- 1School of Life Sciences, Peking University, Beijing, China
- 2RDFZ XiShan School, Beijing, China
- 3Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
- 4Department of Medicine, University of Alberta, Edmonton, AB, Canada
- 5BGI-Shenzhen, Beishan Industrial Zone, Shenzhen, China
- 6University Herbarium and Department of Integrative Biology, University of California, Berkeley, CA, USA
Seeds are one of the most significant innovations in the land plant lineage, critical to the diversification and adaptation of plants to terrestrial environments. From perspective of seed evo-devo, the most crucial developmental stage in this innovation is seed maturation, which includes accumulation of storage reserves, acquisition of desiccation tolerance, and induction of dormancy. Based on previous studies of seed development in the model plant Arabidopsis thaliana, seed maturation is mainly controlled by the LAFL regulatory network, which includes LEAFY COTYLEDON1 (LEC1) and LEC1-LIKE (L1L) of the NF-YB gene family, and ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 (LEAFY COTYLEDON2) of the B3-AFL gene family. In the present study, molecular evolution of these LAFL genes was analyzed, using representative species from across the major plant lineages. Additionally, to elucidate the molecular mechanisms of the seed maturation program, co-expression pattern analyses of LAFL genes were conducted across vascular plants. The results show that the origin of AFL gene family dates back to a common ancestor of bryophytes and vascular plants, while LEC1-type genes are only found in vascular plants. LAFL genes of vascular plants likely specify their co-expression in two different developmental phrases, spore and seed maturation, respectively, and expression patterns vary slightly across the major vascular plants lineages. All the information presented in this study will provide insights into the origin and diversification of seed plants.
Introduction
Seeds, as propagules and dispersal units, play very important roles in the adaptation of seed plants to terrestrial environments (Kenrick and Crane, 1997; Becker and Marin, 2009; Radoeva and Weijers, 2014). Seed development is an intricate process, which can be divided into two conceptually distinct phases: embryo morphogenesis and seed maturation (Goldberg et al., 1994; Harada, 1997; Gutierrez et al., 2007). Seed maturation, which includes all of the events occurring after cell division has ceased within the embryo (following Harada, 1997), can be considered as a developmental module that is added after embryogenesis. It is accomplished with the accumulation of nutrient reserves, the acquisition of desiccation tolerance, the desiccation of seeds, the suppression of precocious germination, and the induction of dormancy (Goldberg et al., 1994; Harada, 1997); these features are each thought to be important in the adaptation of plants to variable and harsh terrestrial environments. Overall, it was considered that seed maturation is a more recently derived adaptation program of land plants (Harada, 1997; Santos-Mendoza et al., 2008).
According to previous studies, especially of Arabidopsis, the seed maturation program involves complex regulatory networks that regulates a large set of genes (Verdier et al., 2013; Righetti et al., 2015). The LAFL network is one of those regulatory networks, which includes LEAFY COTYLEDON1 (LEC1) and LEC1-LIKE (L1L) of the NF-YB gene family, and ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 (LEAFY COTYLEDON2) of the B3-AFL gene family (Suzuki et al., 1997; Luerßen et al., 1998; Santos-Mendoza et al., 2008; Suzuki and McCarty, 2008; Swaminathan et al., 2008; Xie et al., 2008; Jia et al., 2013; Kirkbride et al., 2013). On a basic level, this network was thought to orchestrate the accumulation of storage compounds and the acquisition of desiccation tolerance in seed maturation (Harada, 1997; Santos-Mendoza et al., 2008; Jia et al., 2013; Radoeva and Weijers, 2014). Meanwhile, the LAFL network also represses the expression of genes required for the transition from embryonic to vegetative developments, i.e., the suppression of precocious germination (Giraudat et al., 1992; Nambara et al., 1992; Stone et al., 2001).
Four conserved protein domains can be recognized in the B3-AFL gene family regulatory factors of Arabidopsis thaliana, designated A, B1, B2, and B3 (Giraudat et al., 1992; Suzuki et al., 1997). The A-domain is a functional acidic activation domain found at the N-terminal (McCarty et al., 1991). The B1-domain consists of about 30 amino acids (AAs) involved in the physical interaction with the bZIP transcription factor, such as ABI5 (ABSCISIC ACID INSENSITIVE5; Nakamura et al., 2001). The B2-domain consists of about 15 AAs, which have been shown to be responsible for the ABA-dependent activation of ABA-regulated genes, through the ABA-response element (ABRE; Hill et al., 1996; Bies-Etheve et al., 1999; Ezcurra et al., 2000). The B3-domain, composed of about 100 AAs, has been shown to act as the DNA binding domain (Suzuki et al., 1997; Nag et al., 2005). For the AFL family genes, ABI3 has all the recognized domains of this gene family (Giraudat et al., 1992; Suzuki et al., 1997). FUS3 contains the A, B2, and B3 domains, but the A-domain in the C-terminal (Lu et al., 2010). LEC2 has only the B2 and B3 domains. In the monocots, there are different names for AFL genes. For example, five AFL gene homologous were found in Oryza sativa, e.g., OsVP1, OsLFL1, and OsIDEFs. OsVP1, which contains A, B1, B2, and B3 domains, is homologous with Arabidopsis AtABI3 (Hattori et al., 1994), and OsLFL1 is homologous with Arabidopsis AtFUS3 (Peng et al., 2008). Another three OsIDEFs are considered to be AtLEC2 type genes, but the relationship among them remains unclear (Kobayashi et al., 2007; Sreenivasulu and Wobus, 2013).
In Arabidopsis, AFL genes are mainly expressed in embryo development, but at different developmental stages. AtLEC2 is expressed at early stages of embryogenesis, while AtABI3 and AtFUS3 are highly expressed at late stages (Stone et al., 2001; Kroj et al., 2003; Gazzarrini et al., 2004; Tsuchiya et al., 2004; To et al., 2006; Santos-Mendoza et al., 2008; Fatihi et al., 2016). According to studies in other plants, the AFL family genes are generally expressed in reproductive organs. For instance, OsLFL1 is expressed exclusively in spikes and young embryos (Peng et al., 2008). In Zea mays, ZmaAFL genes are preferentially expressed in pollen and caryopses (Grimault et al., 2015), and in Chamaecyparis nootkatensis, a gymnosperm species, its CnABI3 was detected in the megagametophytes and mature dormant embryos (Zeng and Kermode, 2004).
LEC1-type (LEC1 and L1L) genes are of the intron-less type of the NF-YB family, which are derived from the intron-rich ones, and their earliest occurrence appears to be in a common ancestor of vascular plants (Yang et al., 2005; Xie et al., 2008). LEC1 and L1L genes are highly expressed in embryonic cells and extra-embryonic tissues during seed development (Lotan et al., 1998; Kwong et al., 2003). Expression and function analyses of LEC1 homologs in other species indicate that LEC1 is essential for seed maturation (Stephenson et al., 2007; Cao et al., 2011; Salvini et al., 2012; Tang et al., 2015). In seedless vascular plants (lycophytes and ferns), the expression of LEC1 is restricted to reproductive structures. In Selaginella moellendorffii (a lycophyte), high expression of SmoLEC1 was found in strobili, where megasporangia and microsporangia are located (Kirkbride et al., 2013). Additionally, the maximal expression of AcaLEC1 was detected in mature sporangia of the fern Adiantum capillus-veneris (Fang et al., unpublished data).
