- 1National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology, Ministry of Science and Technology (Government of India), Mohali, India
- 2Department of Biotechnology, Panjab University, Chandigarh, India
Phytoene synthase (PSY) is a key regulatory enzyme of carotenoid biosynthesis pathway in plants. The present study examines the role of PSY in carotenogenesis and stress management in banana. Germplasm screening of 10 Indian cultivars showed that Nendran (3011.94 μg/100 g dry weight) and Rasthali (105.35 μg/100 g dry weight) contained the highest and lowest amounts of β-carotene, respectively in ripe fruit-pulp. Nendran ripe pulp also showed significantly higher antioxidant activity as compared to Rasthali. Meta-analysis of three banana PSY genes (MaPSY1, MaPSY2, and MaPSY3) was performed to identify their structural features, subcellular, and chromosomal localization in banana genome. The distinct expression patterns of MaPSY1, MaPSY2, and MaPSY3 genes were observed in various tissues, and fruit developmental stages of these two contrasting cultivars, suggesting differential regulation of the banana PSY genes. A positive correlation was observed between the expression of MaPSY1 and β-carotene accumulation in the ripe fruit-peel and pulp of Nendran. The presence of stress responsive cis-regulatory motifs in promoter region of MaPSY genes were correlated with the expression pattern during various stress (abscisic acid, methyl jasmonate, salicylic acid and dark) treatments. The positive modulation of MaPSY1 noticed under abiotic stresses suggested its role in plant physiological functions and defense response. The amino acid sequence analysis of the PSY proteins in contrasting cultivars revealed that all PSY comprises conserved domains related to enzyme activity. Bacterial complementation assay has validated the functional activity of six PSY proteins and among them PSY1 of Nendran (Nen-PSY1) gave the highest activity. These data provide new insights into the regulation of PSY expression in banana by developmental and stress related signals that can be explored in the banana improvement programs.
Introduction
Carotenoids are the natural compounds that are synthesized by vascular plants, some fungi and bacteria, but not by animals including humans (Ruiz-Sola and Rodríguez-Concepción, 2012). Carotenoids are essential for plant growth and development, and human nutrition. They are predominantly synthesized through plastid localized 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in chloroplasts and chromoplasts (Flores-Perez et al., 2008; Rodriguez-Concepcion, 2010; Jarvis and López-Juez, 2013; Kaur et al., 2016) (Supplementary Figure S1). Chromoplasts can differentiate from the pre-existing chloroplasts during fruit ripening or directly from proplastids, leucoplasts and amyloplasts in non-green tissues to facilitate carotenoid storage (Sandmann et al., 2006; Maass et al., 2009; Jarvis and López-Juez, 2013; Lado et al., 2015).
Phytoene synthase (PSY) regulates carotenoid metabolic flux in the downstream enzymatic steps for the biosynthesis of lycopene, α-carotene and β-carotene (Sandmann et al., 2006; Cazzonelli and Pogson, 2010; Grassi et al., 2013; Fu et al., 2014). Many plant species are known to have multiple copies of PSY gene (Gallagher et al., 2004; Clotault et al., 2008; Giorio et al., 2008; Li et al., 2008; Fu et al., 2014; Ampomah-Dwamena et al., 2015). The presence of multiple PSY in plants is a result of gene duplication events, which have significance for function and modulation of carotenogenesis (Gallagher et al., 2004; Ampomah-Dwamena et al., 2015). A number of PSY genes have been overexpressed in various plants, such as rice (Ye et al., 2000), tobacco (Busch et al., 2002), tomato (Fraser et al., 2002) and arabidopsis (Maass et al., 2009) leading to enhanced biosynthesis of carotenoids in a specific tissue.
Carotenoids play protective roles against biotic/abiotic stresses in plants. They are involved in signaling process and scavenging of free radicals during the stress management (El-Agamey et al., 2004; Aquino et al., 2016; García-Ruiz et al., 2017). Oxidized carotenoids (apocarotenoids) lead to produce abscisic acid (ABA), strigolactone, β–cyclocitral and β-ionone which subsequently assist plants to adapt in different stress conditions (Pogson et al., 2008; Havaux, 2014; Hou et al., 2016).
Abscisic acid (ABA), salicylic acid (SA), and jasmonic acid (JA) are the major phytohormones which play important roles in the regulation of plant growth and development, and management of variety of abiotic (water, cold, desiccation, and salinity), and biotic (pathogen) stresses (Clarke et al., 2009; Brossa et al., 2011; Rivas et al., 2011; Wang et al., 2011; Khan et al., 2015). They are also key factors in signaling pathway as well as important regulators of defense gene expression and immune response mechanisms (Adie et al., 2007; Koornneef and Pieterse, 2008; Solano and Gimenez-Ibanez, 2013). The slight change in ABA concentration can alter the antioxidative defense mechanism which lead to variation in oxidative damage intensity (Jiang and Zhang, 2001; Nayyar et al., 2005; Nayyar and Gupta, 2006; Duan et al., 2007; Rivas et al., 2011). The PSY is reported for feedback regulation of ABA biosynthesis (Welsch et al., 2008). Stress tolerance conferred by PSY alleles through regulation of ABA in rice, maize and cassava suggested their role in stress control mechanism (Li et al., 2008; Welsch et al., 2008; Arango et al., 2010). The β–cyclocitral produces by oxidation of the β-carotene has role in SA accumulation during the excess light stress (Lv et al., 2015). SA regulates the plant physiological functions and defense mechanism against abiotic stresses (Loake and Grant, 2007; Bari and Jones, 2009; Vlot et al., 2009; An and Mou, 2011). JA and its derivatives such as methyl jasmonate (MeJA) are reported to be involved in various plant biological activities including cell division, fruit growth and ripening (Creelman and Mullet, 1997). It also plays an important role in plant adaptation during abiotic stresses (Clarke et al., 2009; Yoon et al., 2009; Brossa et al., 2011; Nitschke et al., 2016).
Herein, we identified two contrasting cultivars of Indian banana i.e., Nendran and Rasthali with different levels of β-carotene in their fruit-pulp. The PSY was selected as a target gene for finding any correlation with their expression and accumulation of β-carotene in ripe fruit-pulp of contrasting cultivars. The present study also highlighted the differential expression patterns of three banana PSY (MaPSY1, MaPSY2, and MaPSY3) genes in different tissues and during various stress conditions.
Materials and Methods
Plant Materials, Experimental Conditions and Reagents
Ten banana accessions were selected from the field germplasm plot at the National Agri-Food Biotechnology Institute (NABI), Mohali for the carotenoids estimation in ripe fruit-pulp (Table 1). The fruit sampling was performed during the summer season. Plant tissues such as pseudostem (outer, and inner layers), leaf (young, and mature stages), bract, fruit finger, fruit-peel and -pulp (unripe, and ripe stages) were collected from the contrasting cultivars, frozen in liquid nitrogen, and stored at −80°C until further use. For abiotic stress treatments, in-vitro grown Rasthali plants were exposed with ABA (100 μM), MeJA (250 μM) and SA (250 μM), as described previously (Sreedharan et al., 2012; Pandey et al., 2016). The control and treated plants were grown in plant growth chambers (Conviron, Canada) at 26°C under 16/8-h light (200 μmol photon m−2 s−1)/dark period with 60 ± 5% relative humidity (RH). To study the effect of dark condition, plants were kept under the dark for 72 h in a plant growth chamber (Conviron, Canada) at 26°C with 60 ± 5% RH. Leaves were collected at different time intervals, frozen immediately in the liquid nitrogen, and stored at −80°C till further use. All biochemical, standards, kits and reagents unless otherwise stated were molecular biology or cell culture grade from Sigma Chemical Company (St. Louis, MO) and Merck (India).
Quantitative Estimation of Carotenoids
Carotenoid extraction was performed as described previously (Matsumoto et al., 2007; Flowerika et al., 2016) with some modifications. The lyophilized 1 g dry weight (DW) sample was homogenized in 50 ml solution containing 40% (v/v) methanol + 0.5 g magnesium carbonate and centrifuged. The insoluble fraction was extracted in diethyl ether and methanol (7:3, v/v) containing 0.1% (w/v) butylated hydroxy toluene (BHT) till the extract turned colorless. The extract was transferred in a separating funnel and 20 ml diethyl ether was added. The ether phase was washed three-times with 10 ml of saturated sodium chloride and 5 ml of 10% anhydrous sodium sulfate solution. The sample was evaporated using Rota vapor (Buchi Labortechnik AG) at 50°C at 100 rpm and 200 atmospheric pressures. The residue was dissolved in methyl tert-butyl ether (MTBE)-methanol (1:1, v/v) containing 0.1% (w/v) BHT, and centrifuged. The supernatant was filtered through 0.45-μm nylon membrane filter (Millipore, MA) and was used for High Performance Liquid Chromatography (HPLC). Analysis was performed in a liquid chromatograph binary gradient module pumps (Waters, Milford, MA, USA) equipped with photodiode array (Waters, 2998) and autosampler (Waters, 2767). Carotenoids were separated by using YMC C30 column (YMC, Kyoto, Japan). The mobile phase was a gradient, prepared from 95% (v/v) methanol in HPLC-grade water (component A) and MTBE (component B). Carotenoid standards (β-carotene, lutein, zeaxanthin and lycopene) were used for calibration curve, comparison of retention time, and quantification of samples. Data was analyzed using MassLynx™ software and quantification was carried out at 450 nm.
