- 1ICAR – National Research Centre on Plant Biotechnology, New Delhi, India
- 2Shaheed Udham Singh College of Engineering & Technology, I. K. Gujral Punjab Technical University, Jalandhar, India
- 3Divisions of Genetics, ICAR – Indian Agricultural Research Institute, New Delhi, India
The origin and domestication of rice has been a subject of considerable debate in the post-genomic era. Rice varieties have been categorized based on isozyme and DNA markers into two broad cultivar groups, Indica and Japonica. Among other well-known cultivar groups Aus varieties are closer to Indica and Aromatic varieties including Basmati are closer to Japonica, while deep-water rice varieties share kinship to both Indica and Japonica cultivar groups. Here, we analyzed haplotype networks and phylogenetic relationships in a diverse set of genotypes including Indian Oryza nivara/Oryza rufipogon wild rice accessions and representative varieties of four rice cultivar groups based on pericarp color (Rc), grain size (GS3) and eight different starch synthase genes (GBSSI, SSSI, SSIIa, SSIIb, SSIIIa, SSIIIb, SSIVa, and SSIVb). Aus cultivars appear to have the most ancient origin as they shared the maximum number of haplotypes with the wild rice populations, while Indica, Japonica and Aromatic cultivar groups showed varying phylogenetic origins of these genes. Starch synthase genes showed higher variability in cultivated rice than wild rice populations, suggesting diversified selection during and after domestication. O. nivara/O. rufipogon wild rice accessions belonging to different sub-populations shared common haplotypes for all the 10 genes analyzed. Our results support polyphyletic origin of cultivated rice with a complex pattern of migration of domestication alleles from wild to different rice cultivar groups. The findings provide novel insights into evolutionary and domestication history of rice and will help utilization of wild rice germplasm for genetic improvement of rice cultivars.
Introduction
Rice is the most important food crop of Asia, and its domestication began at least 9,000 years ago, although geographical location(s) of its origin has become controversial (Zeder, 2011; Zou et al., 2015). Archeological evidence shows rice domestication beginning 8,000–9,000 years ago in the Yangtze valley (Liu et al., 2007; Fuller et al., 2010) as well as an independent origin of ancestral Indica or proto-Indica rice in the Ganges valley (Fuller, 2011; Prasad et al., 2011). Molecular dating based on synonymous substitution rates suggest that Indica and Japonica rice have evolved from separate wild rice populations that diverged 0.4 to 0.2 million years ago (Vitte et al., 2004; Zhu and Ge, 2005). During the process of domestication and continuous selection for desirable traits, traditional farmers have slowly transformed wild rice into cultivated rice varieties, which is now a pivotal cereal crop for large segment of the human population. Rice has two genetically divergent broad groups of cultivars, namely Indica and Japonica, based on genome wide SSR markers and selected chloroplast genes (Garris et al., 2005). Rice cultivars have been subdivided based on 15 isozyme loci into six genetically distinct sub-populations namely Aus, Indica, Japonica, Aromatic, Rayada and Ashina (Glaszmann, 1987). The Aus cultivars are also known as ‘Aush’ (in Bengal) and ‘Ahu’ (in Assam), meaning ‘early,’ these are sown in summer with the pre-monsoonal showers and harvested in autumn. Aus cultivars are grown primarily in India and Bangladesh during the short growing season under upland conditions. They are known to carry genes for abiotic stress tolerance including drought, heat and salinity. A clear understanding of the evolutionary relationships of rice progenitor species is essential in directing our efforts to search for agronomically useful genes in the wild rice germplasm (Doebley et al., 2006). Two major hypotheses have been proposed on the origin and domestication of rice: (i) single (monophyletic) origin model suggesting that Indica and Japonica cultivars were domesticated at a single location from the wild rice O. rufipogon (Molina et al., 2011; Huang et al., 2012). (ii) Independent domestication at more than one location (polyphyletic) model suggesting that the major groups of rice cultivars were domesticated separately in different parts of Asia from separate wild rice ancestors (Vitte et al., 2004; Londo et al., 2006; Civan et al., 2015; Singh et al., 2015). Most recent study on 286 diverse O. rufipogon accessions originating from 15 countries using 113,739 nuclear single nucleotide polymorphisms (SNPs) and 25 polymorphic sites in chloroplast sequence has confirmed that three separate wild rice sub-populations were genetically and geographically closely related to Indica, Aus, and Japonica cultivar groups of O. sativa (Kim et al., 2016).
Uneven maturity and seed shattering make wild rice unsuitable for cultivation, although human tribes are known to harvest wild rice seeds from natural populations in innovative ways. Rice was domesticated for cultivation making use of one or more of non-shattering mutations in the SH4 (Zhang et al., 2009), qSH1 (Konishi et al., 2006) sh-h (Ji et al., 2010), and SHAT1 (Zhou et al., 2012) genes, which subsequently have spread to wider geographic areas. During rice domestication selection pressure would have favored high seed producing populations and increased inbreeding leading to fixation of domestication alleles. Introgressions from wild rice into domesticated rice could readily have occurred enabling domesticated rice to adapt to new environments. Most of the wild rice germplasm in the gene banks have been collected from human-made habitats such as roadside, ditches, and irrigation channels in the areas where such introgression may have occurred. In Asia, repeated introgression from wild rice to cultivated rice is more likely to have occurred in Indica varieties than Japonica varieties because of the locations where these two varietal groups are grown (Sweeney and Mccouch, 2007).
In recent years, several agronomically important rice genes have been cloned and characterized at the molecular level, which has led to an enhanced understanding of rice domestication. An ideal rice cultivar should have high grain yield potential with desirable grain size, quality, nutritional value, disease resistance and abiotic stress tolerance. Some of the important domestication related genes cloned in rice include SH4 and qSH1genes for reduction in grain shattering (Konishi et al., 2006; Li et al., 2006), Rc gene for red pericarp color (Sweeney et al., 2006), Bh4 gene for brown hull color (Vigueira et al., 2013) GS3 gene for grain size (Fan et al., 2006), An2 gene for awn length (Gu et al., 2015), GAD1 gene for grain number, grain length and awn development (Jin et al., 2016), BADH2 gene for grain fragrance (Bradbury et al., 2005), CKX1 gene for grain number (Ashikari et al., 2005), GW2 grain weight (Song et al., 2007) and GBSS1 gene for amylose content and cooked rice stickiness (Yamanaka et al., 2004). Studies on sequence variation in these genes have aided in tracing the origin and domestication of rice (Civan et al., 2015). In addition to human selection for specific traits, the environment where crops are grown also may have played a role in selection and changes in the genetic diversity of rice (Choudhury et al., 2014). Domestication related genes have experienced severe genetic bottleneck due to selection pressure by tribal farmers and modern plant breeding whereas, enormous genetic diversity exists in their wild ancestors (Doebley et al., 2006; Zhu et al., 2007). Although some alleles exist only within specific rice cultivar groups, as would be expected if multiple domestications occurred independently, other major domestication alleles are claimed to be common to diverse cultivar groups of O. sativa (Kovach et al., 2007). Most of the earlier studies have traced the origin and domestication of rice based on genome wide SNPs, but gene based approach offers means of inferring the origin and dispersal of specific traits that have been favored over the course of rice domestication and breeding.
Pericarp color controlled by Rc gene located on rice chromosome 7 is an important hallmark trait to examine rice domestication (Gross and Olsen, 2010). The vast majority of rice cultivars have white (non-pigmented) pericarp whereas wild rice species mostly possess dark red pericarp (Sweeney et al., 2006, 2007). The genetic basis of white pericarp in O. sativa has been identified as loss of function mutations of the Rc gene, a regulatory protein in the proanthocyanidin synthesis pathway. The loss of function mutation in this gene prevents development of a pigmented pericarp layer (Sweeney et al., 2006). Another crucial trait involved in rice domestication is grain size, which in addition to other minor genes is controlled by a major gene GS3 located on rice chromosome 3. GS3 was one of the first cloned genes underlying a major QTL controlling grain length and weight. It codes for a protein with several conserved domains including a phosphatidylethanolamine-binding protein (PEBP)-like domain and a trans-membrane region and mutations in this gene leading to both increased and decreased grain length as compared to the wild rice have been reported in the cultivated rice (Fan et al., 2006; Anand et al., 2015). Grain starch quality is another important trait in cereals that has been the target of selection during domestication due to its direct impact on cooking and eating quality (Whitt et al., 2002). Starch, accounts for more than 90% of the dry milled rice grains and is a major determinant of both rice yield and quality (Mohapatra et al., 1993). The waxy locus functionally known as GBSSI gene is one of the extensively studied genes of rice and is highly associated with the taste and texture of cooked rice. This locus provides much information regarding how an important mutation spreads in a crop gene pool. It also provides information related to the impact of human selection particularly on rice quality. Starch synthesis pathway is well characterized in plants and until now more than 20 genes involved in the starch synthesis have been identified in the cereal crops. Among these, six genes are known to play major role in rice endosperm starch synthesis: Shrunken2 (Sh2), Brittle2 (Bt2), Waxy (Wx), Starch synthase IIa (SSIIa), Starch branching enzyme IIb (SbeIIb), and Isoamylase1 (Iso1) (James et al., 2003). GBSSI and SSIIa are major genes involved in important grain quality attributes of rice such as amylose content and gelatinization temperature. Starch synthase genes SSI, SSII, SSIII, and SSIV are responsible for amylopectin chain elongation and their distribution between the granular and soluble fractions (Ball and Morell, 2003; Li et al., 2003). The origin of starch synthase subfamilies is clearly ancient and will provide important clues to rice domestication, given the conservation of their orthologs from Chlamydomonas through to the dicots and monocots (Ball and Morell, 2003).
In the present study we investigated the SNP haplotype variation in Rc gene for pericarp color, GS3 gene for grain size and eight different starch synthase genes in a large set of wild rice accessions together with representative varieties of major rice cultivar groups to understand the process of rice evolution and domestication by employing haplotype network and phylogenetic analysis.
Materials and Methods
Plant Materials
A set of 202 diverse genotypes was used for the analysis of gene haplotype network and phylogenetic relationships among Indian Oryza nivara/Oryza rufipogon wild rice accessions and different rice cultivar groups. The set included a 182 wild rice accessions collected from different geographical regions of India; 91 from Mid-Gangetic Plains (MGP), 23 from West-Himalayan Region (WHR), 18 from West Coastal Plains (WCP), 17 from East Plateau Hills (EPH), 9 from Gujarat Plains and Hills (GPH), 7 from Lower Gangetic Plains (LGP), 6 from East Coastal Plains (ECP), 6 from East Himalayan Region (EHR), and 5 from Upper Gangetic Plains (UGP). These wild rice accessions represented admixture-free genotypes representing three different pure model based sub-populations (Pro-Indica, Pro-Aus, and Mid-Gangetic) of Indian O. nivara/O. rufipogon wild rice accessions (Fst values ≥ 0.9, our unpublished results). In addition five of each genotype of Indica, Japonica, Aromatic, and Aus cultivar groups were also included in the analysis. Details of the wild rice accessions and rice cultivars used in present study are provided in Supplementary Table S1 and our online wild rice database1.
DNA Extraction and SNP Array Hybridisation
Genomic DNA was extracted from young green leaves using the method of Murray and Thompson, 1980 with minor modifications, quantified using Nano-drop spectrophotometer absorbance at 260/280 nm and quality checked by electrophoresis in 1% agarose gel. For SNP genotyping an Affymetrix 50K genic SNP chip ‘OsSNPnks’ described earlier was used (Singh et al., 2015). For target probe preparation, 20 μL of genomic DNA was used for each sample, with DNA concentration of 10 ng/μL (200 ng DNA in 20 μL) according to Affymetrix Axiom® 2.0 Assay Manual. DNA amplification, fragmentation, chip hybridisation, single-base extension through DNA ligation and signal amplification were performed using Affymetrix Axiom® 2.0 Assay Manual Target Prep Protocol QRC (P/N 702990). Staining and scanning were performed on the GeneTitan® Multi-Channel instrument according to manufacturer’s instructions2.
SNP Calling and Data Analysis
SNP genotypes were called using Affymetrix Genotyping ConsoleTMv4.1 and AxiomTM Suite. SNPs with low call rates across all samples were removed from the dataset, and high-performing SNPs with a DQC of >0.85 and call rates of >95.0% were used for analyses. To evaluate the data quality the dataset was imported in other compatible formats, such as PLINK (Purcell et al., 2007) and text, was examined using two different softwares, APT with R package and SNPolisher3. Out of the large number of SNPs on the chip we extracted SNPs in 10 agronomically important domestication related rice genes for haplotype and phylogenetic analysis (Table 1).

