- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Shaanxi, China
- 2Key Laboratory of Protected Horticulture Engineering in Northwest China, Ministry of Agriculture, Shaanxi, China
Plant-specific TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTORS (TCP) transcription factors play versatile functions in multiple processes of plant growth and development. However, no systematic study has been performed in strawberry. In this study, 19 FvTCP genes were identified in the diploid woodland strawberry (Fragaria vesca) accession Heilongjiang-3. Phylogenetic analysis suggested that the FvTCP genes were classified into two main classes, with the second class further divided into two subclasses, which was supported by the exon-intron organizations and the conserved motif structures. Promoter analysis revealed various cis-acting elements related to growth and development, hormone and/or stress responses. We analyzed FvTCP gene transcript accumulation patterns in different tissues and fruit developmental stages. Among them, 12 FvTCP genes exhibited distinct tissue-specific transcript accumulation patterns. Eleven FvTCP genes were down-regulated in different fruit developmental stages, while five FvTCP genes were up-regulated. Transcripts of FvTCP genes also varied with different subcultural propagation periods and were induced by hormone treatments and biotic and abiotic stresses. Subcellular localization analysis showed that six FvTCP-GFP fusion proteins showed distinct localizations in Arabidopsis mesophyll protoplasts. Notably, transient over-expression of FvTCP9 in strawberry fruits dramatically affected the expression of a series of genes implicated in fruit development and ripening. Taken together, the present study may provide the basis for functional studies to reveal the role of this gene family in strawberry growth and development.
Introduction
TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTORS (TCP) transcription factors constitute a small family of plant-specific transcription factors that play versatile functions in regulating diverse plant growth and development processes by controlling cell proliferation (Martin-Trillo and Cubas, 2010). TCP transcription factors were discovered in 1999 and named after the first three characterized family members: TEOSINTE BRANCHED 1 (TB1) in maize (Zea mays), CYCLOIDEA (CYC) in snapdragon (Antirrhinum majus), and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) in rice (Oryza sativa; Cubas et al., 1999). This class of transcription factors is characterized by a highly conserved 59-residue-long non-canonical basic helix-loop-helix (bHLH) structure at the N-terminus called the TCP domain, which is involved in DNA binding, protein nuclear localization, and protein–protein interactions (Cubas et al., 1999; Kosugi and Ohashi, 2002). According to the homology of the TCP domains, the members of the TCP family can be divided into two classes: class I (also named PCF or TCP-P) and class II (also named TCP-C; Navaud et al., 2007). The most striking difference between these two classes is a four-amino-acid deletion in the basic region of the TCP domain of class I relative to class II proteins (Martin-Trillo and Cubas, 2010). The members of class II are quite heterogeneous and can be further subdivided into the CIN and CYC/TB1 subclades (Martin-Trillo and Cubas, 2010). Outside the TCP domain, several class II members are present in an 18-20-residue arginine-rich motif called the R domain with an unknown function, which is speculated to facilitate protein–protein interactions (Cubas et al., 1999).
TCP proteins play a versatile function in multiple biological processes during plant growth and development. It has been reported that many TCP transcription factors participate in the regulation of diverse physiological and biological processes, such as branching (Takeda et al., 2003; Aguilar-Martinez et al., 2007), leaf development (Kieffer et al., 2011), flower development (Kieffer et al., 2011), hormone pathways (Aguilar-Martinez et al., 2007), seed germination (Tatematsu et al., 2008), gametophyte development (Pagnussat et al., 2005), mitochondrial biogenesis (Hammani et al., 2011), and regulation of the circadian clock (Giraud et al., 2010) in various plants. The CIN-like clade genes are involved in lateral organ development, and the CYC/TB1 clade genes control axillary meristem development. In Arabidopsis, functional analysis of two homologs of TB1, BRANCHED1 (BRC1, AtTCP18) and BRANCHED2 (BRC2, AtTCP12), demonstrated that these genes were involved in suppressing axillary bud outgrowth (Aguilar-Martinez et al., 2007). Five CIN-like TCP genes (TCP2, TCP3, TCP4, TCP10, and TCP24) in Arabidopsis were all targeted by miR319 and have been implicated in regulating leaf morphogenesis (Nath et al., 2003; Schommer et al., 2008). By contrast, during plant development, class I TCP genes mainly promote cell growth and proliferation (Kosugi and Ohashi, 2002; Danisman et al., 2012). Recently, experimental evidence has shown that TCP proteins could be involved in fruit development and ripening (Parapunova et al., 2014).
Recently, a number of TCP proteins have been identified in various plants due to completion of their whole genome sequence, such as Arabidopsis thaliana (Riechmann et al., 2000), rice (O. sativa; Yao et al., 2007), tomato (Solanum lycopersicum; Parapunova et al., 2014), apple (Malus domestica; Xu et al., 2014), cotton (Gossypium raimondii; Ma et al., 2014), and watermelon (Citrullus lanatus; Shi et al., 2016). However, among the largest and most diverse gene families, the TCP gene family has not been systematically identified in the strawberry genome. To date, only the strawberry FaTCP11 gene has been shown to play a role in ripening and in the regulation of flavan-3-ols synthesis (Pillet et al., 2015).
The cultivated strawberry (Fragaria × ananassa Duch.), which has great nutritive and commercial value, is one of the important horticultural crops grown worldwide for the production of fresh fruit and juice, among other products, and is also an excellent model plant for fleshy fruit development. F. × ananassa has a relatively complex octoploid genome, harboring 56 chromosomes (2n = 8x = 56) that were likely derived from four diploid ancestors (Kang and Liu, 2015). Thus, the sequenced diploid woodland strawberry Fragaria vesca accession Hawaii-4 with a small genome (240 Mb genome, 2n = 2x = 14) offers the possibility of a genome-wide analysis of TCP genes (Shulaev et al., 2011). ‘Heilongjiang-3’ strawberry, from the Heilongjiang province in China, was identified as the diploid woodland strawberry F. vesca (Lei et al., 1997).
In the present study, 19 TCP genes were identified in the diploid woodland strawberry (F. vesca) accession Heilongjiang-3, and a systematic bioinformatics analysis was performed, including determination of the linkage group location, phylogenetic relationships, gene structure, protein motifs and cis-acting elements. We further characterized the transcript accumulation patterns of FvTCP genes in diverse tissues, different stages of fruit development and ripening, different periods of strawberry subcultural propagation, as well in response to hormones and stress treatment. Additionally, we determined the subcellular localization of six FvTCP proteins in Arabidopsis mesophyll protoplasts and transiently over-expressed FvTCP9 in strawberry fruits via agro-infiltration. This study provides details regarding the TCP gene family and facilitates the further functional characterization of TCP genes in strawberry.
Materials and Methods
Plant Materials, Growth Conditions, and Stress Treatments
The wild diploid strawberry F. vesca accession Heilongjiang-3 was obtained from the strawberry germplasm resource greenhouse of the College of Horticulture, Northwest A&F University, Shaanxi, Yangling, China (34° 20′ N 108° 24′ E). The potted strawberry plants were grown at 22°C with 75% relative humidity and no supplemental light. ‘Heilongjiang-3’ strawberry organs/tissues (roots, stems, runners, leaves, floral buds, flowers, mature flowers with partially withered petals, mature green receptacles, white receptacles with green achenes, half white and half red fruits, and fully ripened fruits) were collected for tissue-specific and different developmental stages of the fruits were collected for stage-specific transcript analysis of the FvTCP genes. The strawberry tissue culture plantlets were transferred to proliferation medium consisting of an MS basal medium supplemented with 30 g L-1 sucrose, 7 g L-1 agar, 0.2 mg L-1 6-benzyladenine (6-BA) and 0.8 mg L-1 indole- 3-butyric acid (IBA) with monthly subculturing for induction and the following five different subcultural propagation periods (P1: original plantlet; P2: plantlet with 1–2 branch crowns, approximately 2 weeks after subculture; P3: plantlets with 3–4 branch crowns, approximately 3 weeks after subculture; P4: plantlets with 5–7 branch crowns, approximately 4 weeks after subculture; P5: plantlets with over 10 branch crowns, approximately 6 weeks after subculture) were harvested. Arabidopsis thaliana ecotype Col-gl was grown at 22°C with 75% relative humidity under short-day (8 h of light at 125 μmol⋅m-2⋅s-1 and 16 h of dark) conditions for 4–5 weeks before transformation.
Six-month-old uniformly developed strawberry plantlets were selected for the stress treatments. Salt stress was simulated by irrigating potted strawberry plants with 300 mM NaCl. Another set of control ‘Heilongjiang-3’ plantlets was similarly treated with distilled water. Cold and heat stress treatments were performed by transferring the plants to a 4°C/42°C chamber for 48 h. Another set of potted ‘Heilongjiang-3’ plantlets was maintained in the control temperature range from 22 to 27°C. Hormone treatments were performed by spraying the strawberry leaves with a solution containing 0.1 mM abscisic acid (ABA), 1 mM salicylic acid (SA), 0.1 mM methyl jasmonate (MeJA), or 0.5 g/L ethephon (Eth), while another set of control ‘Heilongjiang-3’ plantlets were similarly sprayed with distilled water. The leaves of all of the above plants treated with salt, cold, heat, and hormone stresses were then collected at 0, 0.5, 2, 4, 8, 12, 24, and 48 h post-treatment (hpt). Drought stress treatment was performed by withholding water, followed by sampling at 0, 24, 48, 72, 96, 120, and 144 hpt. The plants were watered again after 144 h of drought stress and sampled after another 24 h. Strawberry plantlets grown without drought stress were used as a control. The powdery mildew (Podosphaera aphanis) treatment experiment was conducted by touching the adaxial epidermis with sporulating colonies on the surface of the strawberry leaves. The inoculated leaves were collected at 0, 24, 48, 72, 96, 120, 144, and 168 h post-inoculation (hpi), and uninfected leaves served as a negative control. At each time point of each treatment, six leaves from six separate plants were combined to form one sample, and all of the treatments were evaluated in triplicate. All of the collected plant samples were immediately frozen in liquid nitrogen and stored at –80°C until use.
Identification of Strawberry TCP Genes
To identify TCP genes in strawberry, we downloaded the full-length amino acid sequences of hypothetical TCP transcription factor in the diploid woodland strawberry (F. vesca) accession Hawaii-4 from the Plant Transcription Factor Database1. Next, the full-length amino acid sequences of the hypothetical TCP proteins were employed as query to perform BLAST-P searches in National Center for Biotechnology Information (NCBI) database2. An E-value of 1e-10 was used as the threshold to ensure the discovery of all potential TCP DNA binding domain-encoding sequences in the strawberry genome (F. vesca Annotation Release 101). Finally, the putative TCP genes were manually verified using the InterProScan program3 and Pfam database4 to validate the presence and completeness of a TCP DNA-binding domain (PF03634). The identified strawberry TCP genes were annotated based on their respective linkage group distribution. Linkage group assignments were retrieved from annotations downloaded from the NCBI Map Viewer5. FvTCP genes were mapped to the linkage groups using MapInspect software. The TCP protein sequences from Arabidopsis thaliana, apple, rice, and tomato were retrieved from the Plant Transcription Factor Database (PlantTFDB6).
