341.1, HsUEV1A = NP_068823.2); Dr, Danio rerio (DrMMS2 = NP_998680.1, DrUEV1 = NP_001032479); At, A. thaliana (AtUEV1A = NP_564191, AtUEV1B = NP_564994, AtUEV1C = NP_850259, AtUEV1D = NP_001190073); Os, Oryza sativa Japonica Group (OsUEV1A = XP_015632881, OsUEV1B = XP_015620277, OsUEV1C = XP_015611980, OsUEV1D = XP_015635908); Bd, B. distachyon (BdUEV1A = KQK13622, BdUEV1B = XP_003577901, BdUEV1C = XP_003577953). (B) Phylogenetic analyses of UEV family CDSs from different organisms. The phylogenetic tree clustering was conducted by using MEGA6.0.
To further investigate the evolution of B. distachyon UEV1s, the CDS sequences of three BdUEV1s were compared with UEVs from six other organisms. The analysis reveals that BdUEV1s and OsUEV1s are closely related (Figure 1B). Hence, BdUEV1s could be another ideal model to study plant UEV1 gene functions, especially to study crop UEV1 gene functions.
Interaction of BdUev1s with BdUbc13s
Since Uevs play an essential role in Ubc13-mediated Lys63-linked polyubiquitination through forming a stable complex with Ubc13 to maintain its unique E2 activity (Hofmann and Pickart, 1999; McKenna et al., 2001, 2003), a yeast two-hybrid assay (Fields and Song, 1989) was used to determine whether BdUev1s interact with BdUbc13s. As seen in Figure 2A, all three BdUev1s interact with both BdUbc13s and the co-transformants are able to grow in the presence of 5 mM 3-AT, while no self-activation is observed in the -His plate without 3-AT. To further confirm the physical interaction between BdUev1s and BdUbc13s in vitro, a GST-affinity pull-down assay was performed. As shown in Figure 2B, recombinant GST-BdUev1 fusion proteins can pull down purified recombinant His6-BdUbc13 proteins, but the GST alone cannot. In the GST-affinity pull-down assay, the strength of interaction with BdUbc13 has no detectable difference. Based on the above observations, we conclude that BdUev1s can form stable heterodimers with BdUbc13s in vitro.
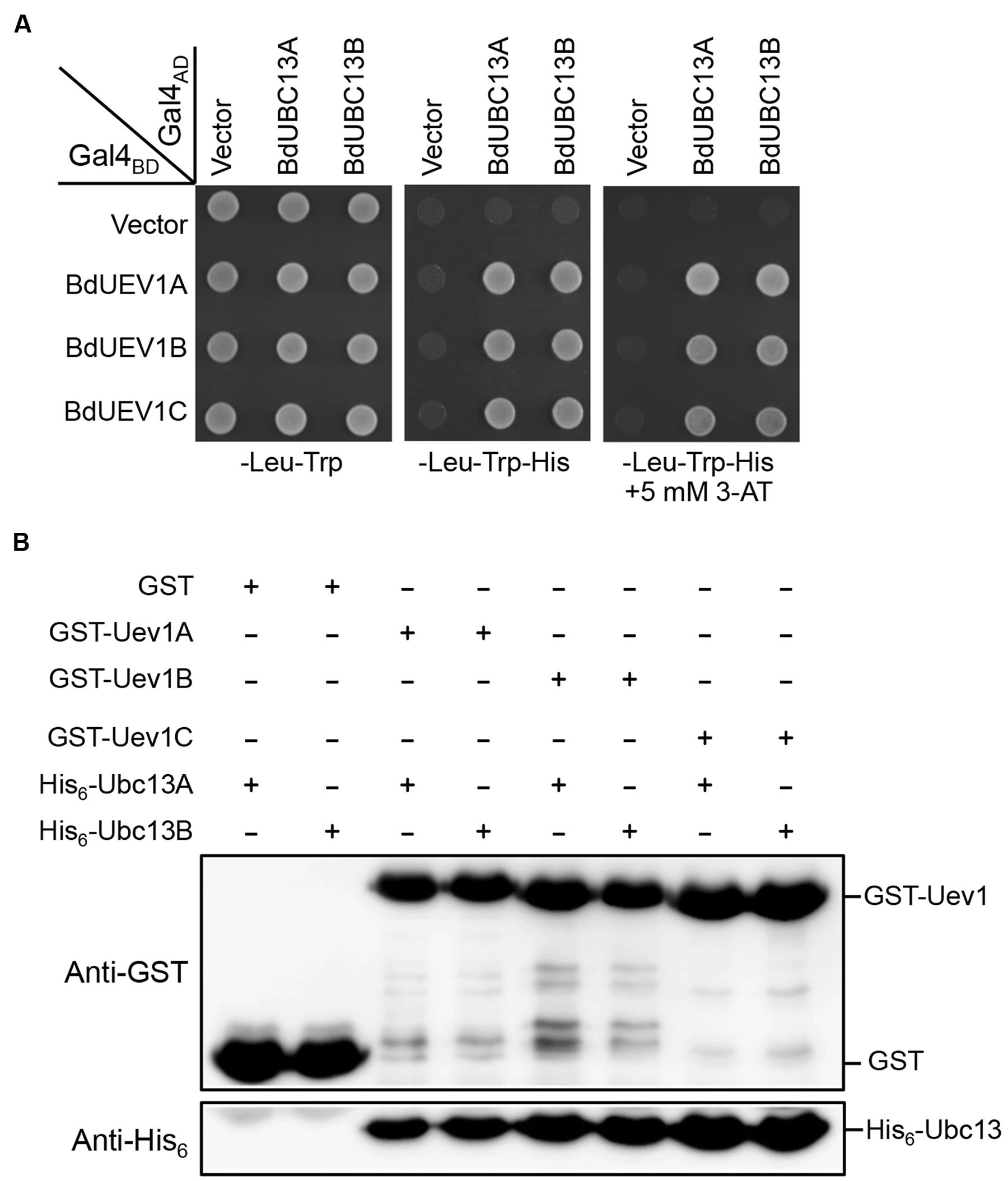
FIGURE 2. BdUev1s physically interact with BdUbc13s. (A) Physical interaction between BdUev1s and BdUbc13s in a yeast two-hybrid assay. PJ69-4a cells were transformed with BdUEV1s and BdUBC13s genes and the transformants carrying one Gal4BD (pGBT9) and one Gal4AD (pGAD424) plasmid were then selected, replicated onto various plates as indicated and incubated for 3 days before being photographed. The result is representative of at least five independent transformants from each treatment. (B) Protein interactions between BdUev1s and BdUbc13s by an affinity pull-down assay. BL21 (DE3) cells were transformed with pGEX-BdUEV1s, and target gene expression was induced by adding 0.2 mM IPTG. Crude cell extracts were loaded on Glutathione SepharoseTM 4B beads and 10 μg of purified His6-BdUbc13.