TABLE 1. Details of genes with number of SNPs, haplotypes and number of rice accessions used for haplotype network and phylogenetic analysis.
Haplotype Network and Phylogenetic Tree Construction
SNP haplotypes for the selected genes were generated using TASSEL 3.2.1 (Bradbury et al., 2007). Based on the SNP haplotype analysis for each gene, a phylogenetic tree was constructed using an improved version of the neighbor-joining algorithm, and visualized using FigTree v1.4.0 (Rambaut, 2009). Haplotype network for each gene was constructed for analysis of genealogical relationship among the haplotypes using Network software4 (Bandelt et al., 1999) and haplotype diversity was calculated with the DnaSP software version 5.105 (Rozas et al., 2003).
Results and Discussion
Rc Gene for Red Pericarp
Haplotype variation in Rc gene was examined in 179 geographically and genetically diverse genotypes, including 159 wild rice accessions and five each of representative varieties from Aus, Indica, Japonica, and Aromatic rice cultivar groups. Even though additional SNP and SSR have been reported in the Rc gene (Sweeney et al., 2006, 2007), present analysis was based on four SNPs in the Rc gene that could be genotyped with high quality score using the 50K rice SNP chip (Singh et al., 2015). Haplotype network analysis revealed four haplotypes (Rc-H1, Rc-H2, Rc-H3, and Rc-H4) with a haplotype diversity (Hd) of 0.7076. Rc-H3 was the most ancestral haplotype located at the root of the NJ phylogenetic tree (Figures 1A,B). Interestingly, Rc-H3 haplotype was present in 62 wild rice accessions all belonging to the MGP region, but it was represented by all three structure based sub-populations of wild rice, viz. ‘Pro-Indica,’ ‘Pro-Aus,’ and ‘Mid-Gangetic types, although predominantly in the Mid-Gangetic sub-population. All wild rice accessions with Rc-H3 haplotype have red pericarp. The other three haplotypes Rc-H1, Rc-H2, and Rc-H4 were derived from Rc-H3 but their origin followed two independent routes, one from Rc-H3 to Rc-H1 while another one was from Rc-H3 to Rc-H4 to Rc-H2 (Figure 1A). Based on NJ phylogenetic tree Rc-H1 was the most recently evolved haplotype, which is shared by Indica, Japonica, and Aromatic rice cultivars, all having white pericarp. In contrast, all the Aus cultivars possessed haplotype Rc-H2 that was shared by 35 wild rice accessions predominantly from the ‘Pro-Aus’ wild rice sub-population. These ‘Pro-Aus’ accessions were collected from diverse geographical regions of India (Supplementary Table S1). Genotypes with Rc-H2 haplotype showed red, green, and white pericarp, suggesting that there are other genes involved in the pericarp color development or there may be further differentiation of the Rc-H2 haplotype based on additional SNP information (Cui et al., 2016). While wild rice accessions with this haplotype showed red or green pericarp, the Aus cultivars showed green or white pericarp (Figures 1A,B). In any case presence of a different Rc allele for white pericarp in Aus cultivars showed independent domestication of Aus cultivars (Civan et al., 2015; Singh et al., 2015). Earlier, a monophyletic origin of cultivated rice has been claimed based on occurrence of common haplotype of Rc gene in Japonica and Indica rice cultivar groups.
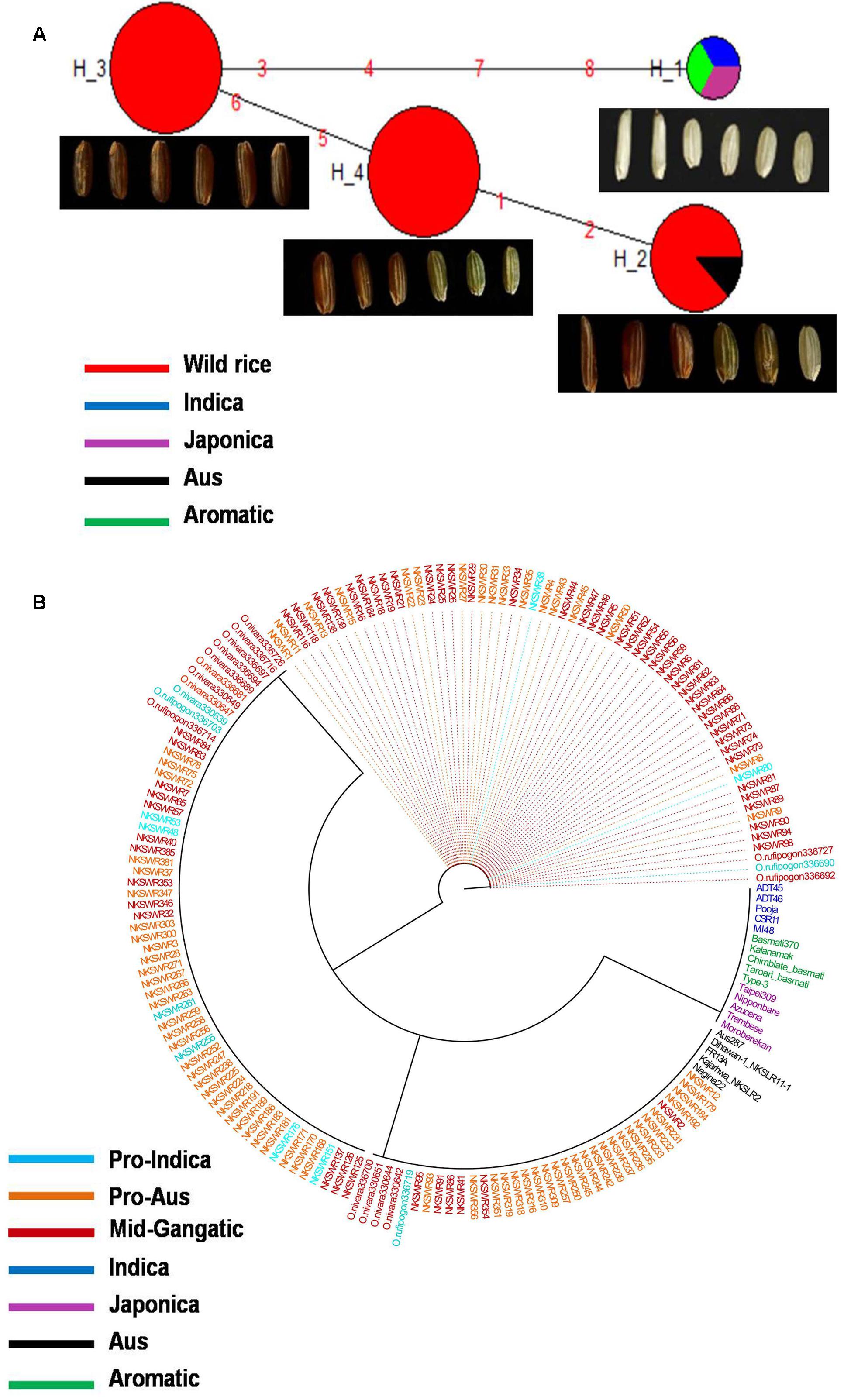
FIGURE 1. (A) A haplotype network of Rc gene along with its pericarp color, in total four haplotypes were formed, Rc-H3 was the ancestral haplotype of wild rice. The size of each circle is proportional to haplotype (allele) frequency. To determine the origin and dispersal of these mutations in Rc gene we examined in total 179 diverse rice genotypes. Color-coding represents different population groups (Red-Wild rice, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica). (B) Haplotype based phylogenetic tree of Rc gene was constructed using 179 diverse rice genotypes and were separated into four major distinct groups. Color-coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
Haplotype analysis of Rc gene conducted earlier by Sweeney et al. (2007) showed that an identical 14 bp deletion was present in 97.9% of the cultivated rice accessions with white pericarp, this deletion was not found in any wild or cultivated rice with red pericarp (Furukawa et al., 2007; Cui et al., 2016). In congruence with our results these studies have also observed, though not highlighted, a second mutation in the Rc gene (Rc-S allele) for white pericarp in the Aus group of rice cultivars. The 14 bp deletion most likely originated in the Japonica-Aromatic cultivar group and then it crossed both geographic and sterility barriers to move into the Indica cultivars perhaps by cultural exchange among ancient farmers over the course of rice domestication with strong positive selection for white pericarp in both Indica and Japonica cultivar groups. The ancestral haplotype Rc-H3 with red color pericarp was present in wild rice accessions along with Rc-H2 and Rc-H4 haplotypes. The recessive rc allele (Rc-H1 haplotype in present study) leading to white pericarp is predominant in the modern rice varieties, but our study confirms that white pericarp selected in the independent domestication of Aus cultivars is due to a different mutation in the Rc gene, a fact not adequately emphasized in the earlier studies. It showed that different mutations leading to same preferred phenotypic trait have arisen in different wild rice sub-populations leading to separate domestication events. Red pericarp wild rice and landraces carrying functional Rc haplotypes are still grown in different parts of India. Wild rice haplotype Rc-H3 was the most ancestral haplotype as most of the red pericarp wild rice clustered within this group (Figures 1A,B). Selection for non-pigmented grains in rice may potentially reflect human selection against seed dormancy, selection for improved taste and cooking qualities and also with the pleasant white appearance. However, the pigment serves as a powerful antioxidant that has been demonstrated to reduce cardiovascular disease, diabetes and help in cancer prevention (Zhang et al., 2010). In another domestication related rice gene BADH2, multiple mutations have been found for the loss of function of gene responsible for grain aroma (Shi et al., 2008). Our results indicate that white pericarp in modern cultivated rice has come from at least two different domestication events in wild rice.
GS3 Gene for Grain Length
GS3 gene located on chromosome 3 is a major gene for grain size in rice, which has also been under strong selection pressure during domestication. Long grain rice varieties are favored in many countries due to their beautiful appearance and high volume expansion upon cooking. There is great diversity of seed size and shape both within and between different sub-populations of rice cultivars, although each sub-population is associated with characteristic seed morphology. Domestication process has resulted in both extra long and extra short rice grains as compared to the wild rice because of different mutations in the GS3 gene (Zhao et al., 2010; Anand et al., 2015). Here we examined the haplotype network and phylogenetic relationship of GS3 alleles to find evidence for the origin and selection of domestication alleles and their association with different sub-populations of wild and cultivated of rice. The SNP variation in GS3 gene was analyzed in 160 diverse wild rice and 20 representative rice cultivars. The haplotype network and phylogenetic tree were constructed based on nine high quality SNPs in the GS3 gene that identified total nine haplotypes with an ‘Hd’ value of 0.6214. There were three major (GS3-H1, GS3-H4, and GS3-H9) and six minor (GS3-H2, GS3-H3, GS3-H5, GS3-H6, GS3-H7, and GS3-H8) haplotypes. It was clear from the phylogenetic analysis that domestication of GS3 gene has followed three independent paths originating from the most ancestral wild rice haplotype GS3-H4 which was shared by the majority 88 wild rice accessions along with four Aus and four Aromatic rice cultivars (Figures 2A,B). The GS3-H4 haplotype was shared by all three wild rice sub-populations, namely ‘Pro-Indica,’ ‘Pro-Aus,’ and ‘Mid-Gangetic’ types indicating its ancient origin. Six minor haplotypes were restricted to cultivated rice, indicating their recent origin under human selection. Two of the three major wild rice haplotypes were also shared by the rice cultivars suggesting that a range of seed sizes have been selected by humans during domestication and breeding while wild environment favored optimum size for efficient seed dispersal and survival. Here, GS3-H1 was the most versatile haplotype comprising of 4 Indica, 2 Aromatic, and 18 wild rice accessions (eight from MGP, four each from LGP and EPH, and two from WCP regions). The third major haplotype GS3-H9 was restricted to 54 wild rice accessions (40 from MGP and 14 from WCP). Five Japonica cultivars possessed three minor haplotyes (GS3-H6, GS3-H7, and GS3-H8), the other three minor haplotypes were represented one each by Indica (GS3H2), Aus (GS3-H3), and Aromatic (GS3-H5) cultivars.
