- Plant Biology Laboratory, Organic and Medicinal Chemistry Division, Council of Scientific and Industrial Research-Indian Institute Chemical Biology, Kolkata, India
Podophylloxin (ptox), primarily obtained from Podophyllum hexandrum, is the precursor for semi-synthetic anticancer drugs viz. etoposide, etopophos, and teniposide. Previous studies established that methyl jasmonate (MeJA) treated cell culture of P. hexandrum accumulate ptox significantly. However, the molecular mechanism of MeJA induced ptox accumulation is yet to be explored. Here, we demonstrate that MeJA induces reactive oxygen species (ROS) production, which stimulates ptox accumulation significantly and up regulates three ROS-responsive ptox biosynthetic genes, namely, PhCAD3, PhCAD4 (cinnamyl alcohol dehydrogenase), and NAC3 by increasing their mRNA stability. Classic uncoupler of oxidative phosphorylation, carbonylcyanide m-chlorophenylhydrazone, as well as H2O2 treatment induced the ROS generation and consequently, enhanced the ptox production. However, when the ROS was inhibited with NADPH oxidase inhibitor diphenylene iodonium and Superoxide dismutase inhibitor diethyldithio-carbamic acid, the ROS inhibiting agent, the ptox production was decreased significantly. We also noted that, MeJA up regulated other ptox biosynthetic pathway genes which are not affected by the MeJA induced ROS. Further, these ROS non-responsive genes were controlled by MeJA through the down regulation of five secondary metabolites biosynthesis specific miRNAs viz. miR172i, miR035, miR1438, miR2275, and miR8291. Finally, this study suggested two possible mechanisms through which MeJA modulates the ptox biosynthesis: primarily by increasing the mRNA stability of ROS-responsive genes and secondly, by the up regulation of ROS non-responsive genes through the down regulation of some ROS non-responsive miRNAs.
Introduction
The mechanism of action of ptox is based on inhibiting the polymerisation of tubulin and arresting of the cell cycle in the metaphase (Ayres and Loike, 1990; Buss and Waigh, 1995). Glycosidic derivatives of ptox, namely, etoposide, etopophos, and teniposide inhibit the cell cycle by inhibiting the DNA topo isomerase II (Fleming et al., 1989; Von Wartburg and Stahelin, 1993). Previous studies have also shown that ptox and its derivatives exhibit biological activity as strong antiviral agents and as antineoplastic drugs and its glycosidic derivatives, etoposide, etopophos (etoposide phosphate), teniposide are thus widely used for the treatment for various types of malignancy (Stahelin and von Wartburg, 1991; Canela et al., 2000). Ptox, is a polyphenolic substance, and formed by the combination of two phenylpropane units. Ptox is one of the most frequently used cytotoxic lignan isolated from podophyllin, which is an ethanolic extract of an endangered medicinal plant, P. hexandrum (Stahelin and von Wartburg, 1991; Gordaliza et al., 2004). This medicinal herb is generally grown at high altitude of the Himalayas and belongs to the Berberidaceae family. In one recent study, the Podophyllum Germination Network (PGN) has been constructed in reference to Arabidopsis thaliana to explore the underlying potential of seed germination mechanisms (Dogra et al., 2015). Previous studies reported that the chemical synthesis of cyclolignan like ptox is yet to be feasible. Thus, various biotechnological techniques like biotransformations, etc. (Kutney, 1999), especially transgenic hairy roots produced by infection of plants with Agrobacterium rhizogenes and cell culture based production of cyclolignan like ptox have been studied extensively (Kadkade, 1981; van Uden et al., 1989; Heyenga et al., 1990; Seidel et al., 2002; Chattopadhyay et al., 2003). It was also reported that MeJA treatment induce the production of lariciresinol, another lignan, in hairy root cultures of Isatis indigotica (Chen et al., 2015).
Previously, the cell suspension cultures of Linum spp. was noted with higher yield of ptox using MeJA and salicylic acid (SA) as elicitors (van Furden et al., 2005; Yousefzadi et al., 2010). MeJA treatment also induced the other secondary metabolie production like paclitaxel in the cell culture of Taxus cuspidate (Lenka et al., 2015). Treatment of the cell culture of L. nodiflorum with coronalon, indanoyl-isoleucine and MeJA induced the accumulation of 6-methoxypodophyllotoxin, a lignan related to ptox (Berim et al., 2005). In our previous studies, we have also reported the 8- to 10-fold higher yield of ptox in MeJA treated old cell culture of P. hexandrum (Bhattacharyya et al., 2012).
Jasmonic acid (JA) and its methyl derivative, MeJA, are well known as plant signaling compound that involves in stress management and development. Wang and Wu (2005) reported that exogenous treatment of JA and MeJA, actually mimic the effects of wounding and these elicitors bring the relative response like production of secondary metabolite as well as ROS. It has also been reported that MeJA induced ROS alter the mitochondrial dynamics that resulted in photosynthetic dysfunction and cell death (Zhang and Xing, 2008). According to Wang and Wu (2005) MeJA induces the ROS, which includes H2O2 and NO, in cell culture of Taxus sp. and the introduction of ROS inhibitor to culture reduce the MeJA induced taxol production. Similar to the changes in taxol from Taxus sp., concentration of other secondary metabolites like phytoalexin were also increased by ROS via cyclopentenone isoprostanes in tomato (Thoma et al., 2003). MeJA induced production of H2O2 also reported in rice leaves. According to Orozco-Cárdenas et al. (2001) H2O2 acts as a second messenger upon treatment with MeJA and induced systemin production to the tomato plant.
Previously, we also did a whole transcriptome analysis of control and MeJA treated cell culture of P. hexandrum and identified ptox specific CAD isoforms namely, PhCAD3 and PhCAD4 (Bhattacharyya et al., 2016, 2013). However, the mechanism through which MeJA regulates ptox biosynthetic pathway genes is yet to be understood. This study revealed the molecular mechanism of MeJA altered ptox accumulation by regulating ptox biosynthetic pathway genes at transcript level. This has shown that MeJA induced ROS alters the mRNA stability of selected pathway genes to regulate their expression at transcript level. In addition to that, it was also noted that MeJA up regulates other ptox biosynthetic pathway genes which were not regulated by ROS. These ROS non-responsive genes may be regulated by the down regulation of particular MeJA responsive miRNAs, which are interfering with ptox pathway genes. Together, this study suggests two possible molecular mechanisms of MeJA induced ptox accumulation in P. hexandrum cell culture.
