- 1Saskatoon Research Centre, Agriculture and Agri-Food Canada, Saskatoon, SK, Canada
- 2Armatus Genetics Inc., Saskatoon, SK, Canada
Seven blackleg resistance (R) genes (Rlm1, Rlm2, Rlm3, Rlm4, LepR1, LepR2 & LepR3) were each introgressed into a common susceptible B. napus doubled-haploid (DH) line through reciprocal back-crossing, producing single-R gene introgression lines (ILs) for use in the pathological and molecular study of Brassica—Leptosphaeria interactions. The genomic positions of the R genes were defined through molecular mapping and analysis with transgenic L. maculans isolates was used to confirm the identity of the introgressed genes where possible. Using L. maculans isolates of contrasting avirulence gene (Avr) profiles, we preformed extensive differential pathology for phenotypic comparison of the ILs to other B. napus varieties, demonstrating the ILs can provide for the accurate assessment of Avr-R gene interactions by avoiding non-Avr dependant alterations to resistance responses which can occur in some commonly used B. napus varieties. Whole-genome SNP-based assessment allowed us to define the donor parent introgressions in each IL and provide a strong basis for comparative molecular dissection of the pathosystem.
Introduction
In a plant-pathogen system that follow gene-for-gene interactions (Flor, 1971), genetically well-defined resistant host germplasm (differential lines) are an essential tool for identifying and characterizing avirulence (Avr) alleles present in pathogen isolates. Ideally, each plant differential line should represent a single resistance (R) gene in an otherwise susceptible host genotype, providing a common background for individual R genes. In addition to being a valuable tool for pathology, producing single R gene differential lines in a common genetic background would allow researchers to dissect and compare the robustness, efficiency and downstream molecular defense pathways for each R gene, free from the interference caused by variation in background genotypic effects.
Resistance against the hemibiotrophic fungal pathogen Leptosphaeria maculans, the causative agent of blackleg disease in of Brassica species (Howlett et al., 2001), is governed largely by race-specific R genes. Nineteen R genes, identified in several Brassica species, have been reported to convey race-specific resistance against L. maculans (reviewed in Raman et al., 2013). Twelve of these R genes have been positioned in the A genome of B. napus or B. rapa (Rlm1, Rlm2, Rlm3, Rlm4, Rlm7, Rlm9, LepR1, LepR2, LepR3, LepR4) or the B genome of B. juncea (LmJR1, LmJR2) via linkage mapping (Delourme et al., 2004; Yu et al., 2005, 2007, 2012; Christianson et al., 2006; Long et al., 2011; Raman et al., 2012a,b; Larkan et al., 2013, 2014, 2016), while two more have been assigned to B genome chromosomes of B. juncea and/or B. nigra (Rlm6, Rlm10) through marker-based assessment of recombinant or chromosome addition lines (Chèvre et al., 1996, 1997; Eber et al., 2011). RlmS, identified in the B. napus variety Surpass 400 (Van De Wouw et al., 2009) likely corresponds to the BlmR2 locus mapped to chromosome A10 (Long et al., 2011; Larkan et al., 2013). The remaining R genes (Rlm5, Rlm8, Rlm11, and RlmJ1) have yet to be genetically characterized in the host Brassica lines in which they reside. However, Rlm8 and Rlm11 are contained within the A genome of B. rapa varieties (Balesdent et al., 2002, 2013), while Rlm5 and RlmJ1 are found in B. juncea (genome AABB), though it has not yet been determined which genome they reside in or if these in fact represent the same gene (Balesdent et al., 2002; van de Wouw et al., 2014). Several other mapping efforts have localized resistance loci (LEM1, LmFr1, LmR1, ClmR1) to chromosome A07 of B. napus (Dion et al., 1995; Ferreira et al., 1995; Mayerhofer et al., 1997, 2005), though it has been assumed that these all correspond to Rlm4 based on map position and phenotypic response (Rouxel et al., 2003b; Rimmer, 2006).
Our understanding of Brassica—Leptosphaeria interactions has expanded tremendously over the last decade, aided greatly by the availability of both pathogen (Rouxel et al., 2011) and host (Wang et al., 2011; Chalhoub et al., 2014; Liu et al., 2014) genomes. To date, eight R gene-interacting effectors have been cloned from L. maculans; AvrLm1, AvrLm2, AvrLm3, AvrLm4-7 & AvrLm7, AvrLm6, AvrLm11, AvrLmJ1 (Gout et al., 2006; Fudal et al., 2007; Parlange et al., 2009; Balesdent et al., 2013; van de Wouw et al., 2014; Ghanbarnia et al., 2015; Plissonneau et al., 2016), along with two of the corresponding host R genes; LepR3 and Rlm2 (Larkan et al., 2013, 2015). Yet despite the rapid expansion of genetic information available for the pathosystem, most characterisations of fungal isolates and host germplasm still rely almost solely on pathological studies for generating Avr and/or R gene hypotheses. The differential host germplasm used in pathology-based studies often varies between labs, can contain multiple known R genes and is sometimes poorly characterized in terms of homozygosity, confirmation of the genetic interactions and the non-specific resistance effects observed during the interaction.
Here we describe the production of single-R gene B. napus introgression lines (ILs) for use in the pathological and molecular study of Brassica—Leptosphaeria interactions. Each IL contains one of seven blackleg resistance (R) genes (Rlm1, Rlm2, Rlm3, Rlm4, LepR1, LepR2 & LepR3) introgressed into a common susceptible B. napus line (Topas DH16516) allowing for the study of Avr-R gene interactions with a minimum of phenotypic ambiguity.
Materials and Methods
Plant Material
B. napus parental lines used in the production of the ILs (Topas, Topas DH16516, Quinta DH24288, Glacier DH24287, Scoop, 1065, 1135, and Surpass 400—Supplementary Table 1) were sourced from collections held by the Rimmer, Lydiate and Borhan labs, AAFC Saskatoon. Doubled-haploid (DH) line Topas DH16516 was originally produced by G. Séguin-Swartz, AAFC Saskatoon. DH lines Quinta DH24288 and Glacier DH24287 were produced by S.R. Rimmer, AAFC Saskatoon, from Quinta and Glacier variety seed originally provided by P.H. Williams, University of Wisconsin. Additional B. napus variety lines were sourced from the Lydiate and Vail Labs, AAFC Saskatoon.
Fungal Material
L. maculans isolates 98-15s, 00-100s, WA30s, 05-31s, and 2367s were produced from single-spore re-isolations of stock isolates contained in the Rimmer Collection, AAFC Saskatoon. Isolate v23.1.3 (Rouxel et al., 2011) was provided by T. Rouxel, INRA-Bioger, France. Isolate B14-13s was isolated from infected field stubble, provided by F.L. Dokken-Bouchard and D.T. Stephens, Saskatchewan Ministry of Agriculture, Canada, collected during the 2014 Saskatchewan Disease Survey (Dokken-Bouchard et al., 2015). Isolate 3R11 was provided by A. Van der Wouw and B. Howlett, University of Melbourne, Australia. Six of the isolates (98-15s, 00-100s, WA30s, 89-12s, 2367s, and 3R11s) have previously been fully-characterized via whole-genome resequencing (Ghanbarnia et al., 2015). Transgenic isolates 3R11:AvrLm1 (Larkan et al., 2013), 2367:AvrLm4-7 and 2367:AvrLm7 (Larkan et al., 2016) have been described previously, as has the genomic AvrLm2 construct used to produce 3R11:AvrLm2 (Ghanbarnia et al., 2015). The remaining transgenic isolates (2367:AvrLm3 and 3R11:AvrLm6) were produced by introducing genomic avirulence allele constructs, including approximately 1 kb 5′ and 500 bp 3′ of the CDS (Supplementary Table 2) using the same Agrobacterium transformation method (Utermark and Karlovsky, 2008) and the fungal transformation vector (Larkan et al., 2013) as described previously.