Multiple Sequence Alignments and Phylogenetic Analysis
Multiple sequence alignments of TCP proteins and domains were performed separately using ClustalX 2.0.12, and the alignment results were presented and manually modified using GeneDoc. Target prediction for miR319 was performed using the psRNATarget online tool7. The full-length amino acid sequences of the putative TCP proteins from Arabidopsis (AtTCP), apple (MdTCP), strawberry (FvTCP), tomato (SlTCP), and rice (OsTCP) were used to generate a phylogenetic tree based on MUSCLE alignment and the unrooted neighbor-joining method with MEGA 5.0. Neighbor-joining analysis with pairwise deletion and bootstrap analysis with 1000 replicates was performed using the p-distance model (Wei et al., 2016).
Conserved Motif Identification and Gene Structure Analysis
TCP protein sequences in F. vesca were submitted to the online Multiple Expectation maximization for Motif Elicitation (MEME8) program to identify conserved protein motifs. The optimized MEME parameters were as follows: minimum motif width, 6; maximum motif width, 100; and maximum number of motifs, 12. The FvTCP genomic sequences and CDS sequences extracted from NCBI were compared using the online program GSDS 2.09 to infer the exon/intron organization of the FvTCP genes.
Putative Promoter cis-Acting Element Analysis
FvTCP nucleotide sequences were used to retrieve whole genomic sequences from NCBI using their gene IDs10. The upstream 1 kb region of the translation start site of the FvTCP genes was used for putative promoter cis-acting element analysis in PlantCARE11. The motifs putatively involved in plant growth and development, phytohormone responses, and abiotic and biotic stress responses are summarized.
Gene Transcript Analysis
To describe the transcript accumulation profiles of TCP genes in F. vesca, total RNA was extracted from tissue samples or treated leaves using an EZNA Plant RNA Kit (R6827-01, Omega Bio-tek, USA), according to the manufacturer’s protocol. Prior to reverse-transcription, RNA was treated with DNase I (RNase free; TaKaRa Biotechnology, Dalian, China) to remove any residual contaminating genomic DNA. Next, 1.5 μg of total RNA isolated from each sample was reverse-transcribed into first-strand cDNA using PrimeScript RTase (TaKaRa Biotechnology, Dalian, China). Gene-specific primers for each FvTCP gene were designed using VECTOR NTI.
Semi-quantitative reverse-transcription (RT) PCR reactions were conducted using the following profile: 95°C for 3 min, 29–33 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, and a final step at 72°C for 5 min. The PCR products were separated in a 1.5% (w/v) agarose gel, stained with ethidium bromide, and imaged under UV light for further gene transcript analysis. Each reaction was performed in triplicate, with three independent analyses for each treatment showing the same trends for each gene. The transcript profiles obtained by semi-quantitative RT-PCR were collated and analyzed using GeneSnap software. The heat maps and hierarchical clustering of gene transcript data were visualized in Multi Experiment Viewer (MeV) 4.9 software (Saeed et al., 2006; Guo et al., 2014). Reverse transcription quantitative PCR (RT-qPCR) was performed as described previously with some modifications (Wei et al., 2016). We performed RT-qPCR in a 21-μl reaction volume containing 10.5 μl of SYBR Green Premix Ex Taq II (TaKaRa Biotechnology), 1.0 μl of cDNA, 0.8 μl of 1.0 μM of each primer, and 7.4 μl of sterile distilled H2O, using an IQ5 real time-PCR machine (Bio-Rad, Hercules, CA, USA). RT-qPCR amplification was performed under the following conditions: 40 cycles at 95°C for 30 s and 58°C for 30 s. After amplification, the samples were maintained at 50°C for 1 min, and the temperature was gradually increased by 0.5°C every 10 s to perform the melting curve analysis. The interspacer 26S-18S strawberry RNA gene (housekeeping gene; Fv18S) was used as an internal control to normalize the expression data (Raab et al., 2006; Hu et al., 2015; Medina-Puche et al., 2015; Paniagua et al., 2016; Wei et al., 2016). Three biological replicates with three technical replicates were assayed for each sample. Reactions for the reference gene were included in each plate. The relative transcript levels of the genes were calculated using the 2-ΔΔCt method, and the standard deviation was calculated from three biological replicates. The gene-specific primers are listed in Supplemental Table S1.
Subcellular Localization of Strawberry TCP Genes
Based on the public NCBI database (F. vesca Annotation Release 101; Darwish et al., 2015), we designed 10 pairs of gene-specific primers to clone the randomly selected 10 TCP genes in the diploid woodland strawberry F. vesca accession Heilongjiang-3. The predicted coding sequences of the selected FvTCP genes were amplified with high-fidelity HS polymerase (TaKaRa Biotechnology, Dalian, China) using the primers listed in Supplementary Table S1. Finally, six FvTCP genes (FvTCP7, FvTCP8, FvTCP9, FvTCP13, FvTCP15, and FvTCP17) belonging to different types were successfully cloned from the cDNA of the leaves of ‘Heilongjiang-3’.
To construct GFP-tagged FvTCP, the six cloned FvTCP sequence-encoding genes were cloned into the XbaI and KpnI sites in the pBI221 vector. The plasmid DNA of pBI221 containing 35S::FvTCPs-GFP was introduced into Arabidopsis mesophyll protoplasts using the PEG-Ca2+-mediated transformation method (Yoo et al., 2007; Wei et al., 2016). The localization of the fusion protein was observed using an Olympus BX-51 inverted fluorescence microscope (Olympus, Japan). GFP fluorescence, the bright field image, and the red autofluorescence of chloroplasts from the protoplast expression assay were imaged simultaneously and merged together. All transient expression assays were repeated at least three times.
Transfection of Strawberry Fruits by Agroinfiltration and Western Blot Analysis
To produce a YFP-tagged FvTCP9, the coding sequence of FvTCP9 was cloned in-frame into the BamHI sites of the binary expression vector C15 to generate plasmid 35S::FvTCP9-YFP. The recombinant plasmids 35S::FvTCP9-YFP and 35S::YFP (C15 empty vector) were transfected into Agrobacterium tumefaciens strain GV3101 via electroporation. C15 empty vector and Agrobacterium tumefaciens strain GV3101 served as the control. Agrobacterium-mediated transient assays were performed as described previously with some modifications (Han et al., 2015). The Agrobacterium suspension was evenly injected throughout the entire Fragaria × ananassa Duch. ‘Toyonoka’ strawberry fruit while it was still attached to the plant, at approximately 12 days after anthesis, using a sterile 1-ml hypodermic syringe. The fruits were harvested for Western blot analysis and ripening-related genes transcript analysis at 3 days after infiltration, and the receptacles were frozen in liquid nitrogen and stored at –80°C until use. For each construct, five uniformly sized fruits were used in the infiltration experiment, and the experiment was repeated three times. The primers used for RT-qPCR analysis of ripening-related genes are listed in Supplemental Table S3 (Han et al., 2015).
For Western blot analysis of FvTCP9 protein, total protein was extracted from infiltrated fruits as described previously with some modifications (Wang et al., 2006). The protein was fractionated by 12% SDS-PAGE and blotted onto PVDF membranes (Millipore) using a semi-dry blotting apparatus as described by the manufacturer (Bio-Rad). GFP was detected using a polyclonal mouse anti-GFP antibody (1:5000 dilution; TransGen Biotech, Beijing, China) and a goat anti-mouse IgG secondary antibody (1:10000 dilution; TransGen Biotech, Beijing, China), according to the manufacturer’s instructions. Proteins separated by SDS-PAGE were detected using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
Statistical Analysis
Statistical significance was determined using a paired Student’s t-test12. The mean ± standard deviations from the mean (SD) of at least three replicates are presented, and significant differences relative to controls are indicated at ∗∗p < 0.05 and ∗p < 0.01.
Results
Identification and Cloning of Strawberry TCP Genes
A total of 21 strawberry TCP members were originally obtained via a BLAST-P search in NCBI using the 18 strawberry TCP amino acid sequences that were predicted in the sequenced diploid woodland strawberry genome accession Hawaii-413 (Shulaev et al., 2011; Darwish et al., 2015). Subsequently, to verify the reliability of the initial results, a survey was conducted to confirm the presence of the conserved TCP domain using the InterProScan database14 and the Pfam database15. The results showed that two putative strawberry TCP genes (XP_011467829.1 and XP_004298742.1) were discarded because they lacked a conserved TCP domain. As a result, 19 non-redundant FvTCP genes were identified in the diploid woodland strawberry and further annotated as FvTCP1 to FvTCP19 according to their locations in strawberry linkage groups 1–7 (Table 1). The 19 FvTCP genes were unevenly distributed in seven linkage groups, with five FvTCP genes located linkage groups 5 and 6, while none of the FvTCP genes mapped to linkage group 1 (Supplementary Figure S1). Detailed characteristics of the TCP transcription factors in F. vesca are provided in Table 1.
Based on the predicted FvTCP coding sequences, we originally cloned the randomly selected 10 homologous TCP genes in the diploid woodland strawberry F. vesca accession Heilongjiang-3. Finally, six FvTCP genes (FvTCP7, FvTCP8, FvTCP9, FvTCP13, FvTCP15, and FvTCP17) belonging to different types were successfully cloned from the cDNA of the leaves of the F. vesca accession Heilongjiang-3 and submitted to NCBI GenBank (GenBank Accession Numbers KX227709, KX227710, KX227711, KX227712, KX227713, and KX227714, respectively). The CDS sequences of the six cloned FvTCP genes from accession Heilongjiang-3 also shared high identities (≥99%, E-value = 0, data not shown) with the corresponding FvTCPs in the strawberry accession Hawaii-4.