Materials and Methods
Plant Growth Condition and Treatment
Callus was derived from mature leaves of P. hexandrum in Murasige and Skoog (MS) medium supplemented with 2.68 μM alpha-Naphthaleneacetic acid (NAA) and 8.88 μM 6-Benzylaminopurine (BAP; Chakraborty et al., 2010) Subculture of callus was done after every 2 weeks in the above mentioned medium. Cell suspension culture was initiated according to Bhattacharyya et al. (2012). In brief, 5 g cells of fresh green callus was inoculated in 50 ml culture containing 60 mM total N2 content, 1.25 mM potassium dihydrogen phosphate, 6% glucose and 11.41 μM 3-Indoleacetic acid (IAA). Cultures were incubated at 22°C for 3 days at 110 rpm before treatment. Three days old cell cultures were treated with MeJA (100 μM); classic uncoupler of oxidative phosphorylation CCCP (Izeradjene et al., 2005; 100 μM); ROS inhibitor, DPI (1 μM); ROS inhibitor, DETC (10 μM), (Sigma-Aldrich, USA) and H2O2 (Merck, USA; 20 mM) in different combination. Two hours was noted as an optimum time point for harvesting the cells after treatment.
Isolation of Protoplast for ROS Determination by Confocal and Flow Cytometry
Protoplast was isolated from 200 mg cells from 3 days old cell suspension culture (Zhang and Xing, 2008). Isolated protoplasts were finally washed in 0.35 M mannitol in liquid MS medium (pH 5.8) and treated with 100 μM MeJA or with ROS inducers or ROS inhibitors as designed and sampled after 2 h for confocal and FACS study.
Determination of ROS by Confocal Microscopy and FACS
For confocal microscopy treated protoplast was incubated with 6-carboxy-2,7-dichlorodihydrofluorescein diacetate (DCFDA; Sigma-Aldrich; 5 mM) in the dark for 5 min (Zhang and Xing, 2008). The intracellular ROS production was visualized under the Andor Spinning Disk Confocal Microscope. For DCFDA, solid state laser excitation was 488 nm and emission was at 525 nm. For ROS generation analysis by FACS, protoplasts were incubated with 40 μM DCFDA for 5 min in the dark after the treatment. DCFDA is deacetylated by intracellular esterase, which is further oxidized by ROS to the fluorescent compound 2,7-dichlorofluorescein (DCF). DCF fluorescence was detected by FACS (Becton–Dickinson), using Cell Quest software. Ten thousand events were recorded for each sample.
Extraction of Ptox by HPLC Analysis
The ptox extraction from various samples was performed as described previously (Bhattacharyya et al., 2012). In brief, 100 mg of harvested cells was homogenized with liquid nitrogen and then extracted with ethyl acetate for over night. For HPLC analysis, the supernatant was decanted after ethyl acetate extraction, evaporated to dryness and dissolved in methanol for further use (Kartal et al., 2004). Column and instrumentation for analysis were performed as standardized (Bhattacharyya et al., 2012).
Extraction of RNA and Quantitative RT-PCR (RT-qPCR) Analysis
Total RNA was extracted from the harvested cells from control and treated culture with Trizol (Invitrogen) according to a standardized protocol (Bhattacharyya et al., 2016) in three replicates. cDNA was synthesized using RevertAid H Minus First Strand cDNA Synthesis kit (Fermentas, USA). The qRT-PCR was performed using the Light Cycler 96 System (Roche Applied Science, USA) with FastStart Essential DNA Green Master (Roche Applied Science). The constitutively expressed actin gene was used as the reference gene. Gene specific primers for performing RT-qPCR are given in Supplemental Table 1. Total 19 ptox biosynthetic pathway genes were selected, including five phenylpropanoid pathway related transcription factors and grouped them in four groups as mentioned below. Group A: four CAD isoforms namely PhCAD1, PhCAD2, PhCAD3, PhCAD4; Group B: five genes upstream of CAD; Group C: five genes downstream of CAD and Group D: five transcription factors related to phenylpropanoid pathway (All genes were submitted under the accession numbers SRX180871 and SRX180389). Here, CAD isoforms were kept in the middle position because CAD is the most important rate limiting gene, which actually controls the trafficking of coniferyl alcohol, a precursor of ptox and lignin biosynthesis, toward ptox and lignin.
mRNA Stability Assay
mRNA stability assay was performed according to Datta et al. (2015). In brief, the control and treated cell cultures were further treated with 200 mM actinomycin D (Sigma-Aldrich) and cells were harvested after 2, 4, 6, 8, 10, and 12 h. Total RNA was isolated for further RT-qPCR by using gene specific primers for PhCAD3, PhCAD4, and NAC3. Cell culture treated with water was taken as mock treated. Prior to the treatment with water, cultures were pre-incubated in actinomycin D for 30 min to allow proper distribution of the antibiotic.
RT-qPCR of miRNA
Stem loop RT-qPCR of miRNA was done according to Varkonyi-Gasic et al. (2007) with minor modifications. In brief, the total miRNA was isolated from the harvested cells of the treated and control cell culture of P. hexandrum using the mirPremier microRNA isolation kit (Sigma-Aldrich) according to the manufacturer’s instructions. Hundred nano-gram miRNA was used for cDNA preparation using stem loop RT primers according to Biswas et al. (2016). The RT-qPCR was performed using Light Cycler 96 System (Roche Applied Science) with FastStart Essential DNA Green Master (Roche Applied Science). The constitutively expressed ubiquitin-6 gene was used as the reference gene. miRNA specific primers for performing RT-qPCR are listed in Supplemental Table 2.
Result
Detection of ROS and Measurement Ptox Content From Treated P. hexandrum Cell Culture
The detection of ROS is concurrent to the enhanced ptox accumulation in 2 h MeJA treated 3 days old P. hexandrum cell culture (Figures 1A–C). Treatment with CCCP and H2O2 also induced the ROS in the cell culture after 2 h of treatment. Amount of ROS produced after H2O2 treatment was maximum (Figures 1A,B). The independent treatments of both CCCP and H2O2 increased the ptox content in the cell culture, where the maximum amount of ptox was accumulated after CCCP treatment for 2 h (Figure 1C). Interestingly, when ROS production was inhibited by the combination of DPI and DETC treatment in MeJA and CCCP treated cultures, decreased ptox content was noted as well (Figures 1A–C).
