Abstract
Drought, salinity, and cold are the major factors limiting wheat quality and productivity; it is thus highly desirable to characterize the abiotic-stress-inducible promoters suitable for the genetic improvement of plant resistance. The sucrose non-fermenting 1-related protein kinase 2 (SnRK2) family genes show distinct regulatory properties in response to abiotic stresses. The present study characterized the approximately 3000-bp upstream sequence (the 313 bp upstream of the ATG was the transcription start site) of the Triticum aestivum TaSnRK2.8 promoter under abscisic acid (ABA) and abiotic stresses. Four different-length 5′ deletion fragments of TaSnRK2.8 promoter were fused with the GUS reporter gene and transformed into Arabidopsis. Tissue expression analysis showed that the TaSnRK2.8 promoter region from position -1481 to -821 contained the stalk-specific elements, and the region from position -2631 to -1481 contained the leaf- and root-specific elements. In the ABA-treated seedlings, the deletion analysis showed that the TaSnRK2.8 promoter region from position -821 to -2631 contained ABA response elements. The abiotic stress responses of the TaSnRK2.8 promoter derivatives demonstrated that they harbored abiotic-stress response elements: the region from position -821 to -408 harbored the osmotic-stress response elements, whereas the region from position -2631 to -1481 contained the positive regulatory motifs and the region from position -1481 to -821 contained the leaf- and stalk-specific enhancers. Further deletion analysis of the promoter region from position -821 to -408 indicated that a 125-bp region from position -693 to -568 was required to induce an osmotic-stress response. These results contribute to a better understanding of the molecular mechanisms of TaSnRK2.8 in response to abiotic stresses, and the TaSnRK2.8 promoter seems to be a candidate for regulating the expression of abiotic stress response genes in transgenic plants.
Introduction
Plant growth is adversely affected by abiotic stresses such as salinity, drought, and low temperature. To cope with environmental stresses, plants have developed intricate signaling networks to protect their cellular activities and maintain whole-plant integrity. Increasing evidence indicates that the SnRK2 (sucrose non-fermenting 1-related protein kinase 2) family plays a crucial role in response to environmental stresses (Boudsocq et al., 2004, 2007; Kobayashi et al., 2004).
The SnRK2 family is a plant-specific gene family that encodes serine/threonine kinases. It is divided into three distinct subclasses (I, II, and III) based on their functional divergence (Halford and Hardie, 1998). In Arabidopsis, 10 SnRK2 members were cloned. Of these, five members (AtSnRK2.2, AtSnRK2.3, AtSnRK2.6, AtSnRK2.7, and AtSnRK2.8) were activated by abscisic acid (ABA). All members, except AtSnRK2.9, responded to drought and salt stresses, and none was induced by cold stress (Boudsocq et al., 2004, 2007). Similarly, in rice, these 10 SnRK2 members, named SAPK1–10, were induced by osmotic stresses, and three (SAPK8–10) were also induced by ABA treatment (Kobayashi et al., 2004). In maize, 10 ZmSnRK2 genes were activated by one or more abiotic stresses (Huai et al., 2008). In wheat, the first identified SnRK2-family gene, PKABA1, was isolated from an ABA-treated embryo cDNA library (Anderberg and Walker-Simmons, 1992). In our recent study, 10 wheat SnRK2 genes were identified, and all responded to multiple stressors (Zhang et al., 2016). Among these, TaSnRK2.1–3 (subclass II) responded weakly to ABA, TaSnRK2.4–7 (subclass I) were not induced by ABA treatment, and TaSnRK2.8–10 (subclass III) were strongly activated by ABA (Zhang et al., 2016). Thus, the SnRK2 members from subclasses I and III have distinct stress signals. Functional analysis of SnRK2 genes indicated that the overexpression of TaSnRK2.3, TaSnRK2.4, TaSnRK2.7, or TaSnRK2.8 in Arabidopsis led to enhanced tolerance to abiotic stresses (Mao et al., 2009; Zhang et al., 2010, 2011; Tian et al., 2013). Various studies have shown that the ABA-activated SnRK2 genes are involved in stress responses through ABA signaling (Melcher et al., 2009; Nishimura et al., 2009; Sheard and Zheng, 2009). However, to date, little research has been conducted on the molecular mechanisms of SnRK2s in response to abiotic stress.
To improve stress tolerance in plants, a large number of genes have been expressed under the control of stress-inducible or constitutive promoters. Constitutive promoters, including the rice Act1 (McElroy et al., 1990), cauliflower mosaic virus CaMV35S (Odell et al., 1985), and maize Ubi1 promoters (Christensen et al., 1992), have been widely used in plant biotechnology applications. However, the high and constitutive expression of transgenes might impose an extra metabolic burden and other adverse effects on plants, finally resulting in their abnormal development (Shelton et al., 2002). Thus, organ-specific and inducible promoters in plant cells have considerable potential for use in plant genetic engineering. Some abiotic-stress-inducible promoters, including Atrd29A (Yamaguchi-Shinozaki et al., 1990), OsABA2 (Rai et al., 2009), and OsNAC6 (Checker et al., 2012), have been characterized in plants. In this study, we characterized and analyzed the wheat TaSnRK2.8 promoter (subclass III) in Arabidopsis. A series of different-length TaSnRK2.8 promoters were fused to the GUS gene to detect the putative regulatory cis-elements that could confer tissue-specific expression and induce stress signaling in Arabidopsis.
