- Department of Agronomy, North China Key Laboratory for Germplasm Resources of Education Ministry, Hebei Agricultural University, Baoding, China
Cotton is one of the most economically important crops, but most cultivated varieties lack adequate innate immunity or resistance to Verticillium wilt. This results in serious losses to both yield and fiber quality. To identify the genetic resources for innate immunity and understand the pathways for pathogen defenses in this crop, here we focus on orthologs of the central Arabidopsis thaliana defense regulator Enhanced Disease Susceptibility 1 (EDS1). The full-length cDNA of GbEDS1 was obtained by screening the full-length cDNA library of Gossypium barbadense combining with RACE strategy. Its open reading frame is 1848 bp long, encoding 615 amino acid residues. Sequence analysis showed that GbEDS1 contains a conserved N-terminal lipase domain and an EDS1-specific KNEDT motif. Expression profiling indicated that the gene is induced by Verticillium dahliae as well as salicylic acid (SA) treatment. Subcellular localization assays revealed that GbEDS1 is located in the cell cytoplasm and nucleus. Overexpression of GbEDS1 in Arabidopsis dramatically up-regulated SA and H2O2 production, resulting in enhanced disease resistance to V. dahliae. Silencing of GbEDS1 in G. barbadense significantly decreased SA and H2O2 accumulation, leading to the cotton more susceptibility. Moreover, combining the gene expression results from transgenic Arabidopsis and silenced-GbEDS1 cotton, it indicated that GbEDS1 could activate GbNDR1 and GbBAK1 expression. These findings not only broaden our knowledge about the biological role of GbEDS1, but also provide new insights into the defense mechanisms of GbEDS1 against V. dahliae in cotton.
Introduction
Cotton (Gossypium sp.) is one of the most economically valuable crops globally. Because its products are used as textile fibers, feed stock, foodstuff, oil, and biofuels. However, its yields have been severely reduced due to increasingly unfavorable environmental conditions, including those associated with biotic and abiotic stresses. Among these, Verticillium wilt, induced by Verticillium dahliae Kleb., is one of the most destructive diseases of G. hirsutum L., and has been detected in most areas where that species is cultivated. Losses to annual yields have risen to more than 1.5 million bales worldwide (Cai et al., 2009).
This soil-borne pathogen penetrates the roots and systemically infects the plant through the xylem (Daayf et al., 1995; Klosterman et al., 2009). Affected cotton plants usually exhibit symptoms of chlorosis or necrosis in the leaves, discoloration of the stem vascular bundles, a decrease in photosynthesis and an increase in respiration, and significant declines in plant biomass and yield (Sink and Grey, 1999). So far, developing resistant cultivars has been regarded as the most effective strategy in combating this stubborn disease. Although the introduction of genetic resistance is considered the best and most sustainable management approach, a lack of information about the molecular mechanisms for cotton resistance against V. dahliae has slowed progress in those research efforts.
Complex molecular mechanisms protect plants from the effects of insects and pathogens. These include constitutive physical and chemical barriers that hinder pathogen entry and infection as well as a powerful defense system that can induce the expression of various stress-related genes (Creelman and Mullet, 1997; Reymond and Farmer, 1998; Glazebrook, 2005; Jones and Dangl, 2006; Howe and Jander, 2008; Fu et al., 2012). The core genetic components that regulate upstream events in the biosynthesis of salicylic acid (SA) include EDS1, PAD4, and SAG101 (Feys et al., 2001; Wiermer et al., 2005; Pieterse et al., 2009; Rietz et al., 2011). In Arabidopsis thaliana, EDS1 is a key positive regulator of basal resistance to biotrophic and hemi-biotrophic pathogens and it participates in effector-triggered immunity that is mediated by a subset of resistance genes (Aarts et al., 1998; Feys et al., 2001; Wiermer et al., 2005; Wirthmueller et al., 2007). EDS1 could control the production of SA required for basal defense and systemic resistance against virulent pathogens (Attaran et al., 2009; Vlot et al., 2009) and is essential for ETI (Feys et al., 2001, 2005; Wirthmueller et al., 2007). In SA signal pathway, EDS1 together with PHYTOALEXIN DEFICIENT4 (PAD4) also needed for many SA-independent gene expression changes in response to infection (Falk et al., 1999; Jirage et al., 1999; Wiermer et al., 2005; Wang L. et al., 2008). EDS1/PAD4 could indirectly up-regulate the expression of isochorismate synthase (SALICYLIC ACID INDUCTIOND EFICIIENT2/ISOCH OR ISMATE SYNTHASE1, SID2/ICS1), an enzyme required for SA synthesis (Wildermuth et al., 2001). Furthermore, the expression of EDS1 could be regulated by calmodulin (CAMTA3), which interacts with the promoter of EDS1 and represses its expression. So EDS1 plays critical role in Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity (Du et al., 2009; Truman et al., 2013; Zhang et al., 2014). In addition, EDS1 plays a crucial role in transducing redox signals in response to biotic and abiotic stresses (Mateo et al., 2004; Straus et al., 2010). Those reactions include the production of SA, reactive oxygen species (Rusterucci et al., 2001), and secondary metabolites, as well as the expression of both SA-dependent and -independent defense genes (Bartsch et al., 2006).
All of these reports suggest that EDS1 plays crucial roles in biotic/abiotic stresses (Attaran et al., 2009; Du et al., 2009; Vlot et al., 2009; Truman et al., 2013; Zhang et al., 2014). Most previous studies of EDS1 gene have emphasized model species such as Arabidopsis and tomato (Rusterucci et al., 2001; Liu et al., 2002; Hu et al., 2005; Rietz et al., 2011). As one of the most important fiber and oil crops, cotton experiences severely impaired growth and yields when exposed to V. dahliae. Therefore, to improve our understanding of the resistance/defense mechanism and identify more resistance genes, we constructed a SSH library and a full- length cDNA library from the tolerance cultival cv. Jimian20 and resistant G. barbadense cv. Pima90-53, respectively (Wang X.F. et al., 2008; Zhang et al., 2013). From both cDNA library, we detected EDS1 gene and found it involved in cotton defense against V. dahliae. However, cotton EDS1 gene has not been studied deeply except Su’ report (2014). In this study, we overexpressed GbEDS1 in Arabidopsis plants and then investigated the effect on V. dahliae invasion and the molecular mechanism of GbEDS1 in resistance to V. dahliae.
Materials and Methods
Plant Growth and Seedling Treatments
Seeds of Gossypium barbadense cv. Pima90–53 (resistant) were germinated in Hoagland solution-saturated vermiculite in a controlled environment chamber [28/25°C (day/night), 16-h photoperiod, and 80% relative humidity]. Plants of the wild-type (WT) A. thaliana Columbia ecotype and transgenic lines (see below) were grown at 22°C, under long-day conditions (16 h light/8 h dark) (Haffner et al., 2010) in the greenhouse.
The highly aggressive defoliating V. dahliae fungus strain, isolated from field-grown upland cotton plants that showed typical symptoms of infection, was cultured on a potato-dextrose agar medium for 7 days at 25°C. Colonies were then incubated for 5 days in Czapek’s medium [NaNO3 (0.3% w/v), MgSO4 (0.1% w/v), KH2PO4 (0.1% w/v), FeSO4 (0.0002% w/v), KCl (0.1% w/v), and sucrose (3% w/v); pH 6.0] at 25°C. Prior to inoculation treatments, the concentration of spores was adjusted to approximately 107 conidia per mL with deionized water. We used a GFP-tagged V. dahliae strain (Vd-gfp77), provided by Professor Dai XF (Chinese Academy of Agricultural Sciences), to observe the process of colonization during the experimental period. Its pathogenicity on cotton is related to that of the WT isolate Vd991 (Xu et al., 2013). When the cotton seedlings were 10 days old, they were treated with 10 mM methyl salicylate (MeSA) (Wang et al., 2007), which is converted to SA in the leaf tissue (Shulaev et al., 1997). For pathogen treatment, 10-day-old seedlings were uprooted gently and their roots rinsed in distilled water. The roots were then dipped for 3 min in the conidia suspension. Control plants were treated similarly with distilled water (Zhang et al., 2011). The root tissues were harvested separately from all the treatments after 0, 12, 24, 48, 72, and 96 h and were frozen in liquid nitrogen immediately.
RNA Preparation and Gene Cloning
Total RNA was extracted from cotton roots with a PlantRNA Kit (Tiangen, China) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 5 μg of total RNA with a StrataScript Kit (Stratagene USA). Based on the candidate sequence of GbEDS1 from the full-length cDNA library, we designed a pair of primers (GbEDS1–F1/GbEDS1–R1) according to the lateral flanking sequence of the open reading frame (ORF) of the candidate sequence. The primers are listed in Supplementary Table S1.
Bioinformatics Analysis
Nucleotide and deduced amino acid sequences were investigated through an NCBI/GenBank/Blast. Sequences were aligned and compared with those of other species via DNAMAN, and a phylogenetic tree was constructed for GbEDS1 and similar genes from other plants. The signal sequences were predicted with SignalP1. Functional regions and activity sites were identified from the PROSITE2 and SMART motif search programs3. Sub-cellular localization was predicted by Psort (Collings, 2013).
Gene Expression Analysis
Total RNA was extracted from cotton as previously described (Zhang et al., 2011). First-strand cDNA was synthesized from 1 μg of total RNA using the iScriptTM cDNA synthesis Kit (Bio-Rad, USA) system. For RT-PCR, gene-specific primers were used to analyze the change in GbEDS1 expression between mock- and V. dahliae-inoculated plants. GhUBQ14 (GenBank: DW505546) from cotton was used as the reference gene to normalize the total amount of cDNA in each reaction (Artico et al., 2010). All qPCRs were performed according to the guidelines of the Minimum Information for Publication of Quantitative Real Time PCR Experiments (Bustin et al., 2009). Diluted cDNA was used with SYBR Green on an ABI 7500 Real Time PCR system (Applied Biosystems, USA). Three biologically independent experiments were run for this analysis. Relative fold-changes were calculated per the 2-ΔΔCt method, as described by Livak and Schmittgen (2001). All primers are shown in Supplementary Table S1.
Subcellular Localization of GbEDS1::GFP Fusion Proteins
To find the cellular localization of GbEDS1, we amplified the coding region of GbEDS1 with GbEDS1–F1/GbEDS1–R2 primers and cloned it into the pCamE-GFP vector to generate a GbEDS1–GFP in-frame fusion with green fluorescence protein (GFP). The plasmid was introduced into onion epidermal cells by particle bombardment, as previously described (Collings, 2013). Afterward, the tissues were incubated for 24 h on an MS agar medium under darkness at 22°C before being examined with a fluorescence microscope.
Generation of Transgenic Arabidopsis Plants
The ORF of GbEDS1 was amplified with primers GbEDS1-F1 and GbEDS1-R1 (Supplementary Table S1), and then inserted into binary vector pBI121 under the control of the CaMV 35S promoter via XbaI and SacI sites. The recombinant construct vector was introduced into Agrobacterium tumefaciens (strain GV3101), and then transferred into eds1 mutant plants of Arabidopsis (Columbia background) by floral-dipping (Clough and Bent, 1998). The transformants were first screened on kanamycin (100 mg L-1) plates and then verified by RT-PCR using primers GbEDS1-F4/GbEDS1-R4 (Supplementary Table S1). Transgenic T3 lines were used for further experiments.