BdUev1 Is Required for Ubc13-Mediated Lys63-Linked Polyubiquitination In vitro
So far, the only defined Uev activity is to promote Lys63-linked polyubiquitination along with Ubc13 (Hofmann and Pickart, 1999; Deng et al., 2000; Hoege et al., 2002). A standard means of characterizing Uev enzymatic activity is by an in vitro Ub conjugation assay (Hofmann and Pickart, 1999). To avoid Ub conjugation to Ubc13 itself (McKenna et al., 2001), we made the corresponding BdUbc13-K94R mutation that does not affect the free Ub chain formation (Guo et al., 2016). As shown in Figure 3, BdUev1s together with BdUbc13A can indeed generate free Ub chains (lanes 3, 6, and 9), while BdUbc13A (lane 1) or BdUev1s (lanes 2, 5, and 8) alone cannot. More importantly, when the Ub-K63 is mutated to R, there are no poly-Ub chains observed in the same reaction (lanes 4, 7, and 10, asterisks indicate the background of UbK63R, see Supplementary Figure S1, lane 3). Hence, we conclude that BdUev1s are indeed required for Lys63-linked poly-Ub chain assembly. Since it has been previously reported that both BdUbc13A and BdUbc13B have the same ability to promote Lys63-linked poly-Ub chains along with AtUevs (Guo et al., 2016), this study did not repeat the reaction with BdUbc13B.
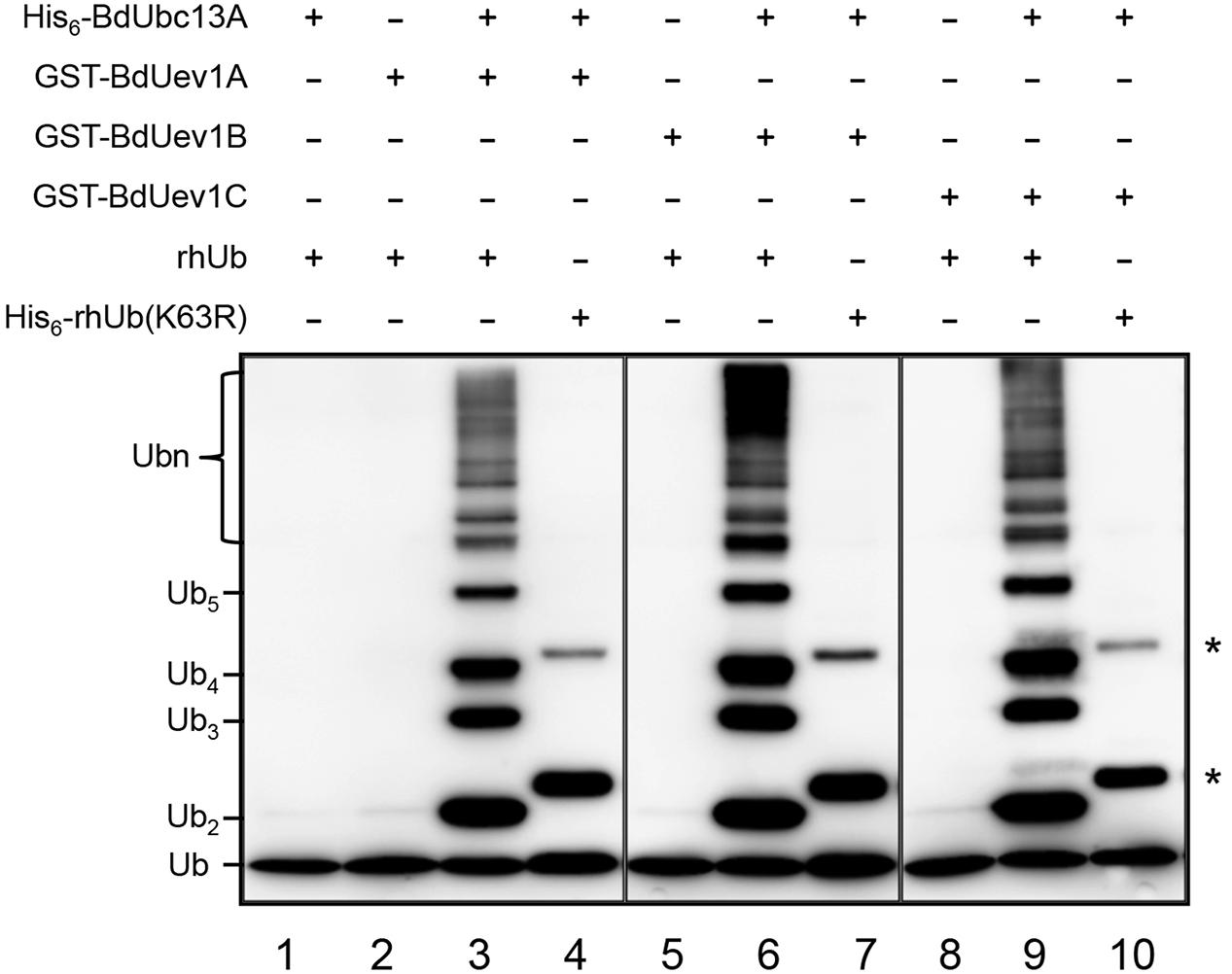
FIGURE 3. In vitro ubiquitin conjugation assay of BdUev1s using purified proteins. After ubiquitination reactions as described, samples were subjected to SDS-PAGE and Western blotting using an anti-Ub antibody to monitor poly-Ub chain formation. rhUb, recombinant human ubiquitin. Asterisks indicate the non-specific bands in lanes 4, 7, and 10.
Functional Complementation of Yeast mms2 by BdUEV1s
Yeast Mms2 is a member of the error-free DDT pathway (Broomfield et al., 1998; Brusky et al., 2000). To determine whether BdUev1s also play critical roles in error-free DDT, we assessed the ability of BdUEV1s to rescue the yeast mms2 mutant phenotypes. As shown in Figure 4A, the expression of any one of the three BdUEV1 genes can protect the mms2 null mutant from killing by MMS, whereas the expression of pGAD424 vector cannot. It is interesting that the ability of BdUEV1A to rescue the mms2 null mutant is slightly less than other two BdUEV1s.