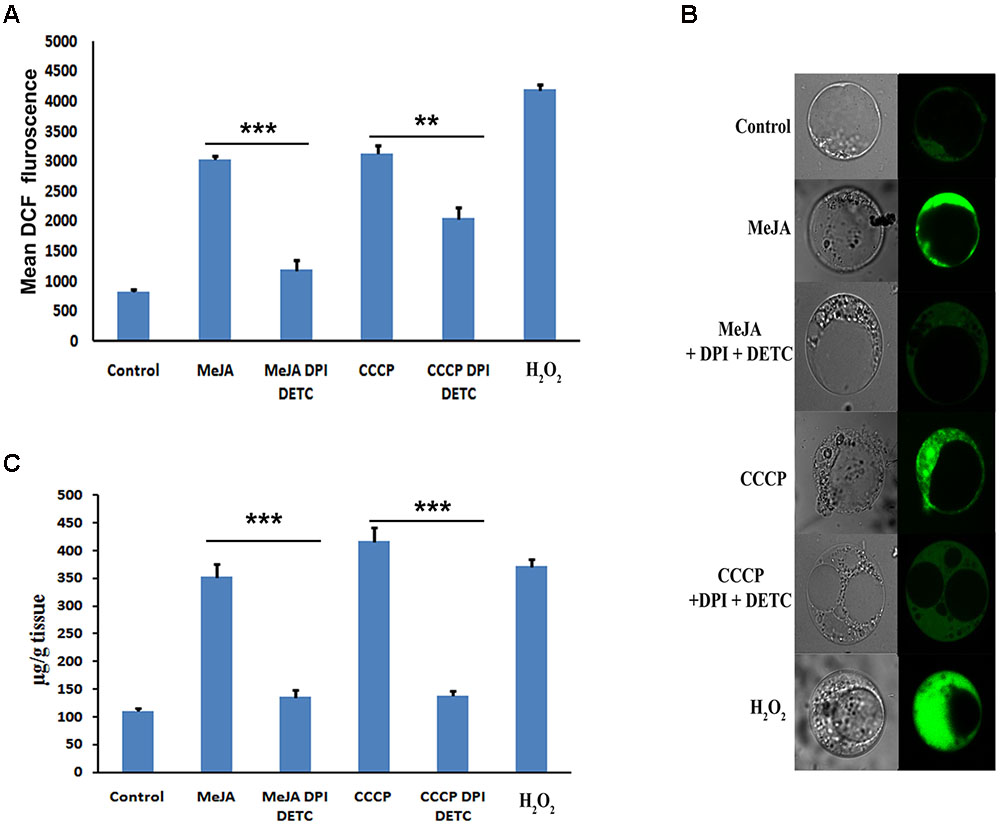
FIGURE 1. (A) Content of ptox after various treatments to the cell culture. (B) Measurement of ROS after various treatments to the cell culture by FACS. (C) Detection of ROS by confocal microscopy after staining with DCFDA of various treated cultures. The statistical analysis of the difference between fold changes was done using GraphPad Instat-3 software. Comparison between groups was done using one-way analysis of variance (ANOVA) followed by Student–Newman–Keuls test. Data were fitted using Sigma plot represented as means ± SD. p < 0.05 was accepted as level of significance; ∗∗∗ highly significant p < 0.001; ∗∗ significant p < 0.01; ∗ less significant p < 0.05; NS not significant for p > 0.05.
Transcript Assay of Selected Ptox Biosynthetic Pathway Genes After Independent MeJa and the Combination of DPI, DETC, and MeJA Treatment
To understand how MeJA induced ROS generation affected the ptox biosynthesis consequently increasing the ptox content, the expression pattern of 19 ptox biosynthetic pathway related genes was checked, in two sets of experiments viz. the first set of treatment was with only MeJA and the second set was a combination of MeJA, DPI, and DETC. Independent MeJA treatment up regulates all 19 selected ptox biosynthetic pathway genes, including five transcription factors related to ptox biosynthesis. Amongst these, PhCAD3 was significantly up regulated, followed by PhCAD4, C4H, SDR, SDG, DPO, NAC3, and so on. Other transcripts like PLR, AdMM, and mTERF1 were comparatively less up regulated after the treatment. Interestingly, when ROS was inhibited in the second set, viz. the combination treatment of MeJA, DPI and DETC, up regulation of only three genes, namely PhCAD3, PhCAD4, and NAC3 were decreased significantly (Figure 2A). Meanwhile, other 16 genes remained unaltered and exhibited almost same expression pattern like independent MeJA treatment (Figure 2A).
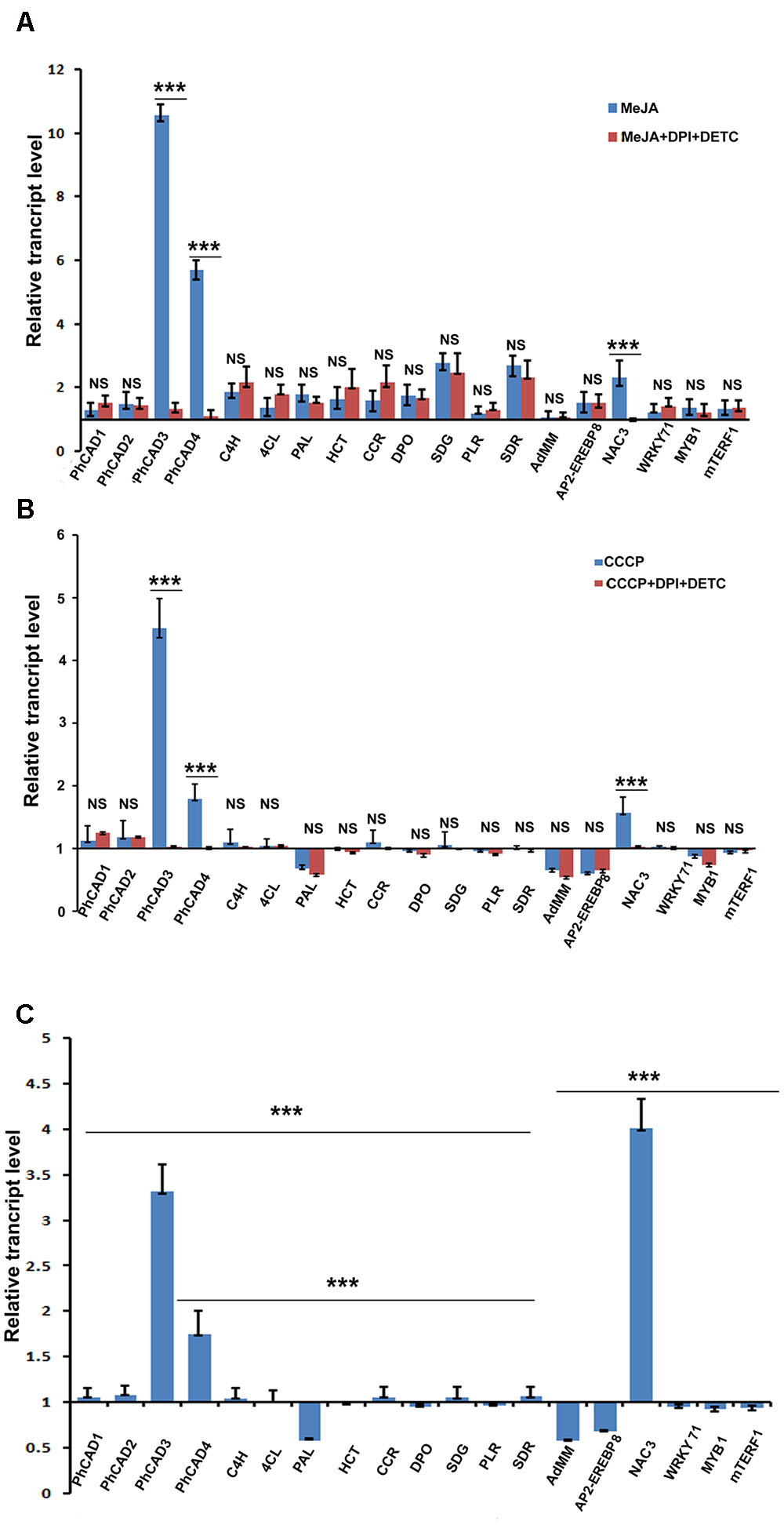
FIGURE 2. Transcript assay of 19 ptox biosynthetic pathway genes after various treatments. (A) Expression pattern after MeJA and MeJA+DPI+DETC ttreatment. (B) Expression pattern after CCCP and CCCP+DPI+DETC treatment. (C) Expression pattern after H2O2 treatment. The statistical analysis of the difference between fold changes was done using GraphPad Instat-3 software. Comparison between groups was done using one-way analysis of variance (ANOVA) followed by Student–Newman–Keuls test. Data were fitted using Sigma plot represented as means ± SD. p < 0.05 was accepted as level of significance; ∗∗∗ highly significant p < 0.001; ∗∗ significant p < 0.01; ∗ less significant p < 0.05; NS not significant for p > 0.05.
Transcript Assay of Selected Ptox Biosynthetic Pathway Genes After Independent CCCP, H2O2, and the Combined DPI, DETC, and CCCP Treatment