Phylogenetic Analysis and Classification of Strawberry TCP Genes
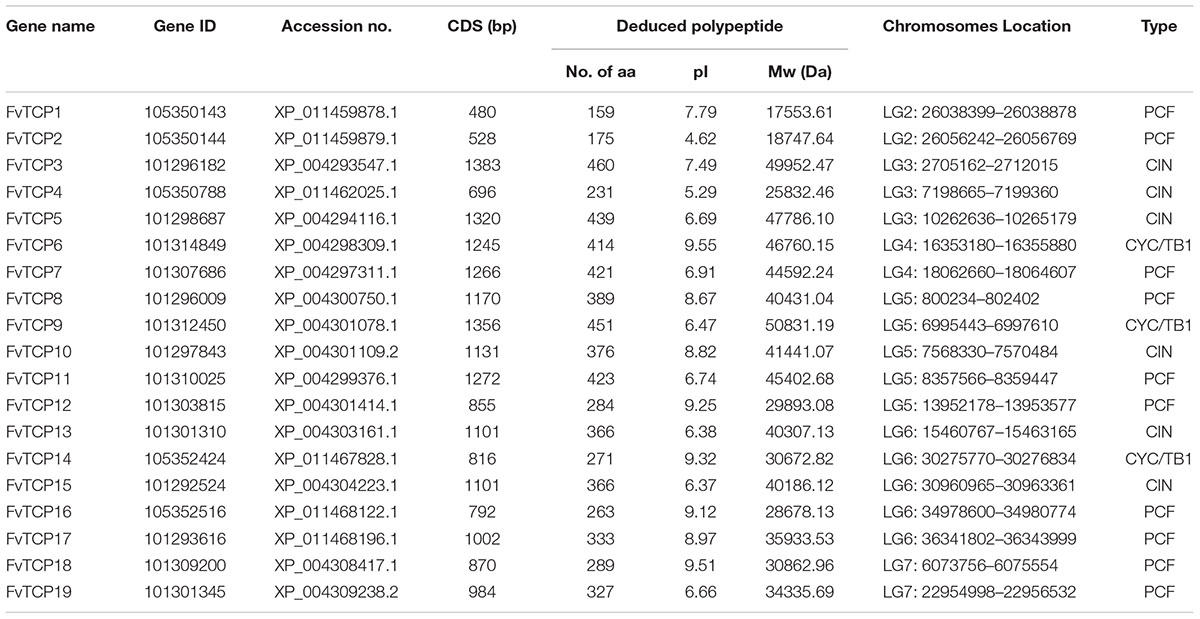
To assess the classification of the FvTCP genes and to gain some insight into the potential function of FvTCP proteins from well-studied TCPs in other plant species, a total of 147 TCP genes, comprising 24 genes from Arabidopsis, 52 from apple, 22 from rice, 19 from strawberry and 30 from tomato, were used to construct a phylogenetic tree (Figure 1). Additionally, to gain a better understanding of the classification of FvTCP members, multiple sequence alignment was performed spanning the approximately 60-amino-acids core TCP domain of all FvTCP amino acid sequences. The phylogenetic analysis and the TCP domain alignment showed that strawberry TCP proteins can be divided into two classes: class I (or PCF) and class II (Figure 1A). The TCP proteins of all five plant species were classified into two major classes (class I and class II; Figure 1B). The most striking difference between these two classes was a four-amino-acid deletion in the basic domain of class I relative to class II proteins (Figure 2A). Further analysis showed that the phylogenetic tree also supported the Arabidopsis and rice described previously in the classification of class II proteins in CIN and CYC/TB1 two subclades (Yao et al., 2007). According to this classification, the CYC/TB1 subclade contains three FvTCP genes (FvTCP6, FvTCP9, and FvTCP14), and the CIN subclade contains six FvTCP genes (FvTCP3, FvTCP4, FvTCP5, FvTCP10, FvTCP13, and FvTCP15; Figure 2A). Outside the TCP domain, the R domain, an approximately 18-residues arginine-rich motif, is conserved and only present in a subset of class II proteins. As shown in Figure 2B, four class II genes (FvTCP6, FvTCP9, FvTCP14, and FvTCP3) contain the R domain, but FvTCP3 is less conserved. Additionally, three CIN subclade genes (FvTCP3, FvTCP5, and FvTCP13) contained the putative miR319 target site and shared the highest sequence similarity with the Arabidopsis and tomato miR319-targeted TCP genes (Figures 1A and 2C).
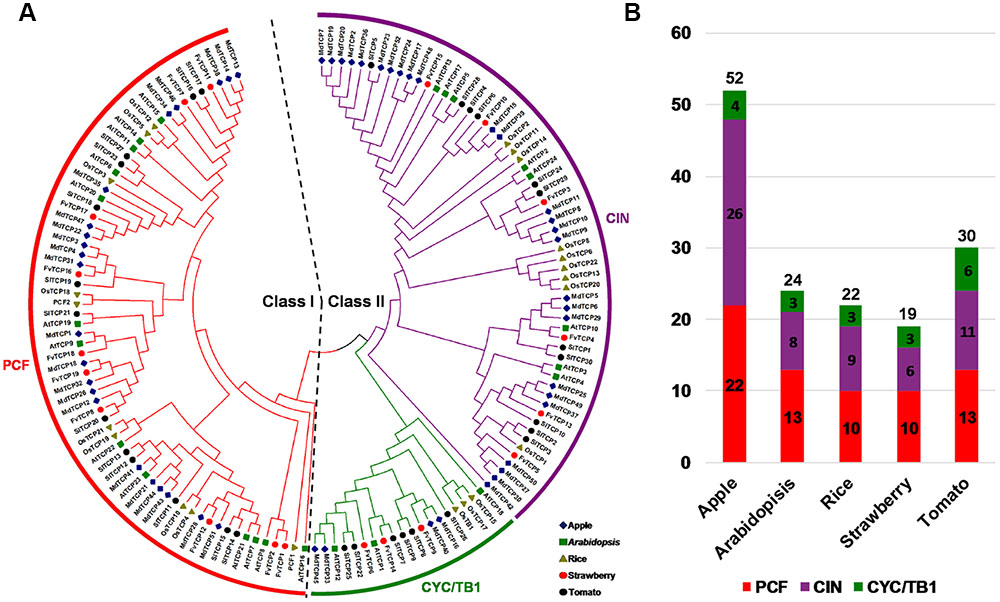
FIGURE 1. Phylogenetic analysis of TCP proteins from strawberry, Arabidopsis, apple, rice, and tomato. (A) The full-length amino acid sequences of TCP proteins from strawberry (FvTCP), Arabidopsis (AtTCP), apple (MdTCP), rice (OsTCP), and tomato (SlTCP) were aligned by ClustalX, and the phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates by MEGA 5.0. The branched lines of the subtrees are colored to indicate different TCP subgroups. (B) TCP family members of apple, Arabidopsis, rice, strawberry, and tomato.
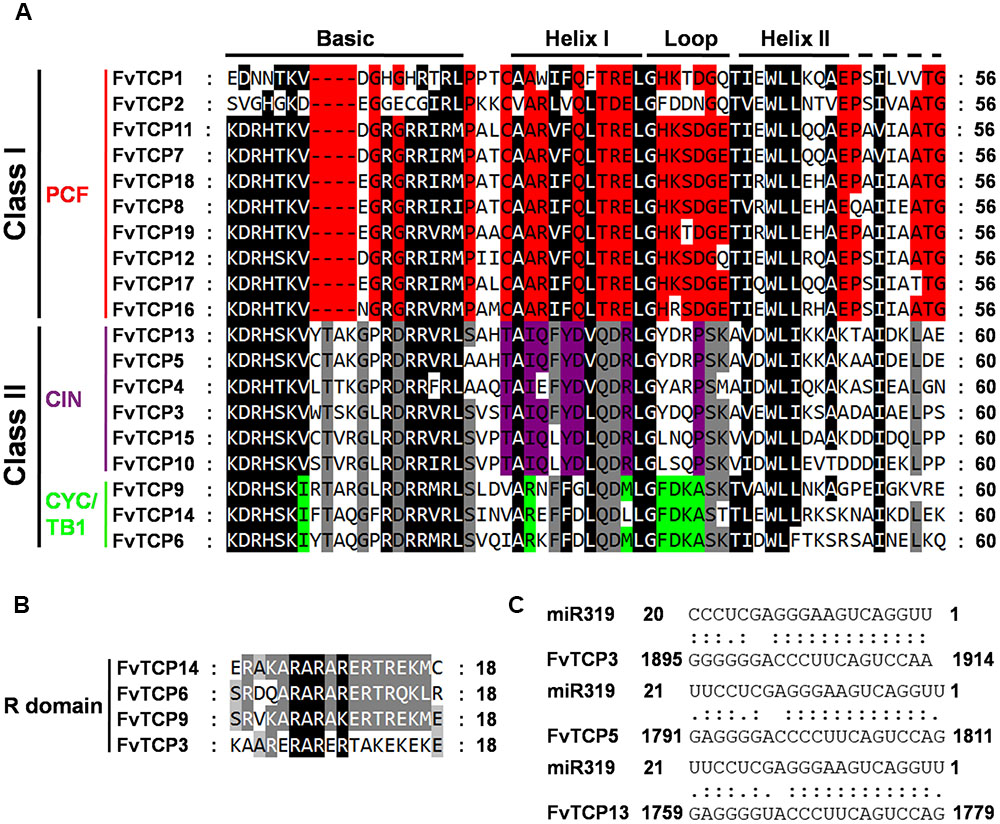
FIGURE 2. Multiple sequence alignment of strawberry TCP transcription factors. (A) Alignment of the TCP domain for the predicted strawberry TCP proteins. Black boxes highlight residues conserved in both TCP classes; red, residues conserved in class I; gray, conserved in class II; purple, conserved in CIN-like proteins; light-green, conserved in CYC/TB1 proteins. The basic, helix I, loop, and helix II regions are indicated. (B) Alignment of the R-domain of Class II subfamily members. The sequences were aligned with ClustalW and visualized with Genedoc. (C) Alignment of putative target areas for miR319.
Conserved Motif Identification and Gene Structure Analysis
To obtain a better understanding of the diversification and evolutionary relationships of the TCP protein in F. vesca, the conserved motifs and exon/intron organization of FvTCPs were analyzed. A new phylogenetic tree was constructed using the protein sequences of FvTCPs, which divided the FvTCP proteins into three subclades. As shown in Figure 3 and Supplementary Figure S2, we used the online MEME tool to predict the conserved FvTCP protein motifs, identifying 12 conserved motifs. As expected, all 19 of the FvTCPs demonstrated the presence of a highly conserved TCP domain (motif 1). The conserved R domain (motif 4) was found in four class II FvTCPs. All of the class II FvTCPs were characterized by motif 2 in the N-terminal TCP domain. By comparison, the C-terminal TCP domain of motif 3 was detected in a subset of the class I proteins. In addition, some motifs were exclusively present in a subset of a particular subclade, suggesting that these motifs may contribute to the specific function of those genes in the subclade. For example, motifs 6, 8, and 10 in PCF, and motifs 9 and 11 in CIN (Figure 3).
FIGURE 3. Structural analysis of strawberry TCP transcription factors. The protein domains of the strawberry TCP genes are shown on the left and are denoted by rectangles with different colors. The exon-intron organization is shown on the right, with exons and introns represented by green round-corner rectangles and red shrunken lines, respectively; untranslated regions (UTRs) are indicated by gray rectangles. The red, green, and purple rectangles are used to cluster the genes into the PCF, CYC/TB1, and CIN classes.
To gain further insight into the evolutionary relationships among FvTCP genes, we investigated the exon/intron organization of individual FvTCP genes by aligning the cDNA sequences and corresponding genomic DNA sequences. Overall, the FvTCP genes exhibited a highly conserved exon-intron organization: 18 of 19 FvTCP genes had no intron, while only FvTCP9 genes possessed one intron. Additionally, most of the FvTCP genes within the nearby paralogous genes demonstrated very similar exon/intron distribution patterns in terms of the exon length and the intron number (Figure 3).