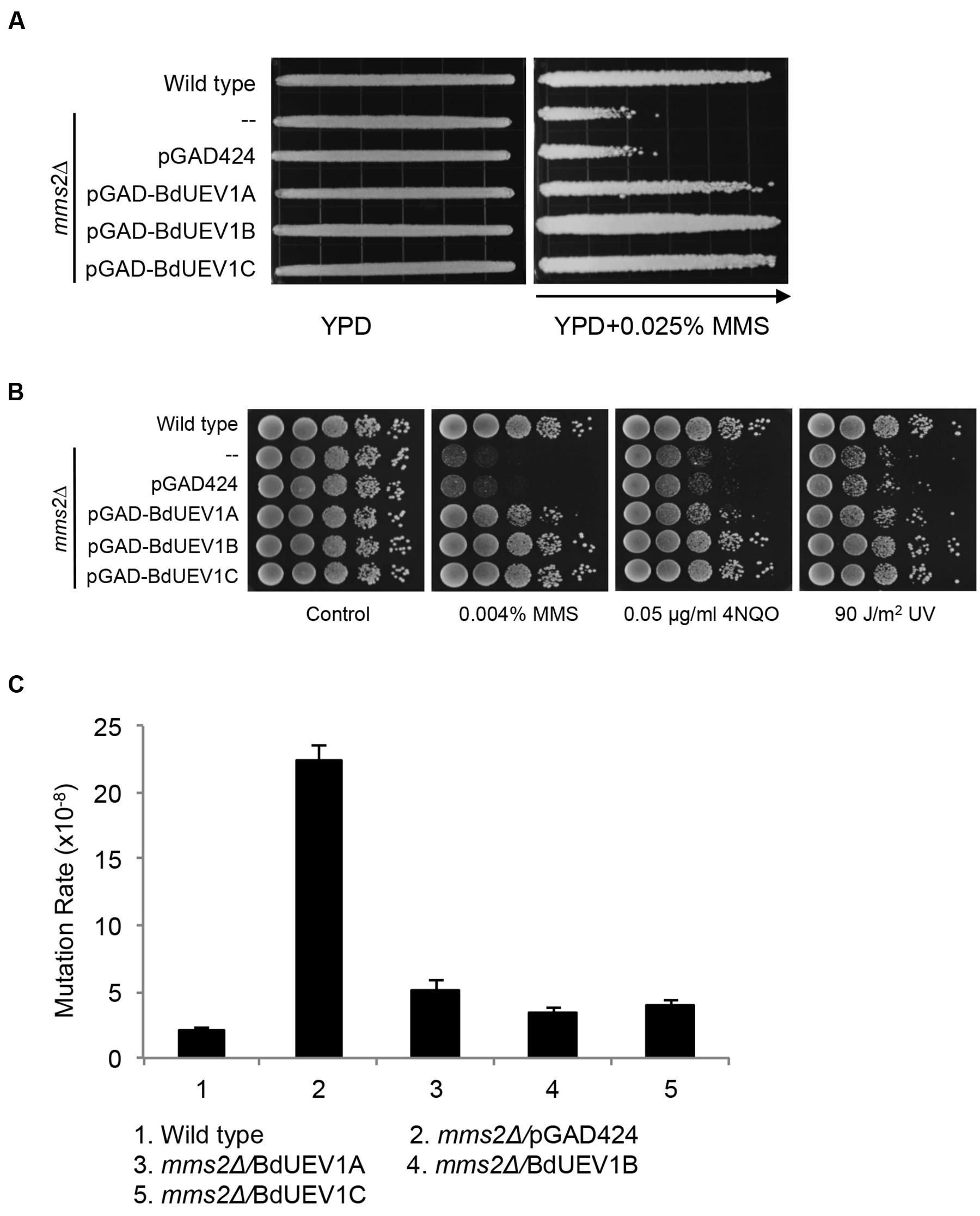
FIGURE 4. Functional complementation of the yeast mms2 null mutant by BdUEV1s. (A) Complementation of the mms2 single mutant by BdUEV1s. Yeast strain HK578-10D (wild type) and the isogenic mms2Δ transformants were grown overnight and then printed onto the gradient plate. The YPD control (left) and YPD+0.025% MMS gradient (right) plates were incubated at 30°C for 3 days. Arrow points to increasing MMS concentrations. (B) Functional complementation of the mms2 null mutant by BdUev1A, BdUev1B and BdUev1C using representative DNA-damaging agents by a serial dilution assay. Yeast strains as indicated were grown overnight in SD selective media, diluted and treated with the DNA-damaging agents. Yeast strains used: HK578-10D (wild type) and WXY942 (mms2Δ). (C) Spontaneous mutation rates of S. cerevisiae mms2 mutants. All strains are isogenic derivatives of DBY747. The results are the average of three independent experiments with standard deviations. All the genes were cloned in pGAD424.
DNA damage induced by MMS, UV irradiation or 4-nitroquinoline 1-oxide (4NQO) is largely considered to cause replication blocks and the mms2 null mutant is hypersensitive to these agents (Broomfield et al., 1998; Xu et al., 2013). To further confirm that BdUEV1s function in the DDT pathway, we performed a serial dilution assay in the presence of several representative DNA-damaging agents as mentioned above. A serial dilution assay is a semi-quantitative method to assess the relative sensitivity of cells to treatment (Xu et al., 2014). As shown in Figure 4B, both BdUEV1B and BdUEV1C can fully protect the yeast mms2 null mutant from killing by MMS, UV or 4-NQO, while the protection provided by BdUEV1A appears to be compromised.
Loss of MMS2 dramatically increases spontaneous mutagenesis, which serves as a hallmark indicating that it is involved in the error-free DDT pathway (Xu et al., 2015). Under our experimental conditions, deletion of MMS2 caused a 10-fold increase in the spontaneous mutation rate. When the mutant cells were transformed with BdUEV1s, the spontaneous mutagenesis rate was reduced to less than twofold (Figure 4C). Again BdUEV1B and BdUEV1C appear to be more effective than BdUEV1A in complementing the yeast MMS2 function. Hence, our results indicate that BdUev1B and BdUev1C can fully replace the function of Mms2 in the yeast error-free DDT pathway, but BdUev1A is partially functional.
The above functional complementation studies require that BdUev1s form stable heterodimers with yeast Ubc13. To further confirm that BdUev1 and BdUbc13 can form a stable heterodimer to replace the function of both yeast Mms2 and Ubc13, we co-transformed yeast ubc13 mms2 double mutant cells with the combination of BdUEV1s and BdUBC13s and assessed their relative tolerance to DNA-damaging agents. As shown in Figure 5 and Supplementary Figure S2, expression of BdUEV1 or BdUBC13 alone cannot rescue the mms2 ubc13 double mutant, whereas co-expression of BdUEV1B or BdUEV1C with BdUBC13 can rescue the ubc13 mms2 double mutant to nearly the wild-type level, regardless of the source of DNA damage. In sharp contrast, BdUBC13A or BdUBC13B co-transformed with BdUEV1A did not effectively rescue the ubc13 mms2 double mutant from killing induced by MMS, UV or 4NQO.