Assay for Disease Resistance
Verticillium wilt resistance was assessed in seedlings that had been gently inoculated with V. dahliae as we previously described (Zhang et al., 2011). All plants were irrigated with Hoagland’s nutrient solution. Disease development was monitored for up to 28 days post-inoculation (dpi). The degree of wilt resistance was evaluated along a scale of disease grades from 0 to 4 (Ma et al., 2000). The disease index was calculated according to the following formula: DI = [(Σ disease grades × number of infected plants) / (total number of checked plants × 4)] × 100 (Zhang et al., 2011). Values obtained for DI represented the status of infection for a population, with lower DI values indicating that the plants were more resistant.
We also quantified the V. dahliae biomass produced in transgenic plants (Jun et al., 2015). After visible symptoms were observed at 14 dpi, all aboveground tissues from each Arabidopsis genotype were harvested and flash-frozen in liquid nitrogen. The samples were ground to powder and approximately 100 mg of each was used for DNA isolation. Three-step Q-PCR was performed on a LightCycler 1.5 Instrument (Roche, Germany) using a SYBR Premix Ex TaqTM II kit (TaKaRa). For measuring the V. dahliae biomass, the internal transcribed spacer (ITS) region of the ribosomal DNA was targeted to generate a 200-bp amplicon by the fungus-specific primers ITS1-F and ST-VE1-R (Gardes and Bruns, 1993; Lievens et al., 2006). The large subunit of the RuBisCo gene from Arabidopsis was used for sample equilibration. The average fungal biomass was determined from at least three Verticillium-inoculated plants per genotype, and quantification was conducted as described by Ellendorff et al. (2009).
Virus-Induced Gene Silencing (VIGS) in Cotton
To investigate the role of EDS1 in Verticillium resistance, we used virus-induced gene silencing (VIGS). The TRV vectors and Agrobacterium tumefaciens for VIGS were prepared according to the method of Gao et al. (2011). Insert fragments to generate TRV::GbEDS1 and the positive control TRV::GbCLA1 were amplified from G. barbadense cv. Pima90-53. Primers to produce the TRV vectors are listed in Supplementary Table S1. The TRV2 plasmid and GbEDS1 fragment were digested with XbaI and SacI, then ligated using T4 DNA ligase (Takara, Japan), and transformed into A. tumefaciens GV3101 by electroporation. The TRV::GbCLA1 was obtained using the same method for TRV::GbEDS1. Subsequently, TRV vectors were agro-infiltrated (Gao et al., 2011) into the cotyledons of 7-day-old seedlings of G. barbadense cv. Pima90-53. The seedlings were then grown at 25°C under a 16-h photoperiod in a controlled environment chamber. A distinct bleaching phenotype for the leaves was observed at 10 days after infiltration in TRV::GbCLA1 plants. Therefore, inoculation with V. dahliae was performed at that time point. Two weeks after inoculation, wilt resistance began assessed. The percentage of diseased plants and a disease index (see below) were determined from about 30 seedlings per treatment and the assessment was repeated at least three times.
Expression Analysis of Defense-Related Genes in Transgenic Arabidopsis Plants
To study whether GbEDS1 overexpression in Arabidopsis influences SA signal transduction and the expression of defense-related genes, we monitored the expression of SA pathway genes (EDS1, NPR1, NDR1) (Shah, 2009; Vlot et al., 2009; Fu and Dong, 2013), a co-receptor and signaling regulator of different pattern recognition receptors—BRI1-associated kinase 1 (BAK1) (Gao et al., 2013), and defense marker genes (PR1, PR5) in transgenic Arabidopsis. For the 20-day transgenic Arabidopsis seedlings were removed gently from vermiculite and their roots were washed in sterile water. Then root part was dipped in the fresh spore suspension (about 107 conidia/mL). After inoculation, the seedlings were re-planted in the pot containing new vermiculite and were irrigated with Hoagland’s nutrient solution. Leaf tissues were sampled separately from transgenic and wild type Arabidopsis after 0, 6, 12, 24 hpi and were frozen in liquid nitrogen. The specificity of the primers in real-time PCR was tested by PCR amplification and sequencing. All PCR procedures were repeated three times and the data were normalized to reference genes according to the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Measurements of Total SA
In Arabidopsis, EDS1 was considered to be one of essential mediators in SA signaling (Falk et al., 1999; Jirage et al., 1999; Wiermer et al., 2005). SA and AtEDS1 were considered to function in a positive feedback regulatory loop (Bartsch et al., 2006). Thus, we want to know whether GbEDS1 could regulate SA level changes. Extraction and measurements of endogenous SA from seedlings were performed as previously described (Nandi et al., 2004). For each sample, 200 mg of leaves was ground and extracted once with 3 mL of 90% methanol and once with 3 mL of 100% methanol. The combined extracts were dried under N2 gas and suspended in 2.5 mL of 5% trichloroacetic acid. Afterward, the samples were acid-hydrolyzed by adding 200 mL of HCl and incubating in a boiling water bath for 30 min. The SA was extracted with 5 mL of a mixture containing cyclohexane:ethylacetate:isopropanol (50:50:1). Samples were dried under N2 gas and dissolved in 0.5 mL of the mobile phase (69:27:4 mix of water:methanol:glacial acetic acid) (Dadgar et al., 1985). They were then passed through a 0.22-mm filter, and 20 to 100 μL was used for HPLC. Concentrations of SA were determined by comparing our samples with available standards.
Determination of H2O2 Content
H2O2 accumulation has been considered to be an important parameter in biotic stresses because it usually regarded as an immunity-associated molecule (Camejo et al., 2016). To further investigation the function of GbEDS1 in defense against V. dahliae, we monitored the levels of H2O2 in transgenic line and GbEDS1-silenced plants. The seedlings leave inoculated with V. dahliae were sampled. Each sample was homogenized in pre-cooled phosphate-buffered saline (PBS), using 1 mL of buffer per 0.1 g of fresh tissue. The homogenate was centrifuged at 10,000 × g for 10 min at 4°C. Freshly isolated supernatant fractions were used immediately for measuring H2O2 with commercial kits (Jiancheng Biotech Inc., Nanjing, China). Adduct formation was measured spectrophotometrically at 405 nm using Thermo Scientific Multiskan FC (Shanghai, China) in strict accordance with the manufacturer’s instructions. Protein contents were determined with an Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China).
Analyses of SOD and POD Activities
The 4-week old Arabidopsis grown in pots were inoculated with V. dahliae according to the method of Zhang et al. (2011). Then the leaves from transgenic and mock plants were sampled at 6, 12, 24, 36, 48 hpi, respectively. The activities of superoxide dismutase (SOD) and peroxidase (POD) in each sample were measured using commercial kits (Jiancheng Biotech Inc., Nanjing, China) according to the manufacturer’s instructions. Protein contents were determined with an Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China).
Statistical Analysis
The experiments were repeated three times. All data were presented as means ± SE. Differences among treatments were evaluated with two-tailed unpaired Student’s t-tests, a value of P < 0.05 or P < 0.01 was considered to be statistically significant.
Results
Cloning and Characterization of the Full-Length GbEDS1 cDNA
Using the full-length cDNA library previously constructed from root samples of G. barbadense cv. Pima90-53 plants that had been inoculated by V. dahliae (Zhang et al., 2013), we obtained a 1706-bp fragment with a poly-A tail. Here, we employed a RACE strategy with GSPs and UPM to amplify the 5′-ends. The complete cDNA of GbEDS1 was assembled and firstly submitted to GenBank (Accession Number EU855795). The full length cDNA of GbEDS1 is 2258 bp, containing an ORF of 1848 bp, as well as a 201-bp 5′-UTR and a 206-bp 3′-UTR. ExPASy-Prosite analysis indicated that GbEDS1 has a conservative lipase-serine active-site signature “IVFTGHSSGG” (Supplementary Figure S1). In addition, this gene displays conserved serine (S), aspartic acid (D), and histidine (H) residues that form a putative hydrolase catalytic triad within its N-terminal lipase-like region (Figure 1) (Falk et al., 1999). In that putative lipase domain, the C-terminal region contains the specific motif KNEDT, which is highly conserved among EDS1 sequences from Populus poplar (Pp), Vitis vinifera (Vv), and A. thaliana (At) (Figure 1; Table 1), suggesting that our GbEDS1 is an ortholog of AtEDS1 and VvEDS1. Furthermore, GbEDS1-homologous sequences from three other Gossypium reference genome and previously reported cotton EDS1 gene from Su’ research were analyzed by BLAST. Sequences blast showed that there was only one copy of EDS1 gene in cotton genome. In particular, our GbEDS1 was more closely related to GrEDS1 (99% identity) than to GhEDS1 (98% identity) or GaEDS1 (97% identity) (Figure 2; Supplementary Figure S2). All above results indicated that EDS1 gene was very conservative across different cotton species. However, why the EDS1 gene from Su’ sequence sharing low identity with GrEDS1, GhEDS1, and GaEDS1 were 75.4, 74.4, and 74.6% is still unclear (Su et al., 2014).

FIGURE 1. Amino acid sequence alignment of EDS1 proteins from different plant species evaluated via ClustalW2. Putative lipase domain and EDS1-specific KNEDT motif are marked with red box and blue box, respectively. Serine (S), aspartate (D), and histidine (H) residues of putative lipase catalytic triad are indicated by asterisks. Pp, Populus poplar; Vv, Vitis vinifera; Gb, Gossypium barbadense; and At, Arabidopsis thaliana.
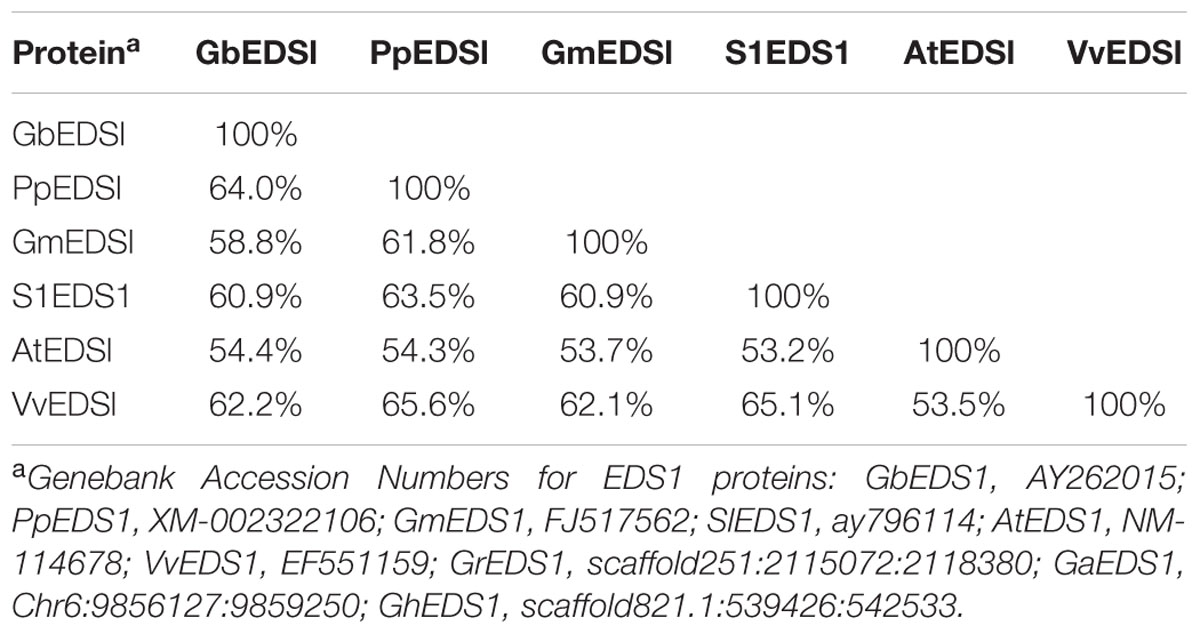
TABLE 1. Sequence homology analysis of GbEDS1 with five other EDS1 proteins, based on their complete ORF amino acid sequences.