To further confirm the ROS-responsiveness of PhCAD3, PhCAD4, and NAC3, P. hexandrum culture was treated independently with CCCP and H2O2 as well as with a combination treatment of CCCP, DPI, and DETC. Only three genes, namely, PhCAD3, PhCAD4, and NAC3 were significantly up regulated after CCCP and H2O2 treatments. All other 16 genes were remained unaffected, although sometimes insignificant down regulation was noted. However, the genes like AdMM, PAL, and AP2-EREBP8 were down regulated insignificantly. While, the ROS in CCCP treated culture was inhibited with the combination treatment of DPI and DETC, only PhCAD3, PhCAD4, and NAC3 were affected and their up regulation was decreased significantly (Figure 2B). However, for other 16 genes the results were similar to those obtained in the case of CCCP treatment alone (Figure 2B). Again, the H2O2 treatment of the cell culture revealed significant up regulation of only PhCAD3, PhCAD4, and NAC3 genes, however, AP2-EREBP8, AdMM, and PAL were down regulated after the treatment (Figure 2C). All other selected genes remain unaffected after the treatment.
mRNA Stability Assay of PhCAD3, PhCAD4, and NAC3
mRNA stability level of 3 ROS-responsive genes, PhCAD3, PhCAD4, and NAC3 was checked after independent treatment with MeJA, CCCP, H2O2 and the combination treatment of MeJA+DPI and DETC and CCCP +DPI and DETC. Three days old cell culture was mock treated with water and considered as a control. Cell treated with actinomycin D alone revealed that remaining mRNA significantly reduced after 4 h of actinomycin D treatment for PhCAD3 whereas for PhCAD4 and NAC3 it significantly reduced after 2 h of treatment. For all three genes after 12 h of treatment there was almost no remaining mRNA (Figure 3). Hence, we check the mRNA stability for all three genes up to 12 h after the respective treatments. MeJA treatment resulted in the higher transcript level of PhCAD3, PhCAD4, and NAC3 after actinomycin D treatment which indicated that MeJA increased the mRNA stability of PhCAD3, PhCAD4, and NAC3 up to 1.61-, 1.9-, and 2.46-fold, respectively, after 2 h. In two separate, independent treatment with CCCP and H2O2 induced ROS, which in turn increased the mRNA stability of PhCAD3, PhCAD4, and NAC3. But when ROS was inhibited with DPI and DETC in MeJA and CCCP treated cultures, then mRNA stability of PhCAD3, PhCAD4, and NAC3 were decreased significantly.
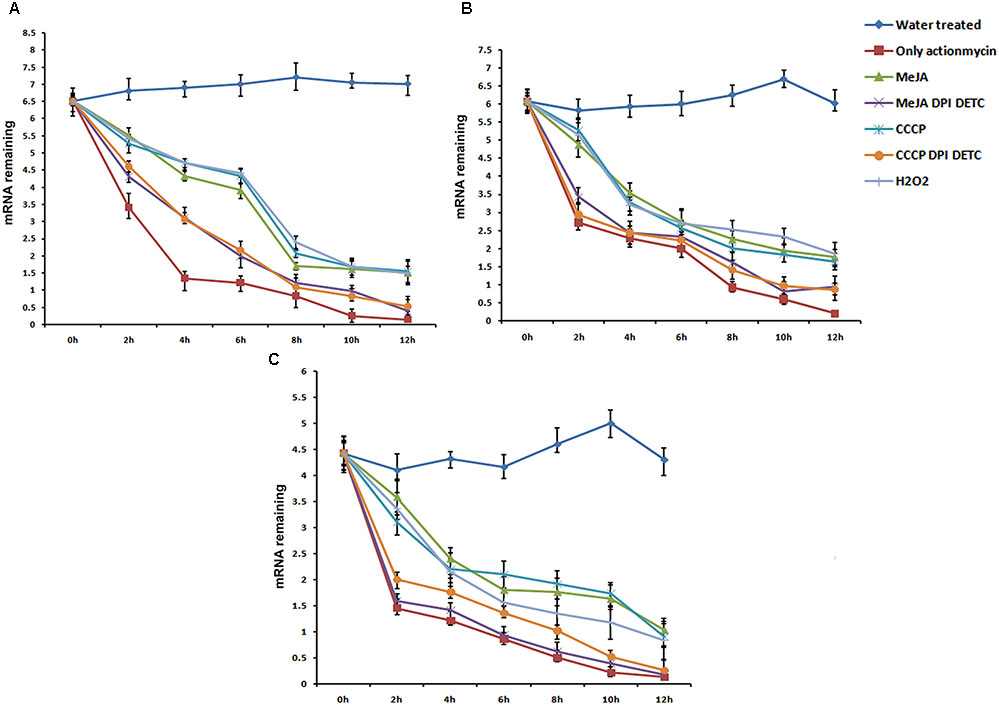
FIGURE 3. mRNA Stability assay of three ROS responsive genes viz, PhCAD3, PhCAD4, and NAC3 after various treatments. (A) mRNA stability of PhCAD3. (B) mRNA stability of PhCAD4. (C) mRNA stability of NAC3. Data are the mean ± SD for three individual experiments.
