- Department of Horticulture, Oregon State University, Corvallis, OR, USA
An understanding of the biology of seeds has been greatly advanced in recent years. The progresses, particularly in the field of seed dormancy and germination research, have been made at a remarkable speed. Some of the possible epigenetic mechanisms, including an involvement of non-coding RNA, which were predicted for DELAY OF GERMINATION1 just a few years ago, have now been demonstrated with strong molecular and genetic evidence. Imprinting, or parent-of-origin-specific gene silencing/expression, which was characterized particularly for developing seeds, was also found in imbibed seeds and suggested for dormancy mechanisms. Hormone biology in seeds, which is the most advanced and almost a traditional area of seed research, also presents a new dimension. Upstream regulators of hormone metabolism and hormone transporters, such as abscisic acid and gibberellin influx/efflux carriers, have been identified. Characterization of the novel posttranslational modification pathways, including the N-end rule and S-nitrosylation pathways, which play a critical role in turnover of the major hormone signal transduction proteins, also expanded our knowledge about the complexity of hormone signaling in seeds. These progresses made at the molecular level are significant steps toward a better understanding of how seeds translate soil and other environmental signals into their internal hormone biology and make an important decision to stay dormant or commence with germination.
Introduction
The previous review article about seed dormancy and germination published in Frontiers in Plant Science (Nonogaki, 2014) summarized the progress made by the international seed research community and highlighted “emerging mechanisms and new hypotheses” at that time. While it has just been a few years since those progresses were reviewed, discoveries continued and more progresses were made, which started to address key biological questions about the mechanisms of seed dormancy and germination. Some of the possible mechanisms predicted by the previous review, such as the regulation of the major seed dormancy genes by long non-coding RNA (lncRNA), have now been demonstrated with convincing evidence. This review will highlight those exciting discoveries to update the current status of our understanding of seed dormancy and germination mechanisms. This article is not intended to provide comprehensive information but will highlight the major discoveries in the relatively unexplored but emerging areas of seed biology research.
New Players in Nitrate and Nitric Oxide Signaling in Seeds
The molecular mechanisms of seed responses to environmental signals, such as light and temperature, have been well characterized (Bae and Choi, 2008; Toh et al., 2008; Seo et al., 2009; Lim et al., 2013; Barrero et al., 2014; Lee and Choi, 2017). Another signal, which is critical for seeds to sense surrounding environments for germination, is the soil components. Nitrate is a major signal in the soil environment for seeds to detect vegetation gaps and germinate in the desirable spots with the likelihood of successful seedling establishment (Bewley et al., 2013). Nitrate signals received by the maternal plants are integrated with temperature signals during seed development and affect performance of mature seeds (He et al., 2016). Seed responses to nitrate, in terms of dormancy release, are well known, however, the mechanisms of nitrate-responsive gene expression in seeds have been elusive.
An understanding of the general mechanisms of nitrate-inducible gene expression in plants was greatly advanced in the past several years. The nitrate-responsive cis-element (NRE) was identified in the promoter region of NITRITE REDUCTASE1 (NIR1) (Konishi and Yanagisawa, 2010). The NRE containing promoter enables efficient gene induction in a nitrate-dependent manner (Konishi and Yanagisawa, 2010) and has been tested for nitrate-inducible gene expression in seeds for a technology development purpose (Nonogaki et al., 2015). The nitrate reductase NIA1, another nitrate-inducible gene, does not appear to contain NRE in the promoter region, however, the 3′-flanking sequence of the NIA1 gene, which is downstream of the transcriptional terminator, contains NREs (Konishi and Yanagisawa, 2011) (Figure 1). Screening for NRE-binding proteins identified Nodule Inception (NIN)-like proteins (NLPs) as NRE-binding factors (Konishi and Yanagisawa, 2013), which significantly advanced our knowledge on nitrate signaling in plants. NLP6 physically interacts with the NREs in NIR1 and NIA1, most likely upon activation of its N-terminal domain by nitrate (Konishi and Yanagisawa, 2013) (Figure 1).
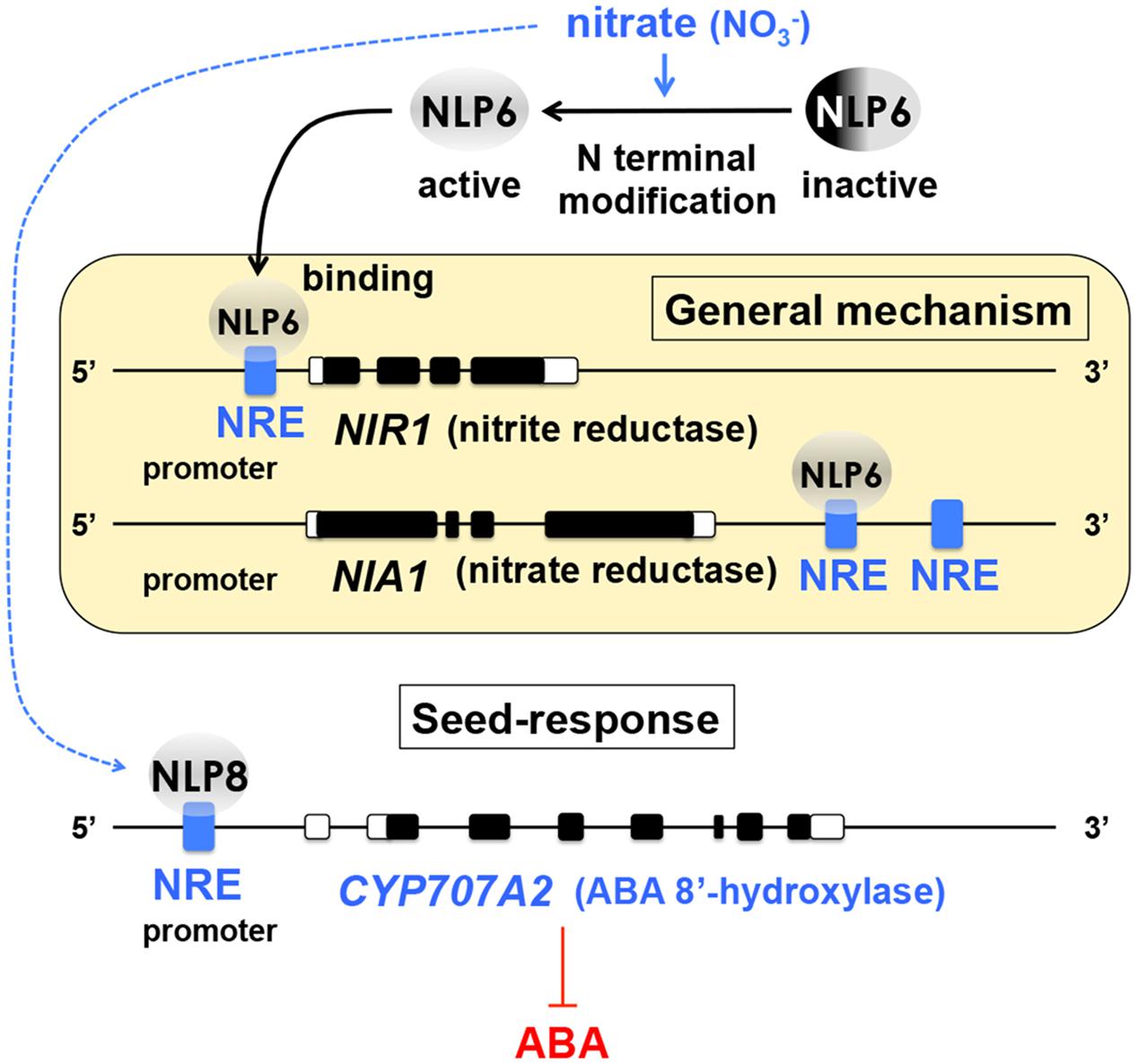
FIGURE 1. Nitrate signaling and ABA catabolism in seeds. An understanding of the general mechanisms of nitrate-induced gene expression in plants has been advanced significantly by the identification of the nitrate responsive cis-element (NRE) in the promoter region of NITRITE REDUCTASE1 (NIR1) and the 3′-flanking sequence of the nitrate reductase NIA1. Nodule Inception (NIN)-like proteins (NLPs) were identified as NRE-binding factors. NLP6 is activated by nitrate possibly through modification of its N-terminal domain (top) and physically interacts with NREs of NIR1 and NIA1 (yellow box). NLP8 is expressed in a narrow window of Phase I of seed imbibition and directly activates CYP707A2, an ABA catabolism gene, which is necessary for seed dormancy release (bottom). This discovery has addressed the missing link between nitrate response and ABA metabolism in seeds and advanced our understanding about how the soil environmental signal can be translated into hormone biology in seeds. Based on Konishi and Yanagisawa (2010, 2011, 2013) and Yan et al. (2016).
In Arabidopsis seeds, nitrate reduces abscisic acid (ABA) levels during imbibition by upregulating the ABA catabolism gene CYP707A2 (Matakiadis et al., 2009), which is required for dormancy release (Kushiro et al., 2004). There was a breakthrough in seed biology research, which has revealed that NLP8 is expressed in a very narrow window during Phase I of imbibition and directly binds to NRE in the promoter region of CYP707A2 to induce its expression (Figure 1). In the nlp8 mutant seeds, both ABA catabolism and germination in response to nitrate are impaired (Yan et al., 2016). This is a significant finding because ABA metabolism is a major determinant of seed germination and therefore identifying its upstream regulators is essential for reaching the core mechanisms of seed dormancy. Factors other than ABA metabolism, such as DELAY OF GERMINATION1 (DOG1) (Bentsink et al., 2006), are also essential for the seed dormancy mechanisms (see below), however, the final “output” of seed dormancy seems to be invariably dependent on expression of ABA biosynthesis genes and concomitant repression of ABA catabolism genes in imbibes seeds (Cadman et al., 2006; Okamoto et al., 2006; Finch-Savage et al., 2007). Application of fluridone, an ABA biosynthesis inhibitor, can induce germination from highly dormant seeds of Arabidopsis Cape Verde Islands (Cvi), from which DOG1 was identified (Ali-Rachedi et al., 2004). This result also demonstrates the essential role of ABA metabolism in imbibed seeds as the output of the dormancy state. An important biological question is: How are the “fates” (differential expression) of ABA biosynthesis and catabolism genes determined and altered in dormant or non-dormant seeds during early imbibition? Identification of NLP8 as a direct regulator of CYP707A2 addresses, at least in part, this important question in seed dormancy and germination research. Besides, uncovering NLP8 as the direct link between nitrate and ABA metabolism is also a significant step toward a better understanding of how the soil environmental signals are translated into hormone biology in seeds.
Nitrate could produce nitric oxide (NO), which also stimulates CYP707A2 expression (Liu et al., 2009; Arc et al., 2013) and seed germination (Bethke et al., 2004, 2007, 2011). However, the NLP8-mediated response is thought to be independent of NO signaling and a direct response to nitrate, because NO-defective mutant seeds still respond to nitrate and germinate in a NLP8-dependent manner (Yan et al., 2016). The nitrate and NO signaling pathways seem to target different transcription factors in seeds.
Nitric oxide targets ABA INSENSITIVE5 (ABI5), a major regulator of ABA signaling, which illustrates the crosstalk between the NO and ABA pathways. NO negatively regulates ABI5 expression by modulating the group VII ethylene response factors (ERFVIIs) through the N-end rule pathway (Gibbs et al., 2014, 2015) (Figure 2A). The N-end rule pathway is a ubiquitin-dependent proteolysis pathway, in which N-terminal residues of proteins serve as degradation signals (N-degrons) and determine half-life of proteins (Bachmair et al., 1986; Tasaki and Kwon, 2007; Tasaki et al., 2012). NO destabilizes ERFVIIs, which are upstream regulators of ABI5, through the N-end rule and 26S proteasome pathways, thereby suppressing ABI5 expression (Gibbs et al., 2014, 2015) (Figure 2A). In this case, NO regulates ABI5 at the level of transcription and indirectly through ERFVIIs.