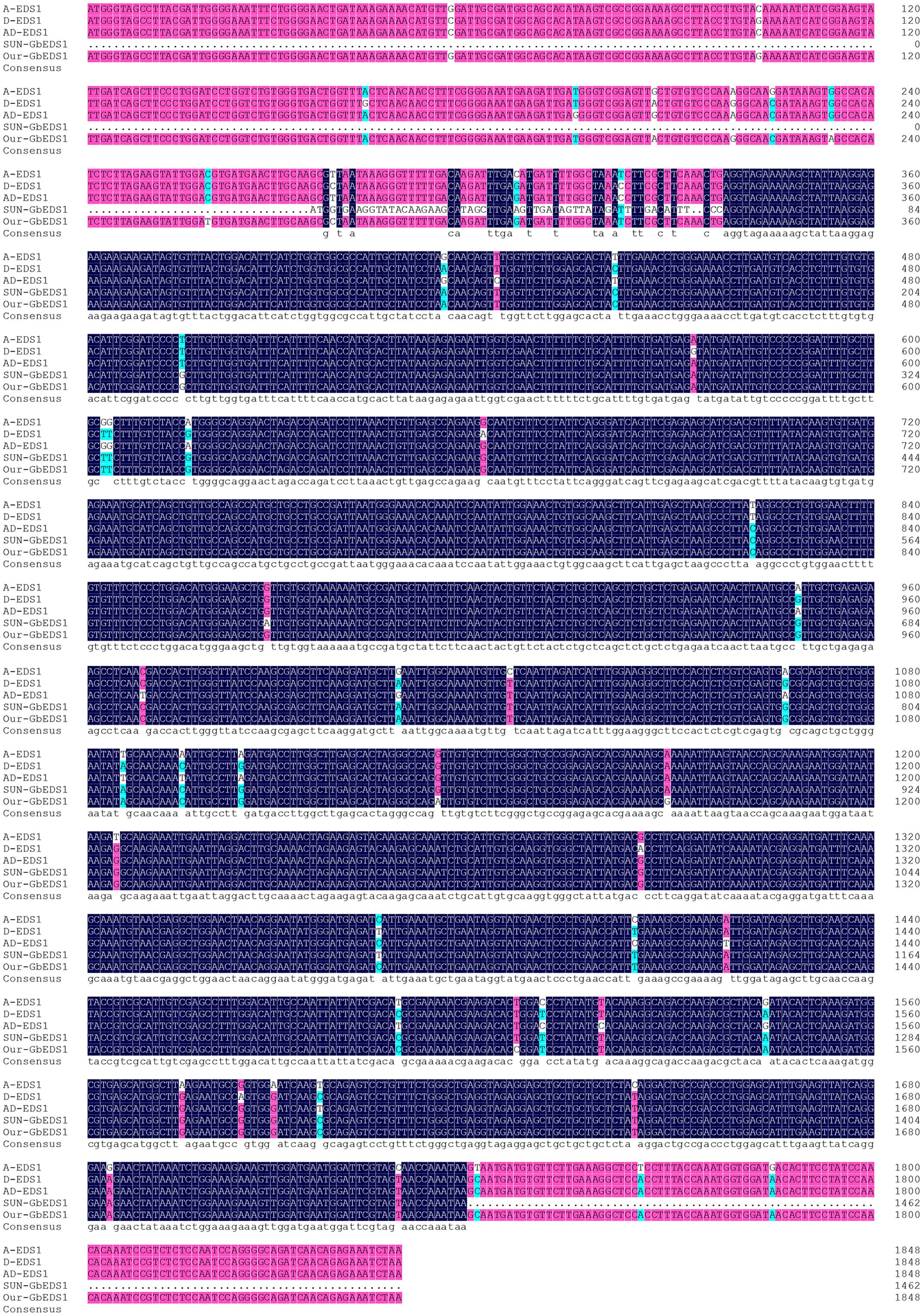
FIGURE 2. Alignment of the sequences of GbEDS1 with its homologous genes in cotton. Comparison of the sequence of EDS1 from G. barbadense, G. hirsutum, G. arboreum, and G. raimondii.
Expression of GbEDS1 was Induced by V. dahliae and SA Treatment
To test the effect of V. dahliae infection on the expression of GbEDS1, we extracted RNA from roots that were harvested at various time points following inoculation. RT-PCR analysis demonstrated that, compared with the uninfected control, GbEDS1 was obviously up-regulated in the infected tissues, with transcript levels being highest between 12 and 96 hpi and peaking at 72 hpi (Figure 3). This result was evidence that GbEDS1 gene is responsive to V. dahliae infection at the transcriptional level, and implied that it might be involved in defense against V. dahliae in G. barbadense.
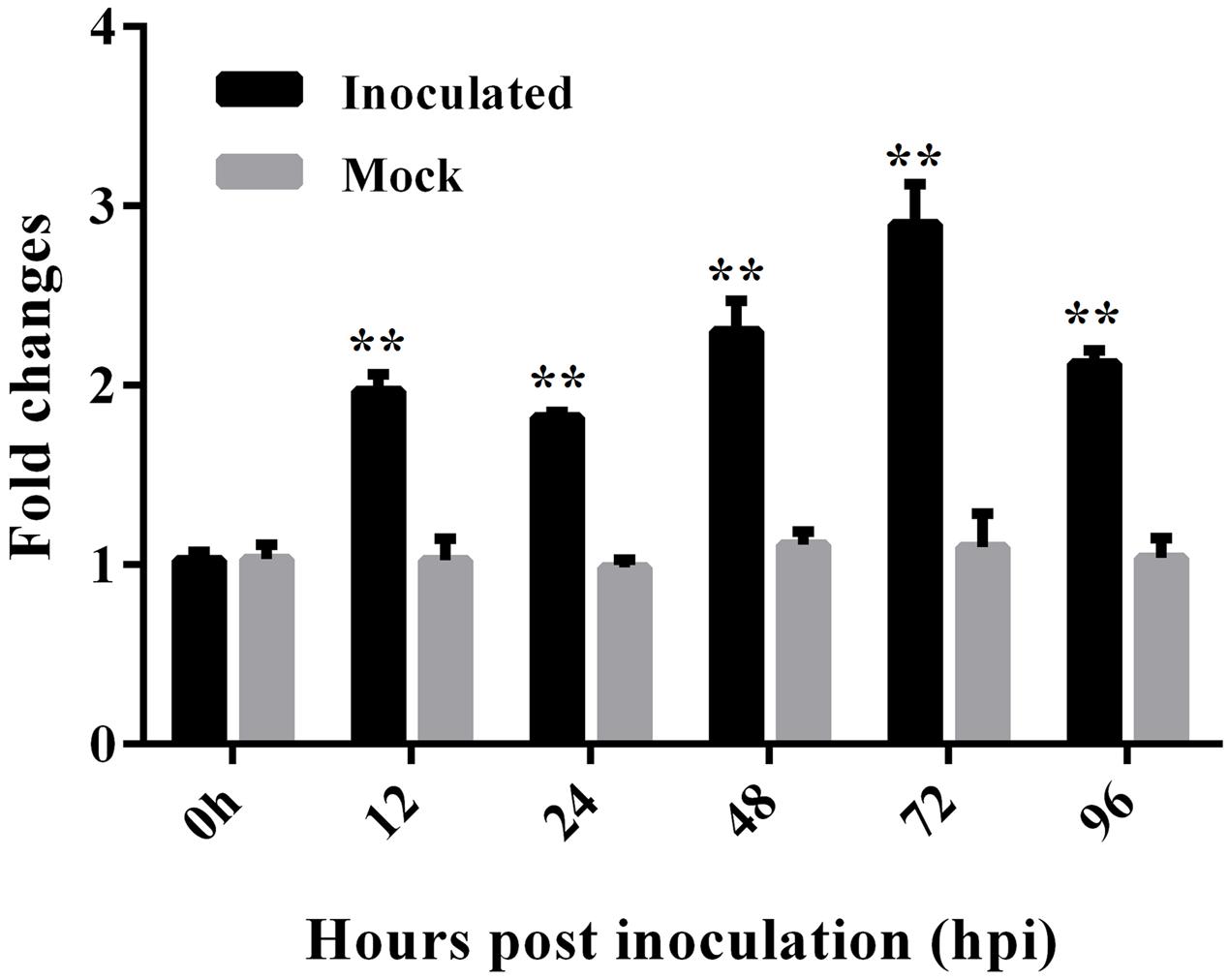
FIGURE 3. Expression analysis of GbEDS1 after induction by V. dahliae in root tissues of Pima90-53. Real-time PCR for time-course tests (12, 24, 48, 72, and 96 hpi) of GbEDS1 expression in response to V. dahliae. Bars represent levels of GbEDS1 transcripts relative to those of cotton GhUBQ14 (for normalization). Seedlings inoculated with water-only served as control. Data are means ± SD of values obtained from triplicate experiments. Asterisks indicate statistical significance (∗∗P < 0.01, Student’s t-test) in comparison with mock control.
Because EDS1 and SA are in a positive feedback loop in Arabidopsis, so we measured transcript level in G. barbadense leaves in response to SA. At 24 h after MeSA treatment, levels of GbEDS1 transcripts were significantly higher (P < 0.01) than the mock treatment (Supplementary Figure S3), suggesting that GbEDS1 is SA-responsive in G. barbadense.
Subcellular Localization of GbEDS1
To determine the cellular localization of GbEDS1, we fused its full-length cDNA, except for the stop codon, in-frame with the GFP-coding sequence to yield a GbEDS1–GFP construct under the control of the CaMV 35S promoter (Figure 4A). After it was introduced into onion epidermal cells, the GFP signal was detected by fluorescence microscopy. The 35SGbEDS1::GFP construct was localized to the cytoplasm and the nucleus (Figure 4B). This was consistent with the targeting of AtEDS1 in plant cells (Zhu et al., 2011). By contrast, the 35S–GFP control construct showed GFP signals throughout the entire cell (Figure 4B).