Complex interactions between the LAFL genes were found in Arabidopsis. For instance, the expression of LEC1 can activate ABI3, FUS3, and LEC2, whereas the ectopic expression of LEC2 up-regulates LEC1 activity in vegetative tissues (Kagaya et al., 2005b; Stone et al., 2007; Guo et al., 2013). The function of LAFL genes involves many aspects of seed maturation including seed storage protein (SSP), late-embryogenesis- abundant (LEA) proteins, hormone metabolism, and signaling pathways (Parcy et al., 1994; Nakamura et al., 2001; Kagaya et al., 2005a,b; Alonso et al., 2009; Yamamoto et al., 2009).
The LAFL network is crucial for seed maturation, and great efforts have been made to investigate the functions of this network genes in Arabidopsis, but little attention was paid to the evolution of the network as a whole. With the increased availability of genomic data and a refined understanding of the distribution of LAFL genes, this work is now feasible. To better understand the origin and evolution of LAFL genes, we performed phylogenetic analyses on an extensive dataset of NF-YB and AFL gene family sequences, focusing particularly on previously underrepresented groups, such as algae, bryophytes, monilophytes, and “early diverging” angiosperms. In addition, we analyzed expression patterns of the LAFL network using online databases and our newly generated qRT-PCR data from S. moellendorffii and A. capillus-veneris (representing lycophytes and monilophytes, respectively). With these data, coupled with aforementioned phylogenetic analyses and cis-element information, we elucidate the evolution of LAFL genes and their association with the seed maturation program.
Materials and Methods
Gene Family Datasets
LAFL genes belong to two gene families: the NF-YB gene family and the AFL gene family, where the latter is a member of the B3 superfamily. To build our dataset of AFL genes, we first queried the Pfam database1 for B3 superfamily genes from three chlorophytes (Volvox carteri, Chlamydomonas reinhardtii, and Chlorella variabilis), one moss (Physcomitrella patens), one lycophyte (S. moellendorffii), and six flowering plants (Brachypodium sylvaticum, Oryza sativa, Zea mays, Populus trichocarpa, Glycine max, and A. thaliana; Supplementary Table S1); this search resulted in 730 sequences. Then, for a better understanding of the evolution of the AFL gene family specifically, we BLASTed the coding sequences of Arabidopsis ABI3, FUS3 and LEC2 against four primary sources: Phytozome2, ConGenIE3, the Klebsormidium flaccidum Genome Project4 (Hori et al., 2014), and the OneKP database5. These queries yielded 253 sequences spanning 68 species representing all major lineages of land plants. The retrieved sequences generally span the complete coding region, but some lack a few AAs at either end. The retrieved sequences range from 200 to 800 AAs in length (Supplementary Table S2).
To obtain sequences of the NF-YB gene family, we BLASTed Arabidopsis LEC1 and L1L coding sequences against five primary sources: NCBI (National Center for Biotechnology Information6), Phytozome2, ConGenIE3, the Klebsormidium flaccidum Genome Project4, and the OneKP project5. In total, 263 sequences spanning 29 species were collected, ranging from 100 to 300 AAs in length (Supplementary Table S3).
Sequence Alignment
All alignments were performed at AA level. For the phylogenetic analysis of the B3 superfamily, only the B3 domain was used for alignment. For the NF-YB and AFL gene families, full-length protein sequences were used. These sequences were aligned with the MAFFT webserver (7Katoh and Standley, 2013). Based on sequence characteristics, we selected an alignment strategy of FFT-NS-i (NF-YB gene family), FFT-NS-1(B3 superfamily), and E-INS-i (AFL gene family), respectively.
Phylogenetic Analysis