Production of Introgression Lines
Initially F1 seed was produced by first crossing the resistant donor parent lines (Supplementary Table 1) to either the susceptible Topas variety line (for Rlm1, Rlm2, and Rlm3 populations) or the DH Topas DH16516 (Rlm4, LepR1, LepR2, and LepR3) once this line became available. F1 seedlings were assayed for resistance to L. maculans isolates previously determined to be avirulent toward the targeted R gene. A single resistant F1 selection was then backcrossed to the susceptible Topas variety or Topas DH16516 line (once this line became available) to produce BC1F1 seed. All backcross generations from BC2F1 onward were produced using Topas DH16516 exclusively, for all lines. Recurrent backcrossing and selection, both for resistance and spring-type growth habit (where applicable), was performed to produce BC5F1 seedlings, which were selfed through three additional generations to produce BC5F4. At this point the lines were tested for homozygous resistance and a final single-plant selection was made. Where the donor parent line contained multiple R genes, duel-isolate tests were performed at either BC1F1 or BC2F1 to select for individuals that only contained the targeted gene.
Genetic Mapping of A07 R Gene Loci
Genetic mapping of three R gene loci (Rlm1, Rlm3, Rlm4) was performed in parallel to the production of the ILs by the previously-described methodology (Larkan et al., 2014). Briefly, large BC1F1 populations (Supplementary Table 1) were produced from the same F1s used for the IL development. Small populations were used to establish linkage of phenotype to simple sequence repeat (SSR) markers positioned on A07 and to assemble draft maps. SSR markers flanking the target genes were then used to screen the full population. Individual BC1F1 seedlings containing recombination events within the targeted R loci regions were reserved for the fine mapping study. The initial genotypes for these ‘recombinants’ were confirmed using DNA from a second round of extractions, and phenotypes were confirmed by examining the segregation of resistance in the BC1F2 generation. Additional mapping using smaller BC1F1 populations was later performed to obtain the genomic positions of Rlm7 and Rlm9 (donor parental lines Roxet and Goéland, respectively) relative to the other A07 R genes (Rlm1, Rlm3, and Rlm4) and to facilitate the future production of additional ILs.
Phenotypic Verification
To demonstrate the phenotypic reaction of the ILs when challenged with L. maculans, seven well-characterized isolates of varying genotypes (Avr complements) where chosen from the lab's collection and used to inoculate wounded cotyledons of each line, after which the pathological interaction was rated on a continuous 1 to 9 scale as previously described (Larkan et al., 2013). In parallel to the IL testing the resistant donor parent lines and several other B. napus lines previously used to assess Brassica—Leptosphaeria interactions (Balesdent et al., 2001, 2002, 2005; Rouxel et al., 2003a,b; Stachowiak et al., 2006; Parlange et al., 2009; Yu et al., 2012; Larkan et al., 2013, 2014, 2015) were also tested. Eight seedlings of each line were used in the test, with one inoculation per cotyledon lobe (4 per seedling). Interactions were scored at 10 and 14 days post-inoculation (dpi) with each lesion being rated independently (32 lesions/line). Phenotyping for the disambiguation of Rlm3 and Rlm4 in B. napus cultivars was performed as described above using 4 seedlings per line (16 inoculations), with phenotypic rating at 14 dpi. Additional confirmation of R gene content in each line was produced using transgenic L. maculans isolates carrying the characterized Avr genes AvrLm1 (Gout et al., 2006), AvrLm2 (Ghanbarnia et al., 2015), AvrLm3 (Plissonneau et al., 2016), AvrLm4-7 and AvrLm7 (Parlange et al., 2009), and AvrLm6 (Fudal et al., 2007).
Whole-Genome SNP Analysis
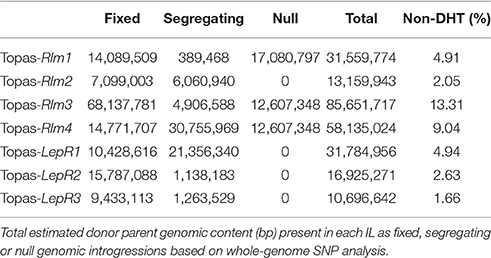
In order to determine the size and genomic location of the donor parent genomic introgressions which harbor each of the targeted R genes, as well as any other significant introgressions elsewhere in the genome of each IL, whole-genome SNP analysis was performed using Infinium SNP arrays (Illumina) containing approx. 52, 000 SNPs for the Brassica A and C genomes (Clarke et al., 2016). This also served to determine the level of polymorphism between Topas DH16516 and the donor parent lines and to assess the overall homogeneity of each IL. To perform the analysis, DNA was extracted as previously described (Larkan et al., 2013) from 218 individual B. napus plants; 20 replicates of each IL line, six replicates of Topas DH16516, three replicates of original Topas variety line and single replicates of each donor parent and several other common blackleg differential lines were also included in the analyses (Supplementary Table 3). Sample were analyzed in two separate Infinium runs. To minimize spurious genotyping, strict criteria were imposed for the selection of high-quality SNP markers. All SNPs were initially clustered using Genome Studio software (Illumina) and markers with 100% heterozygous calls (33) were removed. Further quality control was performed by the removal of markers determined to be skewed, potentially detecting SNPs at more than one locus, based on (a) the polar co-ordinates of genotypic clusters (AA T Mean > 0.35; BB T Mean <0.65; AB T Mean <0.2, >0.8) and (b) cluster separation (<0.2). All markers with a call frequency > 0.091 (at least two samples called) were exported to Microsoft Excel for further sorting. Approximately 600 additional markers were removed due to inconsistencies between Infinium runs [usually low signal markers called as heterozygous (AB) for one run, null for the other]. In total 43,211 SNP markers (82.8%) were retained for use in assessing polymorphism between B. napus lines and to define the donor parent introgressions in each IL.
Results
Genetic Mapping
Genetic mapping of five R gene loci (Rlm1, Rlm3, Rlm4, Rlm7, and Rlm9) was performed to confirm the genes being introgressed (in the case of Rlm1, Rlm3, and Rlm4) were consistent with the originally-described chromosome A07 R loci (Delourme et al., 2004), to determine the physical positions of the genes in the B. napus genome (Chalhoub et al., 2014), and to complement our own previously-described fine mapping of Rlm2 (Larkan et al., 2014), LepR1 and LepR2 (Yu et al., 2012) and LepR3 (Larkan et al., 2013). All five genes were determined to reside within the mapped region of chromosome A07, which was defined by SSR markers spanning approximately 8 Mb of the B. napus “Darmor-bzh” genome (Figure 1). Four of the R genes (Rlm3, Rlm4, Rlm7, and Rlm9) were found to be tightly clustered as previously described (Delourme et al., 2004); all four genes co-segregated with the SSR marker sR7018, which corresponds to chrA07:16027226.16027883 of the B. napus Darmor-bzh genome (nearest gene BnaA07g20490D). Rlm3 and Rlm4 were both localized within an interval of approximately 1.2 Mb containing 230 predicted genes. The mapping of Rlm7 and Rlm9 placed these R loci within intervals of 1.85 Mb (369 genes) and 4.31 Mb (788 genes), respectively. The remaining A07 R gene, Rlm1, was localized to a region 3.8 Mb below the Rlm3/Rlm4 map interval, positioned in an interval of approximately 920 kb (163 predicted genes) between the markers sN9539 and Ind07-02 (Figure 1). The physical location for Rlm3 and Rlm4 are in agreement with our own previous lower-resolution mapping of these genes (Larkan et al., 2016).
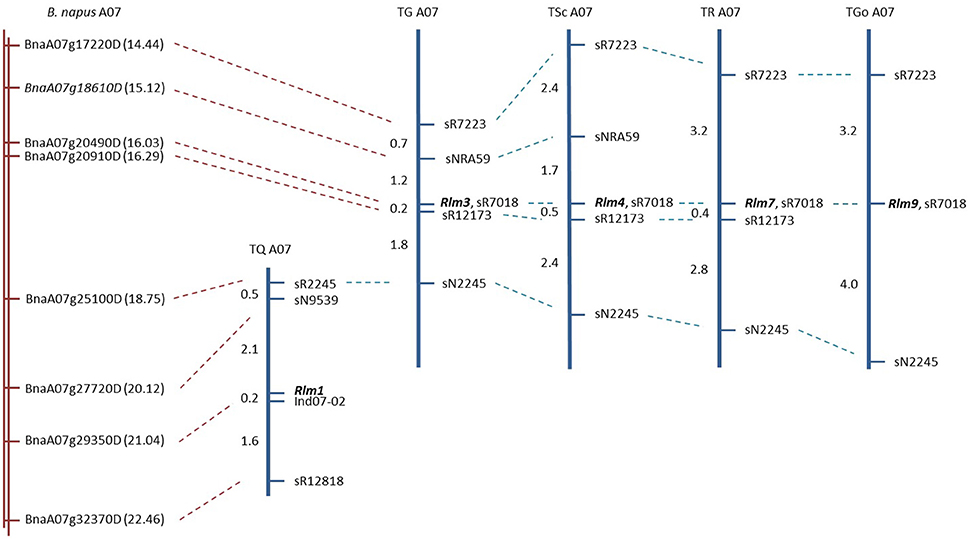
Figure 1. Physical and genetic maps of five A07 blackleg R genes. Portion of reference B. napus Darmor-bzh genome (Chalhoub et al., 2014) corresponding to mapped locations of R genes shown in red. Gene names (BnaA07g…) represent closest gene to corresponding marker, indicated by dashed red line. Gene positions (Mb) given in brackets. Dashed blue lines indicate common markers, mapped R gene names in bold. Genetic distance between markers given in centiMorgans.