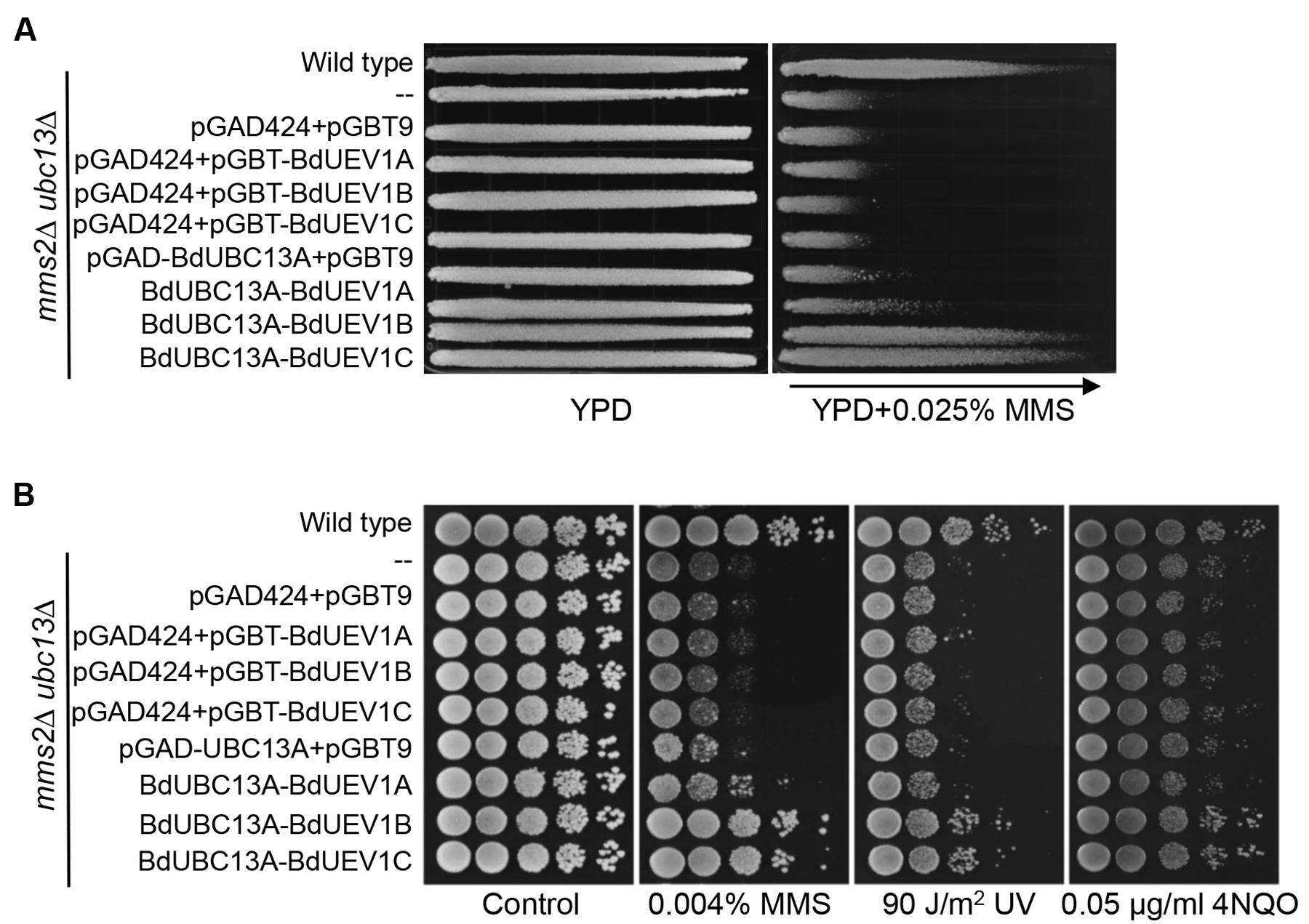
FIGURE 5. Functional complementation of the yeast ubc13 mms2 null mutant by BdUEV1s and BdUBC13A. (A) Complementation of the ubc13 mms2 double mutant by BdUEV1s and BdUBC13A. Yeast strain HK578-10D (wild type) and the isogenic ubc13Δ mms2Δ transformants were grown overnight and then printed onto the gradient plate. The YPD control (left) and YPD+0.025% MMS gradient (right) plates were incubated at 30°C for 3 days. Arrow points to increasing MMS concentration. (B) Functional complementation of the ubc13 mms2 double mutant by BdUBC13A and BdUEV1s using representative DNA-damaging agents by a serial dilution assay. Yeast strains as indicated were grown overnight in SD selective media, diluted and treated with the DNA-damaging agents.
The C-terminal Tail of BdUev1A Determines Its Function in Yeast
Among the three BdUev1s, an obvious sequence difference is the additional 18-amino acid tail found at the C terminus of BdUev1A (Figure 1A). We hypothesized that the lack of complementation of the yeast mms2 mutant by BdUEV1A is due to its encoded C-terminal tail. To test this hypothesis, we created a BdUEV1A-ΔCT construct in which the C-terminal 18 amino-acid coding sequence is removed from BdUEV1A. Figure 6 shows that BdUEV1A-ΔCT provides stronger protection against DNA damage than BdUEV1A and is comparable with that of BdUEV1B. Hence, the C-terminal tail of BdUev1A determines its efficacy in complementing the yeast DDT defect.