The final alignments were analyzed using Protest (Abascal et al., 2005) to choose the best-fitting AA model; the JTT + I + G substitution model was selected for all alignments according to the AIC and BIC selection criteria. Maximum likelihood (ML) phylogenetic analyses were performed with RaxML (Stamatakis et al., 2008) and evaluated by the bootstrap method using 1000 replicates. Trees were observed and edited for presentation using FigTree v1.4.2.8 Based on phylogenetic reconstruction of the B3 superfamily (Supplementary Figure S1), we re-built a dataset with an ingroup sample of 253 AFL genes, and an outgroup of 11 B3 genes from four algal species for further phylogenetic analysis of AFL gene family (Figures 1A, 2A, Table 1, and Supplementary Table S2). For bryophytes and vascular plants, further phylogenetic analyses were carried out, respectively (Figures 1B, 2B). In addition, phylogeny reconstruction of the NF-YB family was performed using the data set containing 263 sequences of 29 species with whole genome sequences (Supplementary Figure S3). To explore the relationship of LEC1-type genes and NF-YB family genes in non-vascular plants, 65 sequences of 26 species were used for further phylogenetic analysis (Table 1 and Figure 4).
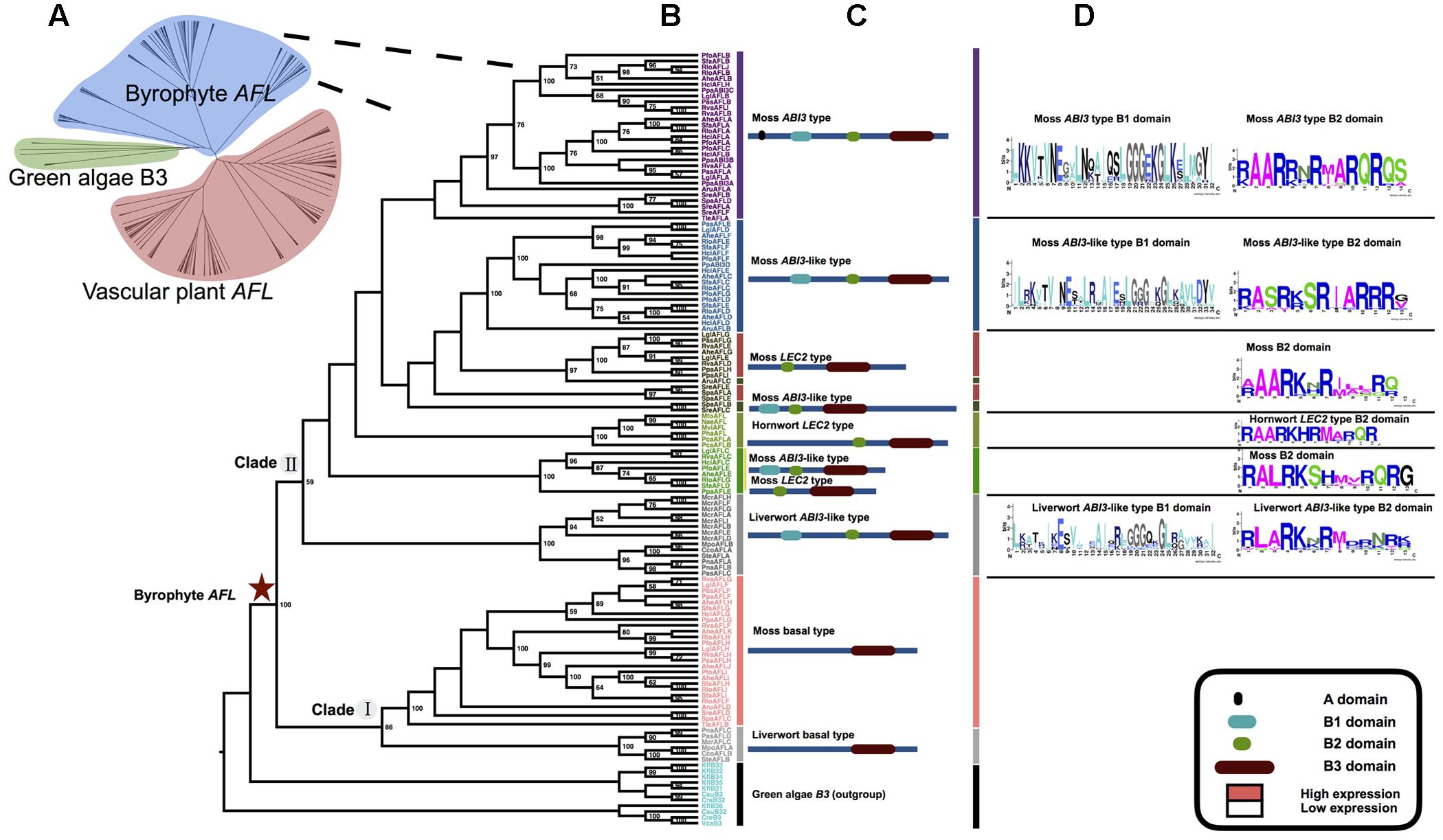
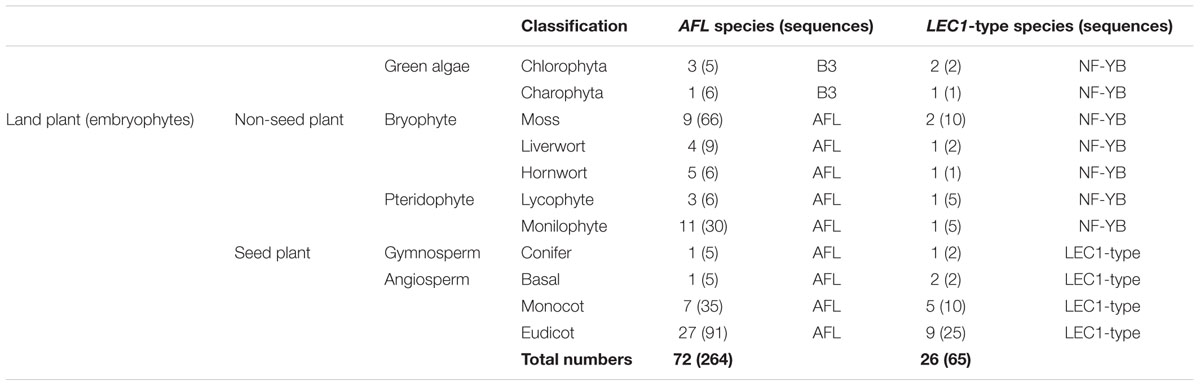
FIGURE 1. Phylogenetic relationships of plant AFL gene family and the details of bryophyte. (A) ML unrooted tree of the plant AFL gene family comprising 264 sequences from 72 taxa (Table 1; for species names see Supplementary Table S2). (B) ML rooted tree of bryophyte AFL using green algae B3 sequences as outgroups (black). Numbers on the branches indicate bootstrap values calculated from 1,000 replicates. Only values higher than 50% are shown. (C) Domain structure of each clade and type, two clades divided into 10 types in total. (D) Amino acids (AAs) characteristic of the B1 and B2 domain. The sequence logo was generated using WebLogo (weblogo.berkeley.edu/).
FIGURE 2. Phylogenetic relationships of plant AFL gene family and the details of the part of vascular plant. (A) ML phylogenetic relationships of plant AFL gene family comprising 264 sequences from 72 taxa (Table 1 for species names see Supplementary Table S2). (B) ML rooted tree of vascular plant AFL using PpaABI3 sequences as outgroups (black). Numbers at the branches indicate bootstrap values calculated from 1,000 replicates. Only values higher than 50% are shown. (C) Domain structure of each clade and type, three clades divided into 10 types in total. (D) AAs characteristic of the B1 and B2 domain. The sequence logo was generated using WebLogo (weblogo.berkeley.edu/). (E) Expression pattern analysis of several AFL genes; expression levels are calculated using the database GENEVESTIGATOR, ConGenIE (Supplementary Figure S3), and data from previous studies (Zeng et al., 2003; Zhang and Xue, 2013).
Gene Structure and Cis-Elements Analysis
For the AFL family genes, we characterized their AA composition and the position of the B1, B2 and B3 domains, because these are known as identification criteria for AFL genes (McCarty et al., 1991; Giraudat et al., 1992; Suzuki et al., 1997; Nag et al., 2005; Lu et al., 2010). The AA composition of B1, B2, and B3 domains was analyzed by the WebLogo online (Figures 2D, 3D9). We performed the intron-exon and position analyses of the NF-YB family genes by using their full-length DNA sequences (Figure 4 and Supplementary Table S3).
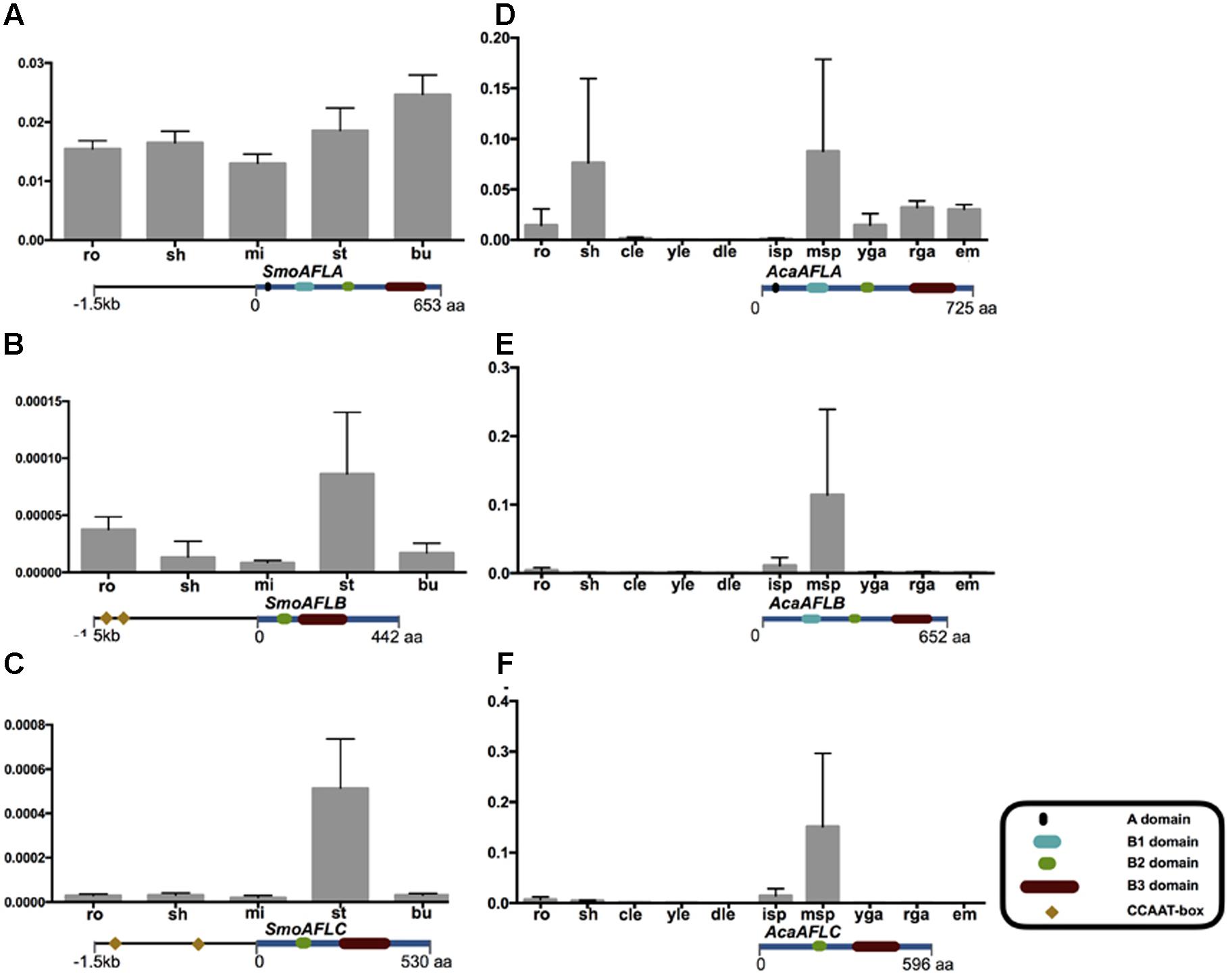
FIGURE 3. AFL mRNA levels in various organs of non-seed plants examined by qRT-PCR. The CCAAT-box cis-elements of the promoter 1.5 kb region of three SmoAFL genes are highlighted in yellow blocks with an arranged number. Selaginella moellendorffii (A–C), ro, roots; sh, shoots; mi, microphylls; st, strobili; bu, bulbils. Adiantum capillus-veneris (D–F), ro, roots; sh, shoots; cle, curled leaves; yle, young leaves; dle, developed leaves; isp, immature sporangia; msp, mature sporangia; yga, young gametophytes; rga, reproductive gametophytes; em, embryos. The detail of each gene domain structure see Supplementary Figure S4.