Whole-Genome SNP Analysis
After sorting of the 43,211 SNP markers to distinguish homozygous (AA or BB), heterozygous (AB) and null (no call) SNP alleles for both Topas DH16516 and R gene donor parent lines, over-all polymorphism relative to Topas DH16516 was analyzed, calculated as a mean of each IL replication, with ILs ranging from 92.9 to 98.9% identical to Topas DH16516 in terms of SNP polymorphism (Supplementary Table 3). Surprisingly, the Topas variety line originally used to establish backcross populations for Topas-Rlm1, Topas-Rlm2 and Topas-Rlm3 was found to share only 82.7% marker identity with Topas DH16516. To further define the genomic arrangements of the ILs, SNPs were ordered based on chromosome position (based on Darmor reference chromosome builds spanning 643,396,264 bp over 19 chromosomes) and surveyed for regions of significant variation from Topas DH16516 (>3 clustered markers) to define the donor parent introgressions and null regions carried in each line. Each IL was found to carry a donor parent chromosomal introgression corresponding to the mapped R locus for each gene, ranging in size from 925 kb to 13.95 Mb, with most R gene introgressions spanning less than 5 Mb (Supplementary Table 4). Several other donor parent introgressions were present as both fixed and segregating genomic segments, including several remnants of the original Topas variety line in the first three ILs. A summary of the total significant donor parent introgressions along with significant null regions is presented in Table 1. The genomic position of each introgression relative to the Darmor-bzh reference genome is presented in Supplementary Table 4. Production of the three ILs carrying A07 R gene introgressions (Topas-Rlm1, Topas-Rlm3, Topas-Rlm4) appeared to be affected by a common A07-C06 reciprocal translocation event as a result of ancestral homology between the Brassica A and C genomes (Osborn et al., 2003). This event is likely carried by the Topas DH16516 parental line, which has resulted in null regions (12.6 Mb) for the affected ILs on lower C06 and corresponding dense marker heterozygosity on the homoeologous region of lower A07.
Pathology
Average ratings for each B. napus line at each time point (10 and 14 dpi) on a scale of 1 to 9 (with 9 being fully susceptible) are presented in Table 2. For the ILs, clear differentials were observed between avirulent (Avr) and virulent (avr) isolates at 10 dpi, with isolates reaching full infection (mean rating score >8) on the susceptible lines by 14 dpi. For incompatible interactions, mean ratings ranged between 2.1 to 4.1 for all ILs except Topas-LepR2, for with scores ranged 3.1 to 4.8. Interactions for Rlm1, Rlm2, Rlm3, Rlm4, and LepR3 were all confirmed using transgenic isolates carrying the corresponding Avr gene (Figure 2). Results were consistent with previous pathological testing conducted with these and other L. maculans isolates over several years.
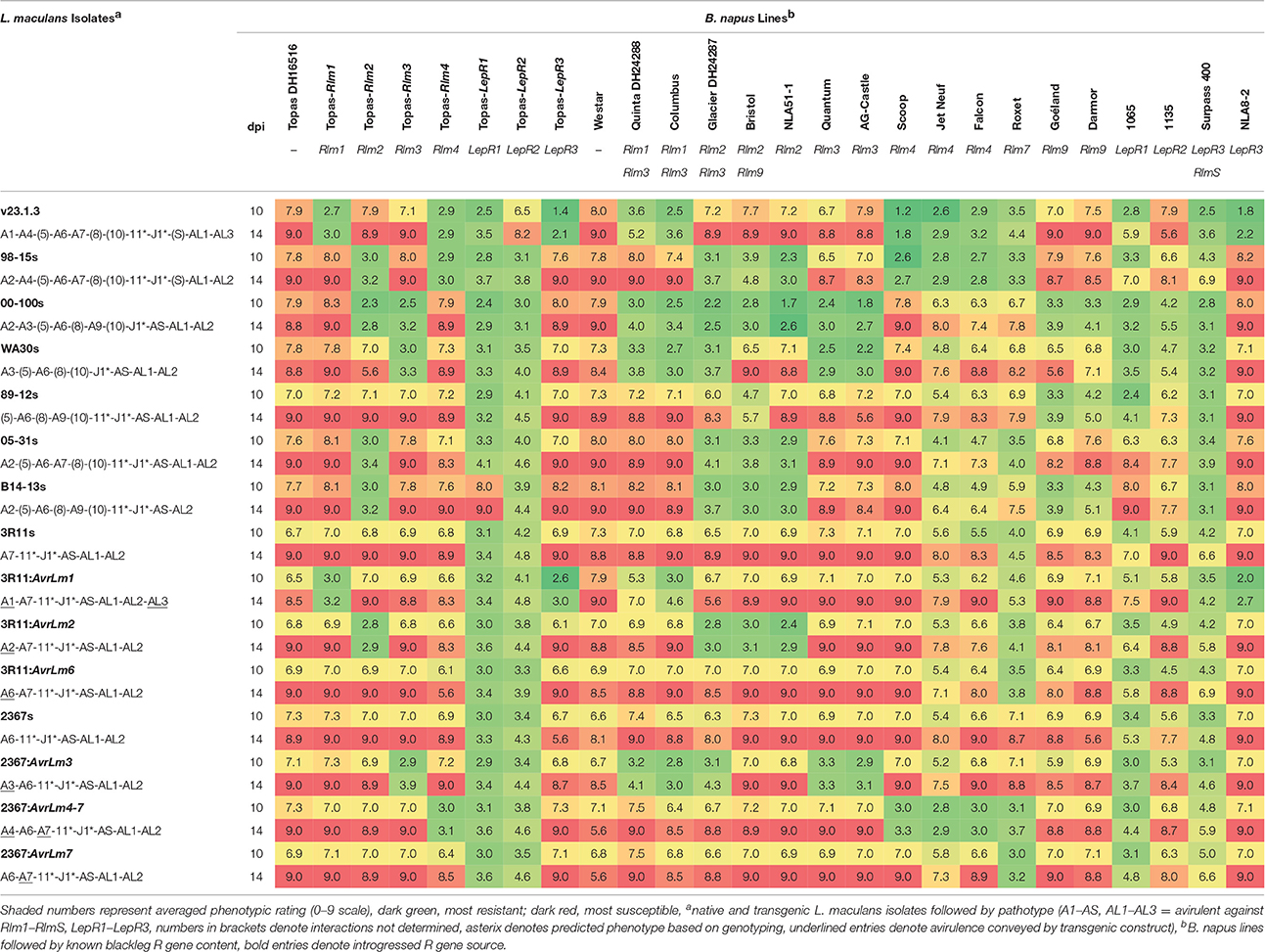
Table 2. Phenotypic characterisation of L. maculans resistance response for ILs, donor parents and other B. napus controls.