Measurement of Antioxidant Activity
To determine antioxidant activity, carotenoid extract was prepared from pulp tissue of Rasthali and Nendran as per procedure described above. Measurement of radical scavenging activity was carried out as per earlier reported 2,2-diphenyl-1-picrylhydrazyl (DPPH) method (Blois, 1958; Awasthi et al., 2016). Briefly, freshly prepared DPPH methanolic solution was added to 200 μl of methanol having different concentration of carotenoid extract. Reaction was incubated in dark for 30 min at room temperature. The absorbance was measured at 517 nm (UV-2700 UV-VIS Spectrophotometer, Shimadzu). Percent inhibition was calculated according to the following equation:
*AbDPPH: Absorbance of DPPH solution at 517 nm,
**AbS: Absorbance of DPPH solution containing extract at a particular concentration at 517 nm.
Experiment was performed in three replicates and ascorbic acid was used as positive control. IC50 values (half maximal inhibitory concentration) were calculated as the concentration of extracts causing 50% inhibition of DPPH radical. Lower IC50 value corresponds to a higher antioxidant activity of extract.
Identification and Sequence Analysis of MaPSY
The whole genome sequence of banana (Musa acuminata) was downloaded from the Banana Genome Hub database (D'Hont et al., 2012; http://banana-genome.cirad.fr/) and used for the identification of PSY gene. BLAST search was performed using annotated PSY sequences of maize (Zea mays), rice (Oryza sativa) and arabidopsis (Arabidopsis thaliana) to explore similar sequences in the banana genome database. The reported PSY gene sequences of banana were also considered in BLAST analysis. The most similar sequences were retrieved from the banana genome database and designated as putative MaPSY genes. The percentage identity of the predicted protein sequences was noted. Selected coding (CDS) and genomic DNA sequences were aligned to identify the gene structures, and numbers of exons and introns in the sequences. Theoretical isoelectric point and molecular weight were predicted using the Compute pI/MW tool on the ExPASy server (Bjellqvist et al., 1994; http://web.expasy.org/compute_pi/). Transmembrane domains were analyzed on the TMHMM server v 2.0 (Krogh et al., 2001; http://www.cbs.dtu.dk/services/TMHMM-2.0/) and subcellular localization of deduced proteins predicted on the TargetP 1.1 server (Emanuelsson et al., 2007; http://www.cbs.dtu.dk/services/TargetP/). The presence of chloroplast transit peptides was predicted using the ChloroP 1.1 server (Emanuelsson et al., 1999). Mapping of the biosynthetic genes on banana chromosome was done by using the locus ID information available on Banana Genome Hub (http://banana-genome.cirad.fr/). The upstream regions (1.5 kbp) of all selected genes were extracted through a custom Perl script for the identification of cis-regulatory elements. This region was considered as the proximal promoter for the individual gene. The promoter sequences were placed in the Plant Care Database (Lescot et al., 2002) in which a brief description of all motifs were extracted and presented.
RNA Isolation, cDNA Preparation, and Expression Analysis
Total RNA was isolated from different tissues (pseudostem, leaf, bract, fruit finger, fruit-peel, and -pulp), and treated samples (ABA, MeJA, SA, and dark) according to previously described protocol (Asif et al., 2006). RNA sample was treated with DNase I Digest kit (Sigma-Aldrich, USA) to eliminate DNA contamination. The cDNA synthesis was performed using Revert Aid First Strand cDNA synthesis Kit (Fermentas, USA) from 3 μg of DNA-free total RNA following the manufacturer's instructions.
The quantitative real-time PCR expression was carried out by following SYBR Green chemistry at ABI PRISM 7700 Fast Real-Time PCR System (Applied Biosystems, USA). The invariant expression under the described experimental conditions was normalized by using two banana housekeeping genes, actin1 (GenBank Accession No. AF246288) and ubiquitin2 (GenBank Accession No. HQ853254). The relative fold expression was calculated by using 2−ΔΔCT method (Schmittgen and Livak, 2008). The list of different primers used in the study is given in Supplementary Table S1.
Cloning, Protein Sequence and Phylogenetic Analyses
Three MaPSY sequences retrieved from Musa acuminata reference genome were used for designing the gene specific primers (Supplementary Table S1). The full-length open reading frames (ORFs) of PSY were amplified from the ripe fruit-pulp of contrasting cultivars (Nendran and Rasthali). The genes isolated from Nendran and Rasthali are designated as NEN-PSY1, NEN-PSY2, NEN-PSY3, and RAS-PSY1, RAS-PSY2, RAS-PSY3 respectively. The amplicons containing BglII and NotI restriction sites were cloned in pBluescript SK+ vector (Stratagene, USA) and confirmed by sequencing. The conserved motifs and sites for PSY functional activity were determined by using NCBI CD-Search (Marchler-Bauer et al., 2011; http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and ScanProsite (Hulo et al., 2008; http://prosite.expasy.org/scanprosite) tools. PSY protein sequence annotation analysis was accelerated using BioEdit sequence alignment editor (Hall, 1999; http://www.mbio.ncsu.edu/bioedit/bioedit). Phylogenetic analysis was conducted in MEGA6 (Tamura et al., 2013) for deduced protein sequences of arabidopsis, rice, maize and banana. The PSY sequences derived from Asupina (high provitamin A) and Cavendish (low provitamin A) banana cultivars (Mlalazi et al., 2012) were also considered in the study. Neighbor-joining method was used to infer homologs and evolution. Bootstrap value was set to 1000.
Functional Complementation Analysis of Six MaPSY in E. coli
A heterologous bacterial complementation system was used to investigate the biological function of six MaPSY proteins derived from Nendran (NEN-PSY1, NEN-PSY2, NEN-PSY3), and Rasthali (RAS-PSY1, RAS-PSY2, and RAS-PSY3). The plasmids pAC-85b and pTrc were used to test the functional activity of PSY genes (Cunningham and Gantt, 2007; Fu et al., 2014). The E. coli cells transformed with these plasmids could not be able to synthesize carotenoid which resulted white colonies. The PCR products of six MaPSY genes comprising BglII and NotI restriction sites (Supplementary Table S1) were cloned into digested pTrc plasmid and named as pTrc-NEN-PSY1, pTrc-NEN-PSY2, pTrc-NEN-PSY3, pTrc-RAS-PSY1, pTrc-RAS-PSY2, and pTrc-RAS-PSY3. The competent E. coli cells were co-transformed with pAC-85b and pTrc plasmid having PSY genes. A single positive colony was inoculated in Luria Broth (LB) medium containing ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) antibiotics and grown at 37°C with 200 rpm. The relative activity of MaPSY proteins could be observed as variation in the color of cultures due to differential accumulation of carotenoid. The β-carotene was extracted from the bacterial cell pellets (0.6 OD) and analyzed by HPLC by the method described earlier (Matsumoto et al., 2007; Flowerika et al., 2016). The extractions were repeated at least three times from independent transformed bacterial cell cultures.
Statistical Analysis
All experiments were repeated with three biological replicates and each experiment consisted of three technical replicates. The data are presented as mean ± SD and analyzed by Student's paired t-test. The mean values under each treatment were compared to determine significance (*P ≤ 0.05; **P ≤ 0.001; ***P ≤ 0.0001). The relationship between gene expression and individual carotenoid accumulation was calculated by Pearson correlation (r) analysis. R2 values were considered significant (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001) correlations. All statistical analyses were performed using Prism GraphPad software Ver. 5.01 (GraphPad Software Inc., San Diego, CA, USA).
Results
Carotenoid Profiling during Banana Fruit Development
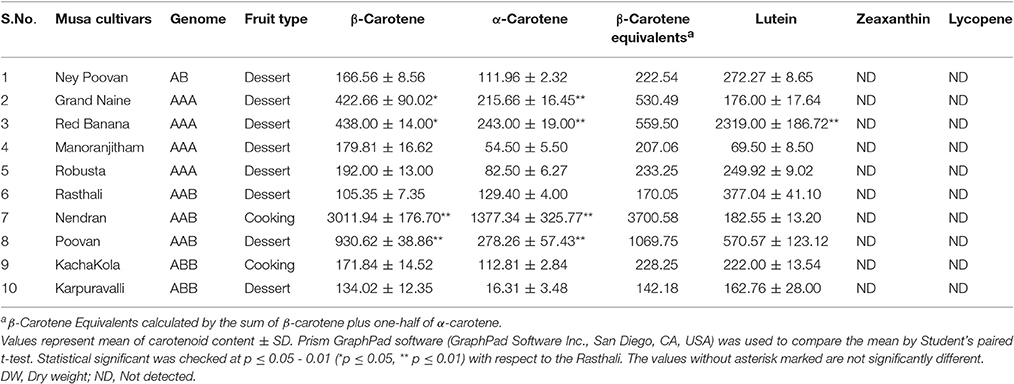
Fruit-pulp collected at the ripening stage of 10 Indian banana cultivars were lyophilized and analyzed for carotenoids. The highest β-Carotene Equivalents (β-carotene + ½ α-carotene) was observed in Nendran (3700.57 μg/100 g DW) while the lowest was found in Rasthali (170.05 μg/100 g DW) (Table 1). The ripe pulp of Rasthali appeared creamy-white and Nendran was deep-yellow in color (Figure 1A). Nendran and Rasthali cultivars were studied for changes in carotenoids in fruit-peel and -pulp as collected at unripe and ripe stages (Figure 1B). Lutein, α-carotene and β-carotene were quantified, whereas lycopene and zeaxanthin were not present in detectable amounts in the tissues. In the pulp tissue, the higher content of all the carotenoids was noticed at the ripening stage. Nearly 28-fold higher β-carotene was present in Nendran (3011.94 ± 176.70 μg/100 g DW) as compared to Rasthali (105.35 ± 7.35 μg/100 g DW) (Figure 1B). Similarly, α-carotene was nearly 10-fold higher in Nendran as compared to Rasthali. Lutein was nearly 2-fold higher in Rasthali as compared to Nendran. Unripe pulp of both Nendran and Rasthali showed significantly low quantities of all carotenoids, compared to the ripe pulp (Figure 1B, Supplementary Figure S2). However, in the peel as compared to the pulp of both the cultivars, higher deposition of all carotenoids (except at the ripening stage of Nendran) was noticed (Figure 1B, Supplementary Figure S3). The greater deposition of α-carotene and β-carotene was found in ripe pulp of Nendran as compared to its ripe peel (Figure 1B).