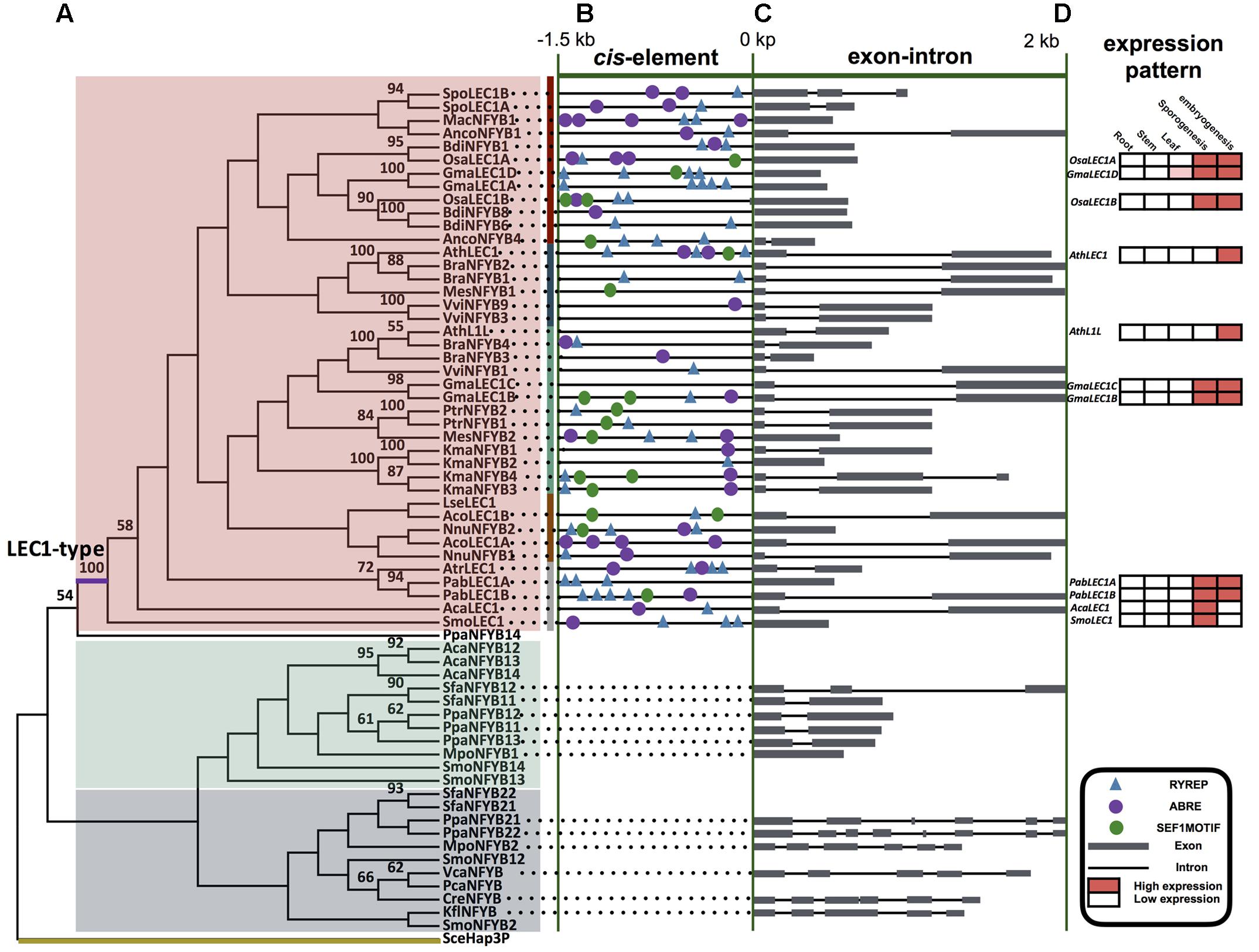
FIGURE 4. LEC1-type and non LEC1-type genes of NF-YB gene family. (A) ML tree of plant LEC1-type and non LEC1-type genes comprising 66 sequences from 27 taxa (for species names see Supplementary Table S3). Numbers on the branches indicate bootstrap values calculated from 1,000 replicates. Only values higher than 50% are shown. Non-seed plant intron-rich clade, non-seed plant intron-less clade, and LEC1-type clade are color-coded with gray, green, and red, respectively. (B) The conserved cis-elements in the promoter 1.5 kb region of LEC1-type genes. Details are shown in Supplementary Table S5 with the number and sequence of motifs. (C) Exon-intron changes at several LEC1-type and non LEC1-type genes. Details are shown in Supplementary Table S5. (D) Expression pattern analysis of several LEC1-type genes; expression levels are calculated using the database GENEVESTIGATOR, ConGenIE (Supplementary Figure S3), and data from experimental studies (Kirkbride et al., 2013; Fang et al., unpublished data).
To characterize cis-elements in the 5′ flanking region of LAFL genes, the 1.5 kb fragment containing promoter and 5′ UTR of six AFL genes and 40 LEC1-type genes were analyzed by PLACE (10this database is temporarily terminated now) and Matlnspector (Genomatix Software Suite11) online (Figures 3, 4 and Supplementary Table S5). Promoter sequences of A. capillus-veneris LEC1 were cloned through genome walking (primers in Supplementary Table S4).
Expression Analysis by qRT-PCR
To investigate the expression of LAFL genes in different vascular plants, the publicly available expression data as well as the expression data of LEC1 homologs in S. moellendorffii and A. capillus-veneris (SmoLEC1 expression data in Kirkbride et al., 2013, AcaLEC1 unpublished expression data) were used to construct an expression heat map, where analyzed species include one monocot (rice), two eudicots (Arabidopsis and soybean), and one gymnosperm (Picea abies; Supplementary Figure S3). In addition, we chose S. moellendorffii and A. capillus-veneris as non-seed plant representatives using qRT-PCR to characterize AFL genes expression patterns at different developmental stages. With respect to sampling for these two species, S. moellendorffii roots, shoots, microphylls, strobili, and bulbils were collected in the field (Sichuan Province, voucher specimen was deposited in Peking University Herbarium, PEY). For A. capillus-veneris, samples were collected from plants cultivated in the greenhouse of Peking University (Voucher specimen was deposited in PEY). We chose roots, shoots, curled leaves, young leaves, fully developed leaves, immature sporangia, mature sporangia, immature gametophytes, reproductive gametophytes, and embryos as materials (Li et al., 2013). Total RNA of plant materials was isolated with Plant RNA Extraction Reagent (Invitrogen, USA) and purified with an RNeasy Mini kit according to the manufacturer’s instructions (Qiagen, Germany). The RNA was then converted to cDNA by reverse transcription with FastQuant RT Kit (Tiangen, China). The qRT-PCR was performed on an Applied Biosystems 7500 Real-Time PCR System (ABI) using cDNA templates mixed with primers (Supplementary Table S4) and SYBR® Premix Ex Tax Mix (Takara, Japan). SmoACTIN and AcaACTIN were selected as the internal standard gene (primer sequences in Supplementary Table S4). Relative expression was calculated via delta-delta threshold method (2-ΔCT; Livak and Schmittgen, 2001). Results were summarized as means ± SE of three biological repeats.