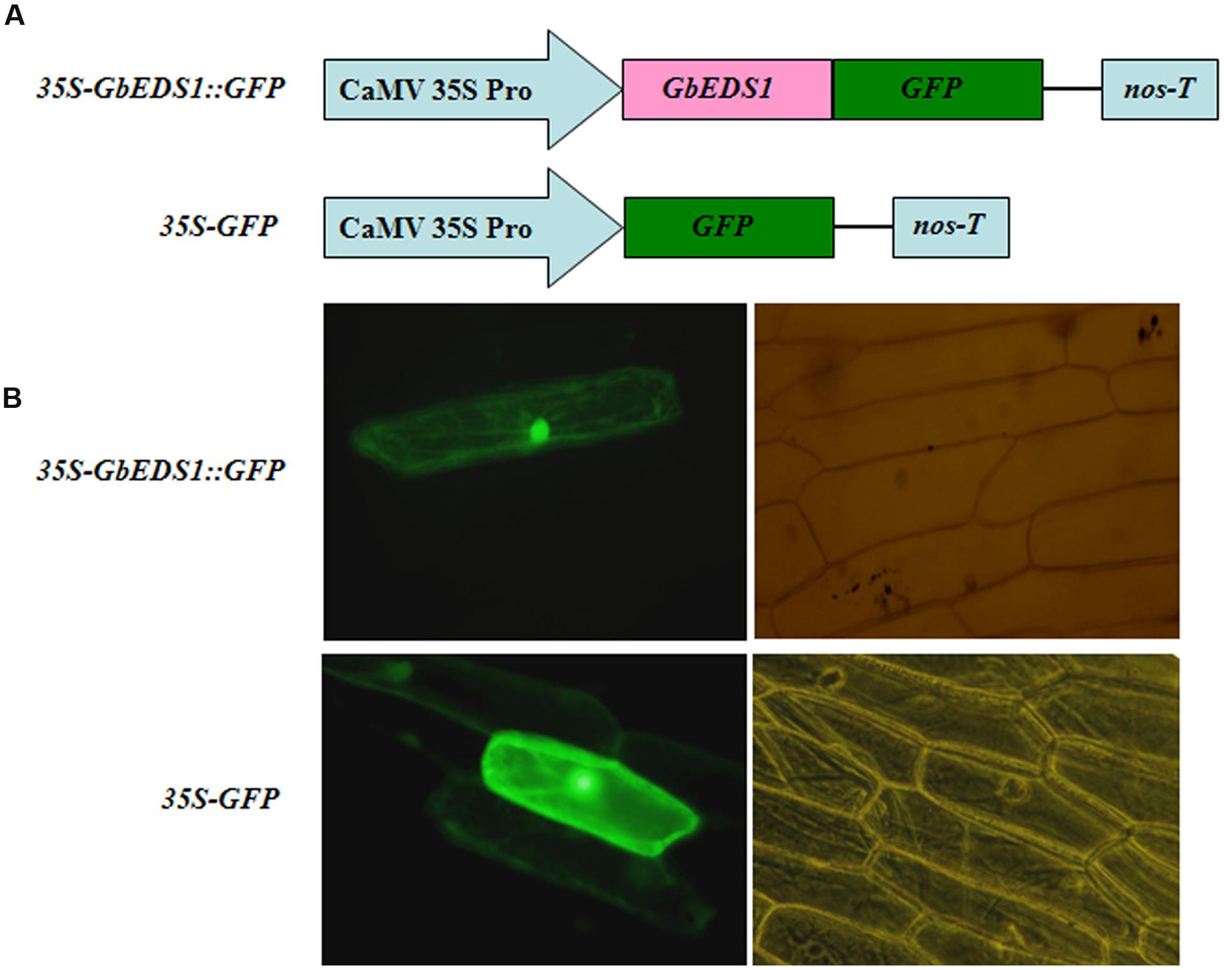
FIGURE 4. Subcellular localization of GbEDS1 in onion epidermal cells. (A), Schematic diagram of 35S–GbEDS1::GFP and 35S–GFP fusion construct. (B), Transient expression of 35S–GbEDS1::GFP and 35S–GFP. Fluorescence images and bright-field images were presented.
Overexpression of GbEDS1 in Arabidopsis Showed Enhanced Defense Response to V. dahliae
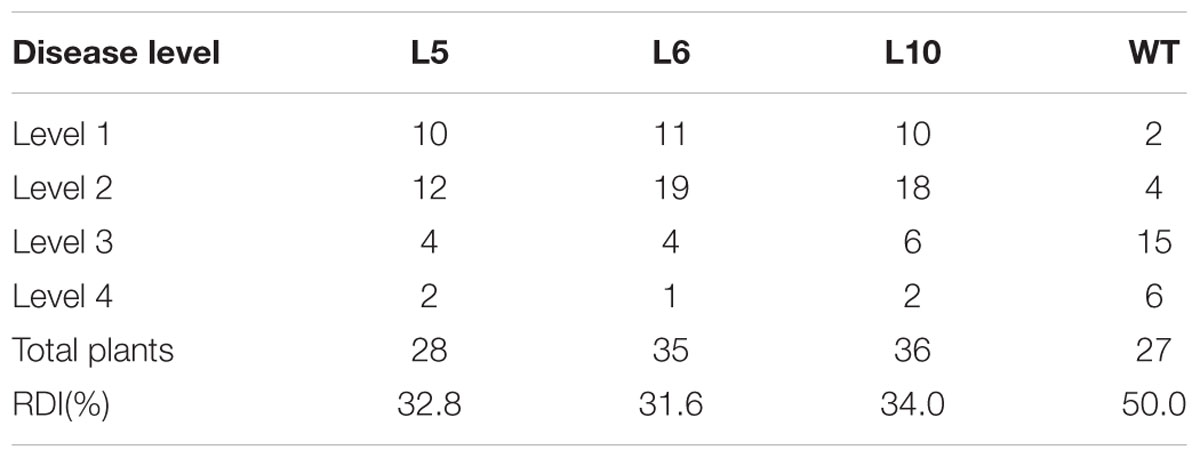
Ecotype ‘Col-0’ Arabidopsis plants were transformed using Agrobacterium tumefaciens to generate transgenic lines overexpressing GbEDS1 driven by the CaMV35S promoter. Of the 13 independent T3 transgenic lines obtained, three (L5, L6, and L10) were selected via genome PCR analysis of the GbEDS1 fragment (Figure 5A). RT-PCR analysis further confirmed that GbEDS1 was successfully expressed in the transgenic plants (Figure 5B). When inoculated with V. dahliae, typical symptoms of vascular disease became apparent in the infected wild-type Col-0 plants but much less pronounced in GbEDS1-overexpressing plants at 20 dpi (Figure 5C). Moreover, the relative disease index values for the wilt-type plants were 50.0, but for L5, L6, and L10 transgenic lines were 32.8, 31.6, and 34.0, respectively (Table 2), indicating that all three transformed lines demonstrated improved wilt resistance. Further evidence that the transgenic lines improved plant resistance were found with regard to the fungal biomass, with significantly less V. dahliae biomass was detected in transgenic lines than that in wild type plants at 20 dpi (Figures 5D,E). Therefore, these results suggested that GbEDS1 in G. barbadense has a crucial function in response to V. dahliae infection. To further investigated the function and potential molecular mechanism of GbEDS1,we choose the best resistance transgenic line –L6 as material to carry out the following related experiments.
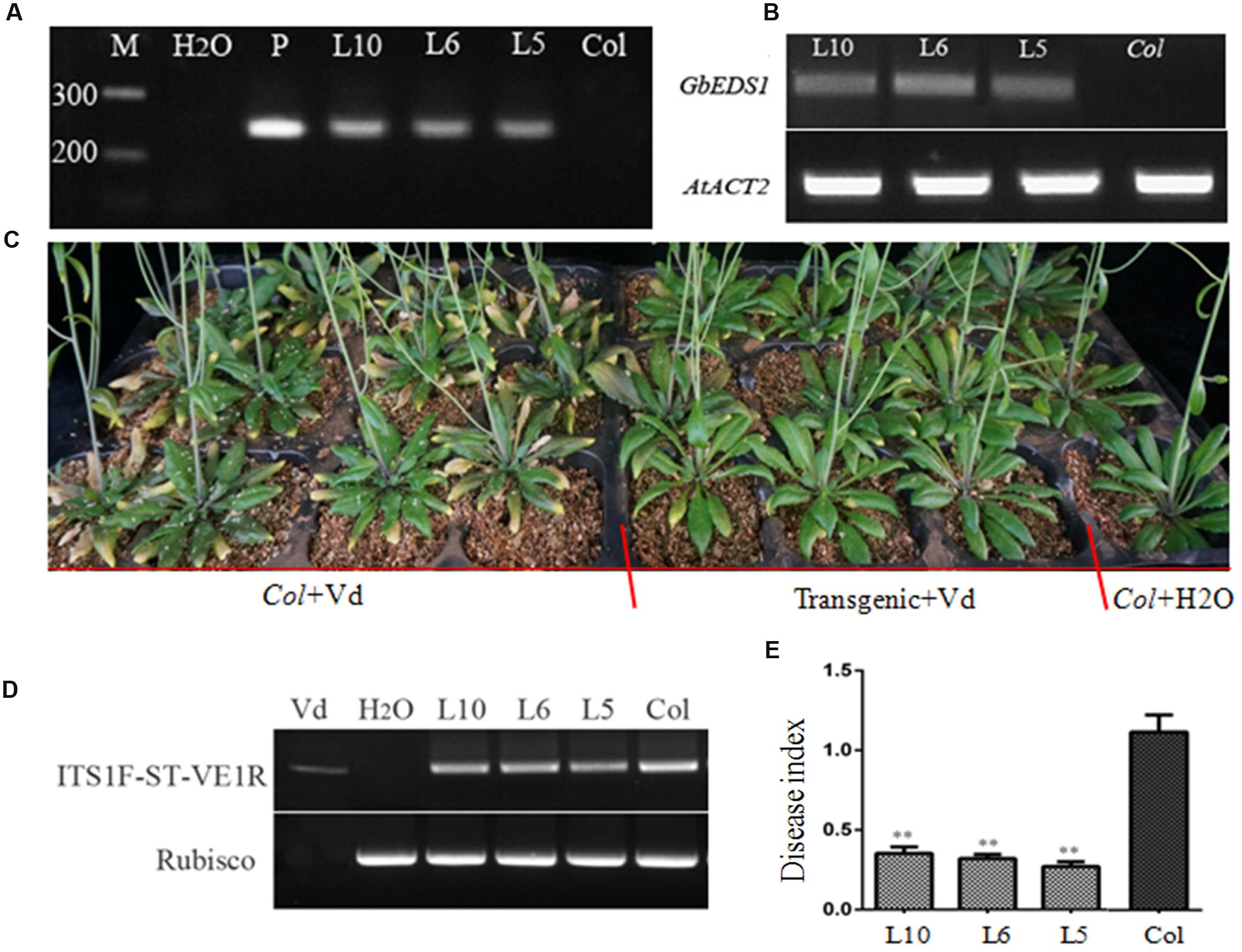
FIGURE 5. Overexpression of GbEDS1 in Arabidopsis improved the resistance to V. dahliae. (A) Direct-PCR analysis of genomic DNA from transgenic lines. (B) RT-PCR analysis the expression of GbEDS1 gene in transgenic lines and wilt type plants. M: Marker, H2O represented negative control in the PCR system. (C) Four-week-old plants were inoculated via root-dipping with 5 × 107 conidia mL-1. Similar results were seen from all transgenic lines. Transgenic and wild type plants showed obvious differences in resistance, transgenic plants showed more resistant than wild type plants. (D) Detection the biomass of the V. dahliae in the transgenic plant with RT-PCR by comparing V. dahliae internal transcribed spacer (ITS) transcript levels (as a measure for fungal biomass) relative to Arabidopsis Rubisco transcript levels (for equilibration) at 14 days post-inoculation. (E) Detection the biomass of the V. dahliae in the transgenic plant with Quantitative real-time PCR. The relative average fungal biomass is shown with standard errors. Asterisks indicate significant differences when compared with colonization of the eds1 mutant. The average fungal biomass was determined using at least three inoculated plants for each genotype. (∗∗P < 0.01).
