- 1College of Horticulture, South China Agricultural University, Guangzhou, China
- 2State key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control, Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
The DnaJ proteins which function as molecular chaperone played critical roles in plant growth and development and response to heat stress (HS) and also called heat shock protein 40 based on molecular weight. However, little was reported on this gene family in pepper. Recently, the release of the whole pepper genome provided an opportunity for identifying putative DnaJ homologous. In this study, a total of 76 putative pepper DnaJ genes (CaDnaJ01 to CaDnaJ76) were identified using bioinformatics methods and classified into five groups by the presence of the complete three domains (J-domain, zinc finger domain, and C-terminal domain). Chromosome mapping suggested that segmental duplication and tandem duplication were occurred in evolution. The multiple stress-related cis-elements were found in the promoter region of these CaDnaJ genes, which indicated that the CaDnaJs might be involved in the process of responding to complex stress conditions. In addition, expression profiles based on RNA-seq showed that the 47 CaDnaJs were expressed in at least one tissue tested. The result implied that they could be involved in the process of pepper growth and development. qRT-PCR analysis found that 80.60% (54/67) CaDnaJs were induced by HS, indicated that they could participated in pepper response to high temperature treatments. In conclusion, all these results would provide a comprehensive basis for further analyzing the function of CaDnaJ members and be also significant for elucidating the evolutionary relationship in pepper.
Introduction
With the increase of global warming, high temperature has become one of the most vital abiotic stresses on crop plants (Glazebrook, 1999). Pepper (Capsicum annuum L.) which originated in the tropical regions of Latin America had been widely cultivated around the world as an important vegetable crop nowadays and is sensitive to high temperature during plant growth and development, especially in reproductive stage (Guo et al., 2014). The optimum temperature of growing pepper is 20–30°C. Over 32°C can bring about serious effects on pollination and fertilization, and results in blossom and fruit dropping which can cause a significant reduction of pepper fruit yield and quality.
In the long-term evolution, plants have evolved a complicated response mechanism to respond to heat stress (HS). Previous researches had reported that heat shock response (HSR) was induced in many plant species under HS condition (Vierling, 1991). Among them, a great deal of ubiquitous and evolutionary-conserved proteins was identified as heat shock proteins (Hsps), one of the main products of the HSR (Vierling, 1991). The Hsp was first discovered in Drosophila melanogaster in response to HS (Ritossa, 1962). In the following years, more Hsps were identified in other plants (Agarwal et al., 2002; Sarkar et al., 2009; Lopes-Caitar et al., 2013; Mulaudzi-Masuku et al., 2015). According to approximate molecular weight and sequence homology, the Hsps were classified into five families, including the Hsp100, Hsp90, Hsp70, Hsp60 and small Hsps (Wang et al., 2004; Kotak et al., 2007; Gupta et al., 2010).
The Hsp40, one of the important plant Hsps, was first identified in Escherichia coli, generally existed in organisms as 41 kDa Hsps (Georgopoulos et al., 1980; Bukau and Horwich, 1998; Craig et al., 2006). The Hsp40s, also known as DnaJ proteins or J-proteins, generally consisted of the J-domain, a proximal G/F-domain, a distal zinc finger (CxxCxGxG) domain, and followed by less conserved C-terminal sequences (Caplan et al., 1993; Silver and Way, 1993). The characteristic feature of the J-proteins was the presence of evolutionarily conserved J-domain which located nearby the N-terminus and composed of approximately 70 amino acids residues (Cyr et al., 1992). The invariant tripeptide (HPD) was the hallmark of J-domain. It stimulated the ATPase activity of Hsp70 and was crucial for keeping J-protein’s function (Kampinga and Craig, 2010). Previously, Cheetham and Caplan (1998) attempted to separate these proteins into three groups. Group I J-proteins were characterized by the J-domain, G/F-domain, and zinc finger domain. Group II would have the J-domain plus either a G/F-domain or zinc finger domain. Group III J-proteins only comprised the J-domain (Ohtsuka and Hata, 2000).
In recent years, it has been found that plant DnaJ proteins played important roles in response to both biotic and abiotic stresses, such as pest, pathogenic bacterium, drought, salt, and heat. In 2007, a J-domain virulence effecter of Pseudomonas syringae remolded host chloroplasts when responded to pathogen (Jelenska et al., 2007). The researchers reported that over-expression of tomato (Solanum lycopersicum) chloroplast-targeted J-protein, LeCDJ1, facilitated heat tolerance in transgenic tomatoes (Kong et al., 2014a) and further found that it also played important role in maintaining photosystem II under chilling stress (Kong et al., 2014b). Subsequently, the study has also demonstrated that this gene could enhance tolerance to drought stress and resistance to P. solanacearum in transgenic tobacco (Wang et al., 2014). In addition, Xia et al. (2014) reported that a putative J-proteins ortholog from Nicotiana tabacum could be involved in drought stress response and its over-expression enhanced drought tolerance possibly through regulating expression of stress-responsive genes.
Up to now, many J-proteins in organisms were identified, such as Arabidopsis thaliana (89) (Miernyk, 2001), yeast (22) (Walsh et al., 2004) and human (41) (Craig et al., 2006). Despite ongoing efforts to characterize the members of J-protein from other organisms (Sarkar et al., 2009; Kong et al., 2014a,b; Xia et al., 2014), none of its from pepper has been identified at the genomic level. Fortunately, the pepper whole genomic sequences were completely available (Kim et al., 2014; Qin et al., 2014), which provided an opportunity for identifying candidate J-protein genes at the genomic level. In the present work, the J-protein gene family members were identified in pepper through bioinformatics method and analyzed by integration of gene structure, conserved motifs, chromosomal localization, cis-element and expression patterns.
Materials and Methods
Genome-Wide Identification of CaDnaJ Genes in Pepper
The genomic sequences of pepper downloaded from the Pepper Genome Database (PGD1) (Kim et al., 2014) were used to build the local database on the software BioEdit 7.0. The Hidden Markov Model (HMM) profile of J-domain (PF00226) downloaded from the Pfam protein family database2 was used as query sequence to search against putative pepper J-protein genes with e-value <10-5. Subsequently, each of all putative pepper J-protein genes was used to identify the presence of J-domain on Pfam3. The protein sequences of identified pepper J-protein gene family members were analyzed with EXPASY PROTOPARAM4 to obtain molecular weight and theoretical isoelectric point (pI).