Results
Phylogenetic Analysis of LAFL Genes
The sequence retrieval and phylogenetic analysis of the B3 gene superfamily showed that no AFL sequences were found in Chlorophytes and Charophytes (Supplementary Figure S1 and Table S1). The B3 domain of AFL genes is highly conserved in seed plants (Supplementary Table S2). According to phylogenetic analysis of AFL genes in land plants, the cluster of bryophytes and vascular plants can be recognized although they were not strongly supported in the tree (Figures 1A, 2A).
Phylogenetic analysis of bryophyte AFL genes showed them to form two clades, clade I and clade II (Figure 1B). Clade I is composed of sequences from liverworts and mosses, while clade II has sequences from liverworts, mosses and hornworts. Mosses have many more AFL gene homologs than do liverworts or hornworts (Figure 1B and Table 1).
In the phylogenetic tree of vascular plant AFL genes, three major clades, clade I (lycophyte AFL clade), clade II, and clade III (ABI3 clade) can be recognized (Table 1 and Figure 2B). Clade I is composed of all lycophyte AFL genes including those of LEC2 type and ABI3 type (Figure 2B). All monilophyte ABI3 genes cluster with seed-plant ABI3 genes as a clade with strong support (clade III) (Figure 2B), while monilophyte LEC2 type genes group with the remaining seed plant LEC2 and FUS3 genes in clade II (Figure 2B). These results indicated that lycophyte AFL clade represents an ancient lineage of AFL gene family, and monilophyte AFL genes are more closely with those of seed plants. In the clade II, we can find five gene types with strong support, i.e., LEC2 type, FUS3 type, LFL1 type, PabFL type, and IDEF type, respectively (Figures 2B,C). In the genome of the gymnosperm P. abies, there were five AFL sequences, and three of them (PabAFLA/B/C) are associated with FUS3 and LFL1 types. In the “early diverging” angiosperm Amborella trichopoda genome (Albert et al., 2013), there were three AFL sequences, none of them of FUS3 type. One ABI3 gene was clustered in the seed plant ABI3 type clade strong support, one was clustered with the LEC2 type sequences and the other was clustered in the IDEF clade.
Phylogenetic analyses of the NF-YB family showed that LEC1-type genes formed a clade and were only present in vascular plants (Figure 4 and Supplementary Figure S3, Table S3), in agreement with previous results (Xie et al., 2008). The number of LEC1-type genes in lycophytes, monilophytes, conifers, “early diverging” angiosperms, monocots, and eudicots averages 1.0, 1.0, 2.0, 1.0, 2.0, and 2.7 per species, respectively (Table 1). AcaLEC1 of the fern A. capillus-veneris was cloned and identified by this study for first time.
Gene Structure Analysis of LAFL Genes
Domain structure analysis of bryophyte AFL genes showed that clade I genes only have one B3 domain in the C-terminal. This structure is similar to B3 genes in green algae. In clade II, ABI3 type genes were easily recognized by the A domain, B1 (about 30 AA), B2 (about 14 AA) and B3 (about 100 AA) domain, while ABI3-like genes lack of the A domain. LEC2 type genes had one unstable B2 domain in middle position and B3 domain in C-terminal. Notably, liverworts only had the clade I and ABI3-like AFL genes. We did not find FUS3 type genes in bryophytes (Figures 1C,D), suggesting that FUS3 may not arise. Domain structure analysis of vascular plant AFL genes showed that there were nine structure types in three clades with specific B2 domain AA characteristics (Figure 2D).
Our analyses of NF-YB gene sequences revealed that: (1) the NF-YB sequences restricted to algae are of the intron-rich type; (2) the liverwort Marchantia polymorpha contains both an intron-rich and an intron-less NF-YB gene; (3) six sequences occur in moss Physcomitrella patens including both intron-rich and intron-less ones; (4) LEC1-type genes, belonging to intron-less NF-YB genes, were only found in vascular plants. All those findings suggest that the intron-less type of NF-YB genes was derived from the intron-rich ones through gene duplication and intron loss in early land plants (Figure 4C, Table 1, and Supplementary Figure S3).
Cis-Element Prediction of LAFL Genes
According to comparative analysis of cis-elements in regulatory region of AFL gene pairs between the seed plant Arabidopsis and the non-seed plant S. moellendorffii, we found that there was CCAAT-box in AthABI3, AthFUS3, AthLEC2, SmoAFLB, and SmoAFLC (Figure 4 and Supplementary Table S5). By cis-elements prediction in the promoter region of 40 LEC1-type genes, we found that (1) LEC1 had more cis-elements than do L1L genes in seed plants; (2) there is not a significant difference in cis-element components in LEC1 genes from seed and non-seed plants: almost all the cis-elements identified in Arabidopsis can be found in the LEC1 promoter of S. moellendorffii and A. capillus-veneris (Figure 4B and Supplementary Table S5).
Expression Pattern Analyses of LAFL Genes
LAFL gene expression was restricted to seed development in Arabidopsis but occurs both in maturing seeds and inflorescences in other species, e.g., soybean, rice, and maize (Figure 4D and Supplementary Figure S4). For LAFL genes of P. abies (a gymnosperm), they were mainly expressed in leaves and cones. SmoLEC1 and AcaLEC1 were each only expressed in strobili and mature sporangia, and were not detected in tissues undergoing embryogenesis (Figure 4D).
The qRT-PCR results showed that the mRNA levels of SmoAFLA (lycophyte ABI3 type) were nearly identical across organs (roots, shoots, microphylls, strobili, and bulbils) of S. moellendorffii. The levels of mRNA of SmoAFLB and SmoAFLC (lycophyte LEC2 type) were higher in strobili than other organs. In A. capillus-veneris (a fern) the levels of mRNA of AcaAFLA (one monilophyte ABI3 type) were higher in shoots and mature sporangia than other organs. The mRNA of AcaAFLB (another monilophyte ABI3 type) and AcaAFLC (monilophyte LEC2 type) were only detected in mature sporangia (Figure 3 and Supplementary Figures S2, S4).
Discussion
LAFL Network and Seed Maturation
Previous studies showed that many genes are involved in seed maturation (Goldberg et al., 1994; Harada, 1997; Radoeva and Weijers, 2014). Among them, the AFL family of B3 transcription factors (TFs) and the LEC1-type of NF-YB TFs, which together form LAFL regulatory network, are considered to play key roles in seed maturation. Although there were studies on the evolution of LEC1-type genes (Xie et al., 2008), AFL genes (Li et al., 2010; Carbonero et al., 2016), this study presents a comprehensive analysis of LAFL genes by integrating their phylogeny, gene structure, cis-elements and expression patterns together for a better understanding of the evolution of seed maturation programs during plant evolution.