miRNA Transcript Assay after Various Treatments
We were also interested to explore the mechanism of MeJA mediated up regulation of upstream and downstream genes of PhCADs which are not regulated by MeJA produced ROS. It was well established that miRNAs play a vital role as a trans-acting regulator of gene expression (Carlborg and Haley, 2004). Here, we selected eight miRNAs from our previously published whole transcriptome data (Bhattacharyya et al., 2013) after KEGG pathway analysis. These eight selected miRNAs were specifically targeted to the secondary metabolite pathway specially phenylpropanoid pathway. Only five miRNAs namely miR172i, miR035, miR1438, miR2275, miR8291 were significantly down regulated after MeJA treatment (Figure 4A). But all these eight miRNAs were completely non-responsive to CCCP and H2O2 induced ROS (Figures 4B,C). Furthermore, these miRNAs also noted with any effect when ROS was inhibited by combining DPI and DETC in the MeJA and CCCP treated cultures (Figures 4A,B). These five miRNAs are interfering with the target gene expression, namely, four coumarate ligage (4CL), Cytochrome P450, Caffeoyl–CoA methyltransferase, Cytochrome P450, and Chalclone synthase and the decline in the level of these miRNAs may bring about an up-regulation of the target genes.
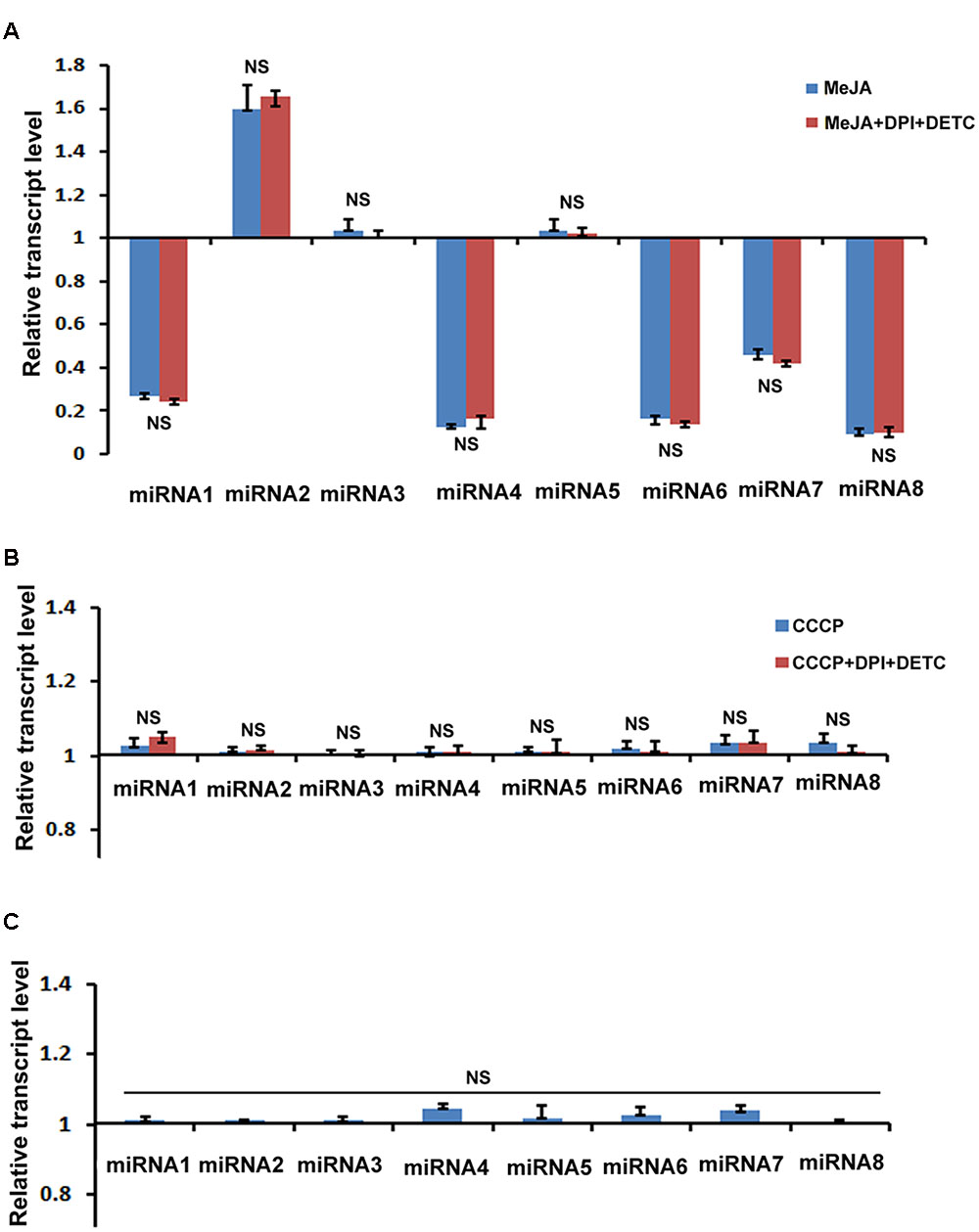
FIGURE 4. Effect of MeJA and other ROS generating compound’s treatment on the expression of eight selected miRNAs. (A) RT-qPCR analysis of MeJA and MeJA+DPI+DETC treatment. (B) RT-qPCR analysis after CCCP and CCCP+DPI+DETC treatment. (C) RT-qPCR analysis after H2O2 treatment. The statistical analysis of the difference between fold changes was done using GraphPad Instat-3 software. Comparison between groups was done using one-way analysis of variance (ANOVA) followed by Student–Newman–Keuls test. Data were fitted using Sigma plot represented as means ± SD. p < 0.05 was accepted as a level of significance; ∗∗∗ highly significant p < 0.001; ∗∗ significant p < 0.01; ∗ less significant p < 0.05; NS not significant for p > 0.05. miRNA1, miR172i; miRNA2, miR1873; miRNA3, miR5538; miRNA4, miR035; miRNA5, miR5532; miRNA6, miR1438; miRNA7, miR2275; miRNA8, miR8291.