Multiple Alignment and Chromosomal Location
In this paper, the J-proteins in pepper were classified based on structural features. In each class, the full amino acid sequences of pepper J-proteins were aligned using the software Clustal X 2.01 (Larkin et al., 2007). Each of the J-protein genes was mapped on chromosomes using MapDraw2.1 (Liu and Meng, 2003) based on information in PGD. Two duplication events, tandem duplication and segmental duplication, were also further elaborated. For tandem duplication, three criteria were adopted. Firstly, two or more pepper DnaJ genes were arrayed within a range of 100 kb distance. Secondly, the multiple alignments of these DnaJ genes had a high coverage rate of the longer gene (more than 70%). Thirdly, the identity of the aligned region in these DnaJ genes was also more than 70% (Li et al., 2009; Huang et al., 2012; Wei et al., 2016). The segmental duplication was investigated according to Plant Genome Duplication Database (PGDD5).
Promoters Analysis of Pepper CaDnaJ Genes
The upstream regions (1.5 kb) of the DnaJ gene sequences were downloaded from PGD, and were used to search for regulating factor such as gibberellins (GA), abscisic acid (ABA), salicylic acid (SA), ethylene, drought, salt and heat.
Tissue-Specific Expression of CaDnaJ Genes Based on RNA-Seq
In this paper, RNA-seq data reported by previous researchers (Kim et al., 2014) were used to investigate expression patterns of putative CaDnaJ genes in pepper. Different tissues were selected: root, stem, leaf and pericarp at 6 days post-anthesis (DPA), 16 DPA, 25 DPA, respectively, and mature green (MG), breaker (B), 5 days post-breaker (B5), B10. RPKM (Reads Per Kilo bases per Million mapped Reads) values of CaDnaJ genes were log2-transformed (Wei et al., 2012). Heat maps of CaDnaJ genes in different tissues were performed using software MultiExperiment Viewer (MeV) (Howe et al., 2010).
Plant Materials and Heat Stress Treatment
A hot pepper hybrid (zhejiao 3#), which developed by Zhejiang Academy of Agricultural Sciences, was selected in the experiments. Seeds were sterilized for 5 min using 10% hypochlorous acid solution and washed three times using distilled water. These seeds were further placed in water-saturated filter paper to germinate, then cultivated in Hoagland solution in a growth chamber which was maintained at a 16 h light at 26°C and 8 h dark at 19°C. At the stage of 6–8 true leaves, plants were treated with 42°C for 4 h and plants grown at 25°C were used as the control group. Young leaves were collected and immediately frozen with liquid nitrogen for total RNA extraction. Each treatment was conducted with three biological replicates, and samples from five plants were collected for each replicate.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted using Total RNA kit (Tiangen Biotech, Beijing, China) and reverse-transcribed using FastQuant RT Kit (Tiangen Biotech, Beijing, China), the operational procedure followed the manufacturer’s procedure.
For quantitative RT-PCR analysis, we amplified PCR products in triplicate using 2 × Taq Master Mix (Vazyme, Nanjing, China) in 20 μL qRT-PCR reactions. PCR was performed using the ABI step-one plus 96-well real-time PCR Detection System (Bio-Rad) and cycling conditions consisted of denaturation at 94°C for 5 min, followed by 33 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s. The UBI gene was used as an internal control (Wan et al., 2011). Gene-specific primers were designed and used for amplification as described in Supplementary Table S1. Analysis of relative gene expression data was performed using the 2-ΔΔct method (Livak and Schmittgen, 2001).
Results
Genome-Wide Identification of CaDnaJ Genes in Pepper
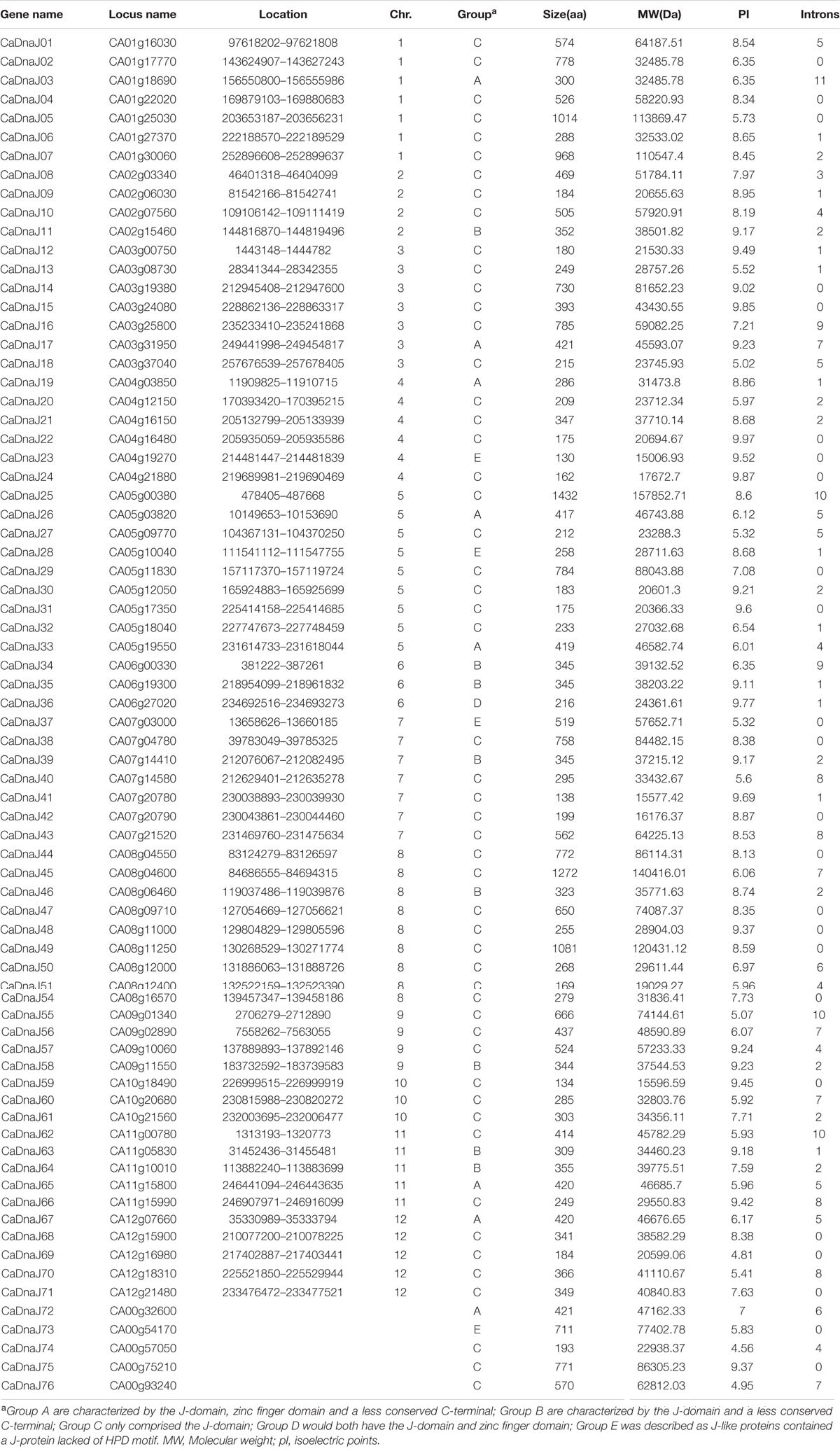
A total of 85 putative sequences of pepper DnaJ genes were gotten from PGD by HMM search. Among them, nine sequences without a complete J-domain (CA03g32700, CA06g22620, CA00g87730, CA12g09540, CA02g21540, CA05g10320, CA11g03100, CA06g00080, and CA05g07590) were removed. The remaining 76 genes were assigned as pepper DnaJ genes. As a matter of convenience, the 76 DnaJ proteins were named as CaDnaJ01 to CaDnaJ76 according to their location on chromosome (Table 1).
