- College of Horticulture, Nanjing Agricultural University, Nanjing, China
Skin color is a key quality attribute of fruits and how to improve fruit coloration has long been a major concern. 5-Aminolevulinic acid (ALA), a natural plant growth regulator, can significantly increase anthocyanin accumulation in fruit skin and therefore effectively improve coloration of many fruits, including apple. However, the molecular mechanism how ALA stimulates anthocyanin accumulation in fruit skin remains unknown. Here, we investigated the impact of ALA on apple skin at the protein and mRNA levels. A total of 85 differentially expressed proteins in apple skins between ALA and water treatment (control) were identified by complementary gel-based and gel-free separation techniques. Most of these differentially expressed proteins were up-regulated by ALA. Function analysis suggested that 87.06% of the ALA-responsive proteins were associated with fruit ripening. To further screen ALA-responsive regulators, we constructed a subtracted cDNA library (tester: ALA treatment; driver: control) and obtained 104 differentially expressed unigenes, of which 38 unigenes were indicators for the fruit ripening-related genes. The differentially changed proteins and transcripts did not correspond well at an individual level, but showed similar regulated direction in function at the pathway level. Among the identified fruit ripening-related genes, the expression of MdMADS1, a developmental transcription regulator of fruit ripening, was positively correlated with expression of anthocyanin biosynthetic genes (MdCHS, MdDFR, MdLDOX, and MdUFGT) in apple skin under ALA treatment. Moreover, overexpression of MdMADS1 enhanced anthocyanin content in transformed apple calli, which was further enhanced by ALA. The anthocyanin content in MdMADS1-silenced calli was less than that in the control with ALA treatment, but higher than that without ALA treatment. These results indicated that MdMADS1 is involved in ALA-induced anthocyanin accumulation. In addition, anthocyanin-related verification in apple calli suggested that the regulation of MdMADS1 on anthocyanin biosynthesis was partially independent of fruit ripening process. Taken together, our findings provide insight into the mechanism how ALA regulates anthocyanin accumulation and add new information on transcriptase regulators of fruit coloration.
Introduction
Skin color is a key quality attribute of apple fruit, and hence one of the most important factors determining apple market acceptance. Generally, well-colored red cultivars are preferred because consumers always associate the red color with some indication of fruit quality, such as maturity, nutrition, taste, and flavor. At commercial apple orchards of southern China, poor red coloration has been an important limiting factor of apple commodity value. Thus, how to promote apple fruit coloration has become a major concern.
Many attempts have been applied to improve red coloration in apple fruits. The traditional fruit production practices contain paper bagging (Ju, 1998) and covering the orchard floor with reflecting films (Meinhold et al., 2010). However, these methods demand a mass of manpower, material resources, and time, or even bring orchard pollution. By contrast, the application of plant growth substances has been proposed as an economically viable alternative. 5-Aminolevulinic acid (ALA), an essential biosynthetic precursor of tetrapyrrole molecules, acts as a new-type plant growth regulator. ALA has gained increasing attention because of its multiple physiological roles in plants, such as increasing plant resistance to various stresses, and improving plant photosynthesis (Akram and Ashraf, 2013; Murooka and Tanaka, 2014). In fruit production, ALA has been demonstrated to be effective for the promotion of fruit coloration in several fruit crops, including apple (Xie et al., 2013; Zhang L. Y. et al., 2015), peach (Guo et al., 2013), pear (Xiao et al., 2012), and litchi (Feng et al., 2015). Importantly, it was also reported that ALA significantly increased fruit interior quality (Gao et al., 2013; Zhang L. Y. et al., 2015). Furthermore, ALA is readily biodegradable and has no adverse effects on animals and humans (Perez et al., 2013). Therefore, ALA can simultaneously improve fruit coloration and fruit interior quality without any detrimental effects, suggesting great application prospect in fruit production.
Red coloration in various plant tissues is predominantly caused by anthocyanin, which accumulates as granules in the vacuole (Bae et al., 2006). This pigment belongs to the diverse group of ubiquitous secondary metabolites collectively known as flavonoids. In plants, two categories of genetic control relate to anthocyanin accumulation. One category is the biosynthetic genes that encode enzymes required for anthocyanin biosynthesis, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glycose: flavonoid 3-O-glucosyl- transferase (UFGT) (An et al., 2015). All of these six genes have been isolated in various plants and their transcription levels are positively correlated with anthocyanin concentration (Han et al., 2010; Feng et al., 2013). Another category is regulatory genes that influence the intensity and pattern of anthocyanin biosynthetic genes. In this category, most studies on the regulation of anthocyanins have focused on transcription factors of R2R3-MYB, basic helix-loop-helix (bHLH), and WD40 classes. These three regulators can form a MYB-bHLH-WD40 protein complex that binds to promoters and then induces transcription of anthocyanin biosynthetic pathway genes. In recent two decades, additional potential regulators have also been reported in model plant Arabidopsis thaliana to affect anthocyanin synthesis, including PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), LONG HYPOCOTYL 5 (HY5), CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), WRKY, WIP domain, MADS-box domain, NAC (NAM, ATAF, CUC), Jasmonate ZIM-domain (JAZ), and the SQUAMOSA promoter-binding protein-like (SPL) (Zhou et al., 2015).
Several studies have been conducted to investigate the regulatory mechanisms behind anthocyanin accumulation in apple. Conserved MYB, bHLH, and WD40 genes in the apple that are homologs of Arabidopsis MYB-bHLH-WD40 protein complex have been demonstrated to be responsible for the accumulation of anthocyanins (Takos et al., 2006; An et al., 2012; Xie et al., 2012). Likewise, new regulators involved in anthocyanin biosynthesis were identified in apple fruits. For example, MdCOP1 has been demonstrated to be involved in the ubiquitination and degradation of the MdMYB1 protein under dark conditions (Li et al., 2012) and MdJAZ2 has been proposed to be involved in the regulation of anthocyanin accumulation during the response of apple fruits to jasmonate (An et al., 2015). Since the regulatory mechanism modulates anthocyanin biosynthesis is highly conserved in higher plants, more research is necessary to develop the anthocyanin regulation network in apple. Research on ALA promoting anthocyanin accumulation in apple fruits has been linked to up-regulating anthocyanin biosynthetic genes and regulatory genes MYB, bHLH, and WD40 (Xie et al., 2013). However, little information is available regarding special regulative effects of ALA on fruit skin and its regulatory mechanisms remain unknown. Current knowledge about the function of ALA on fruit is derived from research on some physiological aspects of fruit growth and ripening. Therefore, an overall molecular framework is needed for better understanding the ALA-associated fruit coloration.
Proteomic and transcriptomic techniques are often used to investigate the molecular mechanisms involved in complex traits. To make a comprehensive understanding of ALA-stimulated fruit coloration, integrated proteomic, and transcriptomic techniques were employed in this study. We identified and analyzed ALA-induced various changes at protein and mRNA levels using gel-free and two-dimensional gel electrophoresis (2-DE) gel-based proteomic techniques and suppression subtractive hybridization (SSH). Based on the results of proteomics and SSH, a candidate biomarker MdMADS1 was selected to explore the molecular mechanism underlying ALA-induced anthocyanin accumulation. Our data offers new molecular evidence elucidating the regulatory mechanism of fruit coloration by ALA, and provides a valuable reference for further research on anthocyanin accumulation in apple fruits.
Materials and Methods
Fruit Source and Apple Flesh Calli Induction
Fruits were collected from apple (Malus × domestica Borkh. cv. Fuji) trees at commercial apple orchards of eastern China, Fengxian County in Jiangsu Province. All fruits were bagged with paper-bags in late May, debagged in early October, and the fruits were commercially harvested in late October. In this study, the debagged fruits which were collected from eight trees were harvested in early October (at onset of fruit coloration) and immediately transported to the laboratory for two different treatments. Solutions containing 0.01% Tween-20 alone (control), or with 200 mg/L ALA (treatment) were sprayed to the surface of debagged fruits. The fruits were then transferred into growth chamber with 150 μmol m−2 s−1 photon flux density (PFD) at 22°C and sampled at 0, 6, 12, 24, 36, 48, and 72 h of light exposure. At each of the sampling points, skins from 15 different fruits were collected and divided randomly into two groups. One group of skin samples was used for measurements of anthocyanin content, and the other was stored at −80°C for RNA and protein isolation after being frozen in liquid nitrogen. Since the time course of anthocyanin accumulation in apple skin showed that the promotion of ALA on anthocyanin accumulation initiated after 24 h light irradiation (See the “Results”), to identify the early upstream regulators of anthocyanin biosynthesis induced by ALA, we chose skins of apples that exposed to light for 24 h for the proteomics and SSH analysis.
Calli from “Fuji” fruit flesh were induced on Murashige and Skoog medium containing 1 mg/L 6-benzylaminopurine (BAP) and 1 mg/L 2,4-dichloropheno-xyacetic acid (2,4-D) at 25°C in the dark, and subcultured at 14-day intervals. For ALA treatment, the calli were transferred to medium containing 100 mg/L ALA. To induce anthocyanin accumulation, transgenic calli were cultured in a culture room under 100 μmol m−2 s−1 PFD at 25°C for 4 days, and then collected for determination of anthocyanin content and RNA isolation.
Measurement of Anthocyanin Content
Anthocyanin content was extracted using 1% (v/v) HCl-methanol for 24 h at room temperature in the dark. The extracts were centrifuged at 15,000 g for 15 min, and the absorbance at 530 nm was then measured with a spectrophotometer. The amount of anthocyanin was expressed as nmol of cyanidin-3-galactoside per g of the sample using a molar extinction coefficient of 3.43 × 104 (Ubi et al., 2006). Mean values were obtained from five independent replicates.
Proteomic Analysis by 2-DE
Protein extraction of apple skin was performed using phenol-based method (Deytieux et al., 2007). The final pellet was dissolved in a solution containing 7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1- propanesulfonate (CHAPS), 1% (w/v) dithiothreitol (DTT), and 0.5% (v/v) immobilized pH gradient (IPG) buffer (pH 4–7; GE Healthcare, USA). The protein concentration was quantified according to the method suggested by Bradford (1976), using bovine serum albumin as standard.