Discussion
It is well established that MeJA is a signal molecule in plant defense responses and an effective inducer of secondary metabolite accumulation in plant cell cultures by means of ROS as a secondary signaling molecule. According to You and Chan (2015) during stress condition ROS plays a vital role in the regulation of complex signaling network of plant stress response. It was also reported by Wang and Wu, 2005 that MeJA also induced the production of H2O2, intracellular malondialdehyde content and lipoxygenase. ROS also regulates the gene expression, translation, metabolism, and turnover (Singh et al., 2016). Exogenous application of MeJA also alters the stomatal closure by ROS where ROS inhibitor DPI prohibited the stomatal closure (Suhita et al., 2004). In this study, we explore the mechanism through which MeJA affects the increased accumulation of ptox. Previously, we have reported that MeJA significantly induces accumulation of ptox by up regulating the genes in the ptox biosynthetic pathway (Bhattacharyya et al., 2013, 2016). However, the molecular mechanism of MeJA-induced expression of ptox biosynthetic pathway genes is still to be elucidated. In this study ROS has been detected by the FACS and confocal microscopy in MeJA treated P. hexandrum cell culture which was simultaneous to the enhanced ptox accumulation (Figures 1A–C). Results also indicate the enhanced ptox accumulation in CCCP and H2O2 treated P. hexandrum cell cultures which may be attributed to the enhanced ROS production (Figures 1A–C). To further confirm the involvement of ROS in ptox accumulation, in both MeJA and CCCP treated cultures, ROS was inhibited by DPI and DETC and that led to the lower degree of ptox accumulation. This observation confirmed the involvement of MeJA induced ROS in increased accumulation of ptox.
It was reported earlier that ROS plays a significant role as a signaling molecule in plant development, abiotic and biotic stress management, photorespiration and photosynthesis, etc. (Noctor and Foyer, 1998; Apel and Hirt, 2004; Mittler et al., 2004).
To understand how MeJA induced ROS affect the ptox biosynthetic pathway genes consequently up accumulating ptox biosynthesis, the expression pattern of 19 ptox biosynthetic pathway genes was checked at transcript level after the MeJA treatment. Results revealed that all these 19 genes were up regulated after the MeJA treatment. Interestingly, when MeJA induced ROS was inhibited by DPI and DETC treatment, consequently, the up regulation of three genes, namely PhCAD3, PhCAD4, and NAC3 was decreased significantly, whereas other 13 genes were noted with the almost same rate of up regulation as obtained after MeJA treatment without ROS scavenging (Figure 2A). Hence, it can be assumed that MeJA induced ROS affect the regulation of PhCAD3, PhCAD4, and NAC3. To confirm the ROS-responsiveness of these three genes, we further treated the culture with CCCP, which induced the ROS generation (Chaudhari et al., 2007; Ji et al., 2016) as well as with H2O2. Results indicated that only three genes, namely, PhCAD3, PhCAD4, and NAC3, were significantly up regulated after the treatment amongst all 19 genes (Figures 2B,C). Furthermore, when ROS generated in CCCP treated culture was inhibited by DPI and DETC treatment, the up regulation of these three genes were deceased significantly. Together, it can be deduced that MeJA induced ROS is the responsible factor for up regulation of these three ROS-responsive genes namely PhCAD3, PhCAD4, and NAC3. In our previous study, we have demonstrated that these two ROS-sensitive isoforms of PhCAD, viz. PhCAD3 and PhCAD4, were specifically responsible for ptox biosynthesis (Bhattacharyya et al., 2016).
We further investigated the underlying mechanism through which ROS-responsive genes of the ptox biosynthetic pathway were regulated by ROS. It is well known that the regulation of mRNA decay is a major checkpoint for controlling the expression level of different genes (Guhaniyogi and Brewer, 2001; Shim and Karin, 2002). According to Chung et al. (2008) the mRNA stability of a particular gene can be altered by ROS. They showed that H2O2 treatment increases the mRNA stability of the SOS1 gene in A. thaliana. In our study, the mRNA stability of PhCAD3, PhCAD4, and NAC3 were increased after independent MeJA, CCCP, and H2O2 treatment. Further, when ROS generated in MeJA and CCCP treated culture, was inhibited by DPI and DETC, the mRNA stability of these three genes was decreased (Figure 3). That observation indicates that MeJA induced ROS up regulates PhCAD3, PhCAD4, and NAC3 by increasing their mRNA stability.
We also proposed the possible mechanism of MeJA mediated up regulation of upstream and downstream genes of PhCADs which are not regulated by MeJA produced ROS. According to Carlborg and Haley, (2004) the differential expression of genes can result from cis- and/or trans-regulatory changes, leading to epistatic effects. miRNA plays a vital role as a trans-acting regulator of gene expression. It was previously reported that miR163 plays important role in plant secondary metabolism (Ng et al., 2011). Robert et al. (2011) also reported that miR163 modulates the secondary metabolite pathway from camalexin toward glucosinolates. It was suggested earlier that biosynthesis of benzylisoquinoline alkaloids were might be regulated by pso-miR13, pso-miR2161, and pso-miR408 in Papaver somniferum (Boke et al., 2015).
In our study, among eight selected miRNAs, which are targeted to the phenylpropanoid pathway genes, only five miRNAs namely miR172i, miR035, miR1438, miR2275, miR8291 were significantly down regulated after MeJA treatment. Heovere, H2O2 as well as CCCP induced ROS, did not show any effect on these eight miRNAs (Figure 4). Hence, these can be concluded that these five down regulated miRNAs were not regulated by the MeJA induced ROS. These five miRNAs are targeted to four coumarate ligage [4CL], Cytochrome P450, Caffeoyl–CoA methyltransferase, Cytochrome P450, and C. synthase as identified by KEGG analysis in our previous report (Bhattacharyya et al., 2016; Biswas et al., 2016). So the down regulation of these ROS non-responsive miRNA in trun bring about the up regulation ptox biosynthetic pathway genes. As we noted here, almost all genes located upstream and downstream of PhCADs, were ROS non-responsive whereas, PhCADs are ROS-responsive. Again, five above mentioned ROS non-responsive miRNAs were also targeted to the genes up- and downstream of PhCADs. These five miRNAs interfere with the target genes expression and the decline in the level of these microRNAs may bring about an up-regulation of the ptox biosynthetic pathway genes. Hence it can be concluded that may be miRNA is just one of the another way to regulate the other sixteen ROS non-responsive genes of the ptox biosynthetic pathway.