The length of CaDnaJ proteins ranged from 130 (CaDnaJ23) to 1272 (CaDnaJ45) amino acids, and the predicted molecular weights were between 15.597 kDa (CaDnaJ59) and 157.85 kDa (CaDnaJ25). The CaDnaJs shared a conserved J-domain comprised about 70 amino acids, in which CaDnaJ57 owned the shortest J-domain with 39 amino acids, while J-domain of CaDnaJ25 was the longest (84 amino acids). The predicted pI-values of CaDnaJ proteins ranged from 4.56 (CaDnaJ74) to 9.87 (CaDnaJ24), indicating acidic and alkaline proteins. Besides, it was also found that 25 (32.89%) of the total 76 CaDnaJ genes had no introns, 11 genes (14.47%) had a single intron, while only CaDnaJ03 had 11 introns (Table 1).
Classification and Sequence Alignment of CaDnaJ Genes
The CaDnaJ protein usually contains conserved J-domain, zinc finger domain, and uncharacterized C-terminal domain (Cheetham and Caplan, 1998). According to the presence of the complete three domains, the CaDnaJ genes were classified into five groups (A, B, C, D, and E), including 9, 8, 53, 1 and 5 members, respectively. Group A CaDnaJ proteins are characterized by the J-domain, zinc finger domain and a less conserved C-terminal. The difference between Group A and B was lack of the zinc finger domain. Group C CaDnaJ proteins only comprised the J-domain, otherwise, Group D would both have the J-domain and zinc finger domain. Group E contains a CaDnaJ protein lacked of HPD motif, which have been described as J-like proteins (Walsh et al., 2004).
Based on the classification above, sequence alignment of CaDnaJ genes was performed separately (Supplementary Table S2). It was found that nine members in group A possessed a conserved HPD motif in J-domain and two zinc finger domains (CxxCxGxG). Eight members in group B were lack of the zinc finger domain but owned conserved HPD motif in J-domain. The largest group (group C) which included 53 members only consisted of complete J-domain. Group D which contained merely one member (CaDnaJ36) had both the conserved HPD motif and zinc finger domain. The last group (group E), which comprised five members, held the least conservation among five groups. All the members in group E possessed J-domains lacked of HPD motif.
Chromosomal Location and Gene Duplication
All these CaDnaJ genes (9 members in group A, 8 members in group B, 53 members in group C, 1 members in group D and 5 members in group E) in pepper were uneven distributed on 12 chromosomes (Figure 1). Among them, eleven and nine genes were located on chromosome 8 and 5, respectively. Seven genes on each of chromosome 1, 3, 7, six genes on chromosome 4, five genes on each of chromosome 11 and 12, four genes on each of chromosome 2, 9, three genes on each of chromosome 6 and 10.
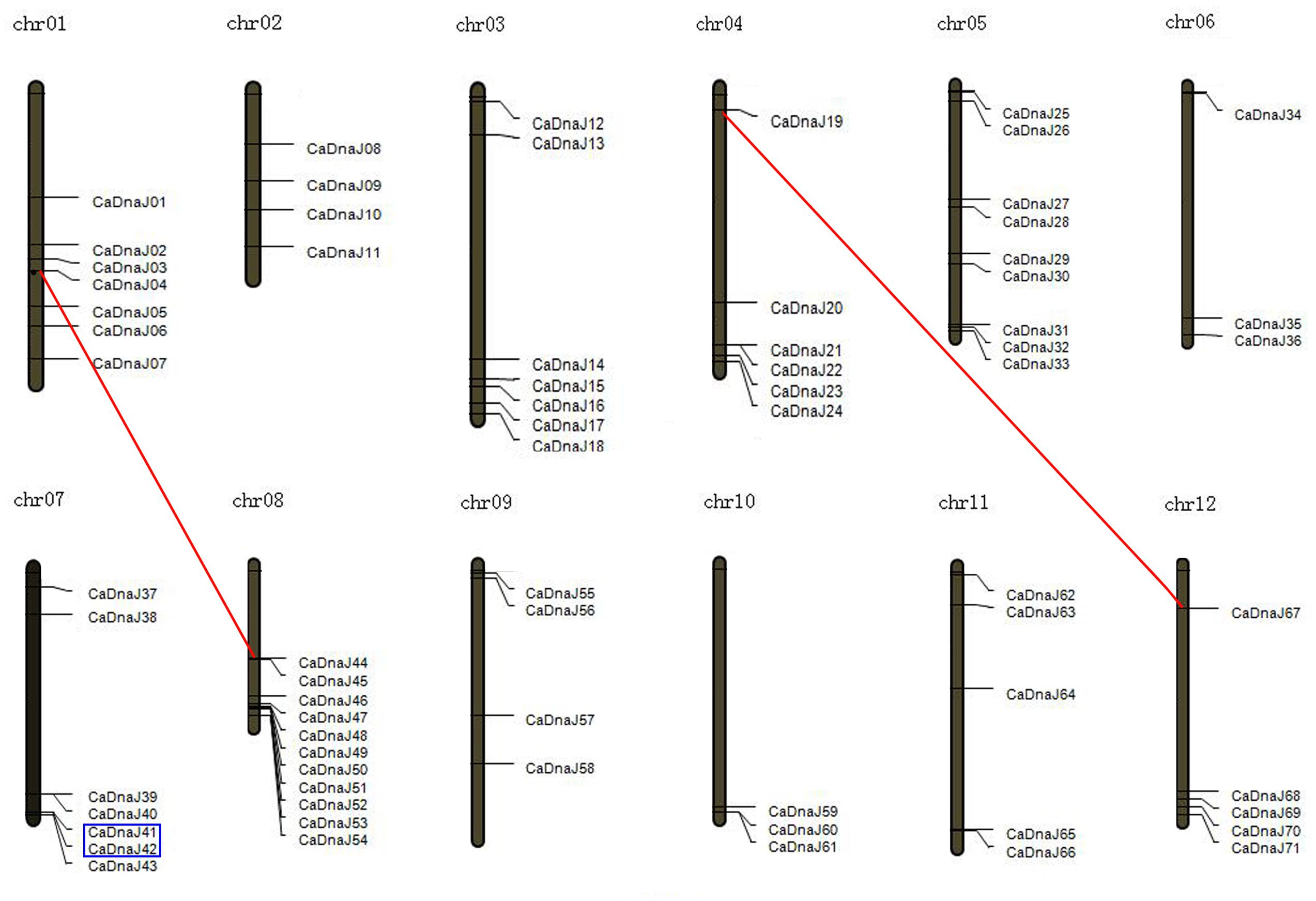
FIGURE 1. Chromosomal map and duplication event coordinates of paralogous CaDnaJ gene candidates. Segmental duplication and tandem duplication were indicated by red lines and blue boxes, respectively.