Identification of cis-Regulatory Elements in the Promoter of FvTCP Genes
The analysis of cis-regulatory elements in promoter sequences is an important feature for understanding gene function and regulation. To identify the likely cis-acting elements of the FvTCPs, the promoter regions (1 kb of genomic DNA sequence upstream of the translation start site) of the FvTCP genes were used to search the PlantCARE database (Postel et al., 2002). A series of cis-acting elements involved in plant growth and development, phytohormone responses, and abiotic and biotic stress responses were identified (Supplementary Table S2). As shown in Figure 4, the Skn-1_motif and GCN4_motifs, cis-acting regulatory elements involved in endosperm expression (Washida et al., 1999), were found in the promoters of 16 and 9 FvTCP genes, respectively. The circadian control element (circadian; Anderson et al., 1994) was found in 10 FvTCP genes. Notably, two leaf development-related cis-acting elements (HD-Zip1 and HD-Zip2; Sessa et al., 1993) were found in the FvTCP15 promoter. Additionally, the zein metabolism regulation element (O2 site), meristem expression and specific activation element (CAT-box and CCGTCC-box), seed-specific regulation element and shoot-specific expression element (RY element and as-2-box; Bobb et al., 1997) were also identified in the promoters of the FvTCP genes. In hormone-related cis-acting elements, the SA-responsive element (TCA element; Goldsbrough et al., 1993), the MeJA-responsive element (CGTCA motif and TGACG motif; Rouster et al., 1997), and the ABA-responsive element (ABRE; Shen and Ho, 1995) were found in the promoters of 11, 8, and 9 FvTCP genes, respectively. The gibberellin-responsive element (GARE motif and P-box; Kim et al., 1992; Washida et al., 1999) and the auxin-responsive element (TGA element, AuxRR core and TGA box; Ulmasov et al., 1997) were observed in 10 and 4 FvTCP genes. Plenty of hormone-responsive elements were identified in the FvTCP promoter sequences, indicating that phytohormones could play crucial roles in the regulation of plant growth and development (Figure 4B). In addition, some stresses-related (e.g., drought, extreme temperatures, salinity, and disease) cis-acting elements were also found in the putative promoter regions of the FvTCP genes (Figure 4).
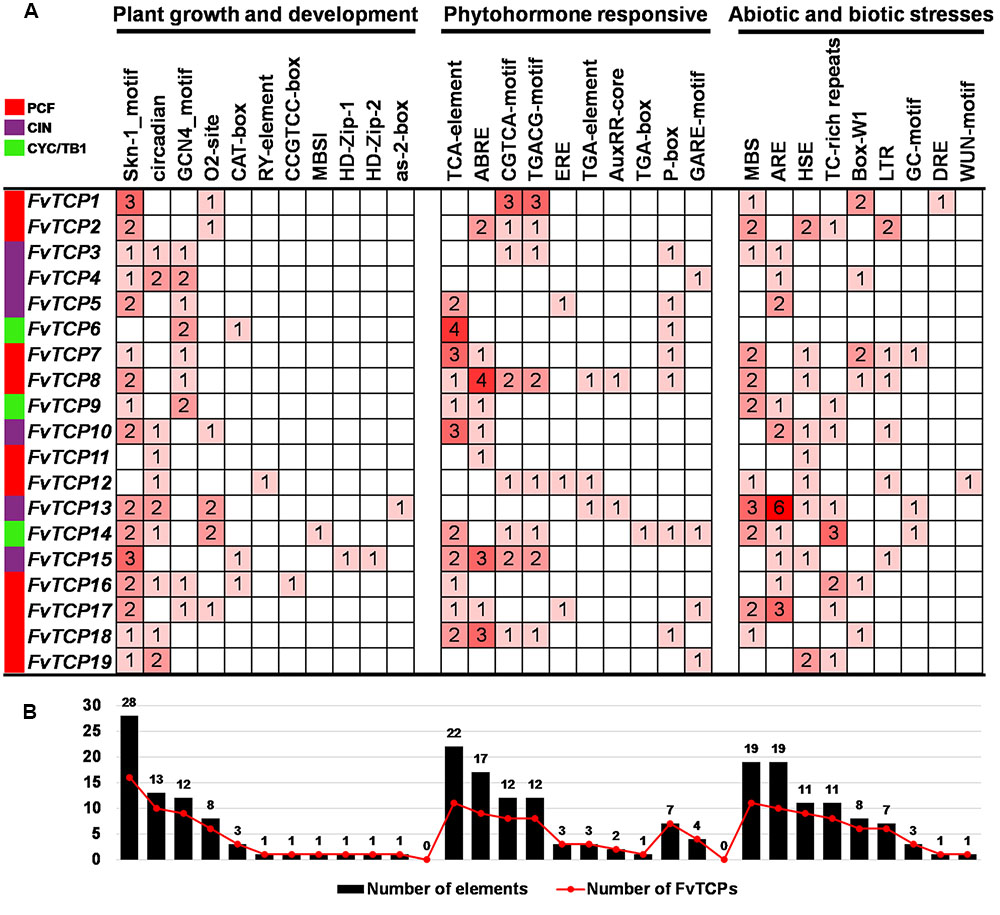
FIGURE 4. Cis-acting element analysis of the promoter regions of strawberry TCP genes. (A) Number of each cis-acting element in the promoter region (1 kb upstream of the translation start site) of FvTCP genes. (B) Statistics for the total number of FvTCP genes, including the corresponding cis-acting elements (red dot) and the total number of cis-acting elements in FvTCP gene family (black box). Based on the functional annotation, the cis-acting elements were classified into three major classes: plant growth and development-, phytohormone responsive-, or abiotic and biotic stresses-related cis-acting elements (detailed results shown in Supplementary Table S2).
Tissue-Specific Transcript Accumulation Patterns in FvTCP Genes
To investigate the tissue-specific transcript accumulation profiles of TCP genes in F. vesca, we analyzed the transcripts of FvTCP genes using semi-quantitative RT-PCR and validated the results via RT-qPCR analysis of different tissues, including roots, stems, runners, leaves, flowers, floral buds, and fruits (fully ripened fruits) from the diploid woodland strawberry accession Heilongjiang-3. As indicated in Figure 5, some FvTCP genes exhibited tissue-specific transcript accumulation patterns, while other FvTCP genes showed similar transcript accumulation patterns in different tissues, potentially indicating the functional divergence of FvTCP genes during strawberry growth and development. For example, FvTCP6, FvTCP8, FvTCP9, and FvTCP15 were constitutively expressed in every tissue tested at relatively high transcript levels, whereas FvTCP1, FvTCP2, and FvTCP18 were expressed at very low level in all tested tissues (Figure 5A). In contrast, the transcript accumulation levels of FvTCP3 and FvTCP5 were very high in runners, young leaves, and floral buds and they were relatively low in flowers, indicating that they might play an important role in the development of runners, young leaves, and floral buds. A similar transcript accumulation pattern was observed for FvTCP13 (Figure 5B). In particular, FvTCP19 displayed extremely high relative transcript levels in stems, which suggested that it might play a role in the development of strawberry stems. FvTCP4 and FvTCP14 were preferentially expressed at high levels in runners, young leaves, or floral buds and at almost undetectable levels in other tissues (Figure 5B). Additionally, the transcript accumulation patterns of certain genes varied among the eight tissues. For example, FvTCP12 and 17 exhibited high transcript levels in roots, runners, and fruits, but relatively low transcript levels in young leaves, mature leaves, flowers, and floral buds, and FvTCP7, which was abundantly expressed in roots, runners, and floral buds, displayed lower levels in other tissues (Figure 5B). Remarkably, RT-qPCR analysis showed that some FvTCP genes (FvTCP9, FvTCP10, and FvTCP17) were highly expressed in fruits (fully ripened fruits), which indicated that they might play an important role in fruit ripening (Figure 5B). These results prompted us to investigate the transcript of FvTCP genes during various fruit development and ripening stages.
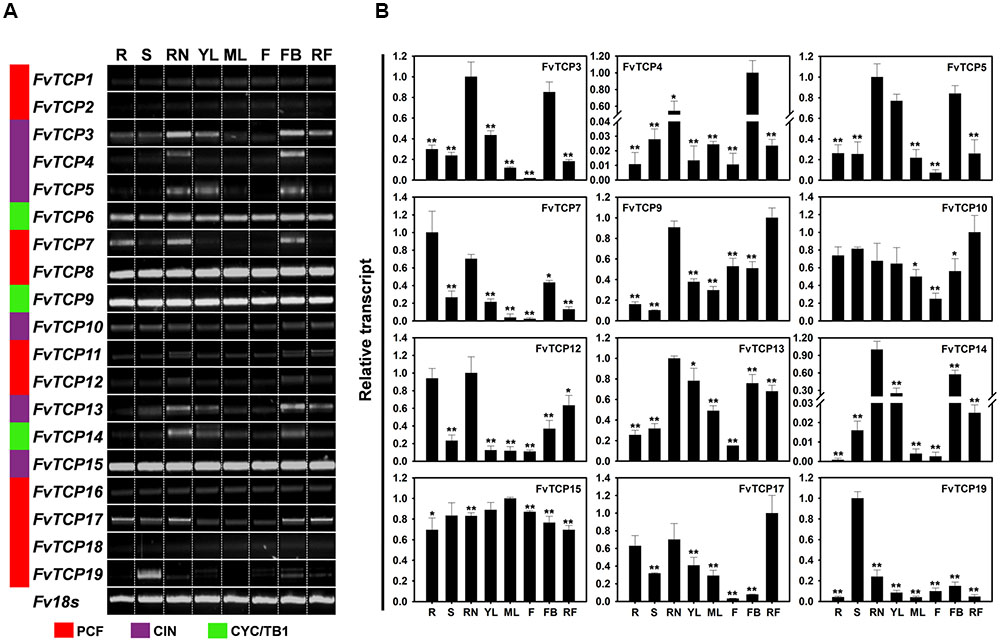
FIGURE 5. Tissue-specific transcript accumulation patterns of 19 TCP genes in the diploid woodland strawberry (F. vesca). (A) Transcript accumulation profiles of 19 FvTCP genes in different tissues using semi-quantitative PCR. Fv18s was used as an internal control. Lanes: R, roots; S, stems; RN, runners; YL, young leaves; ML, mature leaves; F, flowers; FB, floral buds; RF, red fruits. (B) Transcript accumulation profiles of 12 selected FvTCP genes in different tissues using RT-qPCR. The analysis results were normalized using Fv18s. The experiments were repeated three times and provided consistent results. The mean values and SDs were obtained from three biological and three technical replicates. Asterisks indicate significant difference compared to the highest transcription level, as determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
Transcript Accumulation Patterns of FvTCP Genes during Different Fruit Developmental Stages
To investigate fruit development and ripening-related strawberry FvTCP genes, we focused on the transcript accumulation patterns of FvTCP genes in fruits during five different developmental stages (mature flowers with partially withered petals, mature green receptacles, white receptacles with green achenes, half white and half red fruits, and fully ripened fruits). Hierarchical clustering was used to describe the various relative levels of FvTCP gene transcripts, which could be differentiated into two distinct groups. As shown in Figure 6A, 11 FvTCP genes were down-regulated during different fruit developmental stages, while five FvTCP genes exhibited up-regulated transcript accumulation patterns. Additionally, three FvTCP genes showed stable transcript accumulation patterns. Among the 16 down-regulated and up-regulated FvTCP genes, we selected 15 relatively dramatically down-regulated or up-regulated FvTCP genes by RT-qPCR to further test their transcript abundance in the various developmental stages of the fruit (Figure 6B). Among them, five TCP genes (FvTCP5, FvTCP7, FvTCP11, FvTCP12, and FvTCP16) exhibited high transcript accumulation patterns in mature flowers with partially withered petals, and three TCP genes (FvTCP3, FvTCP13, and FvTCP17) showed high accumulation patterns in mature flowers with partially withered petals and mature green receptacles in comparison with the other stages (Figure 6B). These data indicate that the eight FvTCP genes may be involved in strawberry fruit development. The transcripts of four genes (FvTCP6, FvTCP10, FvTCP14, and FvTCP19) were gradually upregulated during the ripening process, which suggested that they might function in strawberry fruit ripening (Figure 6B).