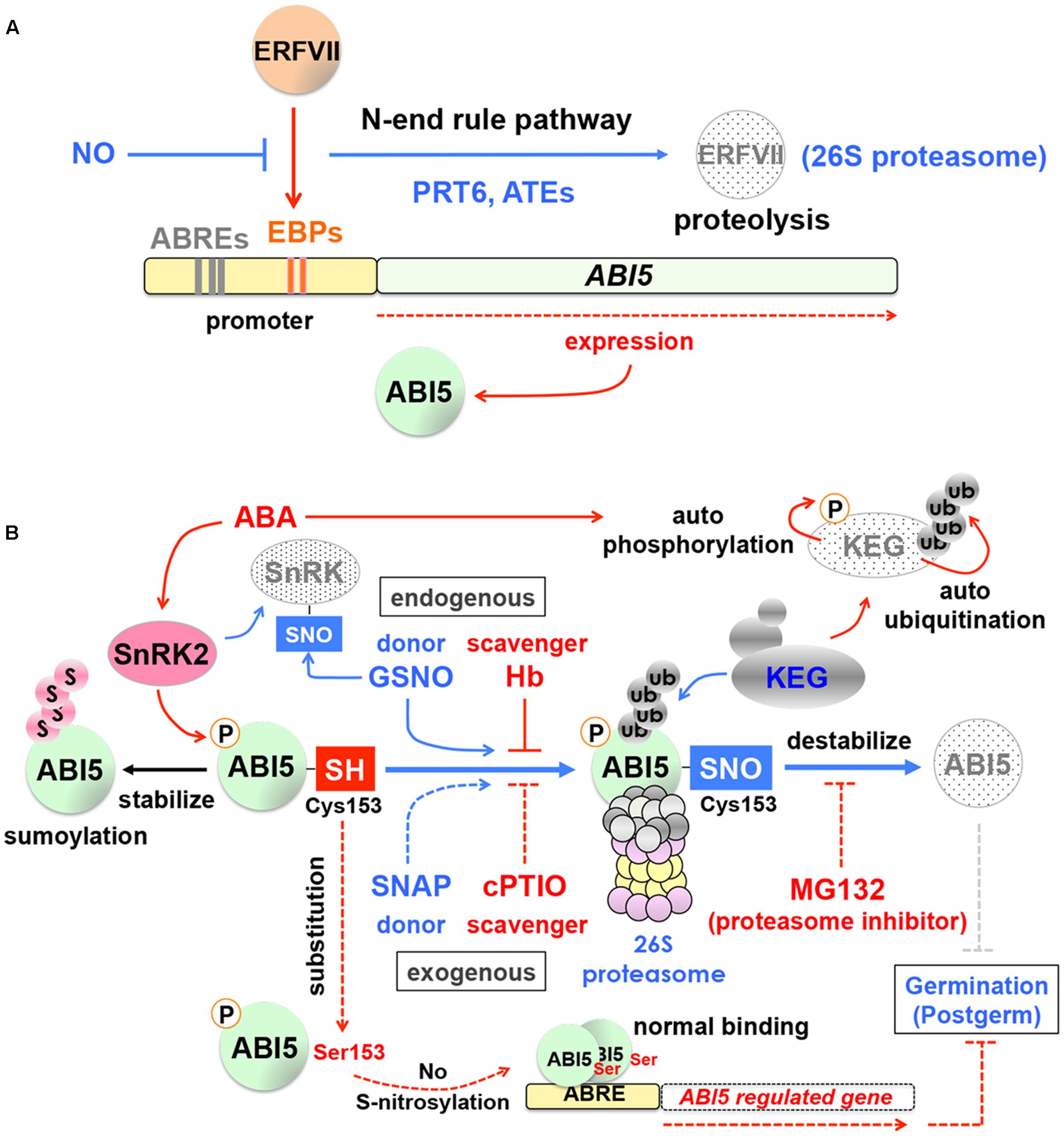
FIGURE 2. Nitric oxide (NO) and ABA signaling in seeds. (A) Indirect regulation of ABA INSENSITIVE5 (ABI5) expression by NO. The group VII ethylene response factors (ERFVIIs) induce expression of ABI5 through EBP-box cis-elements (EBPs) in its promoter region. NO promotes the N-end rule proteolysis pathway and degrade ERFVIIs, thereby reducing ABI5 expression. ABRE, ABA responsive element; PRT6, PROTEOLYSIS 6; ATE, arginyl-tRNA:protein arginyltransferase. Based on Garzon et al. (2007), Holman et al. (2009) and Gibbs et al. (2014, 2015). (B) Direct regulation of ABI5 protein stability by NO. NO counteracts with ABA through the turnover of ABI5 and positively affects seed germination, which is mediated through S-nitrosylation. The thiol side chain (-SH) of the cysteine 153 (Cys153) of ABI5 (ABI5-SH) is subject to S-nitrosylation by NO, which can be provided by the endogenous donor S-nitrosoglutathione (GSNO). In this way, ABI5-SH is converted to ABI5 nitrosothiol (ABI5-SNO), which is destabilized through ubiquitination (ub) by the RING-type E3 ligase KEEP ON GOING (KEG) and degradation by the 26S proteasome pathway. This process reduces ABA signaling and positively affects seed germination and postgermination. In contrast, relatively high levels of ABA promote autophosphorylation of KEG by its own kinase domain (top-right), which destabilizes KEG through autoubiquitination and in turn stabilizes ABI5. ABI5 can also be stabilized by sumoylation (s, top-left), which prevents ubiquitination, although it makes ABI5 inactive. When Cys 153 of ABI5 is substituted by Ser 153 (bottom-left), ABI5 still functions normally, in terms of dimer formation and ABRE binding, however, ABI5 S-nitrosylation is abolished, which negatively affects germination and postgermination. Note that SNF1-related protein kinase 2 (SnRK2), which phosphorylates and activates ABI5, is also subject to S-nitrosylation (top-left). The endogenous NO scavenger hemoglobins (Hb) and the exogenous NO donor S-nitroso-N-acetyl-DL-penicillamine (SNAP) and scavenger 2-(4-carboxyphenyl)-4,4,5,5 tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) are also shown with the proteasome inhibitor MG132. Based on Bethke et al. (2004, 2007), Perazzolli et al. (2004), Stone et al. (2006), Miura et al. (2009), Liu and Stone (2010, 2014), Ragni et al. (2011), Hill (2012), Albertos et al. (2015) and Wang et al. (2015).
In contrast, a novel pathway, in which NO directly affects ABI5 protein stability, has been identified. While the NO-dependent ERFVII degradation by the N-end rule pathway is mediated through PROTEOLYSIS 6 (PRT6), a RING-type E3 ligase (and arginyl-tRNA:protein arginyltransferase [ATE]) (Holman et al., 2009; Gibbs et al., 2014) (Figure 2A), the direct regulation of ABI5 protein by NO is mediated by KEEP ON GOING (KEG), another RING-type E3 ligase. KEG destabilizes ABI5 and acts as a negative regulator of ABA signaling (Stone et al., 2006) (Figure 2B). In turn, ABA causes destabilization of KEG through autoubiquitination, which is possibly caused by autophosphorylation of KEG by its own kinase domain (Liu and Stone, 2010) (Figure 2B). Therefore, in the presence of relatively high levels of ABA, KEG is unable to remove ABI5, which exerts negative effects on seed germination and postgermination. The regulatory mechanism of ABI5 turnover by KEG was well established, however, it was not known how KEG targets ABI5 for ubiquitination. Phosphorylation plays a critical role for ABI5 activity (Lopez-Molina et al., 2001; Piskurewicz et al., 2008) (Figure 2B) and therefore one could speculate that dephosphorylation triggers ubiquitination of ABI5 by KEG. However, phosphorylation status of ABI5 does not affect its turnover by KEG (Liu and Stone, 2014). Thus, it was not known what triggers KEG to target ABI5. This question has been addressed by recent research on the molecular mechanisms of NO-promoted seed germination and seedling growth. It was found that the thiol side chain (-SH) of the cysteine 153 (Cys153) in the ABI5 protein is subject to S-nitrosylation by NO, which results in the modified Cys153 with nitrosothiol (ABI5-SNO) (Figure 2B) and this modification is the trigger of ABI5 ubiquitination by KEG and its subsequent destabilization by the 26S proteasome pathway (Albertos et al., 2015).
In the native system, NO can be supplied through the endogenous donor S-nitrosoglutathione (GSNO) and scavenged by hemoglobins (Hb) (Perazzolli et al., 2004; Hill, 2012; Albertos et al., 2015) (Figure 2B). Similar effects to increase or decrease NO can be achieved by applying the NO donor S-nitroso-N-acetyl-DL-penicillamine (SNAP) or the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5 tetramethylimidazoline-1-oxyl-3-oxide (cPTIO), respectively (Figure 2B). Application of the NO donors GSNO and SNAP promotes ABI5 degradation in dormant Arabidopsis seeds (Albertos et al., 2015), providing evidence for NO-dependent ABI5 turnover. Destabilization of ABI5 is prevented by the proteasome inhibitor MG132 despite the presence of NO donors (Albertos et al., 2015), which verifies that NO-dependent ABI5 degradation is mediated through the 26S proteasome pathway (Figure 2B).
When a mutation is introduced to the ABI5 protein to substitute Cys153 with Ser153, it does not affect the ABI5 function, such as dimer formation and ABA responsive element (ABRE) binding, however, S-nitrosylation and destabilization of ABI5 are abolished by this mutation (Albertos et al., 2015). Thus, ABI5 serves as a NO sensor in seeds and seedlings. Interestingly, SNF1-related protein kinase 2 (SnRK2) that phosphorylates and activates ABI5 is also subject to S-nitrosylation (Wang et al., 2015) (Figure 2B). It appears that NO antagonizes ABA through more than one layer of regulatory mechanism to promote seed germination and early seedling growth. The native system contains the mechanism to stabilize ABI5 through small ubiquitin-related modifier (SUMO) conjugation (sumoylation), which prevents ubiquitination and degradation (Kerscher et al., 2006; Miura et al., 2007) (Figure 2B), although it makes ABI5 inactive (Miura et al., 2009; Liu and Stone, 2014).
Hormone Transport – Interplay Between Seed Tissues
Hormone levels in seeds are determined mainly by its metabolism – biosynthesis and catabolism. Another critical factor, which could significantly affect hormone responses in seeds, is transport of hormones and its precursors from/to different tissues in a seed. Mapping hormone transport between different tissue domains in the embryonic axis, such as the vascular, cortex and endodermis, is critical for a better understanding of the interplay between the distinct cell layers in the embryo, which generates growth potential for germination. It is conceivable that active hormone transport, rather than (or in addition to) diffusion, is involved in the interaction between the embryo and the endosperm. Our knowledge about the mechanisms of hormone transport in seeds is limited. There is little information about ABA or gibberellin (GA) maxima and gradient in the seed cells during imbibition. Localization of hormone transporters in dormant and germinating seeds needs to be characterized.
There were several breakthroughs in the area of hormone transport in seeds. Arabidopsis thaliana ATP-binding cassette (ABC) transporter G family member 25 (AtABCG25), which is a plasma membrane-localized ABA transporter, was found by screening the transposon-tagging lines for mutants exhibiting ABA-sensitivity phenotypes during seed germination and seedling growth (Kuromori et al., 2010). Experiments using isotope-labeled ABA showed that ABA was imported into the AtABCG25-expressing “inside-out” membrane vesicles, which were prepared from insect cells, in an ATP-dependent manner, demonstrating that AtABCG25 is an ABA exporter. When AtABCG25 is overexpressed in plants, it reduces ABA inhibition of seedling growth, which supports the idea that AtABCG25 is an efflux carrier of ABA (Kuromori et al., 2010). A separate study identified AtABCG40 (or Pleiotropic drug resistance transporter 12 [PDR12]) as a plasma membrane-localized ABA transporter, however, in this case AtABCG40 functions as an influx carrier of ABA. Expression of AtABCG40 in yeast and tobacco BY cells increases their ABA uptake. The mesophyll protoplasts isolated from the atabcg40 mutants exhibit slower ABA uptake compared to wild type. Consistently, seeds of atabcg40 exhibit reduced ABA sensitivity in germination (Kang et al., 2010).