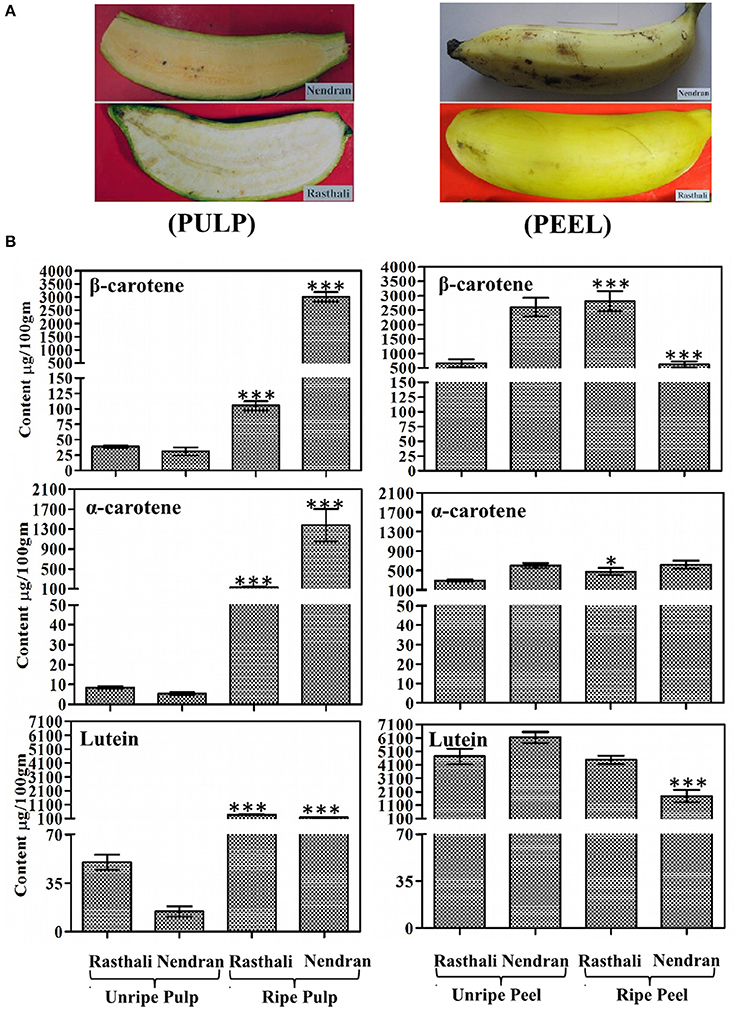
Figure 1. Qualitative and quantitative assessment of carotenoids in fruit tissues of Nendran and Rasthali banana cultivars. (A) Visual observation of ripe fruit-pulp and -peel. (B) Carotenoid profiling in ripe and unripe fruit tissues. Bars denote mean of carotenoid content ± SD. Compounds were quantified by separating extracts from the peel and pulp using HPLC. HPLC chromatograms are given in Supplementary Figures S2, S3. Statistical analysis was performed using Student's paired t-test. Statistical significance was checked at P ≤ 0.05–0.001 (*P ≤ 0.05; ***P ≤ 0.0001) with respect to the unripe- versus ripe-pulp/peel of Rasthali and Nendran.
Antioxidant Activity
In the present study, Nendran unripe/ripe pulp extracts showed the lower IC50 value in comparison to Rasthali unripe/ripe extract (Table 4). Ripe pulp extract of both Nendran and Rasthali had lower IC50 value as compared to the unripe pulp extract. The IC50 values for the extracts of unripe and ripe pulp of Nendran and Rasthali are given in Table 4. The IC50 value of all the extracts was lower than that of ascorbic acid (5.66 ± 0.86), which was used as positive control in the experiment.
In-silico Analysis of PSY in Banana
Three homologs of PSY (MaPSY1-3) were identified in Banana Genome Hub database using known proteins from arabidopsis, maize and rice. Percentage identity of the predicted protein sequences of the banana PSY genes showed high homology with those in arabidopsis, maize and rice varies from 55 to 80% (Supplementary Table S2). Structural analysis of exons and introns of PSY from banana and other species is presented in Supplementary Table S3. The PSY homologs from banana, maize and rice had same number of exons (6) and introns (5) however, arabidopsis PSY was found to be without intron. The accession numbers of the genes, their positions on banana chromosome, predicted polypeptide length, isoelectric point, molecular mass, subcellular localization and transmembrane helices (TMH) are given in Supplementary Table S4. MaPSY1 and MaPSY2 were located on the chromosomes 6 and 9, respectively, while MaPSY3 was present on the un_random chromosome (uncharacterized scaffold). The MaPSY1 and MaPSY2 proteins had similar length of 397 amino acids whereas MaPSY3 was larger in length and contained 426 amino acids. The isoelectric point (9.38), and molecular weight (47.76 kDa) of MaPSY3 was higher than MaPSY1 (9.22, and 45.03 kDa), followed by MaPSY2 (9.12, and 44.35 kDa). All the MaPSY showed to be localized in chloroplast. MaPSY1 has shown the presence of transmembrane helix.
Gene Expression in Different Tissues
Expression analysis of three PSY homologs (MaPSY1, MaPSY2, and MaPSY3) in different tissues of contrasting cultivars (Rasthali and Nendran) was performed (Figures 2, 3). The normalization of transcripts was carried out against actin1 gene that showed most stable expression in various tissues. In general, the expression pattern of MaPSY homologs was similar in both cultivars, but they expressed at varying levels in different tissues. In the pseudostem of both cultivars, higher expression of MaPSY3 was detected in inner and outer layer (Figures 2A,B). The lower expression of MaPSY1 and MaPSY2 was observed in inner and outer layer of both cultivars, respectively. Two leaf developmental stages i.e., young and mature leaves were taken for the analysis (Figures 2A,B). The higher transcript level of MaPSY3 and MaPSY1 was noted in young leaf of Nendran and Rasthali, respectively. Mature leaf of both cultivars has shown higher expression of MaPSY1. The lower expression of MaPSY2 was observed in young and mature leaf of both cultivars. Bract and fruit fingers of both cultivars showed higher expression of MaPSY3. The lower expression of MaPSY2 and MaPSY1 was noted in bracts and fruit fingers of both cultivars, respectively (Figures 2A,B). These results suggest differential expression of MaPSY genes among different tissues as well as between the contrasting cultivars. The analysis indicated that all MaPSY gene homologs were highly expressed in leaf tissue of both cultivars (Figures 2A,B).
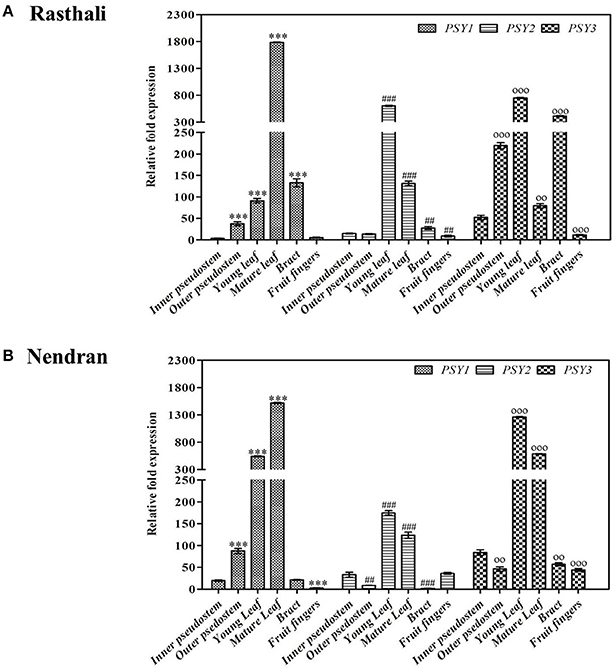
Figure 2. Spatiotemporal real-time PCR expression analysis of PSY genes in banana vegetative tissues. Transcript expression profiles are presented in pseudostem (inner, and outer), leaf developmental stages (young, and mature), bract, and fruit fingers of Rasthali (A), and Nendran (B). The gene expression was normalized with reference to actin1 taken as internal control. Bars denote mean fold expression as compared to the lowest expressing gene in the group of samples ± SD. Statistical analysis was performed using Student's paired t-test. Statistical significance was checked at P ≤ 0.05–0.001 and the symbols on the top of bars represents significance levels (##, oo, P ≤ 0.001; and ***, ###, ooo, P ≤ 0.0001) with respect to inner pseudostem in each group.
Figure 3. Spatiotemporal real-time PCR expression analysis of PSY genes in banana fruit tissues. Transcript expression profiles are presented in unripe/ripe -pulp (A) and -peel (B) of Rasthali and Nendran cultivars. The gene expression was normalized with reference to actin1 taken as internal control. Bars denote mean fold expression as compared to the lowest expressing gene in the group of samples ± SD. Statistical analysis was performed using Student's paired t-test. Statistical significance was checked at P ≤ 0.05–0.001 (**P ≤ 0.001; ***P ≤ 0.0001) with respect to the unripe- versus ripe-pulp/peel of Rasthali and Nendran.
To understand the transcriptional regulation in the carotenogenesis in fruit tissues of two contrasting cultivars, expression of MaPSY homologs was studied in fruit-pulp (Figure 3A) and -peel (Figure 3B) at two developmental stages (unripe and ripe). The homologs were expressed in all fruit tissues of both cultivars, except MaPSY2. The transcript of MaPSY2 was not detected in unripe peel of Rasthali. In the unripe pulp, higher expression of MaPSY1 and MaPSY2 was noticed in Rasthali and Nendran, respectively. Whereas in ripe pulp, high transcript level of MaPSY2 and MaPSY1 was observed in Rasthali and Nendran, respectively (Figure 3A). In the peel at unripe stage, higher expression of MaPSY3 was recorded in Rasthali as well as in Nendran (Figure 3B). The higher expression of MaPSY1 was observed at the ripe stage of peel in both cultivars.