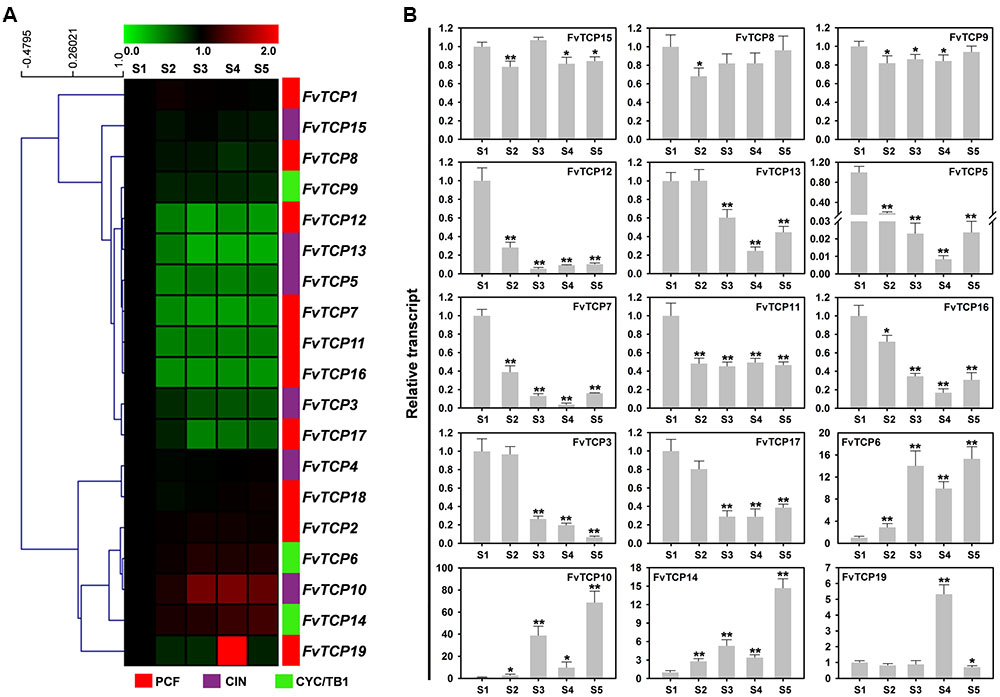
FIGURE 6. Transcript accumulation pattern of 19 TCP genes in the diploid woodland strawberry (F. vesca) during different fruit developmental stages. (A) Hierarchical clustering of the transcript accumulation profiles of 19 FvTCP genes during different fruit developmental stages (S1: mature flowers with partially withered petals, S2: mature green receptacles, S3: white receptacles with green achenes, S4: half white and half red fruits, S5: fully ripened fruits; original results shown in Supplementary Figure S4). Fv18s was used as an internal control. The transcript accumulation profiles were generated by semi-quantitative PCR and were visualized as heat maps. The color scale represents the relative transcript levels with increased (red) or decreased (green) transcript abundance. Genes were hierarchically clustered based on average Pearson’s distance metric and ‘average linkage’ method. (B) RT-qPCR transcript analysis of 15 selected FvTCP genes during different fruit developmental stages. The results were normalized to Fv18s. The experiments were repeated three times and provided consistent results. The mean values and SDs were obtained from three biological and three technical replicates. Asterisks indicate significant difference compared to S1, as determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
Transcript Accumulation Patterns of FvTCP Genes during Different Strawberry Subcultural Propagation Periods
To provide additional information on the growth and developmental functions of TCP genes in strawberry, we investigated their transcript accumulation patterns during five different periods of subcultural propagation in strawberry ‘Heilongjiang-3’ using semi-quantitative RT-PCR (Supplementary Figure S3A). In general, the transcript accumulation patterns obtained for the FvTCP genes could be classified into two types (Supplementary Figure S3B). The majority of the FvTCP genes, specifically FvTCP2, FvTCP4, FvTCP6, FvTCP10, FvTCP11, FvTCP12, and FvTCP14, exhibited downregulation from P1 to P5 stage. The remaining FvTCP genes were only upregulated during specific propagation periods. For example, FvTCP8, FvTCP9, FvTCP13, FvTCP18, and FvTCP19 were upregulated at P2 stage, which indicated that they might promote growth and development during early subcultural propagation. Additionally, the transcript levels of FvTCP1, FvTCP9, and FvTCP18 revealed an upregulation at P5 stage, suggesting that these genes might be involved in rooting and bud germination after subculture. FvTCP17 displayed high transcript levels from P2 to P4 stage, implying that FvTCP17 might play vital biological roles in the subcultural propagation processes of whole strawberry.
Transcript Analysis of FvTCP Genes in Response to Stresses and Hormone Treatments
To determine the potential roles of the FvTCP genes during plant responses to various environmental stresses, semi-quantitative RT-PCR was performed for the 19 FvTCP genes in the leaves of F. vesca plantlets exposed to cold, heat, drought, salt, and powdery mildew treatments. Overall, the FvTCP genes responded to drought and salt treatment to a greater extent than to temperature and biotic treatment (Figure 7). Among them, 10 FvTCP genes were upregulated in response to drought stress, and 13 FvTCP genes responded to salt treatment. In contrast, a minority of the FvTCP genes were only slightly upregulated in response to temperature and biotic treatment (Figure 7). Notably, FvTCP14 and FvTCP19 showed different degrees of upregulation in response to cold, heat, drought, salt, and powdery mildew treatments. The transcript levels of FvTCP4, 7, and 10 responded to at least three treatments (Figure 7).
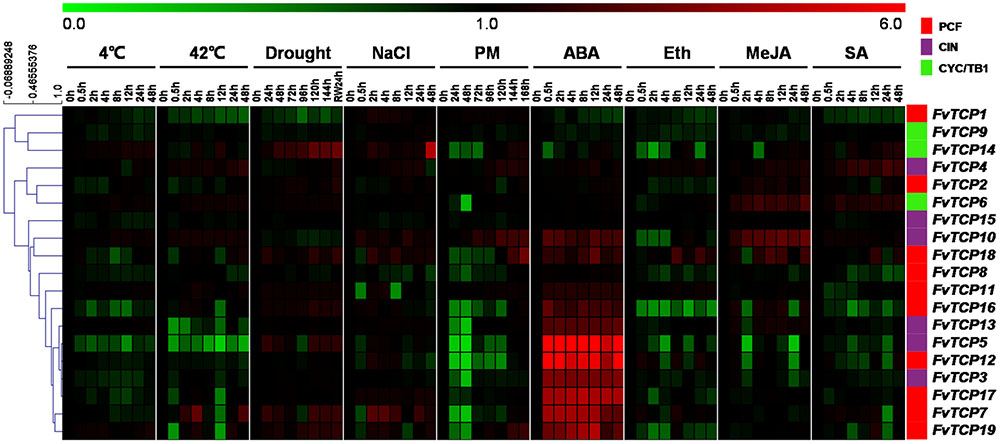
FIGURE 7. Hierarchical clustering of the transcript accumulation profiles of 19 TCP genes in the diploid woodland strawberry (F. vesca) in response to different treatments. The transcript accumulation profiles of 19 FvTCP genes in response to cold, heat, drought, NaCl, powdery mildew infection, ABA, Eth, MeJA, and SA treatments were generated by semi-quantitative PCR and visualized as heat maps (original results shown in Supplementary Figures S5 and S6). The color scale represents the relative transcript levels with increased (red) or decreased (green) transcript abundance. Genes were hierarchically clustered based on average Pearson’s distance metric and ‘average linkage’ method. The experiments were repeated three times with consistent results.
Plant hormones such as SA, MeJA, ethylene (Eth) and ABA have well-established roles in plant stress signaling networks and developmental processes (Bari and Jones, 2009). To understand how FvTCP gene transcripts accumulate in response to plant hormone treatment, semi-quantitative RT-PCR and RT-qPCR were used to analyze FvTCP transcripts in response to ABA, ethephon (Eth), MeJA and SA in the leaves. For ABA treatment, the transcript levels of 12 FvTCP genes were prominent and rapidly increased to significantly high levels at 0.5 hpt, which were maintained throughout the entire treatment period (Figure 7). Notably, the highest transcript levels of FvTCP5, 7, 12, and 17 reached extremely high levels compared with the basal transcript levels after ABA treatment (Figure 8). Conversely, almost all FvTCP was downregulated or remained nearly unchanged relative to basal transcript levels in response to Eth treatment (Figure 7). Additionally, seven and four FvTCP genes were upregulated in response to MeJA and SA, respectively, while the other TCP members were downregulated or exhibited no significant change (Figure 7). It is worth noting that FvTCP4, FvTCP6, FvTCP10, FvTCP13, FvTCP16, and FvTCP18 were upregulated in response to at least two of the hormone treatments (Figure 7).