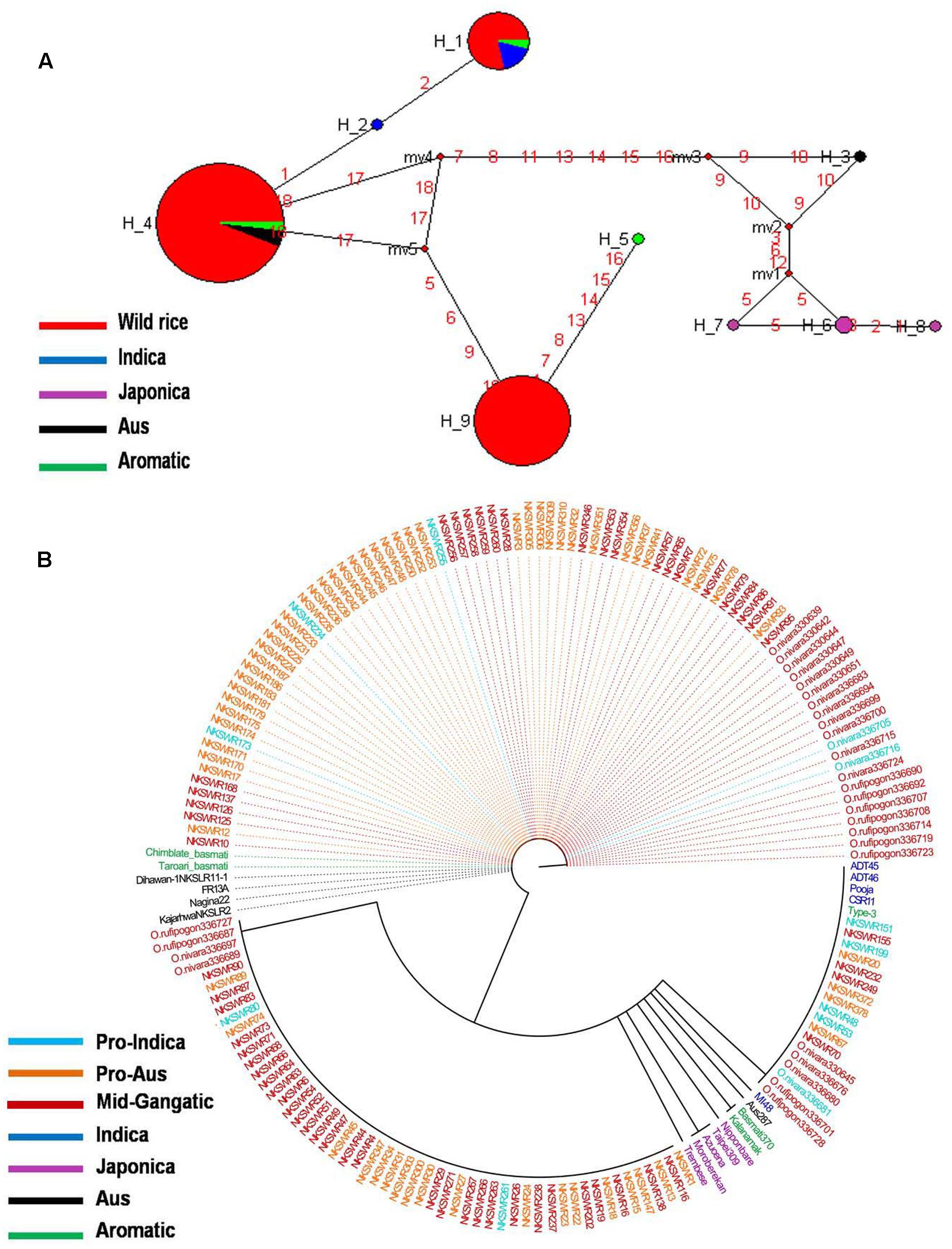
FIGURE 2. (A) Haplotype networks of GS3 gene, in total nine haplotypes were generated, GS3-H4 was the ancestral haplotype representing wild rice along with Aus and Aromatic cultivars. (B) Haplotype based phylogenetic tree of GS3 gene was constructed using 180 diverse rice genotypes and were separated into nine distinct groups. Color-coding represents different sub-populations or varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
We also calculated the average grain length (GL) and grain width (GW) for the three major haplotypes. The average GL for haplotype GS3-H4 was 3.71 mm, average GW 1.39 mm and GL/GW ratio of 2.66 mm. The GS3-H9 haplotype comprising entirely of 54 wild rice accessions mostly from the MGP region has longer average GL, GW, and GL/GW ratio of 4.12, 1.44, and 2.86 mm, respectively. The GS3-H1 haplotype showed the highest average GL of 3.97 mm, GW of 1.33 mm, and GL/GW of 2.98 mm comprising long slender grains. It shows that short grain Aus and Aromatic rice cultivars were domesticated first then subsequent selective breeding by generations of farmers, diversified it into several different grain sizes based on selection of random mutants and recombination with other genes in the domesticated rice, for improved yield, quality and other agronomic or cultural value traits (Deb, 2005). Five Japonica cultivars known for their short, round shape and sticky cooked grains showed most diverse haplotypes derived from mutations in the ancestral wild rice haplotype. Similarly, Indica and Aus cultivars are known for long slender grains while Aromatic rice varieties have both long and short grains, which seem to be directly associated with their GS3 haplotype. In a previous study on the haplotype analysis of 54 diverse rice accessions of 86 SNPs and 28 indels were identified in a 6.57 kb GS3 DNA, suggesting origin of long grain allele in a Japonica type ancestor and subsequent introgression into the Indica gene pool (Takano-Kai et al., 2009). But GS3 haplotype network and phylogenetic analysis here shows independent path of the evolution of Japonica and Indica haplotypes, while major Aus, Indica, and Aromatic rice haplotypes were shared with the wild rice accessions. Earlier, haplotype analysis of 282 diverse rice cultivars has also indicated multiple independent origins of short seeded GS3 alleles suggesting that farmers and early breeders forced artificial selection favoring short seeds (Takano-Kai et al., 2013; Anand et al., 2015). Intercrossing of short and long grain varieties has resulted into evolution of intermediated grain size varieties and modern superior varieties are the result of domestication and artificial selection on GS3 and other grain size genes, including qGL7.1 and qGL1.1 (Amarawathi et al., 2008; Bai et al., 2010; Anand et al., 2015). A GS3 haplotype conferring extra short grain was selected by traditional Indian farmers (Anand et al., 2015).
The preference for grain size and shape varies from one group of consumers to another. Generally long slender grains are preferred in Southeast Asia while Chinese consumers prefer short and bold rice grains. Indian consumers like rice with diverse grain size ranging from very short to extra-long grains with shapes ranging from bold to slender. Consumers in Indonesia and Bangladesh prefer medium slender grains. Japanese, Taiwanese, and South Koreans consume short and bold rice grains, whereas in several states of India, and Sri Lanka, both short and medium grain types are popular (Calingacion et al., 2014). All the short and medium grains genotypes carry the C-allele and the long and extra-long grains carry the A-allele of the GS3 gene (Fan et al., 2009). Further, the grain size and shape have direct impact on yield, head rice recovery during milling. Although popular Basmati rice fetching premium price in the international market has long slender grains, other indigenous Indian aromatic rice varieties have short or medium seed size.
GBSSI (waxy) Gene for Amylose Starch Synthesis
Starch is a major component of the human diet and constitutes about 90% of the total dry weight of polished rice. It occurs in two forms: amylose (15–30%), consisting of predominantly linear chains of glucose monomers linked by α 1–4 glycosidic bonds, and amylopectin (70–85%), in which the chains are branched by the addition of α1–6 glycosidic bonds. Starch property has been under strong selection pressure in rice domestication as it directly affects the cooking and eating quality of the rice. Rice genome has five sub-families of starch synthases, namely granule bound starch synthase (GBSS) and soluble starch synthase I, II, III, and IV (SSI, SSII, SSIII, and SSIV). Each class of soluble starch synthase plays a distinct role in the synthesis of amylopectin. GBSSI also kwon as Waxy (Wx) gene located on rice chromosome 6 encodes a granule-bound starch synthase that is required for amylose synthesis in rice grains has played a crucial role in rice domestication (Hirano et al., 1998; Isshiki et al., 1998). However, other minor genes such as dull and amylose extender (ae) are also reported to play important roles in modification of amylose in rice (Isshiki et al., 2000). The Wx genes have been cloned in O. sativa (Wang et al., 1990; Hirano and Sano, 1991; Okagaki, 1992) and O. glaberrima (Umeda et al., 1991). Two wild-type alleles, Wxa and Wxb, were found at the waxy locus in cultivated rice. Wxa is characteristic of Indica rice, while Wxb is found in Japonica rice (Sano, 1984). Mutation in this gene drastically reduced the amylose synthesis in glutinous rice varieties. Glutinous rice contains a G to T mutation at the 59 splice site of Wx intron 1, which leads to incomplete post-transcriptional processing of the pre-mRNA (Wang et al., 1995; Frances et al., 1998; Hirano et al., 1998; Isshiki et al., 1998). Rice cultivars are categorized based on amylose content as waxy (0–5%), very low (5–12%), low (12–20%), intermediate (20–25%), and high (25–33%). Wild rice does not have glutinous starch which was selected only after domestication, and evolution of glutinous rice may have occurred in various stages (Vaughan et al., 2008). Japonica rice varieties having low amylose content (2–8%) are sticky after cooking as compared to long-grained Indica and Aus varieties. Expression of GBSSI gene is developmentally regulated as it is highly expressed during seed development, which is crucial for amylose synthesis (Sano, 1984; Hirose and Terao, 2004; Ohdan et al., 2005; Fujita et al., 2011). The waxy mutation creating a SNP in exon four of the GBSSI gene reduces the binding of GBSSI enzyme to starch granules resulting in low amylose synthesis (Liu et al., 2009).
Haplotype network and phylogenetic analysis of GBSSI gene was performed using 8 SNPs in a set of 182 wild rice and 20 diverse rice cultivars resulting in eight haplotypes with low ‘Hd’ value of 0.2994. There were two major haplotypes designated GBSSI-H1 and GBSSI-H2 and six minor haplotypes designated GBSSI-H3 to GBSSI-H8. Similar to that with the GS3, wild rice accessions showed significantly less haplotype diversity than rice cultivars. GBSSI-H2 was by far the most predominant and also most ancient haplotype from which the remaining seven haplotypes were derived. It was highly conserved in 164 of the 182 wild rice accessions collected from diverse geographical regions of India (97 MGP, 26 EPH, 14 WHR, 12 UGP, 10 LGP, and 5 WCP) and was also shared by five Aus cultivars, once again showing that Aus cultivars have the most ancient of the four rice cultivar groups. All three sub-populations of wild rice, ‘Pro-Indica,’ ‘Pro-Aus,’ and ‘Mid-Gangetic’ were represented by the ancestral GBSSI-H4 haplotype. Evolution of GBSS1 gene in rice cultivars has taken three independent paths with Aus cultivars having the most ancient GBSS1-H2 haplotype, which was also conserved in 89.1% of the wild rice accessions. The GBSSI-H1 haplotype represented 13 wild rice accessions from wide geographical regions (6 EHR, 3 MGP, 2 each from EPH and WCP) together with all five Indica varieties. Of the 13 wild rice accessions sharing GBSSI-H1 haplotype with the Indica rice cultivars, nine were from ‘Pro-Aus’ and three from ‘Mid-Gangetic’ sub-populations, but none were from Pro-Indica group, suggesting that the GBSS1 haplotype in the Indica rice cultivars has its origin in non-Pro-Indica wild rice ancestors. The five Indica rice cultivars used here possess superior eating quality which is also reported for several wild rice populations harvested by the tribal in different parts of India. Three minor haplotypes GBSSI-H3, GBSSI-H4, and GBSSI-H5 of Japonica varieties were derived from the ancestral haplotype GBSSI-H2. Interestingly, all the five aromatic rice cultivars both short grain and long grain basmati type share a common haplotype GBSSI-H6, which was derived from Japonica haplotype GBSSI-H3. Two more minor haplotypes GBSSI-H7 and GBSSI-H8 were found uniquely in the wild rice accessions collected from the WCP region (Figures 3A,B).