For 2-DE, protein samples (1 mg) were brought to 450 μL of isoelectric focusing (IEF) rehydration solution (7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 1% (w/v) DTT, 0.5% (v/v) IPG buffer, and 0.01% bromphenol blue). The whole volume was transferred into a well of the Immobiline DryStrip Reswelling Tray and IPG strips (24 cm, pH 4–7; GE Healthcare) were rehydrated overnight at 20°C. The strips were then loaded onto an Ettan IPGphor 3 instrument (GE Healthcare), and IEF was performed according to the following steps: 100 V for 1 h, 500 V for 1 h, followed by 8 h gradient from 1000 to 10000 V, and finally focused for 65,000 Vh at 10,000 V. The maximum current per strip was set at 50 μA. After two-step equilibration, the IPG strips were loaded on a 12% w/v sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels using the Ettan Daltsix system (GE Healthcare). The gels were run at 150 V until the dye front reached the bottom of the gel. The gels were visualized using the Coomassie Brilliant Blue G-250 stain and scanned using Image Scanner software (GE Healthcare). Afterward, gel images were processed using the PDQuest 2-DE analysis software (Version 8.0.1, Bio-Rad, USA) in a three-step manner: spot detection, volumetric quantification, and matching. Differences in protein content were analyzed using a t-test and calculated as the fold ratio in three biological replicates with two technical replicates. A threshold of P < 0.05 and fold change of >1.5 or <0.67 was used to identify significant differentially expressed protein spots.
Spots from 2-DE were excised from the gel and digested with trypsin. Then, samples were re-suspended with 5 μL 0.1% TFA followed by mixing in 1:1 ratio with a matrix consisting of a saturated solution of α-cyano-4-hydroxy-trans-cinnamic acid in 50% ACN containing 0.1% TFA. One microliter mixture was spotted on a stainless steel sample target plate. Peptide mass spectrometry (MS) and MS/MS were performed on an ABI 5800 MALDI-TOF/TOF Plus mass spectrometer (Applied Biosystems, USA). Data were acquired in the positive MS reflector using a CalMix5 standard to calibrate the instrument (ABI5800 Calibration Mixture). Both the MS and MS/MS data were integrated and processed using GPS Explorer V3.6 software (Applied Biosystems, USA) with default parameters. Based on the combined MS and MS/MS spectra, proteins were successfully identified based on 95% or higher confidence interval of their scores in MASCOT V2.3 search engine (Matrix Science Ltd, London, U.K.), using the following search parameters: the apple expressed sequence tag (EST) database (32,768 entries, Jan. 14th 2014) downloaded from the Genome Database for Rosaceae (GDR) (https://www.rosaceae.org/species/malus/malus_x_domestica/genome_v1.0); trypsin as the digestion enzyme; one missed cleavage site; fixed modifications of Carbamidomethyl (C); partial modifications of Acetyl (Protein N-term), Deamidated (NQ), Dioxidation (W), Oxidation (M); 100 ppm for precursor ion tolerance, and 0.5 Da for fragment ion tolerance.
The functional annotation of the identified proteins was based on UniProt, GDR, NCBInr protein database, and the literature.
Proteomic Analysis by Label-Free
For shotgun analysis, apple skin proteins (100 μg) dissolved in 6 M urea and 50 mM Tris-HCl (pH 8.0) were reduced by added 1 M DTT until at final concentration of 4 mM for 1 h at 60°C, and then added with 1 M iodoacetamide until at a final concentration of 25 mM for 45 min at 25°C in the dark. The 6 M urea was removed by ultrafiltration in case it influenced digestion. Proteins were dissolved in 50 mM NH4HCO3 (pH 7.8) and then treated with trypsin (2 μg, Promega, USA) at 37°C for 12 h. Finally, protein desalted using a C18 column (Empore) and freeze-dried before sample injection.
A liquid chromatography-MS (LC-MS) system consisting of a Dionex Ultimate 3000 nano-LC system (nano UHPLC, Sunnyvale, CA, USA), connected to a linear quadrupole ion trap Orbitrap (LTQ Orbitrap XL) mass spectrometer (Thermo Electron, Bremen, Germany), and equipped with a nanoelectrospray ion source. For LC separation, an Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex, Sunnyvale, CA, USA) capillary with a 15 cm bed length was used with a flow rate of 300 nL/min. Two solvents, A (0.1% formic acid) and B (aqueous 80% acetonitrile in 0.08% formic acid), were used to elute the peptides from the nanocolumn. The gradient went from 5 to 40% B in 32 min and from 40 to 95% B in 1 min, with a total run time of 60 min. The mass spectrometer was operated in the data-dependent mode so as to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Electrospray voltage and the temperature of the ion transfer capillary were 2.2 kV and 200°C, respectively. Survey full scan MS spectra (from m/z 350 to 1800) were acquired in the Orbitrap with a resolution r = 60,000 at m/z 400, allowing the sequential isolation of the top ten signal intensity ions for collision-induced dissociation at a collision energy of 35 V. A dynamic exclusion mode was enabled to exclude the previously selected ions during the repeated cycle of 60 s. The external mass calibration of the Orbitrap was performed once every 3 days to ensure a working mass accuracy <5 ppm. For each run, 1.5 μg of the digest was injected on a reverse-phase C18 column.
The obtained MS/MS spectra were searched against the apple EST database using SEQUEST algorithm in Proteome Discoverer 1.3 software (Thermo Scientific, San Jose, CA, USA). Search results were filtered for a False Discovery rate of 1% employing a decoy search strategy utilizing a reverse database.
For quantitative proteome analysis, four MS raw files from each group were analyzed using SIEVE software (Version 2.0, Thermo Scientific, USA). Eight MS raw files were performed the SIEVE experimental workflow of “two sample differential analysis,” where ALA–treated samples were compared to control samples. For the alignment step, the chromatographic peaks detected by Orbitrap were aligned by their retention time (± 2.5 min) and mass (± 0.02 unit) among all sample runs. After alignment, the feature detection and integration (or framing) process were performed using the MS level data with a feature called “Frames from MS2 Scans” only. The parameters consisted of a frame m/z width of 10 ppm and a retention time width of 2.5 min. Then, the peak integration was performed for each frame and these values were used for statistical analysis. Next, peptide sequences obtained from the database search were imported into SIEVE. A frame filter rule which was defined as “PRRoot > 0 and GoodID > 0 and CV_ALA treatment <25 and CV_control <25” was applied to obtain confident overall protein ratio. Peptides were grouped into proteins and a protein ratio and P-value were calculated. Only proteins observed in all two groups were quantified. The quantified proteins were considered as significantly different expressed proteins if they matched at least two peptides, and changed over 1.5-fold cutoff (ratio above 1.5 or below 1/1.5) with P-value < 0.05.
Suppression Subtractive Hybridization (SSH)
Total RNA was isolated from apple skins by CTAB-LiCl methods (Jaakola et al., 2001). Isolation of poly A+ RNA from total RNA was performed by PolyATtract mRNA Isolation Systems Kit (Promega, USA) according to the manufacturer's instructions. The integrity of RNA was ascertained by electrophoresis on 1% agarose gel. The concentration of total RNA and poly A+ RNA was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific, USA).
The SSH library was carried out using the PCR-Select™ cDNA Subtraction Kit (Clontech, USA) according to the manufacturer's instructions, starting with 2 μg poly A+ RNA from the tester (ALA) and driver (control) samples. After checking subtraction efficiency, subtracted second PCR products were cloned into the pMD-19T vector (Takara, Dalian, China) and then transformed into Trans1-T1 phage resistant chemically competent cells (TransGen Biotech, China). Subsequently, white colonies from Luria-Bertani (LB) solid medium plates containing ampicillin, 5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside (X-Gal), and isopropyl-β-D-1- thiogalactopyranoside (IPTG) were selected for colony polymerase chain reaction (PCR). The PCR amplification used the primer set of Nested PCR primer 1 and Nested PCR primer 2R provided along the SSH kit. Then, clones which PCR products were single inserts and longer than 100 bp insertion fragment size were sequenced with the universal M13 sequencing primer. After removing vector and adaptor sequences from the raw EST sequences, the resulting ESTs were assembled into unigenes using iAssembler (Zheng et al., 2011). Annotation and function analysis of the resulting unigenes were compared against the apple EST database.
Transfection of Apple Fruit Calli by Agroinfiltration
In order to overexpress MdMADS1 (Accession No. U78947), the open reading frame was amplified by PCR from cDNA of Fuji apple fruits skin with primers MdMADS1(OE)-F and MdMADS1(OE)-R (Supplementary Table S1), followed by ligation to the linearized plant transformation vector pBI121 based on homologous recombination method using the NovoRec® PCR Seamless Cloning Kit (Novoprotein Scientific Inc, China).
To silence MdMADS1, a 259 base pair (bp) fragment including 58 bp sequence homologous to the RNA-interference (RNAi) vector pHELLSGATE2 and 201 bp of MdMADS1 coding sequence was amplified by RT-PCR using primers MdMADS1(i)-F and MdMADS1(i)-R (Supplementary Table S1). Amplified fragments were transferred to the RNAi vector through Gateway BP reaction generating RNAi construct in which the sense and antisense MdMADS1 RNA sequences would be linked in tandem separated by the PDK intron.
The resulting recombinant plasmids and empty plasmids were transformed into Agrobacterium strain EHA105. One single separated colony containing the target gene was grown at 28°C in LB broth supplemented with appropriate antibiotics. When the optical density at 600 nm (OD600) of the culture liquid reached approximately 0.5, Agrobacterium cells were harvested and resuspended in a modified MacConkey agar (MMA) medium (Murashige and Shoog salts, 10 mM morpholine ethane sulphonic acid pH 5.6, 20 g/L sucrose). The fresh calli were soaked into the Agrobacterium solution for 15 min. The calli were co-cultured on Murashige and Skoog medium containing 1 mg/L 2, 4-D and 1 mg/L BAP for 2 d at 25°C in the dark. Subsequently, the calli were washed 5 times with sterile water and transferred to MS medium supplemented with 250 mg/L of carbenicillin and 30 mg/L kanamycin for transgenic selection.
Quantitative Real-Time Reverse Transcription-PCR
The cDNA synthesis was performed using the PrimeScript First-Stand cDNA Synthesis Kit (TaKaRa Bio, China). Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed with SYBR® Premix Ex Taq™ (TaKaRa Bio, China) as described by the manufacturer's instructions. Using specific primers (Supplementary Table S1), the transcript levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), where MdACTIN gene was used as an internal reference. In this study, the relative expression levels of corresponding genes were presented as values relative to corresponding indicated samples. Data were derived from 3 independent replicates.
Statistical Analysis
All data were obtained from at least three independent experiments. Statistical and correlation analysis was performed using SPSS statistical computer package (version 19.0 SPSS Inc. Chicago. IL). Data was compared with the control or among treatments by analysis of variance (ANOVA) to discriminate significant differences at P < 0.05 followed by least significant difference tests (LSD).
Results
ALA Promoted Anthocyanin Accumulation in the Skin of Debagged Apple Fruits
To confirm the effect of ALA on apple fruit coloration and provide more detailed information on this process, we investigated the time course of anthocyanin accumulation in apple skin after ALA treatment. Results showed that ALA treatment notably promoted fruit coloration within 72 h of light irradiation (Figure 1A). Anthocyanin determination showed that ALA did not stimulate anthocyanin accumulation within 24 h, but significantly increased anthocyanin content after 24 h, compared with the control (Figure 1B). Anthocyanin content in ALA-treated apple skin was about 23, 50, and 45%, respectively, higher than that in control at 36, 48, and 72 h. These results indicated that ALA-induced upstream regulatory factors of anthocyanin accumulation probably function before 36 h. Therefore, to identify the early upstream regulators of anthocyanin biosynthesis induced by ALA, skins of apples that exposed to light for 24 h were used for the following proteomics and SSH analysis.