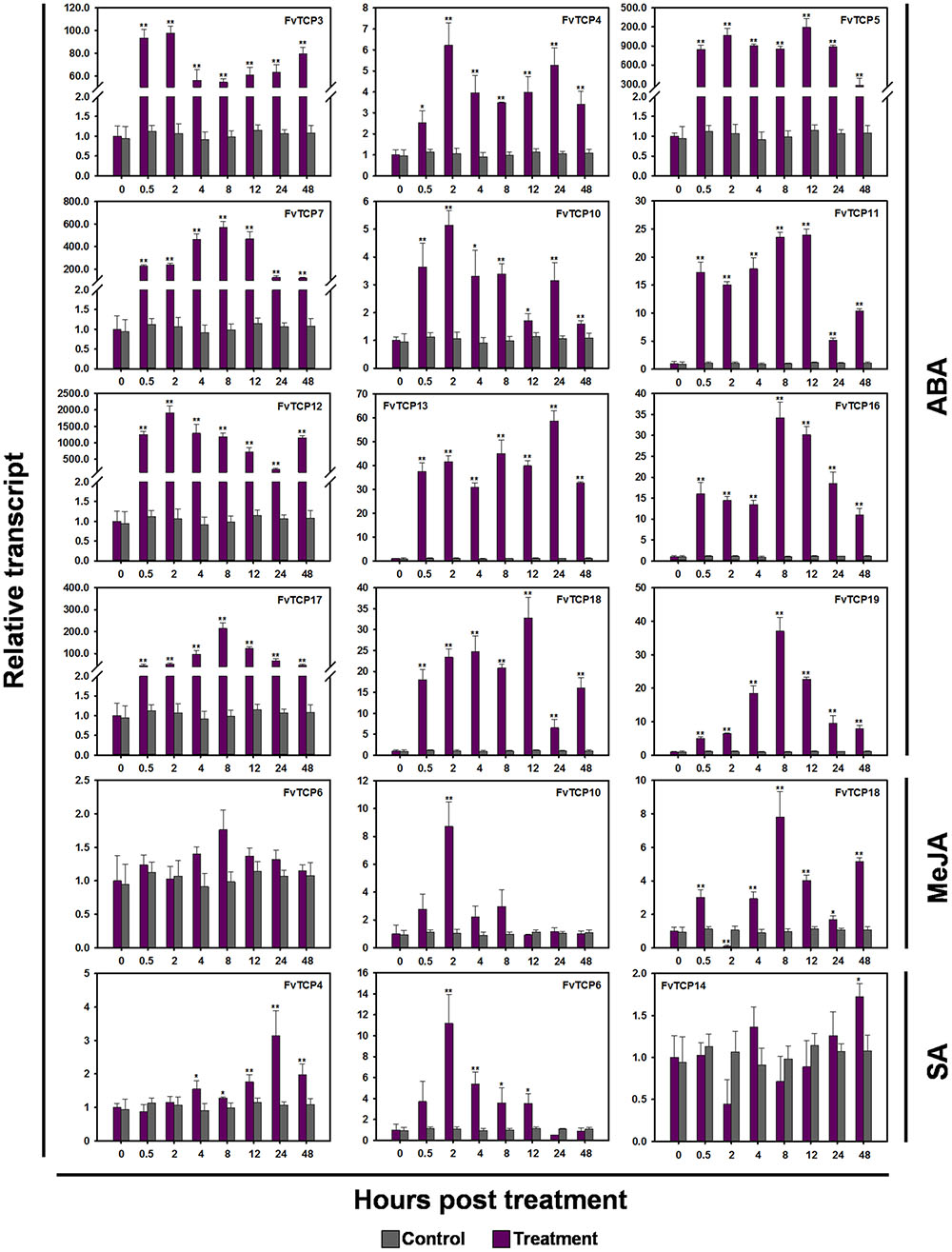
FIGURE 8. RT-qPCR analysis of several TCP genes in the diploid woodland strawberry (F. vesca) in response to ABA, MeJA, and SA treatments. The detailed transcript levels of several FvTCP genes revealed unusual transcript accumulation patterns in response to ABA, MeJA, and SA treatments. The results were normalized to Fv18s. The experiments were repeated three times and provided consistent results. The mean values and SDs were obtained from three biological and three technical replicates. The asterisks indicate that the corresponding gene was significantly up or down-regulated in response to treatment, as determined by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
Subcellular Localization of FvTCPs
It is known that TFs regulate the transcription of target genes by binding to specific cis-elements in their promoters and that this binding occurs in the nucleus. To assess the subcellular localization of the FvTCP TFs, the full-length open reading frames (ORFs) without the stop codon of six cloned FvTCP genes were cloned into a vector in-frame with green fluorescence protein (GFP) under the control of the CaMV 35S promoter. The resulting constructs and empty (control) vector were transiently expressed in Arabidopsis mesophyll protoplasts. Fluorescence microscopy revealed that six FvTCP fusion proteins were clearly localized in the nucleus (Figure 9). Notably, FvTCP7-GFP (ii) and FvTCP17-GFP (ii) were also localized in the nucleus and cytoplasm (Figure 9). To confirm the nuclear and cytoplasm localization of FvTCP7 (ii) and FvTCP17 (ii), we also examined the subcellular localization of FvHsfC1a, a strawberry heat shock transcription factor that has been reported to localize to both the nucleus and cytoplasm of Arabidopsis mesophyll protoplasts (Hu et al., 2015). FvTCP7 (ii), FvTCP17 (ii), and FvHsfC1a exhibited similar subcellular localizations (Figure 9). Additionally, FvTCP7-GFP (iii) and FvTCP17-GFP (iii) fluorescent signals showed a punctate pattern in the cytoplasm that resembled mitochondria (Figure 9).
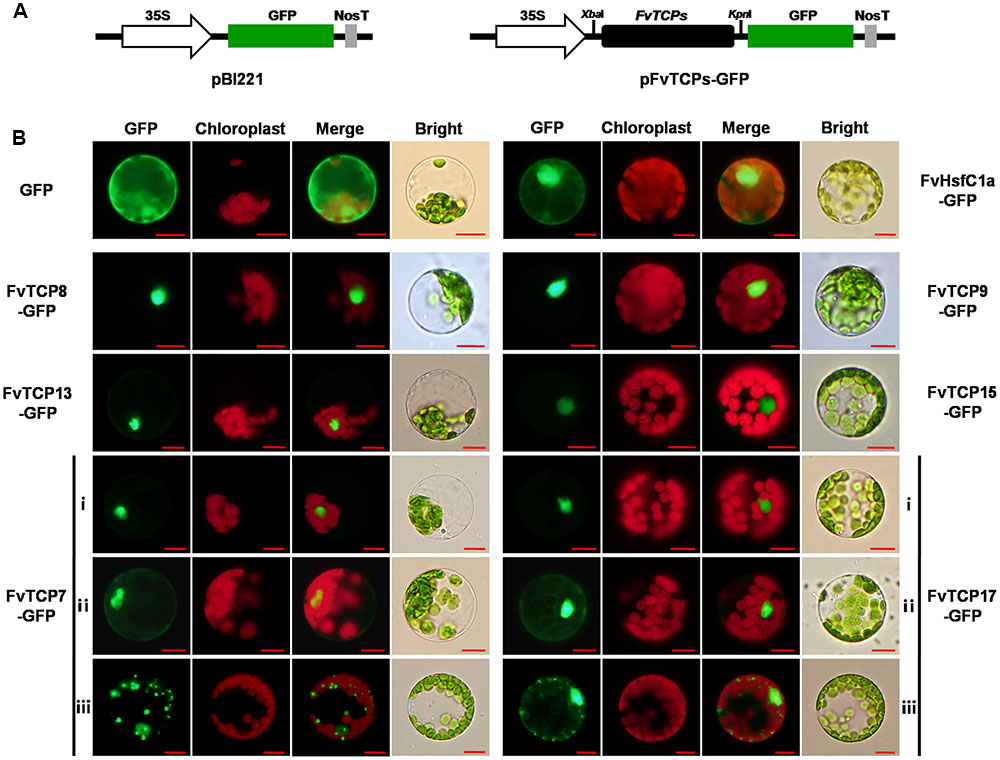
FIGURE 9. Subcellular localization of six strawberry TCP genes. (A) Schematic illustration of vectors pBI221 and FvTCPs. The selected TCP genes were cloned from a diploid woodland strawberry (F. vesca) and used to construct the CaMV35S::TCPs-GFP vectors, in which GFP was fused at the C-terminus. (B) The six FvTCP-GFP fusion proteins (FvTCP7-GFP, FvTCP8-GFP, FvTCP9-GFP, FvTCP13-GFP, FvTCP15-GFP, and FvTCP17-GFP), the FvHsfC1a-GFP marker protein, and GFP as a control were transiently expressed in Col-gl Arabidopsis protoplasts and observed under a fluorescence microscope. The merged images were constructed in the green fluorescence channel (first panels) and the chloroplast autofluorescence channel (second panels). The corresponding bright field images are shown on the right. Bar = 10 μm.
Transient Over-Expression of the FvTCP9 Gene in Strawberry Fruits
Agrobacterium-mediated transient gene expression is a rapid and powerful tool for the analysis of gene function in plants. For a more detailed analysis of the biological roles of the FvTCP genes during strawberry fruit development and ripening, we cloned the FvTCP9 gene into the BamHI sites of the binary expression vector C15 and transiently over-expressed it in Fragaria × ananassa Duch. ‘Toyonoka’ attached fruits (12 days after anthesis) (Figure 10A). The Western blot results revealed single protein bands of the expected size (∼75 kDa for FvTCP9-YFP and ∼25 kDa for YFP) in the agroinfiltrated strawberry fruits, whereas antigen-specific bands were not detected in protein extracts from agroinfiltrated Agrobacterium strain GV3101 control fruits (Figure 10B). These results clearly demonstrated the stable integration and protein expression of FvTCP9 in agroinfiltrated strawberry fruits. Additionally, several strawberry fruit ripening-related genes were analyzed by RT-qPCR in the injected fruits. As shown in Figure 10C, transient over-expression of FvTCP9 markedly up-regulated the expression of a series of genes implicated in fruit color and aroma metabolism, including FaCHS (chalcone synthase), FaF3H (flavanone 3-hydroxylase), FaUFGT (UDP-glycose flavonoid 3-O-glycosyltransferase), and FaQR (quinine oxidoreductase). Simultaneously, transient over-expression FvTCP9 dramatically affected the expression of a series of genes implicated in fruit softening, such as FaPE (pectinesterase), FaPG (polygalacturonase), FaCEL (cellulose), FaGAL1/2 (b-galactosidase1/2), FaXYL1 (b-xylosidase1), and FaEXP1/2/5 (expansin1/2/5).
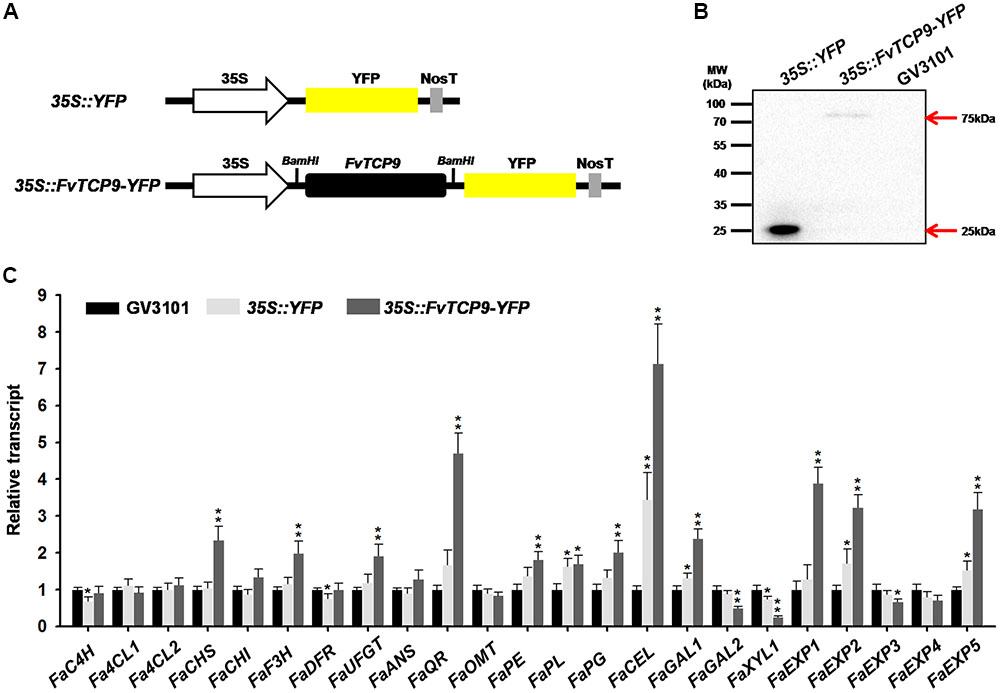
FIGURE 10. Transient over-expression of FvTCP9 in Fragaria × ananassa Duch. ‘Toyonoka’ fruits. (A) Schematic illustration of vectors 35S::YFP and 35S::FvTCP9-YFP. Black filled boxed denote the FvTCP9 gene. Yellow filled boxed indicate yellow fluorescence protein (YFP). (B) Western blot analysis of FvTCP9 in transiently over-expressing FvTCP9 strawberry fruits. Samples were harvested at 3 days after agroinfiltration, and total soluble proteins were extracted. (C) RT-qPCR transcript analysis of ripening-related genes during the transient over-expression of FvTCP9 in Fragaria × ananassa Duch. ‘Toyonoka’ fruits. The strawberry 26S-18S RNA gene (housekeeping gene) was used as an internal control to normalize the expression data. The ripening-related gene primers are listed in Supplementary Table S3 (Han et al., 2015). Asterisks indicate significant differences compared with the control sample, as determined by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
Discussion
The TCP gene family encodes plant-specific transcription factors that are involved in plant growth and development (Manassero et al., 2013). To date, the features and functions of the TCP gene family have been identified and investigated in several plant species, including Arabidopsis (Riechmann et al., 2000), rice (Yao et al., 2007), tomato (Parapunova et al., 2014), apple (Xu et al., 2014), cotton (Ma et al., 2014), and watermelon (Shi et al., 2016). However, no comprehensive analyses of the TCP gene family in F. vesca, an important Rosaceae model plant that is widely used in fruit research, have been conducted. In this study, we conducted a broad analysis of the TCP genes in strawberry by investigating their linkage group organization, evolutionary relationships, gene structure, protein motifs, cis-acting elements, and expression profiles in different tissues and developmental stages and under various stress conditions, subcellular localizations, and during transient over-expression. Genome-wide analysis of the TCP genes in F. vesca will facilitate a better understanding of the role of this gene family during strawberry growth and development.