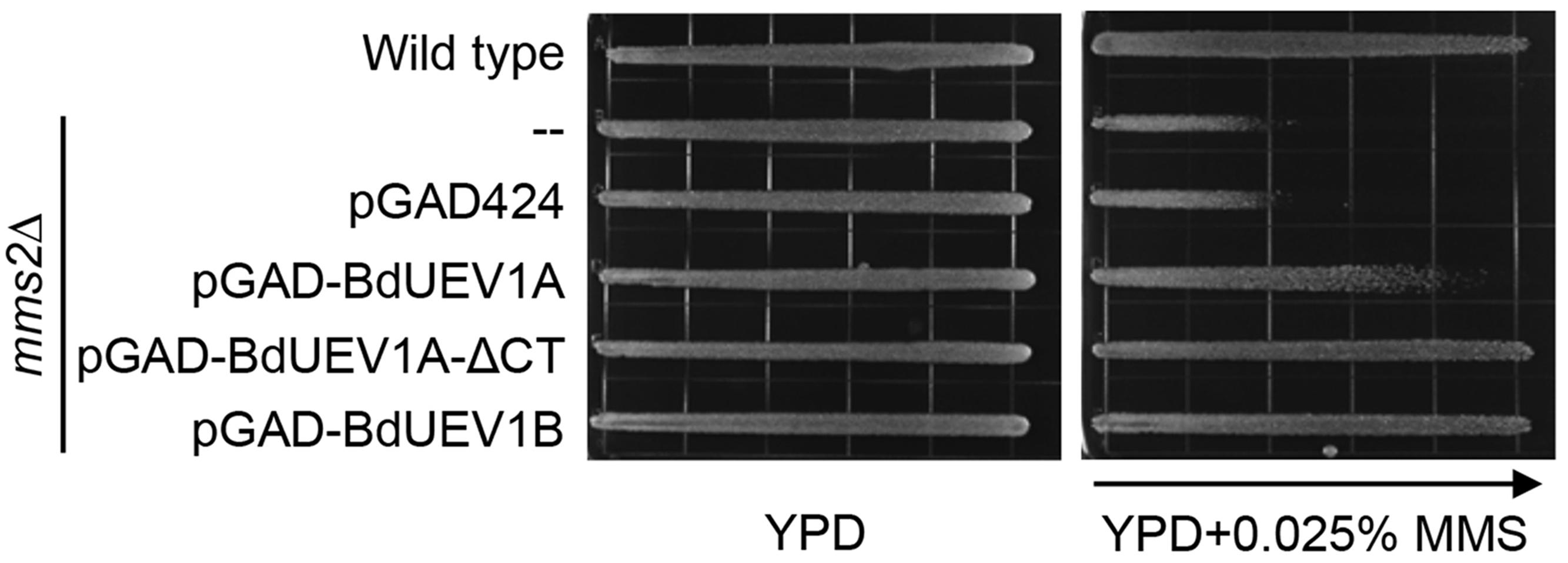
FIGURE 6. The C-terminal tail of BdUev1A affects functional complementation of the yeast mms2 null mutant. mms2 null mutant (WXY942) transformants with plasmids expressing BdUev1A, BdUev1A-ΔCT or BdUev1B were assessed by an MMS gradient plate assay. The plates were incubated at 30°C for 3 days. Arrow points to gradually increasing MMS concentration.
Differential Subcellular Locations of BdUev1s
To further address the difference between BdUEV1A and BdUEV1B/C in complementing the yeast mms2 mutation, we entertained a hypothesis that their encoded proteins may have different subcellular distributions in plants. To test this hypothesis, three GFP-BdUEV1 fusion constructs were made, transiently transfected to tobacco (N. benthamiana) leaves by A. tumefaciens and their subcellular localization was monitored by fluorescence microscopy. As shown in Figure 7, both GFP-BdUev1B and GFP-BdUev1C were primarily found in the nucleus. In sharp contrast, GFP-BdUev1A was excluded from the nucleus, while truncation of its C-terminal 18 amino acids (BdUev1A-ΔCT) results in its redistribution to the nucleus, reminiscent of the subcellular localization patterns of GFP-BdUev1B and GFP-BdUev1C. Since the DDT function must occur in the nucleus, the above observations further suggest that BdUev1B and BdUev1C are involved in plant DNA-damage response while BdUev1A is not. Meanwhile, this observation indicates that the C-terminal tail of BdUev1A plays a critical role in its subcellular localization and physiological functions.
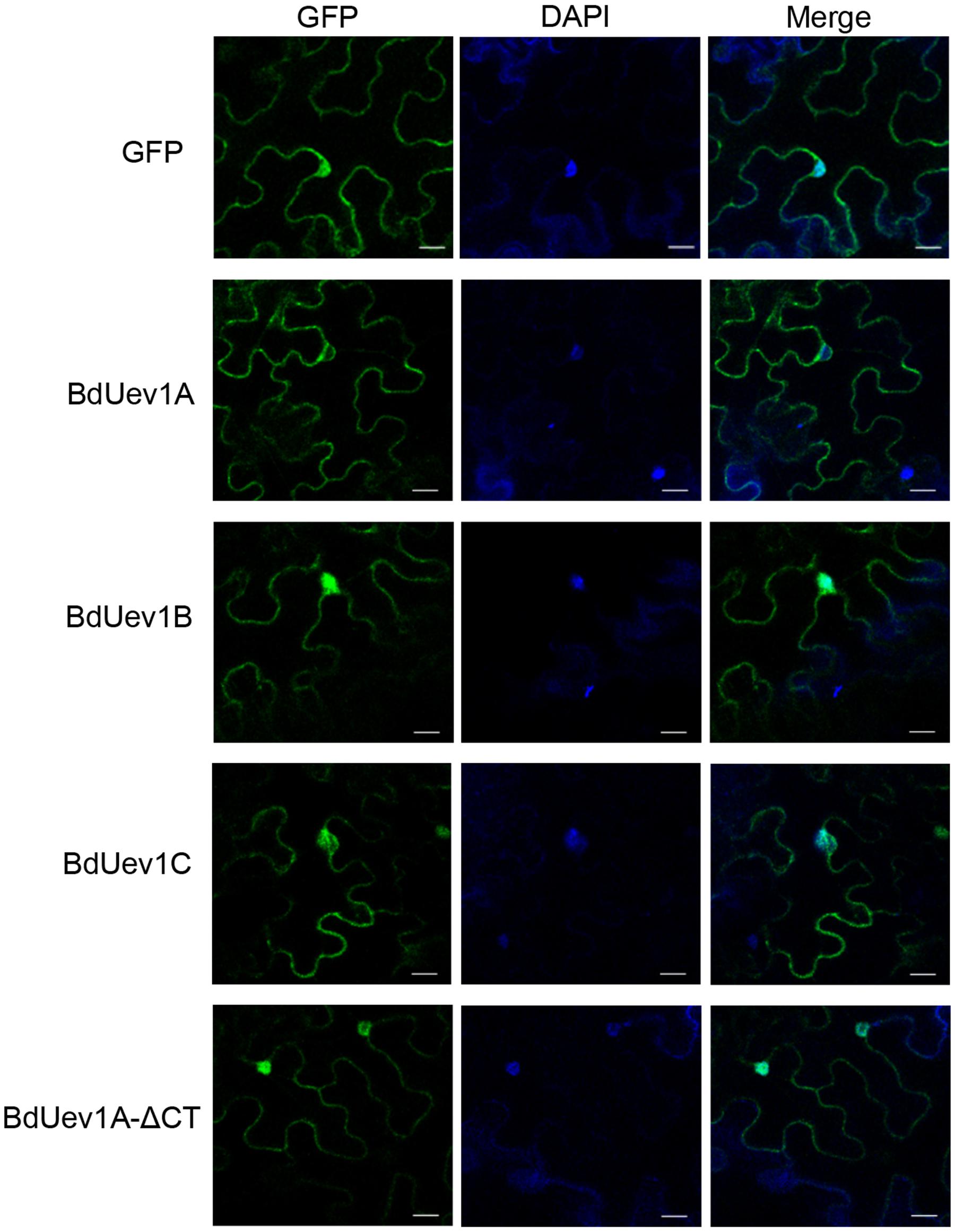
FIGURE 7. Subcellular localization of GFP-BdUev1s. Left panels, transient expression of GFP and GFP-tagged BdUev1s in tobacco epidermal cells visualized by epifluorescence microscopy. Middle panels, DAPI staining to visualize nuclei. Right panels, merge of images from corresponding fluorescence and DAPI staining. Bars = 20 μm.