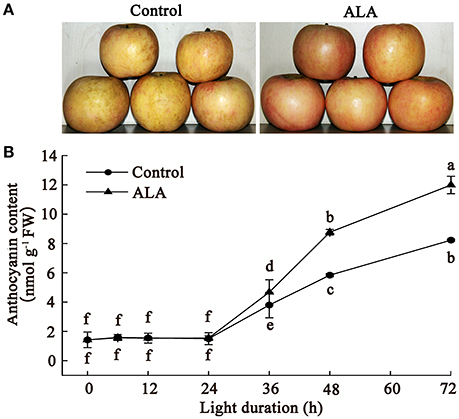
Figure 1. ALA induces anthocyanin accumulation in apple skin. The bagged fruits from Fuji apple trees were collected in early October and immediately transported to the laboratory for 200 mg/L ALA treatment and water treatment (control). Then, fruits coloration was induced in a growth chamber with 150 μmol m−2 s−1 photon flux density at 22°C. (A) A photo displays the color difference between ALA treatment and the control in apple skin at 72 h light duration. (B) Time courses of anthocyanin accumulation after ALA application. The different small letters represent significant differences at P = 0.05 level.
Protein Identification Using Gel-Based and Gel-Free Proteomics
To explore the mechanism how ALA regulated anthocyanin accumulation, we measured changes in the abundance of proteins and compared them between control and ALA-treated fruit skin using 2-DE gel-based proteomics. Analysis of the 2-DE pattern revealed that nearly 500 resolved spots were detected after ignoring very faint spots and spots with undefined shapes and areas (Figure 2). To analyze ALA-responsive proteins, the changes in spot intensity between ALA-treated and control were quantified by PDQuet software. Quantitative analysis of spot intensity by integration of the staining signal for each gel revealed that levels of 57 proteins changed in an ALA-dependent manner (ratio > 1.5 or ratio <0.67) in three independent replications. Using MALDI-TOF/TOF MS/MS, 56 protein spots representing 50 differently expressed proteins were then successfully identified according to the GDR database (Supplementary Table S2).
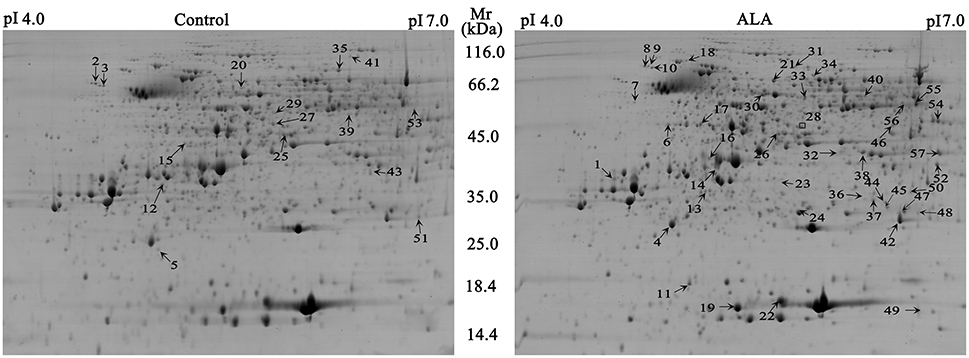
Figure 2. Representative 2-DE gel images of ALA treatment and the control in apple skin. Nearly 500 resolved spots were detected with Mr ranging from 116.0 to 14.4 kDa and pI ranging from 4.0 to 7.0. A total of 57 spots (marked with arrow and number) were identified as differentially expressed and of these, 15 proteins were down-regulated (left), whereas 42 proteins were up-regulated in ALA treatment (right). One spot was unsuccessfully identified (marked with square).
To obtain a more comprehensive understanding of proteins affected by ALA, we further performed label-free shotgun proteomics of apple skin. Data acquired by nano-ESI-MS/MS on a Q-Exactive spectrometer were processed with SIEVE software to reveal up- or down-regulated proteins by ALA. A total of four runs per group were analyzed and the quality of the alignment of the chromatographic peaks between ALA treatment group and control group was high (Supplementary Figure S1). Significant differences in protein abundance were considered with a ratio of ALA-treated/control higher than 1.5 or lower than 0.67 (P < 0.05). As a result, a total of 47 proteins were identified (Supplementary Table S3).
The identified proteins by gel-based and gel-free proteomics were further categorized into different classes (Figure 3A), where the 50 proteins identified using gel-based proteomics were mainly involved in stress response and defense, carbohydrate metabolism and energy, amino acid metabolism, nucleotide metabolism, lipid metabolism, nucleotide metabolism, and secondary metabolism. Similarly, the 47 proteins identified using gel-free proteomics were predominantly related to stress response and defense, carbohydrate metabolism and energy, amino acid metabolism and secondary metabolism.
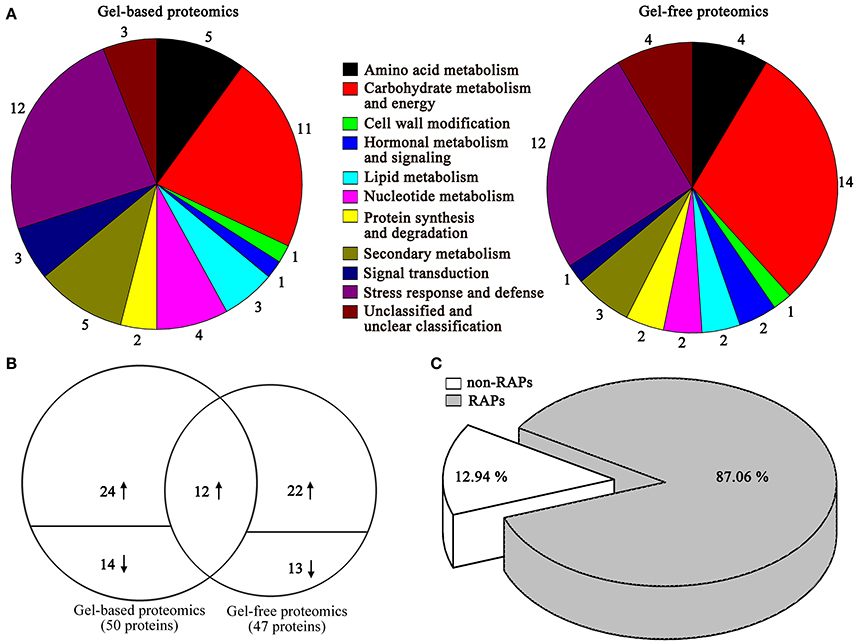
Figure 3. Proteomic analyses of differentially expressed proteins between ALA treatment and the control in apple skin using gel-free and gel-based techniques. (A) Primary functional distribution of 50 and 47 regulated gene products as identified by gel-free and gel-based techniques. (B) Venn diagram of the distribution of differentially expressed proteins identified by gel-free and gel-based techniques. The number above or below the horizontal line in each portion indicated the number of up-regulated or down-regulated proteins. The overlapping regions indicated the number of proteins commonly identified by the two proteomics techniques. (C) Further classification was carried out to identify proteins related to previous studies orthologous ripening associated proteins (RAPs).
Combined the results of gel-based and gel-free proteomics, a total of 85 differently expressed proteins were identified (Table 1, Figure 3B). Among these proteins, 72.82% of changed proteins were up-regulated by ALA, and 12 proteins were commonly identified by the two proteomics techniques (Supplementary Table S4). Moreover, among the total ALA-responsive proteins, about 87.06% of changed proteins are related to orthologous fruit ripening associated genes (Figure 3C), indicating that ALA might increase anthocyanin accumulation by regulating fruit ripening process.
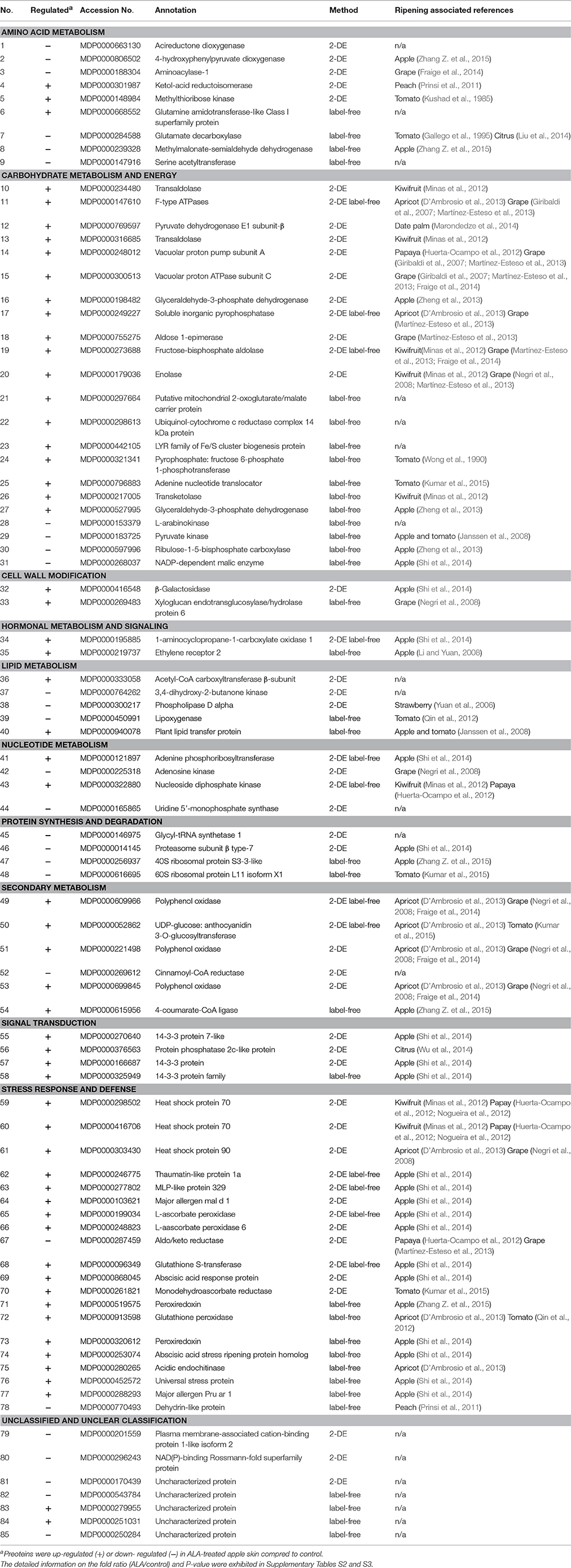
Table 1. Differently expressed proteins obtained by 2-DE and label-free analysis in ALA-treated apple skin.