We further analyzed the gene duplication of CaDnaJ genes in pepper. As shown in Figure 1, one tandem duplication event (CaDnaJ41/CaDnaJ42) was identified on chromosome 7. The chromosome location of this pair of CaDnaJ genes was close in distance and was inserted by less than one gene. In addition, two segmental duplication events were detected. CaDnaJ04 on chromosome 1 presented synteny to CaDnaJ44 localized on a duplicated segment of chromosome 8. Similar scenario was observed for CaDnaJ19 on chromosome 4 and CaDnaJ67 on chromosome 12. These results suggested that these duplication events made contributions to expansion of pepper CaDnaJ gene family.
Analysis of Stress-Related cis-Elements in Pepper CaDnaJ Promoters
The upstream regions (1.5 kb) of the CaDnaJ sequences were searched for regulating factor on different stress conditions. The stress-related cis-elements were not been found in the promoter region of two CaDnaJ genes (CaDnaJ26 and 27), and the rest of 74 CaDnaJ genes possessed multiple cis-elements. Among them, the CaDnaJ 28 and 50 possessed the maximum types of stress-related cis-elements (10). On the contrary, only two types of stress-related cis-elements were held on CaDnaJ10. Besides, CaDnaJ 34 has the most stress-related cis-elements (23), including twelve methyl jasmonic acid (MeJA) related, six drought and indoleacetic acid (IAA) related, four ABA related and one GA related (Figure 2 and Supplementary Table S3).
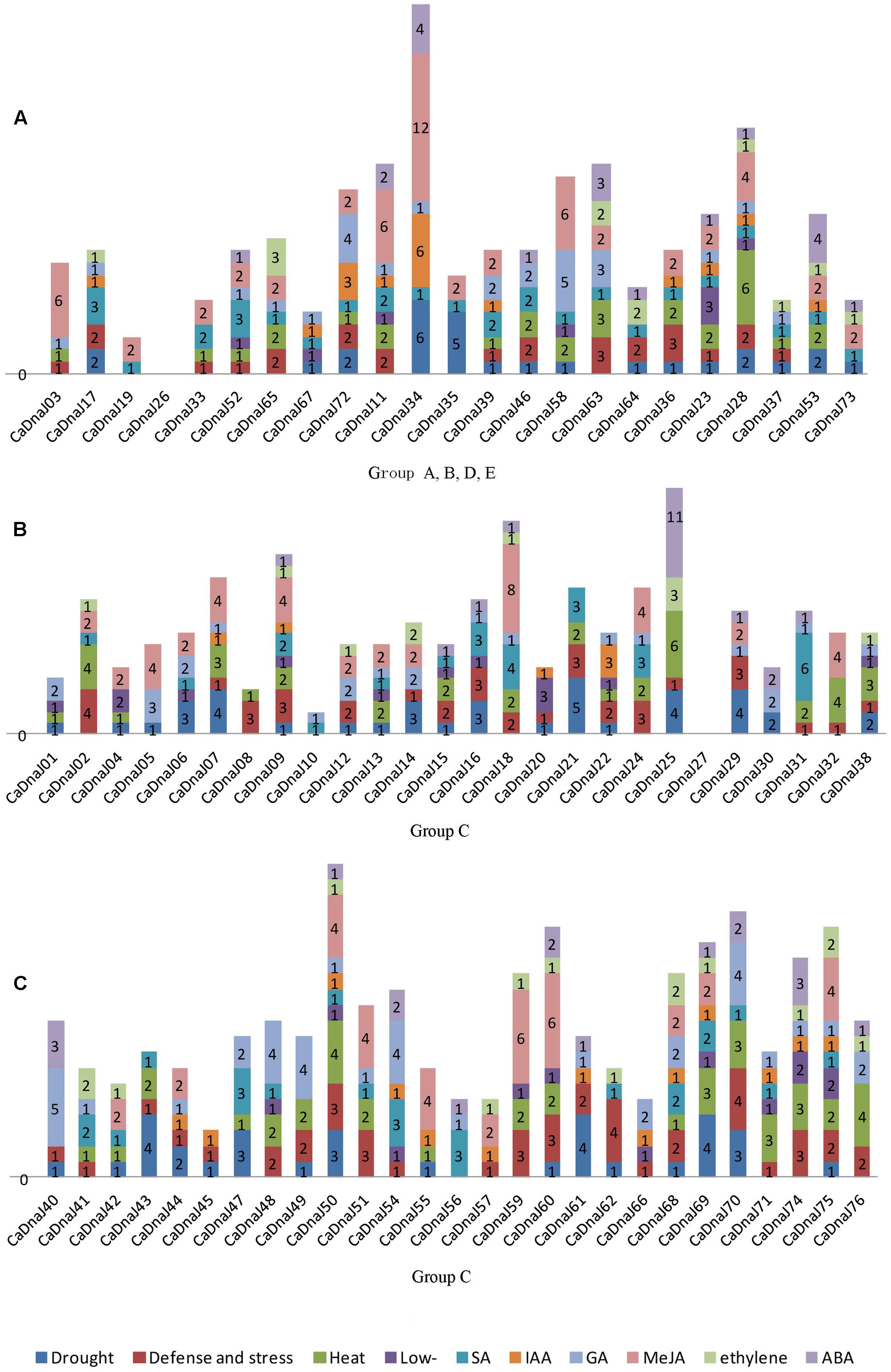
FIGURE 2. Predicted cis-elements in the promoter regions of CaDnaJ genes. Promoter sequences (–1500 bp) for 74 CaDnaJ genes (promoter regions of CaDnaJ26 and 27 were absent) are analyzed. The names of the promoters of CaDnaJ genes are shown on the bottom of the figure. Different cis-elements with the common functions are marked with same color. (A) Predicted cis-elements in the promoter regions of group A, B, D, E. (B) Predicted cis-elements in the promoter regions of group C (26 of 53 CaDnaJs). (C) Predicted cis-elements in the promoter regions of group C (27 of 53 CaDnaJs).