EDS1 Is Necessary for Conferring Resistance to Verticillium Wilt in Cotton
Using VIGS, we monitored the silencing effect with CLA1, a gene involved in chloroplast development. Two weeks after Agrobacterium infiltration in cotton, silencing of CLA1 led to an albino phenotype on newly developing true leaves and stem (Figure 6a), suggesting that the VIGS approach was successful. Meanwhile, RT-PCR results showed that levels of EDS1 transcripts were significantly reduced in TRV::GbEDS1 plants when compared with TRV::00 plants (Figure 6b). It indicated that GbEDS1 was effectively silenced in cotton. After inoculation with V. dahliae by 14 d, the TRV::00 plants displayed good Verticillium wilt resistance (Figure 6c) whereas TRV::GbEDS1 seedlings were stunted growth, wilting, chlorosis, and defoliation (Figure 6d). With the infection development, the disease symptom became obviously at 28 dpi (Figures 6e,f). Values calculated for the rate of diseased plants and DI showed that suppressing the expression of GbEDS1 in cotton resulted in decreased V. dahliae resistance (Figures 6g,h). All above results implied that GbEDS1 played an important role in defense against V. dahliae.
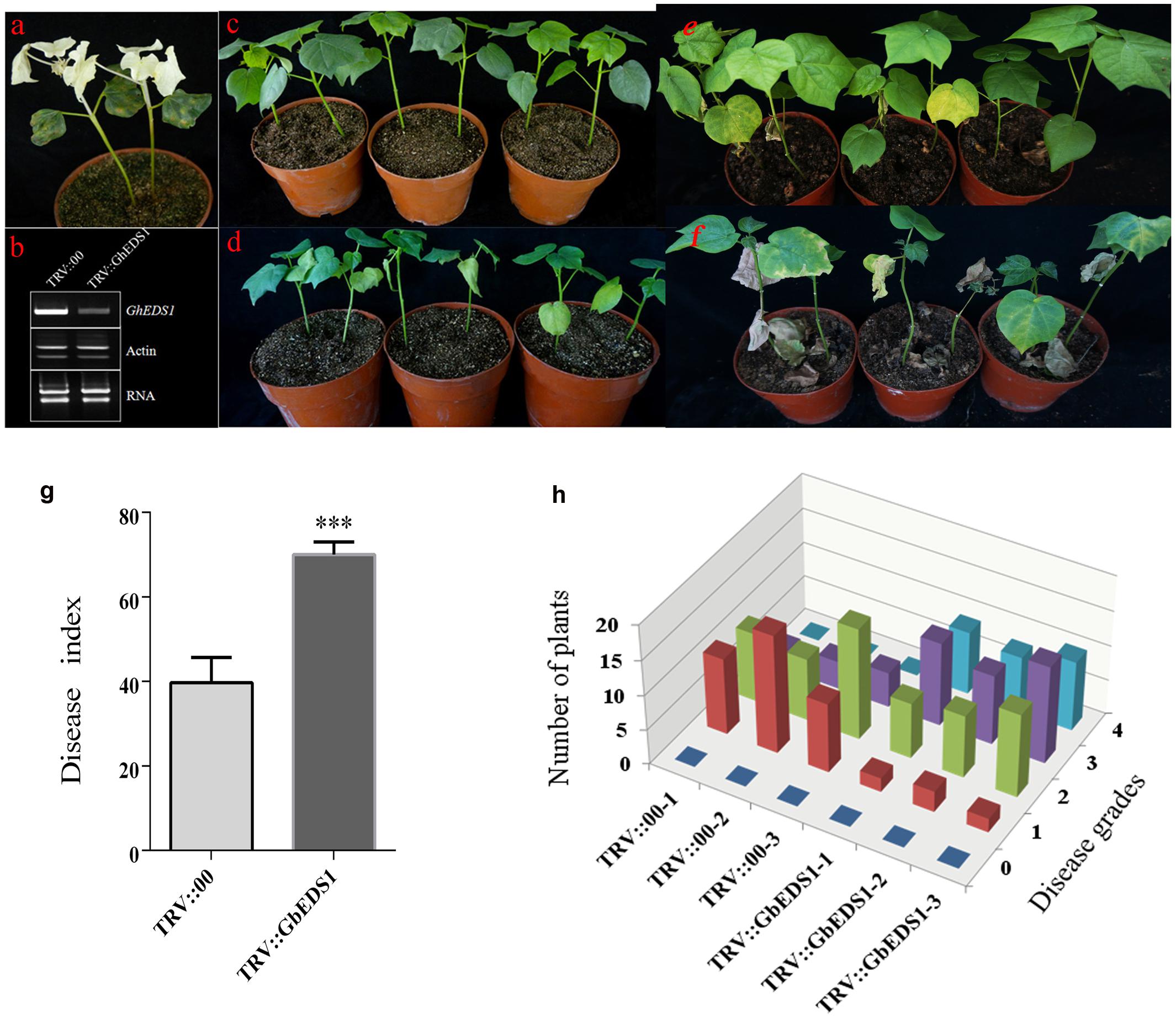
FIGURE 6. Silencing of GbEDS1 resulted in enhancing susceptibility in cotton against V. dahliae. (a) Disease symptoms induced on TRV:00, TRV:GbEDS1 seedlings after inoculation with V. dahliae. Ten-day-old G. barbadense cv. Pima90-53 seedlings were hand-infiltrated with Agrobacterium carrying target gene in the VIGS vector. Two weeks after infiltration, the seedlings were dip-inoculated with V. dahliae. Photos were taken at 14 and 28 days post inoculation. (b) qPCR analysis of the GbEDS1 transcripts in control and silencing seedlings. Disease symptoms inoculated with V. dahliae strain on TRV::00 (c,e) and TRV::GbEDS1 (d,f) plants. (g,h) The rate of diseased plants and disease index were measured at 20 dpi. Error bars represent the standard deviation of three biological replicates (n ≥ 30); asterisks indicate statistically significant differences, as determined by the Student t-test (∗∗∗P < 0.001).
GbEDS1 Influences Verticillium Wilt Resistance by Regulating SA Level
To learn whether GbEDS1 regulates the SA signal pathway, we determined the SA levels both in transgenic line and TRV::GbEDS1 plants. After inoculated with V. dahliae, the contents of SA were increased both in wild type and transgenic plants. Furthermore, the SA levels in transgenic plants were significantly higher than those in the wild type except for the point at 48 hpi (Figure 7A). In contrast, the decreased SA level were detected in GbEDS1-silenced plants comparing to the control (Figure 7B), indicating that suppressing the expression of GbEDS1 gene resulted declining SA accumulation in cotton. These results suggested that SA signaling pathway involved in cotton defense against V. dahliae, and GbEDS1 played a positive role in Verticillium wilt resistance by regulating the levels of SA.
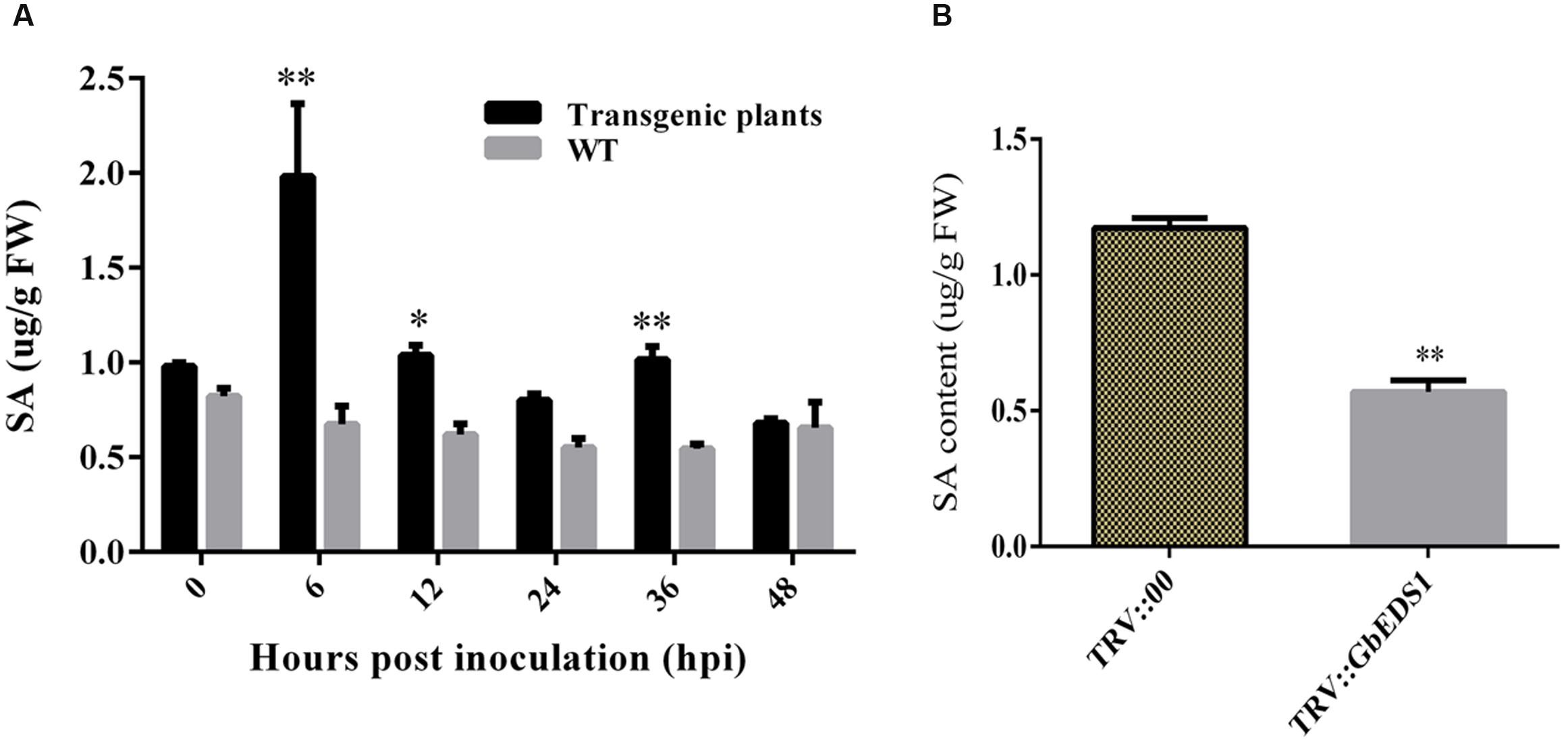
FIGURE 7. Detection of SA content in Arabidopsis and cotton. (A) Measurement of SA accumulation in transgenic Arabidopsis in response to V. dahliae infection. (B) Measurement of SA accumulation in silenced cotton. Error bars of SA levels represent the SD of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student’s t-test (∗P < 0.05; ∗∗P < 0.01).