Materials and Methods
Plant Materials
For the evolutionary analysis of the TaSnRK2.8 promoter sequences, 10 wheat species were used: hexaploid wheat (Triticum aestivum, AABBDD) cultivars “Hanxuan 10” and “Chinese Spring”; tetraploid wheat (AABB) PS9 (Triticum persicum) and DM 50 (Triticum dicoccum); diploid wheat Triticum urartu (AA) UR201 and UR204; diploid wheat Aegilops speltoides (BB) Y2001 and Y2041; and diploid wheat Aegilops tauschii (DD) Y85 and Ae46. Leaves were harvested for DNA isolation.
Sequence Isolation and Analysis
To isolate TaSnRK2.8 promoter, its genomic sequence (Zhang et al., 2013) was used as a query to screen the Wheat Genome Browser1. The approximately 3000-bp upstream sequence of the translation start site was considered to be the putative TaSnRK2.8 promoter region. It was polymerase chain reaction (PCR)-amplified using the primers listed in supplementary Supplementary Table S1.
The SnRK2.8 promoter sequences from wheat (MF351624), rice (LOC_Os03g55600), and maize (AC206916.1) were aligned for element prediction using MegAlign in the DNAStar software, and then predicted using the PlantCARE software2. To gain insight into the evolution and origin of the TaSnRK2.8 promoter sequences, the hexaploid wheat varieties and their relatives were used to construct a phylogenetic tree using MEGA 7.0.
The transcriptional start site was determined by 5′ rapid amplification of cDNA ends (RACE) using the GeneRacer Kit (Invitrogen, China). Total RNA was isolated using TRIzol reagent (Sangon, China). Total RNA (3.5 μg) was used as a template for 5′-RACE. The gene-specific primers used for the nested PCR are listed in Supplementary Table S1. The nested PCR products were cloned into the pGEM-T-Easy vectors and sequenced.
Transgenic Plant Generation
Successive 5′ truncations of the TaSnRK2.8 promoter were PCR-amplified using the primers shown in Supplementary Table S1. Nine promoter fragments (-2631 to ATG, Dp2947; -1481 to ATG, Dp1797; -821 to ATG, Dp1137; -774 to ATG, Dp1090; -693 to ATG, Dp1009; -568 to ATG, Dp884; -463 to ATG, Dp779; -408 to ATG, Dp724; Dp724 carrying the 125-bp fragment from position -693 to -568, named as Δ125) were separately fused with the GUS reporter gene in pCAMBIA1391, yielding the PTaSnRK2:GUS constructs (Figure 1). These expression constructs and the pCAMBIA1391 vector (CaMV35S:GUS) were transformed into Agrobacterium, and then transferred into the wild-type Arabidopsis plants by floral infiltration. The transgenic plants were initially screened on hygromycin plates and subsequently characterized by PCR (Supplementary Table S1) and GUS (beta-glucuronidase) staining. The T3-generation plants were selected for subsequent functional analyses.
FIGURE 1
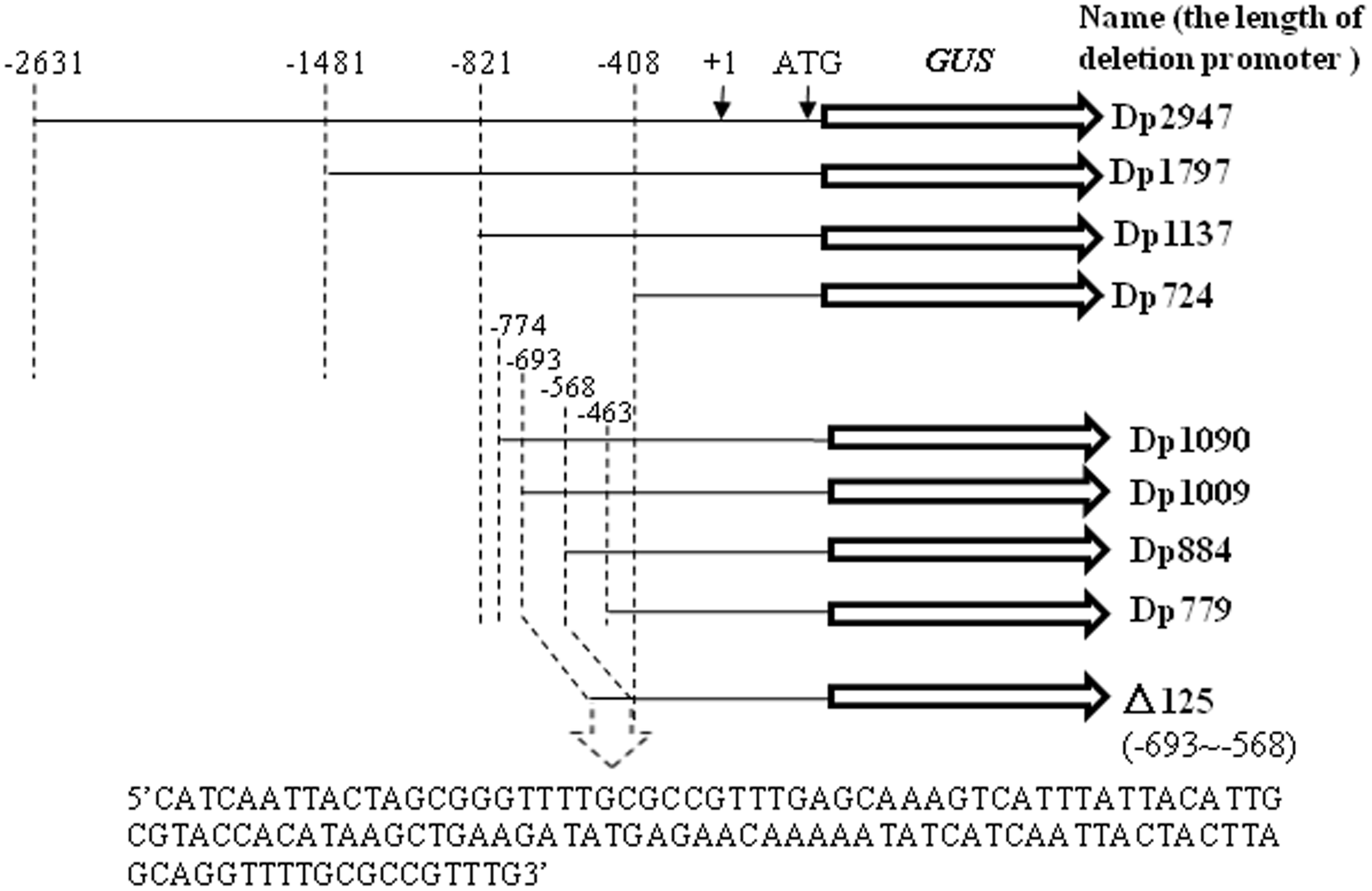
Schematic depiction of deletion fragments generated in the study. Successive 5′ promoter truncations were fused to a GUS reporter. The putative transcription start site was designated as “+1”.