Evolution and Function Differentiation of AFL Genes
According to our extensive phylogenic and gene structure analyses, LEC2 type and ABI3 type genes evolved in a common ancestor of bryophytes and vascular plants, and their gene structure is very conservative. However, FUS3 type genes were only found in seed plants (Figures 1, 2), suggesting that FUS3 genes originate relatively late in the AFL family.
In embryophytes, LEC2 type genes had one B2 domain in a middle position and a B3 domain in the C-terminal. In the seedless species S. moellendorffii (lycophyte) and A. capillus-veneris (fern), the expression pattern of LEC2 type genes (SmoAFLB, SmoAFLC, and AcaAFLC) was restricted to shoots (S. moellendorffii) and maturing spores (both S. moellendorffii and A. capillus-veneris; Figure 3). In the “early diverging” angiosperm Amborella trichopoda, there were three AFL genes. One of them is of ABI3 type, and the other two are LEC2 type and IDEF type, respectively. Interestingly, IDEF type genes were identified only from monocots, and have only B3 domain in C-terminal (Kobayashi et al., 2007), which is different from LEC2 gene structure (Figure 2C). In rice, OsaIDEF transcripts are constitutively present in roots, leaves, inflorescences, and seeds. In eudicots, LEC2 plays central roles in seed embryogenesis and morphogenesis (Figure 2 and Supplementary Figure S4). All these data suggest that LEC2 and IDEF type genes diverged very early, and LEC2 type genes may be lost in monocots.
During the review of this manuscript, Carbonero et al. (2016) published their work on the AFL family. In agreement with our results, they suggest that the origin of the AFL family traces back to a common ancestor of bryophytes and vascular plants, and that this family has expanded in the angiosperms. However, due to different sampling regimes and sequence coverage, there are some different results between these two studies, especially relating to the evolution of LEC2 genes. According to Carbonero et al. (2016), seven LEC2 genes were described from three monocots, Oryza sativa, Brachypodium distachyon and Hordeum vulgare (all grasses), but the relationship of those seven genes with other AFL homologs needs to be verified; differences in gene structure, phylogenetic position, and expression pattern suggests that these may not be LEC2 genes.
Considering ABI3 genes of land plants, there is a clear evolutionary trajectory according to our study. Phylogenetically, monilophyte ABI3 genes are more closely related to those of seed plants, rather than to lycophyte ABI3 types. In P. abies (gymnosperm) and Amborella trichopoda (“early diverging” angiosperm), there was only one PabABI3 and AtrABI3 sequence, respectively. This may be due to the lack of a lineage-specific whole genome duplication (WGD) in these species (Albert et al., 2013; Nystedt et al., 2013). Expression patterns of SmoAFLA (S. moellendorffii, one lycophyte ABI3 type) are more similar to those of bryophyte ABI3 type genes, which are only expressed in vegetative tissues (Figure 3; Khandelwal et al., 2010). The expression of AcaAFLA and AcaAFLB (A. capillus-veneris, two monilophyte ABI3 type genes) are found in shoots and spore maturation, which are consistent with that of PabABI3 (P. abies) (Figures 2B,C,E, 3). This suggest the expression pattern of ABI3 genes has slightly differentiated across major land plant lineages.
FUS3 type genes appear to have originated relatively late because they are restricted to the seed plant clade. Three PabAFL sequences (PabAFLA, B, and C) from the gymnosperm P. abies belong to Pab-FL (FUS3 and LFL) type clade, which is associated with FUS3 type and LFL type. These finding, coupled with expression patterns of PabAFLA/B/C genes suggest that the Pab-FL type may represent ancestral FUS3/LFL gene function. There is no FUS3 type member in Amborella trichopoda, which suggests that FUS3 type genes likely originated in a common ancestor of seed plants and were subsequently lost in Amborella. In eudicots and monocots, FL genes are divided to FUS3 type and LFL type, respectively. OsLFL1, involved in the photoperiodic flowering of rice and expressed exclusively in spikes and young embryos, is functionally similar to AthFUS3 in Arabidopsis (Peng et al., 2008; Tiedemann et al., 2008). The FUS3 type (found only in eudicots) and the LFL type (restricted to monocots) are clustered together with strong bootstrap support, and they have similar domain structure and functions (Figures 2B,C,E, 3).
Evolution of the LEC1-Type Genes
As members of the LAFL network, LEC1-type genes are CCAAT-binding factors (CBFs), which are present in all eukaryotes (Forsburg and Guarente, 1989; Mantovani, 1999; Matuoka and Chen, 2002; Siefers et al., 2009; Dolfini et al., 2012). There is no clear correlation between expression patterns and the classification of NF-YB family genes with an exception of the LEC1-type genes, which are considered seed-specific (Stephenson et al., 2007; Salvini et al., 2012). Arabidopsis LEC1-type genes (AthLEC1 and AthL1L) have significant functions at late stages of embryogenesis (Lotan et al., 1998; Kwong et al., 2003). Our phylogenetic analyses of the NF-YB gene family support some findings of previous studies, e.g., only one intron-rich type of NF-YB genes occurs in chlorophytes, the intron-less genes are derived from the intron-rich ones, and LEC1-type genes are restricted to vascular plants (Xie et al., 2008; Cagliari et al., 2014; Table 1 and Figures 4A,C).
In addition, there are some new findings, e.g., only one copy of the intron-rich type of NF-YB genes is found in the alga Klebsormidium flaccidum, which is considered to be one of the closest relatives of land plants (Hori et al., 2014). The liverwort Marchantia polymorpha, one of the earliest diverged land plants (Rövekamp et al., 2016), has two copies of NF-YB genes in its genome, one of which is intron-rich and the other intron-less. The six copies found in the moss Physcomitrella patens, have been proven to originate from duplication events (Yang et al., 2005; Rensing et al., 2008; Xie et al., 2008). In addition, our analyses demonstrate that there is only one copy of LEC1-type genes in the genome of S. moellendorffii (lycophyte), A. capillus-veneris (fern), P. abies (gymnosperm), and Amborella trichopoda (“early diverging” angiosperm). These data support that LEC1 and L1L genes result from the duplication of LEC1-type genes likely occurring after the origin of extant angiosperms (Table 1 and Figures 4A,C).
The Cis-Element Prediction and Co-expression of LAFL Genes
The LAFL network has been considered to play central roles in seed maturation, and LAFL genes regulate different facets of this developmental process by their interactions with up- and down-stream genes (Harada, 1997; Santos-Mendoza et al., 2008; Fatihi et al., 2016; González-Morales et al., 2016). The cis-element prediction shows that LEC1 genes of seed plants and non-seed plants have similar cis-elements, suggesting the LEC1-type genes could be regulated by similar regulators (Figure 4). Among the cis-elements of LEC1, RYREPART and ABRE are thought to be very important for LEC1 activity. The RYREPEAT is considered to be a RY-like element, and the binding site of the B3 domain (Braybrook et al., 2006; Mönke et al., 2012; Wang and Perry, 2013; Tang et al., 2015). The ABRE is functionally important in many ABA-regulated genes (Fan et al., 2015). Additionally, LEC1, as a subunit of the CCAAT-box binding factor (CBF), activates its downstream genes by the CCAAT-box element (Junker et al., 2012). According to the CCAAT-box element prediction of AFL genes in S. moellendorffii, there is a CCAAT-box element in the regulatory region of its AFL genes, e.g., SmoAFLB and SmoAFLC (Figure 3).