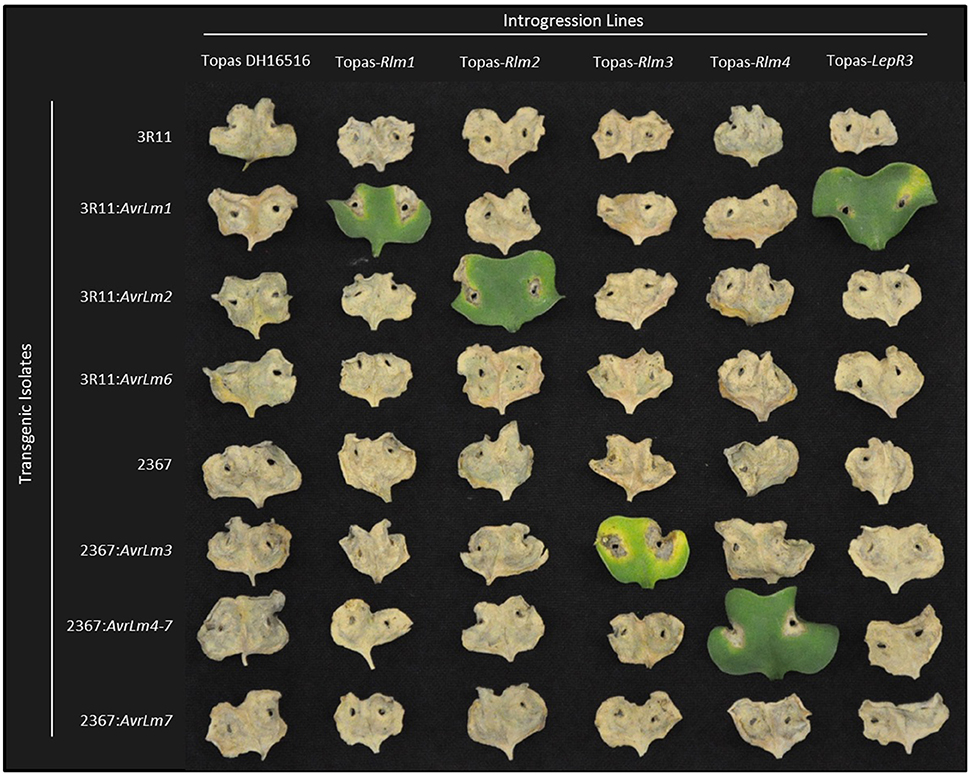
Figure 2. Confirmation of introgression line R gene content though transgenic complementation of avirulence determinants. Cotyledon reactions of susceptible B. napus line Topas DH16516 and five ILs challenged with two native L. maculans isolates (3R11 & 2367) and six transgenic isolates carrying additional Avr gene constructs. Cotyledons shown at 14 days post-infection.
The clear differential between susceptible and resistant reactions observed for the ILs was not true for several of the other control lines tested, with incompatible interactions resulting in intermediate to mildly susceptible mean rating scores. For example, the Rlm1 line Quinta DH24288 (donor parent of the Topas-Rlm1 IL) produced an intermediate score of 5.2 when challenged with the AvrLm1 isolate v23.1.3, and ratings progressed from 5.3 to 7.0 between 10 and 14 dpi when inoculated with the transgenic isolate 3R11:AvrLm1. This weak response was not mirrored by another Rlm1 variety, Columbus, which resulted in mean scores of 3.6 and 4.6 when challenged with v23.1.3 and 3R11:AvrLm1, respectively. While all isolates, with the exception of B14-13s, were clearly avirulent on the IL Topas-LepR1, several isolates produced intermediate or susceptible scores (5.3 to 8.4) on the LepR1 donor line 1065. The most dramatic difference between IL and donor line and was observed for Topas-LepR2 and 1135. While all isolates except v23.1.3 were avirulent on Topas-LepR2 at 14 dpi (3.1 to 4.8), all isolates were rated as intermediate to susceptible (5.4 to 9) on the LepR2 donor parent 1135. Though clear resistance reactions (scores <5) were observed for at least a few individual inoculation points on 1135 cotyledons for most isolates, a wide distribution of rating scores was observed, usually resulting in an overall susceptible rating. Even isolates that produced intermediate averaged rating scores, such as 00-100s, a broad range of individual lesion ratings, including several highly-infected lesions (score 7–8), were observed. This volatility was not observed for Topas-LepR2 which displayed a much more consistent resistance response (Figure 3).
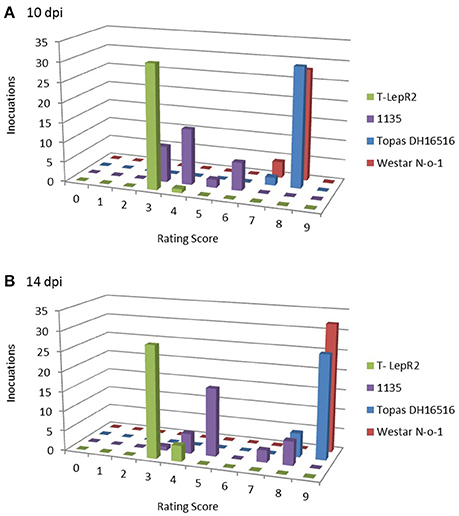
Figure 3. Distribution of cotyledon lesion ratings for LepR2 lines Topas-LepR2 and 1135. Vertical bars represent number of lesions scored for each rating scale graduation (0, no infection; 9, complete tissue collapse), total 32 lesions per line, at (A) 10 days post-inoculation (dpi) and (B) 14 dpi, when challenged with L. maculans isolate 00–100.
In phenotypic comparisons between ILs and the other B. napus control lines, restriction of lesions in some B. napus lines was observed which were not associated with known Avr-R gene interactions, particularly for the variety line Jet Neuf carrying Rlm4. All Rlm4-virulent (avrLm4) isolates were slower to establish infection on Jet Neuf than on other susceptible lines (10 dpi scores 4.1–6.3) and some weak HR was observed around the infection sites. These lesions failed to reach full infection (<8) by 14 dpi. Similar, though less severe restriction of infection was also observed for the varieties Falcon and Roxet (Supplementary Figure 1).
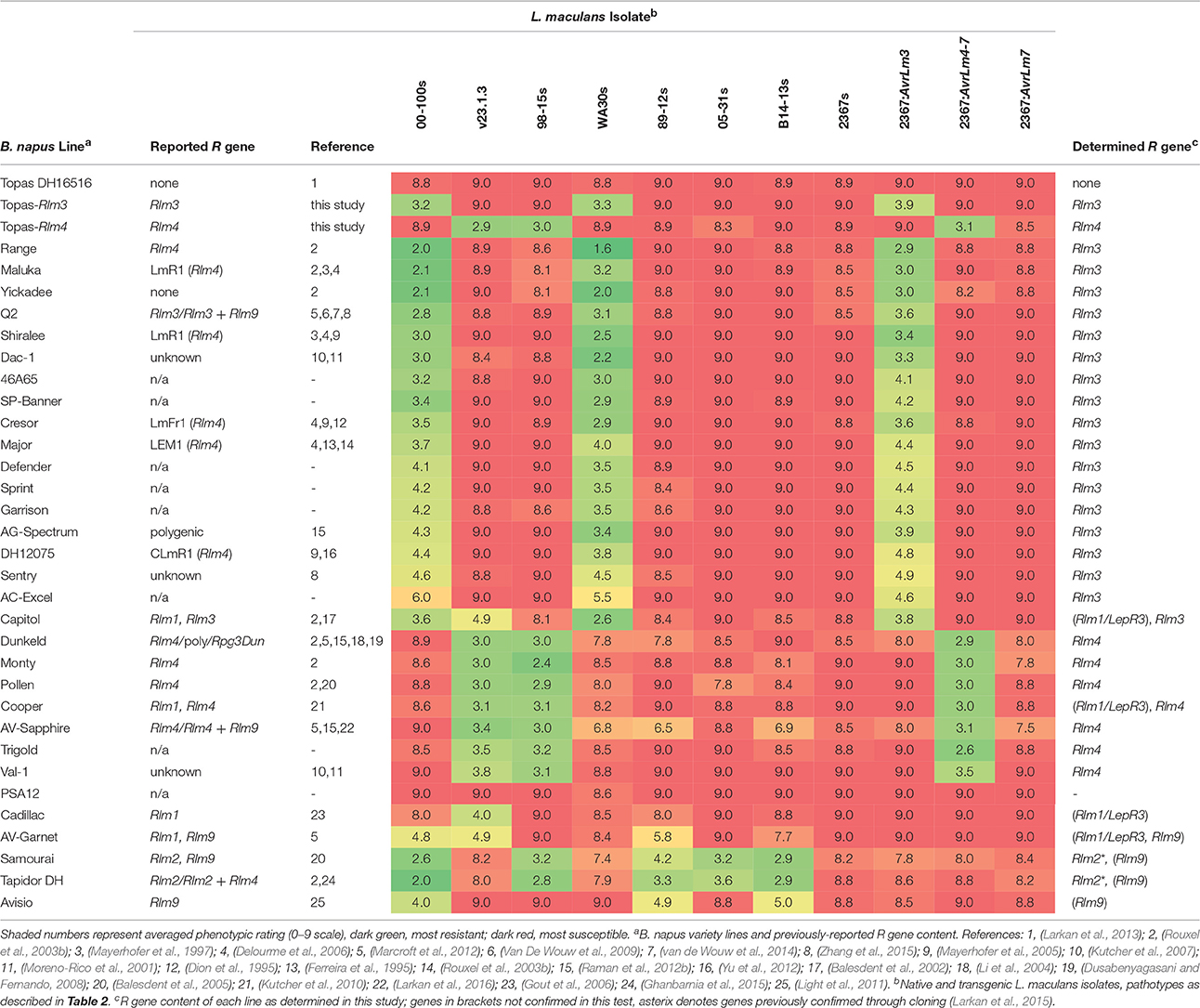
In additional testing, 31 B. napus variety lines held within AAFC collections, many of which had previously been assessed for use in differential L. maculans pathology testing and designated as Rlm3 or Rlm4 varieties, were assayed for the presence of Rlm3 and Rlm4 using the same isolates used to define the IL interactions (v23.1.3, 98-15s, 00-100s, WA30s, 89-12s, 05-31s, B14-13s, and 2367s), along with the transgenic isolates 2367:AvrLm3, 2367:AvrLm4-7 and 2367:AvrLm7. Topas DH16516, Topas-Rlm3 and Topas-Rlm4 were included as controls. Eighteen lines were confirmed as carrying Rlm3 by their interactions with both native AvrLm3 isolates (00-100s and WA30s) and the transgenic isolate 2367:AvrLm3 (Table 3). These 18 lines included six lines previously designated as Rlm4 varieties (Major, Crésor, Shiralee, Maluka, DH12075 and Range) and 10 lines which had not previously been characterized or had been deemed as “susceptible” varieties. Seven lines were confirmed to carry Rlm4, including two varieties not previously defined as Rlm4 varieties. Of particular note is the Australian variety Trigold, which is included as a susceptible control in some field-based disease nurseries (Raman et al., 2012b; Larkan et al., 2016). No lines were detected to carry both Rlm3 and Rlm4, though Tapidor DH, which we have previously shown to carry Rlm2 (Larkan et al., 2015), appears to additionally contain Rlm9. Similar to observations made during the characterisation of the ILs, B. napus variety lines carrying the same R gene (in this case Rlm3) displayed wide variance in the amplitude of HR after infection with AvrLm3 isolates. For example, variety Range produced highly resistant averaged ratings (1.6 to 2.9) while the average scores for AC-Excel were graded as intermediate to mildly susceptible (4.6 to 6.0). Rating scores for the Rlm4 varieties appeared to be more consistent (2.4 to 3.8) though several lines, including Dunkeld and AV-Sapphire, showed similar non-Avr associated restriction of lesions as previously observed for Jet Neuf. This was also apparent in the Rlm2, Rlm9 lines Samourai and Tapidor DH (Table 3).
Discussion