AtABCG25 exports ABA from the vascular bundles and AtABCG40 imports it to guard cells (Kang et al., 2010; Kuromori et al., 2010, 2014), which is important for translocation of ABA in a plant body. ABA transport has been suggested to occur also in seeds, based on the characterization of AtABCG30 and AtABCG31, which were found to be ABA importer and exporter, respectively. The exporters AtABCG31 and AtABCG25 localize mainly in the endosperm of Arabidopsis seeds while the importers AtABCG30 and AtABCG40 localize mainly in the embryo, suggesting that ABA produced in the endosperm is transported to, and function in, the embryo (Kang et al., 2015; Figure 3A). While the hypothesis seems reasonable, there are more questions to be answered. For example, it is not known whether the ABA exporters in the Arabidopsis endosperm specifically localize at the plasma membranes facing the embryo. In the case of auxin transporters PIN-FORMEDs (PINs) in developing embryos, their cellular polarity and the consequential auxin flow in a unidirectional manner have been well documented (Bowman and Floyd, 2008). The endosperm and the embryo in a mature seed are not connected by cells but separated by spatial gaps. Therefore, endosperm cells might just secrete ABA uniformly, which could reach the embryo just by passive diffusion. It is not known whether the ABA importers in the embryo of imbibed seeds localize at the specific cells (e.g., epidermal cells adjacent to the endosperm or inner layers). Are there any specific roles for ABA exported from the endosperm versus ABA produced in the embryo itself? It is conceivable that the endospermic and embryonic origin of ABA target distinct cell layers of the embryo, which can be precisely controlled by specific localization of different ABA transporters, although it is not hard to imagine that the two sources of ABA are inevitably blended in the embryo. ABA production by the endosperm might affect its production in the embryo, or vice versa (i.e., positive feedback or feedforward regulation). Alternatively, ABA exporters might just function to avoid accumulation of ABA in undesirable cell layers by pumping out the hormone from them and the importers might just function to retain ABA in the site of biosynthesis (see below). These strategies could function to contain ABA in specific cell layers, rather than actively transporting the hormone to other locations. Information about the ABA transporters at the cellular level, including an involvement of endocytosis and abiotic stress responses, in roots (not the radicle) is emerging (Park Y. et al., 2016). Similar characterization can be performed to describe the role of ABA transporters in imbibed seeds. While there are still many questions to be answered, the findings about ABCG transporters in seeds suggest an interesting possibility of active transport of ABA between the endosperm and the embryo and opened a new area of seed dormancy and germination research.
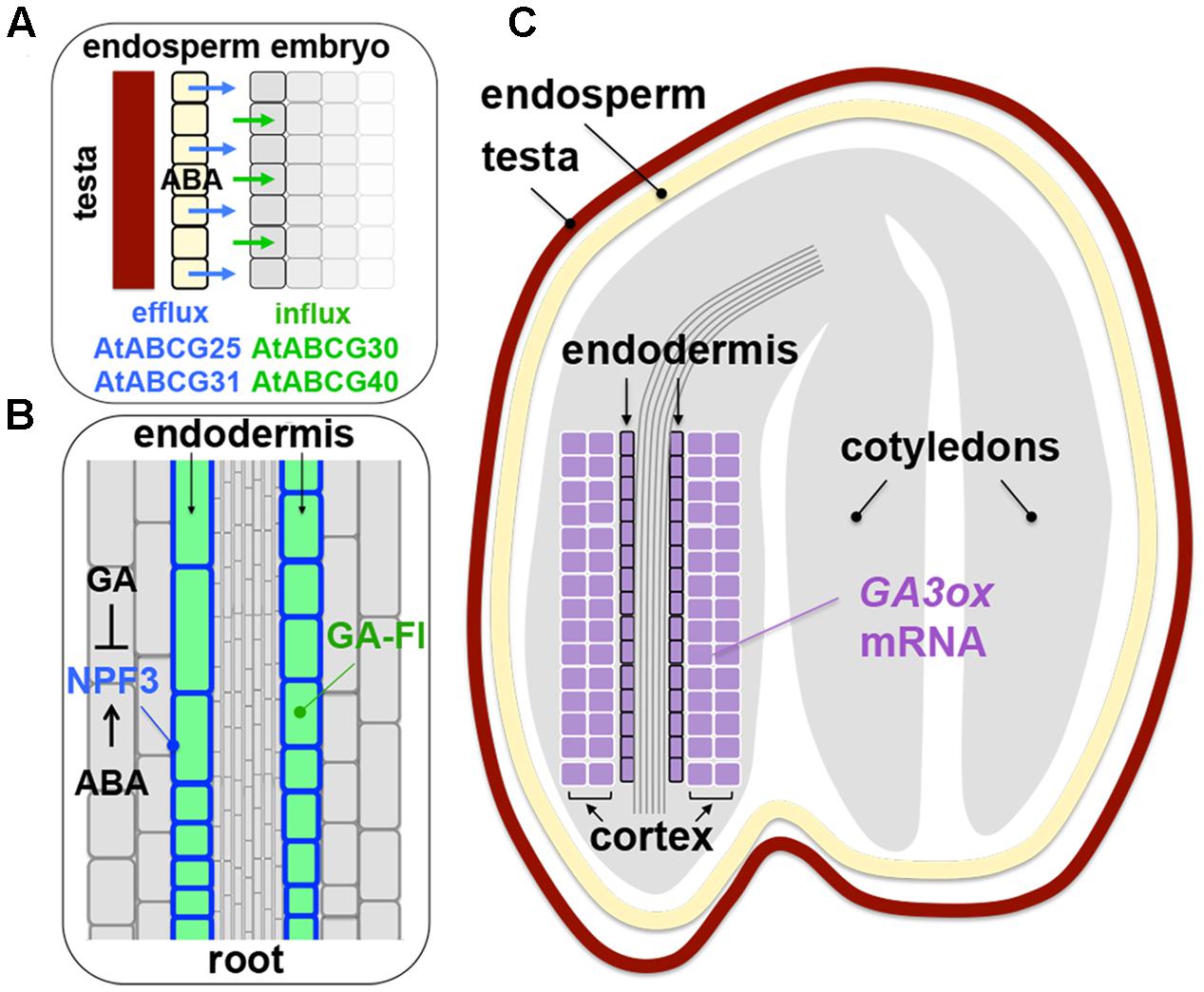
FIGURE 3. Hormone transporters in seeds and roots. (A) Localization of the ABA efflux carriers Arabidopsis thaliana ATP-binding cassette (ABC) transporter G family member 25 (AtABCG25) and AtABCG31 (blue arrows) in the endosperm (yellow layer) and the influx carriers AtABCG30 and AtABCG40 (green arrows) in the embryo (gray layers) of Arabidopsis seeds, which implies ABA transport from the endosperm to the embryo. Whether the ABA efflux carriers specifically localize at the embryonic side of the plasma membranes of endospermic cells is not known. Details of the localization of the ABA influx carriers in the embryo (e.g., epidermal, endodermal cells) remain to be examined. Based on Kang et al. (2015). (B) Localization of the GA influx carrier NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER (NRT1/PTR) family (NPF) member 3 (NPF3) in the plasma membrane of the endodermal cells (blue) of Arabidopsis root, which causes accumulation of fluorescently labeled GA (GA-Fl, green) in those cells. NPF3 expression is reduced by GA and enhanced by ABA. See text for details. Based on Shani et al. (2013) and Tal et al. (2016). (C) Localization of GA3ox transcripts (purple) in the endodermis and cortex of the embryonic axis of Arabidopsis seeds. While little information is available for the localization of GA transporters in seed tissues, it is possible that active transport of GA occurs in a seed also. Based on Yamaguchi et al. (2001).
In addition to the ABC transporters ABCGs, a different type of ABA transporter, has been identified from the NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER (NRT1/PTR) family (NPF) (Leran et al., 2014). Arabidopsis thaliana NPF4.6 (AtNPF4.6), which had been characterized as a low-affinity nitrate transporter NRT1.2, was identified as ABA-IMPORTING TRANSPORTER1 (AIT1) (Kanno et al., 2012). NPF4.6/NRT1.2/AIT1 (called AIT1 hereafter) was found by an elegant screening, which employed a modified yeast two-hybrid system. This system takes advantage of the well-characterized interaction between the ABA receptor and protein phosphatase 2C (PP2C) (Cutler et al., 2010), which occurs only in the presence of a sufficient level of ABA in the cell. The ABA receptor (PYRABACTIN RESISTANCE1 [PYR1]), which is fused to the DNA binding domain (BD-PYR1), and the PP2C (ABA INSENSITIVE1 [ABI1]), which is fused to the activation domain (AD-ABI1), co-induce expression of a reporter (selection marker) gene only when ABA is actively imported into the cell and triggers interaction between the receptor (BD-PYR1) and PP2C (AD-ABI1). Using this system, cDNAs encoding for ABA importers were searched for. The screening identified AIT1 (and other AITs) as a high-affinity, plasma membrane-localized ABA transporter. Overexpression of AIT1 makes germination of transgenic seeds more sensitive to ABA compared to wild type while ait1 mutant seeds are insensitive to exogenous ABA (Kanno et al., 2012), both of which support the function of AIT1 as an ABA influx carrier. Detailed localization of AIT1 in imbibed seed cells is not clear. However, the AIT1 promoter is activated specifically in the vascular tissues of inflorescence stems, which is the same localization as the expression of the major ABA biosynthesis genes. Therefore, it has been proposed that AIT1 function is to maintain the ABA pool size in the site of ABA production (Kanno et al., 2012) (see below for a similar role of the GA influx carrier). It should be noted that AIT1 does not import GA, indole-3-acetic acid (IAA) and jasmonic acid (JA) into the cell, unlike some other NPFs, which transport more than one hormone (Chiba et al., 2015). Thus, the efficient screening of ABA transporters using the ABA receptor as a sensor, further advanced ABA transport studies. More research on the role of active transport of ABA between distinct tissues or its retention by certain cell layers will highlight different functions of seed tissues and cell layers during dormancy.
Information about a GA transporter is also emerging, which may be relevant to ABA transport as well (see below). GA is a mobile hormone, which was clearly demonstrated by grafting experiments (Ragni et al., 2011). Inactive GAs are also subject to long-distance transport (Regnault et al., 2015). Some GA biosynthesis genes, such as copalyl diphosphate synthase and GA 3-oxidase (GA3ox), are expressed in different tissues (provascular vs. cortex/endodermis) in the embryonic axis of Arabidopsis seeds (Yamaguchi et al., 2001). Therefore, it is possible that GA and its precursors are actively transported inside the embryo and a seed by transporters. There is a classical example of GA secretion by cereal embryos to stimulate amylase gene expression in the aleurone layers (Jones and Armstrong, 1971). Likewise, GA produced by the tomato embryo is thought to induce mannanase expression in the endosperm (Groot and Karssen, 1987; Nonogaki et al., 2000; Martinez-Andujar et al., 2012), although involvement of GA transporters in these events has not been demonstrated.
There was a new finding of a GA transporter NPF3 in Arabidopsis. NPF3 has been characterized mainly for roots (not the radicle), however, the findings about the general function of NPF3 in roots provide significant implications for possible roles of GA transporters in seeds. It was known that a fluorescently labeled GA (GA-Fl) accumulates in the endodermal cells in the elongation zone of Arabidopsis roots (Shani et al., 2013). Screening of the T-DNA insertion mutants of the ABC and NPF transporters identified the npf3 mutants, which were defective in GA-Fl accumulation in the endodermis. NPF3 is an influx carrier, which localizes at the plasma membrane and transports GA-Fl into the root endodermal cells (Tal et al., 2016) (Figure 3B). The npf3 mutants do not show phenotypes in seed germination, probably due to redundancy. NPF3-overexpressing seeds exhibit delayed germination (Tal et al., 2016), although more analysis should be performed on the role of NPF3 (and other GA transporters) specifically for sensu stricto germination.