Quantitative Real-Time PCR analysis
Ten similarly sized 10-day-old seedlings from each transgenic line were used for total RNA isolation using the TRIzol reagent (Sangon) and then treated with RNase-free DNase (Takara, China). The quantitative real-time PCR (qRT-PCR) assays were performed in triplicate with the SYBR Premix Ex Taq (Takara, Shiga, Japan) using an ABI PRISM 7000 system (Applied Biosystems, Foster City, CA, United States). The relative transcript levels of GUS gene were determined using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The actin transcript was used to quantify the expression level.
Southern Blotting
Total genomic DNA was extracted from 10 similarly sized 10-day-old seedlings collected from each transgenic line using the Qiagen DNeasy Mini Kit (Qiagen, Valencia, CA, United States). Approximately 10 μg DNA was digested with EcoRI, segregated in 0.8% agarose gels, and then blotted onto nylon membranes (Hybond N+; Amersham Biosciences, Little Chalfont, United Kingdom). A 993-bp GUS construct labeled with digoxigenin (DIG) was used as a probe. Probe hybridization was performed using the AlkPhos Direct Labeling and Detection System (Amersham Biosciences) according to the manufacturer’s protocol.
Abiotic Stress Treatments
Seeds were germinated on Murashige and Skoog (MS) medium solidified with 1.0% agar, and cultured in a growth chamber (25°C, 14 h light/10 h dark cycle, and 150 μMM-2 S-1 light intensity). After 7 days, the seedlings were planted on MS medium containing 150 mM mannitol, 150 mM NaCl, or 50 μM ABA, which were shown to constitute a significant stress in our pilot experiments. In cold stress treatment, seedlings were planted on MS medium and cultured in a cold growth chamber (4°C). Control plants were grown under non-stress conditions. After 3 days, twenty 10-day-old seedlings of each transgenic line were collected and subjected to GUS activity assays. All experiments were performed in triplicate.
GUS Activity Assays
Histochemical and fluorometric analyses of the GUS activity were performed as described previously (Jefferson, 1987). Seedlings were incubated in GUS reaction buffer at 37°C for 24 h, and then rinsed in an ethanol series to remove chlorophyll. Tissue observations were performed using a Leica MZ12 binocular microscope and images were captured using a Leica DC300 camera (Leica Microsystems, Wetzlar, Germany).
For the fluorometric assay, plants were homogenized in GUS extraction buffer. The homogenate was centrifuged at 12,000 × g for 10 min at 4°C, and then the supernatant activity was detected via an assay buffer containing 1 mM 4-methylumbelliferyl-β-glucuronide (Sigma, United States) at 37°C. The reaction was terminated by the addition of 200 mM Na2CO3 to a final concentration of 180 mM. Fluorometric estimations were performed using a PerkinElmer LS45 spectrofluorimeter (PerkinElmer).
Statistical Analysis
The Student’s t-test, variance analysis, and Chi-square test, were performed using the statistical software SAS 9.2 to determine significant differences (P < 0.05) between the means.
Results
Regulatory Element Analysis
The putative transcription start site was determined by two independent 5′-RACE experiments, indicating that the nucleotide position 313 bp upstream of the ATG start codon was the putative transcription start site of TaSnRK2.8 (Supplementary Figure S1).
The possible cis-elements were predicted by the multiple alignments of the SnRK2.8 promoter sequences from wheat, rice, and maize, and then determined using the PlantCARE software. As shown in Table 1, 11 predicted elements, including one ABA response element (ABRE), two binding sites for MYB transcription factors, and one low-temperature response element (LTRE), were identified in the TaSnRK2.8 promoter.
Table 1
| N | Element name | Conserved sequence/strand | Start position |
|---|---|---|---|
| 1 | ABRE | CACGTG(+) | -1165 |
| 2 | MBS | GTCAAT(-) TAACTG(+) | -2539 -1576 |
| 3 | LTR | CCGAAA(+) CCGAAA(+) | -393 +24 |
| 4 | Unknown | CTACTCC(+) | -11 |
| 5 | Unknown | AAAAAAT(+) | -2462, -2375 |
| 6 | Unknown | AATTTT(+) | -2645, -1188 |
| 7 | Unknown | AATCTA(+) | -2661, -2650, -2124, -1586, -1260, -903, -264 |
| 8 | Unknown | ATGTAT(+) | -2165, -2134, -1753, -1648, -305, -274 |
| 9 | Unknown | TGTATC(+) | -2145, -285 |
| 10 | Unknown | TTTTTAGGCA(+) | -80 |
| 11 | Unknown | CATTTATGAC(+) | +7 |
Predictions of cis-elements present in the TaSnRK2.8 promoter.