To further explore the possible regulation mechanism of CaDnaJ genes to HS, the heat stress responsiveness elements (HSEs) were also searched in the promoter region of all these CaDnaJ genes. The result showed that 66.21% (49 out of 74) CaDnaJ genes have HSEs in the promoter regions. The maximum numbers of HSEs (6) were identified in CaDnaJ25 and CaDnaJ28. Only one HSEs was found in fifteen CaDnaJ genes (CaDnaJ01, 03, 04, 08, 22, 33, 37, 39, 41, 42, 47, 52, 55, 68, and 72). In addition, other stress-related cis-elements were also detected. There were 107 TC-rich repeats in 55 genes, 99 MBS in 49 genes, 33 LTR in 26 genes, 79 TCA-element in 47 genes, 36 TGA-element in 27 genes, 87 GARE-motif in 49 genes, 138 CGTCA-motif in 41 genes, 37 ERE in 27 genes and 55 ABRE in 28 genes.
Expression Patterns of CaDnaJ Genes in Different Tissues
Based on RNA-seq data of different pepper tissue (root, stem, leaf, and pericarp) published previously (Kim et al., 2014), expression profiles of CaDnaJ genes were revealed (Figure 3). A total of seven different stages of pericarp [6 DPA, 16 DPA, 25 DPA, MG, breaker (B), 5 days post-breaker (B5), B10] were selected for expression analysis in the present study. As shown in Figure 3, 29 out of 76 CaDnaJ genes were barely expressed in the tested tissues, including CaDnaJ01, 03, 05, 09, 12,14, 15, 16, 19, 22, 26, 29, 30, 38, 39, 48, 49, 51, 57, 58, 59, 63, 64, 65, 67, 69, 71, 72 and 75. The left 47 CaDnaJ genes could be detected at least in one tissue. Ten genes (CaDnaJ06, 08, 21, 27, 28, 47, 50, 55, 56 and 76) were expressed in all tested tissues. Of them, four genes (CaDnaJ06, 21, 28 and 76) were expressed at relatively high levels. Besides, tissue-specific expression was also found in some CaDnaJ genes. The CaDnaJ66 was specifically expressed in leaf, CaDnaJ07 and 13 were specifically expressed in root. During the stage of pericarp development, the expression of CaDnaJ02 was significantly up-regulated, while CaDnaJ74 and CaDnaJ45was obviously down-regulated.
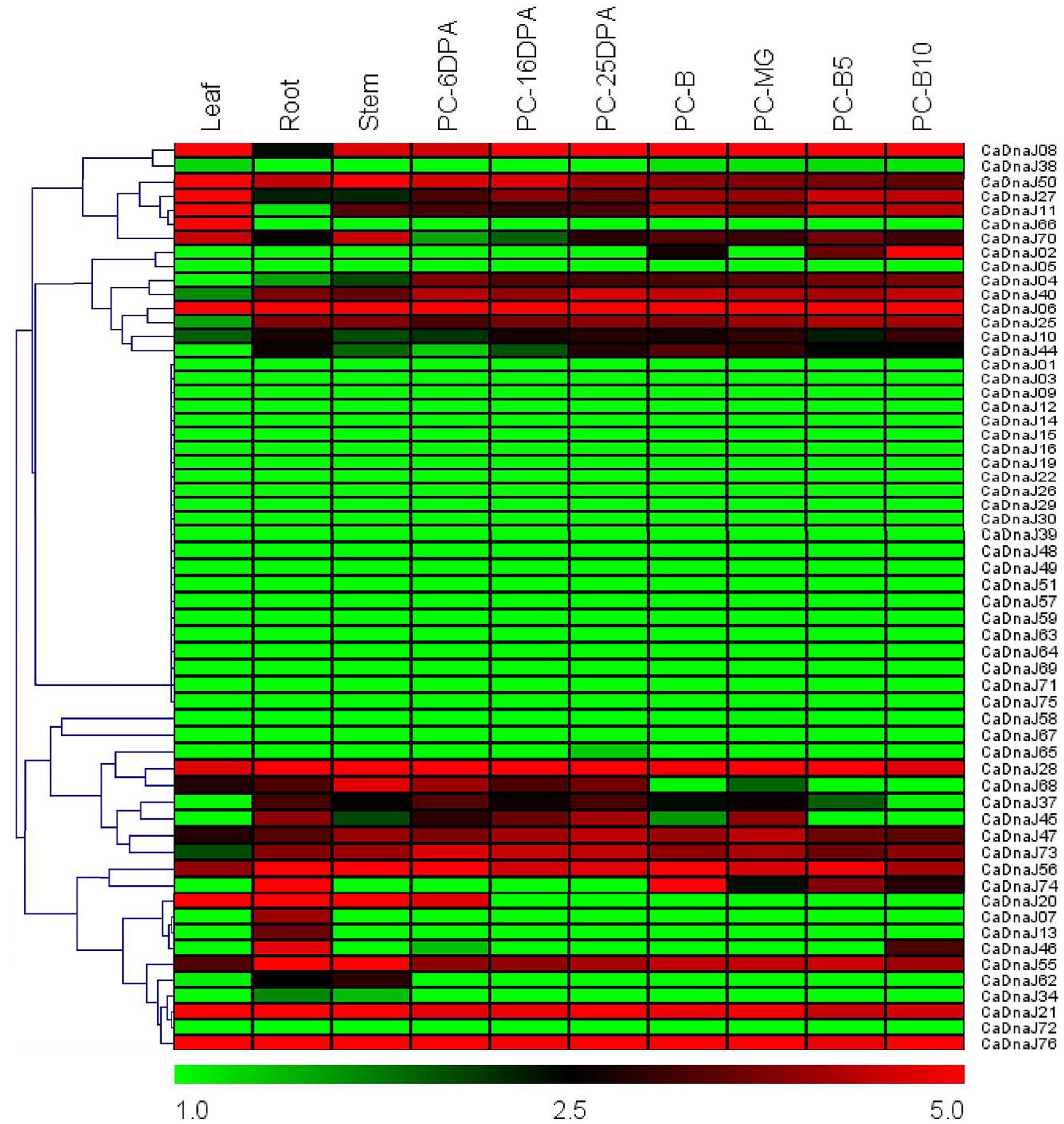
FIGURE 3. Heat map of the expression profiles of CaDnaJ genes in various tissues. The tested tissues are root, leaf, stem, and pericarp. PC, pericarp; MG, mature green; B, breaker; B5, 5 days post-breaker; B10, 10 days post-breaker; 6 DPA, 6 days post-anthesis; 16 DPA, 16 days post-anthesis; 25 DPA, 25 days post-anthesis. Fragments per kilobase of exon model per million mapped (FPKM) values were log2-transformed and heat maps with hierarchical clustering were exhibited using the software Mev 4.9.0.