NPF3 expression in roots is repressed by GA and promoted by ABA (Tal et al., 2016) (Figure 3B). A plausible interpretation of this transcriptional control is that NPF3 repression by GA is a negative-feedback mechanism for GA homeostasis, which prevents excessive entry of GA into the cells, while NPF3 promotion by ABA could be a cell response to increase GA transport into the cells and counteract ABA to maintain a certain GA-ABA balance in the root endodermis. However, the mechanism might not be that simple because interestingly, NPF3 could serve as an ABA importer also; that is, GA and ABA antagonize each other at the level of transport (Tal et al., 2016), in addition to the well-known antagonism at the levels of metabolism and signal transduction (Seo et al., 2009). This is an intriguing mechanism, which has not been explored in terms of hormonal regulation of seed dormancy and germination, and adds a new dimension to GA-ABA antagonism in seeds.
The endodermis (and cortex) is probably the production site of active GA in the embryo during germination, because the transcripts of the rate-limiting GA biosynthesis enzymes GA3ox1 and GA3ox2, which catalyze the final conversion of inactive GAs (GA20, GA9) to the active forms (GA1, GA4), are detected in the endodermis (and cortex) (Yamaguchi et al., 2001; Figure 3C). It is interesting to examine localization of NPF3 and other GA transporters in the embryonic axis (and the endosperm) of imbibed seeds before testa rupture. If GA importers localize at the endodermis of the elongation zone of the radicle as observed for NPF3 in roots, they probably retain GA in the site of biosynthesis, which is analogous to ABA retention by AIT1 (NPF4.6) in the vascular tissues discussed above. In fact, ectopic expression of NPF3 in roots with the 35S promoter caused the GA influx carrier to import and trap GA-Fl in the epidermal cells (misguided localization), which caused delayed germination, probably due to reduced transport of GA into the endodermis (targeted growing tissue) (Tal et al., 2016). These results exemplify the importance of precise control of hormone transport and retention in specific tissues and present the complexity of hormonal regulation in roots, which may be applied to seeds also. Comprehensive information about the localization of all ABA and GA metabolism enzymes and transporters (and precursor transporters if any) in different seed tissues will draw a clear picture of hormone production, transport and antagonism during seed dormancy and germination (see EPIGENETICS below for auxin transport).
Making A Seed Dormancy Gene “Dormant”
Does it make sense to maintain a seed dormancy gene “dormant”? Yes, it does but it seems to be done through antisense. Before entering this topic, it is probably important to touch on the significance of the repression of developmental programs in seeds. A number of lessons have been learned about the biological significance of repression and de-repression of seed developmental programs. During seed development, the embryogenesis program occurs in the embryo proper and part of the suspensor (West and Harada, 1993) while this program is strictly repressed in the rest of suspensor cells. When this control is lost by a mutation, such as leafy cotyledon1 (lec1), aberrant cell divisions occur in the suspensor cells, and a secondary embryo could be formed from these cells, which result in abnormal seeds containing double-embryos (Lotan et al., 1998). These results demonstrate the importance of well-coordinated spatial and temporal repression of certain developmental programs to allow others. Similar repression and de-repression occur also for testa development. The developmental program of the testa, which is precisely repressed in the integuments by the Polycomb Repressive Complex 2 (PRC2) pathway, is de-repressed upon fertilization through auxin signals from the endosperm (Figueiredo et al., 2016). In addition to these examples during seed development, repression and de-repression events play a central role also in the regulation of seed germination. This is well exemplified by the suppression of seed germination by the DELLA proteins during seed dormancy. GA-inducible genes, which are important for seed germination, are repressed by DELLAs (Cao et al., 2006). Upon GA perception by the receptor and its interaction with DELLAs, the repressor proteins are ubiquitinated and subject to degradation or inactivation by the 26S proteasome pathway (McGinnis et al., 2003; Dill et al., 2004; Ueguchi-Tanaka et al., 2005, 2007). In this case, the repression and de-repression at the posttranslational level play a role in the transition from the dormant to germinable state of seeds. The significance of repression of specific transcription factors by small RNAs at the posttranscriptional level has also been demonstrated for hormonal regulation of seed germination (Liu P.-P. et al., 2007; Reyes and Chua, 2007; Nonogaki, 2010).
Seed dormancy itself is a suppressive mechanism, which prevents mature seeds from germinating under conditions otherwise favorable for germination (Bewley et al., 2013). Interestingly, the dormancy mechanisms are also subject to repression when seeds need to become the germinative mode. The previous review focused intensively on the possible epigenetic mechanisms to repress the major dormancy genes (e.g., DOG1, ABI3) through chromatin remodeling, such as histone and DNA methylation by the PRC2 and KRYPTONITE (KYP) pathways (Nonogaki, 2014). While there were more developments in research, which reinforced the idea of PRC2 and KYP involvement in DOG1 regulation (Footitt et al., 2015), this topic will not be repeated here. However, it should be stressed that detailed mechanisms of DOG1 and ABI3 silencing by the PRC2 and KYP pathways are still unknown. It is necessary to elucidate the regulatory mechanisms controlling expression of the major dormancy genes, including possible involvement of lncRNAs in their silencing, which was predicted by the previous review (Nonogaki, 2014).
Information about the triggers of DOG1 and ABI3 silencing by the PRC2 and KYP pathways is still missing. However, recent studies on the regulatory mechanisms of DOG1 expression started to decode how the seed dormancy gene could be repressed. When the DOG1 gene was first identified, the presence of several splicing variants was reported (Bentsink et al., 2006). Further analysis provided more detailed information about the five transcript variants (α, β, γ, δ, 𝜀), which produce three different proteins (Nakabayashi et al., 2015) (Figure 4A). Among them, DOG1-𝜀 is the predominant form in the developing Arabidopsis seeds (Nakabayashi et al., 2015) (although DOG1-𝜀 is not exactly a splicing variant; see below). One could speculate that alternative splicing differentiates function of proteins, including their subcellular localization and potential to impose dormancy. However, all the three proteins are transported to the nucleus (Nakabayashi et al., 2015), which is critical for the predicted function of DOG1 as a regulatory protein (Nakabayashi et al., 2012). Overexpression analysis suggests that all the three isoforms are functional in terms of seed dormancy induction, although they are more stable when co-expressed. DOG1 is thought to function as a homodimer (in a protein complex) (Nakabayashi et al., 2015). Therefore, formation of heterodimers may not explain the better stability of DOG1 proteins. The mechanisms underlying the positive role of co-expression of DOG1 isoforms for their stability is unknown.
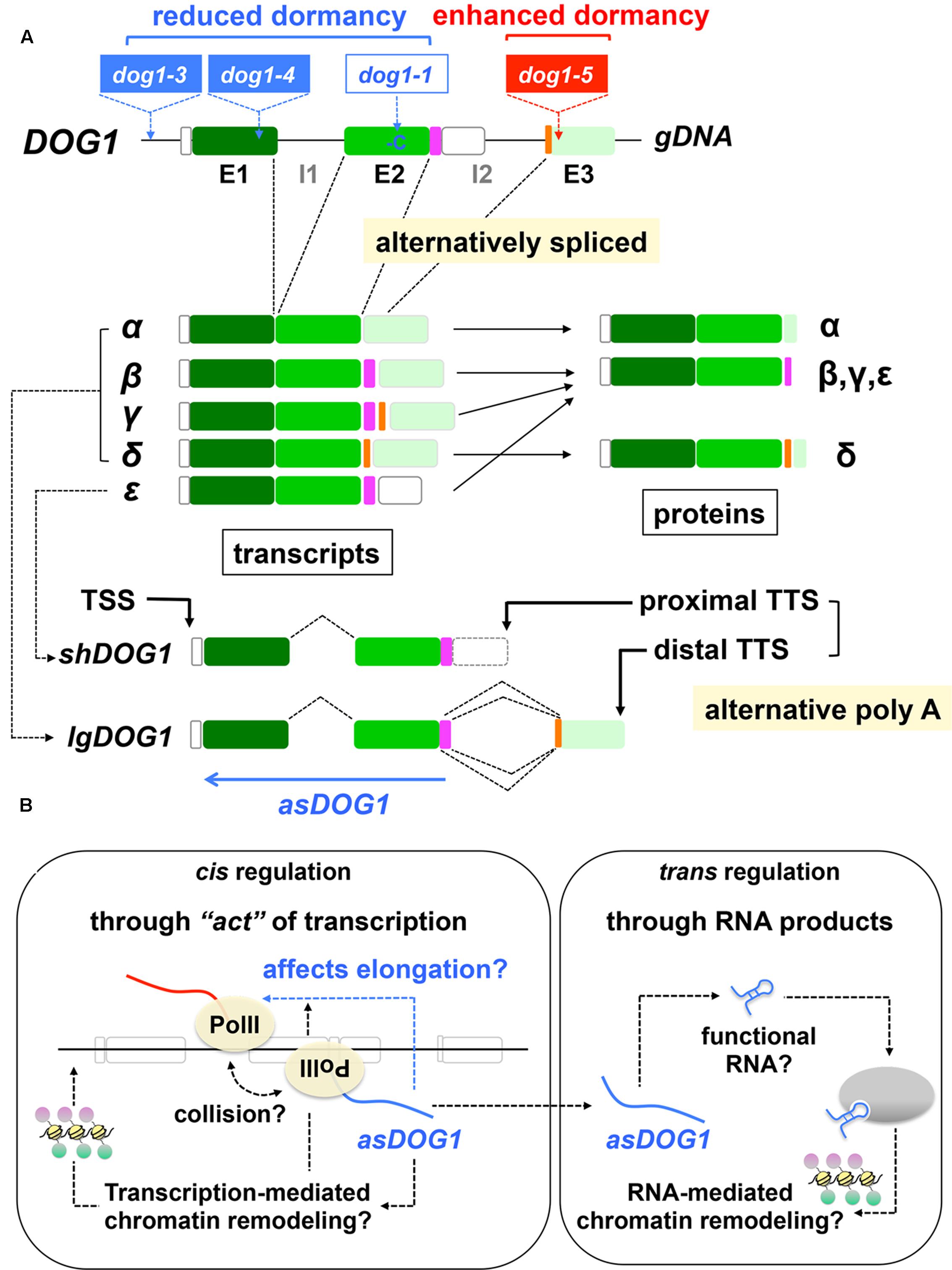
FIGURE 4. Regulation of DOG1 expression and function. (A) Structures of the DOG1 gene. Top, DOG1 gDNA with exons (E1, E2, E3) and introns (I1, I2). Alternatively spliced regions are highlighted in pink and orange. Approximate positions of the dog1 mutations (T-DNAs in dog1-3, dog1-4, dog1-5 and a single-base deletion [-C] in dog1-1) are also indicated. Middle, alternative DOG1 transcripts (α, β, γ, δ, 𝜀) and the corresponding proteins. Note that DOG1-𝜀 is not exactly an alternative splicing product. See text for details. Bottom, alternatively polyadenylated short DOG1 (shDOG1), which is identical to DOG1-𝜀 and long (lgDOG1) transcripts, which comprises DOG1-α, -β, -γ and -δ. The transcriptional start (TSS) and termination (TTS) sites are indicated. Approximate position and the orientation of antisense DOG1 (asDOG1) are shown as a blue arrow. Based on Bentsink et al. (2006), Dolata et al. (2015), Nakabayashi et al. (2015), Cyrek et al. (2016), Fedak et al. (2016). (B) Possible mechanisms of asDOG1 function. Relatively stable asDOG1 RNA could function as a regulatory RNA, in a sequence-specific manner or through its secondary structure, for RNA-mediated chromatin remodeling (right panel, trans regulation). However, allele-specific asDOG1 expression has indicated that asDOG1 functions in cis (left panel). The “act” of transcription itself, rather than its product (RNA), exerts the negative effects of asDOG1 expression to DOG1 expression and dormancy. Antisense expression could cause transcriptional interference and affect transcription elongation, which is known to be important for DOG1 expression and seed dormancy while transcription-mediated chromatin remodeling is also possible. Based on Shearwin et al. (2005), Hongay et al. (2006), Geisler and Coller (2013), Kornienko et al. (2013), Pelechano and Steinmetz (2013), Fedak et al. (2016), Quinn and Chang (2016).