GbEDS1 Overexpression Enhanced the Expression of NDR1 and BAK1
To learn how GbEDS1 regulates the SA signal pathway and defense response, we used qPCR to analyze EDS1, NPR1, NDR1 BAK1, PR1, and PR5 genes that participate in signal transduction or defense response. As shown in Figure 8, the transcripts of AtEDS1, AtNDR1, and AtBAK1 were significantly up-regulated in transgenic line than in Col after infection by V. dahliae. However, the transcripts of AtNPR1 and AtPR1 and AtPR5 were lower in transgenic lines than that in Col. The opposite trends of above genes expression were obtained in GbEDS1-silenced plants (Figure 9). Combination the results from overexpression transgenic line and GbEDS1-silenced plants, it deduced that GbEDS1 could enhance the expression of NDR1 and BAK1 genes, which were reported to mediated the Verticillium wilt resistance in cotton (Gao et al., 2011, 2013)
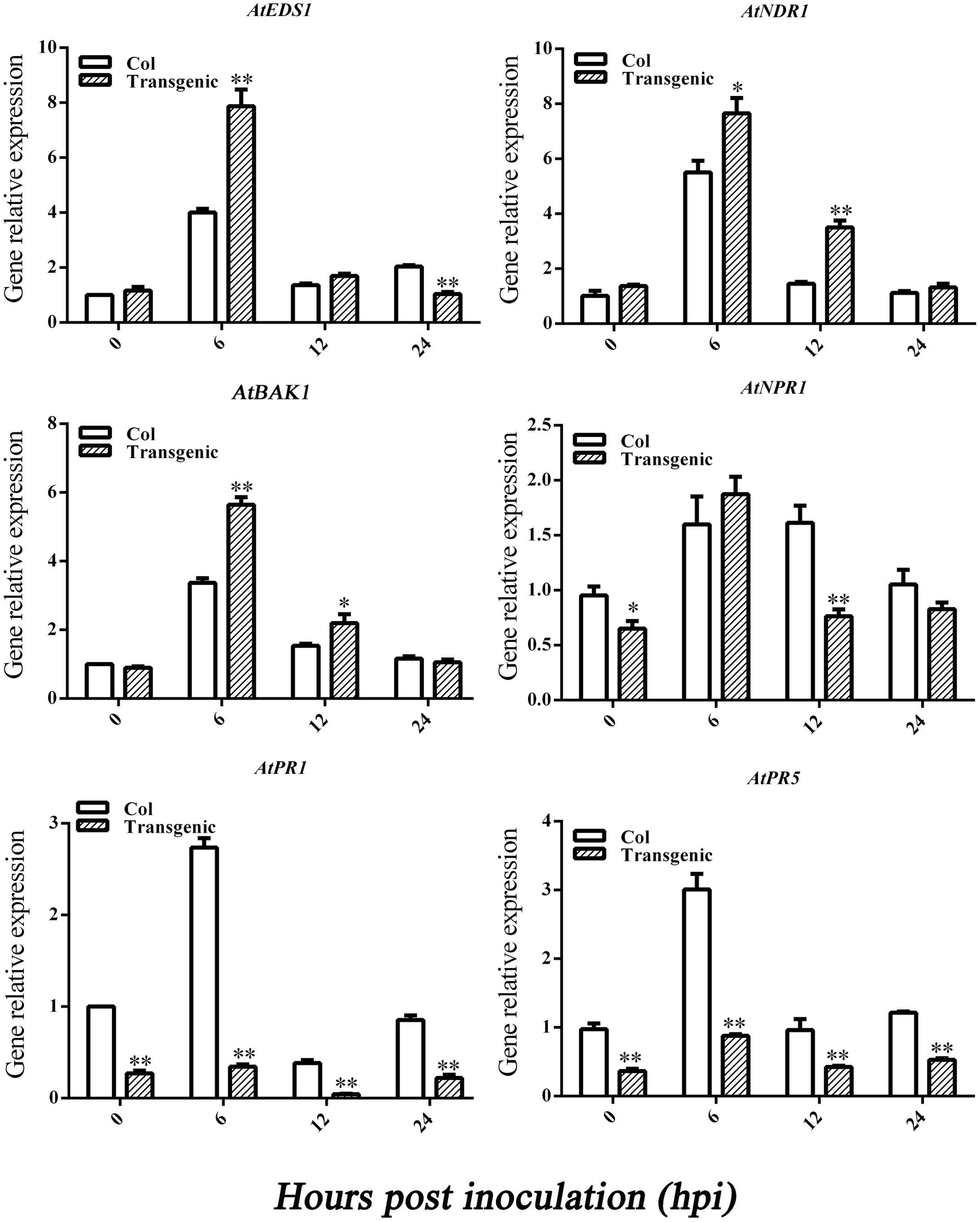
FIGURE 8. Detection of SA signaling pathway-related genes and defense related genes. qRT-PCR analysis the expression of genes (EDS1, NPR1, NDR1, BAK1, PR1, and PR5) in transgenic plants. Error bars represent the standard deviation for three independent experiments, and three technical replicates were analyzed; asterisks indicate statistically significant differences, as determined by the Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
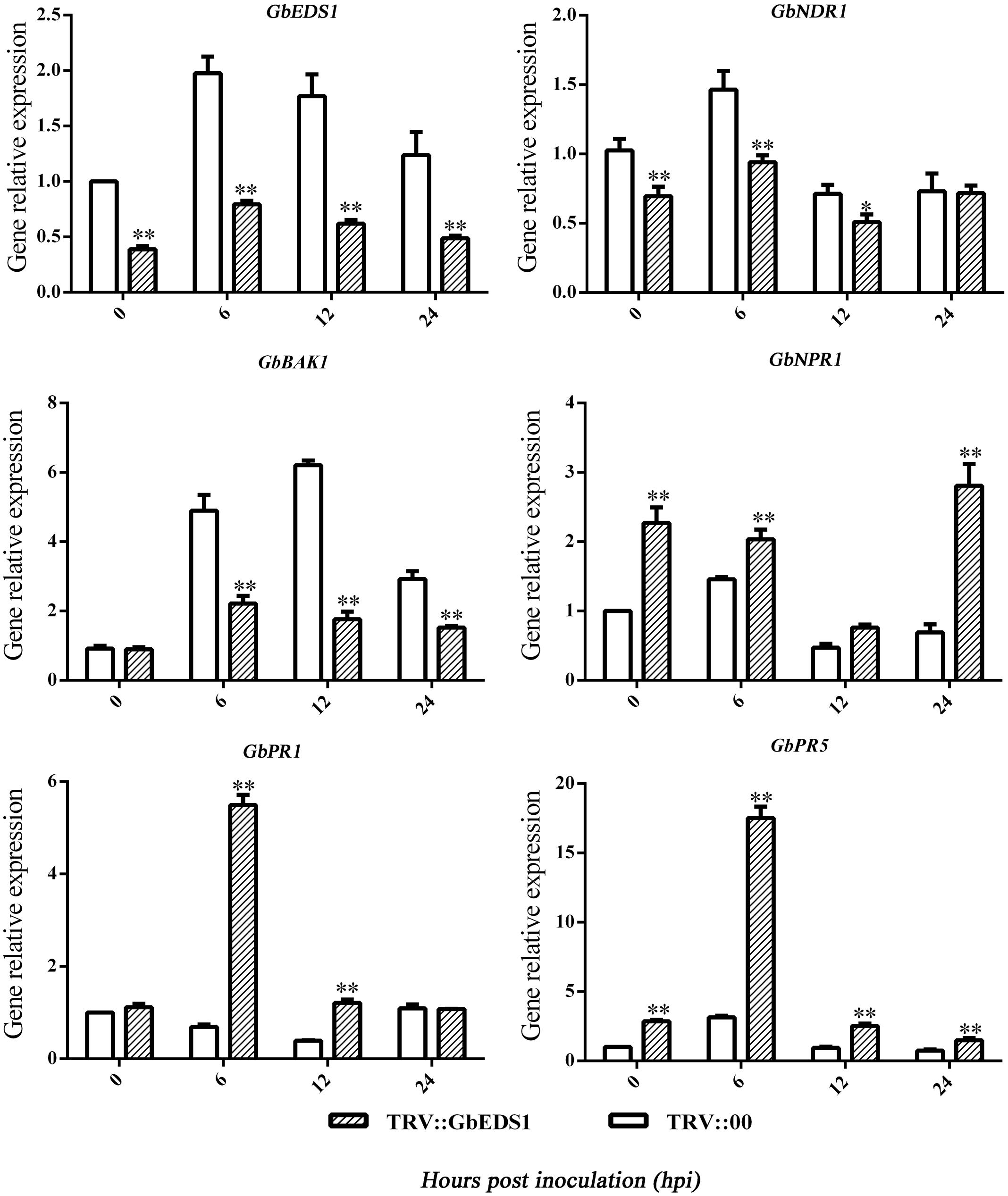
FIGURE 9. Detection of SA signaling pathway-related genes and defense related genes. qRT-PCR analysis the expression of genes (EDS1, NPR1, NDR1, BAK1, PR1, and PR5) in silenced cotton. Error bars represent the standard deviation for three independent experiments, and three technical replicates were analyzed; asterisks indicate statistically significant differences, as determined by the Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
GbEDS1 Regulated H2O2 Content
H2O2 accumulation has been considered to be an important parameter in biotic stresses because it usually regarded as an immunity-associated molecule (Camejo et al., 2016). To further investigation the function of GbEDS1 in defense against V. dahliae, we monitored the levels of H2O2 in transgenic line and GbEDS1-silenced plants. As shown in Figure 10, comparing to the wild type, levels of H2O2 in transgenic plants was higher, especially at 0, 6, and 12 hpi (Figure 10A), while the levels of H2O2 were lower than that in mock plants within 48 h except 36 hpi in GbEDS1-silenced plants (Figure 10B). Changes of H2O2 levels prompted us to assay the major antioxidant enzyme (SOD and POD) activities. According to the results of SOD and POD activities, only several time points exhibited changes comparing to the wild type, however, the whole trend did not obvious, especially in POD activity (Figure 11). All above results indicated that overexpression of GbEDS1 promoted the production of H2O2 in Arabidopsis upon V. dahliae infection.