Expression of BdUEV1s during Development and in Response to Abiotic Stresses
In a recent study, we found no obvious fluctuation in the expression of BdUBC13 genes in different tissues or during plant development (Guo et al., 2016). This expression pattern is consistent with UBC13 genes from Arabidopsis (Wen et al., 2006) and rice (Zang et al., 2012). In contrast, the expression of Arabidopsis UEV1 genes appears to fluctuate in different tissues and during development (Wen et al., 2008). To investigate the BdUEV1 gene expression during development, total RNA was extracted from 7-day seedlings as well as different tissues and the BdUEV1 transcript levels were determined by ddPCR. As shown in Figure 8A, the three BdUEV1 genes have a relatively uniform expression in different tissues except for BdUEV1C in L2 (leaves from transition phase) and Sp8 (spikes from 8-week old) phases. The BdUEV1B expression also has a significant but moderate increase during L2.
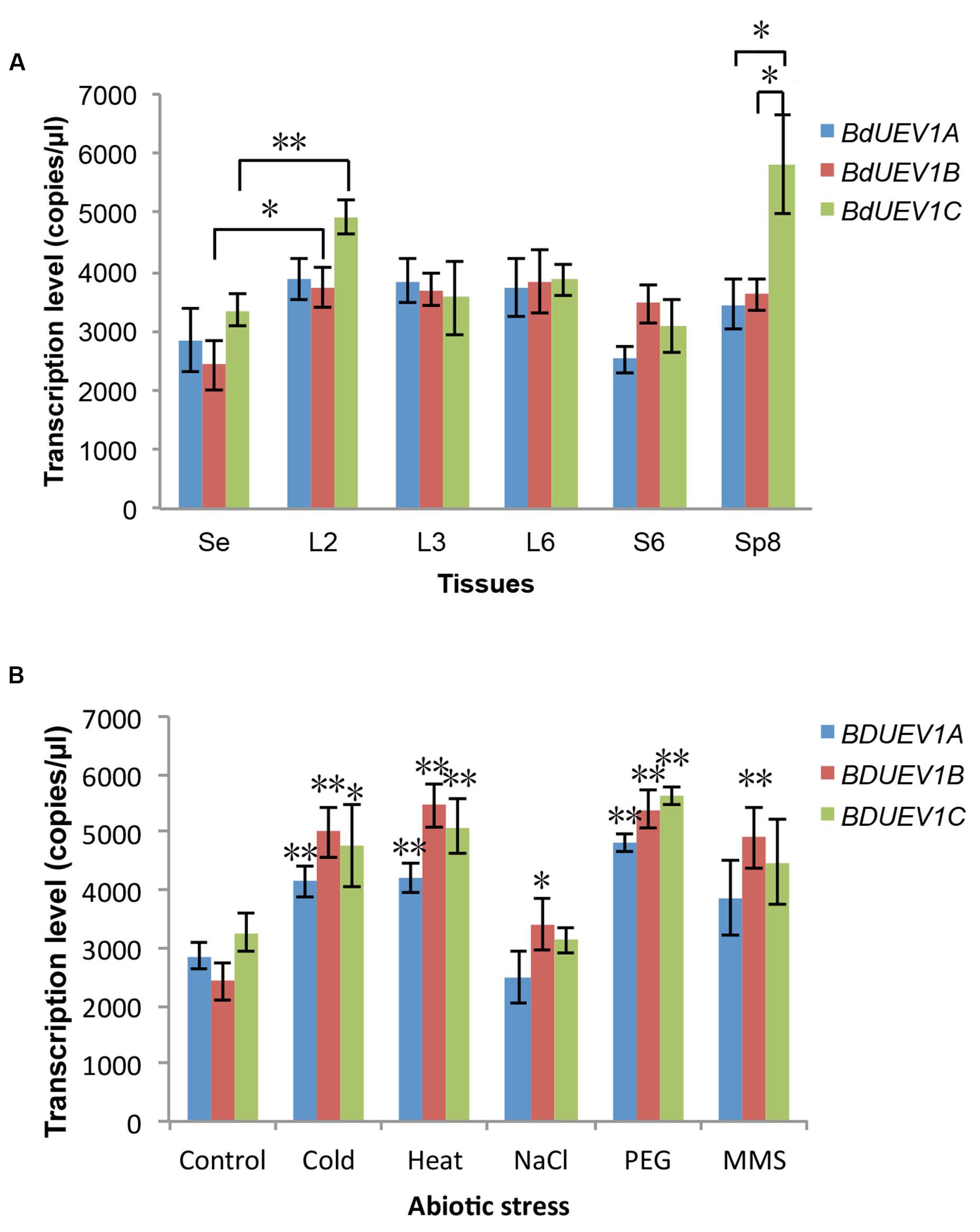
FIGURE 8. Quantitative analysis of expression of BdUEV1s. (A) Expression levels of BdUEV1s in different tissues during B. distachyon development. Samples were taken from different developmental stages and tissues as indicated. Se: 7-day seedling; L2: leaves from 2-week old plants; L3: leaves from 3-week old plants; L6 and S6: leaves and stems from 6-week old plants; Sp8: spikes from 8-week old plants. (B) The expression of BdUEV1s under different abiotic stresses. Control: 7-day old seedlings; Cold: 7-day old seedlings were treated at 4°C for 12 h; Heat: 7-day old seedlings were treated at 42°C for 12 h; PEG: 7-day old seedlings were treated with 25% PEG for 12 h; MMS: 7-day old seedlings were treated with 0.01% MMS for 12 h. ∗P < 0.05; ∗∗P < 0.01.
We also analyzed BdUEV1 expression in response to abiotic stress using ddRT-PCR (Figure 8B). It is of great interest to note that cold stress induces the expression of all three BdUEV1s reminiscent of cold-induced BdUBC13 expression (Guo et al., 2016). The transcription level of all BdUEV1s also significantly increased after heat and PEG treatments. On the other hand, only BdUEV1B expression has a twofold increase after MMS treatment, suggesting the involvement of BdUEV1B in DNA-damage response.