When the Arabidopsis ortholog of the yeast spliceosomal component NineTeen complex-Related proteins 1 (AtNTR1) is mutated, it causes the major defects of intron retention and exon skipping in DOG1. This misregulation of DOG1 splicing reduces seed dormancy, although this phenotype may not be caused by alternative splicing itself but is probably the consequence of reduced DOG1 expression level in this mutant (Dolata et al., 2015). As focused in the previous review, efficiency of transcription elongation is a critical factor for DOG1 expression and seed dormancy (Liu Y. et al., 2007; Liu et al., 2011; Mortensen and Grasser, 2014; Nonogaki, 2014). AtNTR1 is thought to control RNA polymerase II (PolII) at the splice sites and serve as the checkpoint of transcription elongation (Dolata et al., 2015).
Another mechanism to produce transcript variants is alternative polyadenylation, which generates transcripts differing in 3′ ends (Di Giammartino et al., 2011; Cyrek et al., 2016). The two forms of DOG1 transcripts, short DOG1 (shDOG1) and long DOG1 (lgDOG1), which are produced by alternative polyadenylation, have been characterized (Cyrek et al., 2016). shDOG1 is identical to DOG1-𝜀 while lgDOG1 comprises DOG1-α, -β, -γ and -δ (Figure 4A). The C-terminus of DOG1 protein is absent or not conserved in many plant species, suggesting that it is not essential for the DOG1 function. In fact, shDOG1 is sufficient to complement the dog1 mutation and recovers seed dormancy (Cyrek et al., 2016). Although there is some discrepancy in published results in terms of the importance of longer versions of DOG1 transcripts, there seems to be a consensus that the short two-exonic DOG1 is functional and the major protein that is necessary for seed dormancy (Nakabayashi et al., 2015; Cyrek et al., 2016; Fedak et al., 2016).
There is little conservation of the exon 3 region of DOG1 genomic DNA in terms of encoded polypeptide sequences. In contrast, this region is highly conserved at the level of DNA, which is extended (back) to intron 2 (Fedak et al., 2016). The conservation of this region of DOG1 sequence at the DNA level, which is contradictory to the low evolutionary pressure for the protein sequences in the same region, implies a possible role of this genomic region of DOG1 as a production site of a regulatory ncRNA. In fact, expression of a lncRNA in an antisense orientation (antisense DOG1 [asDOG1]) from this region (and the vicinity) has been found (Fedak et al., 2016) (Figure 4A). Its expression is not spurious transcriptional noise but is regulated by a transcriptionally active promoter in an antisense orientation, which has been experimentally verified (Fedak et al., 2016). Expression of asDOG1 negatively affects expression of shDOG1, suggesting that asDOG1 is a negative regulator of DOG1 expression and seed dormancy. The mutations in the (sense) DOG1 promoter (dog1-3 [T-DNA]), exon 1 (dog1-4 [T-DNA]) and exon 2 (dog1-1 [1-bp deletion]) cause reduced or little seed dormancy (Bentsink et al., 2006; Cyrek et al., 2016; Fedak et al., 2016). In contrast, a mutation in the exon 3 (asDOG1 promoter) region (dog1-5 [T-DNA]) rather enhances seed dormancy (Cyrek et al., 2016; Fedak et al., 2016) (Figure 4A), providing convincing evidence for the role of asDOG1 as a negative regulator of seed dormancy.
Antisense DOG1 is a relatively stable RNA (a half-life of approximately 46 min) (Fedak et al., 2016), which is typical of regulatory RNAs, and therefore it is possible that the asDOG1 function depends on the RNA molecule at the posttranscriptional level. However, the detailed analysis by allele-specific asDOG1 expression has concluded that asDOG1 is unable to function in trans but does function in cis (Fedak et al., 2016). That is, the products of asDOG1 transcription (RNA molecules themselves) may not be important but the “act” of transcription itself (Kornienko et al., 2013; Pelechano and Steinmetz, 2013) is probably the cause of DOG1 repression (Fedak et al., 2016). The co-transcriptional effects of antisense expression, rather than posttranscriptional regulation by antisense RNA molecules, are known to cause transcriptional interference (Pelechano and Steinmetz, 2013). Transcriptional interference could be mediated by various mechanisms including direct collision of RNA polymerases and promoter competition (Shearwin et al., 2005; Pelechano and Steinmetz, 2013; Quinn and Chang, 2016) (Figure 4B). In yeast, an antisense-mediated transcriptional interference blocks transcription elongation of the IME4 gene (Hongay et al., 2006; Pelechano and Steinmetz, 2013). Therefore, it is possible that asDOG1 expression affects transcription elongation of DOG1, which has been demonstrated to be a critical factor for seed dormancy (Liu Y. et al., 2007; Liu et al., 2011; Mortensen and Grasser, 2014; Nonogaki, 2014) (Figure 4B).
Since asDOG1 is a repressor of seed dormancy, it is reasonable to hypothesize that asDOG1 might play a regulatory role in germination induction. However, both DOG1 and asDOG1 expression is reduced during seed imbibition, and therefore the asDOG1 function may be restricted to seed maturation (Fedak et al., 2016). Modification of DOG1 protein function, rather than its transcriptional control, may be critical for germination induction through after-ripening (Nakabayashi et al., 2012; Nee et al., 2016). Expression levels of asDOG1 during the maturation stage might determine the depth of seed dormancy in mature seeds, although more analyses are necessary to conclude the precise role of this interesting mechanism of asDOG1 in seed dormancy biology.
Here, only DOG1, one of the best-characterized dormancy genes, in terms of alternative splicing, alternative polyadenylation and the role of antisense RNA, was focused on. However, other dormancy genes, such as ABI3 and its ortholog in wheat Viviparous 1 (Vp-1), also produce transcript variants, which are developmentally regulated and important for dormancy (McKibbin et al., 2002; Sugliani et al., 2010). Therefore, more discoveries are anticipated from extended studies of the biological roles of alternative splicing, alternative polyadenylation and antisense transcription in regulation of other dormancy genes, including those in different species.
Epigenetics – Updates and New Developments
The topic of epigenetic regulation of seed dormancy and germination, which was intensively covered by the previous review, was bypassed above. However, as witnessed through the asDOG1 study, novel mechanisms associated with epigenetic regulation are emerging. Therefore, some updates and new developments in epigenetic studies on seed dormancy and germination will be briefly examined here.
As summarized in the previous review, deacetylation of histone H3 at lysines 9 and 18 (H3K9/18), which is a repressive mark of gene expression, plays a critical role in regulation of seed germination through the GA, ethylene and ABA pathways (Nonogaki, 2014) (Figure 5, shaded background). HISTONE DEACETYLASE 2B (HD2B) promotes GA accumulation in seeds by suppressing the GA catabolism gene GA2ox2 and (indirectly) enhancing GA3ox1 and GA3ox2, which positively affect germination (Yano et al., 2013). In contrast, SWI-INDEPENDENT3 (SIN3)-LIKEs (SNLs) in the histone deacetylase (HDAC) complex exert negative effects on germination by reducing ethylene levels and signals in seeds through repression of the ethylene biosynthesis genes 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASEs (ACOs) and signaling genes ERFs (Wang et al., 2013). The negative effects of the SNLs on germination are exerted also through repression of the ABA catabolism genes CYP707A1 and CYP707A2, which results in ABA accumulation in seeds (Wang et al., 2013) (Figure 5).
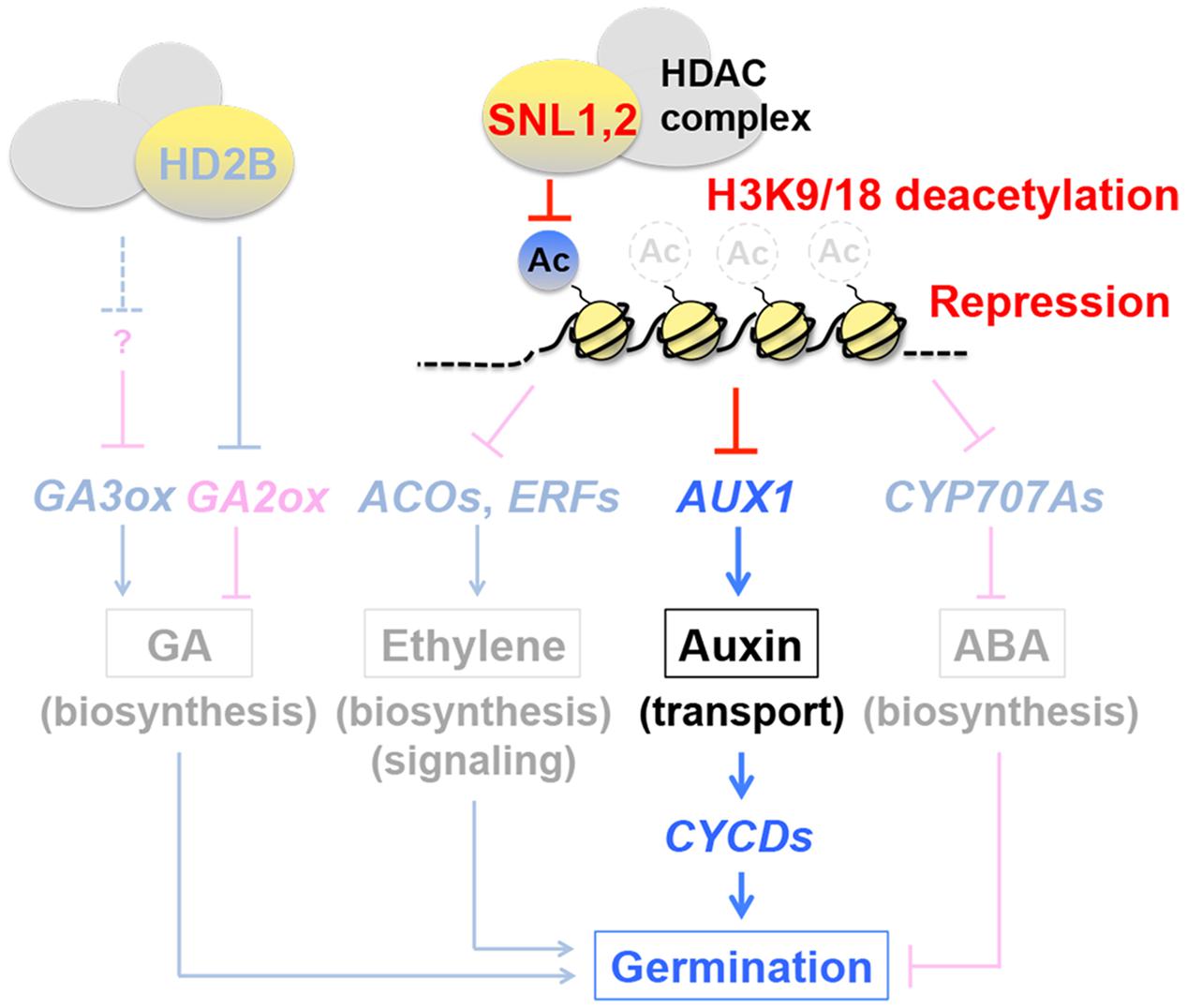
FIGURE 5. Regulation of hormone synthesis, signaling and transport in seeds by histone deacetylation. Previous studies had indicated that histone deacetylation modulates the GA, ethylene and ABA pathways (shaded background). HISTONE DEACETYLASE 2B positively affects GA levels in seeds and germination by repressing the GA catabolism gene GA2ox and enhancing the GA biosynthesis genes GA3ox1 and GA3ox2. SWI-INDEPENDENT3 (SIN3)-LIKEs (SNLs) in the histone deacetylase (HDAC) complex exert negative effects on ethylene levels in seeds and germination by repressing the ethylene biosynthesis genes 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASEs (ACOs) and signaling genes ETHYLENE RESPONSE FACTORs (ERFs), through deacetylation of histone H3 at lysines 9 and 18 (H3K9/18). SNLs repress CYP707A1 and CYP707A2 also, which results in ABA accumulation in seeds and negative regulation of germination. A new study has revealed that the negative role of SNLs in seed germination (speed) is also mediated through the auxin pathway. Unlike the other hormonal pathways where metabolism or signaling genes are the targets of histone deacetylation, SNLs repress AUXIN RESISTANT 1 (AUX1), an auxin influx carrier, thereby affecting auxin transport (and synthesis). Repression of AUX1 reduces the expression of the downstream factors D-type cyclin genes CYCDs, which are positive regulators of germination. The auxin transporter appears to play an important role for (the synthesis and) proper distribution of auxin in the radicle tip. Based on Wang et al. (2013, 2016), Yano et al. (2013), Nonogaki (2014).