Screening of ALA-Induced Genes from Apple Skins by Suppression Subtractive Hybridization
To further determine how ALA stimulates fruit coloration, a forward SSH library was constructed with mRNA isolated from ALA-treated to water-treated (control) apple peel (Figure 4A). Subtraction was performed between ALA-treated cDNA (tester) and the control cDNA (driver). Positive clones were selected for sequencing, and the vector and other uninformative sequences were removed. A total of 125 ESTs were successfully sequenced and identified in the apple database and then these ESTs were further assembled into 104 unigenes (Supplementary Table S5). The unigenes present in the forward SSH library were classified into 13 major functional groups, using information from various sources (Figure 4B). Among that, 10 out of 12 functional groups (not including unknown category) were regulated at both protein and mRNA levels. Genes involved in structural component and transcription factor were only detected at mRNA levels. In addition, 1-aminocyclopropane-1-carboxylate oxidase 1 (MDP0000195885), abscisic acid stress ripening protein (MDP0000253074), universal stress protein (MDP0000452572), glutamine amidotransferase-like class I superfamily protein (MDP0000668552), and an uncharacterized protein (MDP0000170439), were detected at both mRNA and protein levels.
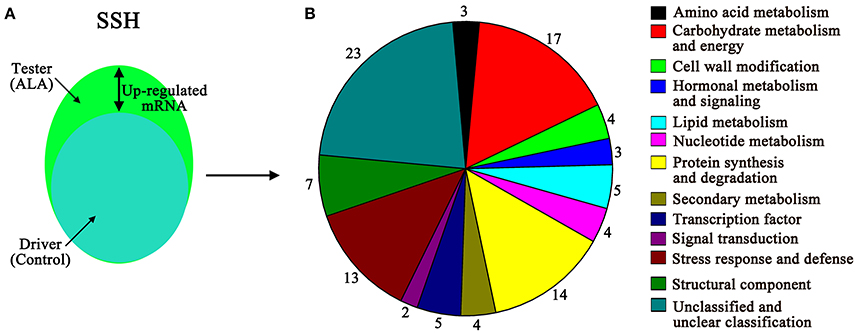
Figure 4. Up-regulated genes in response to ALA treatment were identified by SSH in apple skin. (A) Visual representation of the experimental plot of suppression subtractive hybridization (SSH). Tester, ALA-treated apple skin; driver, water-treated apple skin (control). (B) The function of unigenes from up-regulated genes pool was classified into 13 functional groups.
Thirty-eight unigenes have been reported to be relevant to fruit ripening (Table 2). The largest category, consisting of 9 genes, was associated with stress response and defense, such as major allergen Mal d 1, a universal stress protein, acidic endochitinase, and dehydrin family protein. Eleven genes were predicted to be associated with primary metabolism, including amino acid metabolism, carbohydrate metabolism and energy, lipid metabolism. Four genes were predicted to be associated with protein synthesis and degradation. In secondary metabolism group, two pigments biosynthesis genes (phytoene dehydrogenase and UFGT) and two aroma production gene (α-farnesene synthase and farnesyl pyrophosphate synthase) were identified. Among the differentially expressed genes, five genes associated with hormonal metabolism and signal transduction. Interestingly, a transcription factor MADS1, also referred to as MADS8 and SEPALLATA1 (Ireland et al., 2013) was identified from the SSH libraries, which accounted for ripening-related genes.
MdMADS1 Expression Is Positively Correlated with Anthocyanin Biosynthesis-Related Genes in ALA Treatment
The above results indicated that ALA-induced anthocyanin accumulation was probably associated with its regulation of fruit ripening process. Recently, MdMADS1 was reported to be involved in regulation of apple fruit ripening and MdMADS1- antisense lines showed inhibited fruit coloration (Ireland et al., 2013). Therefore, we speculated that the up-regulation of MdMADS1 expression might play an important role in ALA-induced anthocyanin accumulation. We confirmed by qRT-PCR that ALA indeed enhanced the expression of MdMADS1 at 12, 24, and 48 h of light irradiation (Figure 5), but decreased it at 72 h. These results indicated that MdMADS1 was induced by ALA application.
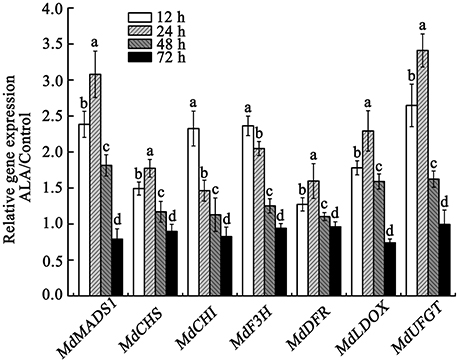
Figure 5. qRT-PCR analysis of the expression of MdMADS1 and anthocyanin biosynthetic genes in apple skin under ALA treatment. The relative expression of MdMADS1 and anthocyanin biosynthetic genes (MdCHS, MdCHI, MdF3H, MdDFR, MdLDOX, MdUFGT) were simultaneously analyzed in apple skin with ALA treatment at 12, 24, 48, and 72 h light duration. The expression level of each gene in control was used as a reference sample at each time point.
To show the potential role of MdMADS1 in ALA-induced anthocyanin accumulation, we simultaneously measured the expression of anthocyanin biosynthetic genes (MdCHS, MdCHI, MdF3H, MdDFR, MdLDOX, and MdUFGT) in response to ALA and investigated the relationship between the expression of MdMADS1 and anthocyanin biosynthetic genes. We found that the expression of MdMADS1 showed similar changing pattern to that of anthocyanin biosynthetic genes (MdCHS, MdDFR, MdLDOX, and MdUFGT) at different light duration (Figure 5). Correlation analysis further showed that the expression of MdMADS1 was significantly positively correlated with anthocyanin biosynthetic genes, except MdCHI and MdF3H (Table 3). These results suggested a positive role of MdMADS1 in ALA-induced improvement of anthocyanin biosynthesis in apple skin.
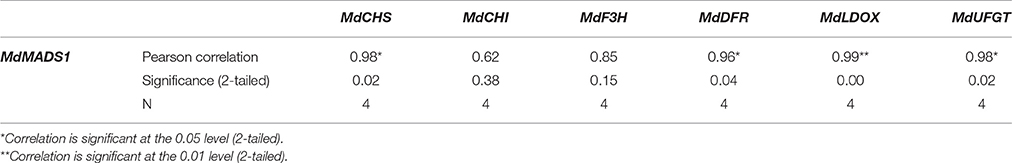
Table 3. Correlations between the relative expressions of MdMADS1 and anthocyanin biosynthetic genes under ALA treatment.
MdMADS1 Is Involved in Anthocyanin Regulation in Apple Calli
Callus was a reliable material for anthocyanin-related research (Lalusin et al., 2006; Xie et al., 2012; An et al., 2015). To confirm the role of MdMADS1 in ALA-induced anthocyanin accumulation, the gene was overexpressed and silenced in apple fruit calli, respectively. In overexpression lines (OE), the expression of MdMADS1 was significantly increased compared with the corresponding empty plasmids control [control(OE)], and in the silenced lines (RNAi), the expression was drastically reduced compared with the corresponding empty plasmids control [control(i)] (Figure 6A). Under light condition, the appearance revealed that OE calli looked redder in color than the control and RNAi calli (Figure 6B), suggesting that MdMADS1 overexpression increased anthocyanin accumulation. Spectrophotometric analysis further confirmed that OE calli contained significantly higher content of anthocyanin than the control and RNAi calli (Figure 6C), suggesting that MdMADS1 play a positive role in anthocyanin accumulation. The expression of the anthocyanin biosynthetic genes were significantly up-regulated and down-regulated in OE and RNAi calli than the control (Figures 6D,E), respectively, indicating that MdMADS1 regulates anthocyanin accumulation by modulating expression of anthocyanin biosynthesis-related genes.
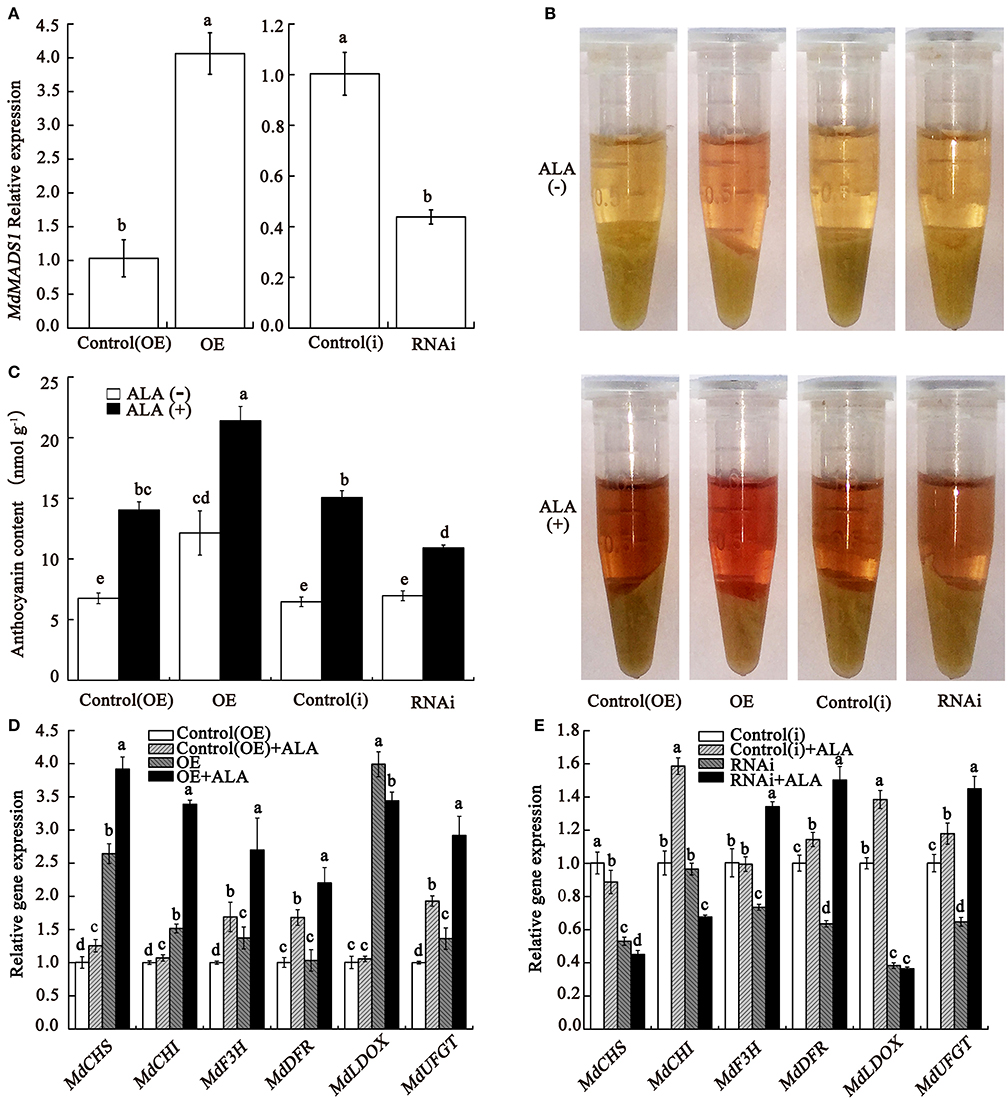
Figure 6. MdMADS1 involves in ALA-induced anthocyanin accumulation. The calli were induced from Fuji fruit on MS medium containing 1 mg/L BAP and 1 mg/L 2, 4-D at 25°C in the dark. (A) qRT-PCR analysis of the relative expression of MdMADS1 in transgenic calli. (B,C) Color (B) and anthocyanin content (C) in transgenic calli. (D,E) ALA regulated the expression of anthocyanin biosynthetic genes in overexpression (D) and RNAi (E) transgenic calli. The expression level of each gene in the control(OE) or the control(i) calli was used as a reference sample. The expression level of each gene in calli was used as a reference sample. OE, calli infiltrated with the plasmid for overexpressing MdMADS1; Control(OE), calli infiltrated with an empty pBI121 vector; RNAi, calli infiltrated with the plasmid for silencing MdMADS1; Control(i), calli infiltrated with an empty pHELLSGATE2 vector.