Possible cis-elements were predicted by multiple alignments using the SnRK2.8 promoter sequences from wheat, rice, and maize.
Evolutionary Analysis of the TaSnRK2.8 Promoter
The hexaploid wheat T. aestivum originated from hybridization between the diploid goat grass (Ae. tauschii, DD) and the tetraploid emmer wheat (Triticum turgidum, AABB) (Dubcovsky and Dvorak, 2007). To gain insight into the origin of the TaSnRK2.8 promoter sequences, the hexaploid wheat varieties and their relatives were used to construct a phylogenetic tree. Consistent with the genomic sequence analysis of TaSnRK2.8 (Zhang et al., 2010), these TaSnRK2.8 promoter sequences were divided into three distinct groups. As shown in Figure 2, two genotypes (PTaSnRK2.8-A and PTaSnRK2.8-B) were present in all hexaploid and tetraploid wheats: PTaSnRK2.8-A and PTaSnRK2.8-B originated from the A and B diploids, respectively. No TaSnRK2.8 promoters were detected in the D diploids. Moreover, no genetic diversity of TaSnRK2.8 promoters existed in the B and D genomes. The TaSnRK2.8 promoter in B genome was further examined through the GUS activity assays.
FIGURE 2
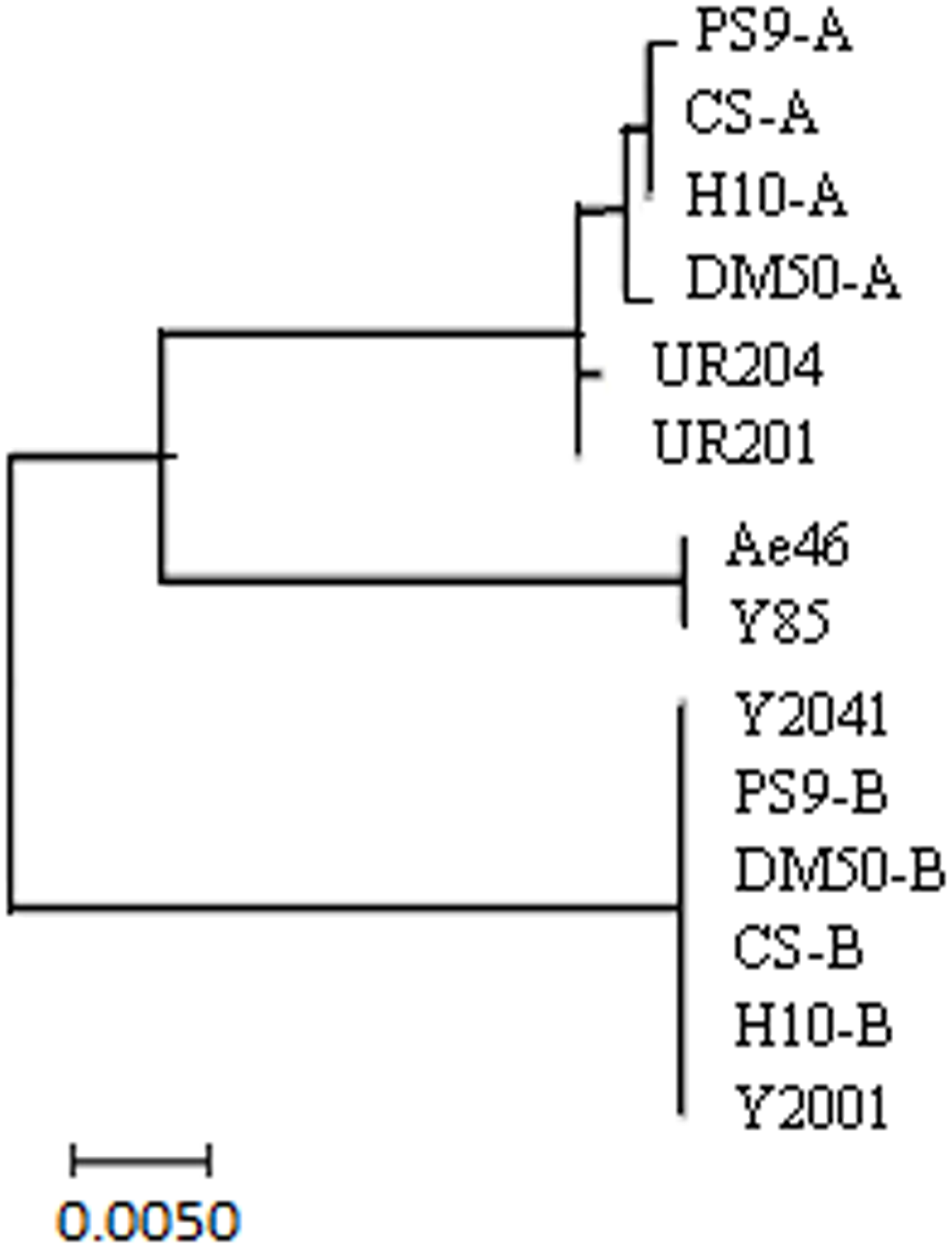
Phylogenetic analyses of TaSnRK2.8 promoters from hexaploid, tetraploid, and diploid wheats. Hexaploid wheat: Chinese Spring (CS) and Hanxuan 10 (H10); tetraploid wheat: PS9 and DM 50; A genome diploid: UR201 and UR204; B genome diploid: Y2001 and Y2041; D genome diploid: Y85 and Ae46. Phylogenetic analysis was conducted in MEGA 7.0.