The findings presented in this study suggest that a partial LAFL network, consisting of ABI3 and LEC2 genes, arose in a common ancestor of land plants, and then became more complex with the occurrence of FUS3 and LEC1 genes. With evolution of vascular plants, LAFL network genes likely specify their co-expression in two different developmental processes, spore and seed maturation, respectively. The co-expression of LAFL genes in these two processes alone or simultaneously, which correspond to two reproductive structures, suggest that the biological process involved in spore maturation is similar to those of seed maturation.
Author Contributions
J-DH analyzed data and drafted the manuscript. XL and C-KJ carried out the experiments. GW and CR provided some samples and analyzed sequences. G-YR designed the research.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC, Grant no. 91231105).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Prof. Ji Yang of Fudan University for discussions, and two reviewers for their critical comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00439/full#supplementary-material
Footnotes
- ^ https://pfam.xfam.org/family/PF02362#tabview=tab0
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ congenie.org
- ^ www.plantmorphogenesis.bio.titech.ac.jp/~algae_genome_project/klebsormidium/index.html
- ^ https://db.cngb.org/blast4onekp/
- ^ www.ncbi.nlm.nih.gov
- ^ http://mafft.cbrc.jp/alignment/server/
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ weblogo.berkeley.edu
- ^ sogo.dna.affrc.go.jp/
- ^ https://www.genomatix.de/solutions/genomatix-software-suite.html
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Albert, V. A., Barbazuk, W. B., Der, J. P., Leebens-Mack, J., Ma, H., Palmer, J. D., et al. (2013). The Amborella genome and the evolution of flowering plants. Science 342:1241089. doi: 10.1126/science.1241089
Alonso, R., Oñate-Sánchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., et al. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21, 1747–1761. doi: 10.1105/tpc.108.062968
Becker, B., and Marin, B. (2009). Streptophyte algae and the origin of embryophytes. Ann. Bot. 103, 999–1004. doi: 10.1093/aob/mcp044
Bies-Etheve, N., da Silva Conceicao, A., Koornneef, M., Léon-Kloosterziel, K., Valon, C., and Delseny, M. (1999). Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 6, 1045–1054. doi: 10.1023/A:1006252512202
Braybrook, S. A., Stone, S. L., Park, S., Bui, A. Q., Le, B. H., Fischer, R. L., et al. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473. doi: 10.1073/pnas.0511331103
Cagliari, A., Turchetto-Zolet, A. C., Korbes, A. P., dos Santos Maraschin, F., Margis, R., and Margis-Pinheiro, M. (2014). New insights on the evolution of Leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 103, 380–387. doi: 10.1016/j.ygeno.2014.03.005
Cao, S., Kumimoto, R. W., Siriwardana, C. L., Risinger, J. R., and Holt, B. F. III (2011). Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS ONE 6:e21805. doi: 10.1371/journal.pone.0021805
Carbonero, P., Iglesias-Fernández, R., and Vicente-Carbajosa, J. (2016). The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J. Exp. Bot. 4, 871–880. doi: 10.1093/jxb/erw458
Dolfini, D., Gatta, R., and Mantovani, R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 47, 29–49. doi: 10.3109/10409238.2011.628970
Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerström, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. doi: 10.1046/j.1365-313x.2000.00857.x
Fan, K., Shen, H., Bibi, N., Li, F., Yuan, S., Wang, M., et al. (2015). Molecular evolution and species-specific expansion of the NAP members in plants. J. Integr. Plant Biol. 57, 673–687. doi: 10.1111/jipb.12344
Fatihi, A., Boulard, C., Bouyer, D., Baud, S., Dubreucq, B., and Lepiniec, L. (2016). Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci. 250, 198–204. doi: 10.1016/j.plantsci.2016.06.013
Forsburg, S. L., and Guarente, L. (1989). Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3, 1166–1178. doi: 10.1101/gad.3.8.1166
Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385. doi: 10.1016/j.devcel.2004.06.017
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., and Goodman, H. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Goldberg, R. B., de Paiva, P., and Yadegari, R. (1994). Plant embryogenesis: zygote to seed. Science 266, 605–614. doi: 10.1126/science.266.5185.605
González-Morales, S. I., Chávez-Montes, R. A., Hayano-Kanashiro, C., Alejo-Jacuinde, G., Rico-Cambron, T. Y., de Folter, S., et al. (2016). Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 113, E5232–E5241. doi: 10.1073/pnas.1610985113
Grimault, A., Gendrot, G., Chaignon, S., Gilard, F., Tcherkezd, G., Thévenine, J., et al. (2015). Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant Sci. 236, 116–125. doi: 10.1016/j.plantsci.2015.03.021
Guo, X., Hou, X., Fang, J., Wei, P., Xu, B., Chen, M., et al. (2013). The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 75, 403–416. doi: 10.1111/tpj.12209
Gutierrez, L., Wuytswinkel, O., Castelain, M., and Bellini, C. (2007). Combined networks regulating seed maturation. Trends Plant Sci. 12, 294–300. doi: 10.1016/j.tplants.2007.06.003
Harada, J. J. (1997). “Seed maturation and control of dormancy,” in Cellular and Molecular Biology of Plant Seed Development, eds B. A. Larkins and I. K. Vasil (Gainesville, FL: University of Florida Press), 545–592. doi: 10.1007/978-94-015-8909-3_15
Hattori, T., Terada, T., and Hamasuna, S. T. (1994). Sequence and functional analysis of the rice gene homologous to maize Vp1. Plant Mol. Biol. 24, 805–810. doi: 10.1007/BF00029862
Hill, A., Nantel, A., Rock, C. D., and Quatrano, R. S. (1996). A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 271, 3366–3374. doi: 10.1074/jbc.271.7.3366
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5:3978. doi: 10.1038/ncomms4978
Jia, H., Suzuki, M., and McCarty, D. R. (2013). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. WIREs Dev. Biol. 3, 135–145. doi: 10.1002/wdev.126
Junker, A., Mönke, G., Rutten, T., Keilwagen, J., Seifert, M., Thi, T. M., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71, 427–442. doi: 10.1111/j.1365-313X.2012.04999.x
Kagaya, Y., Okuda, R., Ban, A., Toyoshima, R., Tsutsumida, K., Usui, H., et al. (2005a). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 46, 300–311. doi: 10.1093/pcp/pci031
Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., and Hattori, T. (2005b). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406. doi: 10.1093/pcp/pci048
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kenrick, P., and Crane, P. (1997). The origin and early evolution of plants on land. Nature 389, 33–39. doi: 10.1038/37918
Khandelwal, A., Cho, S. H., Marella, H., Sakata, Y., Perroud, P. F., Pan, A., et al. (2010). Role of ABA and ABI3 in desiccation tolerance. Science 327, 546–546. doi: 10.1126/science.1183672
Kirkbride, R. C., Fischer, R. L., and Harada, J. J. (2013). LEAFY COTYLEDON1, a key regulator of seed development, is expressed in vegetative and sexual propagules of Selaginella moellendorffii. PLoS ONE 8:e67971. doi: 10.1371/journal.pone.0067971
Kobayashi, T., Ogo, Y., Itai, R. N., Nakanishi, H., Takahashi, M., Mori, S., et al. (2007). The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 19150–19155. doi: 10.1073/pnas.0707010104
Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130, 6065–6073. doi: 10.1242/dev.00814
Kwong, R. W., Bui, A. Q., Lee, H., Kwong, L. W., Fischer, R. L., Goldberg, R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18. doi: 10.1105/tpc.006973