Conclusion
Taken together, it can be concluded that MeJA can up regulate the ptox biosynthetic genes by two possible ways, one through increasing the mRNA stability of PhCAD3, PhCAD4, and NAC3 by MeJA induced ROS and another through the down regulating some miRNAs which are targeted to other important genes of the ptox biosynthetic pathway.
Author Contributions
SH carried out the experimental work, analyzed the data, and draft the manuscript. DB helps to analyze additional data and performed initial experiments. SC conceived and designed the experiments and prepared the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. P. S. Ahuja, Ex-Director, CSIR-IHBT, Palampur, and Ex-Director-General, CSIR, New Delhi, for the generous gift of fresh P. hexandrum. The authors would like to express their gratitude to the Director, CSIR-IICB, Kolkata. This work received financial support from the Council of Scientific and Industrial Research (CSIR), SH acknowledges UGC for fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00164/full#supplementary-material
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Ayres, D. C., and Loike, J. D. (1990). Lignans. Chemical, Biological and Clinical Properties. Cambridge: Cambridge University Press.
Berim, A., Spring, O., Conrad, J., Maitrejean, M., Boland, W., and Petersen, M. (2005). Enhancement of lignan biosynthesis in suspension cultures of Linum nodiflorum by coronalon, indanoyl-isoleucine and methyl jasmonate. Planta 222, 769–776. doi: 10.1007/s00425-005-0019-9
Bhattacharyya, D., Hazra, S., Banerjee, A., Datta, R., Kumar, D., Chakrabarti, S., et al. (2016). Transcriptome-wide identification and characterization of CAD isoforms specific for podophyllotoxin biosynthesis from Podophyllum hexandrum. Plant Mol. Biol. 92, 1–23. doi: 10.1007/s11103-016-0492-5
Bhattacharyya, D., Sinha, R., Ghanta, S., Chakraborty, A., Hazra, S., and Chattopadhyay, S. (2012). Proteins differentially expressed in elicited cell suspension culture of Podophyllum hexandrum with enhanced podophyllotoxin content. Proteome Sci. 10:34. doi: 10.1186/1477-5956-10-34
Bhattacharyya, D., Sinha, R., Hazra, S., Datta, R., and Chattopadhyay, S. (2013). De novo transcriptome analysis using 454 pyrosequencing of the Himalayan Mayapple, Podophyllum hexandrum. BMC Genomics 14:748. doi: 10.1186/1471-2164-14-748
Biswas, S., Hazra, S., and Chattopadhyay, S. (2016). Identification of conserved miRNAs and their putative target genes in Podophyllum hexandrum (Himalayan Mayapple). Plant Gene 6, 82–89. doi: 10.1016/j.plgene.2016.04.002
Boke, H., Ozhuner, E., Turktas, M., Parmaksiz, I., Ozcan, S., and Unver, T. (2015). Regulation of the alkaloid biosynthesis by miRNA in Opium poppy. Plant Biotechnol. J. 13, 409–420. doi: 10.1111/pbi.12346
Buss, A. D., and Waigh, R. D. (1995). “Natural products as leads for new pharmaceuticals,” in Burger’s Medicinal Chemistry and Drug Discovery Principles and Practice, ed. M. E. Wolff (New York, NY: Wiley), 983–1033.
Canela, C., Moraesb, R. M., Dayana, F. E., and Ferreira, D. (2000). Podophyllotoxin. Phytochemistry. 54, 115–120. doi: 10.1016/S0031-9422(00)00094-7
Carlborg, O., and Haley, C. S. (2004). Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 5, 618–625. doi: 10.1038/nrg1407
Chakraborty, A., Bhattacharya, D., Ghanta, S., and Chattopadhyay, S. (2010). An efficient protocol for in vitro regeneration of Podophyllum hexandrum, a critically endangered medicinal plant. Indian J. Biotechnol. 9, 217–220.
Chattopadhyay, S., Mehra, R. S., Srivastava, A. K., Bhojwani, S. S., and Bisaria, V. S. (2003). Effect of major nutrients on podophyllotoxin production in Podophyllum hexandrum suspension cultures. Appl. Microbiol. Biotechnol. 60, 541–546. doi: 10.1007/s00253-002-1149-7
Chaudhari, A. A., Seol, J. W., Kim, S. J., Lee, Y. J., Kang, H. S., Kim, I. S., et al. (2007). Reactive oxygen species regulate Bax translocation and mitochondrial transmembrane potential, a possible mechanism for enhanced TRAIL-induced apoptosis by CCCP. Oncol. Rep. 18, 71–76.
Chen, R., Li, Q., Tan, H., Chen, J., Xiao, Y., Ma, R., et al. (2015). Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front. Plant Sci. 6:952. doi: 10.3389/fpls.2015.00952
Chung, J. S., Zhu, J. K., Bressan, R. A., Hasegawa, P. M., and Shi, H. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53, 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Datta, R., Kumar, D., Sultana, A., Hazra, S., Bhattacharyya, D., and Chattopadhyay, S. (2015). Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via wrky33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169, 2963–2981. doi: 10.1104/pp.15.01543
Dogra, V., Bagler, G., and Sreenivasulu, Y. (2015). Re-analysis of protein data reveals the germination pathway and up accumulation mechanism of cell wall hydrolases during the radicle protrusion step of seed germination in Podophyllum hexandrum- a high altitude plant. Front. Plant Sci. 6:874. doi: 10.3389/fpls.2015.00874
Fleming, R. A., Miller, A. A., and Steward, C. F. (1989). Etoposide: an update. Clin. Pharm. 8, 274–293.