We present here the development and characterisation of single-blackleg R gene introgression lines for the study of the Brassica–Leptosphaeria pathosystem, allowing for the accurate assessment of Avr-R gene interactions in a highly homogenous genomic background free from ambiguous, phenotype-altering effects present in many other B. napus lines. Assessing the R gene content of B. napus material with varied genomic backgrounds poses several challenges. We noted several varieties which produced mild and inconsistent HR in a non-Avr dependent fashion, restricting the spread of infection in some tests. This phenomenon has been noted several times in the past (Rouxel et al., 2003b; Balesdent et al., 2005; Dilmaghani et al., 2009). Specifically, Dilmaghani et al. (2009) noted atypical reactions for tests performed using Jet Neuf which could not be explained by the presence of AvrLm4 in the affected isolates. Restriction of cotyledon lesions, resulting in less than full susceptibility, has also been observed and associated with adult-plant resistance and field-derived quantitative trait loci (QTL) in several studies (Delourme et al., 2004; Raman et al., 2012b; Huang et al., 2014; Larkan et al., 2016). While inoculation of variety lines affected by this minor restriction of infection still produced 14 dpi rating scores that were regarded as susceptible interactions (>6 on the 0–9 scale), using these lines as differentials under non-ideal conditions, or when using less vigorous inoculum, could lead to an incorrect assessment of either the virulence of the isolate being tested or the R gene content of the host line. Phenotyping of Brassica—Leptosphaeria interactions can also be affected by growth conditions, including temperature and moisture (Huang et al., 2006) and these appear to affect different R gene-mediated resistance responses unequally. It should be noted that we did not subject our seedlings to additional humidity to establish infection during the pathological assays, which is a common practice for L. maculans pathology. In our experience, L. maculans inoculum that fails to reach full infection on Topas DH16516 will also perform poorly on Westar and other susceptible varieties. Inoculum that is sub-optimal may then be subject to further inhibition by non-specific cotyledon resistance found in many lines, leading to a false classification of specific avirulence. The judicious use of susceptible controls is vital to evaluating the interaction of isolates with resistant material. Our results reflect as close to “ideal” conditions for pathotyping as we can achieve.
We also demonstrate here that a range of phenotypic responses can be observed for B. napus varieties carrying the same R gene. The most striking example of this phenomenon can be seen in the comparison between the IL Topas-LepR2 and 1135, the LepR2 donor line (Table 2). 1135 was produced from an initial B. napus x B. rapa ssp. “sylvestris” (BRS) F1, were the B. napus parent was DH12075, a Crésor × Westar F1-derived DH line (Mayerhofer et al., 2005). 1135 was produced from the DH12075 × BRS F1 following successive backcrossing to the DH line Westar N-o-1 before selection at BC4 (Yu et al., 2012), so the LepR2 gene is present in a largely Westar-derived genomic background in this line. In our testing, six of the eight non-transgenic isolates used in the characterisation of the ILs were able to produce mild to severe infection on 1135 cotyledons (averaged ratings of 7.3–9) while inoculation with the remaining two isolates (00-100s and WA30s) resulted in intermediate interactions (5.5. and 5.4, respectively). In contrast, seven of the eight isolates proved to be avirulent on the IL Topas-LepR2 (average scores 3.1 to 4.8) and only the reference isolate v23.1.3 proved to be truly virulent against LepR2 (8.2). Variation in resistance response was also clearly shown for Rlm3, in which we show a wide range of averaged lesion scores (2.0–6.0 for interactions with AvrLm3 isolate 00-100s) over 23 variety lines harboring the gene (Tables 2, 3). We speculate that this phenomenon can be attributed to variation in the genomic background of individual varieties, whereby the amplitude of HR is affected, either positively or negatively, by other host loci involved in recognition or signaling during pathogen invasion. The recognition of the L. maculans effectors AvrLm1 (Gout et al., 2006) and AvrLm2 (Ghanbarnia et al., 2015) by the R genes LepR3 (Larkan et al., 2013) and Rlm2 (Larkan et al., 2015), respectively, is known to involve at least two additional partner proteins in the membrane-bound signaling complex; SOBIR1 and BAK1 (Larkan et al., 2015; Ma and Borhan, 2015). Other host proteins may also be involved in the signaling complex, based on other plant interactions with apoplastic fungal pathogens (van der Hoorn and Kamoun, 2008; Stotz et al., 2014; Misas-Villamil et al., 2016). Subtle variation in either the expression or sequence of these partner proteins may account for the range of resistance responses observed. Indeed, in each case where we compared an R gene present in a largely Westar-derived genomic background; 1065 (LepR1), 1135 (LepR2), DH12075 (Rlm3) and the Rlm3 variety AC Excel (a selection of Karat x Westar Rakow, 1993), against the Topas DH16516-derived genomic background of the ILs (Topas-LepR1, Topas-LepR2 and Topas-Rlm3) we observed significantly stronger HR and much lower variance in IL cotyledon ratings across the test (Tables 2, 3, Figure 3). This would suggest that the Westar genomic background is somewhat deficient in its ability to fully express R gene-mediated cotyledon resistance. Another notable discrepancy was the comparatively weak reaction of Rlm1 in the donor parent line Quinta DH24288 compared to Topas-Rlm1 or Columbus when challenged with isolate v23.1.3 (AvrLm1) or the transgenic isolate 3R11:AvrLm1 (Table 2), again suggesting that substitution of the genomic background can affect the amplitude of the R gene response and hence the ability of the observer to accurately infer the presence or absence of corresponding Avr genes in the pathogen or the R gene present in the respective host plant.