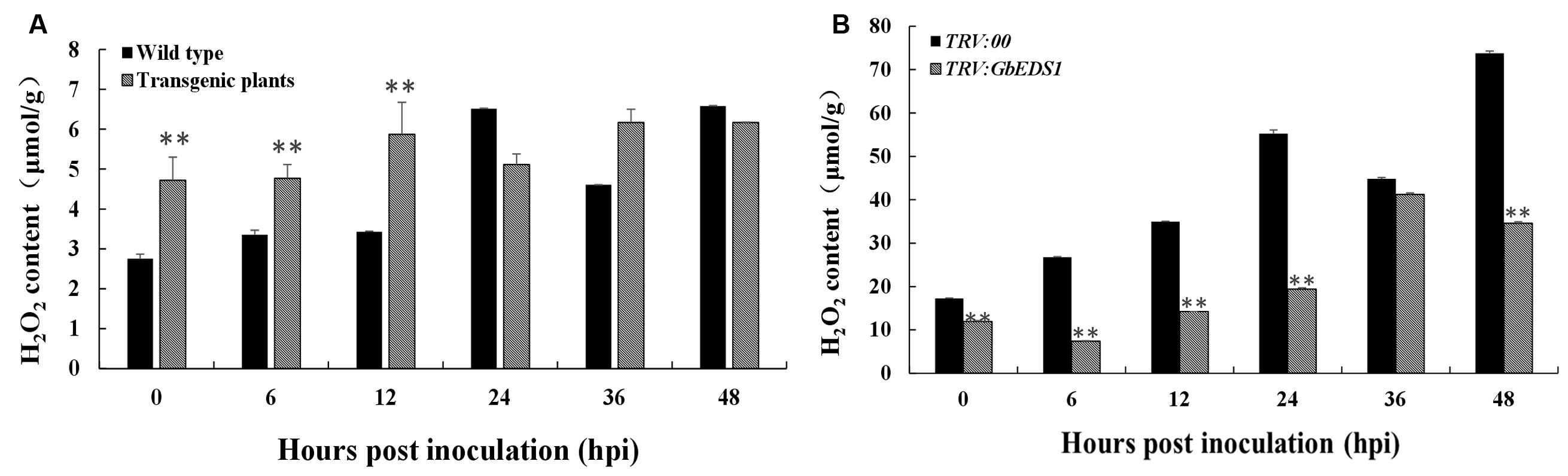
FIGURE 10. Measurement of H2O2 in response to V. dahliae infection. Detection of H2O2 accumulation in transgenic Arabidopsis (A) and silenced cotton (B), respectively. Error bars of H2O2 levels represent the standard deviation of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student’s t-test (∗∗P < 0.01).
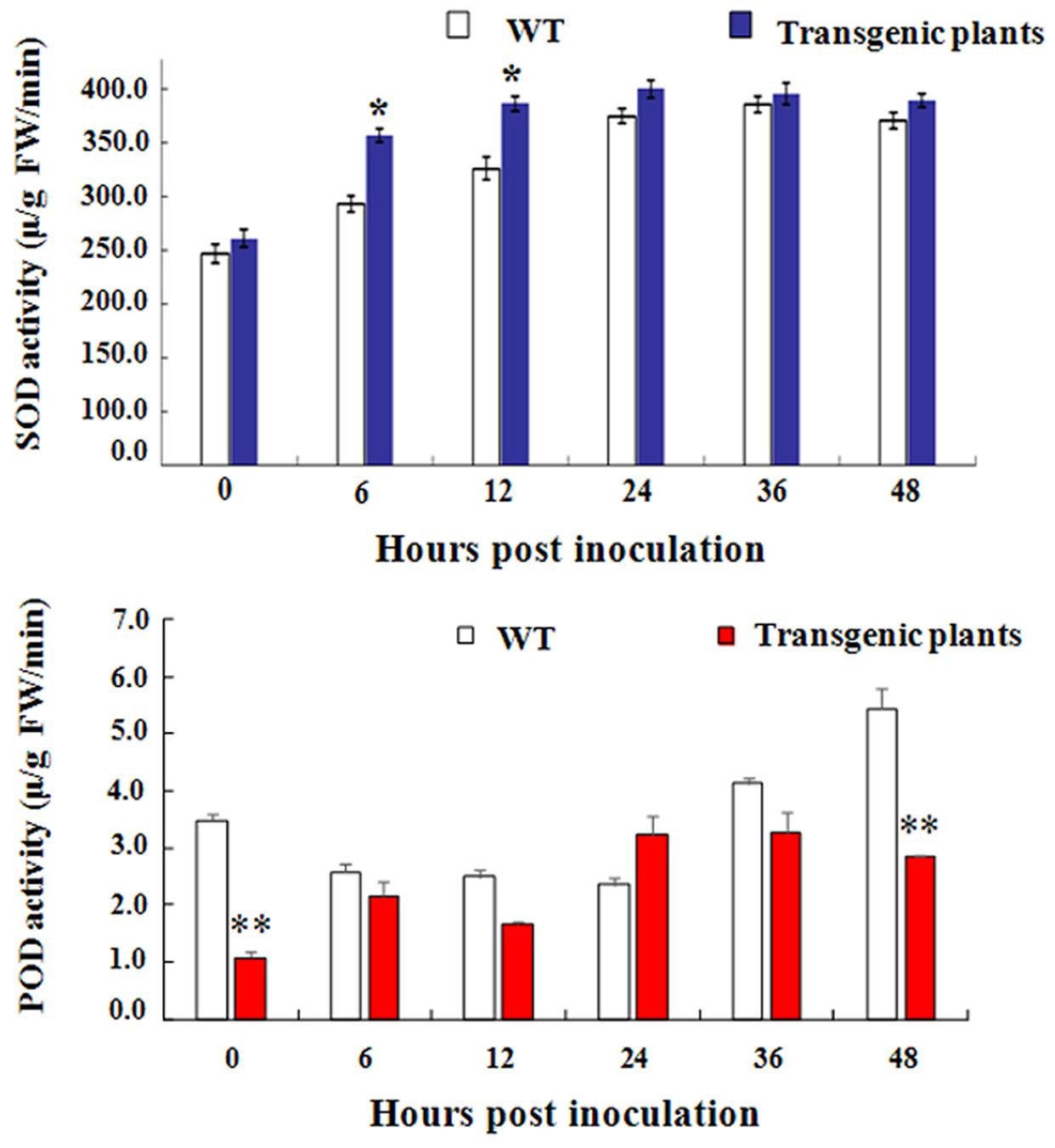
FIGURE 11. Detection of SOD and POD activity in transgenic Arabidopsis, respectively. The total activities of the antioxidant enzymes SOD (A) and POD (B) in Arabidopsis plants within 48 h after inoculated with V. dahliae. The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01, Student’s t-test).
Discussion
We characterized and analyzed the function of GbEDS1, a lipase-like protein gene from G. barbadense. The results suggested that GbEDS1 played a crucial role in Verticillium wilt resistance responses by regulating the accumulation of SA, H2O2. These findings not only broaden our knowledge about the biological role of GbEDS1, but also provide new insights into the defense mechanisms of GbEDS1 against V. dahliae in cotton.
Consistent with previous alignments made between GbEDS1 and eukaryotic lipases, its C-terminal region contains a specific motif, KNEDT, which is highly conserved among other EDS1 genes, e.g., NbEDS1 and AtEDS1. In addition, this motif is an important tool for distinguishing EDS1 from PAD4, another lipase-like gene required for TIR–NB–LRR-type R protein-mediated resistance. Therefore, we propose that GbEDS1 has biochemical properties and physiological functions similar to those of the previously identified AtEDS1 and NbEDS1. The ORF of GbEDS1 in this studied is 1,848 bp long and encodes for a protein with 615 aa. These data differ from those previously reported by Su et al. (2014), who cloned a GbEDS1 gene with a 1647-bp ORF and 548 amino acid residues. According to the sequence analysis of EDS1 genes from different cotton species, including GrEDS1, GaEDS1, GhEDS1, GbEDS1, indicated that EDS1 gene is very conservative in cotton. However, the EDS1 sequence reported by Su et al. (2014) was only about 75% identify with different cotton species reference genomes, and it seems like that this sequence is not a correct sequence of EDS1 itself in cotton.
To investigate the gene function, we examined the subcellular localization of GbEDS1 protein and found gfp-fluorescence signal both in the nucleus and at the cytomembrane. This is similar to the reported localizations of AtEDS1 and GmEDS1, which were all located to the nucleus and the cytomembrane (Zhu et al., 2011; Wang et al., 2014). Previous study have reported that AtEDS1 forms a complex with PAD4 as well as SAG101, which is preferentially localized in the nucleus and could be translocated to the cytoplasm if an extranuclear form of AtEDS1 existed (Zhu et al., 2011). Our data revealing the nucleus- and cytoplasm-localized GbEDS1 is consistent with the targeting of AtEDS1 in plant cells, also suggesting the functional similarity between the GbEDS1 and AtEDS1.
The genetic basis and molecular mechanisms of EDS1 and analogous genes have been studied in Arabidopsis and other model plant species, including Lycopersicon esculentum and Nicotiana tabacum (Liu et al., 2002; Hu et al., 2005). However, there was seldom report about EDS1 in cotton (Su et al., 2014), and how cotton EDS1 gene function is remain unclear. In this research, we first study the GbEDS1 function simultaneously by overexpression Arabidopsis and silenced cotton. Both the transgenic Arabidopsis displaying enhanced resistance and silenced-GbEDS1 cotton showing enhanced susceptibility coincident indicated that GbEDS1 played a positive role in Verticillium wilt resistance. In model plants, EDS1 regulates the accumulation of SA as part of a positive feedback loop through which it promotes its own expression and the expression of downstream pathway genes (NPR1 and PR1) leading to defense amplification (Feys et al., 2001; Eulgem et al., 2004). In this study, we found that the SA content was higher in transgenic Arabidopsis than in wild type, which promoted the expression of AtEDS1 and AtBAK1, however, the expression of NPR1, PR1, and was not. Previous studies had shown that cotton NDR1 and BAK1 mediated the Verticillium wilt resistance (Gao et al., 2011, 2013), thus we deduced that GbEDS1 positively mediated cotton defense through the enhance the expression of NDR1 and BAK1. This special side of GbEDS1 that differented from other homologous EDS1 genes was firstly discovered in our study. It was reported that EDS1 had a‘master’role in coordinating SA and ROS activities in response to abiotic and biotic stress stimuli (Straus et al., 2010). In this study, it proved that GbEDS1 could regulate SA and H2O2 content, which were important to defense against V. dahliae. Furthermore, the regulating network of GbEDS1 in coordinating SA and H2O2 levels remains deeply research.