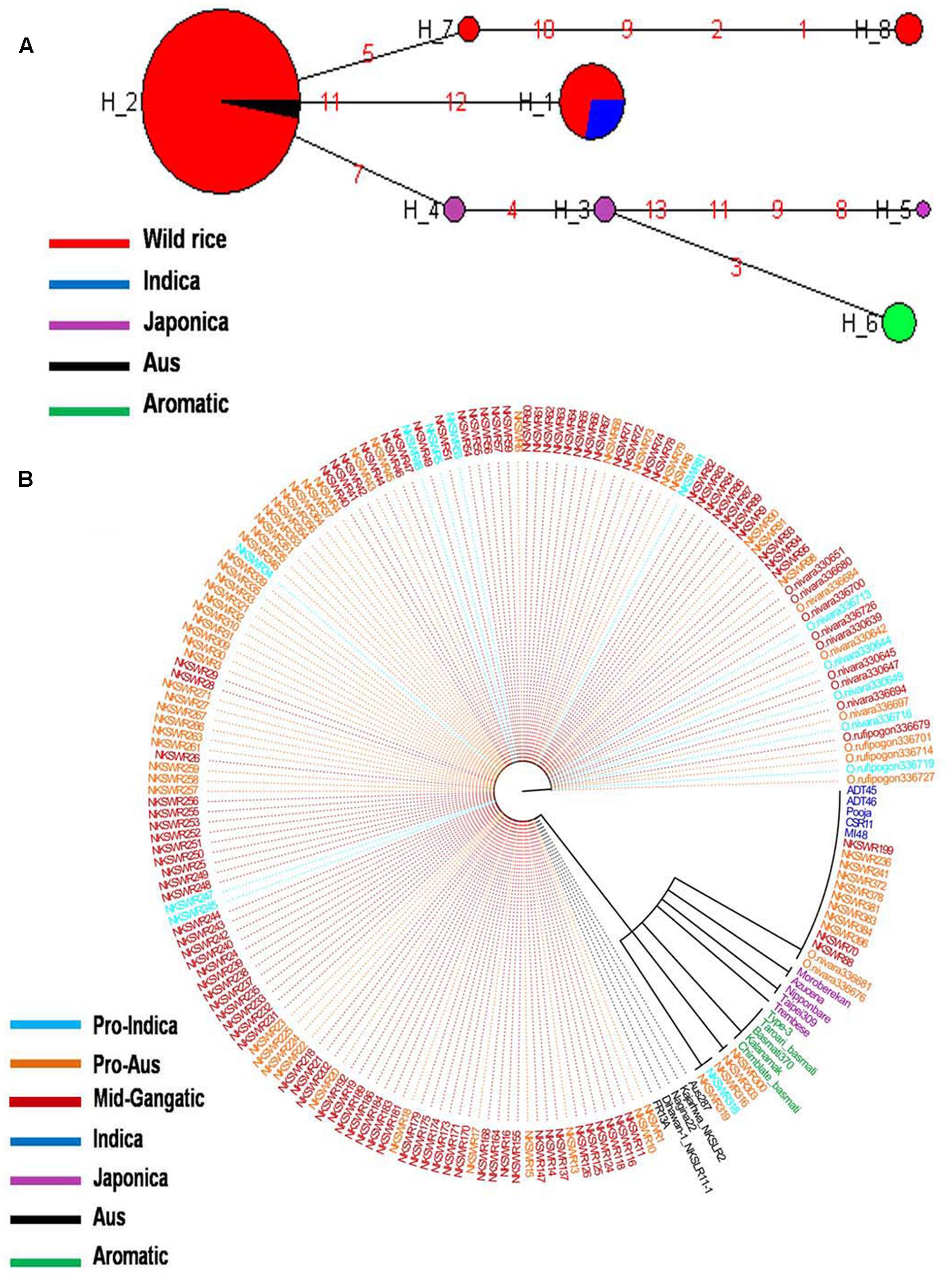
FIGURE 3. (A) Haplotype networks of GBSSI gene, in total eight haplotypes were formed, GBSS1-H2 was the major ancestral haplotype of wild rice sub-populations, shared with Aus cultivars. (B) Haplotype based phylogenetic tree of GBSSI gene was constructed using 202 diverse rice genotypes and were separated into eight distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
It was exciting to see that similar to the grain size gene GS3, GBSSI gene was also more conserved in wild rice than in cultivated rice. While 182 wild rice genotypes showed only four haplotypes, only 20 rice cultivars possessed six different haplotypes two of which were shared by wild rice. The highly conserved haplotype GBSS1-H2 present in 89.1% of the wild rice accessions must have selective advantage for survival in natural environment. On the other hand large number of haplotypes have been selected during the domestication and breeding of rice cultivars for various user preferences of cooking and eating quality. These results also showed three independent origins of cultivated rice, the most ancestral haplotype is shared by the Aus cultivars which have intermediate amylose content, the second haplotype is common to high amylose Indica cultivars that is shared by another small group of mostly Pro-Aus wild rice accessions and the third clade is that of Japonica and Aromatic rice cultivars which have low to intermediate amylose content. Japonica cultivars showed the maximum number of three haplotypes in just five varieties and Aromatic cultivars including both short grain and basmati rice have single haplotype originating from one of the Japonica haplotypes. These results are consistent with recent genome wide phylogenetic studies showing tri-phyletic origin of domesticated rice (Civan et al., 2015; Singh et al., 2015; Kim et al., 2016).
The three main properties that determine rice eating and cooking quality are, amylose content, gel consistency, and gelatinization temperature, which also correlate with one another. Grain amylose content is the major determinant of rice eating and cooking qualities. High amylose rice is mainly found in tropical countries while low amylose rice is preferred in temperate countries. High amylose rice varieties are less sticky, have low glycemic index which is beneficial for diabetics. Rice with intermediate amylose content is preferred in Iran, Laos, Pakistan, Malaysia, Philippines, India, and some provinces of China, Vietnam, Indonesia, and Uruguay. High amylose and low amounts of amylopectin varieties are popular in Myanmar, Sri Lanka, provinces of Indonesia, and many states of India (Calingacion et al., 2014).
SSSI Gene for Amylopectin Synthesis
Soluble Starch Synthase I (SSSI gene) located very close to the GBSS1 gene on rice chromosome 6 is responsible for the formation of α 1–4 glycosidic bonds in amylopectin. The expression of SSSI gene in Indica rice cultivars is lower than that of Japonica rice which reduces the synthesis of short chains in Indica rice amylopectin (Takemoto-Kuno et al., 2006). Haplotype network and phylogenetic analysis of SSSI gene was performed based on 46 SNPs in a set of 66 diverse rice genotypes including 46 wild rice accessions and 20 representative rice cultivars. Total 17 haplotypes were generated, one major (SSSI-H14), the remaining 16 were minor haplotypes with overall ‘Hd’ value of 0.6466. SSSI-H14 was the most ancestral haplotype present in 39 wild rice accessions, all from the MGP region. Indica and Aus cultivars were represented by three haplotypes each, SSSI-H1-SSSI-H3 and SSSI-H9-SSSI-H11, respectively. Five Japonica cultivars were represented by two minor haplotypes (SSSI-H12 and SSSI-H13). Each of the five aromatic cultivars possessed a different haplotype (SSSI-H4-SSSI-H8). There were also three minor haplotypes (SSSI-H15-SSSI-H17) of wild rice derived from SSSI-H14 (Figure 4 and Supplementary Figure S1). The results showed that Japonica, Indica, Aus, and Aromatic rice cultivars have evolved from separate ancestral wild rice groups and then further separated into minor haplo-groups, supporting polyphyletic origin of rice.
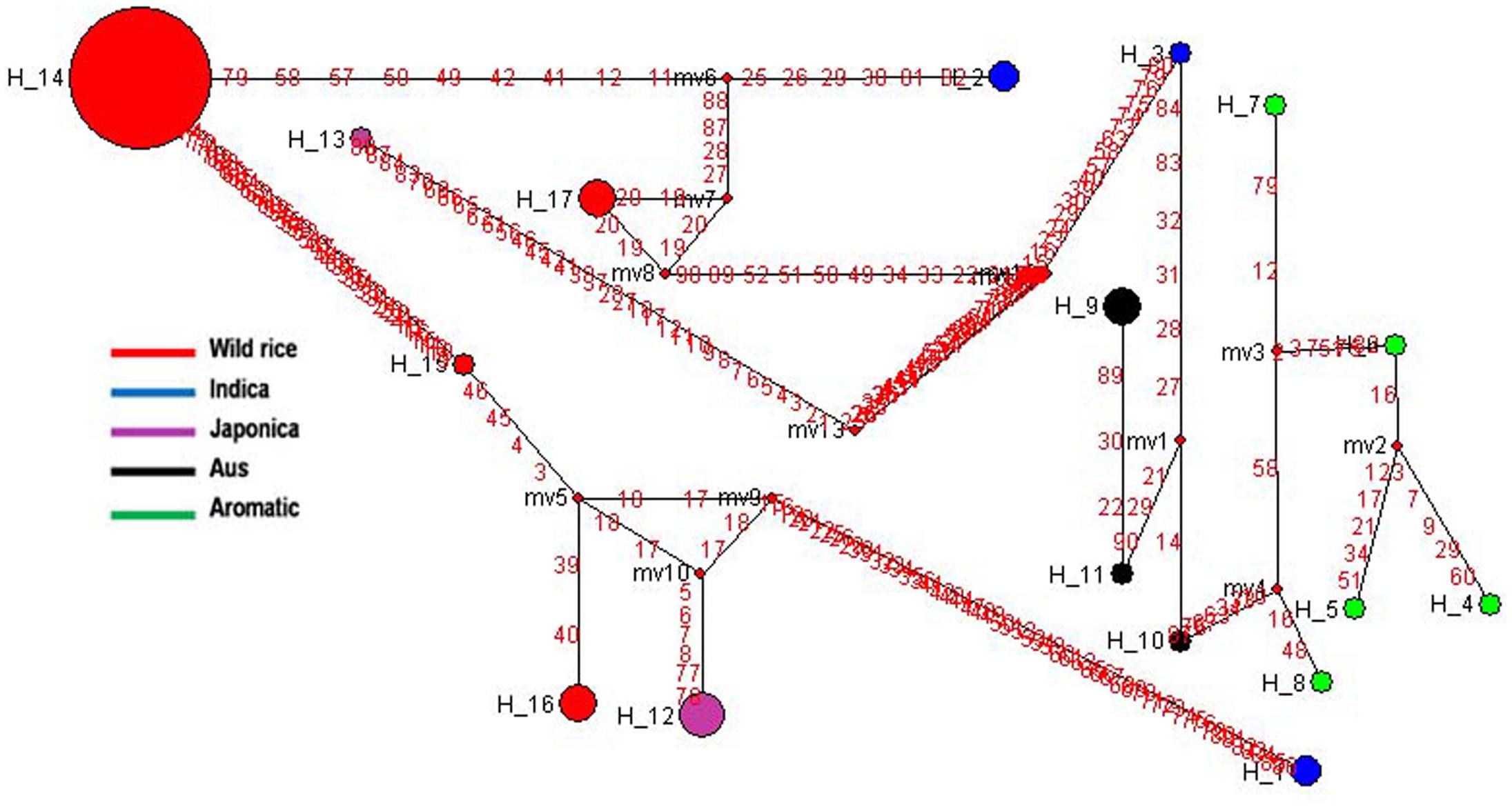
FIGURE 4. Haplotype networks of SSSI gene, in total 17 haplotypes were generated, and SSS1-H14 was the ancestral haplotype.
SSIIa and SSIIb Genes for Amylopectin Synthesis
SSIIa is known to have a major affect on starch quality and located at the alk locus on the short arm of chromosome 6 in the rice genome (Umemoto et al., 2002). This gene is predominantly expressed in the endosperm at very high level and presumably affects amylopectin structure (Morell et al., 2003). The effect of this gene on cooking quality and starch texture has clearly been revealed (Umemoto et al., 2004, 2008). Gelatinization temperature of rice starch is mainly determined by SSIIa gene (Luo et al., 2015). Previously genotyped 60 rice cultivars for two SNPs of SSIIa gene and also incorporated three other SNPs of this gene generated five haplotyes. These SNPs alter the branch-length distribution of amylopectin and gelatinization properties and alkali spreading value of rice (Umemoto and Aoki, 2005). In the present study, total five SNPs of SSIIa gene were genotyped for the construction of haplotype network and phylogenetic tree of 175 diverse genotypes including 155 wild rice and 20 rice cultivars. Total seven haplotypes were generated of which four were major (SSIIa-H1, SSIIa-H2, SSIIa-H5, and SSIIa-H6) and three were minor (SSIIa-H3, SSIIa-H4, and SSIIa-H7) with ‘Hd’ value of 0.7387. The SSIIa-H5 haplotype present in the wild rice accessions collected from MGP and UGP regions was the ancestral haplotype for the other six haplotypes. However, SSIIa-H2 and SSIIa-H6 were closest to SSIIa-H5 with their allele flow in two opposite directions. SSIIa-H2 haplotype was shared by all three groups of wild rice, all five Japonica cultivars and two Indica cultivars, while SSIIa-H6 was represented by wild rice only. SSIIa-H1 was shared by wild rice, all of the Aus and most of the Indica cultivars (Figure 5 and Supplementary Figure S2). The aromatic rice varieties showed two minor haplotypes, SSIIa-H3 and SSIIa-H4, whereas another minor haplotype SSIIa-H7 was present in three wild rice accessions, collected 1 from MGP and 2 from EPH regions. The phylogenetic analysis of SSIIa gene also showed a polyphyletic origin of cultivated rice because rice cultivar groups have followed three independent evolutionary pathways. Recently, nucleotide diversity and molecular evolution based on 93 SNPs in this gene was analyzed in 199 Chinese wild rice accessions and 122 cultivated accessions, also reporting three different haplotypes in both wild and cultivated rice accessions. Their hypothesis was also supported that cultivated rice was independently domesticated from multiple domestication and has undergone balancing selection on separate haplotypes in multiple populations in China (Zhou et al., 2016).