Identification of Transgenic Plants
To characterize the function of the TaSnRK2.8 promoter, a series of 5′ promoter deletions were performed and four derivatives of TaSnRK2.8 promoter fragments were fused to the GUS (beta-glucuronidase) reporter gene (Figure 1). Hygromycin segregation in the T1 and T2 seeds was analyzed through the Chi-square tests. The T3 homozygous lines were used for performing the GUS activities assays. The integration of DNA in transformed plants was identified via PCR (Figure 3A). The qRT-PCR performed to determine the transcript levels of GUS under non-stress conditions showed that different promoter constructs have different abilities to regulate gene expression, and no GUS transcription was detected in the Dp724 and Dp1137 plants (Figure 3B). Southern blotting was used for selecting two independent transgenic lines with a single gene copy, which were further used in the GUS activity assays (Figure 3C). Expression patterns of GUS in various tissues showed that GUS was strongly expressed in all tissues of 35S:GUS seedlings, weakly in Dp2947 seedlings, and marginally in seedling stalks of Dp1797 plants under non-stress conditions. For Dp2947 plants, the transcript levels of GUS in stalks and roots were higher than those in leaves (Figure 3D).
FIGURE 3
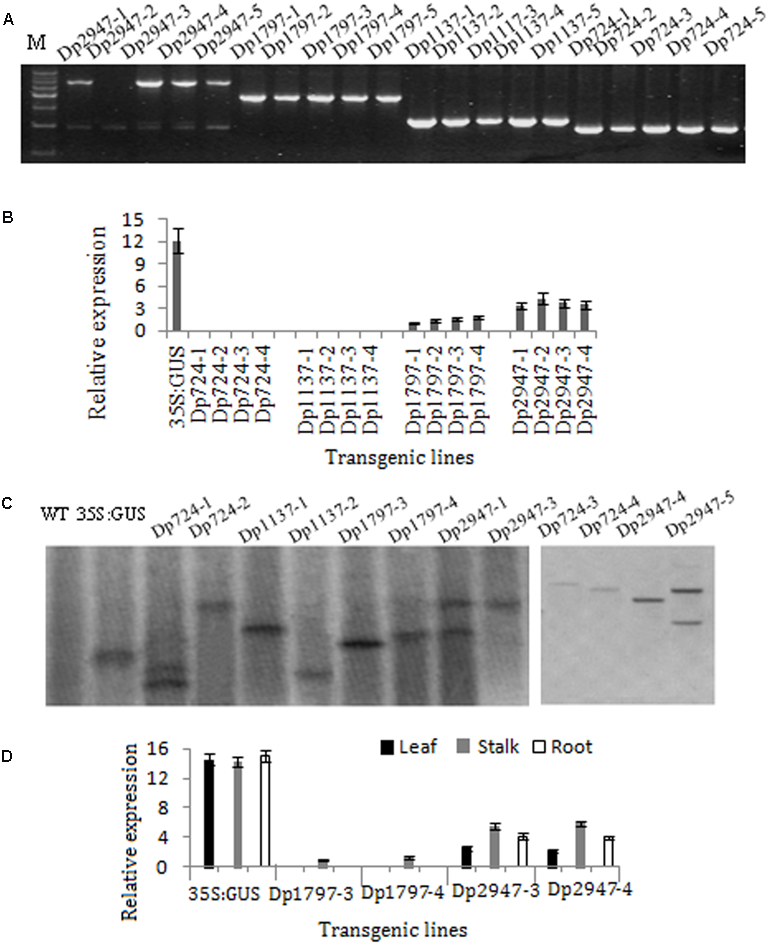
Identification of transformed Arabidopsis plants. (A) DNA-PCR analysis of transgenic plants. (B,D) Expression levels of GUS under the control of TaSnRK2.8 promoter deletion segments. Values are means ± SD (n = 3). (C) Southern blot analysis of transgenic plants. Total genomic DNA from the transgenic Arabidopsis leaves was digested with EcoRI. A DIG-labeled DNA fragment of GUS was used as the hybridization probe.
Deletion Analysis of the TaSnRK2.8 Promoter in Transgenic Seedlings under Non-stress Conditions
Under non-stress conditions, the CaMV35S plants displayed the highest GUS expression intensity in all tissues, whereas only a slight GUS expression was observed in the controls of the TaSnRK2.8 promoters (Dp2947). Moreover, different expression patterns and activities were observed among the deletion segments of the TaSnRK2.8 promoter(Figure 4).