Expression Patterns of CaDnaJ Genes in Response to Heat Treatments
To gain more insight into the role of CaDnaJ genes under HS condition, expression profiles of CaDnaJ genes in pepper response to high temperature based on qRT-PCR technique were performed. In this study, a total of 67 CaDnaJ genes were used to design successfully specific primers for expression analysis. As shown in Figure 4, expression of these tested CaDnaJ genes was significantly changed under high temperature stress treatments. Expression levels of five CaDnaJ genes (CaDnaJ4, 50, 59, 63, and 72) were down-regulated, and 49 CaDnaJ genes were up-regulated. For the remaining 13 CaDnaJ genes (CaDnaJ1, 17, 27, 31, 43, 48, 49, 53, 54, 58, 60, 73 and 75), no difference was observed. Notably, among the CaDnaJ genes up-regulated, the expression levels of 71.4% (35/49) were increased to three folds. In total, expression of most of CaDnaJ genes was significantly altered under HS, indicating that the CaDnaJ genes were involved in plants response to high temperature stress.
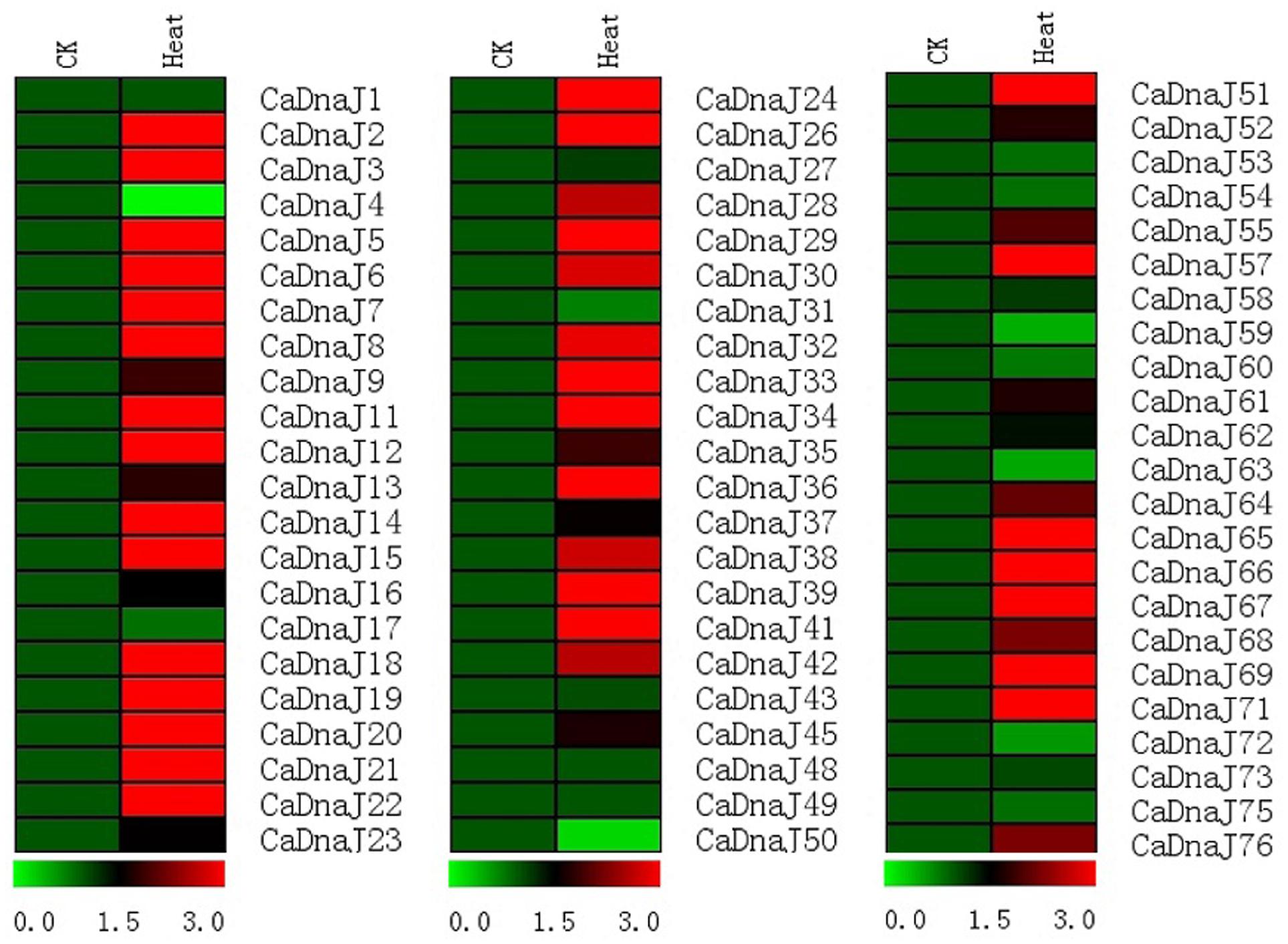
FIGURE 4. Gene expression profiles of CaDnaJ genes in response to heat stress treatment. The expression levels of these CaDnaJs under high temperature stress treatment were tested using qRT-PCR. Green, black, and red elements indicate low, regular, and high signal intensity, respectively.
Discussion
As an important vegetable crop all over the world, pepper is deeply loved by a large population since its major ingredient in cuisines, essential vitamins and other healthy nutrients (Kim et al., 2014). High temperature has become one of the important environmental stresses and affected seriously the growth and development in pepper. The DnaJs, one of the significant Hsps, was produced in the process of plant responding to HS. Up to now, functional identification of DnaJ has been reported in many plant species (Miernyk, 2001; Qiu et al., 2006; Bekh-Ochir et al., 2013; Fristedt et al., 2014; Kong et al., 2014a,b). At the whole genome level, it has been found that Arabidopsis has 89 members and encoded multiple gene family (Miernyk, 2001). Recently, the whole pepper genome has released and provided an opportunity for identifying putative DnaJ homologous. In the current paper, a systematic analysis of CaDnaJ gene family was performed using bioinformatics methods, which focused on gene structure, chromosomal localization, stress-related cis-elements, and expression profiles in different tissues. The results would be significant for further analyzing the function of CaDnaJ members and illuminating the evolutionary relationship in pepper.