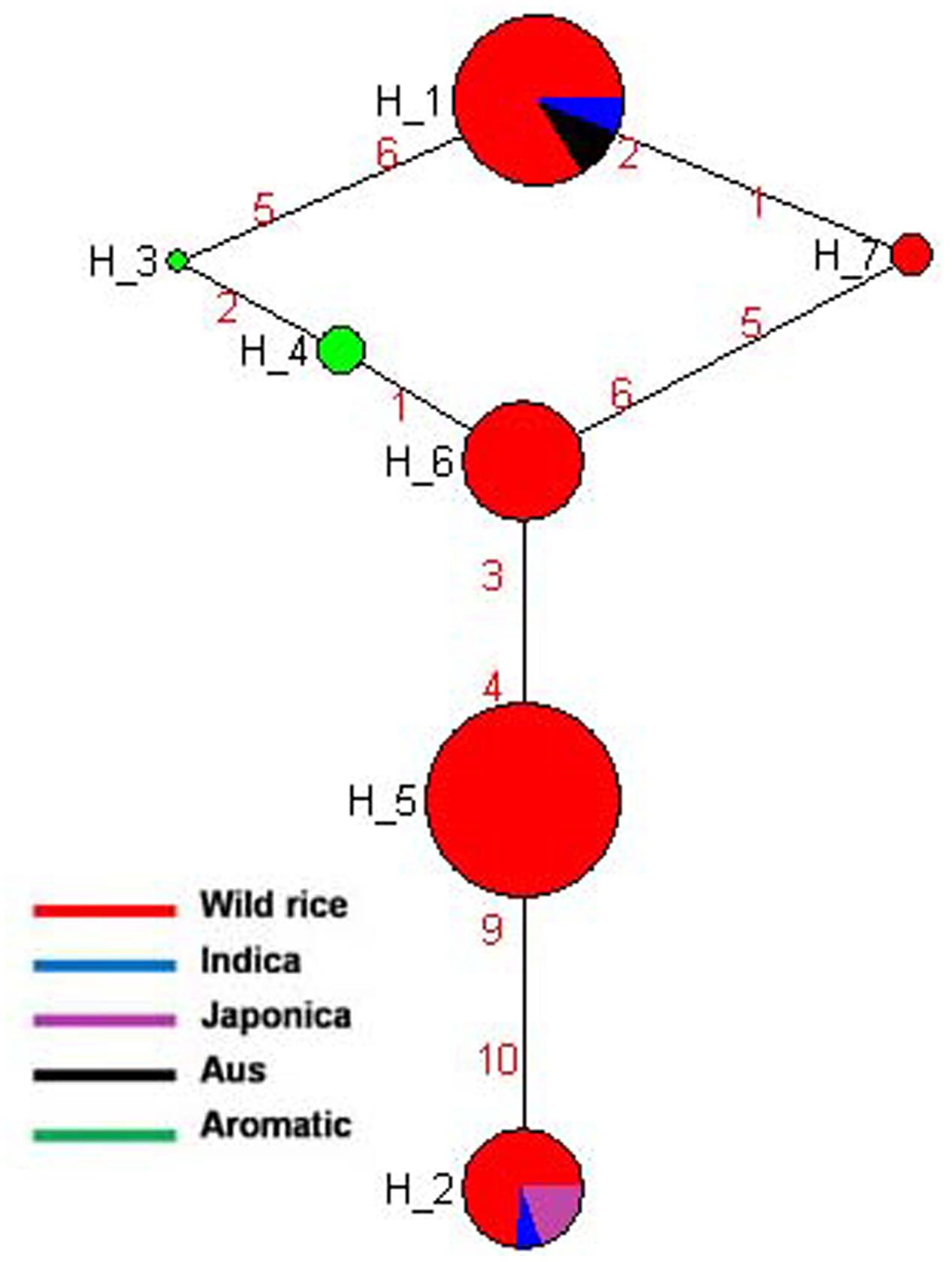
FIGURE 5. Haplotype networks of SSIIa gene, in total seven haplotypes were generated, and SSIIa-H5 was the ancestral haplotype.
SSIIb gene located on rice chromosome 2 is expressed primarily in leaf blades and sheaths (leaf specific) at early stage of grain filling (Hirose and Terao, 2004). A total of 188 diverse rice genotypes were analyzed based on four SNPs in the SSIIb gene. Both haplotype network and phylogenetic analysis showed similar pattern of gene diversification. In total six haplotypes were generated, three major (SSIIb-H1, SSIIb-H2, and SSIIb-H5) and three minor (SSIIb-H3, SSIIb-H4, and SSIIb-H6) haplotypes with ‘Hd’-0.7256. SSIIb-H2 was the ancestral haplotype shared by majority of the wild rice accessions from diverse geographical regions (EHR, MGP, UGP, and WCP) along with four Aus and three Aromatic cultivars. The other five haplotypes were derived from this ancestral haplotype. Here also we can see a tri-phyletic origin of the SSIIb haplotypes in rice cultivars the ancestral SSIIb-H2 was shared by Aus and Aromatic cultivars, SSIIb-H1 was present in all Indica rice cultivars, while in another evolutionary route, comprising of 4 different haplotypes (SSIIb-H3 to SSIIb-H6), two haplotypes (SSIIb-H5 and SSIIb-H6) were solely represented by wild rice genotypes. Wild rice accessions belong to MGP and UGP regions were clustered in SSIIb-H5 group while SSIIb-H6 haplotype was specific to the GPH region. Japonica cultivars showed two haplotypes (SSIIb-H1 and SSIIb-H3) but SSIIb-H3 was the major haplotype present in wild rice, four Japonica and one Aromatic cultivars (Figure 6 and Supplementary Figure S3). These results showed that Indica and Japonica cultivars groups have originated from wild rice genotypes through independent phylogenetic routes while haplotype of Aus cultivars was the most ancient and still shared by diverse wild rice genotypes.
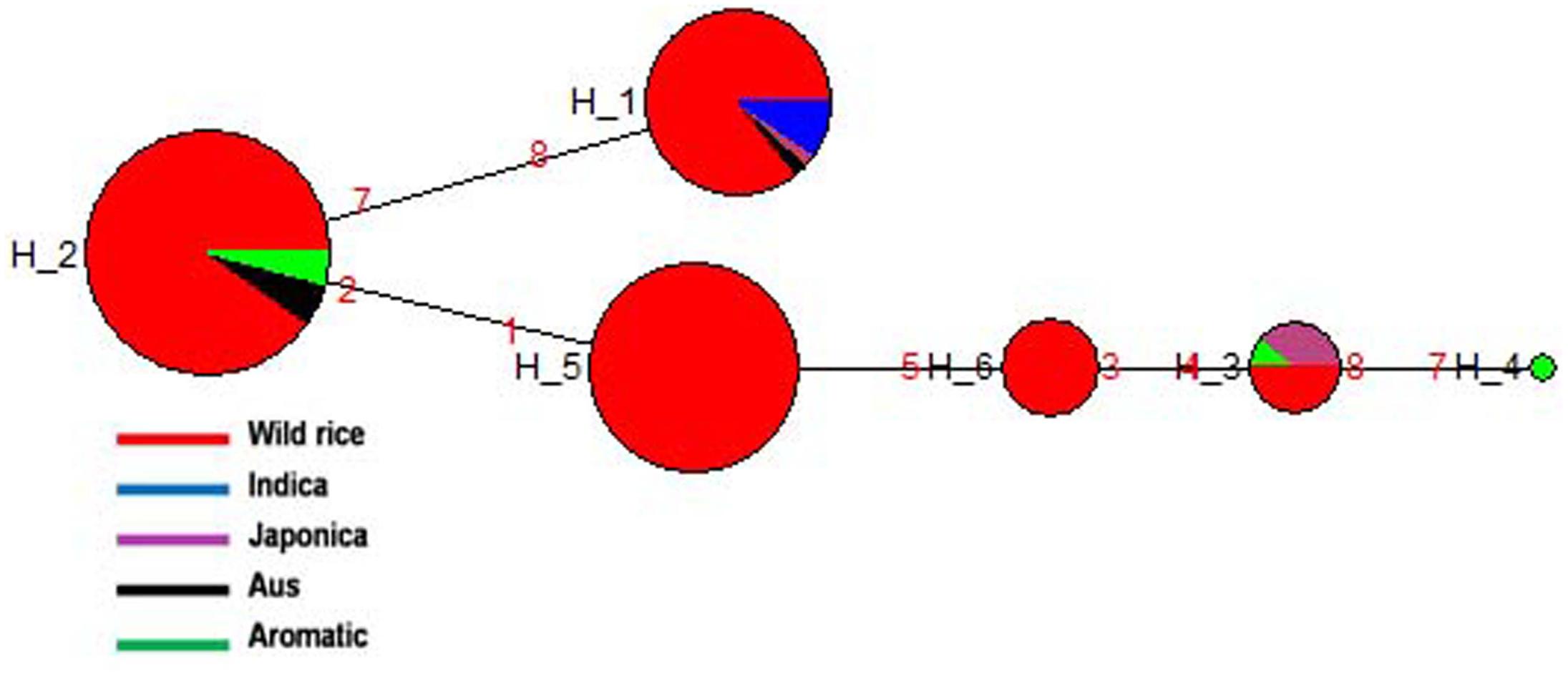
FIGURE 6. Haplotype networks of SSIIb gene six haplotypes were generated, SSIIb-H2 was the ancestral haplotype comprising large number of wild rice genotypes, shared with Aus and Aromatic cultivar groups.
SSIIIa and SSIIIb Genes for Amylopectin Synthesis
The SSIIIa gene located on chromosome 8 is highly expressed in endosperm and green tissues. Haplotype network and phylogenetic analysis was done based on 20 SNPs of SSIIIa gene in 84 diverse rice genotypes, which was nicely classified into 18 distinct haplotypes with a haplotype diversity ‘Hd’ value of 0.8153. Of these SSIIIa-H15 and SSIIIa-H16 were the major haplotypes present entirely in the wild rice accessions. Here, SSIIIa-H16 was the most ancestral haplotype comprising of wild rice accessions from EHR, UGP, WCP, and EPH region. Five haplotypes of wild rice (SSIIIa-H13-SSIIIa-H15, SSIIIa-H17- and SSIIIa-H18) belonged to different geographical regions of India (Figure 7 and Supplementary Figure S4). Three haplotypes were formed in Aromatic, Aus, and Indica. Similarly, Japonica cultivars also carried three minor haplotypes, SSIIIa-H4-SSIIIa-H6. Here also the cultivated rice showed more variations than wild rice, suggesting selection for diverse end-usage. Aus and Indica cultivars were derived from the same ancestral wild rice but grouped into separate haplotypes. Hence, this gene also supports the hypothesis of polyphyletic origin of cultivated rice, because all the four cultivar groups show multiple independent origins.