Li, X., Fang, Y. H., Yang, J., Bai, S. N., and Rao, G. Y. (2013). Overview of the morphology, anatomy and ontogeny of Adiantum capillus-veneris: an experimental system to study the development of ferns. J. Syst. Evol. 51, 499–510. doi: 10.1111/jse.12034
Li, Y., Jin, K., Zhu, Z., and Yang, J. (2010). Stepwise origin and functional diversification of the AFL subfamily B3 genes during land plant evolution. J. Bioinform. Comput. Biol. 8, 33–45. doi: 10.1142/S0219720010005129
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lotan, T., Ohto, M., Yee, K. M., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lu, Q. S., Paz, J., Pathmanathan, A., Chiu, R. S., Tsai, A., and Gazzarrini, S. (2010). The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 64, 100–113. doi: 10.1111/j.1365-313X.2010.04307.x
Luerßen, H., Kirik, V., Herrmann, P., and Miséra, S. (1998). FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313X.1998.00259.x
Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. doi: 10.1016/S0378-1119(99)00368-6
Matuoka, K., and Chen, K. Y. (2002). Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res. Rev. 1, 639–651. doi: 10.1016/S1568-1637(02)00026-0
McCarty, D. R., Hattori, T., Carson, C. B., Vasil, V., Lazar, M., and Vasil, I. K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. doi: 10.1016/0092-8674(91)90436-3
Mönke, G., Seifert, M., Keilwagen, J., Mohr, M., Grosse, I., Hähnel, U., et al. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254. doi: 10.1093/nar/gks594
Nag, R., Maity, M. K., and DasGupta, M. (2005). Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol. Biol. 59, 821–838. doi: 10.1007/s11103-005-1387-z
Nakamura, S., Lynch, T. J., and Finkelstein, R. R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. doi: 10.1046/j.1365-313x.2001.01069.x
Nambara, E., Naito, S., and McCourt, P. (1992). A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 2, 435–441. doi: 10.1111/j.1365-313X.1992.00435.x
Nystedt, B., Street, N. R., Wetterbom, A., Zuccolo, A., Lin, Y. C., Scofield, D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. doi: 10.1038/nature12211
Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. doi: 10.1105/tpc.6.11.1567
Peng, L. T., Shia, Z. Y., Lia, L., Shen, G. Z., and Zhang, J. L. (2008). Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J. Plant Physiol. 165, 876–885. doi: 10.1016/j.jplph.2007.07.010
Radoeva, T., and Weijers, D. (2014). A roadmap to embryo identity in plants. Trends Plant Sci. 19, 709–716. doi: 10.1016/j.tplants.2014.06.009
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Righetti, K., Vu, J., Pelletier, S., Vu, B., Glaab, E., Lalanne, D., et al. (2015). Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell 27, 2692–2708. doi: 10.1105/tpc.15.00632
Rövekamp, M., Bowman, J. L., and Grossniklaus, U. (2016). Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 26, 1–8. doi: 10.1016/j.cub.2016.05.028
Salvini, M., Sani, E., Fambrini, M., Pistelli, L., Pucciariello, C., and Pugliesi, C. (2012). Molecular analysis of a sunflower gene encoding an homologous of the B subunit of a CAAT binding factor. Mol. Biol. Rep. 39, 6449–6465. doi: 10.1007/s11033-012-1463-9
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., and Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Siefers, N., Dang, K. K., Kumimoto, R. W., Bynum, W. E., Tayrose, G., and Holt, B. F. (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. doi: 10.1104/pp.108.130591
Sreenivasulu, N., and Wobus, U. (2013). Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu. Rev. Plant Biol. 64, 189–217. doi: 10.1146/annurev-arplant-050312-120215
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stephenson, T. J., McIntyre, C. L., Collet, C., and Xue, G. P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65, 77–92. doi: 10.1007/s11103-007-9200-9
Stone, S., Braybrook, S., Paula, S., Kwong, L., Meuser, J., Pelletier, J., et al. (2007). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3151–3156. doi: 10.1073/pnas.0712364105
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Suzuki, M., Kao, C., and McCarty, D. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. doi: 10.1105/tpc.9.5.799
Suzuki, M., and McCarty, D. R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11, 548–553. doi: 10.1016/j.pbi.2008.06.015
Swaminathan, K., Peterson, K., and Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 12, 647–655. doi: 10.1016/j.tplants.2008.09.006
Tang, G., Xu, P., Liu, W., Liu, Z., and Shan, L. (2015). Cloning and characterization of 5’ flanking regulatory sequences of AhLEC1B gene from Arachis hypogaea L. PLoS ONE 10:e0139213. doi: 10.1371/journal.pone.0139213
Tiedemann, J., Rutten, T., Mönke, G., Vorwieger, A., Rolletschek, H., Meissner, D., et al. (2008). Dissection of a complex seed phenotype: novel insights of FUSCA3 regulated developmental processes. Dev. Biol. 317, 1–12. doi: 10.1016/j.ydbio.2008.01.034
To, A., Valon, C., Savino, G., Guilleminot, J., Devic, M., Giraudat, J., et al. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18, 1642–1651. doi: 10.1105/tpc.105.039925
Tsuchiya, Y., Nambara, E., Naito, S., and McCourt, P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37, 73–81. doi: 10.1046/j.1365-313X.2003.01939.x
Verdier, J., Torres-Jerez, I., Wang, M., Andriankaja, A., Allen, S., He, J., et al. (2013). Establishment of the lotus japonicus gene expression atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 74, 351–362. doi: 10.1111/tpj.12119
Wang, F., and Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi: 10.1104/pp.112.212282
Xie, Z., Li, X., Glover, B. J., Bai, S. N., Rao, G. Y., Luo, J., et al. (2008). Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 25, 1581–1592. doi: 10.1093/molbev/msn105
Yamamoto, A., Kagaya, Y., Toyoshima, R., Kagaya, M., Takeda, S., and Hattori, T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856. doi: 10.1111/j.1365-313X.2009.03817.x
Yang, J., Xie, Z., and Glover, B. J. (2005). Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol. 165, 623–632. doi: 10.1111/j.1469-8137.2004.01260.x
Zeng, Y., and Kermode, A. R. (2004). A gymnosperm ABI3 gene functions in a severe abscisic acid-insensitive mutant of Arabidopsis (abi3-6) to restore the wild-type phenotype and demonstrates a strong synergistic effect with sugar in the inhibition of post-germinative growth. Plant Mol. Biol. 56, 731–746. doi: 10.1007/s11103-004-4952-y
Zeng, Y., Raimondi, N., and Kermode, A. R. (2003). Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant Mol. Biol. 51, 39–49. doi: 10.1023/A:1020762304937
Keywords: seed maturation program, LAFL network, gene structure, expression analysis, phylogenetic analysis
Citation: Han J-D, Li X, Jiang C-K, Wong GK-S, Rothfels CJ and Rao G-Y (2017) Evolutionary Analysis of the LAFL Genes Involved in the Land Plant Seed Maturation Program. Front. Plant Sci. 8:439. doi: 10.3389/fpls.2017.00439
Received: 19 January 2017; Accepted: 14 March 2017;
Published: 04 April 2017.
Edited by:
Zhong-Jian Liu, The Orchid Conservation & Research Center of Shenzhen, ChinaReviewed by:
Pablo Daniel Jenik, Franklin & Marshall College, USAMarie Monniaux, Max Planck Institute for Plant Breeding Research, Germany
Copyright © 2017 Han, Li, Jiang, Wong, Rothfels and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang-Yuan Rao, cmFvQHBrdS5lZHUuY24=
 Jing-Dan Han
Jing-Dan Han Xia Li
Xia Li Chen-Kun Jiang
Chen-Kun Jiang Gane K.-S. Wong
Gane K.-S. Wong Carl J. Rothfels6
Carl J. Rothfels6 Guang-Yuan Rao
Guang-Yuan Rao