FIGURE 4
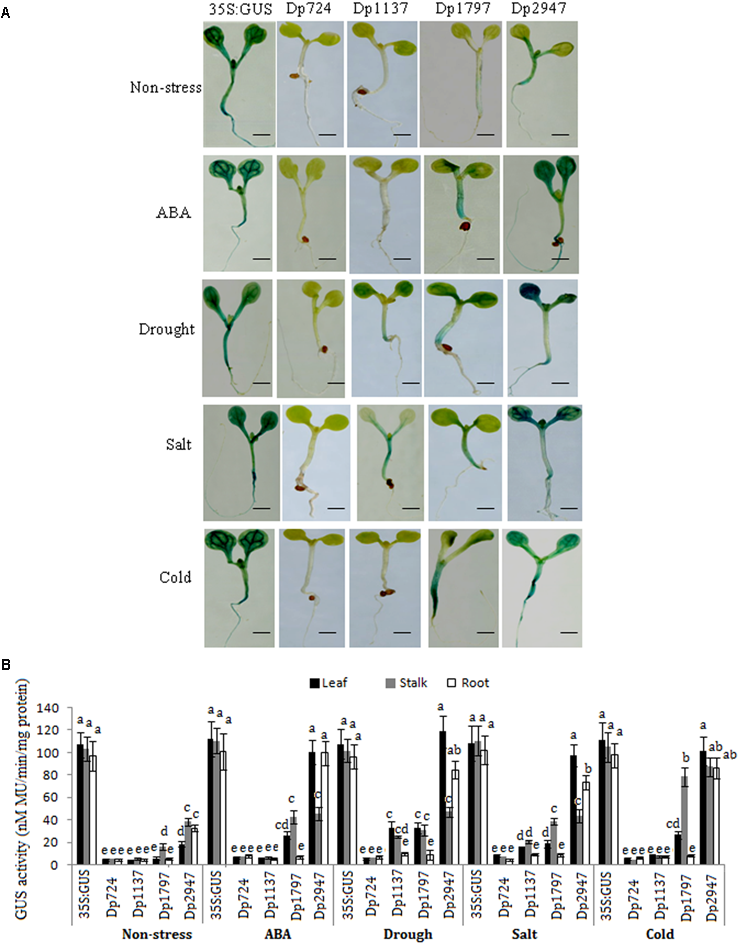
Effects of various abiotic stresses on the GUS activity under the control of different TaSnRK2.8 promoter deletion segments in the transgenic Arabidopsis seedlings. (A)GUS staining of transgenic Arabidopsis seedlings containing promoter deletion constructs under normal and stress conditions. (B)GUS activity under the control of different TaSnRK2.8 promoter deletion segments. The 7-day-old seedlings were planted on the MS medium containing 150 mM NaCl (salt), 150 mM mannitol (drought), or 50 μM ABA. Cold, cultured in a cold growth chamber (4°C). The plants used for the analysis were two independent lines carrying the promoter deletion constructs. GUS activity was expressed as nM MU/min/mg protein. Values are the mean ± SE (n = 20). a-eValues followed by the different letter are significantly different (P < 0.05). Dp724, Dp1137, Dp1797, and Dp2947 indicate the TaSnRK2.8 promoter regions from –408 to +316, –821 to +316, –1481 to +316, and –2631 to +316, respectively. Bar, 2 mm.
No GUS activity was observed in the Dp724 and Dp1137 plants, and only a faint expression was detected in the stalks of the Dp1797 seedlings under non-stress conditions. Although Dp2947 induced GUS expression in all tissues, weak GUS activity was observed in the leaves. These results indicated that the 660-bp segment (from position -1481 to -821) might harbor the stalk-specific expression elements under non-stress conditions, and that some enhancers were conserved in the region from position -2631 to -1481.
Activity of the Promoter Fragments in Response to ABA and Abiotic Stresses
Under ABA treatment, the GUS expression was detected in Dp1797, but not in Dp724 and Dp1137 plants. When the promoter length was increased to position -2631, stronger GUS activities were detected in the leaves and roots of the Dp2947 seedlings (Figure 4). This finding suggested that the region from position -821 to -2631 might harbor some ABREs.
In the abiotic stress assays, no GUS activity was observed in the Dp724 (-408) seedlings. When the promoter length was increased to position -821 (Dp1137), a weak GUS expression was observed in the leaves and stalks in response to drought and salt stresses (Figure 4), suggesting the likely presence of osmotic stress-response elements in the region from position -821 to -408. The Dp1797 (-1481) seedlings showed relatively higher GUS expression levels in the leaves and stalks under various stresses, whereas no GUS activity was observed in the roots. Under stress treatments, the longest promoter, Dp2947, conferred the highest levels of GUS expression in all tissues, predominantly in the leaves. The results indicated that the fragment from position -1481 to -821 contained positive regulatory elements that enhanced gene expression in the leaves and stalks. Furthermore, the TaSnRK2.8 promoter (Dp2947) conferred the highest gene expression, suggesting the likely presence of positive regulatory motifs in the region from position -2631 to -1481.
A 125-bp Region was Required for Osmotic Stress Response
The promoter region from position -821 to -408 was used for further deletion analysis (Figure 2). The integration of DNA in the transformed plants was identified via PCR (Figure 5A). Under drought and salt stresses, no GUS activity was observed in the Dp779 and Dp884 seedlings. Increasing the length of the promoter to position -693 (Dp1009) or -774 (Dp1090) led to the detection of GUS activity in the leaves and stalks in response to osmotic stress in a pattern similar to the Dp1137 seedlings (Figures 5B,C). This finding suggested that some osmotic stress-response elements might exist in the 125-bp region from position -639 to -568. To further verify the osmotic stress response element, the 125-bp region was fused to Dp724 through an overlapping PCR, which cannot induce the GUS transcription under non-stress and stress conditions. It was found that the promoter fragment Dp724 carrying the 125-bp region could induce the expression of GUS in the leaves and stalks in response to osmotic stress, very similar to what was observed in the Dp1009, Dp1090, and Dp1137 seedlings (Figures 5B,C). Collectively, these results suggest that the 125-bp TaSnRK2.8 promoter region from position -693 to -568 was required for inducing an osmotic stress response in Arabidopsis plants. The effects of different TaSnRK2.8 promoter deletion segments in response to ABA and abiotic stresses were shown in Supplementary Figure S2.