Additionally, we have also observed what appears to be an effect of heterozygosity on the amplitude of HR. In particular, during the mapping of Rlm7, it was very difficult to determine the resistance status of individual BC1F1 seedlings, with most appearing intermediate to susceptible by 14 dpi. Only after assessment of the resulting BC1F2 progeny, where individuals harboring homozygous Rlm7 could be observed, was it possible to confidently assign resistance or susceptibility to each member of the mapping population (data not shown). This suggests a co-dominant phenotype for Rlm7. Additional testing for potential co-dominant phenotypic effects is required for blackleg R genes, particularly in regards to the accurate assessment of R gene content in modern hybrid (heterozygous) varieties.
Our results for the detection of Rlm3 and Rlm4 in B. napus cultivars conflict with several other reports in the literature. The blackleg R gene Rlm4 was first identified in the varieties Jet Neuf and Quinta V, a single-plant selection of cv. Quinta (Balesdent et al., 2001). However, in determining the Rlm4 resistance phenotype for Quinta V selection, the authors remarked on the heterogeneous nature of the Quinta variety seed, noting the isolate used to characterize the Rlm4 interaction showed virulence on the original seed lot obtained, which appeared to segregate for both Rlm1 and Rlm4 (Balesdent et al., 2001). As demonstrated in the present study and noted in previous reports (Kutcher et al., 2010; Larkan et al., 2013), the DH line Quinta DH24288, produced at AAFC Saskatoon from a Quinta seed lot that was originally obtained as breeder seed (Mengistu et al., 1991; P. H. Williams, pers. comm), is homozygous for Rlm1 and Rlm3. Genetic characterisation of the A07 blackleg R gene locus has a long history. A single dominant R gene was originally mapped from the French cultivar Major and termed LEM1 (Ferreira et al., 1995). LEM1 was later shown to induce resistance to the L. maculans PG2 isolate PHW1245 carrying the avirulence gene alm1 (Pongam et al., 1998). LEM1 from Major was later assumed to be Rlm4, as were mapped resistance loci LmFr1 from French cv. Crésor (Dion et al., 1995) and LmR1 from the Australian cultivars Shiralee and Maluka (Mayerhofer et al., 1997), based on map location and/or differential pathology tests (Rouxel et al., 2003b; Rimmer, 2006). The Crésor resistance, present in DH12075, a DH line produced from a Crésor × Westar F1, was again mapped (as CLmR1) and shown to localize to the same chromosomal position as LmR1 from Shiralee (Mayerhofer et al., 2005). Our tests suggest that all of these lines in fact carry Rlm3, not Rlm4. This is supported in part by previous work in which cultivars Shiralee, Maluka, Quinta, and Major were all shown to be susceptible to an isolate carrying AvrLm4-7 (Raman et al., 2012b) and also by the pedigree of the Rlm3 varieties Quantum and Q2, which contain L. maculans resistance derived from Maluka and Shiralee, respectively (Stringam et al., 1995a,b; 1999). However, the B. napus variety lines tested here have been selected over several generations for homozygous phenotypes when challenged with L. maculans, with several of the selections coming from source seed that was clearly segregating for R gene content on first assessment (data not shown). The variety lines used in the testing therefore do not necessarily represent the cultivar as it was upon commercial release, particularly in regards to the homozygosity of the R genes carried by the variety, nor do they necessarily represent the variety lines of the same name held by other institutions. Disparities in either R gene classification of B. napus varieties or pathotyping of L. maculans isolates, due to either error or variation in germplasm, only highlights the need for unambiguous, standardized material for pathological studies.
Brassica napus is an allotetraploid species, consisting of two highly-homologous diploid genomes known to undergo inter-genomic homoeologous recombination which can result in complex genomic rearrangements (Nicolas et al., 2007; Cai et al., 2014; Mason et al., 2014). Three of the ILs appear to be missing the lower portion of chromosome C06, likely due to the parental lines containing non-compatible A07-C06 genomic arrangements (Osborn et al., 2003). The whole-genome SNP analysis indicated that these lines carry two copies of lower A07; one copy from the R gene donor parent and a second copy replacing lower C06, derived from Topas DH16516. This event affects C06 from approximately 12.6 Mb to the end of the lower chromosome arm, with the duplication occurring at approximately 16.85 Mb on A07, around 550 kb below the mapped Rlm3 and Rlm4 interval, leaving these genes unaffected. However, the Rlm1 region of A07, positioned approximately 4 Mb below this point (Figure 1) is represented twice in the Topas-Rlm1 IL. We infer that Topas-Rlm1 carries the Quinta DH24288 Rlm1 allele on A07, while the Topas DH16516 rlm1 allele is carried on C06. This duplication of lower A07 does not appear to affect the phenotypic expression of the R genes in question, with all three ILs producing consistent resistance responses equal to or better than other B. napus varieties harboring the same R genes. However, this genomic event is something that should be taken into consideration when analyzing these lines in complex, whole-genome molecular studies. Significant homoeologous translocation events were also detected in several of the other B. napus varieties surveyed, the most notable resulting in very large null regions detected for lower C01 (22.92 Mb) in Goéland and for upper C02 (33.68 Mb) in Scoop (Supplementary Table 5). In these cases no marker heterozygosity was detected for the corresponding homoeologous A genome regions, suggesting these varieties are affected by non-reciprocal translocations, where the varieties carry two highly-monomorphic copies of the corresponding A01 and A02 chromosomal regions, respectively, with total loss of the affected the C genome regions.
Our hope is that the ILs described here will be widely distributed amongst the Leptosphaeria research community and will provide a strong basis for comparison of Leptosphaeria—Brassica interactions between labs. By introgressing the R genes into a common genomic background we have produced highly standardized material for Leptosphaeria pathology and related studies of host gene expression, and largely eliminated both negative and positive background variation which could interfere with the unambiguous pathotyping of L. maculans isolates and determination of R gene- specific defense responses. Work to add to the IL set through the production of new ILs (Rlm7, Rlm9 etc.) is on-going.
Author Contributions
NL, FY, DL, SR, and MB designed experiments. NL and FY carried out production of ILs and genetic mapping of R loci. NL performed all final pathological and genomic characterisations of lines. NL and MB drafted manuscript. All authors read and approved final manuscript with the exception of SR (deceased).