Relationship between MaPSY Expression and Carotenoid Accumulation in Fruit Tissues
Pearson correlation analysis was carried out between relative fold expression of MaPSY transcripts and individual carotenoid content in fruit-pulp (Table 2) and -peel (Table 3) in the contrasting cultivars. In the Nendran pulp, significant positive correlation was observed for NEN-PSY1 (r = 0.9995, P < 0.05) and NEN-PSY2 (r = 0.9978, P < 0.05) with β-carotene and α-carotene, respectively. No significant correlation was noted for lutein in Nendran pulp. In Rasthali pulp, significant positive correlation was noted for RAS-PSY2 (r = 1.00, P < 0.005) and RAS-PSY1 (r = 0.9973, P < 0.05) with β-carotene. RAS-PSY2 (r = 0.9969, P < 0.05) showed significant positive correlation with α-carotene. Lutein accumulation in Rasthali showed significant positive correlation with both RAS-PSY1 (r = 0.9983, P < 0.05) and RAS-PSY2 (r = 0.9998, P < 0.05). In the Nendran peel, significant positive correlation was established for NEN-PSY1 (r = 1.00, P < 0.005) with β-carotene whereas, negative correlation was observed for NEN-PSY1 (r = −0.9997, P < 0.05) with lutein. No correlation was observed for α-carotene in Nendran. In the Rasthali peel, significant positive correlation was established for RAS-PSY1 with lutein (r = 0.9994, P < 0.05). However, with β-carotene significant negative correlation was noted for RAS-PSY2 (r = −0.999, P < 0.05). Besides, no significant correlation (r = 0.8262 to 0.9945, P ≥ 0.1031 to ≤ 0.381) was found between carotenoids and PSY3 homolog of both cultivars in pulp and peel (Table 2).
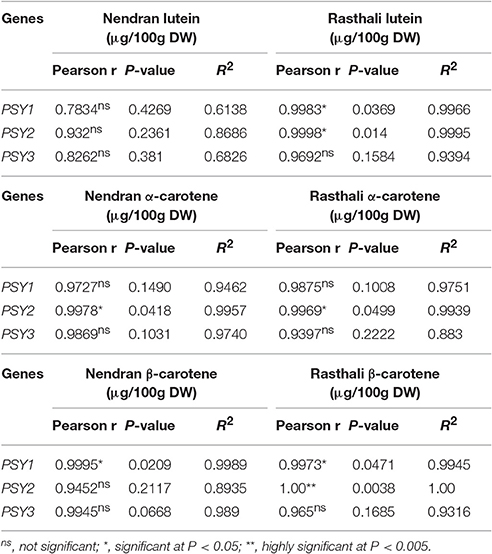
Table 2. Correlation of PSY expression with the accumulation of individual carotenoids in fruit-pulp of Nendran and Rasthali.
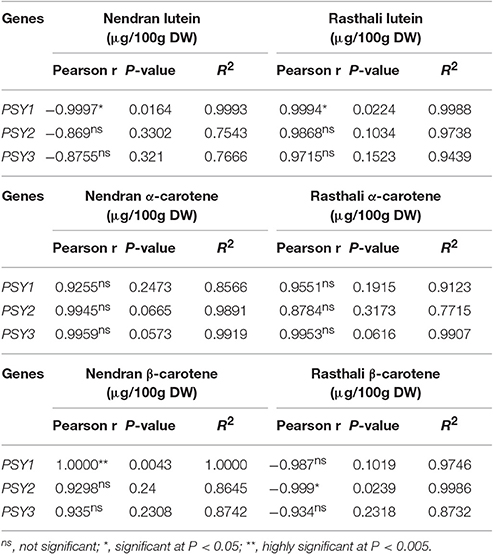
Table 3. Correlation of PSY expression with the accumulation of individual carotenoids in fruit-peel of Nendran and Rasthali.
Promoter Analysis and MaPSY Expression under Stress Conditions
Various cis-regulatory elements were detected in the proximal promoter regions of three MaPSY genes (Supplementary Table S5). The cis-elements such as Skn-1 motif related to growth and development, Box4, and Sp1 motifs related to light response were present in all three MaPSY genes. The light responsive elements were mostly found in MaPSY1 and MaPSY3 while higher number stress responsive motifs were detected in MaPSY1 and MaPSY2, which suggested their possible role in these conditions.
To understand the effect of various environmental factors on transcriptional regulation of MaPSY genes, expression analysis was performed following different treatments in Rasthali plants. The differential expression of all three MaPSY genes was observed during the dark exposure to the plants. The analysis reveals that the dark exposure leads to significantly down-regulated expression of MaPSY1 and MaPSY3 genes (Figure 4A). Interestingly, nearly 26- and 7.5- fold up-regulated expression of MaPSY2 was noted after 12 and 48 h, respectively of incubation under the dark as compared to control (0 h) plants (Figure 4A). The expression of MaPSY2 was comparable to control (0 h) at 72 h. The expression patterns suggest a positive correlation with the presence of high number light-responsive cis-elements in MaPSY1 and MaPSY3 promoter regions.
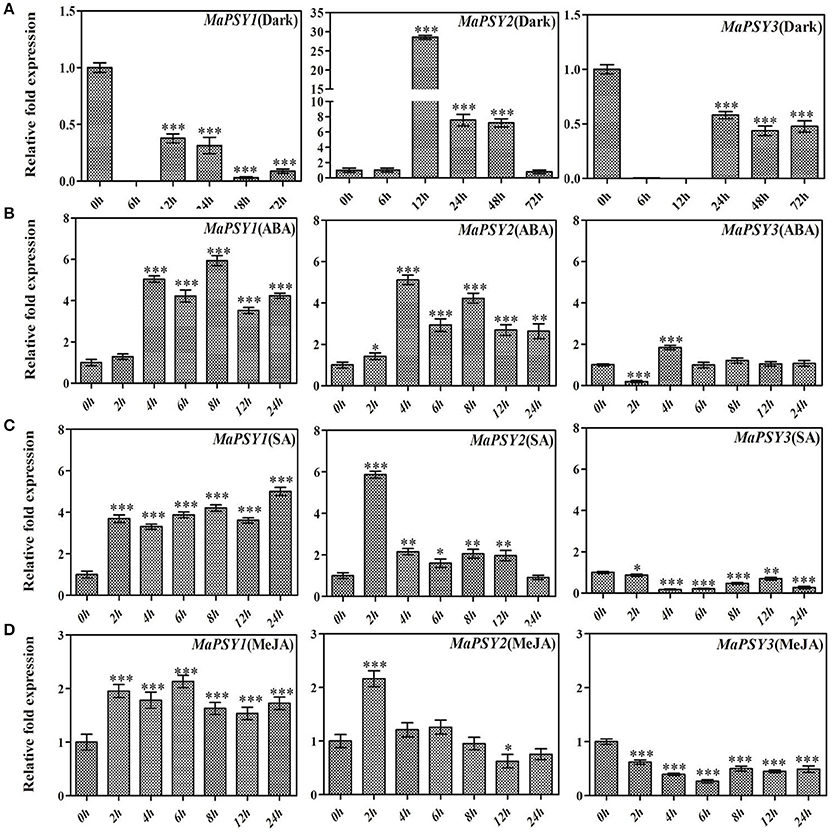
Figure 4. Real-time PCR expression analysis of PSY genes under stress conditions. Transcript expression profiles are presented in in-vitro grown Rasthali plants under (A) dark, (B) methyl jasmonate, (C) abscisic acid, and (D) salicylic acid exposures. The gene expression was normalized with reference to actin1 taken as internal control. Bars denote mean fold expression as compared to the control (0 h sample). Statistical analysis was performed using Student's paired t-test. Statistical significance was checked at P ≤ 0.05–0.001 (*P ≤ 0.05; **P ≤ 0.001; ***P ≤ 0.0001) with respect to the 0 h sample.
The exogenous applications of ABA, SA and MeJA have modulated expression of MaPSY genes (Figures 4B–D). ABA exposure led to the significant enhancement in the expression of all three MaPSY genes at 4 h, but the differential modulation was noted for different genes at different time points (Figure 4B). MaPSY1 and MaPSY2 showed a significant up-regulation in the expression at 4 h which continued till 72 h. In case of MaPSY3, biphasic expression pattern was observed (Figure 4B). ABRE motif responsible for ABA response was solely present in the promoter region of MaPSY2 gene.
Exposure of SA led to the significant up-regulation in expression of MaPSY1 at 2 h and continued till 24 h. On the other hand, MaPSY2 transcript was found to be up-regulated at 2 h and then reduced to comparable levels of control (Figure 4C). MaPSY3 showed significant down-regulation in expression at different time points after treatment. Interestingly, TCA element that is responsible for SA response has found in three copies at the proximal promoter region of MaPSY1 whereas it was absent in MaPSY2 promoter. A single copy of TCA element was present in MaPSY3 promoter region.
The exogenous exposure of MeJA showed nearly 2-fold up-regulation in expression of MaPSY1 at 2 h which continued till 24 h (Figure 4D). MaPSY2 expression was up-regulated at 2 h and afterwards expression was comparable to the control (Figure 4D). The significant down-regulation in expression of MaPSY3 was found at different time points as compare to control. MeJA responsive elements (CGTCA and TGACG motifs) were present in the proximal promoter regions of MaPSY1 and MaPSY2, but absent in MaPSY3. These results suggest that MaPSY genes expression in response to different treatments largely correlates with the presence of cis-regulatory elements in their promoter regions.