To determine whether MdMADS1 functioned in ALA-induced anthocyanin accumulation, we compared the anthocyanin content in transgenic calli which were treated with or without ALA. In Figure 6B, results showed that OE calli looked redder and accumulated the markedly higher level of anthocyanin after ALA treatment; ALA-treated RNAi calli were less red than ALA-treated control but much redder than the control without ALA. Spectrophotometric assay showed that ALA significantly promoted the anthocyanin accumulation in OE and RNAi calli (Figure 6C). In addition, we found that the expression levels of anthocyanin biosynthetic genes were higher in OE calli with ALA treatment than that in control(OE) calli with ALA treatment and OE calli, except MdLDOX (Figure 6D). Interestingly, ALA reversed the expression levels of three anthocyanin biosynthetic genes (MdF3H, MdLDOX, and MdUFGT) in RNAi transgenic calli (Figure 6E). These results indicated that MdMADS1 was involved in ALA-induced anthocyanin accumulation, but the latter was not completely dependent on the former.
Discussion
ALA has shown to be effective in promoting fruit coloration, yield and quality, so that it has great application potential in horticulture. However, little information is available on regulatory mechanisms behind ALA-induced fruit coloration. Here, two proteomic techniques and SSH were employed to identify the early responses of apple skin coloration to ALA treatment at protein and mRNA levels.
The difference in pretreatment, depth of proteome coverage, analyses of isoforms and quantification statistical power often result in poor correspondence between the proteins identified by different proteomic techniques (Scherp et al., 2011). For example, Yin et al. (2014) found only 9 of 115 in soybean root tips were commonly detected by gel-based and gel-free proteomics under flooding stress. Majeran et al. (2005) reported that among the 125 chloroplast proteins quantified in the three methods (2-DE, ICAT, and label-free), only 20 proteins were quantified in common. Moreover, the SDS-PAGE-based and gel-free-based proteomic techniques were combined to explore the molecular mechanism responsible for low silk production, a total of 17 of 174 changed proteins were common between the two techniques (Wang et al., 2014). Similarly, here, we identified 85 changed proteins using gel-based and gel-free techniques, among which only 12 proteins were commonly identified by two methods. Although most of the proteins were not identified simultaneously by different proteomic techniques, they showed similar altered trends in biological function (Figure 3A, Yin et al., 2014). Our data together with the above previous studies indicate that utilization of different proteomic approaches can lead to a more comprehensive proteome profiling, providing complementary information and hence a better understanding of the mechanisms.
Only five ALA-responsive genes were identified at both protein and mRNA levels, indicating that the expression levels of individual proteins were not strictly correlated with the up-regulated transcripts. Similar results have been reported in several previous studies (Lan et al., 2012; Arcondéguy et al., 2013; Wang et al., 2014). The regulation of mRNA synthesis, the post-transcriptional regulation of mRNA splicing and supervision mechanism, and the post-translational modification of the mRNA translated product can all result in altered protein levels (Arcondéguy et al., 2013). In addition, the different expression time course between mRNA and proteins also can lead to the divergence of mRNA and protein expression levels (Lan et al., 2012; Wang et al., 2014). Although the differentially changed proteins and transcripts did not correspond well at an individual level, the pathway analysis showed that proteome and transcriptome in the present study were well-matched. A total of 10 out of 12 transcriptome-involved pathways were found in the proteome-involved pathways, suggesting that the changed direction of proteins and transcripts were coordinated. Therefore, the proteomics and SSH techniques were mutually complementary and verified in this study, and provide valuable information on mechanisms involved in ALA-regulated apple skin.
Higher levels of anthocyanins are closely related to higher expression of anthocyanin biosynthetic genes. Previous studies have reported that exogenous ALA up-regulated the expression level of key genes in the pathway of anthocyanin biosynthesis (Xie et al., 2013; Feng et al., 2015). At protein level, a 4-coumarate-CoA ligase (No. 54 in Table 1), which catalyses the last step of the phenylpropanoid pathway leading to either lignins or flavonoids (Dixon et al., 2002), was induced by ALA treatment. UFGT is the last enzyme in anthocyanin biosynthetic pathway and has widely been considered as the key enzyme determining apple coloration (Li et al., 2002; Kim et al., 2003). Ju et al. (1995) found that anthocyanin biosynthesis in apple peel was most strongly correlated with UFGT activity. Here, the expression levels of UFGT (No. 50 in Table 1 and UN007 in Table 2) were up-regulated at both transcript and protein level under ALA treatment. These results indicate that up-regulation of biosynthetic genes contributes greatly to ALA-promoted anthocyanin accumulation. Except up-regulation of anthocyanin biosynthetic genes, ALA also repressed cinnamoyl-CoA reductase (No. 52 in Table 1) at protein level in the lignin biosynthetic pathway. Ludwig et al. (2013) reported that there was a competition between lignin and anthocyanin biosynthetic pathways for their common precursors. These results indicate that ALA helps divert the metabolic flux from lignin to anthocyanin pathway, which also contribute to the anthocyanin accumulation. Our results add new evidence supporting the positive regulation of ALA on anthocyanin accumulation.
ALA, as a potential plant growth regulator, is known to be effective in improving plant tolerance to various stresses (Akram and Ashraf, 2013). In this study, ALA alters the expression of numerous genes at mRNA and protein levels associated with various biological processes in apple skin. One of the outstanding abundant classes is protein and genes involved in stress response, indicating the positive role of ALA in defense response of apple skin to stresses. Anthocyanin itself, is a secondary metabolite which shows antioxidant activity (Alessio et al., 2011), and plays essential roles in ameliorating environmental stresses, such as UV-B radiation, drought, and cold temperatures (Chalker-Scott, 1999). In the red-fleshed apples, a recent study suggested that anthocyanin was both associated with the red coloration and the stress tolerance (Wang et al., 2015). Here, the effects of ALA on apple skin included anthocyanin accumulation as well as a stress response. Thus, it is reasonable to speculate that ALA might promote anthocyanins accumulation, participating in stress resistance of apple skin.
Another more abundant functional class in ALA treatment is related to primary metabolites involved in the metabolism of amino acid, sugar, and fatty acid. Li et al. (2014) found that the differences between anthocyanin concentrations in the pear peel of green “Anjou” and its bud mutation “Red Anjou” were accompanied with up-regulated of sorbitol metabolism and altered amino acid metabolism in the peel of “Red Anjou.” It is demonstrated that the manipulation of primary metabolism can change the production of secondary metabolites (Henkes et al., 2001; Dauwe et al., 2007). In this study, change of primary metabolism in apple skin under ALA treatment indicates that they may be crucial at the initial stage of ALA-induced anthocyanin accumulation. Therefore, we attempted to capture the potential links between primary metabolism and anthocyanin accumulation in ALA treatment (Figure 7). The first key step of the anthocyanin biosynthesis, catalyzed by chalcone synthase, involves two reaction substrates malonyl-CoA and 4-coumaroyl-CoA (Winkel-Shirley, 2001). In plants, the formation of malonyl-CoA from the carboxylation of acetyl-CoA in fatty acid chain elongation pathway is catalyzed by acetyl-CoA carboxylase, which is an ATP-dependent biotinylated protein complex (Sasaki and Nagano, 2014). The β subunit of this protein (No. 36 in Table 1) was identified here, and it appears positively regulated by ALA. Another substrate 4-coumaroyl-CoA is converted by phenylalanine. The formation of phenylalanine in shikimate pathway is connected to two intermediate metabolites, phosphoenolpyruvate of glycolysis pathway and erythrose 4-phosphate of the pentose phosphate pathway in carbohydrate metabolism (Tzin and Galili, 2010). Here, we found that genes related to glycolysis, such as pyrophosphate: fructose 6-phosphate 1-phosphotransferase (No. 24), glyceraldehyde-3-phosphate dehydrogenase (No. 16 and 27), enolase (No. 20), fructose-bisphosphate aldolase (No. 19), pyruvate kinase (No. 29), and pyruvate dehydrogenase E1 subunit β (No. 12) were regulated and at protein level (Table 1) while 6-phosphofructokinase (UN092) and pyruvate phosphate dikinase (UN016) was up-regulated at mRNA level (Table 2) under ALA treatment. Moverover, plastidial and cytosolic enolase, respectively, have specific functions in metabolism (Voll et al., 2009). In the present study, the subcelluar locations of enolase was predicted using an online tool Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant/. Chou and Shen, 2010), suggesting that glycolytic enzyme is a cytosolic protein. This data supported the suggestion that, cytosolic enolase plays a central role to modulate the synthesis of aromatic amino acids and secondary phenylpropanoid compounds, even in the absence of a complete glycolysis pathway in the plastids (Voll et al., 2009; Eremina et al., 2015; Negri et al., 2015). Therefore, our results suggest that carbon flux into anthocyanin biosynthesis is activated by ALA during fruit coloration period. Transaldolase links the pentose phosphate pathway to glycosis and it was up-regulated by ALA at both protein and mRNA levels (No. 10, 13, 26 in Table 1 and UN088 in Table 2). The high expression level of this gene could result in an accumulation of erythrose 4-phosphate. These results together indicate that the promotion of ALA-induced anthocyanin synthesis may also be associated with an ALA-enhanced supply of precursors from primary metabolism to secondary metabolism.