Although these findings broaden our knowledge about the biological role of GbEDS1, further investigation is required. As an important crop plant, our understanding about the molecular biology and functional genomics of cotton lags behind that of other systems, such as Oryza sativa, Triticum aestivum, and Zea mays, largely because molecular tools and resources have, to date, been limited (Gao et al., 2011). It is not as easy to obtain direct genetic evidence and analyze gene functioning because genetic transformation is more difficult with cotton than with other plants. Fortunately, Agrobacterium-mediated VIGS can be utilized to silence genes of interest for loss-of-function assays. In addition, unlike more conventional methods for cotton transformation, transient assays based on the VIGS approach are time-saving and are not limited to only a few select cotton cultivars. Therefore, we believe that applying these tools will allow us to obtain a wealth of information about the molecular mechanisms of cotton resistance to fungi, bacteria, nematodes, and viruses. In summary, we characterized GbEDS1, a lipase-like protein from G. barbadense. Expression of GbEDS1 was up-regulated under V. dahliae stress and SA treatment. Overexpression of GbEDS1 enhanced resistance to Verticillium wilt in Arabidopsis. Furthermore, silencing of GbEDS1 in cotton led to enhanced susceptibility to V. dahliae. These results indicate that GbEDS1 plays an important role in the defense response in cotton. Further experiments that obtain stable knockout or overexpression lines in cotton will allow us to elucidate more fully the resistance mechanism of GbEDS1 in defense against V. dahliae.
Author Contributions
The experiments were designed by WX and MZ, and were conducted by ZY, RW, and YJ. ZY also performed the data analysis and prepared the manuscript along with MZ. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Yule Liu (College of Life Science, Tsinghua University, Beijing) for providing the TRV-VIGS vectors. This work was financially supported by the Natural Science Foundation of China (31301371), and Foundation for Ph.D. Granting Disciplines from the Education Ministry (20131302120002).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01830/full#supplementary-material
Abbreviations
DPI, days post-inoculation; GbEDS1, Gossypium barbadense enhanced disease susceptibility 1; GFP, green fluorescence protein; Hpi, hours post-inoculation; ORF, open reading frame; SA, salicylic acid; VIGS, virus-induced gene silencing.
Footnotes
- ^ http://www.cbs.dtu.dk/services/SignalP-3.0/
- ^ http://expasy.hcuge.ch/sprot/prosite.html
- ^ http://smart.embl-heidelberg.de/
References
Aarts, N., Metz, M., Holub, E., Staskawicz, B. J., Daniels, M. J., and Parker, J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 10306–10311. doi: 10.1073/pnas.95.17.10306
Artico, S., Nardeli, S. M., Brilhante, O., Grossi-de-Sa, M. F., and Alves-Ferreira, M. (2010). Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 10:49. doi: 10.1186/1471-2229-10-49
Attaran, E., Zeler, T. E., Griebel, T., and Zeler, J. (2009). Methyl salicylate production and jasmonate signaling are essential for systemic acquired resistance in Arabidopsis. Plant Cell 21, 954–971. doi: 10.1105/tpc.108.063164
Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J. L., Bautor, J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18, 1038–1051. doi: 10.1105/tpc.105.039982
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, Y. F., He, X. H., Mo, J. C., Sun, Q., Yang, J. P., and Liu, J. G. (2009). Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr. J. Biotechnol. 8, 7363–7372.
Camejo, D., Guzman-Cedeno, A., and Moreno, A. (2016). Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 103, 10–23. doi: 10.1016/j.plaphy.2016.02.035
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collings, D. A. (2013). Subcellular localization of transiently expressed fluorescent fusion proteins. Methods Mol. Biol. 1069, 227–258. doi: 10.1007/978-1-62703-613-9-16
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Daayf, F., Nicole, M., and Geiger, J. P. (1995). Differentiation of Verticillium dahliae populations on the basis of vegetative compatibility and pathogenicity on cotton. Eur. J. Plant Pathol. 101, 69–79. doi: 10.1007/BF01876095
Dadgar, D., Climax, J., Lambe, R., and Darragh, A. (1985). High-performance liquid chromatographic determination of certain salicylates and their major metabolites in plasma following topical administration of a liniment to healthy subjects. J. Chromatogr. 342, 315–321. doi: 10.1016/S0378-4347(00)84522-3
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S., et al. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Ellendorff, U., Fradin, E. F., de Jonge, R., and Thomma, J. (2009). RNA silencing is required for Arabidopsis defense against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. doi: 10.1093/jxb/ern306
Eulgem, T., Weigman, V. J., Chang, H. S., McDowell, J. M., Holub, E. B., Glazebrook, J., et al. (2004). Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 135, 1129–1144. doi: 10.1104/pp.104.040444
Falk, A., Feys, B. J., Frost, L. N., Jones, J. D., Daniels, M. J., and Parker, J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S.A. 96, 3292–3297. doi: 10.1073/pnas.96.6.3292
Feys, B. J., Moisan, L. J., Newman, M. A., and Parker, J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. doi: 10.1093/emboj/20.19.5400
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gao, X., Li, F., Li, M., Kianinejad, A. S., Dever, J. K., Wheeler, T. A., et al. (2013). Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55, 586–596. doi: 10.1111/jipb.12064
Gao, X. Q., Wheeler, T., Li, Z. H., Kenerley, C. M., He, P., and Shan, L. B. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. doi: 10.1111/j.1365-313X.2011.04491.x
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes, application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Haffner, E., Karlovsky, P., and Diederichsen, E. (2010). Genetic and environmental control of the Verticillium syndrome in Arabidopsis thaliana. BMC Plant Biol. 10:235. doi: 10.1186/1471-2229-10-235
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Hu, G., deHart, A. K., Li, Y., Ustach, C., Handley, V., Navarre, R., et al. (2005). EDS1 in tomato is required for resistance mediated by TIR-class R genes and the receptor-like R gene Ve. Plant J. 42, 376–391. doi: 10.1111/j.1365-313X.2005.02380.x
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588. doi: 10.1073/pnas.96.23.13583
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jun, Z., Zhang, Z. Y., Gao, Y. L., Zhou, L., Fang, L., Chen, X. D., et al. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5:15048. doi: 10.1038/srep15048
Klosterman, S. J., Atallah, Z. K., Vallad, G. E., and Subbarao, K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. doi: 10.1146/annurev-phyto-080508-081748
Lievens, B., Brouwer, M., Vanachter, A. C. R. C., Cammue, B. P. A., and Thomma, B. P. H. J. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171, 155–165. doi: 10.1016/j.plantsci.2006.03.009
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Z. Y., Wang, X. F., Zhang, G. Y., Liu, S. Q., Liu, J. L., and Sun, J. Z. (2000). Genetic studies of Verticillium wilt resistance among different types of Sea Island cottons. Acta Agron. Sin. 26, 315–321.
Mateo, A., Muhlenbock, P., Rusterucci, C., Chang, C. C. C., Miszalski, Z., Karpinska, B., et al. (2004). Lesion simulating disease 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. doi: 10.1104/pp.104.043646
Nandi, A., Welti, R., and Shah, J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16, 465–477. doi: 10.1105/tpc.016907
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Reymond, P., and Farmer, E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. doi: 10.1016/S1369-5266(98)80264-1
Rietz, S., Stamm, A., Malonek, S., Wagner, S., Becker, D., Medina-Escobar, N., et al. (2011). Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. doi: 10.1111/j.1469-8137.2011.03675.x
Rusterucci, C., Aviv, D. H., Holt, B. F., Dangl, J. L., and Parker, J. E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. doi: 10.1105/tpc.010085
Shah, J. (2009). Plants under attack: systemic signals in defence. Curr. Opin. Plant Biol. 12, 459–464. doi: 10.1016/j.pbi.2009.05.011
Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. doi: 10.1038/385718a0
Sink, K. C., and Grey, W. E. (1999). A root-injection method to assess verticillium wilt resistance of peppermint and its use in identifying resistant somaclones of cv. Black Mitcham. Euphytica 106, 223–230. doi: 10.1023/A:1003591908308
Straus, M. R., Rietz, S., van Themaat, E., Bartsch, M., and Parker, J. E. (2010). Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 62, 628–640. doi: 10.1111/j.1365-313X.2010.04178.x
Su, X. F., Qi, X. L., and Cheng, H. M. (2014). Molecular cloning and characterization of enhanced disease susceptibility 1 (EDS1) from Gossypium barbadense. Mol. Biol. Rep. 41, 3821–3828. doi: 10.1007/s11033-014-3248-9
Truman, W., Sreekanta, S., Lu, Y., Bethke, G., Tsuda, K., Katagiri, F., et al. (2013). The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163, 1741–1751. doi: 10.1104/pp.113.227108
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, J. L., Shine, M. B., Gao, Q. M., Navarre, D., Jiang, W., Liu, C. Y., et al. (2014). EDS1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 165, 1269–1284. doi: 10.1104/pp.114.242495
Wang, L., Mitra, R. M., Hasselmann, K. D., Sato, M., Lenarz-Wyatt, L., Cohen, J. D., et al. (2008). The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol. Plant Microbe Interact. 21, 1408–1420. doi: 10.1094/MPMI-21-11-1408
Wang, M. M., Zhang, Y., Wang, J., Wu, X. L., and Guo, X. Q. (2007). A novel MAP kinase gene in cotton (Gossypium hirsutum L.), GhMAPK, is involved in response to diverse environmental stresses. J. Biochem. Mol. Biol. 40, 325–332.
Wang, X. F., Tian, H. Y., Ma, Z. Y., Xuan, Z. L., Wang, W. S., and Sun, Y. X. (2008). SSH library construction of upland cotton resistant cultivar under the stress of Verticillium dahliae. Cotton Sci. 20, 3–8.
Wiermer, M., Feys, B. J., and Parker, J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. doi: 10.1016/j.pbi.2005.05.010
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wirthmueller, L., Zhang, Y., Jones, J. D. G., and Parker, J. E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17, 2023–2029. doi: 10.1016/j.cub.2007.10.042
Xu, M., Gui, Y. J., Qin, W. Y., Liu, S. Y., Chen, J. Y., and Dai, X. F. (2013). Verticillium dahliae labeled with green fluorescent protein gene. Plant Protect. 39, 128–133. doi: 10.3969/j.issn.0529-1542.2013.05.018
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca(2+)-calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Zhang, Y., Wang, S. F., Yang, S., Chi, J. N., Zhang, G. Y., and Ma, Z. Y. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. doi: 10.1007/s00299-011-1115-x
Zhang, Y., Wang, X. F., Ding, Z. G., Ma, Q., Zhang, G. R., Zhang, S. L., et al. (2013). Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics 14:637. doi: 10.1186/1471-2164-14-637
Keywords: cotton (Gossypium barbadense), EDS1, Verticillium wilt resistant, transgenic Arabidopsis, virus-induced gene silence, SA level, H2O2 accumulation
Citation: Yan Z, Xingfen W, Wei R, Jun Y and Zhiying M (2016) Island Cotton Enhanced Disease Susceptibility 1 Gene Encoding a Lipase-Like Protein Plays a Crucial Role in Response to Verticillium dahliae by Regulating the SA Level and H2O2 Accumulation. Front. Plant Sci. 7:1830. doi: 10.3389/fpls.2016.01830
Received: 15 July 2016; Accepted: 21 November 2016;
Published: 15 December 2016.
Edited by:
Vincenzo Lionetti, Sapienza University of Rome, ItalyCopyright © 2016 Yan, Xingfen, Wei, Jun and Zhiying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ma Zhiying, bXpoeUBoZWJhdS5lZHUuY24=
 Zhang Yan
Zhang Yan Wang Xingfen
Wang Xingfen Rong Wei
Rong Wei Yang Jun
Yang Jun Ma Zhiying
Ma Zhiying