Full-Length Cloning and Phylogenetic Analysis of PSY from Contrasting Cultivars
Full-length gene sequences of Nendran (NEN-PSY1, NEN-PSY2, and NEN-PSY3), and Rasthali (RAS-PSY1, RAS-PSY2, and RAS-PSY3) were amplified for characterization. The sequencing analysis revealed that all the homologs of PSY in both the cultivars shares considerable (≥95%) homology at both nucleotide and protein levels. The phylogenetic relationship of MaPSY proteins with known banana and other plant species was constructed using neighbor joining method. Three MaPSY identified from each of Rasthali (RAS-PSY1, RAS-PSY2, and RAS-PSY3), and Nendran (NEN-PSY1, NEN-PSY2, and NEN-PSY3) showed general trend of clustering with other monocots (Figure 5). MaPSY homologs shared three distinct clades with other known banana PSY sequences. RAS-PSY1 and NEN-PSY1 were grouped into a clade with PSY2 proteins of Asupina (APSY2b) and Ladyfinger (LfPSY2). On the other hand, RAS-PSY2 and NEN-PSY2 grouped in a clade with PSY2a proteins of Asupina and Cavendish banana varieties. RAS-PSY3 and NEN-PSY3 were clustered in a different clade with PSY1 proteins of Asupina, Cavendish and Ladyfinger.
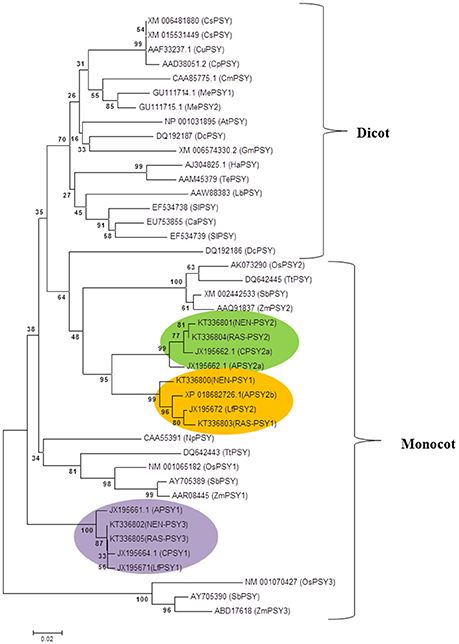
Figure 5. Phylogenetic tree of PSY proteins. Neighbor joining based phylogenetic tree of the deduced PSY protein sequences of Nendran (NEN-PSY1 - 3) and Rasthali (RAS-PSY1 - 3) inferred with different plant species. The tree was bootstrapped to 10,000 trials, and bootstrapping values are shown at each node of the rooted rectangular cladogram. The GenBank accession numbers for different PSY sequences were given in the Supplementary Table S6.
Motif Analysis for MaPSY Functional Activity
The deduced amino acid sequences of Nendran (NEN-PSY1-3) and Rasthali (RAS-PSY1-3) were compared to determine if differences in sequences could account for the differences in fruit carotenoids content between these two cultivars of banana (Figure 6). All PSY proteins contain a predicted chloroplast transient peptide at the N-terminal region. Conserved domain analysis revealed that all the six MaPSY proteins belong to class 1 superfamily of isoprenoid biosynthesis enzymes containing the conserved trans-isoprenyl diphosphate synthases, head-to-head (trans-IPPS_HH) domain. This domain has the binding sites for substrate and cofactor that catalyzes the production of phytoene by condensation of two molecules of GGPP (Geranyl geranyl pyrophosphate). ScanProsite analysis showed the presence of squalene/phytoene synthase signatures 1 and 2 (SQS-PSY 1 and 2) in all MaPSY proteins for PSY activity. The SQS-PSY1 motif (YCyyVAGTVGlmSvpV) and aspartate-rich motifs (ARM) (DELVD and DVGED) were found conserved in each PSY protein of Nendran and Rasthali (Figure 6). This observation was further confirmed by alignments with PSY sequences of several other plant species (Supplementary Figure S4). We noted that PSY1 of both the cultivars were 95% similar and differed by a total of 19 amino acid residues which included 4 in the transit peptide and 15 in the mature protein. Among these amino acid substitutions, four located in evolutionarily conserved domains of the mature protein. The substitutions from threonine to asparagine (N149T) and tyrosine to cysteine (C398Y) in the active site lid motifs (YAKTF and RAYV), valine to isoleucine (I292V) in the SQS-PSY2 (LGianQltNIlRDVgeDarrgRiYlP), and the tyrosine to aspartic acid (D169Y) in the GGPP substrate binding pocket were detected in the RAS-PSY1 (Figure 6). PSY2 and PSY3 of both the cultivars showed nearly 99% similarities and differed by 5 and 6 amino acids, respectively. The alanine (A307) and isoleucine (I312) residues were present in SQS-PSY2 motif of the PSY1 while these were changed by serine (S307A) and valine (V312I) in PSY2 of both the cultivars. The changes of lysine to glutamine (Q148K) in active site lid-motif and isoleucine to leucine (L292I) in SQS-PSY2 motif were found in the NEN-PSY3 protein (Figure 6).

Figure 6. Analysis of banana PSY protein sequences. Multiple sequence alignment of putative protein sequences of PSY from Rasthali (RAS-PSY1 - 3) and Nendran (NEN-PSY1 - 3) cultivars.
Functional Complementation Analysis
To determine whether the six MaPSY (NEN-PSY1, NEN-PSY2, NEN-PSY3, RAS-PSY1, RAS-PSY2, and RAS-PSY3) proteins have functional activity, the ORF of each gene was cloned into the pTrc plasmid and co-transformed along with the pAC-85b into E. coli TOP10F. The visual observation suggested that co-transformed cells accumulated intense yellow color as compared to the control vector pAC-85b (no color) (Figure 7A), indicating that all six MaPSY accelerate the accumulation of β-carotene but with different levels of enzymatic activity. The pigments accumulated in the transformed E. coli cells were extracted and analyzed by HPLC. The expected product, β-carotene was confirmed by matching spectra and retention time. The highest β-carotene content was found in the bacteria transformed with pTrc-NEN-PSY1 (410 ± 15.45 μg/100 ml culture) while the lowest was recorded with pTrc-RAS-PSY3 (187 ± 9.59 μg/100 ml culture) (Figure 7B). The β-carotene was also not detected in individually transformed pTrc-NEN-PSY1–3 and pTrc-RAS-PSY1–3 plasmid controls. These results established that all six MaPSY proteins were enzymatically functional and could accelerate the biosynthesis of β-carotene at different levels.
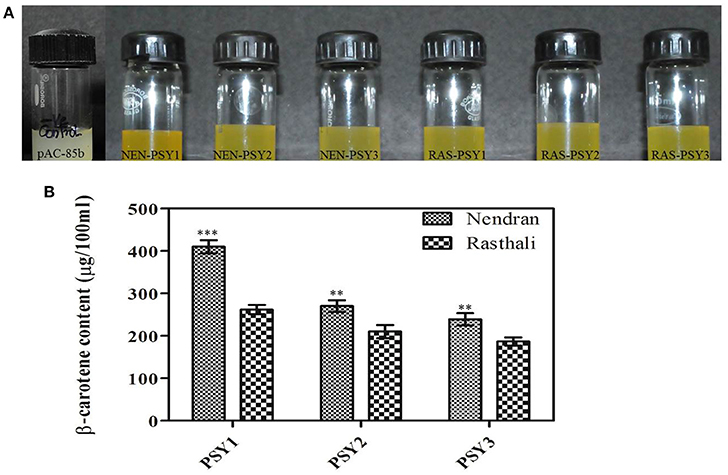
Figure 7. Functional complementation analysis. (A) Color phenotype of E. coli cultures consisting of pAC-85b (control) and complemented with plasmids containing PSY homologs (NEN-PSY1-3 and RAS-PSY1-3). (B) Concentration of β-carotene in E. coli cells after complementation with plasmids pAC-85b and pTrc containing PSY1-3 of Nendran and Rasthali. Bars denote mean of β-carotene content ± SD. Statistical significance was checked at P ≤ 0.05–0.001 (** significant; *** highly significant) with respect to the Rasthali (PSY1-3).
Discussion
Banana is a lucrative target to be developed as a rich source of dietary carotenoid with improved agronomic traits through intervention of genetic tools. The role of PSY in the carotenoid biosynthesis (Sandmann et al., 2006; Cazzonelli and Pogson, 2010; Grassi et al., 2013; Fu et al., 2014) and stress tolerance (Li et al., 2008; Welsch et al., 2008; Arango et al., 2010) has been studied in several plants species. Therefore, an insight into the PSY genes involved in carotenoid biosynthesis and stress management in banana is essential for strategically selecting suitable homolog that could be utilized in the precise metabolic engineering program. In the present study, we analyzed the expression pattern of MaPSY homologs in different vegetative and fruit tissues. The expression of MaPSY in response to different environmental stimuli was also correlated with the presence of cis-acting elements in their promoter. The correlation between the levels of expression of MaPSY genes and the carotenoid content was examined to predict the homolog likely to elevate β-carotene in banana fruit pulp.
The homology and gene structure analyses suggested that the identified MaPSY genes have functional homologs of closely related monocots and arabidopsis. The multiple paralogs for PSY were found at various locations on different chromosomes in the banana genome (Supplementary Table S4). The existence of gene paralogs in banana is mainly attributed to duplication events during evolution and suggests distinct as well as overlapping roles (D'Hont et al., 2012). The banana lineage, after separation from the common ancestors of banana and poales might have experienced three whole genome duplication events, as opposed to the two in the case of poales lineage (Pandey et al., 2016).