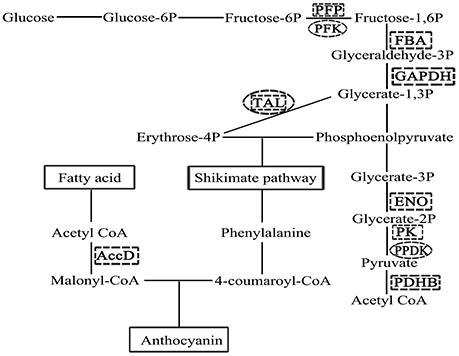
Figure 7. Schematic representation of the supply of precursors for the anthocyanin biosynthetic pathway. The formation of two reaction substrates in anthocyanin biosynthesis, malonyl-CoA and 4-coumaroyl-CoA, were linked to glycolysis, pentose phosphate pathway, shikimate pathway, and fatty acid metabolism. Enzymes identified by proteomics (marked with a dotted square) or SSH (marked with a dotted circle) were shown in bold uppercase letters (AccD, acetyl-CoA carboxyltransferase, β subunit; PFP, pyrophosphate: fructose 6-phosphate 1- phosphotransferase; PFK, 6-phosphofructokinase; FBA, fructose-bisphosphate aldolase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ENO, enolase; PK: pyruvate kinase; PPDK: pyruvate phosphate dikinase; PDHB, pyruvate dehydrogenase E1 subunit β; TAL: transaldolase).
It is well-documented that the expression of genes related to stress responses, primary and secondary metabolism, cell wall metabolism, and hormonal metabolism contribute to fruit ripening process (Shi et al., 2014; Kumar et al., 2015). Previous studies have suggested that ALA enhanced fruit maturation with a promotion of many ripening-related biological events (Watanabe et al., 2006). However, to date, there is little evidence elucidating the molecular mechanism how ALA regulates fruit ripening. In this study, about 85% of changed proteins and 38% of up-regulated mRNA in ALA-treated apple skin have a secondary function associated with fruit ripening (Figure 3C and Table 2). This result is not surprising because most fruits accumulate anthocyanin only in their ripening phase (Jaakola et al., 2002; Whale and Singh, 2007; Negri et al., 2008; Bureau et al., 2009). Our results demonstrate that ALA regulates the expression of ripening-related genes and therefore provide more evidence on the regulation of fruit ripening by ALA.
Our results suggest a role of ALA in the modulation of apple skin physiology by regulating fruit ripening-related processes. Transcription factors play important roles in controlling the switch to the ripening phase in fruits (Giovannoni, 2007; Karlova et al., 2014). Since studies on ripening inhibitor (Rin) mutation in tomato revealed SEPALLATA (SEP) subfamily of MADS-box genes play a key role in the developmental control of fruit ripening (Vrebalov et al., 2002), there is evidence that members of SEP-like orthologous gene families are involved in fruit development in other species, such as strawberry (Seymour et al., 2011), and banana (Elitzur et al., 2010). In fact, some MADS box TF from the AP1/SQUA class have been implicated in controlling of anthocyanin biosynthesis. IbMADS10 expression was correlated with red pigmentation in sweet potato, and ectopic expression resulted in anthocyanin accumulation in transgenic sweet potato calli and transgenic Arabidopsis (Lalusin et al., 2006). VmTDR4 expression was linked with color development and anthocyanin-related gene expression in bilberry (Vaccinium myrtillus), while silencing of this gene reduced anthocyanin levels (Jaakola et al., 2010). Meanwhile, SEP-like genes have also been reported to be associated with the accumulation of anthocyanin in pear (Wu et al., 2013). In strawberry, suppression of SEP1/2-like FaMADS9 resulted in delayed ripening with respect to anthocyanin accumulation. Therefore, SEP-like gene regulated fruit ripening process including skin color change. Here, we identified an ALA-responsive gene, MdMADS1 (UN038 in Table 2), which belongs to the SEP subgroup (Sung and An, 1997). In apple, fruits of the MdMADS1 antisense lines do not ripen in terms of both developmentally controlled ripening characters, such as starch degradation and background color change of fruit skin (Ireland et al., 2013). However, the authors did not investigate anthocyanin biosynthesis and accumulation in the transgenic fruits, and the exact relationship between MdMADS1 and fruit coloration remains unknown. Our SSH result showed that ALA up-regulated the expression of MdMADS1, and this regulation was further confirmed by real-time quantitative PCR. Furthermore, under ALA treatment, the expression of MdMADS1 was significantly positively correlated with that of anthocyanin biosynthetic genes, including MdCHS, MdDFR, MdLDOX, and MdUFGT. These results indicate that MdMADS1 may play an important role in ALA-promoted anthocyanin accumulation. To prove the role of MdMADS1, we transformed and successfully obtained transgenic fruit calli. Overexpression of MdMADS1 led to more anthocyanin accumulation. The six anthocyanin biosynthetic genes were up-regulated in overexpression transgenic calli and down-regulated in RNAi transgenic calli. Anthocyanin accumulated in overexpression and RNAi transgenic calli treated by ALA was higher not only than their corresponding control but also than that in transgenic calli without ALA. These results indicate that MdMADS1 involves in ALA-induced anthocyanin accumulation, but the latter is not completely dependent on the former.
In apple fruit, MdMADS1 is a master regulator that controls fruit ripening process including the initiation of ethylene biosynthesis (Ireland et al., 2013; Schaffer et al., 2013). The central role of ethylene in apple fruit ripening has been well-studies (Dandekar et al., 2004; Schaffer et al., 2007). Of these, ethylene appeared to be a key factor regulating anthocyanin biosynthesis and color development in apple fruit (Whale and Singh, 2007). In the present study, we also identified 1-aminocyclopropane-1-carboxylate oxidase 1 (ACO1, No. 34 in Table 1 and UN095 in Table 2) at protein and mRNA levels, a key enzyme in ethylene biosynthesis, and ethylene receptor 2 (ERS2, No. 35 in Table 1) at protein level, an ethylene signal transduction gene, suggesting that MdMADS1 might regulate ALA-induced anthocyanin accumulation by its regulation of ethylene metabolism and action. However, the role of ethylene in apple cultivar “Fuji” fruit may be quiet limited, because the ethylene production in “Fuji” fruit was significant lower than that in other cultivars (Harada et al., 2000; Tatsuki et al., 2007). In addition, the skin color change in MdMADS1 antisense fruits was not completely compensated for by exogenous ethylene (Ireland et al., 2013), suggesting a non-ethylene-dependent pathway exists in MdMADS1-regulated fruit coloration. In this study, the function of MdMADS1 was tested in fruit calli, proving that the regulation of MdMADS1 on anthocyanin biosynthesis is at least partly independent of fruit ripening process. Our data firstly reveal a positive regulation role of MdMADS1 in anthocyanin biosynthesis. Further study is needed to reveal the mechanisms behind ripening-related and non-ripening-related anthocyanin accumulation mediated by MdMADS1. Except MdMADS1, ALA promotes anthocyanin accumulation has been linked to regulatory genes MYB, bHLH, and WD40 (Xie et al., 2013). Therefore, the interaction between the MdMADS1 and MYB–bHLH–WD40 complexes need further study.
Conclusion
In summary, the integrated proteomics and SSH techniques, in this study, provide us a comprehensive understanding of biological events that are relevant to ALA-improved fruit coloration. Based on the results, we identified a positive regulator, MdMADS1, and verified its role in ALA-induced anthocyanin accumulation by further functional characterization. In apple fruits, the expression of MdMADS1 was induced by ALA, which was significantly positively correlated to that of anthocyanin biosynthetic genes under ALA treatment. In fruit calli, overexpressed MdMADS1 enhanced anthocyanin content, and the accumulation was further enhanced by ALA treatment. However, silenced MdMADS1 cannot completely repress anthocyanin accumulation in ALA-treated calli. The results indicate synergistic or additive responses between ALA and MdMADS1 exists for regulation of apple skin anthocyanin accumulation. In addition, verification in apple calli, a non-fruit test system, suggested the regulation of MdMADS1 on anthocyanin biosynthesis is partially independent of fruit ripening process. Our results contribute to the understanding of ALA-stimulated fruit coloration and expand the existing information on transcription regulation of anthocyanin accumulation in fruit. Further study is needed to elucidate how MdMADS1 regulates the genes encoding anthocyanin biosynthetic pathway enzymes.
Author Contributions
XF, YA conceived and designed research. XF, JZ, MS carried out all the experiments. XF, YA analyzed the data. YA, LW wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the Natural Science Foundation of Jiangsu Province, China (BK20140702), the National Natural Science Foundation of China (31401820), and the Fundamental Research Funds for the Central Universities (KJQN201538). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Meixiang Zhang from Nanjing Agricultural University for his valuable comments and suggestions that help improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01615/full#supplementary-material
References
Aguilar, M., Osuna, D., Caballero, J. L., and Munoz, J. (1997). Isolation of a cDNA encoding a metallothionein-like protein (Accession No. U81041) from strawberry fruit. Plant Physiol. 113, 663.
Akram, N. A., and Ashraf, M. (2013). Regulation in plant stress tolerance by a potential plant growth regulator, 5-aminolevulinic acid. J. Plant Growth Regul. 32, 663–679. doi: 10.1007/s00344-013-9325-9
Alessio, F., Cecilia, B., Martina, D. F., Francesco, F., and Massimiliano, T. (2011). Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 6, 709–711. doi: 10.4161/psb.6.5.15069
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., and Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Arcondéguy, T., Lacazette, E., Millevoi, S., Prats, H., and Touriol, C. (2013). VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 41, 7997–8010. doi: 10.1093/nar/gkt539
Bae, R. N., Kim, K. W., Kim, T. C., and Lee, S. K. (2006). Anatomical observations of anthocyanin rich cells in apple skins. HortScience 41, 733–736.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bregoli, A. M., Scaramagli, S., Costa, G., Sabatini, E., Ziosi, V., Biondi, S., et al. (2002). Peach (Prunus persica) fruit ripening: aminoethoxyvinylglycine (AVG) and exogenous polyamines affect ethylene emission and flesh firmness. Physiol. Plant. 114, 472–481. doi: 10.1034/j.1399-3054.2002.1140317.x
Bureau, S., Renard, C. M. G. C., Reich, M., Ginies, C., and Audergon, J. M. (2009). Change in anthocyanin concentrations in red apricot fruits during ripening. LWT Food Sci. Technol. 42, 372–377. doi: 10.1016/j.lwt.2008.03.010
Burns, J. K., Hartmond, U., and Kender, W. J. (1999). Acetolactate synthase inhibitors increase ethylene production and cause fruit drop in citrus. HortScience 34, 908–910.