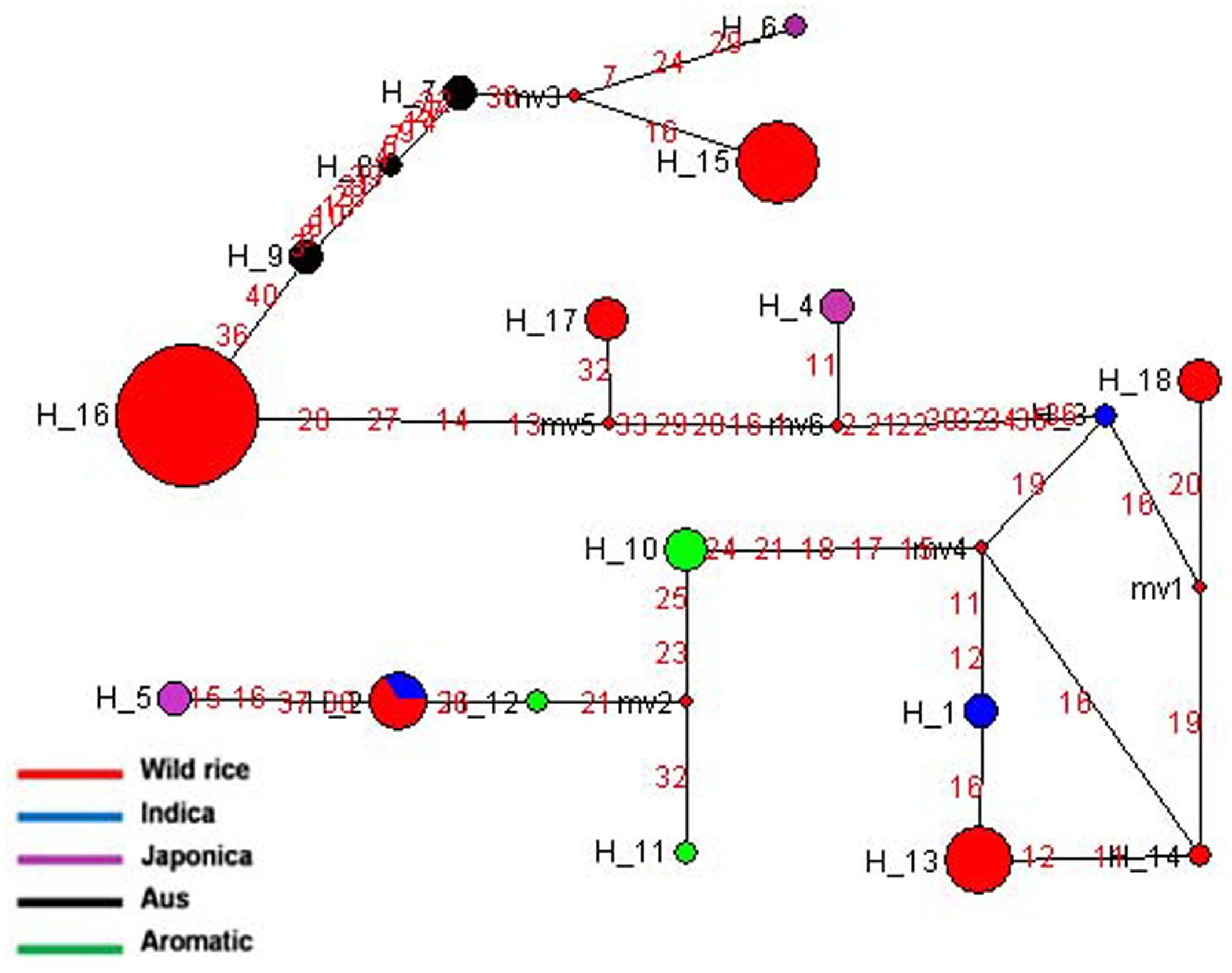
FIGURE 7. Haplotype networks of SSIIIa gene, in total 18 haplotypes were generated, and SSIIIa-H16 was the ancestral haplotype of wild rice.
SSIIIb gene located on rice chromosome 4 is expressed mainly in endosperm but also in leaf sheaths. It showed 11 haplotypes based on 7 SNPs in 192 diverse rice genotypes. SSIIIb-H2 and SSIIIb-H9 were the two major haplotypes along with nine minor haplotypes with ‘Hd’ value of 0.7275. SSIIIb-H9 was the most predominant ancestral haplotype present in 77 wild rice accessions and was the source of origin for remaining 10 haplotypes. The ancestral wild rice haplotype was prevalent in the MGP, WHR, and WCP regions, and other haplotypes derived from this followed three separate evolutionary routes. Another major haplotype SSIIIb-H2 originated directly from SSIIIb-H9 and was shared by 57 wild rice accessions along with two Indica cultivars. Wild rice accessions were represented by total eight haplotypes, three of which were present exclusively in wild rice accessions (SSIIIb-H9 to SSIIIb-H11). Indica cultivars were represented by three different haplotypes, (SSIIIb-H1 to SSIIIb-H3) where they were nested with the wild rice progenitors. Aus cultivars also showed three haplotypes (SSIIIb-H3 to SSIIIb-H5), shared by all other cultivar groups including Indica, Japonica, and Aromatic. SSIIIb-H3, was shared by wild rice and Indica cultivars, SSIIIb-H4 by wild rice and Aromatic, whereas SSIIIb-H5 by Aus Japonica. Similar to the Indica and Aus cultivars, Japonica cultivars were distributed in all three evolutionary routes and showed four different haplotypes (SSIIIb-H2, SSIIIb-H5-SSIIIb-H6, SSIIIb-H7 and SSIIIb-H8, Figure 8 and Supplementary Figure S5). Aromatic rice cultivars including both short grain and Basmati type showed single haplotype (SSIIIb-H4), which was shared by two wild and one Aus cultivar. Thus, SSIIIb gene showed mixed phylogenic patterns among the cultivated rice.
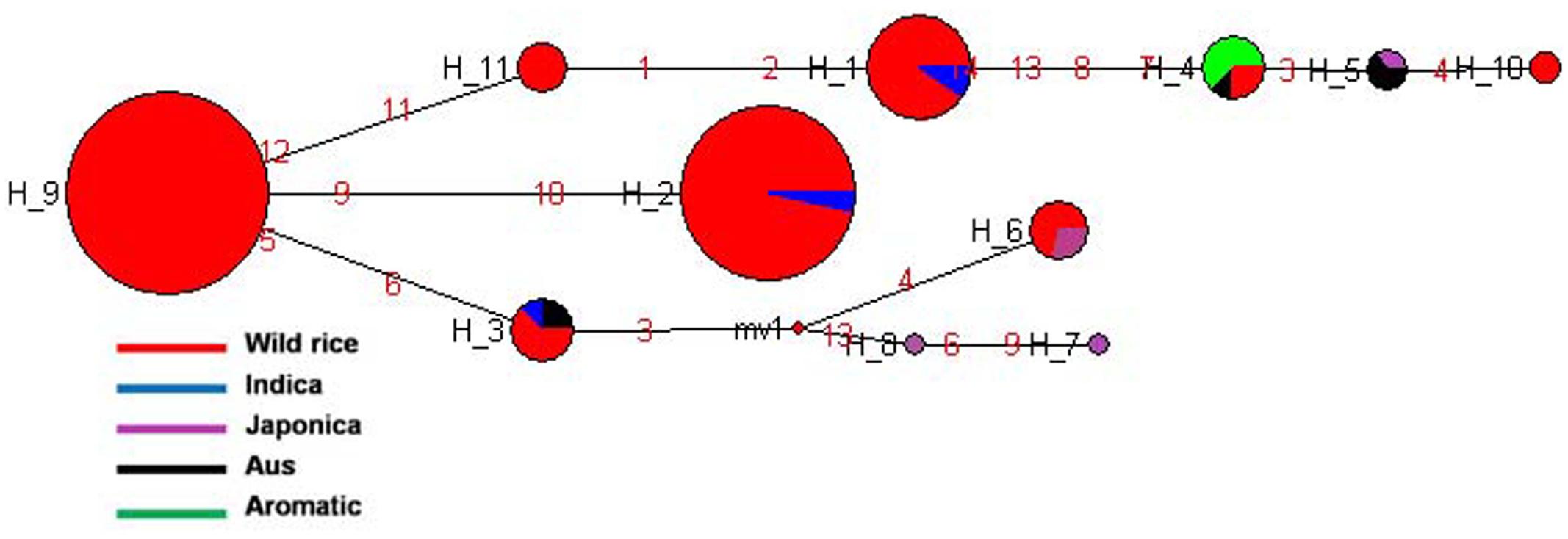
FIGURE 8. Haplotype networks of SSIIIb gene, 11 haplotypes were generated, and SSIIIb-H9 was the ancestral haplotype of wild rice.
SSIVa and SSIVb Genes for Amylopectin Synthesis
SSIVa gene located on rice chromosome 1 is one of the least characterized starch synthase genes in plants and is involved in amylopectin biosynthesis. SSIVa and SSIVb genes are expressed during grain filling in pericarp and endosperm and contribute significantly to grain chalkiness (Hirose and Terao, 2004; Yamakawa et al., 2008). The haplotype network and phylogenetic tree were generated based on 19 SNPs in the SSIVa gene from 110 diverse rice genotypes. In total 12 haplotypes were observed, including three major and nine minor haplotypes with ‘Hd’ value of 0.6455 (Figure 9 and Supplementary Figure S6). Here, SSIVa-H3 was the most primitive and predominant haplotype shared by 60 wild rice accessions collected from MGP, GPH, and EPH regions and also by a flood tolerant Aus cultivar FR13A. The other two major haplotypes, SSIVa-H1 and SSIVa-H2 were represented by Indica and Aus cultivars nested in 11 and 15 different wild rice accessions, respectively. SSIVa-H1 was also shared by one Japonica cultivar Moroberekan. There were three minor haplotypes each for Japonica and Aromatic cultivars, which were not present in any wild rice accession (SSIVa-H5 to SSIVa-H7) and (SSIVa-H8 to SSIVa-H10). Another haplotype SSIVa-H4 was represented by a single Aus cultivar. Thus, analysis of SSIVa gene also showed that Aus cultivars have the most ancient origin as they carry the most ancestral haplotype common to diverse groups of wild rice ancestors. Once again we notice that like other starch synthase genes cultivated rice has more haplotype diversity than wild rice, suggesting selection for diversified end-usage after domestication.
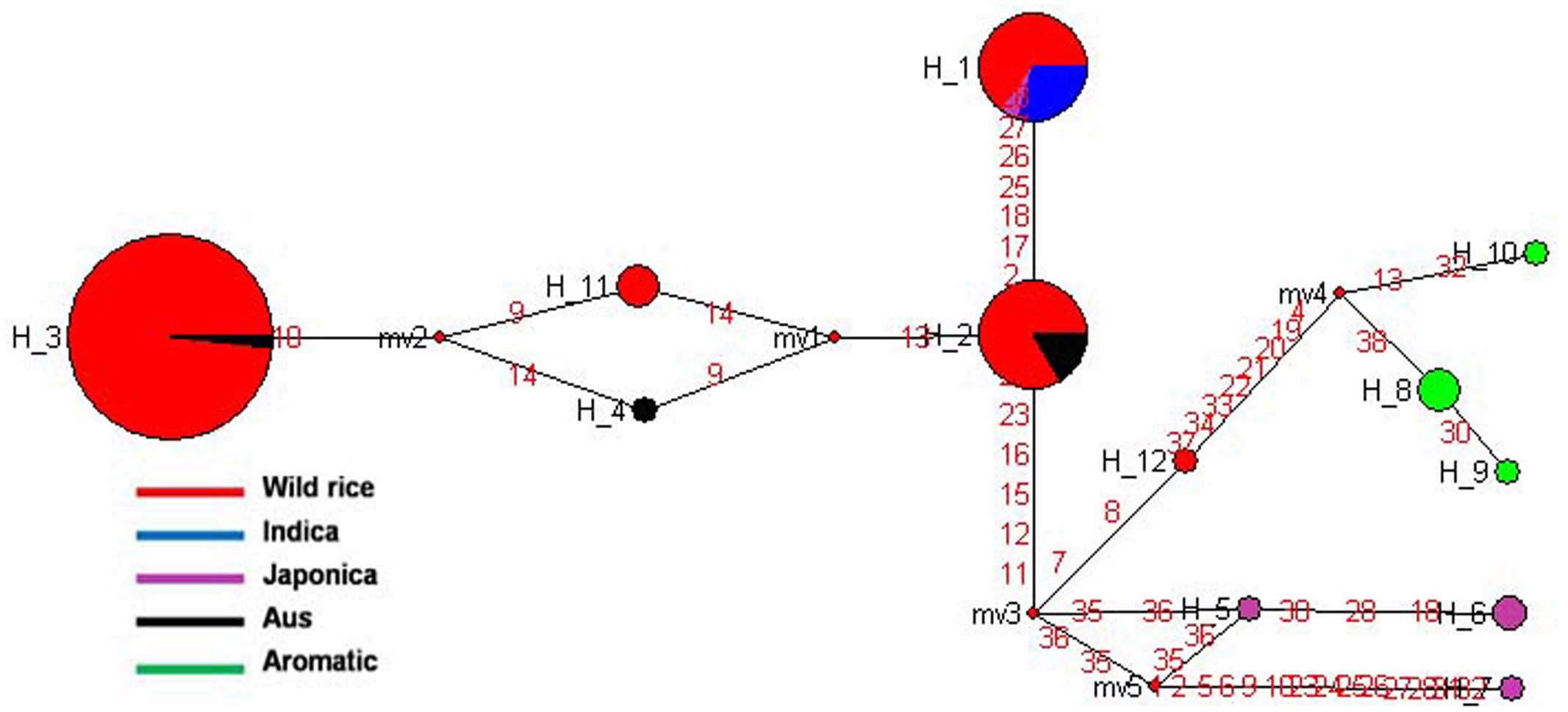
FIGURE 9. Haplotype networks of SSIVa gene, in total 12 haplotypes were formed, and SSIVa-H3 was the ancestral haplotype of wild rice along with single Aus FR13A.