Funding
This work was funded by the Western Grains Development Fund, the AAFC-Industry Blackleg Consortium II (members include Agriculture Victoria Services Pty Ltd on behalf of the State of Victoria, Australia, Bayer, Crop Production Services (Canada), Dow AgroSciences, Lantmännen and Monsanto Canada) and the AAFC Canadian Agri-Science Cluster.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Sarah Kuzmicz, Kyla Horner, Christine Hammond, Helen Lui, Catherine Guenther, Stephanie McMillan, and Justin Skurdal for assistance with technical aspects of the project, Sally Vail (AAFC, Saskatoon) for providing several B. napus variety lines, Ginette Seguin-Swartz (AAFC, Saskatoon) for providing Topas DH16516, and Raymond Gadoua (Western Grains/Canola Council of Canada) for initial input on the project. We would also like to thank Thierry Rouxel (INRA-Bioger, France) for providing the reference L. maculans isolate v23.1.3 and Angela Van de Wouw & Barbara Howlett (University of Melbourne, Australia) for providing the isolate 3R11.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01771/full#supplementary-material
References
Balesdent, M. H., Attard, A., Ansan-Melayah, D. A., Delourme, R., Renard, M., and Rouxel, T. (2001). Genetic control and host range of avirulence toward Brassica napus cultivars Quinta and Jet Neuf in Leptosphaeria maculans. Phytopathology 91, 70–76. doi: 10.1094/PHYTO.2001.91.1.70
Balesdent, M. H., Attard, A., Kühn, M. L., and Rouxel, T. (2002). New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology 92, 1122–1133. doi: 10.1094/PHYTO.2002.92.10.1122
Balesdent, M. H., Barbetti, M. J., Li, H., Sivasithamparam, K., Gout, L., and Rouxel, T. (2005). Analysis of Leptosphaeria maculans race structure in a worldwide collection of isolates. Phytopathology 95, 1061–1071. doi: 10.1094/PHYTO-95-1061
Balesdent, M. H., Fudal, I., Ollivier, B., Bally, P., Grandaubert, J., Eber, F., et al. (2013). The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol. 198, 887–898. doi: 10.1111/nph.12178
Cai, G., Yang, Q., Yi, B., Fan, C., Edwards, D., Batley, J., et al. (2014). A complex recombination pattern in the genome of allotetraploid Brassica napus as revealed by a high-density genetic map. PLoS ONE 9:e109910. doi: 10.1371/journal.pone.0109910
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chèvre, A. M., Barret, P., Eber, F., Dupuy, P., Brun, H., Tanguy, X., et al. (1997). Selection of stable Brassica napus-B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor. Appl. Genet. 95, 1104–1111. doi: 10.1007/s001220050669
Chèvre, A. M., Eber, F., This, P., Barret, P., Tanguy, X., Brun, H., et al. (1996). Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B. nigra addition lines. Plant Breed. 115, 113–118. doi: 10.1111/j.1439-0523.1996.tb00884.x
Christianson, J. A., Rimmer, S. R., Good, A. G., and Lydiate, D. J. (2006). Mapping genes for resistance to Leptosphaeria maculans in Brassica juncea. Genome 49, 30–41. doi: 10.1139/g05-085
Clarke, W. E., Higgins, E. E., Plieske, J., Wieseke, R., Sidebottom, C., Khedikar, Y., et al. (2016). A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 129, 1887–1899. doi: 10.1007/s00122-016-2746-7
Delourme, R., Chevre, A. M., Brun, H., Rouxel, T., Balesdent, M. H., Dias, J. S., et al. (2006). Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur. J. Plant Pathol. 114, 41–52. doi: 10.1007/s10658-005-2108-9
Delourme, R., Pilet-Nayel, M. L., Archipiano, M., Horvais, R., Tanguy, X., Rouxel, T., et al. (2004). A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94, 578–583. doi: 10.1094/PHYTO.2004.94.6.578
Dilmaghani, A., Balesdent, M. H., Didier, J. P., Wu, C., Davey, J., Barbetti, M. J., et al. (2009). The Leptosphaeria maculans - Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 58, 1044–1058. doi: 10.1111/j.1365-3059.2009.02149.x
Dion, Y., Gugel, R. K., Rakow, G. F. W., Séguin-Swartz, G., and Landry, B. S. (1995). RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et de Not.] in canola (Brassica napus L.) Theor. Appl. Genet. 91, 1190–1194. doi: 10.1007/BF00220928
Dokken-Bouchard, F. L., Anderson, K., Bassendowski, K. A., Bauche, C., Bergsveinson, M., and Bruce, S. (2015). “Survey of Canola diseases in Saskatchewan, 2014,” in Canadian Plant Disease Survey, ed R. A. A. Morrall (Canadian Phytopathological Society). Available online at: http://phytopath.ca/wp-content/uploads/2016/04/CPDS_2016vol96.pdf
Dusabenyagasani, M., and Fernando, W. G. D. (2008). Development of a SCAR marker to track canola resistance against blackleg caused by Leptosphaeria maculans pathogenicity group 3. Plant Dis. 92, 903–908. doi: 10.1094/PDIS-92-6-0903
Eber, F., Lourgant, K., Brun, H., Lodé, M., Huteau, V., Coriton, O., et al. (2011). “Analysis of Brassica nigra chromosomes allows identification of a new effective Leptosphaeria maculans resistance gene introgressed in Brassica napus,” in 13th International Rapeseed Congress, (Prague).
Ferreira, M. E., Rimmer, S. R., Williams, P. H., and Osborn, T. C. (1995). Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 85, 213–217. doi: 10.1094/Phyto-85-213
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Fudal, I., Ross, S., Gout, L., Blaise, F., Kuhn, M. L., Eckert, M. R., et al. (2007). Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Mol. Plant Microbe Interact. 20, 459–470. doi: 10.1094/MPMI-20-4-0459
Ghanbarnia, K., Fudal, I., Larkan, N. J., Links, M. G., Balesdent, M.-H., Profotova, B., et al. (2015). Rapid identification of the Leptosphaeria maculans avirulence gene AvrLm2 using an intraspecific comparative genomics approach. Mol. Plant Pathol. 16, 699–709. doi: 10.1111/mpp.12228
Gout, L., Fudal, I., Kuhn, M.-L., Blaise, F., Eckert, M., Cattolico, L., et al. (2006). Lost in the middle of nowhere: the AvrLm1 avirulence gene of the dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60, 67–80. doi: 10.1111/j.1365-2958.2006.05076.x
Howlett, B. J., Idnurm, A., and Pedras, M. S. C. (2001). Leptosphaeria maculans, the causal agent of blackleg disease of Brassicas. Fungal Genet. Biol. 33, 1–14. doi: 10.1006/fgbi.2001.1274
Huang, Y. J., Evans, N., Li, Z. Q., Eckert, M., Chèvre, A. M., Renard, M., et al. (2006). Temperature and leaf wetness duration affect phenotypic expression of Rlm6-mediated resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 170, 129–141. doi: 10.1111/j.1469-8137.2006.01651.x
Huang, Y. J., Qi, A., King, G. J., and Fitt, B. D. L. (2014). Assessing quantitative resistance against Leptosphaeria maculans (phoma stem canker) in Brassica napus (oilseed rape) in young plants. PLoS ONE 9:e84924. doi: 10.1371/journal.pone.0084924
Kutcher, H. R., Balesdent, M. H., Rimmer, S. R., Rouxel, T., Chevre, A. M., Delourme, R., et al. (2010). Frequency of avirulence genes in Leptosphaeria maculans in western Canada. Can. J. Plant Pathol. 32, 77–85. doi: 10.1080/07060661003594109
Kutcher, H. R., Keri, M., Mclaren, D. L., and Rimmer, S. R. (2007). Pathogenic variability of Leptosphaeria maculans in western Canada. Can. J. Plant Pathol. 29, 388–393. doi: 10.1080/07060660709507484
Larkan, N. J., Lydiate, D. J., Parkin, I. A. P., Nelson, M. N., Epp, D. J., Cowling, W. A., et al. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. doi: 10.1111/nph.12043
Larkan, N. J., Lydiate, D. J., Yu, F., Rimmer, S. R., and Borhan, M. H. (2014). Co-localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol. 14:1595. doi: 10.1186/s12870-014-0387-z
Larkan, N. J., Ma, L., and Borhan, M. H. (2015). The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 13, 983–992. doi: 10.1111/pbi.12341
Larkan, N. J., Raman, H., Lydiate, D. J., Robinson, S. J., Yu, F., Barbulescu, D. M., et al. (2016). Multi-environment QTL studies suggest a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus. BMC Plant Biol. 16:183. doi: 10.1186/s12870-016-0877-2
Li, H., Sivasithamparam, K., Barbetti, M. J., and Kuo, J. (2004). Germination and invasion by ascospores and pycnidiospores of Leptosphaeria maculans on spring-type Brassica napus canola varieties with varying susceptibility to blackleg. J. Gen. Plant Pathol. 70, 261–269. doi: 10.1007/s10327-004-0125-8
Light, K. A., Gororo, N. N., and Salisbury, P. A. (2011). Usefulness of winter canola (Brassica napus) race-specific resistance genes against blackleg (causal agent Leptosphaeria maculans) in southern Australian growing conditions. Crop Pasture Sci. 62, 162–168. doi: 10.1071/CP10187
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. P., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. doi: 10.1038/ncomms4930
Long, Y., Wang, Z., Sun, Z., Fernando, D. W. G., Mcvetty, P. B. E., and Li, G. (2011). Identification of two blackleg resistance genes and fine mapping of one of these two genes in a Brassica napus canola cultivar ‘Surpass 400’. Theor. Appl. Genet.122, 1223–1231. doi: 10.1007/s00122-010-1526-z
Ma, L., and Borhan, M. H. (2015). The receptor-like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1-triggered immunity. Front. Plant Sci. 6:933. doi: 10.3389/fpls.2015.00933
Marcroft, S. J., Elliott, V. L., Cozijnsen, A. J., Salisbury, P. A., Howlett, B. J., and Van De Wouw, A. P. (2012). Identifying resistance genes to Leptosphaeria maculans in Australian Brassica napus cultivars based on reactions to isolates with known avirulence genotypes. Crop Pasture Sci. 63, 338–350. doi: 10.1071/CP11341
Mason, A. S., Batley, J., Bayer, P. E., Hayward, A., Cowling, W. A., and Nelson, M. N. (2014). High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. New Phytol. 202, 964–974. doi: 10.1111/nph.12706
Mayerhofer, R., Bansal, V. K., Thiagarajah, M. R., Stringam, G. R., and Good, A. G. (1997). Molecular mapping of resistance to Leptosphaeria maculans in Australian cultivars of Brassica napus. Genome 40, 294–301. doi: 10.1139/g97-041
Mayerhofer, R., Wilde, K., Mayerhofer, M., Lydiate, D., Bansal, V. K., Good, A. G.a.P., et al. (2005). Complexities of chromosome landing in a highly duplicated genome: toward map-based cloning of a gene controlling blackleg resistance in Brassica napus. Genetics 171, 1977–1988. doi: 10.1534/genetics.105.049098
Mengistu, A., Rimmer, S. R., Koch, E., and Williams, P. H. (1991). Pathogenicity grouping of isolates of Leptosphaeria maculans on Brassica napus cultivars and their disease reaction profiles on rapid-cycling Brassicas. Plant Dis. 75, 1279–1282. doi: 10.1094/PD-75-1279
Misas-Villamil, J. C., Van Der Hoorn, R. A. L., and Doehlemann, G. (2016). Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 212, 902–907. doi: 10.1111/nph.14117
Moreno-Rico, O., Frias-Trevi-o, A. G., Luna-Ruiz, J. J., Manzano-Flores, D. E., Romero-Cova, S., and Séguin-Swartz, G. (2001). Characterization and pathogenicity of isolates of Leptosphaeria maculans from Aguascalientes and Zacatecas, Mexico. Can. J. Plant Pathol. 23, 270–278. doi: 10.1080/07060660109506940
Nicolas, S. D., Mignon, G. L., Eber, F., Coriton, O., Monod, H., Clouet, V., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175, 487–503. doi: 10.1534/genetics.106.062968
Osborn, T. C., Butrulle, D. V., Sharpe, A. G., Pickering, K. J., Parkin, I. A. P., Parker, J. S., et al. (2003). Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165, 1569–1577.