Evolution and Structure of the TCP Gene Family in F. vesca
A total of 19 FvTCPs were identified based on the diploid woodland strawberry genome (accession Hawaii-4; Shulaev et al., 2011; Darwish et al., 2015). This number is highly conserved among Arabidopsis (24 members) and rice (22 members; Figure 1B) (Riechmann et al., 2000; Yao et al., 2007). However, it is significantly lower than that present in apple (52 members; Figure 1B), which is consistent with the genome sizes of apple (∼742.3 Mb in M. domestica; Velasco et al., 2010), indicating that TCP genes in different plants have expanded to differing degrees. Sequence alignment and phylogenetic analysis of FvTCP proteins has resulted in their classification into three major subclasses (I, II, and III), with FvTCP genes distributed across all three subclasses. In addition, each subclass contains TCP genes from Arabidopsis, tomato, apple, and rice. FvTCP genes are more closely related to genes from apple TCP genes, demonstrating that apple and strawberry are Rosaceae and evolved more recently from a common ancestor. These results indicate that although plant TCP genes may be derived from a common ancestor, many have undergone distinct patterns of differentiation with the divergence of different lineages. Moreover, the consistency of the motif compositions of FvTCP proteins as well as the exon/intron structures of most FvTCP genes with phylogenetic subclasses further supported the close evolutionary relationships among FvTCPs as well as the dependability of our phylogenetic analysis, as described previously, in cotton and apple (Ma et al., 2014; Xu et al., 2014).
Potential Roles of FvTCP Genes in Plant Growth and Development
Accumulating evidence suggests that TCP transcription factors are involved in the regulation of cell growth and proliferation, performing diverse functions in multiple aspects of plant growth and development (Martin-Trillo and Cubas, 2010). The CYC/TB1 clade includes genes that are mainly involved in the development of axillary meristems that give rise to either flowers or lateral shoots. AtTCP1, the closest homolog of CYC in Arabidopsis, is involved in the longitudinal elongation of petioles, rosette leaves, and inflorescent stems. The expression pattern of AtTCP1 is strong in the lower part of the inflorescence stem, the distal region of expanding rosette leaves, and the midrib of the blade and petiole during leaf development (Koyama et al., 2010). Additionally, Arabidopsis gain-of-function tcp1-1D mutant plants show elongated leaves and petioles, whereas TCP1-SRDX plants display rounded and epinastic leaves, shortened petioles, and reduced statures (Guo et al., 2010). FvTCP14, which is closely related to AtTCP1, was transcribed at high levels in runners, floral buds, or young leaves and was almost undetectable in other tissues (Figures 1A and 5B). This result is consistent, in part, with the expression profile of AtTCP1 and implies that the FvTCP14 gene in F. vesca functions in runners and in inflorescence and leaf development. BRANCHED1 (BRC1, AtTCP18) and BRANCHED2 (BRC2, AtTCP12), two homologs of TB1 in Arabidopsis, were transcribed at high levels in tissues that mainly contained axillary buds, such as leaf bases and stem, inflorescences, and siliques (Aguilar-Martinez et al., 2007). AtTCP18 acts downstream of auxin and strigolactone to coordinate axillary bud outgrowth, and mutants with reduced activity of either gene show an increased number of rosette branches. In contrast, the up-regulation of AtTCP18 results in an inhibition of lateral branching. AtTCP12 exhibits a weaker or no mutant phenotype compared with AtTCP18 (Aguilar-Martinez et al., 2007; Finlayson, 2007). The phylogenetically close gene of AtTCP18 in strawberry is FvTCP9 (Figure 1A), which displays higher homology with AtTCP18 by NCBI BLAST-P and a significantly higher relative transcript level in runners and fruits (Figure 5). These transcript similarities suggest that FvTCP9 is likely to perform roles similar to AtTCP18 in axillary bud tissue development in strawberry.
Additionally, CIN-like clade genes could be more ancient than the CYC/TB1 clade TCPs and are important for leaf growth and development. In Arabidopsis, five of the CIN subclade members (AtTCP2, 3, 4, 10, and 24) are post-transcriptionally regulated by miRNA319, and miR319 modulates jasmonate biosynthesis, negative leaf curvature, and crinkly leaves, while it positively regulates leaf senescence and affects petal development (Nath et al., 2003; Palatnik et al., 2007; Schommer et al., 2008; Nag et al., 2009). In the present study, the closest strawberry homologs of these Arabidopsis genes are the four FvTCP genes, FvTCP3, FvTCP4, FvTCP5, and FvTCP13, all of which, excluding FvTCP4, carry a putative binding site for miR319 (Figures 1A and 2C). The transcript accumulation levels of FvTCP3, FvTCP5, and FvTCP13 were very high in runners, young leaves, and floral buds. These results indicated that the regulation of leaf growth and development by miRNA-targeted TCP TFs that are homologous to those in strawberry may be consistent in Arabidopsis.
By contrast, Class I FvTCP genes showed more widespread and less tissue-specific transcript accumulation patterns, such as in roots, runners, floral buds, and fruits (Figure 5). These results implied that Class I FvTCP genes might play diverse regulatory roles at multiple growth and development stages. In Arabidopsis, AtTCP14 regulates embryonic growth potential during seed germination, which is related to ABA and GA responses (Tatematsu et al., 2008). In our study, FvTCP7, 11, and 17 were phylogenetically close to AtTCP14 and had similarly high transcript accumulation levels after ABA treatment (Figures 1A and 8). However, FvTCP7, 11, and 17 were down-regulated during different developmental stages of the fruit (Figure 6), suggesting that they were also down-regulated during embryo and seed development. These findings indicate that FvTCP7, 11, and 17 might have functions that differ from those of ABA-mediated embryo and seed development in response to AtTCP14.
FvTCP is Likely to Play a Role in Fruit Development and Ripening
Fruit development and ripening is a complex and highly controlled biological process that is controlled by transcriptional regulatory networks involving many transcription factors, such as MADS-box, NAC, and EIN3/EIL (Seymour et al., 2013). In strawberry, the endosperm and seed coat play a crucial role in the fruit set and early stage fruit development (Kang et al., 2013). Promoter analysis showed that most of the FvTCP genes harbored Skn-1_motif and GCN4_motif cis-regulatory elements involved in endosperm expression in their promoters (Figure 4). These results imply that FvTCP genes are likely to play an important role in strawberry fruit development and ripening. In tomato, several SlTCP genes, such as SlTCP12, SlTCP15, and SlTCP18, are preferentially expressed in tomato fruit. Moreover, these genes are regulated by RIN (RIPENING INHIBITOR), CNR (COLORLESS NON-RIPENING), and SlAP2a (APETALA2a) proteins, which are transcription factors with key roles in ripening, suggesting a role during tomato fruit development or ripening (Parapunova et al., 2014). In strawberry, FvTCP12 and FvTCP17, which are homologs of SlTCP15 and SlTCP18, respectively, also showed high transcript accumulation levels in fruits, indicating that FvTCP12 and 17 might play a role in strawberry fruit development or ripening (Figures 1A and 5). Additionally, ethylene is a key regulator during fleshy fruit ripening (Kumar et al., 2014). The current results demonstrate that the majority of FvTCP genes detected herein were downregulated or nearly unchanged following Eth treatment (Figure 7), suggesting that TCP transcription factors regulate strawberry fruit ripening independently of the Eth pathway.
Agro-infiltration of maturing strawberry fruit has been a useful tool for defining the gene contributions to fruit development and ripening (Hoffmann et al., 2006). However, few TCPs have been well characterized in terms of their potential roles in this process. In this study, agro-infiltrated fruits overexpressing FvTCP9 revealed that the expression of a series of ripening-related genes was distinctly up-regulated (more than 3.0-fold), such as FaQR (quinine oxidoreductase), FaCEL (cellulose), and FaEXP1/2/5 (expansin1/2/5). FaQR, an enzyme involved in the biosynthesis of 4-hydroxy-2,5-dimethyl-3(2H)-furanone (HDMF; Furaneol), is a key flavor compound in strawberries (Raab et al., 2006). FaCEL is involved in strawberry the fruit softening process via the regulation of cellulose degradation (Woolley et al., 2001). Expansins are proteins that have been demonstrated to induce cell wall extension in vitro, and six FaEXP genes have demonstrated that expansions from ripening strawberry fruit are able to catalyze extension (Harrison et al., 2001). Therefore, we speculate that FvTCP9 might play an important role in strawberry fruit development and ripening. However, we must note that FvTCP9 is the only gene that contains an intron in strawberry (Figure 3), and it was quite highly and consistently expressed during strawberry fruit development (Figure 5). Thus, it would be interesting to investigate whether other FvTCP or FvTCP9 homologs in other plants have similar roles in fruit development and ripening.
Conclusion
In this study, 19 FvTCP genes were identified in the diploid woodland strawberry Fragaria vesca and placed in an evolutionary context based on phylogenetic and structural feature analyses. Numerous cis-acting elements were found in the FvTCP promoter sequences, suggesting that FvTCP gene transcripts are controlled by a complex regulatory regime. We characterized FvTCP gene transcripts in different tissues and developmental stages and under various stress conditions, which suggested that FvTCP genes could play important roles in strawberry growth and development. In addition, we examined the subcellular localization of six FvTCP-GFP fusion proteins, which provided additional insights into their functions. Notably, transient over-expression of FvTCP9 in strawberry fruits up-regulated several fruit ripening-related genes, indicating that FvTCP9 might be involved in the regulation of strawberry fruit development and ripening. Taken together, genome-wide analysis of the TCP genes in F. vesca might lay the foundation for further studies unraveling the functions of strawberry TCP genes during strawberry growth and development.
Author Contributions
The experiments were conceived and designed by J-YF. The experiments were performed by WW, YH, M-YC, and Y-TH. WW and J-YF analyzed the data. KG provided the ‘Heilongjiang-3’ tissue culture plantlets. WW and J-YF contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present study was financially supported by grants from the National Natural Science Foundation of China (Grant No. 31201657), the Shaanxi province science and technology research and development program (2014K02-02-02), and the major program of Yangling Agricultural Hi-tech Industry Demo Zone synergy innovation for cooperation in production, study, and research (2016CXY-11). We would like to thank the reviewers for their comments on the manuscript. We thank Dr. Ke Duan of the Shanghai Academy of Agricultural Sciences for generously providing the wild diploid woodland strawberry Fragaria vesca plants.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01937/full#supplementary-material
FIGURE S1 | Linkage group distributions of FvTCP genes. Linkage group numbers are provided at the top of each linkage group. The names on the left side of each linkage group correspond to the approximate location of each FvTCP gene. The scale is in megabases (Mb).