Cao, Y., Tang, X., Giovannoni, J., Xiao, F., and Liu, Y. (2012). Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. BMC Plant Biol. 12:211. doi: 10.1186/1471-2229-12-211
Chalker-Scott, L. (1999). Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 70, 1–9. doi: 10.1111/j.1751-1097.1999.tb01944.x
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5:e11335. doi: 10.1371/journal.pone.0011335
D'Ambrosio, C., Arena, S., Rocco, M., Verrillo, F., Novi, G., Viscosi, V., et al. (2013). Proteomic analysis of apricot fruit during ripening. J. Proteomics 78, 39–57. doi: 10.1016/j.jprot.2012.11.008
Dandekar, A. M., Teo, G., Defilippi, B. G., Uratsu, S. L., Passey, A. J., Kader, A. A., et al. (2004). Effect of down-regulation of ethylene biosynthesis on fruit flavor complex in apple fruit. Transgenic Res. 13, 373–384. doi: 10.1023/B:TRAG.0000040037.90435.45
Dauwe, R., Morreel, K., Goeminne, G., Gielen, B., Rohde, A., Beeumen, J. V., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52, 263–285. doi: 10.1111/j.1365-313X.2007.03233.x
Deytieux, C., Geny, L., Lapaillerie, D., Claverol, S., Bonneu, M., and Donèche, B. (2007). Proteome analysis of grape skins during ripening. J. Exp. Bot. 58, 1851–1862. doi: 10.1093/jxb/erm049
Dixon, R. A., Achnine, L., Kota, P., Liu, C. J., Reddy, M. S. S., and Wang, L. J. (2002). The phenylpropanoid pathway and plant defence - a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Elitzur, T., Vrebalov, J., Giovannoni, J. J., Goldschmidt, E. E., and Friedman, H. (2010). The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J. Exp. Bot. 61, 1523–1535. doi: 10.1093/jxb/erq017
Eremina, M., Rozhon, W., Yang, S., and Poppenberger, B. (2015). ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1. Plant J. 81, 895–906. doi: 10.1111/tpj.12775
Feng, F., Li, M., Ma, F., and Cheng, L. (2013). Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 69, 54–61. doi: 10.1016/j.plaphy.2013.04.020
Feng, S., Li, M. F., Wu, F., Li, W. L., and Li, S. P. (2015). 5-Aminolevulinic acid affects fruit coloration, growth, and nutrition quality of Litchi chinensis Sonn. cv. Feizixiao in Hainan, tropical China. Sci. Hortic. 193, 188–194. doi: 10.1016/j.scienta.2015.07.010
Fraige, K., González-Fernández, R., Carrilho, E., and Jorrín-Novo, J. V. (2014). Metabolite and proteome changes during the ripening of Syrah and Cabernet Sauvignon grape varieties cultured in a nontraditional wine region in Brazil. J. Proteomics 113, 206–225. doi: 10.1016/j.jprot.2014.09.021
Gallego, P. P., Whotton, L., Picton, S., Grierson, D., and Gray, J. E. (1995). A role for glutamate decarboxylase during tomato ripening: the characterisation of a cDNA encoding a putative glutamate decarboxylase with a calmodulin-binding site. Plant Mol. Biol. 27, 1143–1151. doi: 10.1007/BF00020887
Gao, J. J., Feng, X. X., Duan, C. H., Jian-Hua, L. I., Shi, Z. X., Gao, F. Y., et al. (2013). Effects of 5-aminolevulinic acid (ALA) on leaf photosynthesis and fruit quality of apples. J. Fruit Sci. 30, 944–951. doi: 10.13925/j.cnki.gsxb.2013.06.015
Giovannoni, J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10, 283–289. doi: 10.1016/j.pbi.2007.04.008
Giribaldi, M., Perugini, I., Sauvage, F. X., and Schubert, A. (2007). Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 7, 3154–3170. doi: 10.1002/pmic.200600974
Guo, L., Cai, Z. X., Zhang, B. B., Xu, J. L., Song, H. F., and Ma, R. J. (2013). The mechanism analysis of anthocyanin accumulation in peach accelerated by ALA. Acta Hortic. Sin. 40, 1043–1050. doi: 10.16420/j.issn.0513-353x.2013.06.004
Han, Y. P., Vimolmangkang, S., Soria-Guerra, R. E., Rosales-Mendoza, S., Zheng, D. M., Lygin, A. V., et al. (2010). Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 153, 806–820. doi: 10.1104/pp.109.152801
Harada, T., Sunako, T., Wakasa, Y., Soejima, J., Satoh, T., and Niizeki, M. (2000). An allele of the 1-aminocyclopropane-1-carboxylate synthase gene (Md-ACS1) accounts for the low level of ethylene production in climacteric fruits of some apple cultivars. Theor. App. Genet. 101, 742–746. doi: 10.1007/s001220051539
Henkes, S., Sonnewald, U., Badur, R., Flachmann, R., and Stitt, M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13, 535–551. doi: 10.1105/tpc.13.3.535
Huerta-Ocampo, J. Á., Osuna-Castro, J. A., Lino-López, G. J., Barrera-Pacheco, A., Mendoza-Hernández, G., De León-Rodríguez, A., et al. (2012). Proteomic analysis of differentially accumulated proteins during ripening and in response to 1-MCP in papaya fruit. J. Proteomics 75, 2160–2169. doi: 10.1016/j.jprot.2012.01.015
Ireland, H. S., Yao, J. L., Tomes, S., Sutherland, P. W., Nieuwenhuizen, N., Gunaseelan, K., et al. (2013). Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 73, 1044–1056. doi: 10.1111/tpj.12094
Jaakola, L., Määttä, K., Pirttilä, A. M., Törrönen, R., Kärenlampi, S., and Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/pp.006957
Jaakola, L., Pirttilä, A. M., Halonen, M., and Hohtola, A. (2001). Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19, 201–203. doi: 10.1385/MB:19:2:201
Jaakola, L., Poole, M., Jones, M. O., Kämäräinen-Karppinen, T., Koskimäki, J. J., Hohtola, A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. doi: 10.1104/pp.110.158279
Janssen, B. J., Thodey, K., Schaffer, R. J., Alba, R., Balakrishnan, L., Bishop, R., et al. (2008). Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol. 8:16. doi: 10.1186/1471-2229-8-16
Ju, Z. G. (1998). Fruit bagging, a useful method for studying anthocyanin synthesis and gene expression in apples. Sci. Hortic. 77, 155–164. doi: 10.1016/S0304-4238(98)00161-7
Ju, Z. G., and Curry, E. A. (2000). Evidence that α-farnesene biosynthesis during fruit ripening is mediated by ethylene regulated gene expression in apples. Postharvest Biol. Technol. 19, 9–16. doi: 10.1016/S0925-5214(00)00078-8
Ju, Z. G., Liu, C. L., and Yuan, Y. B. (1995). Activities of chalcone synthase and UDPGal: flavonoid-3-o-glycosyltransferase in relation to anthocyanin synthesis in apple. Sci. Hortic. 63, 175–185. doi: 10.1016/0304-4238(95)00807-6
Karlova, R., Chapman, N., David, K., Angenent, G. C., Seymour, G. B., and de Maagd, R. A. (2014). Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. doi: 10.1093/jxb/eru316
Kim, S. H., Lee, J. R., Hong, S. T., Yoo, Y. K., An, G., and Kim, S. R. (2003). Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 165, 403–413. doi: 10.1016/S0168-9452(03)00201-2
Kumar, V., Irfan, M., Ghosh, S., Chakraborty, N., Chakraborty, S., and Datta, A. (2015). Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma 253, 1–14. doi: 10.1007/s00709-015-0836-z
Kushad, M. M., Richardson, D. G., and Ferro, A. J. (1985). 5′-Methylthioadenosine nucleosidase and 5-methylthioribose kinase activities and ethylene production during tomato fruit development and ripening. Plant Physiol. 79, 525–529. doi: 10.1104/pp.79.2.525
Lalusin, A. G., Nishita, K., Kim, S. H., Ohta, M., and Fujimura, T. (2006). A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Mol. Genet. Genomics 275, 44–54. doi: 10.1007/s00438-005-0080-x
Lan, P., Li, W., and Schmidt, W. (2012). Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11, 1156–1166. doi: 10.1074/mcp.M112.020461
Law, R. D., and Plaxton, W. C. (1997). Regulatory phosphorylation of banana fruit phosphoenolpyruvate carboxylase by a copurifying phosphoedpyruvate carboxylase kinase. Eur. J. Biochem. 247, 642–651. doi: 10.1111/j.1432-1033.1997.00642.x
Li, J., and Yuan, R. (2008). NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. J. Plant Growth Regul. 27, 283–295. doi: 10.1007/s00344-008-9055-6
Li, P., Zhang, Y., Einhorn, T. C., and Cheng, L. (2014). Comparison of phenolic metabolism and primary metabolism between green ‘Anjou’ pear and its bud mutation, red ‘Anjou’. Physiol. Plant. 150, 339–354. doi: 10.1111/ppl.12105
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Li, Z., Sugaya, S., Gemma, H., and Iwahori, S. (2002). Flavonoid biosynthesis and accumulation and related enzyme activities in the sin of ‘Fuji’ and ‘Oorin’ apples during their development. J. Jpn. Soc. Hortic. Sci. 71, 317–321. doi: 10.2503/jjshs.71.317
Liu, J. H., Zhang, J., Xu, B. Y., Zhang, J. B., Jia, C. H., Wang, J. S., et al. (2012). Expression analysis of banana MaECHI1 during fruit ripening with different treatments. Afr. J. Biotechnol. 11, 12951–12957. doi: 10.5897/AJB12.763
Liu, X., Hu, X. M., Jin, L. F., Shi, C. Y., Liu, Y. Z., and Peng, S. A. (2014). Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD1 and CsGAD2, reveal the strong relationship between CsGAD1 and citrate utilization in citrus fruit. Mol. Biol. Rep. 41, 6253–6262. doi: 10.1007/s11033-014-3506-x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ludwig, R., Su-Ying, Y., Stephanie, H., Thomas, H., Rosario, B. P., Mathieu, F., et al. (2013). Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 163, 43–60. doi: 10.1104/pp.113.222778
Majeran, W., Yang, C., Qi, S., and Wijk, K. J. V. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17, 3111–3140. doi: 10.1105/tpc.105.035519
Marondedze, C., Gehring, C., and Thomas, L. (2014). Dynamic changes in the date palm fruit proteome during development and ripening. Hortic. Res. 1, 14039. doi: 10.1038/hortres.2014.39
Martínez-Esteso, M. J., Vilella-Antón, M. T., Pedreño, M. Á., Valero, M. L., and Bru-Martínez, R. (2013). iTRAQ-based protein profiling provides insights into the central metabolism changes driving grape berry development and ripening. BMC Plant Biol. 13:167. doi: 10.1186/1471-2229-13-167
Meinhold, T., Richters, J. P., Damerow, L., and Blanke, M. M. (2010). Optical properties of reflection ground covers with potential for enhancing fruit colouration. Biosys. Eng. 107, 155–160. doi: 10.1016/j.biosystemseng.2010.07.006
Minas, I. S., Tanou, G., Belghazi, M., Job, D., Manganaris, G. A., Molassiotis, A., et al. (2012). Physiological and proteomic approaches to address the active role of ozone in kiwifruit post-harvest ripening. J. Exp. Bot. 63, 2449–2464. doi: 10.1093/jxb/err418
Murooka, Y., and Tanaka, T. (2014). “5-Aminolevulinic acid (5-ALA) - a multifunctional amino acid as a plant growth stimulator and stress tolerance factor,” in Plant Adaptation to Environmental Change: Significance of Amino Acids and their Derivatives, eds N. A. Anjum, S. S. Gill, and R. Gill (Wallingford: CABI Publishing), 18–34.