SSIVb gene located on rice chromosome 5 plays important role in determining starch granule morphology and in maintaining amyloplast envelope structure. The haplotype network and phylogenetic tree were constructed based on nine SNPs in this gene genotyped in 150 diverse accessions including 130 wild rice accessions and 20 cultivars. There were nine haplotypes, two major and seven minor haplotypes with ‘Hd’ value of 0.5550. SSIVb-H1 was the most abundant and ancient haplotype representing 86 wild rice accessions along with five Indica and two Aus cultivars. The 130 wild rice accessions possessed four distinct haplotypes, SSIVb-H1 and SSIVb-H2 were shared by Aus and Indica cultivars while SSIVb-H8 and SSIVb-H9 were present exclusively in wild rice accessions (Figure 10 and Supplementary Figure S7). Five Japonica cultivars were grouped into three different haplotypes (SSIVb-H4 to SSIVb-H6), whereas five Aromatic cultivars showed a separate unique haplotype SSIVb-H7. Aus cultivars were also showed three haplotypes(SSIVb-H1 to SSIVb-H3) two of which were shared with wild rice accessions and Indica where SSIVb-H3 was a unique Aus haplotype. Haplotype SSIVb-H9 was unique to wild rice accessions collected from MGP region. These results indicate that Japonica rice cultivars originated later from wild rice progenitor and made a separate group. Once again we see three separate phylogenetic routes for the evolution of SSIVb gene. There was higher haplotype diversity in Japonica and Aus cultivars as compared to Indica and Aromatic cultivars. These results reveal that evolution of SSIVb gene in rice cultivars has a tri-phyletic origin with Aus and Indica cultivars possessing ancient haplotypes.
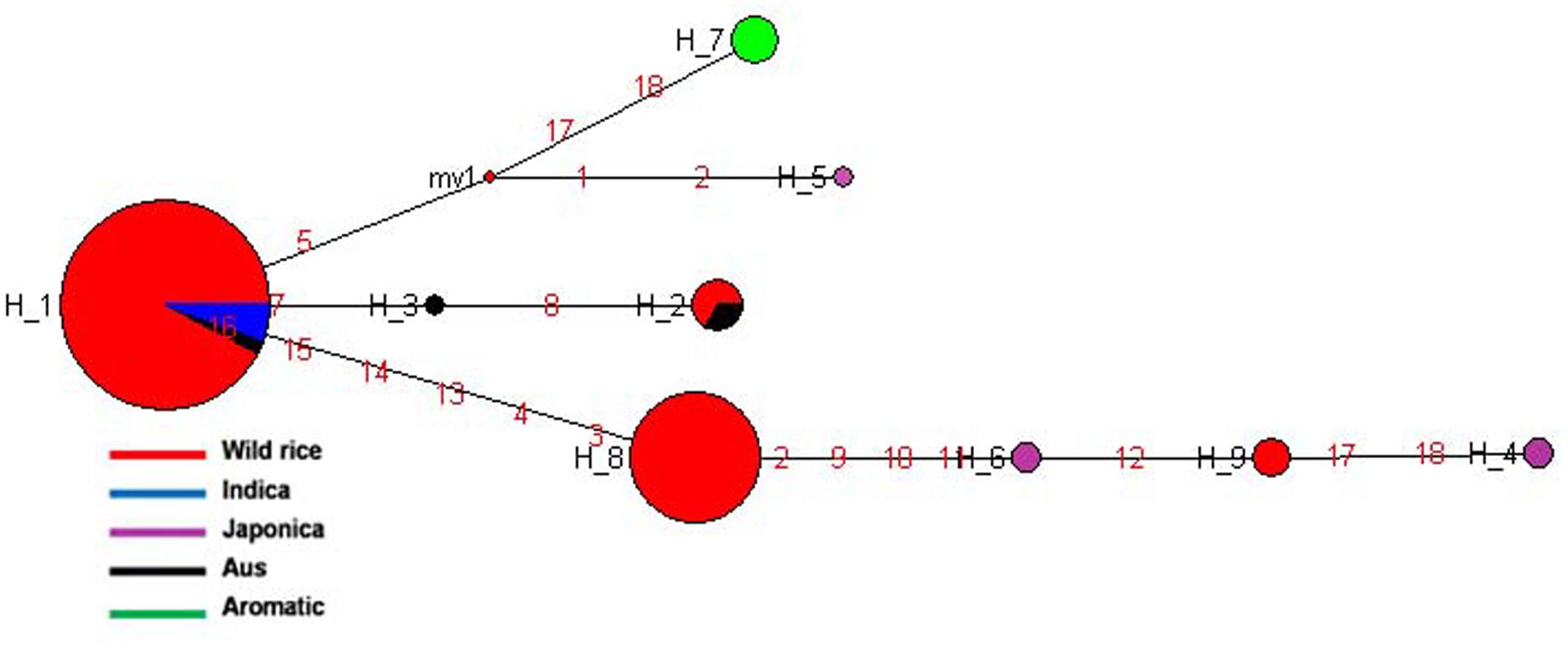
FIGURE 10. Haplotype networks of SSIVb gene, in total nine haplotypes were generated, and SSIVb-H1 was the ancestral haplotype combinations of wild rice genotypes and Indica and Aus cultivar groups.
Conclusion
The haplotype and phylogenetic analysis of 10 agronomically important key genes for pericarp color, seed size, and starch synthase enzymes suggested a polyphyletic origin of cultivated rice. Here, Rc, SSSI, SSIIa, SSIIb, SSIIIa, and SSIVa genes showed the biphyletic origin and GS3, GBSSI, SSIIIb, and SSIVb genes showed the tri-phyletic origin of cultivated rice from their ancestral wild rice. A recent study based on ∼8 million SNPs employing a large data set of 1,083 O. sativa and 446 wild rice accessions showed that there were three independent domestications of rice in different parts of Asia (Civan et al., 2015). It has been shown that the Yangtze valley of China is the source of the Japonica rice gene pool, and populations in Indochina and the Brahmaputra valley are the source of the Indica rice gene pool (Civan et al., 2015). Our study showed that the sources of these genes in major rice cultivar groups are genetically diverse wild rice sub-populations. Aus and Indica cultivars have haplotypes common with wild rice populations, whereas haplotypes of Aromatic cultivars including Basmati were closer to Japonica except for GS3 and SSIIa, genes and seem to have evolved more recently. The ancestral haplotypes were shared by wild rice accessions collected from diverse geographical regions including Mid-Gangetic plains, Upper-Gangetic plains, West-Coast plains, and Eastern-Hills of India and three sub-populations of wild rice, namely ‘Pro-Indica,’ ‘Pro-Aus,’ and ‘Mid-Gangetic’ elucidated using a genome wide 48-plex SNP assay (Singh et al., unpublished). Here we showed that Aus cultivars have the most ancient origin as they shared most haplotypes with ancestral wild rice haplotypes, which subsequently may have been diversified into several modern elite varietal groups as confirmed by previous studies (Hazarika, 2006; Kim et al., 2016). These findings will be useful for rice researchers to understand the evolution and domestication of rice and utilization of diverse wild rice germplasm for introgression of novel traits into cultivated rice. Because cultivated rice has less variability as a result of selection during domestication, and the diversity within wild rice genotypes is of immense value for rice improvement especially for impending climate changes.
Author Contributions
NS conceptualized, designed and conducted the experiments and prepared the manuscript. BS collected maintained and genotyped wild rice. VR, SS, and AS co-supervised the study and NKS conceptualized, supervised the study, edited and finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Indian Council of Agricultural Research “National Professor BP Pal chair” for the financial support. NS is also thankful to I.K.G. Punjab Technical University, Jalandhar for the technical support of Ph.D. work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00972/full#supplementary-material
FIGURE S1 | Haplotype based phylogenetic tree of SSSI gene was constructed using 66 diverse rice genotypes and were separated into 17 different groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S2 | Haplotype based phylogenetic tree of SSIIA gene was constructed using 175 diverse rice genotypes and were separated into seven distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S3 | Haplotype based phylogenetic tree of SSIIB gene was constructed using 188 diverse rice genotypes and were separated into six groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S4 | Haplotype based phylogenetic tree of SSIIIA gene was constructed using 84 diverse rice genotypes and was separated into 18 distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S5 | Haplotype based phylogenetic tree of SSIIIB gene was constructed using 192 diverse rice genotypes and were separated into 11 major distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S6 | Haplotype based phylogenetic tree of SSIVA gene was constructed using 110 diverse rice genotypes and were separated into 12 distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
FIGURE S7 | Haplotype based phylogenetic tree of SSIVB gene was constructed using 150 diverse rice genotypes and were separated into nine major distinct groups. Color coding represents different varietal groups (Cyan-Pro-Indica, Orange-Pro-Aus, Red-Mid-Gangetic, Blue-Indica, Black-Aus, Green-Aromatic, and Magenta-Japonica).
Footnotes
- ^ http://nksingh.nationalprof.in:8080/iwrdb/index.jsp
- ^ http://www.medsci.uu.se/digitalAssets/160/c_160695-l_1-k_axiom2.pdf
- ^ http://www.affymetrix.com
- ^ http://www.fluxus-engineering.com
- ^ http://www.ub.es/dnasp
References
Amarawathi, Y., Singh, R., Singh, A. K., Singh, V. P., Mohapatra, T., Sharma, T. R., et al. (2008). Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 21, 49–65. doi: 10.1007/s11032-007-9108-8
Anand, D., Baunthiyal, M., Krishnan, S. G., Singh, N. K., Prabhu, K. V., and Singh, A. K. (2015). Novel InDel variation in GS3 locus and development of InDel based marker for marker assisted breeding of short grain aromatic rices. J. Plant Biochem. Biotechnol. 24, 120–127. doi: 10.1007/s13562-013-0243-5
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Bai, X. F., Luo, L. J., Yan, W. H., Kovi, M. R., Zhan, W., and Xing, Y. Z. (2010). Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11:16. doi: 10.1186/1471-2156-11-16
Ball, S. G., and Morell, M. K. (2003). From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. doi: 10.1146/annurev.arplant.54.031902.134927
Bandelt, H. J., Forster, P., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bradbury, L. M., Fitzgerald, T. L., Henry, R. J., Jin, Q., and Waters, D. L. (2005). The gene for fragrance in rice. Plant Biotechnol. J. 3, 363–370. doi: 10.1111/j.1467-7652.2005.00131.x
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Calingacion, M., Laborte, A., Nelson, A., Resurreccion, A., Concepcion, J. C., Daygon, V. D., et al. (2014). Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS ONE 9:e85106. doi: 10.1371/journal.pone.0085106
Choudhury, B. I., Khan, M. L., and Dayanandan, S. (2014). Patterns of nucleotide diversity and phenotypes of two domestication related genes (OsC1 and Wx) in indigenous rice varieties in Northeast India. BMC Genet. 15:71. doi: 10.1186/1471-2156-15-71
Civan, P., Craig, H., Cox, C. J., and Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nat. Plants 1:15164. doi: 10.1038/nplants.2015.164
Cui, Y., Song, B. K., Li, L. F., Li, Y. L., Huang, Z., Caicedo, A. L., et al. (2016). Little white lies: pericarp color provides insights into the origins and evolution of Southeast Asian weedy rice. G3, 4105–4114. doi: 10.1534/g3.116.035881
Deb, D. (2005). Seeds of Tradition, Seeds of Future, Folk Rice Varieties of Eastern India. New Delhi: Research Foundation for Science Technology & Ecology, doi: 10.1093/aobpla/plt032
Doebley, J., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Fan, C. C., Yu, S. B., Wang, C. R., and Xing, Y. Z. (2009). A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet 118, 465–472. doi: 10.1007/s00122-008-0913-1
Frances, H., Bligh, J., Larkin, P. D., Roach, P. S., Jones, C. A., Fu, H., et al. (1998). Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol. Biol. 38, 407–415. doi: 10.1023/A:1006021807799
Fujita, N., Satoh, R., Hayashi, A., Kodama, M., Itoh, R., Aihara, S., et al. (2011). Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 62, 4819–4831. doi: 10.1093/jxb/err125
Fuller, D. Q. (2011). Finding plant domestication in the Indian subcontinent. Curr. Anthropol. 52, S347–S362.