Our phytochemical screening of different cultivars suggested that Nendran and Rasthali are the contrasting cultivars in terms of β carotene in fruit pulp (Table 1). The deep-yellow color observed in the ripe pulp of Nendran also suggested high level accumulation of carotenoids. The regulatory mechanisms leading to carotenoid synthesis and accumulation can be affected by transcriptional and/or post-transcriptional levels. The carotenoid profiling and gene expression analysis in fruit-peel and -pulp at two developmental stages (unripe and ripe) presented here gave information into transcriptional regulation of carotenoid biosynthesis in banana. The ripening induced biosynthesis of carotenoid was observed in pulp tissue in both the cultivars. However, such pattern was not noticed in the peel. Hence, their regulation in different tissues was controlled by different developmental cues. Numerous reports have highlighted antioxidant property of carotenoids (El-Agamey et al., 2004; Das and Roychoudhury, 2014; Aquino et al., 2016; García-Ruiz et al., 2017). Radical scavenging assay (inhibition of DPPH) showed ripe pulp extract has significantly higher antioxidant activity as compared to the unripe pulp. This observation is in agreement with Aquino et al. (2016). Our results suggested that the content of carotenoids in ripe pulp of Nendran is directly correlated with higher antioxidant activity (Table 4). Transcriptional regulation is an important mode to control carotenoid biosynthesis in banana fruit, as noticed by us and also in agreement with previous studies on other fruit crops such as watermelon (Grassi et al., 2013), grape (Young et al., 2012), and tomato (Giorio et al., 2008; Smita et al., 2013) where carotenoid content increases during ripening stage. The PSY encoding enzymatic step has been characterized as the first committed and rate-limiting step toward carotenoid biosynthesis (Bouvier et al., 2005; Sandmann et al., 2006; Cazzonelli and Pogson, 2010; Fu et al., 2014; Liu et al., 2014). Differential transcript expression of the three PSY genes in pulp of Nendran and Rasthali may explain the difference in carotenoids content in these two cultivars. The expression analysis revealed that PSY1 was highly expressed in the ripe pulp of Nendran. This could be responsible for high carotenoids accumulation similar to the findings of Giorio et al. (2008) who reported the same in tomato ripening.
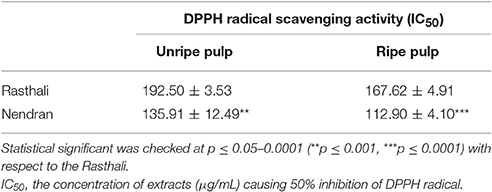
Table 4. Measurement of antioxidant activity in extracts prepared from pulp of Nendran and Rasthali.
The expression analysis in different tissues indicated that MaPSY homologs were expressed at varying levels which suggest their tissue specific role in banana. The similar trend of PSY expression was reported in other plant species (Grassi et al., 2013; Smita et al., 2013). The up-regulation in expression of MaPSY1 in developmental leaf stages suggests its role in growth and development.
Cis-acting elements known to participate in various functions in promoter region of genes suggest that they could be coordinately regulated by multiple transcription factors and appears to be modulated by multiple exogenous and endogenous factors (Pandey et al., 2016). The promoter analysis indicated that the light responsive elements such as AE box and ATCT motif were exclusively absent in MaPSY2 that is correlated with a positive modulation in the absence of light. Such up-regulation of MaPSY2 expression suggests it salient role during the dark condition as earlier reported for the Maize PSY1 (Li et al., 2008).
ABA is a cis-carotenoid metabolite that can alter the production of carotenoids (Kachanovsky et al., 2012). We found that ABRE, myb binding site (MBS) and G-box are responsible for ABA response were located in the promoter region of MaPSY2 suggested it possible role in defense mechanism. Enhanced expression of MaPSY1 and MaPSY2 genes was observed during ABA treatment, although ABRE element was absent in proximal promoter of PSY1. This might be due to ABA induced production of ethylene (Zhang et al., 2009a,b) and many other metabolites (Jeong et al., 2004; Giribaldi et al., 2010) those have significant role in fruit ripening. Moreover, ABA is a cis-carotenoid metabolite can alter the production of carotenoids which are precursors for the ABA biosynthesis (Kachanovsky et al., 2012).
SA protects the chloroplast membrane and enhances the antioxidant capacity of the plant (Huang et al., 2016) under stresses. Furthermore, MeJA has been reported to enhance the β-carotene content in broccoli sprouts (Natella et al., 2016) and in mango (Muengkaew et al., 2016). Impositions of SA and MeJA on banana plantlets showed positive modulation of MaPSY1 with respect to other genes, delineating its role in plant development, physiology and antioxidant defense mechanism.
The functional protein encoded by PSY should include the Lid, ARM, and SQS-PSY evolutionarily conserved domains which are responsible for substrate binding and the catalytic activity (Mlalazi et al., 2012; López-Emparán et al., 2014). Mutation of critical amino acid residues in PSY changes the functionality of the mature protein (Howitt et al., 2009; Welsch et al., 2010; Gady et al., 2012; Mlalazi et al., 2012; Shumskaya et al., 2012; Nogueira et al., 2013), thus impacting on carotenoid biosynthesis.
All three PSY1-3 sequences of Nendran and Rasthali contain these conserved domains. However, in-depth sequence analysis revealed that NEN-PSY1, NEN-PSY2 and NEN-PSY3 are differed from RAS-PSY1, RAS-PSY2, and RAS-PSY3, in 19, five and six amino acids, respectively. These differences may play an important role in the carotenoid content between the two cultivars. Threonine and tyrosine are reported to be more conserved amino acids in functional active domains of PSY in several plant systems (Supplementary Figure S4). The change in active site lid motif threonine to asparagine (N149T) and tyrosine to cysteine (C398Y), SQS-PSY2 motif valine to isoleucine (I292V) and the replacement of tyrosine residue with aspartic acid (D169Y) involved in the catalytic activity as well as substrate binding might result in alteration of catalytic activity and substrate binding efficiency of the enzyme encoded by RAS-PSY1. Substitutions in PSY2 and PSY3 in their SQS-PSY2 and active site lid motif may also lead to variation in the carotenoid content found in Nendran and Rasthali. The similar observation was noticed by Mlalazi et al. (2012) where the differences in the 10 amino acid sequences of Cavendish and Asupina PSY2a showed upto 20-fold higher β-carotene content in Asupina fruit. Moreover, differences in amino acids at different positions in the putative peptide may affect the structural properties of protein by altering interactions between the residues adjacent to each other. However, the impact of different amino acid substitutions on PSY activity requires further characterization.
We analyzed and compared the functional activities of all six MaPSY using a bacterial complementation system. Similar system has already been used to demonstrate the function of different carotenoid pathway genes from hazel (Wang et al., 2007), papaya (Devitt et al., 2010), jatropha (Lin et al., 2010), grapevine (Young et al., 2012), and loquat (Fu et al., 2014). We observed significantly higher activity with NEN-PSY1 compared to the other homologs suggesting its role in the carotenoid biosynthesis (Figure 5). Differential accumulation of β-carotene in bacterial system transformed with PSY could be affected by the variety of factors including catalytic activity, protein localization, stability, folding, solubility and/or the differences in amino acid sequence found between the different homologs. Hence, it would be needed to confirm the functional activity of banana PSY in plant. Further, it also remains to be seen if carotenoid degrading enzymes such as carotenoid cleavage dioxygenases (CCDs), β-ring hydroxylases (BCHs) and 9-cis carotenoid cleavage dioxygenases (NCEDs) play any critical role in carotenoid accumulation in banana fruit.
Accession Numbers
Sequence data from this article have been deposited in the GenBank data libraries under accession numbers: NEN-PSY1 (KT336800), NEN-PSY2 (KT336801), NEN-PSY3 (KT336803), RAS-PSY1 (KT336804), RAS-PSY2 (KT336805), and RAS-PSY3 (KT336807).
Author Contributions
NK, AP, and ST conceived the idea and designed research experiments. ST supervised the research. NK, AP, SS, PK, and PP performed the experiments related to plants. NK, AP, PK, and AK performed phytochemical analysis. NK and PA performed antioxidant activity. NK, AP, and SM performed bioinformatics analysis. NK, AP, and ST analyzed data, prepared the figures and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors express their gratitude to National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology (Government of India) and Biotechnology Industry Research Assistance Council (BIRAC) (A Government of India Enterprise) for the research support on provitamin A biofortification work in Indian banana. NK and SS are thankful to Department of Biotechnology, Panjab University, Chandigarh for Ph.D. support and facility. We thank Dr. M. Sabu (University of Calicut) and Dr. K. Soorianatha Sundaram (Tamil Nadu Agricultural University, Coimbatore) for providing banana germplasm for establishment at NABI.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00462/full#supplementary-material
Supplementary Figure S1. Schematic representation of pathways for biosynthesis of MVA, MEP and carotenoids in higher plants. Enzymatic reactions are represented by arrows; dashed lines represent multiple enzymatic steps. Enzymes: HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy3-methylglutaryl-CoA reductase; DXS, 1- deoxy-D-xylulose-5-phosphate synthase; DXR, 1- deoxy-D-xylulose-5-phosphate reductase; HDR, 1-hydroxy-2-methyl-2-(E)-butenyl-4diphosphate reductase; GGPP synthase, geranylgeranyl diphosphate synthase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; LCYB, lycopene β-cyclase; LCYE, lycopene ε-cyclase; CRTISO, carotenoid isomerase; BCH, β-ring hydroxylase, ECH, ε-ring hydroxylase. Compounds: HMG-CoA, 3-hydroxy-3methylglutaryl-CoA; MVA, mevalonic acid; GAP, glyceraldehyde-3-phosphate; DOXP, 1deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; HMBPP, 1hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGPP, geranylgeranyl diphosphate.