FIGURE S2 | Motif sequences of FvTCP proteins identified using MEME tools.
FIGURE S3 | Transcript accumulation pattern of 19 TCP genes in the diploid woodland strawberry (F. vesca) during different periods of subcultural propagation. (A) Photos of strawberry subcultural propagation during the five different periods assessed (P1: original plantlet; P2: plantlet with 1–2 branch crowns, approximately 2 weeks after subculture; P3: plantlets with 3–4 branch crowns, approximately 3 weeks after subculture; P4: plantlets with 5–7 branch crowns, approximately 4 weeks after subculture; P5: plantlets with over 10 branch crowns, approximately 6 weeks after subculture). Bar = 1 cm. (B) Hierarchical clustering of the transcript accumulation profiles of 19 FvTCP genes during different strawberry subcultural propagation periods (original results shown in Supplementary Figure S4). The transcript accumulation profiles were generated by semi-quantitative PCR and were visualized as heat maps. The color scale represents relative transcript levels with increased (red) or decreased (green) transcript abundance. Genes were hierarchically clustered based on average Pearson’s distance metric and ‘average linkage’ method. Fv18s was used as an internal control. The experiments were repeated three times and provided consistent results.
FIGURE S4 | Transcript accumulation patterns of the 19 FvTCP genes during different fruit developmental stages (A) and strawberry subcultural propagation stages (B) analyzed by semi-quantitative RT-PCR. Fv18s was used as an internal control. Lanes: A: S1: mature flowers with partially withered petals, S2: mature green receptacles, S3: white receptacles with green achenes, S4: half white and half red fruits, S5: fully ripened fruits. B: P1: original plantlet; P2: plantlet with 1–2 branch crowns, approximately 2 weeks after subculture; P3: plantlets with 3–4 branch crowns, approximately 3 weeks after subculture; P4: plantlets with 5–7 branch crowns, approximately 4 weeks after subculture; P5: plantlets with over 10 branch crowns, approximately 6 weeks after subculture.
FIGURE S5 | Transcript accumulation patterns of the 19 FvTCP genes under abiotic (cold, heat, drought, and NaCl) and biotic treatments (powdery mildew infection) analyzed by semi-quantitative RT-PCR. Fv18s was used as an internal control.
FIGURE S6 | Transcript accumulation patterns of the 19 FvTCP genes exposed to hormone treatments (ABA, Eth, MeJA, and SA) and analyzed by semi-quantitative RT-PCR. Fv18s served as an internal control.
Footnotes
- ^http://planttfdb.cbi.pku.edu.cn/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://www.ebi.ac.uk/Tools/pfa/iprscan/
- ^http://pfam.xfam.org/search/
- ^http://www.ncbi.nlm.nih.gov/mapview/
- ^http://planttfdb.cbi.pku.edu.cn/
- ^http://plantgrn.noble.org/psRNATarget/
- ^http://meme-suite.org/tools/meme
- ^http://gsds.cbi.pku.edu.cn/
- ^http://www.ncbi.nlm.nih.gov/
- ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^http://www.physics.csbsju.edu/stats/
- ^http://planttfdb.cbi.pku.edu.cn/
- ^http://www.ebi.ac.uk/interpro/search/sequence-search
- ^http://pfam.xfam.org/search/
References
Aguilar-Martinez, J. A., Poza-Carrion, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Anderson, S. L., Teakle, G. R., Martino-Catt, S. J., and Kay, S. A. (1994). Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 6, 457–470. doi: 10.1046/j.1365-313X.1994.6040457.x
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bobb, A. J., Chern, M. S., and Bustos, M. M. (1997). Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucleic Acids Res. 25, 641–647. doi: 10.1093/nar/25.3.641
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Danisman, S., Van Der Wal, F., Dhondt, S., Waites, R., De Folter, S., Bimbo, A., et al. (2012). Arabidopsis Class I and Class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Darwish, O., Shahan, R., Liu, Z. C., Slovin, J. P., and Alkharouf, N. W. (2015). Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genomics 16:29. doi: 10.1186/s12864-015-1221-1
Finlayson, S. A. (2007). Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 48, 667–677. doi: 10.1093/pcp/pcm044
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Goldsbrough, A. P., Albrecht, H., and Stratford, R. (1993). Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 3, 563–571. doi: 10.1046/j.1365-313X.1993.03040563.x
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Guo, Z. X., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X. P., and Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
Hammani, K., Gobert, A., Hleibieh, K., Choulier, L., Small, I., and Giege, P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23, 730–740. doi: 10.1105/tpc.110.081638
Han, Y., Dang, R. H., Li, J. X., Jiang, J. Z., Zhang, N., Jia, M. R., et al. (2015). SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an Ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 167, 915–930. doi: 10.1104/pp.114.251314
Harrison, E. P., Mcqueen-Mason, S. J., and Manning, K. (2001). Expression of six expansin genes in relation to extension activity in developing strawberry fruit. J. Exp. Bot. 52, 1437–1446. doi: 10.1093/jexbot/52.360.1437
Hoffmann, T., Kalinowski, G., and Schwab, W. (2006). RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J. 48, 818–826. doi: 10.1111/j.1365-313X.2006.02913.x
Hu, Y., Han, Y. T., Wei, W., Li, Y. J., Zhang, K., Gao, Y. R., et al. (2015). Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 6:736. doi: 10.3389/fpls.2015.00736
Kang, C., Darwish, O., Geretz, A., Shahan, R., Alkharouf, N., and Liu, Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25, 1960–1978. doi: 10.1105/tpc.113.111732
Kang, C. Y., and Liu, Z. C. (2015). Global identification and analysis of long non-coding RNAs in diploid strawberry Fragaria vesca during flower and fruit development. BMC Genomics 16:815. doi: 10.1186/s12864-015-2014-2
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kim, J. K., Cao, J., and Wu, R. (1992). Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pI alpha-amylase gene. Mol. Gen. Genet. 232, 383–393. doi: 10.1007/BF00266241
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Koyama, T., Sato, F., and Ohme-Takagi, M. (2010). A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 74, 2145–2147. doi: 10.1271/bbb.100442
Kumar, R., Khurana, A., and Sharma, A. K. (2014). Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 65, 4561–4575. doi: 10.1093/jxb/eru277
Lei, J. J., Yang, G., Dai, H. P., Wu, L. P., and Deng, M. Q. (1997). The wild strawberry gemplasm resources in China. J. Fruit Sci. 14, 198–200.
Ma, J., Wang, Q. L., Sun, R. R., Xie, F. L., Jones, D. C., and Zhang, B. H. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. doi: 10.1038/srep06645
Manassero, N. G., Viola, I. L., Welchen, E., and Gonzalez, D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. Concepts 4, 111–127. doi: 10.1515/bmc-2012-0051
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Medina-Puche, L., Molina-Hidalgo, F. J., Boersma, M., Schuurink, R. C., Lopez-Vidriero, I., Solano, R., et al. (2015). An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol. 168, 598–614. doi: 10.1104/pp.114.252908
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Nath, U., Crawford, B. C. W., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Herve, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Palatnik, J. F., Wollmann, H., Schommer, C., Schwab, R., Boisbouvier, J., Rodriguez, R., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis MicroRNAs miR159 and miR319. Dev. Cell 13, 115–125. doi: 10.1016/j.devcel.2007.04.012
Paniagua, C., Blanco-Portales, R., Barcelo-Munoz, M., Garcia-Gago, J. A., Waldron, K. W., Quesada, M. A., et al. (2016). Antisense down-regulation of the strawberry beta-galactosidase gene FabetaGal4 increases cell wall galactose levels and reduces fruit softening. J. Exp. Bot. 67, 619–631. doi: 10.1093/jxb/erv462
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. doi: 10.1186/1471-2229-14-157
Pillet, J., Yu, H. W., Chambers, A. H., Whitaker, V. M., and Folta, K. M. (2015). Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 66, 4455–4467. doi: 10.1093/jxb/erv205
Postel, D., Vanlemmens, P., Gode, P., Ronco, G., and Villa, P. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Raab, T., Lopez-Raez, J. A., Klein, D., Caballero, J. L., Moyano, E., Schwab, W., et al. (2006). FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18, 1023–1037. doi: 10.1105/tpc.105.039784
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rouster, J., Leah, R., Mundy, J., and Cameron-Mills, V. (1997). Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 11, 513–523. doi: 10.1046/j.1365-313X.1997.11030513.x
Saeed, A. I., Hagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 microarray software suite. Methods Enzymol. 411, 134–193.
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Sessa, G., Morelli, G., and Ruberti, I. (1993). The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12, 3507–3517.
Seymour, G. B., Ostergaard, L., Chapman, N. H., Knapp, S., and Martin, C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Shen, Q., and Ho, T. H. (1995). Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307. doi: 10.1105/tpc.7.3.295
Shi, P. B., Guy, K. M., Wu, W. F., Fang, B. S., Yang, J. H., Zhang, M. F., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. doi: 10.1186/s12870-016-0765-9
Shulaev, V., Sargent, D. J., Crowhurst, R. N., Mockler, T. C., Folkerts, O., Delcher, A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. doi: 10.1038/ng.740
Takeda, T., Suwa, Y., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., Matsuoka, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant Cell Physiol. 44, S146–S146.
Tatematsu, K., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Wang, W., Vignani, R., Scali, M., and Cresti, M. (2006). A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27, 2782–2786. doi: 10.1002/elps.200500722
Washida, H., Wu, C. Y., Suzuki, A., Yamanouchi, U., Akihama, T., Harada, K., et al. (1999). Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol. Biol. 40, 1–12. doi: 10.1023/A:1026459229671
Wei, W., Hu, Y., Han, Y. T., Zhang, K., Zhao, F. L., and Feng, J. Y. (2016). The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 105, 129–144. doi: 10.1016/j.plaphy.2016.04.014
Woolley, L. C., James, D. J., and Manning, K. (2001). Purification and properties of an endo-beta-1,4-glucanase from strawberry and down-regulation of the corresponding gene, cel1. Planta 214, 11–21. doi: 10.1007/s004250100577
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Yao, X., Ma, H., Wang, J., and Zhang, D. B. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897. doi: 10.1111/j.1744-7909.2007.00509.x
Keywords: Strawberry (Fragaria vesca), TCP transcription factors, plant growth and development, transcript accumulation profiles, subcellular localization, transient over-expression
Citation: Wei W, Hu Y, Cui M-Y, Han Y-T, Gao K and Feng J-Y (2016) Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front. Plant Sci. 7:1937. doi: 10.3389/fpls.2016.01937
Received: 05 September 2016; Accepted: 06 December 2016;
Published: 22 December 2016.
Edited by:
Stefan De Folter, CINVESTAV, MexicoReviewed by:
Erin E. Sparks, Duke University, USAEmma Larsson, Swedish University of Agricultural Sciences (SLU), Sweden
Kevin Michael Folta, University of Florida, USA
Copyright © 2016 Wei, Hu, Cui, Han, Gao and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-Yue Feng, ZmVuZ2p5MTkxNTFAbndzdWFmLmVkdS5jbg==
 Wei Wei
Wei Wei Yang Hu
Yang Hu Meng-Yuan Cui1,2
Meng-Yuan Cui1,2 Jia-Yue Feng
Jia-Yue Feng