Fuller, D. Q., Sato, Y. I., Castillo, C., Qin, L., Weisskopf, A. R., KingwellBanham, E. J., et al. (2010). Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthropol. Sci. 2, 115–131. doi: 10.1007/s12520-010-0035-y
Furukawa, T., Maekawa, M., Oki, T., Suda, I., Iida, S., Shimada, H., et al. (2007). The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49, 91–102. doi: 10.1111/j.1365-313X.2006.02958.x
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., and McCouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi: 10.1534/genetics.104.035642
Glaszmann, J. C. (1987). Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 74, 21–30. doi: 10.1007/BF00290078
Gross, B. L., and Olsen, K. M. (2010). Genetic perspectives on crop domestication. Trends Plant Sci. 15, 529–537. doi: 10.1016/j.tplants.2010.05.008
Gu, B., Zhou, T., Luo, J., Liu, H., Wang, Y., Shangguan, Y., et al. (2015). An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant 8, 1635–1650. doi: 10.1016/j.molp.2015.08.001
Hazarika, M. (2006). Neolithic culture of Northeast India: a recent perspective on the origins of pottery and agriculture. Ancient Asia 1, 25–44.
Hirano, H., and Sano, Y. (1991). Molecular characterization of the WAXY locus of rice (Oryza sativa). Plant Cell Physiol. 32, 989–997. doi: 10.1093/oxfordjournals.pcp.a078186
Hirano, H. Y., Eiguchi, M., and Sano, Y. (1998). A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 15, 978–987. doi: 10.1093/oxfordjournals.molbev.a026013
Hirose, T., and Terao, T. (2004). A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220, 9–16. doi: 10.1007/s00425-004-1314-6
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Isshiki, M., Morino, K., Nakajima, M., Okagaki, R. J., Wessler, S. R., Izawa, T., et al. (1998). A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 15, 133–138. doi: 10.1046/j.1365-313X.1998.00189.x
Isshiki, M., Nakajima, M., Satoh, H., and Shimamoto, K. (2000). dull: rice mutants with tissue-specific effects on the splicing of the waxy pre-mRNA. Plant J. 23, 451–460. doi: 10.1046/j.1365-313x.2000.00803.x
James, M. G., Denyer, K., and Myers, A. M. (2003). Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6, 215–222. doi: 10.1016/S1369-5266(03)00042-6
Ji, H., Kim, S. R., Kim, Y. H., Kim, H., Eun, M. Y., Jin, I. D., et al. (2010). Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 61, 96–106. doi: 10.1111/j.1365-313X.2009.04039.x
Jin, J., Hua, L., Zhu, Z., Tan, L., Zhao, X., Zhang, W., et al. (2016). GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28, 2453–2463. doi: 10.1105/tpc.16.00379
Kim, H., Jung, J., Singh, N., Greenberg, A., Doyle, J. J., Tyagi, W., et al. (2016). Population dynamics among six major groups of the Oryza rufipogon species complex, wild relative of cultivated Asian rice. Rice 9:56. doi: 10.1186/s12284-016-0119-0
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Kovach, M. J., Sweeney, M. T., and McCouch, S. R. (2007). New insights into the history of rice domestication. Trends Genet. 23, 578–587. doi: 10.1016/j.tig.2007.08.012
Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Li, Z., Sun, F., Xu, S., Chu, X., Mukai, Y., Yamamoto, M., et al. (2003). The structural organisation of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthases in plants. Funct. Integr. Genomics 3, 76–85. doi: 10.1007/s10142-002-0072-4
Liu, L., Lee, G. A., Jiang, L., and Zhang, J. (2007). Evidence for the early beginning (c. 9000 cal. BP) of rice domestication in China: a response. Holocene 17, 1059–1068. doi: 10.1177/0959683607085121
Liu, L., Ma, X., Liu, S., Zhu, C., Jiang, L., Wang, Y., et al. (2009). Identification and characterization of a novel waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71, 609–626. doi: 10.1007/s11103-009-9544-4
Londo, J. P., Chiang, Y. C., Hung, K. H., Chiang, T. Y., and Schaal, B. A. (2006). Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, O. sativa. Proc. Natl. Acad. Sci. U.S.A. 103, 9578–9583. doi: 10.1073/pnas.0603152103
Luo, J., Jobling, S. A., Millar, A., Morell, M. K., and Li, Z. (2015). Allelic effects on starch structure and properties of six starch biosynthetic genes in a rice recombinant inbred line population. Rice 8:15. doi: 10.1186/s12284-015-0046-5
Mohapatra, P. K., Patel, R., and Sahu, S. K. (1993). Time of flowering affects grain quality and spikelet partitioning within the rice panicle. Aust. J. Plant Physiol. 20, 231–241. doi: 10.1071/PP9930231
Molina, J., Sikora, M., Garud, N., Flowers, J. M., Rubinstein, S., Reynolds, A., et al. (2011). Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. U.S.A. 108, 8351–8356. doi: 10.1073/pnas.1104686108
Morell, M. K., Kosar-Hashemi, B., Cmiel, M., Samuel, M. S., Chandler, P., Rahman, S., et al. (2003). Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 34, 172–184. doi: 10.1046/j.1365-313X.2003.01712.x
Murray, M., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Ohdan, T., Francisco, P. B., Sawada, T., Hirose, T., Terao, T., Satoh, H., et al. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56, 3229–3244. doi: 10.1093/jxb/eri292
Okagaki, R. J. (1992). Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol. Biol. 19, 513–516. doi: 10.1007/BF00023402
Prasad, V., Stromberg, C. A. E., Leache, A. D., Samant, B., Patnaik, R., Tang, L., et al. (2011). Late Cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nat. Commun. 2:480. doi: 10.1038/ncomms1482
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Rambaut, A. (2009). Molecular Evolution, Phylogenetics and Epidemiology. Available at: http://tree.bio.ed.ac.uk/software/figtree/ [accessed December 5, 2012].
Rozas, J., Sanchez-Del Barrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1093/bioinformatics/btg359
Sano, Y. (1984). Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68, 467–473. doi: 10.1007/BF00254822
Shi, W., Yang, Y., Chen, S., and Xu, M. (2008). Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 22, 185–192. doi: 10.1007/s11032-008-9165-7
Singh, N., Jayaswal, P. K., Panda, K., Mandal, P., Kumar, V., Singh, B., et al. (2015). Single-copy gene based 50 K SNP chip for genetic studies and molecular breeding in rice. Sci. Rep. 5:11600. doi: 10.1038/srep11600
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Sweeney, M., and Mccouch, S. (2007). The complex history of the domestication of rice. Ann. Bot. 100, 951–957. doi: 10.1093/aob/mcm128
Sweeney, M. T., Thomson, M. J., Cho, Y. G., Park, Y. J., Williamson, S. H., Bustamante, C. D., et al. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3:e133. doi: 10.1371/journal.pgen.0030133
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and McCouch, S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi: 10.1105/tpc.105.038430
Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., Kanamori, H., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1323–1334. doi: 10.1534/genetics.109.103002
Takano-Kai, N., Jiang, H., Powell, A., McCouch, S., Takamure, I., Furuya, N., et al. (2013). Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 63, 77–85. doi: 10.1270/jsbbs.63.77
Takemoto-Kuno, Y., Suzuki, K., Nakamura, S., Satoh, H., and Ohtsubo, K. (2006). Soluble starch synthase I effects differences in amylopectin structure between indica and japonica rice varieties. J. Agric. Food Chem. 54, 9234–9240. doi: 10.1021/jf061200i
Umeda, M., Ohtsubo, H., and Ohtsubo, E. (1991). Diversification of the rice Waxy gene by insertion of mobile DNA elements into introns. Jpn. J. Genet. 66, 569–586. doi: 10.1266/jjg.66.569
Umemoto, T., and Aoki, N. (2005). Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 32, 763–768. doi: 10.1071/FP04214
Umemoto, T., Aoki, N., Lin, H., Nakamura, Y., Inouchi, N., Sato, Y., et al. (2004). Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 31, 671–684. doi: 10.1071/FP04009
Umemoto, T., Horibatac, T., Aoki, N., Hiratsukab, M., Yanod, M., and Inouchi, N. (2008). Effects of variations in starch synthase on starch properties and eating quality of rice. Plant Prod. Sci. 11, 472–480. doi: 10.1626/pps.11.472
Umemoto, T., Yano, M., Satoh, H., Shomura, A., and Nakamura, Y. (2002). Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 104, 1–8. doi: 10.1007/s001220200000
Vaughan, D. A., Lu, B. R., and Tomooka, N. (2008). The evolving story of rice evolution. Plant Sci. 174, 394–408. doi: 10.1016/j.plantsci.2008.01.016
Vigueira, C. C., Li, W., and Olsen, K. M. (2013). The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 2, 1738–1749. doi: 10.1111/jeb.12171
Vitte, C., Ishii, T., Lamy, F., Brar, D., and Panaud, O. (2004). Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol. Gen. Genet. 272, 504–511.
Wang, Z. Y., Wu, Z. I., Xing, Y. Y., Zheng, F. G., Guo, X. I., Zhang, W. G., et al. (1990). Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 18:5898. doi: 10.1093/nar/18.19.5898
Wang, Z. Y., Zheng, F. Q., Shen, G. Z., Gao, J. P., Sunstad, D. P., Li, M. G., et al. (1995). The amylose content in rice endosperm is related to the post transcriptional regulation of the WAXY gene. Plant J. 7, 613–622. doi: 10.1046/j.1365-313X.1995.7040613.x
Whitt, S. R., Wilson, M., Tenaillon, M. I., Gaut, B. S., and Buckler, E. S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 12959–12962. doi: 10.1073/pnas.202476999
Yamakawa, H., Ebitani, T., and Terao, T. (2008). Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breed. Sci. 58, 337–343. doi: 10.1186/1471-2156-15-49
Yamanaka, S., Nakamura, I., Watanabe, K. N., and Sato, Y. I. (2004). Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 108, 1200–1204. doi: 10.1007/s00122-003-1564-x
Zeder, M. A. (2011). The origins of agriculture in the Near East. Curr. Anthropol. 52, S221–S236. doi: 10.1086/659307
Zhang, L. B., Zhu, Q. H., Wu, Z. Q., Ross-Ibarra, J., Gaut, B. S., Ge, S., et al. (2009). Selection on grain shattering genes and rates of rice domestication. New Phytol. 184, 708–720. doi: 10.1111/j.1469-8137.2009.02984.x
Zhang, M. W., Zhang, R. F., Zhang, F. X., and Liu, R. H. (2010). Phenolic profiles and antioxidant activity of black rice bran of different commercially available varieties. J. Agric. Food Chem. 58, 7580–7587. doi: 10.1021/jf1007665
Zhao, K., Wright, M., Kimball, J., Eizenga, G., McClung, A., Kovach, M., et al. (2010). Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE 5:e10780. doi: 10.1371/journal.pone.0010780
Zhou, Y., Lu, D., Li, C., Luo, J., Zhu, B. F., Zhu, J., et al. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24, 1034–1048. doi: 10.1105/tpc.111.094383
Zhou, Y., Zheng, H., Wei, G., Zhou, H., Han, Y., Bai, X., et al. (2016). Nucleotide diversity and molecular evolution of the ALK gene in cultivated rice and its wild relatives. Plant Mol. Biol. Rep. 34, 923–930. doi: 10.1007/s11105-016-0975
Zhu, Q., and Ge, S. (2005). Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 167, 249–265. doi: 10.1111/j.1469-8137.2005.01406.x
Zhu, Q., Zheng, X., Luo, J., Gaut, B. S., and Ge, S. (2007). Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol. Biol. Evol. 24, 875–888. doi: 10.1093/molbev/msm005
Keywords: domestication, haplotype, grain size, red pericarp, starch synthase, wild rice
Citation: Singh N, Singh B, Rai V, Sidhu S, Singh AK and Singh NK (2017) Evolutionary Insights Based on SNP Haplotypes of Red Pericarp, Grain Size and Starch Synthase Genes in Wild and Cultivated Rice. Front. Plant Sci. 8:972. doi: 10.3389/fpls.2017.00972
Received: 10 March 2017; Accepted: 23 May 2017;
Published: 09 June 2017.
Edited by:
Marion S. Röder, Institute of Plant Genetics and Crop Plant Research (LG), GermanyReviewed by:
Manish Kumar Pandey, International Crops Research Institute for the Semi-Arid Tropics, IndiaUmesh K. Reddy, West Virginia State University, United States
Keisuke Nagai, Nagoya University, Japan
Copyright © 2017 Singh, Singh, Rai, Sidhu, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nagendra K. Singh, bmtzaW5naDRAZ21haWwuY29t; bmtzaW5naEBucmNwYi5vcmc=
 Nisha Singh
Nisha Singh Balwant Singh
Balwant Singh Vandna Rai
Vandna Rai Sukhjeet Sidhu
Sukhjeet Sidhu Ashok K. Singh3
Ashok K. Singh3 Nagendra K. Singh
Nagendra K. Singh