Parlange, F., Daverdin, G., Fudal, I., Kuhn, M.-L., Balesdent, M.-H., Blaise, F., et al. (2009). Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. doi: 10.1111/j.1365-2958.2008.06547.x
Plissonneau, C., Daverdin, G., Ollivier, B., Blaise, F., Degrave, A., Fudal, I., et al. (2016). A game of hide and seek between avirulence genes AvrLm4-7 and AvrLm3 in Leptosphaeria maculans. New Phytol. 209, 1613–1624. doi: 10.1111/nph.13736
Pongam, P., Osborn, T. C., and Williams, P. H. (1998). Genetic analysis and identification of amplified fragment length polymorphism markers linked to the alm1 avirulence gene of Leptosphaeria maculans. Phytopathology 88, 1068–1072. doi: 10.1094/PHYTO.1998.88.10.1068
Raman, H., Raman, R., and Larkan, N. (2013). “Genetic dissection of blackleg resistance loci in rapeseed (Brassica napus L.),” in Plant Breeding from Laboratories to Fields, ed S. B. Andersen (Rijeka: InTech), 85–120.
Raman, R., Taylor, B., Lindbeck, K., Coombes, N., Barbulescu, D., Salisbury, P., et al. (2012a). Molecular mapping and validation of Rlm1 gene for resistance to Leptosphaeria maculans in canola (Brassica napus L.). Crop Pasture Sci. 63, 1007–1017. doi: 10.1071/CP12255
Raman, R., Taylor, B., Marcroft, S., Stiller, J., Eckermann, P., Coombes, N., et al. (2012b). Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.). Theor. Appl. Genet. 125, 405–418. doi: 10.1007/s00122-012-1842-6
Rimmer, S. R. (2006). Resistance genes to Leptosphaeria maculans in Brassica napus. Can. J. Plant Pathol. 28, S288–S297. doi: 10.1080/07060660609507386
Rouxel, T., Grandaubert, J., Hane, J. K., Hoede, C., Van De Wouw, A. P., Couloux, A., et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat. Commun. 2, 202. doi: 10.1038/ncomms1189
Rouxel, T., Penaud, A., Pinochet, X., Brun, H., Gout, L., Delourme, R., et al. (2003a). A 10-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape. Eur. J. Plant Pathol. 109, 871–881. doi: 10.1023/A:1026189225466
Rouxel, T., Willner, E., Coudard, L., and Balesdent, M. H. (2003b). Screening and identification of resistance to Leptosphaeria maculans (stem canker) in Brassica napus accessions. Euphytica 133, 219–231. doi: 10.1023/A:1025597622490
Stachowiak, A., Olechnowicz, J., Jedryczka, M., Rouxel, T., Balesdent, M. H., Happstadius, I., et al. (2006). Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur. J. Plant Pathol. 114, 67–75. doi: 10.1007/s10658-005-2931-z
Stotz, H. U., Mitrousia, G. K., De Wit, P. J. G. M., and Fitt, B. D. L. (2014). Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500. doi: 10.1016/j.tplants.2014.04.009
Stringam, G. R., Bansal, V. K., Thiagarajah, M. R., Degenhardt, D. F., and Tewari, J. P. (1995a). Development of an agronomically superior blackleg resistant canola cultivar in Brassica napus L. using doubled haploidy. Can. J. Plant Sci. 75, 437–439. doi: 10.4141/cjps95-072
Stringam, G. R., Degenhardt, D. F., Thiagarajah, M. R., and Bansal, V. K. (1995b). Quantum summer rape. Can. J. Plant Sci. 75, 903–904. doi: 10.4141/cjps95-151
Stringam, G. R., Degenhardt, D. F., Thiagarajah, M. R., and Bansal, V. K. (1999). Q2 summer rape. Can. J. Plant Sci. 79, 597–598. doi: 10.4141/P98-136
Utermark, J., and Karlovsky, P. (2008). Genetic transformation of filamentous fungi by Agrobacterium tumefaciens. Nat. Protoc. Exch. 10.1038/nprot.2008.83 Available online at: http://www.nature.com/protocolexchange/protocols/427
van der Hoorn, R. A. L., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
van de Wouw, A. P., Lowe, R. G. T., Elliott, C. E., Dubois, D. J., and Howlett, B. J. (2014). An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. doi: 10.1111/mpp.12105
Van De Wouw, A. P., Marcroft, S. J., Barbetti, M. J., Hua, L., Salisbury, P. A., Gout, L., et al. (2009). Dual control of avirulence in Leptosphaeria maculans towards a Brassica napus cultivar with ‘sylvestris-derived’ resistance suggests involvement of two resistance genes. Plant Pathol. 58, 305–313. doi: 10.1111/j.1365-3059.2008.01982.x
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Yu, F., Lydiate, D. J., Gugel, R. K., Sharpe, A. G., and Rimmer, S. R. (2012). Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus. Mol. Breed. 30, 1495–1506. doi: 10.1007/s11032-012-9735-6
Yu, F., Lydiate, D. J., Hahn, K., Kuzmicz, S., Hammond, C., and Rimmer, S. R. (2007). “Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus x B. rapa subsp. Sylvestris,” in 12th International Rapeseed Congress, (Wuhan).
Yu, F., Lydiate, D. J., and Rimmer, S. R. (2005). Identification of two novel genes for blackleg resistance in Brassica napus. Theor. Appl. Genet. 110, 969–979. doi: 10.1007/s00122-004-1919-y
Keywords: Brassica napus, Leptosphaeria maculans, introgression lines, resistance genes, blackleg, differentical lines
Citation: Larkan NJ, Yu F, Lydiate DJ, Rimmer SR and Borhan MH (2016) Single R Gene Introgression Lines for Accurate Dissection of the Brassica - Leptosphaeria Pathosystem. Front. Plant Sci. 7:1771. doi: 10.3389/fpls.2016.01771
Received: 26 September 2016; Accepted: 10 November 2016;
Published: 28 November 2016.
Edited by:
Dirk Balmer, Syngenta Crop Protection, SwitzerlandReviewed by:
Jens Staal, Ghent University, BelgiumZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2016 Larkan, Yu, Lydiate, Rimmer and Borhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Hossein Borhan, SG9zc2Vpbi5Cb3JoYW5AYWdyLmdjLmNh
 Nicholas J. Larkan
Nicholas J. Larkan Fengqun Yu1
Fengqun Yu1 Derek J. Lydiate
Derek J. Lydiate M. Hossein Borhan
M. Hossein Borhan