FIGURE 5
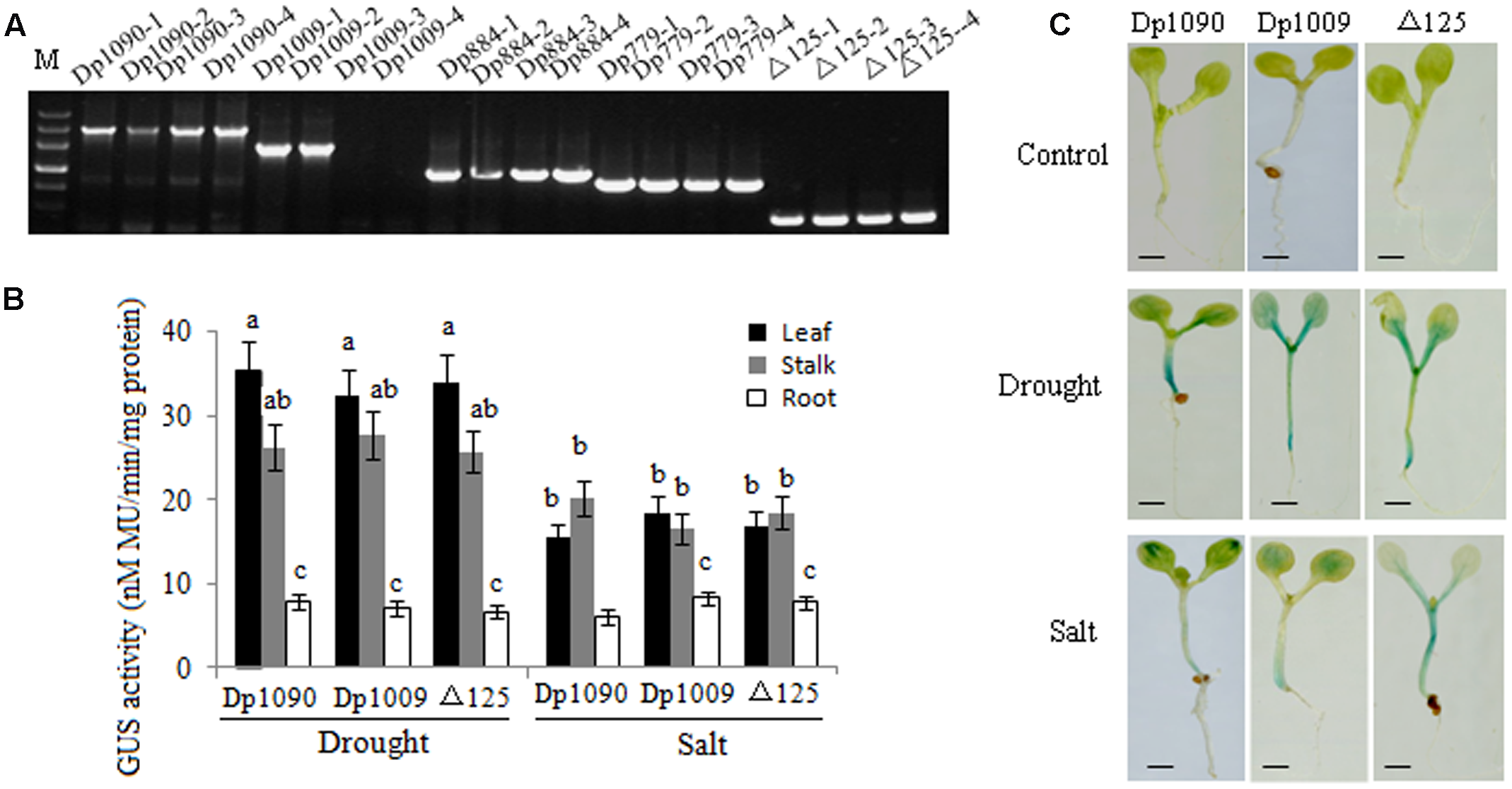
TaSnRK2.8 promoter region from –693 to –568 (125 bp) is required for osmotic stress response. (A) DNA-PCR analysis of transgenic plants. (B) Effects of osmotic stress on GUS activity under the control of different TaSnRK2.8 promoter deletion segments. The plants used for the analysis were two independent lines carrying the promoter deletion constructs. GUS activity was expressed as nM MU/min/mg protein. Values are the mean ± SE (n = 20). a–cValues followed by the different letter are significantly different (P < 0.05). (C) The GUS staining of transgenic Arabidopsis seedlings containing promoter deletion constructs under normal and osmotic stress conditions. The 7-day-old seedlings were planted on the MS medium containing 150 mM NaCl (salt) or 150 mM mannitol (drought). –774 to ATG, Dp1090; –693 to ATG, Dp1009; –568 to ATG, Dp884; –463 to ATG, Dp779; –408 to ATG, Dp724; Dp724 carrying the 125-bp fragment from –693 to –568 bp, Δ125. Bar, 2 mm.
Discussion
Different Promoter Fragments of TaSnRK2.8 Harbored Different Elements for Tissue-Preferential Expression
Previous studies found that TaSnRK2.8 (subclass III) was strongly expressed in the roots, and faintly in the stems and leaves. The over-expression of TaSnRK2.8 in Arabidopsis enhanced its tolerance to abiotic stresses through different stress-signaling routes (Zhang et al., 2010, 2011, 2016). To more thoroughly investigate the signaling and regulation of TaSnRK2.8 under different stresses, we characterized the spatial expression pattern of different TaSnRK2.8 promoter derivatives under ABA and environmental stress conditions.
Gene expression patterns might be a direct indication of a promoter’s function in stress and developmental processes. In Arabidopsis, the GUS gene driven by AtSnRK2.7 promoter was specifically expressed in the roots, whereas the AtSnRK2.8 promoter-mediated GUS activity was observed in all tissues (Umezawa et al., 2004; Mizoguchi et al., 2010). Furthermore, AtSnRK2.6 was confirmed to play a key role in stomatal closure under stress conditions (Mustilli et al., 2002). These data indicate that the SnRK2 family members play different roles in different tissues. The promoter deletion analyses of TaSnRK2.8 indicated that different promoter fragments harbored different elements for tissue-preferential expression. In general, the region from position -1481 to -821 contained the stalk-specific elements, whereas the region from position -2631 to -1481 contained the leaf- and root-specific elements. The spatial variation in expression levels was closely related to the different physiological stages of the plant lines and to the duration of stress treatment.