The previous findings had revealed the inhibitory roles of SNLs in seed germination through the ethylene and ABA pathways and their antagonism. A new development in research suggests that SNLs exert negative effects on germination through the auxin pathway also. SNL1 and SNL2 repress AUXIN RESISTANT 1 (AUX1) (Maher and Martindale, 1980) through H3K9/18 deacetylation (Wang et al., 2016) (Figure 5). Unlike the SNL targets in the ethylene and ABA pathways, which are hormone metabolism and signaling genes, AUX1 is an auxin influx carrier (Bennett et al., 1996; Yang et al., 2006), suggesting that SNLs modulate seed germination through auxin transport. Here also, the importance of hormone transport in seeds, which was discussed above, is emerging. Auxin could affect seed germination positively and negatively at low (0.03–3 nM) and high (0.3–1 μM) concentrations, respectively (Hsueh and Lou, 1947; Liu P.-P. et al., 2007; He et al., 2012). Application of the auxin synthesis inhibitor aminoethoxyvinylglycine (AVG) and transport inhibitors 2,3,5-triidobenzoid acid (TIBA) and 1-naphthoxyacetic acids (1-NOA) negatively affects germination speed, suggesting that certain levels of auxin synthesis and transport are necessary for normal seed germination. AUX1 is thought to affect germination through the synthesis and distribution of auxin (possibly at low concentrations) in the radicle tip (Wang et al., 2016). AUX1 is not essential for radicle emergence, however, it plays a significant role for germination speed through the activation of the D-type cyclin genes CYCDs (Wang et al., 2016), which are known to play a role in germination (Masubelele et al., 2005) (Figure 5). The function of AUX1 as a transporter in seeds still needs to be investigated into more details. However, the SNL-AUX1 study has made dual impacts on epigenetics and hormone transport in seeds. Seed vigor, including germination speed, is an important aspect of seed quality in agriculture. The SNL-type of regulation of seed germination by epigenetics, which could be affected by seed production conditions and kept as a “memory” in seeds, may be an important constituent of seed vigor. Therefore, this area of research should also be expanded for applied aspects of seed biology.
Another new development of epigenetic research, which is relevant to the seed dormancy and germination mechanisms, is a finding of imprinting in imbibed seeds. Imprinting, or parent-of-origin-specific gene silencing/expression, is known for both animals and plants (Haig and Westoby, 1989, 1991; Moore and Haig, 1991; Feil and Berger, 2007). In imprinting, either the maternal or paternal origin of gene is specifically silenced, which results in preferential expression of the counterpart, independently of Mendelian genetics (Feil and Berger, 2007; Pignatta and Gehring, 2012). The biological significance of imprinting is explained by parental conflicts over resource allocation. Sibling offspring of one mother, which carry genes from different fathers, compete with each other for available resources while the mother favors equal distribution of resources to all offspring. This creates a conflict between the mother and each offspring, which is a “manifestation” of the conflict between the mother and the father (Haig and Westoby, 1989).
The “parental tug-of-war” (Moore and Haig, 1991), between males favoring collective resource acquisition by their own offspring and a female favoring equal resource allocation to all offspring, is exactly what could happen during seed development (Figure 6A). Individual seeds compete with each other, which creates the conflicts between the maternal plant and developing seeds. The parent-of-origin effects on seed size, which are also regulated by chromatin remodeling, have been well documented for crossing between plants in different ploidy and the mutants defective in chromatin remodeling (Scott et al., 1998; Ohto et al., 2007). Imprinting during endosperm development has been well characterized (Hsieh et al., 2011; Gehring, 2013; Tonosaki and Kinoshita, 2015). Unlike mammals where de novo methylation causes allele-specific gene silencing, DNA methylation and silencing in both alleles are the default state of the imprinted genes in the endosperm (central cell). The allele-specific DNA demethylation, which removes the repressive marks specifically from the maternal allele, drives maternal-specific gene expression in the central cell (Kinoshita et al., 2004; Penterman et al., 2007; Zhu et al., 2007; Jullien and Berger, 2009; Hsieh et al., 2011; Park K. et al., 2016) (Figure 6B). While imprinting has been characterized for seed development, it is not clear whether imprinting plays a role in mature seeds, particularly for dormancy and germination.
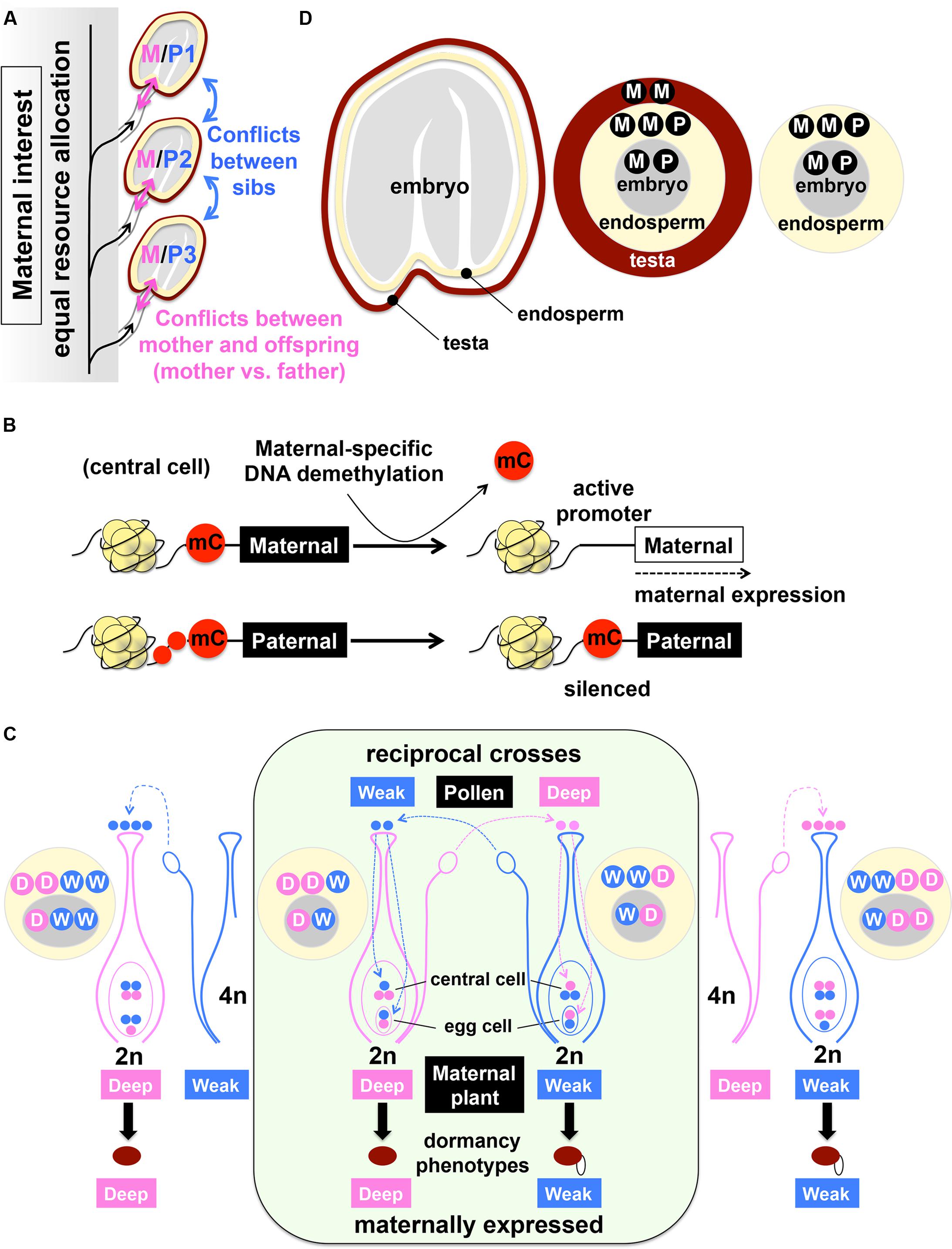
FIGURE 6. Imprinting and maternal expression of dormancy traits and genes. (A) Illustration of parental conflict over resource allocation in developing seeds. Imprinting, or parent-of-origin-specific gene expression, is explained by parental conflict over resource allocation. Each offspring competes with siblings for available resources (blue double-headed arrows) while the mother favors equal resource allocation to all offspring (black arrows). The conflict between the mother and each offspring (pink double-headed arrows) is a “manifestation” of the conflict between the mother (M, maternal genome) and the father (P, paternal genome). Based on Haig and Westoby (1989, 1991). (B) Maternal-specific activation of gene expression by DNA demethylation (mC removal) in the endosperm, which results in paternal-specific gene silencing. Based on Kinoshita et al. (2004), Ikeda and Kinoshita (2009), Jullien and Berger (2009). (C) Reciprocal crosses between deeply (Deep) and weakly (Weak) dormant accessions, using diploid (2n, inside green panel) or tetraploid (4n, outside green panel) plants as pollen donors. Pink and blue dots indicate genomic information and dosage from Deep (D) and Weak (W) dormant accessions, respectively. The F-1 hybrid seeds from reciprocal crosses between D and W tend to phenocopy the maternal seed dormancy traits, which are not explained by dosage effects. The genotypes of the embryo (gray circle) and the endosperm (yellow circle) are shown. See text for details. Based on Piskurewicz et al. (2016). (D) Contribution of the maternal (M) and paternal (P) genome to the embryo (gray), endosperm (yellow) and testa (brown).
Recent studies focusing on the molecular mechanisms of maternal expression of seed dormancy traits started to shed light on a possible role of imprinting in seed dormancy. When reciprocal crosses are performed between a deeply dormant accession of Arabidopsis (Cvi) and a relatively weak dormant accession (C24), the two populations of F-1 hybrid seeds from these reciprocal crosses exhibit distinct levels of dormancy, which tend to phenocopy the maternal traits (Piskurewicz et al., 2016): the F-1 seeds produced from the deeply (D) dormant accession as the mother are more dormant compared to those produced from weakly (W) dormant accession as the mother (Figure 6C). The genetic information in the embryo, which comes from the maternal (M) and paternal (P) genome (M/P), is essentially the same (D/W or W/D) between the two populations of F-1 seeds (Figures 6C,D). The maternally expressed dormancy phenotypes can be caused by the testa, which is derived from the integuments, a maternal (MM) tissue (DD or WW). The maternal effects of testa properties on seed dormancy phenotypes have been well demonstrated by the reciprocal crosses between wild type and transparent testa (tt) mutants, which exhibit defects in testa pigmentation and dormancy (Debeaujon and Koornneef, 2000; Debeaujon et al., 2000, 2007). However, both Cvi (DD) and C24 (WW) have the pigmented testa. Therefore, it is more likely that the differential expression of dormancy between the reciprocal crosses was caused by the endosperm. The endosperm contains two doses from the maternal genome while the paternal genome contributes a single dose (MM/P) to this tissue (Figure 6D). Therefore, the two populations of F-1 seeds differ in their endosperm genotypes (DD/W or WW/D), which could cause dosage effects (Figure 6C). However, when this possibility is tested by using tetraploid as pollen donors, which breaks off the genomic imbalance in the endosperm (DD/WW or WW/DD), the maternal expression of the dormancy phenotypes is still observed (Piskurewicz et al., 2016) (Figure 6C). Therefore, dosage effects in the endosperm do not explain the maternal expression of dormancy phenotypes. The remaining possibility – imprinting in the endosperm – has been suggested as an alternative explanation (Piskurewicz et al., 2016). Indeed, expression of 71 maternally expressed genes (MEG) and 5 paternally expressed genes (PEG) in the endosperm of dormant seeds and 50 MEG and 8 PEG expression in non-dormant seeds have been detected, with MEG expression showing close correlations with seed dormancy levels (Piskurewicz et al., 2016). Since imprinting occurs during seed development as mentioned above and it could be carried over through dormant seeds, more studies may be necessary to conclude the causal effects (cause or consequence?) of imprinting in imbibed seeds on the maintenance of seed dormancy. Nonetheless, possible involvement of imprinting in seed dormancy and germination regulation is very interesting. Maternal gene expression and histone modification are also emerging as a likely mechanism of heterosis in hybrid seeds (Alonso-Peral et al., 2017; Zhu et al., 2017), which has been a long-lasting question in basic science and is also an important foundation of seed business. Expanding the area of seed epigenetics will advance both basic and applied seed biology.