Since the first DnaJ proteins were isolated from E. Coli as 41 kDa Hsps (Georgopoulos et al., 1980), many DnaJ proteins have been subsequently reported in other life species (Bukau and Horwich, 1998; Miernyk, 2001; Walsh et al., 2004; Craig et al., 2006). Generally, DnaJ proteins contained one to four domains (J-domain, G/F-domain, zinc finger domains and less conserved C-terminal) (Silver and Way, 1993). Initially, the DnaJ proteins were classified into three groups (A, B, C) based on domain composition (Cheetham and Caplan, 1998; Miernyk, 2001). In this study, more complex structure of DnaJ genes was observed in pepper. For example, the HPD tripeptide which was crucial for J-domain function (Kampinga and Craig, 2010) was not found in CaDnaJ23, 28, 37, 53 and 73. Therefore, given the high diversity of CaDnaJ genes, a more systematic classification was proposed in our study. A total of 76 CaDnaJ genes were classified into five groups (A, B, C, D and E). Group A was characterized by the J-domain, zinc finger domains and a less conserved C-terminal. Group B were lack of the zinc finger domains but contained the J-domain and a less conserved C-terminal. Group C only comprised the J-domain. Group D would both have the J-domain and zinc finger domains but lack of C-terminal, and group E which have been described as J-like proteins with the J-domain lack of HPD motif (Table 1 and Supplementary Table S2).
It was reported that gene duplication was become one of the primary evolution forces during the processes of genetic systems and genomes (Moore and Purugganan, 2003). In our study, chromosome location showed that all these CaDnaJ genes were mapped unevenly on 12 pepper chromosomes (Figure 1). Six of them were involved in gene duplication, including tandem duplication and segmental duplication (Figure 1). One tandem duplication event (CaDnaJ41/CaDnaJ42) and two segmental duplication events (CaDnaJ04/CaDnaJ44, and CaDnaJ19/CaDnaJ67) were observed. These results indicated that both tandem duplication and segmental duplication played role in expansion of the CaDnaJ gene family in pepper.
It has been demonstrated that cis-elements participated in responding multiple abiotic and biotic stresses. For instance, several cis-elements, such as ABRE, DRE, CRT, SARE and SURE, had been identified for responding to ABA, dehydration, cold, SA, and sulfur, respectively (Sakuma et al., 2002; Maruyama-Nakashita et al., 2005; Shi et al., 2010; Osakabe et al., 2014; Feng et al., 2016). In this paper, we identified cis-elements in the promoter regions of CaDnaJ genes using the PlantCARE server (Lescot et al., 2002). Two major groups of cis-elements were observed, including stress-responsive and hormone-responsive. The former contained HSE, LTR, and TC-rich cis-elements, which was responsive to heat, low-temperature, and defense, respectively. The latter was composed of TCA-element, TGA-element, GARE-motif, CGTCA-motif, ABRE and ERE, which was responsive to SA, IAA, GA, MeJA, ABA and ethylene, respectively (Figure 2 and Supplementary Table S3). These results implied that CaDnaJ genes could be involved in the process of plant respond to multiple stresses. Especially, one of the most important cis-element, HSEs, which kept AAAAAATTTC as the core sequence, accounted for 13.64% of all the cis-elements. As we all known that the expression of Hsp was controlled by heat shock transcription factors that bind to HSEs in the promoter region of the Hsp genes (Hancock et al., 2009). A total of 49 (66.21%) CaDnaJ genes have HSEs in the promoter region. Thus, the results will contribute to further understand the vital function role of CaDnaJ genes under HS condition in the further.
To obtain more insights into the expression profiles of CaDnaJ genes in different tissues, RNA-seq data were acquired from leaf, root, stem and different stage of pericarp (Kim et al., 2014). Based on RNA-seq, we found that all these CaDnaJ genes exhibited three different expression patterns: (1) barely expression or too low expression level to detect; (2) constitutive expression; (3) tissue-specific expression patterns. The expression of these CaDnaJ genes differed in tissues tested, indicating that the CaDnaJ proteins may play different functional roles. Expression of some CaDnaJ genes showed tissue- and development-specific in root, stem, leaf and pericarp, suggested that they may participated in growth and development of pepper. In addition, we also found that 10 genes (CaDnaJ06, 08, 21, 27, 28, 47, 50, 55, 56 and 76) were highly expressed in all the tested tissues, implying that they might be involved in specific housekeeping action under normal growth conditions in pepper (Figure 3).
It has been known that plant growth and development were frequently affected by biotic and abiotic stresses under natural conditions (Xia et al., 2014). Previous researchers had reported that the DnaJ protein was involved in plant response to heat and drought stresses (Xia et al., 2014; Wang et al., 2015). To further comprehend the putative roles of CaDnaJ genes in pepper response to HS, expressions patterns of CaDnaJ genes under HS treatment conditions were analyzed by qRT-PCR. Expression analysis revealed that most CaDnaJ genes were changed in response to HS (Figure 4). Among them, almost half of these CaDnaJ genes were up-regulated for three folds. We also found that there are multiple types and numbers of cis-elements in these CaDnaJs promoter, which is reported to involved in abiotic stresses (Feng et al., 2016), including TC-rich, HSEs, MBS motif. All these results suggested that these CaDnaJ genes could involved in plant response to HS.
Author Contributions
Conceived and designed the experiments: FF and YK. Performed the experiments: XY, XC, and YC. Analyzed the data: FF and YK. Wrote the paper: FF. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research is supported by National Natural Science Foundation of China (No. 31301767), Foundation for Distinguished Young Talents in Higher Education of Guangdong, China (No. YQ2015028), and Modern agricultural industry technology system construction of Guangdong, China (No. 2016LM1109).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00689/full#supplementary-material
TABLE S1 | Specific primer for qRT-PCR of each CaDnaJ gene.
TABLE S2 | Multiple sequence alignment of the pepper CaDnaJs. The sequences of 76 CaDnaJs were aligned according to the classification. Names of all the 76 members are showed on the left side of the figure. Conserved amino acid residues are highlighted in black. The sequences which boxed in red rectangle represented the conserved HPD motif, and sequences in yellow rectangle were zinc finger domains.
TABLE S3 | Stress-related cis-elements in pepper CaDnaJ promoters.
Footnotes
- ^ http://peppergenome.snu.ac.kr/
- ^ http://pfam.xfam.org/family/PF00226.29
- ^ http://pfam.xfam.org/
- ^ http://www.expasy.org/tools/protparam.html
- ^ http://chibba.agtec.uga.edu/duplication/
References
Agarwal, M., Katiyar-Agarwal, S., and Grover, A. (2002). Plant Hsp100 proteins: structure, function and regulation. Plant Sci. 163, 397–405.
Bekh-Ochir, D., Shimada, S., Yamagami, A., Kanda, S., Ogawa, K., Nakazawa, M., et al. (2013). A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 237, 1509–1525. doi: 10.1007/s00425-013-1859-3
Caplan, A. J., Cyr, D. M., and Douglas, M. G. (1993). Eukaryotic homologues of Escherichia coli DnaJ: a diverse protein family that functions with Hsp70 stress proteins. Mol. Biol. Cell 4, 555–563.