Discussion
In this study, we isolated and characterized three B. distachyon UEV1 genes encoding Ubc-like proteins. Our observations collectively indicate that there are two classes of UEV genes in B. distachyon as well as other plant species. First of all, genome database analysis shows that all known plant species contain several highly conserved UEV genes, in which at least one (e.g., BdUEV1B and BdUEV1C) encodes a short version of Uev1 and one (e.g., BdUEV1A) encodes one with an additional C-terminal tail. Secondly, phylogenetic analysis groups all plant Uevs with the additional C-terminal tail in one branch, indicating that the two UEV classes were independently evolved early in higher plants. Thirdly, it appears that BdUEV1B/C can fully rescue yeast mms2 null mutant phenotypes while BdUEV1A is only partially functional, and this difference becomes more obvious when BdUBC13 is used to replace yUBC13 in yeast cells. Fourthly, we found in this study that BdUev1A and BdUev1B/C have different subcellular distribution patterns in cultured tobacco cells. While BdUev1B/C proteins are primarily located in the nucleus, BdUev1A is excluded from the nucleus, suggesting that their physiological functions are different. Finally, we surprisingly found that when the C-terminal tail is deleted from BdUev1A, it behaves like BdUev1B/C with respect to both functional complementation of the yeast mms2 mutant and subcellular localization. The above observations along with a previous report on the AtUEV1D function (Wen et al., 2008) collectively allow us to speculate that BdUev1B/C are involved in nuclear functions including DNA-damage response, while BdUev1A plays roles in non-nuclear functions. Hence, the reduced ability for BdUEV1A to complement the yeast mms2 mutant is perhaps due to product exclusion from the yeast nucleus. It is interesting to note that plant UBC13 genes are expressed at a high level and do not fluctuate (Wen et al., 2006; Zang et al., 2012; Guo et al., 2016), and BdUbc13 is distributed to both nucleus and cytoplasm, as well as the cytoplasmic membrane (Guo et al., 2016), implying that UBC13 may serve as a housekeeping gene and different UEV1 genes encode regulatory subunits to modulate the heterodimer activity through spatial distribution and differential expression. The above envisioned plant UEV functions are reminiscent of the mammalian UEVs, in which UEV1A encodes a Uev with a unique 25-aa N-terminal sequence (Xiao et al., 1998). While both Uev1A and Mms2 are able to form stable complexes with Ubc13 and promote Lys63-linked polyubiquitination, they have distinct in vivo functions: Mms2 is involved in DNA-damage response but not NF-κB signal transduction, whereas Uev1A is required for the NF-κB activation but not for DNA-damage response (Andersen et al., 2005). Furthermore, expression of UEV1C, a UEV1 splicing variant lacking the UEV1A sequence coding for its N-terminal unique region, failed to confer NF-κB activation (Wu et al., 2014).
Lys63-linked polyubiquitination is found to be involved in several important cell signaling pathways, including DDT (Brusky et al., 2000), DNA double-strand break repair (Thorslund et al., 2015), inflammation and immunity (Deng et al., 2000; Chang et al., 2012), protein endocytosis (Marchal et al., 2000; Soetens et al., 2001), mitochondrial inheritance (Fisk and Yaffe, 1999) and in ribosome function (Spence et al., 2000). Ubc13 is the only known E2 dedicated to catalyzing Lys63-linked Ub chain assembly and this activity absolutely requires Uev as a cofactor (Hofmann and Pickart, 1999; McKenna et al., 2001). While plant Ubc13 has been shown to be involved in several cellular processes, in the majority of cases, its cognate E3 is unclear and whether it requires Uev as a cofactor is not known. We anticipate that if Lys63-linked polyubiquitination of the target protein is required for the Ubc13 function, it must need a Uev as a cofactor. In this respect, it is of great interest to determine which UEV1 gene(s) is required for these processes.
Author Contributions
Experimental design: HG, RW, and WX; Experiments: HG, RW, and QW; manuscript preparation: HG and WX; Supervision, funding, and reagents: WX and RD.
Funding
This work is supported by the National Natural Science Foundation of China (31270823) and Beijing Municipal Commission of Education to WX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Zhi Liu, Qian Wang, Yiran Zhang and Yuepeng Zang for reagents and technical assistance and Michelle Hanna for proofreading the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01551
References
Andersen, P. L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., et al. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170, 745–755. doi: 10.1083/jcb.200502113
Bartel, P. L., and Fields, S. (1995). Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254, 241–263. doi: 10.1016/0076-6879(95)54018-0
Blackwell, S., Hanna, M. D., and Xiao, W. (2014). Spontaneous mutagenesis assay. Methods Mol. Biol. 1163, 193–199. doi: 10.1007/978-1-4939-0799-1_14
Broomfield, S., Chow, B. L., and Xiao, W. (1998). MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5678–5683. doi: 10.1073/pnas.95.10.5678
Brusky, J., Zhu, Y., and Xiao, W. (2000). UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37, 168–174. doi: 10.1007/s002940050515
Catalan, P., Chalhoub, B., Chochois, V., Garvin, D. F., Hasterok, R., Manzaneda, A. J., et al. (2014). Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends Plant Sci. 19, 414–418. doi: 10.1016/j.tplants.2014.05.002
Chang, J. H., Xiao, Y., Hu, H., Jin, J., Yu, J., Zhou, X., et al. (2012). Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat. Immunol. 13, 481–490. doi: 10.1038/ni.2267
Chen, Z. J., and Sun, L. J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 33, 275–286. doi: 10.1016/j.molcel.2009.01.014
Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., et al. (2000). Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361. doi: 10.1016/S0092-8674(00)00126-4
Draper, J., Mur, L. A., Jenkins, G., Ghosh-Biswas, G. C., Bablak, P., Hasterok, R., et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. doi: 10.1104/pp.010196
Eddins, M. J., Carlile, C. M., Gomez, K. M., Pickart, C. M., and Wolberger, C. (2006). Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920. doi: 10.1038/nsmb1148
Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. doi: 10.1038/340245a0
Fisk, H. A., and Yaffe, M. P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199–1208. doi: 10.1083/jcb.145.6.1199
Guo, H., Wen, R., Liu, Z., Datla, R., and Xiao, W. (2016). Molecular cloning and functional characterization of two Brachypodium distachyon UBC13 genes whose products promote K63-linked polyubiquitination. Front. Plant Sci. 6:1222. doi: 10.3389/fpls.2015.01222
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. doi: 10.1146/annurev.biochem.67.1.425
Hochstrasser, M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439. doi: 10.1146/annurev.genet.30.1.405
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141. doi: 10.1038/nature00991
Hofmann, R. M., and Pickart, C. M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653. doi: 10.1016/S0092-8674(00)80575-9
Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168.
James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436.