Supplementary Figure S2. Representative HPLC profiles illustrating the accumulation of carotenoids in fruit-pulp of banana. Peaks in chromatograms are representing 1: lutein, 2: zeaxanthin, 3: α-carotene, 4: β-carotene and 5: lycopene. Extract was separated with HPLC coupled with UV and PDA detector.
Supplementary Figure S3. Representative HPLC profiles illustrating the accumulation of carotenoids in fruit-peel of banana. Peaks in chromatograms are representing 1: lutein, 2: zeaxanthin, 3: α-carotene, 4: β-carotene and 5: lycopene. Extract was separated with HPLC coupled with UV and PDA detector.
Supplementary Figure S4. Analysis of PSY protein sequences. Multiple sequence alignment of putative protein sequences of PSY1 of Rasthali and Nendran along with different monocots PSY sequences. The GenBank accession numbers for different PSY sequences were given in the Supplementary Table S6.
Supplementary Table S1. List of the primers used in the present study.
Supplementary Table S2. Percentage identity of the predicted protein sequence of MaPSY.
Supplementary Table S3. Gene families and the exons and introns numbers in MaPSY homologs.
Supplementary Table S4. Structural features of three PSY homologs in banana.
Supplementary Table S5. Cis-regulatory elements in the promoter of MaPSYgenes.
Supplementary Table S6. PSY protein sequences used for phylogenetic tree analysis in Figure 5.
References
Adie, B. A., Pérez-Pérez, J., Pérez-Pérez, M. M., Godoy, M., Sánchez-Serrano, J. J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defences in Arabidopsis. Plant Cell 19, 1665–1681. doi: 10.1105/tpc.106.048041
Ampomah-Dwamena, C., Driedonks, N., Lewis, D., Shumskaya, M., Chen, X., Wurtze, E. T., et al. (2015). The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol. 15:185. doi: 10.1186/s12870-015-0573-7
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integ. Plant. Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
Aquino, C. F., Salomã, O. L. C. C., Ribeiro, S., Rocha, M., Siqueira, D. L. D., and Cecon, P. R. (2016). Carbohydrates, phenolic compounds and antioxidant activity in pulp and peel of 15 banana cultivars. Rev. Bras. Fruticultura. 38:e-090. doi: 10.1590/0100-29452016090
Arango, J., Wüst, F., Beyer, P., and Welsch, R. (2010). Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 232, 1251–1262. doi: 10.1007/s00425-010-1250-6
Asif, M., Trivedi, P., Solomos, T., and Tucker, M. (2006). Isolation of high-quality RNA from apple (Malus domestica) fruit. J. Agric. Food Chem. 54, 5227–5229. doi: 10.1021/jf053137n
Awasthi, P., Mahajan, V., Jamwal, V. L., Kapoor, N., Rasool, S., Bedi, Y. S., et al. (2016). Cloning and expression analysis of chalcone synthase gene from Coleus forskohlii. J. Genet. 95, 647–657. doi: 10.1007/s12041-016-0680-8
Bari, R., and Jones, J. D. G. (2009). Role of plant hormones in plant defence response. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Bjellqvist, B., Basse, B., Olsen, E., and Celis, J. E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539. doi: 10.1002/elps.1150150171
Blois, M. S. (1958). Antioxidant determinations by the use of a stable free radical. Nature 181, 1199–1200.
Bouvier, F., Rahier, A., and Camara, B. (2005). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44, 357–429. doi: 10.1016/j.plipres.2005.09.003
Brossa, R., López-Carbonell, M., Jubany-Mar,í, T., and Alegre, L. (2011). Interplay between abscisic acid and jasmonic acid and its role in water-oxidative stress in wild-type, ABA-deficient, JA-deficient, and ascorbate-deficient Arabidopsis plants. J. Plant Growth Regul. 30, 322–333. doi: 10.1007/s00344-011-9194-z
Busch, M., Seuter, A., and Hain, R. (2002). Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol. 128, 439–453. doi: 10.1104/pp.010573
Cazzonelli, C. I., and Pogson, B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15, 266–274. doi: 10.1016/j.tplants.2010.02.003
Clarke, S. M., Cristescu, S. M., Miersch, O., Harren, F. J., Wasternack, C., and Mur, L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182, 175–187. doi: 10.1111/j.1469-8137.2008.02735.x
Clotault, J., Peltier, D., Berruyer, R., Thomas, M., Briard, M., and Geoffriau, E. (2008). Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 59, 3563–3573. doi: 10.1093/jxb/ern210
Creelman, R. A., and Mullet, J. E. (1997). Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9, 1211–1223.
Cunningham, F. X. Jr., and Gantt, E. (2007). A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosyn. Res. 92, 245–259. doi: 10.1007/s11120-007-9210-0
D'Hont, A., Denoeud, F., Aury, J. M., Baurens, F. C., Carreel, F., Garsmeur, O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. doi: 10.1038/nature11241
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
Devitt, L. C., Fanning, K., Dietzgen, R. G., and Holton, T. A. (2010). Isolation and functional characterization of a lycopene β-cyclase gene that controls fruit colour of papaya (Carica papaya L.). J. Exp. Bot. 61, 33–39. doi: 10.1093/jxb/erp284
Duan, B., Yang, Y., Lu, Y., Korpelainen, H., Berninger, F., and Li, C. (2007). Interactions between water deficit, ABA, and provenances in Picea asperata. J. Exp. Bot. 58, 3025–3036. doi: 10.1093/jxb/erm160
El-Agamey, A., Lowe, G. M., McGarvey, D. J., Mortensen, A., Phillip, D. M., Truscott, T. G., et al. (2004). Carotenoid radical chemistry and antioxidant/pro-oxidant properties. Arch. Biochem. Biophys. 430, 37–48. doi: 10.1016/j.abb.2004.03.007
Emanuelsson, O., Brunak, S., Von, Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoco. 2, 953–971. doi: 10.1038/nprot.2007.131
Emanuelsson, O., Nielsen, H., and VonHeijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Prot. Sci. 8, 978–984.
Flowerika Alok, A., Kumar, J., Thakur, N., Pandey, A., Pandey, A. K., Upadhyay, S. K., et al. (2016). Characterization and expression analysis of phytoene synthase from bread wheat (Triticum aestivum L.). PLoS ONE 11:e0162443. doi: 10.1371/journal.pone.0162443
Flores-Perez, U., Perez-Gil, J., Rodriguez-Villalon, A., Gil, M. J., Vera, P., and Rodriguez-Concepcion, M. (2008). Contribution of hydroxymethylbutenyl diphosphate synthase to carotenoid biosynthesis in bacteria and plants. Biochem. Biophys. Res. Commun. 371, 510–514. doi: 10.1016/j.bbrc.2008.04.115
Fraser, P. D., Romer, S., Shipton, C. A., Mills, P. B., Kiano, J. W., Misawa, N., et al. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U.S.A 99, 1092–1097. doi: 10.1073/pnas.241374598
Fu, X., Feng, C., Wang, C., Yin, X., Lu, P., Grierson, D., et al. (2014). Involvement of multiple phytoene synthase genes in tissue- and cultivar-specific accumulation of carotenoids in loquat. J. Exp. Bot. 65, 4679–4689. doi: 10.1093/jxb/eru257
Gady, A. L., Vriezen, W. H., Van de Wal, M. H., Huang, P., Bovy, A. G., Visser, R. G., et al. (2012). Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breed. 29, 801–812. doi: 10.1007/s11032-011-9591-9
Gallagher, C. E., Matthews, P. D., Li, F., and Wurtzel, E. T. (2004). Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 135, 1776–1783. doi: 10.1104/pp.104.039818
García-Ruiz, A., Baenas, N., Benítez-González, A. M., Stinco, C. M., Meléndez-Martínez, A. J., Moreno, D. A., et al. (2017). Guayusa (Ilex guayusa L.) new tea: phenolic and carotenoid composition and antioxidant capacity. J. Sci. Food Agr. doi: 10.1002/jsfa.8255
Giorio, G., Stigliani, A. L., and D'Ambrosio, C. (2008). Phytoene synthase genes in tomato (Solanum lycopersicum L.)–new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 275, 527–535. doi: 10.1111/j.1742-4658.2007.06219.x
Giribaldi, M., Gény, L., Delrot, S., and Schubert, A. (2010). Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. J. Exp. Bot. 61, 2447–2458. doi: 10.1093/jxb/erq079
Grassi, S., Piro, G., Lee, J. M., Zheng, Y., Fei, Z., Dalessandro, G., et al. (2013). Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genomics 14:781. doi: 10.1186/1471-2164-14-781
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98.
Havaux, M. (2014). Carotenoid oxidation products as stress signals in plants. Plant J. 79, 597–606. doi: 10.1111/tpj.12386
Hou, X., Rivers, J., León, P., McQuinn, R. P., and Pogson, B. J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 21, 792–803. doi: 10.1016/j.tplants.2016.06.001
Howitt, C. A., Cavanagh, C. R., Bowerman, A. F., Cazzonelli, C., Rampling, L., Mimica, J. L., et al. (2009). Alternative splicing, activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Funct. Integr. Genomics 9, 363–376. doi: 10.1007/s10142-009-0121-3
Huang, C., Wang, D., Sun, L., and Wei, L. (2016). Effects of exogenous salicylic acid on the physiological characteristics of Dendrobium officinale under chilling stress. J. Plant Growth Regul. 79, 199–208. doi: 10.1007/s10725-015-0125-z
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., Cuche, B. A., De Castro, E., et al. (2008). The 20 years of PROSITE. Nucleic Acids Res. 36, D245–D249. doi: 10.1093/nar/gkm977
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 15, 147–147. doi: 10.1038/nrm3744
Jeong, S. T., Goto-Yamamoto, N., Kobayashi, S., and Esaka, M. (2004). Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 167, 247–252. doi: 10.1016/j.plantsci.2004.03.021
Jiang, M., and Zhang, J. (2001). Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 42, 1265–1273. doi: 10.1093/pcp/pce162
Kachanovsky, D. E., Filler, S., Isaacson, T., and Hirschberg, J. (2012). Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc. Natl. Acad. Sci. U.S.A. 109, 19021–19026. doi: 10.1073/pnas.1214808109
Kaur, N., Shivani., Pandey, A., and Tiwari, S. (2016). “Provitamin A enrichment for tackling malnutrition,” in Banana: Genomics and Transgenic Approaches for Genetic Improvement, eds S. Mohandas and K. V. Ravishankar (Singapore: Springer-Verlag Ltd.), 277–299.