Gordaliza, M., Garcia, P. A., del Corral, J. M. M., Castro, M. A., and Gomez-Zurit, M. A. (2004). Podophyllotoxin: distribution, sources, applications and new cytotoxic derivatives. Toxicon 44, 441–459. doi: 10.1016/j.toxicon.2004.05.008
Guhaniyogi, J., and Brewer, G. (2001). Regulation of mRNA stability in mammalian cells. Gene 265, 11–23. doi: 10.1016/S0378-1119(01)00350-X
Heyenga, A. G., Lucas, J. A., and Dewick, P. M. (1990). Production of tumour-inhibitory lignans in callus cultures of Podophyllum hexandrum. Plant Cell Rep. 9, 382–385. doi: 10.1007/BF00232404
Izeradjene, K., Douglas, L., Tillman, D. M., Delaney, A. B., and Houghton, J. A. (2005). Reactive oxygen species regulate caspase activation in tumor necrosis factor–related apoptosis-inducing ligand–resistant human colon carcinoma cell lines. Cancer Res. 65, 7436–7445. doi: 10.1158/0008-5472.CAN-04-2628
Ji, W., Wei, S., Hao, P., Xing, J., Yuan, Q., Wang, J., et al. (2016). aldehyde dehydrogenase 2 has cardioprotective effects on myocardial ischaemia/reperfusion injury via suppressing mitophagy. Front. Pharmacol. 7:101. doi: 10.3389/fphar.2016.00101
Kadkade, P. G. (1981). Formation of podophyllotoxins by Podophyllum peltatum tissue cultures. Naturwissenschaften 68, 481–482. doi: 10.1007/BF01047526
Kartal, M., Kartal, B., Indrayanto, G., and Alfermann, A. W. (2004). Comparison of different extraction methods for the determination of podophyllotoxin and 6-methoxypodophyllotoxin in Linum species. J Pharm. Biomed. Anal. 35, 441–447. doi: 10.1016/j.jpba.2004.01.016
Kutney, J. P. (1999). Biotechnology and synthetic chemistry—routes to clinically important compounds. Pure Appl. Chem. 71, 1025–1032. doi: 10.1351/pac199971061025
Lenka, S. K., Nims, N. E., Vongpaseuth, K., Boshar, R. A., Roberts, S. C., and Walker, E. (2015). Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 6:115. doi: 10.3389/fpls.2015.00115
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Ng, D. W., Zhang, C., Miller, M., Palmer, G., Whiteley, M., Tholl, D., et al. (2011). Cis- and trans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23, 1729–1740. doi: 10.1105/tpc.111.083915
Noctor, G., and Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Orozco-Cárdenas, M. L., Narváez, V. J., and Ryan, C. A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. doi: 10.2307/3871162
Robert, S. A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., et al. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67, 218–231. doi: 10.1111/j.1365-313X.2011.04591.x
Seidel, V., Windhövel, J. G., Eaton, A. W., Alfermann, R. R., Arroo, M., Medarde, M., et al. (2002). Biosynthesis of podophyllotoxin in Linum album cell cultures. Planta 215, 1031–1039. doi: 10.1007/s00425-002-0834-1
Shim, J., and Karin, M. (2002). The control of mRNA stability in response to extracellular stimuli. Mol. Cells 14, 323–331.
Singh, R., Singh, S., Parihar, P., Mishra, R. K., Tripathi, D. K., Singh, V. P., et al. (2016). Reactive Oxygen Species (ROS): beneficial companions of plants’ developmental processes. Front. Plant Sci. 7:1299. doi: 10.3389/fpls.2016.01299
Stahelin, H. F., and von Wartburg, A. (1991). The chemical and biological route from podophyllotoxin glucoside to etoposide. Cancer Res. 5l, 5–15.
Suhita, D., Raghavendra, A. S., Kwak, J. M., and Vavasseur, A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate and abscisic acid-induced stomatal closure. Plant Physiol. 134, 1536–1545. doi: 10.1104/pp.103.032250
Thoma, I., Loeffler, C., Sinha, A. K., Gupta, M., Krischke, M., Steffan, B., et al. (2003). Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 34, 363–375. doi: 10.1046/j.1365-313X.2003.01730.x
van Furden, B., Humburg, A., and Fuss, E. (2005). Influence of methyl jasmonate on podophyllotoxin and 6-methoxypodophyllotoxin accumulation in Linum album cell suspension cultures. Plant Cell Rep. 24, 312–317. doi: 10.1007/s00299-005-0954-8
van Uden, W., Pras, N., Visser, J. F., and Malingré, T. M. (1989). Detection and identification of podophyllotoxin produced by cell cultures derived from Podophyllum hexandrum royle. Plant Cell Rep. 8, 165–168. doi: 10.1007/BF00716832
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., and Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi: 10.1186/1746-4811-3-12
Von Wartburg, A., and Stahelin, H. F. (1993). “Etoposide,” in Chronicles of Drug Discovery, ed. D. Lednicer (Washington, DC: American Chemical Society), 349–380.
Wang, J. W., and Wu, J. Y. (2005). Nitric Oxide is involved in methyl jasmonate-induced defense responses and secondary metabolism activities of Taxus cells. Plant Cell Physiol. 46, 923–930. doi: 10.1093/pcp/pci098
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Yousefzadi, M., Sharifi, M., Behmanesh, M., Ghasempour, A., Moyano, E., and Palazon, J. (2010). Salicylic acid improves podophyllotoxin production in cell cultures of Linum album by increasing the expression of genes related with its biosynthesis. Biotechnol. Lett. 32, 1739–1743. doi: 10.1007/s10529-010-0343-4
Keywords: mRNA decay, MeJA, miRNA, Podophyllum hexandrum, ptox, ROS
Citation: Hazra S, Bhattacharyya D and Chattopadhyay S (2017) Methyl Jasmonate Regulates Podophyllotoxin Accumulation in Podophyllum hexandrum by Altering the ROS-Responsive Podophyllotoxin Pathway Gene Expression Additionally through the Down Regulation of Few Interfering miRNAs. Front. Plant Sci. 8:164. doi: 10.3389/fpls.2017.00164
Received: 30 November 2016; Accepted: 26 January 2017;
Published: 14 February 2017.
Edited by:
Fumiya Kurosaki, University of Toyama, JapanReviewed by:
Guzel Kudoyarova, Institute of Biology (RAS), RussiaChunhua Fu, Huazhong University of Science and Technology, China
Copyright © 2017 Hazra, Bhattacharyya and Chattopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharmila Chattopadhyay, c2hhcm1pbGFAaWljYi5yZXMuaW4=; Y2hhdHRvcGFkaHlheTYyQGdtYWlsLmNvbQ==
†Present address: Dipto Bhattacharyya, Division of Biotechnology, Chonbuk National University, Iksan-si, South Korea
 Saptarshi Hazra
Saptarshi Hazra Sharmila Chattopadhyay
Sharmila Chattopadhyay