Cheetham, M. E., and Caplan, A. J. (1998). Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36.
Craig, E. A., Huang, P., Aron, R., and Andrew, A. (2006). The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156, 1–21.
Cyr, D. M., Lu, X., and Douglas, M. G. (1992). Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267, 20927–20931.
Feng, K., Yu, J., Cheng, Y., Ruan, M., Wang, R., Ye, Q., et al. (2016). The SOD gene family in tomato: identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 7:1279. doi: 10.3389/fpls.2016.01279
Fristedt, R., Williams-Carrie, R., Merchant, S. S., and Barkan, A. (2014). A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J. Biol. Chem. 289, 30657–30667. doi: 10.1074/jbc.M114.587758
Georgopoulos, C. P., Lundquist-Heil, A., Yochem, J., and Feiss, M. (1980). Identification of the E. coli DnaJ gene product. Mol. Gen. Genet. 178, 583–588. doi: 10.1007/BF00337864
Glazebrook, J. (1999). Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 2, 280–286. doi: 10.1016/S1369-5266(99)80050-8
Guo, M., Yin, Y. X., Ji, J. J., Ma, B. P., Lu, M. H., and Gong, Z. H. (2014). Cloning and expression analysis of heat-shock transcription factor gene CaHsfA2 from pepper (Capsicum annuum L.). Genet. Mol. Res. 13, 1865–1875. doi: 10.4238/2014.March.17.14
Gupta, S. C., Sharma, A., Mishra, M., Mishra, R. K., and Chowdhuri, D. K. (2010). Heat shock proteins in toxicology: how close and how far? Life Sci. 86, 377–384. doi: 10.1016/j.lfs.2009.12.015
Hancock, M. K., Xia, M., Frey, E. S., Sakamuru, S., and Bi, K. (2009). HTS-compatible β-lactamase transcriptional reporter gene assay for interrogating the heat shock response pathway. Curr. Chem. Genomics 3, 1–6. doi: 10.2174/1875397300903010001
Howe, E., Holton, K., Nair, S., Schlauch, D., Sinha, R., and Quackenbush, J. (2010). “Mev: multi experiment viewer,” in Biomedical Informatics for Cancer Research, ed. M. F. Ochs (New York, NY: Springer), 267–277.
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Jelenska, J., Yao, N., Vinatzer, B. A., Wright, C. M., Brodsky, J. L., and Greenberg, J. T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. doi: 10.1016/j.cub.2007.02.028
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. doi: 10.1038/nrm2941
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kong, F., Deng, Y., Wang, G., Wang, J., Liang, X., and Meng, Q. (2014a). LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. J. Integr. Plant Biol. 56, 63–74. doi: 10.1111/jipb.12119
Kong, F., Deng, Y., Zhou, B., Wang, G., Wang, Y., and Meng, Q. (2014b). A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 65, 143–158. doi: 10.1093/jxb/ert357
Kotak, S., Larkindale, J., Lee, U., von Koskull-Doring, P., Vierling, E., and Scharf, K. D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. doi: 10.1016/j.pbi.2007.04.011
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Z. W., Li, X., Yu, Q. Y., Xiang, Z. H., Kishino, H., and Zhang, Z. (2009). The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insects HSP genes. BMC Evol. Biol. 9:215. doi: 10.1186/1471-2148-9-215
Liu, R. H., and Meng, J. L. (2003). MapDraw: a Microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopes-Caitar, V. S., de Carvalho, M. C., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14:577. doi: 10.1186/1471-2164-14-577
Maruyama-Nakashita, A., Nakamura, Y., Watanabe-Takahashi, A., Inoue, E., Yamaya, T., and Takahashi, H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42, 305–314. doi: 10.1111/j.1365-313X.2005.02363.x
Miernyk, J. A. (2001). The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209–218.
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Mulaudzi-Masuku, T., Mutepe, R. D., Mukhoro, O. C., Faro, A., and Ndimba, B. (2015). Identification and characterization of a heat-inducible Hsp70 gene from Sorghum bicolor which confers tolerance to thermal stress. Cell Stress Chaperones 20, 793–804. doi: 10.1007/s12192-015-0591-2
Ohtsuka, K., and Hata, M. (2000). Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5, 98–112.
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Qiu, X. B., Shao, Y. M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. doi: 10.1007/s00018-006-6192-6
Ritossa, F. (1962). A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18, 571–573. doi: 10.1007/BF02172188
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sarkar, N. K., Kim, Y. K., and Grover, A. (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10:393. doi: 10.1186/1471-2164-10-393
Shi, Z., Maximova, S. N., Liu, Y., Verica, J., and Guiltinan, M. J. (2010). Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis. BMC Plant Biol. 10:248. doi: 10.1186/1471-2229-10-248
Silver, P. A., and Way, J. C. (1993). Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 74, 5–6.
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 42, 579–620. doi: 10.1146/annurev.pp.42.060191.003051
Walsh, P., Bursac, D., Law, Y. C., Cyr, D., and Lithgow, T. (2004). The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571. doi: 10.1038/sj.embor.7400172
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416, 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, G. D., Cai, G. H., Kong, F. Y., Deng, Y. S., Ma, N. N., and Meng, Q. W. (2014). Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol. Biochem. 82, 95–104. doi: 10.1016/j.plaphy.2014.05.011
Wang, G. D., Kong, F. Y., Zhang, S., Meng, X., Wang, Y., and Meng, Q. W. (2015). A tomato chloroplast-targeted DnaJ protein protects Rubisco activity under heat stress. J. Exp. Bot. 66, 3027–3040. doi: 10.1093/jxb/erv102
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wei, K. F., Chen, J., Chen, Y. F., Wu, L. J., and Xie, D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wei, Y., Wan, H., Wu, Z., Wang, R., Ruan, M., Ye, Q., et al. (2016). A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol. Biol. Rep. 34, 512–523. doi: 10.1007/s11105-015-0943-1
Keywords: DnaJ, heat shock protein 40, chromosomal localization, stress-related cis-elements, expression patterns
Citation: Fan F, Yang X, Cheng Y, Kang Y and Chai X (2017) The DnaJ Gene Family in Pepper (Capsicum annuum L.): Comprehensive Identification, Characterization and Expression Profiles. Front. Plant Sci. 8:689. doi: 10.3389/fpls.2017.00689
Received: 21 January 2017; Accepted: 13 April 2017;
Published: 01 May 2017.
Edited by:
Jie Zhou, Zhejiang University, ChinaReviewed by:
Sergey Morozov, Moscow State University, RussiaHao Li, Northwest A&F University, China
Copyright © 2017 Fan, Yang, Cheng, Kang and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunyan Kang, a2FuZ3l1bnlhbkBzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work.
 FangFei Fan1†
FangFei Fan1† Yunyan Kang
Yunyan Kang