Negri, A. S., Prinsi, B., Failla, O., Scienza, A., and Espen, L. (2015). Proteomic and metabolic traits of grape exocarp to explain different anthocyanin concentrations of the cultivars. Front. Plant Sci. 6:603. doi: 10.3389/fpls.2015.00603
Negri, A. S., Prinsi, B., Rossoni, M., Failla, O., Scienza, A., Cocucci, M., et al. (2008). Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9:378. doi: 10.1186/1471-2164-9-378
Nogueira, S. B., Labate, C. A., Gozzo, F. C., Pilau, E. J., Lajolo, F. M., and Oliveira do Nascimento, J. R. O. (2012). Proteomic analysis of papaya fruit ripening using 2DE-DIGE. J. Proteomics 75, 1428–1439. doi: 10.1016/j.jprot.2011.11.015
Perez, M. H., Rodriguez, B. L., Shintani, T. T., Watanabe, K., Miyanari, S., and Harrigan, R. C. (2013). 5-Aminolevulinic acid (5-ALA): analysis of preclinical and safety literature. Food Nutr. Sci. 4, 1009–1013. doi: 10.4236/fns.2013.410131
Prinsi, B., Negri, A. S., Fedeli, C., Morgutti, S., Negrini, N., Cocucci, M., et al. (2011). Peach fruit ripening: a proteomic comparative analysis of the mesocarp of two cultivars with different flesh firmness at two ripening stages. Phytochemical 72, 1251–1262. doi: 10.1016/j.phytochem.2011.01.012
Qin, G., Wang, Y., Cao, B., Wang, W., and Tian, S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70, 243–255. doi: 10.1111/j.1365-313X.2011.04861.x
Sasaki, Y., and Nagano, Y. (2014). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Schaffer, R. J., Friel, E. N., Souleyre, E. J. F., Bolitho, K., Thodey, K., Ledger, S., et al. (2007). A genomics approach reveals that aroma production in apple is controlled by ethylene predominantly at the final step in each biosynthetic pathway. Plant Physiol. 144, 1899–1912. doi: 10.1104/pp.106.093765
Schaffer, R. J., Ireland, H. S., Ross, J. J., Ling, T. J., and David, K. M. (2013). SEPALLATA1/2-suppressed mature apples have low ethylene, high auxin and reduced transcription of ripening-related genes. Aob Plants 5:pls047. doi: 10.1093/aobpla/pls047
Scherp, P., Ku, G., Coleman, L., and Kheterpal, I. (2011). Gel-based and gel-free proteomic technologies. Methods Mol. Biol. 702, 163–190. doi: 10.1007/978-1-61737-960-4_13
Seymour, G. B., Ryder, C. D., Cevik, V., Hammond, J. P., Popovich, A., King, G. J., et al. (2011). A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 62, 1179–1188. doi: 10.1093/jxb/erq360
Shi, Y., Jiang, L., Zhang, L., Kang, R., and Yu, Z. (2014). Dynamic changes in proteins during apple (Malus × domestica) fruit ripening and storage. Hortic. Res. 1:6. doi: 10.1038/hortres.2014.6
Sunchung, P., Nobuko, S., Larson, M. D., Randy, B., and Steven, V. N. (2006). Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol. 141, 811–824. doi: 10.1104/pp.106.080994
Sung, S. K., and An, G. H. (1997). Molecular cloning and characterization of a MADS-box cDNA clone of the Fuji apple. Plant Cell Physiol. 38, 484–489. doi: 10.1093/oxfordjournals.pcp.a029193
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tatsuki, M., Endo, A., and Ohkawa, H. (2007). Influence of time from harvest to 1-MCP treatment on apple fruit quality and expression of genes for ethylene biosynthesis enzymes and ethylene receptors. Postharvest Biol. Technol. 43, 28–35. doi: 10.1016/j.postharvbio.2006.08.010
Turner, W. L., and Plaxton, W. C. (2003). Purification and characterization of pyrophosphate- and ATP-dependent phosphofructokinases from banana fruit. Planta 217, 113–121. doi: 10.1007/s00425-002-0962-7
Tzin, V., and Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3, 956–972. doi: 10.1093/mp/ssq048
Ubi, B. E., Honda, C., Bessho, H., Kondo, S., Wada, M., Kobayashi, S., et al. (2006). Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci. 170, 571–578. doi: 10.1016/j.plantsci.2005.10.009
Voll, L. M., Hajirezaei, M. R., Czogalla-Peter, C., Lein, W., Stitt, M., Sonnewald, U., et al. (2009). Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol. 184, 607–618. doi: 10.1111/j.1469-8137.2009.02998.x
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, N., Zheng, Y., Duan, N., Zhang, Z., Ji, X., Jiang, S., et al. (2015). Comparative transcriptomes analysis of red- and white-fleshed apples in an F1 population of malus sieversii f. niedzwetzkyana crossed with M. domestica ‘Fuji’. PLoS ONE 10:e0133468. doi: 10.1371/journal.pone.0133468
Wang, S. H., You, Z. Y., Ye, L. P., Che, J., Qian, Q., Nanjo, Y., et al. (2014). Quantitative proteomic and transcriptomic analyses of molecular mechanisms associated with low silk production in silkworm Bombyx mori. J. Proteome Res. 13, 735–751. doi: 10.1021/pr4008333
Watanabe, K., Nishihara, E., Watanabe, S., Tanaka, T., Takahashi, K., and Takeuchi, Y. (2006). Enhancement of growth and fruit maturity in 2-year-old grapevines cv. Delaware by 5-aminolevulinic acid. Plant Growth Regul. 49, 35–42. doi: 10.1007/s10725-006-0024-4
Weiss, J., and Egea-Cortines, M. (2009). Transcriptomic analysis of cold response in tomato fruits identifies dehydrin as a marker of cold stress. J. Appl. Genet. 50, 311–319. doi: 10.1007/BF03195689
Whale, S. K., and Singh, Z. (2007). Endogenous ethylene and color development in the skin of ‘Pink lady’ apple. J. Am. Soc. Hort. Sci. 132, 20–28.
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wong, J. H., Kiss, F., Wu, M. X., and Buchanan, B. B. (1990). Pyrophosphate fructose-6-P 1-phosphotransferase from tomato fruit: evidence for change during ripening. Plant Physiol. 94, 499–506. doi: 10.1104/pp.94.2.499
Wu, J., Xu, Z., Zhang, Y., Chai, L., Yi, H., and Deng, X. (2014). An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 65, 1651–1671. doi: 10.1093/jxb/eru044
Wu, J., Zhao, G., Yang, Y. N., Le, W. Q., Khan, M. A., Zhang, S. L., et al. (2013). Identification of differentially expressed genes related to coloration in red/green mutant pear (Pyrus communis L.). Tree Genet. Genomes 9, 75–83. doi: 10.1007/s11295-012-0534-3
Xiao, C. C., Zhang, S. L., Hu, H. J., Tian, R., Wu, J., Yang, Z. J., et al. (2012). Effects of bagging and exogenous 5-aminolevulinic acid treatment on coloration of ‘Yunhongli 2’. J. Nanjing Agricult. Univ. 35, 25–29. doi: 10.7685/j.issn.1000-2030.2012.06.005
Xie, L., Wang, Z. H., Cheng, X. H., Gao, J. J., Zhang, Z. P., and Wang, L. J. (2013). 5-Aminolevulinic acid promotes anthocyanin accumulation in Fuji apples. Plant Growth Regul. 69, 295–303. doi: 10.1007/s10725-012-9772-5
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Yin, X., Sakata, K., Nanjo, Y., and Komatsu, S. (2014). Analysis of initial changes in the proteins of soybean root tip under flooding stress using gel-free and gel-based proteomic techniques. J. Proteomics 106, 1–16. doi: 10.1016/j.jprot.2014.04.004
Yuan, H., Chen, L., Paliyath, G., Sullivan, A., Murr, D. P., and Novotna, Z. (2006). Immunohistochemical localization of phospholipase D in strawberry (Fragaria ananassa Duch.) fruits. Sci. Hortic. 109, 35–42. doi: 10.1016/j.scienta.2006.02.023
Zhang, L. Y., Feng, X. X., Gao, J. J., An, Y. Y., Tian, F., Li, J., et al. (2015). Effects of rhizosphere-applied 5-aminolevulinic acid(ALA) solutions on leaf physiological characteristics and fruit quality of apples. Jiangsu J. Agric. Sci. 31, 158–165. doi: 10.3969/j.issn.1000-4440.2015.01.025
Zhang, Z., Jiang, S., Wang, N., Li, M., Ji, X., Sun, S., et al. (2015). Identification of differentially expressed genes associated with apple fruit ripening and softening by suppression subtractive hybridization. PLoS ONE 10:e0146061. doi: 10.1371/journal.pone.0146061
Zheng, Q., Song, J., Campbell-Palmer, L., Thompson, K., Li, L., Walker, B., et al. (2013). A proteomic investigation of apple fruit during ripening and in response to ethylene treatment. J. Proteomics 93, 276–294. doi: 10.1016/j.jprot.2013.02.006
Zheng, Y., Zhao, L., Gao, J., and Fei, Z. (2011). iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinform. 12:453. doi: 10.1186/1471-2105-12-453
Keywords: anthocyanin, apple, 5-aminolevulinic acid (ALA), proteomics, suppression subtractive hybridization (SSH), MdMADS1
Citation: Feng X, An Y, Zheng J, Sun M and Wang L (2016) Proteomics and SSH Analyses of ALA-Promoted Fruit Coloration and Evidence for the Involvement of a MADS-Box Gene, MdMADS1. Front. Plant Sci. 7:1615. doi: 10.3389/fpls.2016.01615
Received: 29 June 2016; Accepted: 12 October 2016;
Published: 07 November 2016.
Edited by:
Maarten Hertog, KU Leuven, BelgiumReviewed by:
Panagiotis Kalaitzis, Mediterranean Agronomic Institute of Chania, GreeceLuca Espen, University of Milan, Italy
Copyright © 2016 Feng, An, Zheng, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangju Wang, d2xqQG5qYXUuZWR1LmNu
†These authors have contributed equally to this work.
 Xinxin Feng
Xinxin Feng Yuyan An
Yuyan An Jie Zheng
Jie Zheng Miao Sun
Miao Sun Liangju Wang
Liangju Wang