Li, W., and Schmidt, W. (2010). A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 62, 330–343. doi: 10.1111/j.1365-313X.2010.04150.x
Marchal, C., Haguenauer-Tsapis, R., and Urban-Grimal, D. (2000). Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem. 275, 23608–23614. doi: 10.1074/jbc.M001735200
McKenna, S., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., Spyracopoulos, L., et al. (2003). An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13. The structural basis for lysine 63 chain catalysis. J. Biol. Chem. 278, 13151–13158. doi: 10.1074/jbc.M212353200
McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., et al. (2001). Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J. Biol. Chem. 276, 40120–40126. doi: 10.1074/jbc.M102858200
Moraes, T. F., Edwards, R. A., McKenna, S., Pastushok, L., Xiao, W., Glover, J. N., et al. (2001). Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat. Struct. Biol. 8, 669–673. doi: 10.1038/90373
Mural, R. V., Liu, Y., Rosebrock., T. R., Brady, J. J., Hamera, S., Connor, R. A., et al. (2013). The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25, 3615–3631. doi: 10.1105/tpc.113.117093
Pastushok, L., Moraes, T. F., Ellison, M. J., and Xiao, W. (2005). A single Mms2 key residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. J. Biol. Chem. 280, 17891–17900. doi: 10.1074/jbc.M410469200
Pastushok, L., Spyracopoulos, L., and Xiao, W. (2007). Two Mms2 residues cooperatively interact with ubiquitin and are critical for Lys63 polyubiquitination in vitro and in vivo. FEBS Lett. 581, 5343–5348. doi: 10.1016/j.febslet.2007.10.028
Pickart, C. M. (2000). Ubiquitin in chains. Trends Biochem. Sci. 25, 544–548. doi: 10.1016/S0968-0004(00)01681-9
Soetens, O., De Craene, J. O., and Andre, B. (2001). Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276, 43949–43957. doi: 10.1074/jbc.M102945200
Spence, J., Gali, R. R., Dittmar, G., Sherman, F., Karin, M., and Finley, D. (2000). Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102, 67–76. doi: 10.1016/S0092-8674(00)00011-8
Thorslund, T., Ripplinger, A., Hoffmann, S., Wild, T., Uckelmann, M., Villumsen, B., et al. (2015). Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393. doi: 10.1038/nature15401
Tsui, C., Raguraj, A., and Pickart, C. M. (2005). Ubiquitin binding site of the ubiquitin E2 variant (UEV) protein Mms2 is required for DNA damage tolerance in the yeast RAD6 pathway. J. Biol. Chem. 280, 19829–19835. doi: 10.1074/jbc.M414060200
VanDemark, A. P., Hofmann, R. M., Tsui, C., Pickart, C. M., and Wolberger, C. (2001). Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720. doi: 10.1016/S0092-8674(01)00387-7
Waadt, R., and Kudla, J. (2008). In Planta visualization of protein interactions using Bimolecular Fluorescence Complementation (BiFC). CSH Protoc. 2008:dbrot4995.
Wen, R., Li, J., Xu, X., Cui, Z., and Xiao, W. (2012). Zebrafish Mms2 promotes K63-linked polyubiquitination and is involved in p53-mediated DNA-damage response. DNA Repair (Amst) 11, 157–166. doi: 10.1016/j.dnarep.2011.10.015
Wen, R., Newton, L., Li, G., Wang, H., and Xiao, W. (2006). Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 61, 241–253. doi: 10.1007/s11103-006-0007-x
Wen, R., Torres-Acosta, J. A., Pastushok, L., Lai, X., Pelzer, L., Wang, H., et al. (2008). Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 20, 213–227. doi: 10.1105/tpc.107.051862
Wen, R., Wang, S., Xiang, D., Venglat, P., Shi, X., Zang, Y., et al. (2014). UBC13, an E2 enzyme for Lys63-linked ubiquitination, functions in root development by affecting auxin signaling and Aux/IAA protein stability. Plant J. 80, 424–436. doi: 10.1111/tpj.12644
Wu, Z., Shen, S., Zhang, Z., Zhang, W., and Xiao, W. (2014). Ubiquitin-conjugating enzyme complex Uev1A-Ubc13 promotes breast cancer metastasis through nuclear factor-small ka, CyrillicB mediated matrix metalloproteinase-1 gene regulation. Breast Cancer Res. 16:R75. doi: 10.1186/bcr3692
Xiao, W., Chow, B. L., Fontanie, T., Ma, L., Bacchetti, S., Hryciw, T., et al. (1999). Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 435, 1–11. doi: 10.1016/S0921-8777(99)00034-8
Xiao, W., Lin, S. L., Broomfield, S., Chow, B. L., and Wei, Y. F. (1998). The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 26, 3908–3914. doi: 10.1093/nar/26.17.3908
Xiao, W., and Samson, L. (1993). In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 90, 2117–2121. doi: 10.1073/pnas.90.6.2117
Xu, X., Ball, L., Chen, W., Tian, X., Lambrecht, A., Hanna, M., et al. (2013). The yeast Shu complex utilizes homologous recombination machinery for error-free lesion bypass via physical interaction with a Rad51 paralogue. PLoS ONE 8:e81371. doi: 10.1371/journal.pone.0081371
Xu, X., Blackwell, S., Lin, A., Li, F., Qin, Z., and Xiao, W. (2015). Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev. mutat. Res. 764, 43–50. doi: 10.1016/j.mrrev.2015.02.001
Xu, X., Lambrecht, A. D., and Xiao, W. (2014). Yeast survival and growth assays. Methods Mol. Biol. 1163, 183–191. doi: 10.1007/978-1-4939-0799-1_13
Yin, X. J., Volk, S., Ljung, K., Mehlmer, N., Dolezal, K., Ditengou, F., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19, 1898–1911. doi: 10.1105/tpc.107.052035
Keywords: Brachypodium distachyon, Uev1, Ubc13, K63-linked polyubiquitination, DNA-damage response
Citation: Guo H, Wen R, Wang Q, Datla R and Xiao W (2016) Three Brachypodium distachyon Uev1s Promote Ubc13-Mediated Lys63-Linked Polyubiquitination and Confer Different Functions. Front. Plant Sci. 7:1551. doi: 10.3389/fpls.2016.01551
Received: 04 August 2016; Accepted: 03 October 2016;
Published: 18 October 2016.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Hongyong Fu, Institute of Plant and Microbial Biology, Academia Sinica, TaiwanWan-sheng Lo, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan
Copyright © 2016 Guo, Wen, Wang, Datla and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Xiao, d2VpLnhpYW9AdXNhc2suY2E=
†These authors have contributed equally to this work.
 Huiping Guo
Huiping Guo Rui Wen
Rui Wen Qianqian Wang
Qianqian Wang Raju Datla
Raju Datla Wei Xiao
Wei Xiao