Concluding Remarks
This review highlighted a limited number of major findings that started to address the key questions in seed biology, by focusing on the relatively less explored and challenging areas, such as hormone metabolism upstream, hormone transport and lncRNA-mediated gene regulation. There were more findings relevant to hormonal signaling (Jiang et al., 2016), seed structures (Graeber et al., 2014; De Giorgi et al., 2015), crop seed dormancy (Nakamura et al., 2016; Sato et al., 2016; Torada et al., 2016) and many others, which were not integrated into this review due to the limited space and the scope of this article. Different dots and lines provided by other findings will probably be connected to each other by more discoveries in the near future. There are still many questions to be answered. For example, how is CYP707A2 activated without nitrate in after-ripened seeds? Is it still NLP8-dependent or independent? There are other important questions in an evolutionary context also. How did DOG1 emerge over the course of evolution and when did asDOG1 appear and become a DOG1 repressor? Is imprinting in seed dormancy genes indeed advantageous for plant survival? If so, how is it managed in gymnosperm seeds, which lack the endosperm? What are the crucial differences between megagametophyte vs. endosperm, in terms of gene silencing and chromatin remodeling machinery? These difficult questions will probably be answered by elegant experiments like those highlighted in this review and also by unexpected discoveries in seed and other plant sciences in the future. There are exciting times ahead for seed biology research.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I am grateful to Khadidiatou Sall, Ryosuke Koyari and Mariko Nonogaki, Oregon State University for critical reading of and helpful suggestions for the manuscript.
References
Albertos, P., Romero-Puertas, M. C., Tatematsu, K., Mateos, I., Sanchez-Vicente, I., Nambara, E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Comm. 6:8669. doi: 10.1038/ncomms9669
Ali-Rachedi, S., Bouinot, D., Wagner, M. H., Bonnet, M., Sotta, B., Grappin, P., et al. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219, 479–488. doi: 10.1007/s00425-004-1251-4
Alonso-Peral, M. M., Trigueros, M., Sherman, B., Ying, H., Taylor, J. M., Peacock, W. J., et al. (2017). Patterns of gene expression in developing embryos of Arabidopsis hybrids. Plant J. 89, 927–939. doi: 10.1111/tpj.13432
Arc, E., Sechet, J., Corbineau, F., Rajjou, L., and Marion-Poll, A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63. doi: 10.3389/fpls.2013.00063
Bachmair, A., Finley, D., and Varshavsky, A. (1986). In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186. doi: 10.1126/science.3018930
Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311. doi: 10.1146/annurev.arplant.59.032607.092859
Barrero, J. M., Downie, A. B., Xu, Q., and Gubler, F. (2014). A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 26, 1094–1104. doi: 10.1105/tpc.113.121830
Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. doi: 10.1126/science.273.5277.948
Bentsink, L., Jowett, J., Hanhart, C. J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047. doi: 10.1073/pnas.0607877103
Bethke, P. C., Gubler, F., Jacobsen, J. V., and Jones, R. L. (2004). Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219, 847–855. doi: 10.1007/s00425-004-1282-x
Bethke, P. C., Libourel, I. G. L., Aoyama, N., Chung, Y.-Y., Still, D. W., and Jones, R. L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143, 1173–1188. doi: 10.1104/pp.106.093435
Bethke, P. C., Libourel, I. G. L., Vitecek, J., and Jones, R. L. (2011). “Nitric oxide methods in seed biology,” in Seed Dormancy: Methods and Protocols, ed. A. R. Kermode (Totowa, NJ: Humana Press), 385–400. doi: 10.1007/978-1-61779-231-1_22
Bewley, J. D., Bradford, K. J., Hilhorst, H. W. M., and Nonogaki, H. (2013). Seeds: Physiology of Development, Germination and Dormancy. New York, NY: Springer. doi: 10.1007/978-1-4614-4693-4
Bowman, J. L., and Floyd, S. K. (2008). Patterning and polarity in seed plant shoots. Annu. Rev. Plant Biol. 59, 67–88. doi: 10.1146/annurev.arplant.57.032905.105356
Cadman, C. S. C., Toorop, P. E., Hilhorst, H. W. M., and Finch-Savage, W. E. (2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46, 805–822. doi: 10.1111/j.1365-313X.2006.02738.x
Cao, D., Cheng, H., Wu, W., Soo, H. M., and Peng, J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142, 509–525. doi: 10.1104/pp.106.082289
Chiba, Y., Shimizu, T., Miyakawa, S., Kanno, Y., Koshiba, T., Kamiya, Y., et al. (2015). Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 128, 679–686. doi: 10.1007/s10265-015-0710-2
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Cyrek, M., Fedak, H., Ciesielski, A., Guo, Y. W., Sliwa, A., Brzezniak, L., et al. (2016). Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 170, 947–955. doi: 10.1104/pp.15.01483
De Giorgi, J., Piskurewicz, U., Loubery, S., Utz-Pugin, A., Bailly, C., Mene-Saffrane, L., et al. (2015). An endosperm-associated cuticle is required for Arabidopsis seed viability, dormancy and early control of germination. PLoS Genet. 11:e1005708. doi: 10.1371/journal.pgen.1005708
Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. doi: 10.1104/pp.122.2.415
Debeaujon, I., Leon-Kloosterziel, K. M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122, 403–414. doi: 10.1104/pp.122.2.403
Debeaujon, I., Lepiniec, L., Pourcel, L., and Routaboul, J. M. (2007). “Seed coat development and dormancy,” in Seed Development, Dormancy and Germination, eds K. J. Bradford and H. Nonogaki (Oxford: Blackwell Publishing), 25–49. doi: 10.1002/9780470988848
Di Giammartino, D. C., Nishida, K., and Manley, J. L. (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866. doi: 10.1016/j.molcel.2011.08.017
Dill, A., Thomas, S. G., Hu, J., Steber, C. M., and Sun, T.-P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405. doi: 10.1105/tpc.020958
Dolata, J., Guo, Y. W., Kolowerzo, A., Smolinski, D., Brzyzek, G., Jarmolowski, A., et al. (2015). NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 34, 544–558. doi: 10.15252/embj.201489478
Fedak, H., Palusinska, M., Krzyczmonik, K., Brzezniak, L., Yatusevich, R., Pietras, Z., et al. (2016). Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. U.S.A. 113, E7846–E7855. doi: 10.1073/pnas.1608827113
Feil, R., and Berger, F. (2007). Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 23, 192–199. doi: 10.1016/j.tig.2007.02.004
Figueiredo, D. D., Batista, R. A., Roszakt, P. J., Hennig, L., and Kohler, C. (2016). Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5:e20542. doi: 10.7554/eLife.20542
Finch-Savage, W. E., Cadman, C. S. C., Toorop, P. E., Lynn, J. R., and Hilhorst, H. W. M. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51, 60–78. doi: 10.1111/j.1365-313X.2007.03118.x
Footitt, S., Muller, K., Kermode, A. R., and Finch-Savage, W. E. (2015). Seed dormancy cycling in Arabidopsis: chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 81, 413–425. doi: 10.1111/tpj.12735
Garzon, M., Eifler, K., Faust, A., Scheel, H., Hofmann, K., Koncz, C., et al. (2007). PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196. doi: 10.1016/j.febslet.2007.06.005
Gehring, M. (2013). Genomic imprinting: insights from plants. Annu. Rev. Genet. 47, 187–208. doi: 10.1146/annurev-genet-110711-155527
Geisler, S., and Coller, J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699–712. doi: 10.1038/nrm3679
Gibbs, D. J., Conde, J. V., Berckhan, S., Prasad, G., Mendiondo, G. M., and Holdsworth, M. J. (2015). Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169, 23–31. doi: 10.1104/pp.15.00338
Gibbs, D. J., Isa, N. M., Movahedi, M., Lozano-Juste, J., Mendiondo, G. M., Berckhan, S., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379. doi: 10.1016/j.molcel.2013.12.020
Graeber, K., Linkies, A., Steinbrecher, T., Mummenhoff, K., Tarkowska, D., Tureckova, V., et al. (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. U.S.A. 111, E3571–E3580. doi: 10.1073/pnas.1403851111
Groot, S. P. C., and Karssen, C. M. (1987). Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 171, 525–531. doi: 10.1007/BF00392302
Haig, D., and Westoby, M. (1989). Parent-specific gene-expression and the triploid endosperm. Am. Nat. 134, 147–155. doi: 10.1086/284971
Haig, D., and Westoby, M. (1991). Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333, 1–13. doi: 10.1098/rstb.1991.0057
He, H., Willems, L. A., Batushansky, A., Fait, A., Hanson, J., Nijveen, H., et al. (2016). Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol. 57, 473–487. doi: 10.1093/pcp/pcv207
He, J. N., Duan, Y., Hua, D. P., Fan, G. J., Wang, L., Liu, Y., et al. (2012). DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24, 1815–1833. doi: 10.1105/tpc.112.098707
Hill, R. D. (2012). Non-symbiotic haemoglobins-What’s happening beyond nitric oxide scavenging? AoB Plants 2012:pls004. doi: 10.1093/aobpla/pls004
Holman, T. J., Jones, P. D., Russell, L., Medhurst, A., Tomas, S. U., Talloji, P., et al. (2009). The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4549–4554. doi: 10.1073/pnas.0810280106
Hongay, C. F., Grisafi, P. L., Galitski, T., and Fink, G. R. (2006). Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127, 735–745. doi: 10.1016/j.cell.2006.09.038
Hsieh, T. F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M. J., et al. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 1755–1762. doi: 10.1073/pnas.1019273108
Hsueh, Y. L., and Lou, C. H. (1947). Effects of 2,4-D on seed germination and respiration. Science 105, 283–285. doi: 10.1126/science.105.2724.283
Ikeda, Y., and Kinoshita, T. (2009). DNA demethylation: a lesson from the garden. Chromosoma 118, 37–41. doi: 10.1007/s00412-008-0183-3
Jiang, Z. M., Xu, G., Jing, Y. J., Tang, W. J., and Lin, R. C. (2016). Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 7:12377. doi: 10.1038/ncomms12377
Jones, R. L., and Armstrong, J. E. (1971). Evidence for osmotic regulation of hydrolytic enzyme production in germinating barley seeds. Plant Physiol. 48, 137–142. doi: 10.1104/pp.48.2.137
Jullien, P. E., and Berger, F. (2009). Gamete-specific epigenetic mechanisms shape genomic imprinting. Curr. Opin. Plant Biol. 12, 637–642. doi: 10.1016/j.pbi.2009.07.004
Kang, J., Hwang, J. U., Lee, M., Kim, Y. Y., Assmann, S. M., Martinoia, E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. doi: 10.1073/pnas.0909222107
Kang, J., Yim, S., Choi, H., Kim, A., Lee, K. P., Lopez-Molina, L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6:8113. doi: 10.1038/ncomms9113
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180. doi: 10.1146/annurev.cellbio.22.010605.093503
Kinoshita, T., Miura, A., Choi, Y. H., Kinoshita, Y., Cao, X. F., Jacobsen, S. E., et al. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303, 521–523. doi: 10.1126/science.1089835
Konishi, M., and Yanagisawa, S. (2010). Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 63, 269–282. doi: 10.1111/j.1365-313X.2010.04239.x
Konishi, M., and Yanagisawa, S. (2011). The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant Cell Physiol. 52, 824–836. doi: 10.1093/pcp/pcr033
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4, 1617. doi: 10.1038/ncomms2621
Kornienko, A. E., Guenzl, P. M., Barlow, D. P., and Pauler, F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. doi: 10.1186/1741-7007-11-59