Khan, M. I. R., Fatma, M., Per, T. S., Anjum, N. A., and Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6:462. doi: 10.3389/fpls.2015.00462
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defence signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lado, J., Zacarías, L., Gurrea, A., Page, A., Stead, A., and Rodrigo, M. J. (2015). Exploring the diversity in Citrus fruit colouration to decipher the relationship between plastid ultrastructure and carotenoid composition. Planta 242, 645–661. doi: 10.1007/s00425-015-2370-9
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van, de., Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, F., Vallabhaneni, R., and Wurtzel, E. T. (2008). PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 146, 1333–1345. doi: 10.1104/pp.107.111120
Lin, J., Jin, Y., Zhou, X., and Wang, J. (2010). Molecular cloning and functional analysis of the gene encoding geranylgeranyl diphosphate synthase from Jatropha curcas. Afr. J. Biotechnol. 9, 3342–3351.
Liu, J. X., Chiou, C. Y., Shen, C. H., Chen, P. J., Liu, Y. C., Jian, C. D., et al. (2014). RNA interference-based gene silencing of phytoene synthase impairs growth, carotenoids, and plastid phenotype in Oncidium hybrid orchid. Springerplus 3, 1. doi: 10.1186/2193-1801-3-478
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
López-Emparán, A., Quezada-Martinez, D., Zúñiga-Bustos, M., Cifuentes, V., Iñiguez-Luy, F., and Federico, M. L. (2014). Functional analysis of the Brassica napus L. phytoene synthase (PSY) gene family. PLoS ONE 9:e114878. doi: 10.1371/journal.pone.0114878
Lv, F., Zhou, J., Zeng, L., and Xing, D. (2015). β-cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J. Exp. Bot. 66, 4719–4732. doi: 10.1093/jxb/erv231
Maass, D., Arango, J., Wüst, F., Beyer, P., and Welsch, R. (2009). Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 4:e6373. doi: 10.1371/journal.pone.0006373
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Matsumoto, H., Ikoma, Y., Kato, M., Kuniga, T., Nakajima, N., and Yoshida, T. (2007). Quantification of carotenoids in citrus fruit by LC-MS and comparison of patterns of seasonal changes for carotenoids among citrus varieties. J. Agric. Food Chem. 55, 2356–2368. doi: 10.1021/jf062629c
Mlalazi, B., Welsch, R., Namanya, P., Khanna, H., Geijskes, R. J., Harrison, M. D., et al. (2012). Isolation and functional characterisation of banana phytoene synthase genes as potential cisgenes. Planta 236, 1585–1598. doi: 10.1007/s00425-012-1717-8
Muengkaew, R., Chaiprasart, P., and Warrington, I. (2016). Changing of physiochemical properties and color development of mango fruit sprayed methyl Jasmonate. Sci. Hort. 198, 70–77. doi: 10.1016/j.scienta.2015.11.033
Natella, F., Maldini, M., Nardini, M., Azzini, E., Foddai, M. S., Giusti, A. M., et al. (2016). Improvement of the nutraceutical quality of broccoli sprouts by elicitation. Food Chem. 201, 101–109. doi: 10.1016/j.foodchem.2016.01.063
Nayyar, H., Bains, T., and Kumar, S. (2005). Low temperature induced floral abortion in chickpea: relationship to abscisic acid and cryoprotectants in reproductive organs. Environ. Exp. Bot. 53, 39–47. doi: 10.1016/j.envexpbot.2004.02.011
Nayyar, H., and Gupta, D. (2006). Differential sensitivity of C 3 and C 4 plants to water deficit stress: association with oxidative stress and antioxidants. Environ. Exp. Bot. 58, 106–113. doi: 10.1016/j.envexpbot.2005.06.021
Nitschke, S., Cortleven, A., Iven, T., Feussner, I., Havaux, M., Riefler, M., et al. (2016). Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient arabidopsis plants. Plant Cell 28, 1616–1639. doi: 10.1105/tpc.16.00016
Nogueira, M., Mora, L., Enfissi, E. M., Bramley, P. M., and Fraser, P. D. (2013). Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell 25, 4560–4579. doi: 10.1105/tpc.113.116210
Pandey, A., Misra, P., Alok, A., Kaur, N., Sharma, S., Lakhwani, D., et al. (2016). Genome-wide identification and expression analysis of homeodomain leucine zipper subfamily IV (HDZ IV) gene family from Musa accuminata. Front. Plant Sci. 7:20. doi: 10.3389/fpls.2016.00020
Pogson, B. J., Woo, N. S., Förster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Rivas, F., Fornes, F., Rodrigo, M. J., Zacarias, L., and Agusti, M. (2011). Changes in carotenoids and ABA content in citrus leaves in response to girdling. Sci. Hort. 127, 482–487. doi: 10.1016/j.scienta.2010.11.010
Rodriguez-Concepcion, M. (2010). Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 504, 118–122. doi: 10.1016/j.abb.2010.06.016
Ruiz-Sola, M. Á., and Rodríguez-Concepción, M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158. doi: 10.1199/tab.0158
Sandmann, G., Romer, S., and Fraser, P. D. (2006). Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab. Eng. 8, 291–302. doi: 10.1016/j.ymben.2006.01.005
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shumskaya, M., Bradbury, L. M., Monaco, R. R., and Wurtzel, E. T. (2012). Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell. 24, 3725–3741. doi: 10.1105/tpc.112.104174
Smita, S., Rajwanshi, R., Lenka, S. K., Katiyar, A., Chinnusamy, V., and Bansal, K. C. (2013). Expression profile of genes coding for carotenoid biosynthetic pathway during ripening and their association with accumulation of lycopene in tomato fruits. J. Genet. 92, 363–368. doi: 10.1007/s12041-013-0275-6
Solano, R., and Gimenez-Ibanez, S. (2013). Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 4:72. doi: 10.3389/fpls.2013.00072
Sreedharan, S., Shekhawat, U. K. S., and Ganapathi, T. R. (2012). MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 80, 503–517. doi: 10.1007/s11103-012-9964-4
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, C., Zhang, S., Wang, P., Hou, J., Qian, J., Ao, Y., et al. (2011). Salicylic acid involved in the regulation of nutrient elements uptake and oxidative stress in Vallisneria natans (Lour.) Hara under Pb stress. Chemosphere 84, 136–142. doi: 10.1016/j.chemosphere.2011.02.026
Wang, Y., Guo, B., Zhang, F., Yao, H., Miao, Z., and Tang, K. (2007). Molecular cloning and functional analysis of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from hazel (Corylusavellana L. Gasaway). BMB Rep. 40, 861–869. doi: 10.5483/BMBRep.2007.40.6.861
Welsch, R., Arango, J., Bär, C., Salazar, B., Al-Babili, S., Beltrán, J., et al. (2010). Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22, 3348–3356. doi: 10.1105/tpc.110.077560
Welsch, R., Wüst, F., Bär, C., Al-Babili, S., and Beyer, P. (2008). A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 147, 367–380. doi: 10.1104/pp.108.117028
Ye, X., Al-Babili, S., Klöti, A., Zhang, J., Lucca, P., Beyer, P., et al. (2000). Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305. doi: 10.1126/science.287.5451.303
Yoon, J. Y., Hamayun, M., Lee, S. K., and Lee, I. J. (2009). Methyl jasmonate alleviated salinity stress in soybean. J. Crop Sci. Biotechnol. 12, 63–68. doi: 10.1007/s12892-009-0060-5
Young, P. R., Lashbrooke, J. G., Alexandersson, E., Jacobson, D., Moser, C., Velasco, R., et al. (2012). The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genomics 15, 13:243. doi: 10.1186/1471-2164-13-243
Zhang, M., Leng, P., Zhang, G. L., and Li, X. X. (2009b). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. Plant Physiol. 166, 1241–1252. doi: 10.1016/j.jplph.2009.01.013
Keywords: antioxidant activity, banana, carotenoid, gene expression, provitamin A, stress
Citation: Kaur N, Pandey A, Shivani, Kumar P, Pandey P, Kesarwani AK, Mantri SS, Awasthi P and Tiwari S (2017) Regulation of Banana Phytoene Synthase (MaPSY) Expression, Characterization and Their Modulation under Various Abiotic Stress Conditions. Front. Plant Sci. 8:462. doi: 10.3389/fpls.2017.00462
Received: 28 November 2016; Accepted: 16 March 2017;
Published: 03 April 2017.
Edited by:
Claudio Bonghi, University of Padua, ItalyReviewed by:
Mahesh Kumar Basantani, Shri Ramswaroop Memorial University, IndiaRajesh Mehrotra, Birla Institute of Technology and Science, India
Copyright © 2017 Kaur, Pandey, Shivani, Kumar, Pandey, Kesarwani, Mantri, Awasthi and Tiwari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siddharth Tiwari, c2lkZGhhcnRoZG5hMjBAeWFob28uY29t; c2lkZGhhcnRoQG5hYmkucmVzLmlu
 Navneet Kaur
Navneet Kaur Ashutosh Pandey
Ashutosh Pandey Shivani
Shivani Prateek Kumar1
Prateek Kumar1 Pankaj Pandey
Pankaj Pandey Praveen Awasthi
Praveen Awasthi Siddharth Tiwari
Siddharth Tiwari