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2361–2366. doi: 10.1073/pnas.0912516107
Kuromori, T., Sugimoto, E., and Shinozaki, K. (2014). Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol. 164, 1587–1592. doi: 10.1104/pp.114.235556
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Lee, N., and Choi, G. (2017). Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 35, 54–60. doi: 10.1016/j.pbi.2016.11.004
Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi: 10.1105/tpc.113
Liu, H., and Stone, S. L. (2014). Regulation of ABI5 turnover by reversible post-translational modifications. Plant Signal. Behav. 9:e27577. doi: 10.4161/psb.27577
Liu, H. X., and Stone, S. L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22, 2630–2641. doi: 10.1105/tpc.110.076075
Liu, P.-P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H., and Carrington, J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146. doi: 10.1111/j.1365-313X.2007.03218.x
Liu, Y., Geyer, R., van Zanten, M., Carles, A., Li, Y., Hörold, A., et al. (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy.PLoS ONE 6:e22241. doi: 10.1371/journal.pone.0022241
Liu, Y. G., Shi, L., Ye, N. H., Liu, R., Jia, W. S., and Zhang, J. H. (2009). Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183, 1030–1042. doi: 10.1111/j.1469-8137.2009.02899.x
Liu, Y., Koornneef, M., and Soppe, W. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19, 433–444. doi: 10.1105/tpc.106.049221
Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lotan, T., Ohto, M.-A., Yee, K. M., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Maher, E. P., and Martindale, S. J. B. (1980). Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem. Genet. 18, 1041–1053. doi: 10.1007/BF00484337
Martinez-Andujar, C., Pluskota, W. E., Bassel, G. W., Asahina, M., Pupel, P., Nguyen, T. T., et al. (2012). Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J. 71, 575–586. doi: 10.1111/j.1365-313X.2012.05010.x
Masubelele, N. H., Dewitte, W., Menges, M., Maughan, S., Collins, C., Huntley, R., et al. (2005). D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 15694–15699. doi: 10.1073/pnas.0507581102
Matakiadis, T., Alboresi, A., Jikumaru, Y., Tatematsu, K., Pichon, O., Renou, J.-P., et al. (2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149, 949–960. doi: 10.1104/pp.108.126938
McGinnis, K. M., Thomas, S. G., Soule, J. D., Strader, L. C., Zale, J. M., Sun, T. P., et al. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130. doi: 10.1105/tpc.010827
McKibbin, R. S., Wilkinson, M. D., Bailey, P. C., Flintham, J. E., Andrew, L. M., Lazzeri, P. A., et al. (2002). Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. U.S.A. 99, 10203–10208. doi: 10.1073/pnas.152318599
Miura, K., Jin, J. B., and Hasegawa, P. M. (2007). Sumoylation, a post-translational regulatory-process in plants. Curr. Opin. Plant Biol. 10, 495–502. doi: 10.1016/j.pbi.2007.07.002
Miura, K., Lee, J., Jin, J. B., Yoo, C. Y., Miura, T., and Hasegawa, P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 5418–5423. doi: 10.1073/pnas.0811088106
Moore, T., and Haig, D. (1991). Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7, 45–49. doi: 10.1016/0168-9525(91)90230-N
Mortensen, S. A., and Grasser, K. D. (2014). The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Lett. 588, 47–51. doi: 10.1016/j.febslet.2013.10.047
Nakabayashi, K., Bartsch, M., Ding, J., and Soppe, W. J. (2015). Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet. 11:e1005737. doi: 10.1371/journal.pgen.1005737
Nakabayashi, K., Bartsch, M., Xiang, Y., Miatton, E., Pellengahr, S., Yano, R., et al. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. doi: 10.1105/tpc.112.100214
Nakamura, S., Pourkheirandish, M., Morishige, H., Kubo, Y., Nakamura, M., Ichimura, K., et al. (2016). Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr. Biol. 26, 775–781. doi: 10.1016/j.cub.2016.01.024
Nee, G., Xiang, Y., and Soppe, W. J. (2016). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. doi: 10.1016/j.pbi.2016.09.002
Nonogaki, H. (2010). MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol. 51, 1840–1846. doi: 10.1093/pcp/pcq154
Nonogaki, H. (2014). Seed dormancy and germination-emerging mechanisms and new hypotheses. Front. Plant Sci. 5:233. doi: 10.3389/fpls.2014.00233
Nonogaki, H., Gee, O. H., and Bradford, K. J. (2000). A germination-specific endo-beta-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 123, 1235–1246. doi: 10.1104/pp.123.4.1235
Nonogaki, M., Sekine, T., and Nonogaki, H. (2015). Chemically inducible gene expression in seeds before testa rupture. Seed Sci. Res. 25, 345–352. doi: 10.1017/S0960258515000240
Ohto, M., Stone, S. L., and Harada, J. J. (2007). “Genetic control of seed development and seed mass,” in Seed Development, Dormancy and Germination, eds K. J. Bradford and H. Nonogaki (Oxford: Blackwell Publishing), 1–24. doi: 10.1002/9780470988848
Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8 ’-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141, 97–107. doi: 10.1104/pp.106.079475
Park, K., Kim, M. Y., Vickers, M., Park, J. S., Hyun, Y., Okamoto, T., et al. (2016). DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. U.S.A. 113, 15138–15143. doi: 10.1073/pnas.1619047114
Park, Y., Xu, Z. Y., Kim, S. Y., Lee, J., Choi, B., Lee, J., et al. (2016). Spatial regulation of ABCG25, an ABA exporter, is an important component of the mechanism controlling cellular ABA levels. Plant Cell 28, 2528–2544. doi: 10.1105/tpc.16.00359
Pelechano, V., and Steinmetz, L. M. (2013). Non-coding RNA gene regulation by antisense transcription. Nat. Rev. Genet. 14, 880–893. doi: 10.1038/nrg3594
Penterman, J., Zilberman, D., Huh, J. H., Ballinger, T., Henikoff, S., and Fischer, R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757. doi: 10.1073/pnas.0701861104
Perazzolli, M., Dominici, P., Romero-Puertas, M. C., Zago, E., Zeier, A., Sonoda, M., et al. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16, 2785–2794. doi: 10.1105/tpc.104.025379
Pignatta, D., and Gehring, M. (2012). Imprinting meets genomics: new insights and new challenges. Curr. Opin. Plant Biol. 15, 530–535. doi: 10.1016/j.pbi.2012.09.004
Piskurewicz, U., Iwasaki, M., Susaki, D., Megies, C., Kinoshita, T., and Lopez-Molina, L. (2016). Dormancy-specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana. eLife 5:e19573. doi: 10.7554/eLife.19573
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Ragni, L., Nieminen, K., Pacheco-Villalobos, D., Sibout, R., Schwechheimer, C., and Hardtke, C. S. (2011). Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 23, 1322–1336. doi: 10.1105/tpc.111.084020
Regnault, T., Davière, J.-M., Wild, M., Sakvarelidze-Achard, L., Heintz, D., Carrera Bergua, E., et al. (2015). The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants 1:15073. doi: 10.1038/nplants.2015.73
Reyes, J. L., and Chua, N.-H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Sato, K., Yamane, M., Yamaji, N., Kanamori, H., Tagiri, A., Schwerdt, J. G., et al. (2016). Alanine aminotransferase controls seed dormancy in barley. Nat. Commun. 7:11625. doi: 10.1038/ncomms11625
Scott, R. J., Spielman, M., Bailey, J., and Dickinson, H. G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341.
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69, 463–472. doi: 10.1007/s11103-008-9429-y
Shani, E., Weinstain, R., Zhang, Y., Castillejo, C., Kaiserli, E., Chory, J., et al. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 110, 4834–4839. doi: 10.1073/pnas.1300436110
Shearwin, K. E., Callen, B. P., and Egan, J. B. (2005). Transcriptional interference - a crash course. Trends Genet. 21, 339–345. doi: 10.1016/j.tig.2005.04.009
Stone, S., Williams, L., Farmer, L., Vierstra, R., and Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Sugliani, M., Brambilla, V., Clerkx, E. J. M., Koornneef, M., and Soppe, W. J. J. (2010). The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 22, 1936–1946. doi: 10.1105/tpc.110.074674
Tal, I., Zhang, Y., Jorgensen, M. E., Pisanty, O., Barbosa, I. C. R., Zourelidou, M., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7:11486. doi: 10.1038/ncomms11486
Tasaki, T., and Kwon, Y. T. (2007). The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem. Sci. 32, 520–528. doi: 10.1016/j.tibs.2007.08.010
Tasaki, T., Sriram, S. M., Park, K. S., and Kwon, Y. T. (2012). The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289. doi: 10.1146/annurev-biochem-051710-093308
Toh, S., Imamura, A., Watanabe, A., Nakabayashi, K., Okamoto, M., Jikumaru, Y., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385. doi: 10.1104/pp.107.113738
Tonosaki, K., and Kinoshita, T. (2015). Possible roles for polycomb repressive complex 2 in cereal endosperm. Front. Plant Sci. 6:144. doi: 10.3389/fpls.2015.00144
Torada, A., Koike, M., Ogawa, T., Takenouchi, Y., Tadamura, K., Wu, J., et al. (2016). A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr. Biol. 26, 782–787. doi: 10.1016/j.cub.2016.01.063
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Ueguchi-Tanaka, M., Nakajima, M., Motoyuki, A., and Matsuoka, M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58, 183–198. doi: 10.1146/annurev.arplant.58.032806.103830
Wang, P., Zhu, J.-K., and Lang, Z. (2015). Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal. Behav. 10:e1031939. doi: 10.1080/15592324.2015.1031939
Wang, Z., Cao, H., Sun, Y., Li, X., Chen, F., Carles, A., et al. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25, 149–166. doi: 10.1105/tpc.112.108191
Wang, Z., Chen, F. Y., Li, X. Y., Cao, H., Ding, M., Zhang, C., et al. (2016). Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat. Commun. 7:13412. doi: 10.1038/ncomms13412
West, M., and Harada, J. J. (1993). Embryogenesis in higher plants: an overview. Plant Cell 5, 1361–1369. doi: 10.1105/tpc.5.10.1361
Yamaguchi, S., Kamiya, Y., and Sun, T. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28, 443–453. doi: 10.1046/j.1365-313X.2001.01168.x
Yan, D., Easwaran, V., Chau, V., Okamoto, M., Ierullo, M., Kimura, M., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7:13179. doi: 10.1038/ncomms13179
Yang, Y. D., Hammes, U. Z., Taylor, C. G., Schachtman, D. P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16, 1123–1127. doi: 10.1016/j.cub.2006.04.029
Yano, R., Takebayashi, Y., Nambara, E., Kamiya, Y., and Seo, M. (2013). Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana. Plant J. 74, 815–828. doi: 10.1111/tpj.12167
Zhu, A., Greaves, I. K., Dennis, E. S., and Peacock, W. J. (2017). Genome-wide analyses of four major histone modifications in Arabidopsis hybrids at the germinating seed stage. BMC Genomics 18:137. doi: 10.1186/s12864-017-3542-8
Keywords: antisense RNA, germination, hormone transport, imprinting, nitrate, nitric oxide
Citation: Nonogaki H (2017) Seed Biology Updates – Highlights and New Discoveries in Seed Dormancy and Germination Research. Front. Plant Sci. 8:524. doi: 10.3389/fpls.2017.00524
Received: 17 February 2017; Accepted: 24 March 2017;
Published: 11 April 2017.
Edited by:
Kimberley Cathryn Snowden, Plant & Food Research Auckland, New ZealandReviewed by:
Richard Macknight, University of Otago, New ZealandSzymon Swiezewski, Institute of Biochemistry and Biophysics – Polish Academy of Sciences, Poland
Copyright © 2017 Nonogaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroyuki Nonogaki, aGlyby5ub25vZ2FraUBvcmVnb25zdGF0ZS5lZHU=
 Hiroyuki Nonogaki
Hiroyuki Nonogaki