TaSnRK2.8 Promoter Could Drive Strong Gene Expression in Response to ABA
ABA is a vital hormone involved in plant abiotic stress responses. Plants can respond to stresses via ABA-dependent or -independent pathways. As a stress hormone, ABA acts through the regulatory pathways that control gene expression in response to stress. Increasing evidence shows that the subclass III SnRK2s respond strongly to ABA, the subclass II SnRK2s are only faintly induced by ABA, and the subclass I SnRK2s are moderately induced by ABA (Kobayashi et al., 2004, 2005; Boudsocq et al., 2007; Huai et al., 2008; Fujita et al., 2009). In the present study, the ability of TaSnRK2.8 promoter to induce GUS expression in Arabidopsis revealed that TaSnRK2.8 could drive transgene expression in response to ABA, and the promoter region from position -821 to -2631 might contain ABREs.
TaSnRK2.8 Promoters Harbored Abiotic-Stress Response Elements
The identification of tissue-specific or inducible promoters in expression analysis would be of considerable practical value because it could eliminate the unnecessary burdens on plants by restricting the genetic expression to specific tissues or in response to specific environmental conditions (Hsieh et al., 2002; Zavallo et al., 2010; Hou et al., 2016). Here, we examined the activation of GUS under the regulation of TaSnRK2.8 promoter fragments in response to abiotic stresses and showed that a typical stress-inducible promoter contains some regions for regulating tissue specificity and transcription levels under stress, leading to different regulation patterns. For example, there might be osmotic stress response elements in the region from position -693 to -568 and positive regulatory motifs from -2631 to -1481, whereas the fragment from -1481 to -821 contained the leaf- and stalk-specific enhancers. Further analysis might enable the identification of essential control elements in these functional regions.
The comparative functional analysis of different stress-responsive promoters has provided useful information, leading to enhancements in the specificity and inducibility of stress responsive genes. Therefore, the TaSnRK2.8 promoter examined in the present study might prove beneficial for the overexpression of foreign genes in the plants subjected to abiotic stresses. Moreover, the findings of the deletion analysis reported here provide the basis for identifying the direct upstream regulators of SnRKs and stress-response elements in TaSnRK2.8 promoter.
Statements
Author contributions
HZ performed the experiments and participated to the data analysis. XM performed the qRT-PCR experiments, RJ projected design and supervision.
Funding
This work was supported by the National Natural Science Foundation of China (31401371).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01198/full#supplementary-material
Footnotes
1.^http://www.ebi.ac.uk/ena/search/
2.^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
1
AnderbergR. J.Walker-SimmonsM. K. (1992). Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases.Proc. Natl. Acad. Sci. U.S.A.8910183–10187. 10.1073/pnas.89.21.10183
2
BoudsocqM.Barbier-BrygooH.LauriereC. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana.Biol. Chem.27941758–41766. 10.1074/jbc.M405259200
3
BoudsocqM.DroillardM. J.Barbier-BrygooH.LauriereC. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid.Plant Mol. Biol.63491–503. 10.1007/s11103-006-9103-1
4
CheckerV. G.ChhibbarA. K.KhuranaP. (2012). Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress.Transgenic Res.21939–957. 10.1007/s11248-011-9577-8
5
ChristensenA. H.SharrockR. A.QuailP. H. (1992). Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation.Plant Mol. Biol.18675–689. 10.1007/BF00020010
6
DubcovskyJ.DvorakJ. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication.Science3161862–1866. 10.1126/science.1143986
7
FujitaY.NakashimaK.YoshidaT.KatagiriT.KidokoroS.KanamoriN.et al (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis.Plant Cell Physiol.502123–2132. 10.1093/pcp/pcp147
8
HalfordN. G.HardieD. G. (1998). SNF1-related protein kinases: global regulators of carbon metabolism in plants?Plant Mol. Biol.37735–748. 10.1023/A:1006024231305
9
HouJ.JiangP.QiS.ZhangK.HeQ.XuC.et al (2016). Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants.PLoS ONE11:e0154041. 10.1371/journal.pone.0154041
10
HsiehT. H.LeeJ. T.CharngY. Y.ChanM. T. (2002). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress.Plant Physiol.130618–626. 10.1104/pp.006783
11
HuaiJ.WangM.HeJ.ZhengJ.DongZ.LvH.et al (2008). Cloning and characterization of the SnRK2 gene family from Zea mays.Plant Cell Rep.271861–1868. 10.1007/s00299-008-0608-8
12
JeffersonR. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system.Plant Mol. Biol. Rep.5387–405. 10.1007/BF02667740
13
KobayashiY.MurataM.MinamiH.YamamotoS.KagayaY.HoboT.et al (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors.Plant J.44939–949. 10.1111/j.1365-313X.2005.02583.x
14
KobayashiY.YamamotoS.MinamiH.KagayaY.HattoriT. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid.Plant Cell16k1163–1177. 10.1105/tpc.019943
15
LivakK. J.SchmittgenT. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ ΔCT method.Methods25402–408. 10.1006/meth.2001.1262
16
MaoX. G.ZhangH. Y.TianS. J.ChangX. P.JingR. L. (2009). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis.J. Exp. Bot.61683–696. 10.1093/jxb/erp331
17
McElroyD.ZhangW.CaoJ.WuR. (1990). Isolation of an efficient actin promoter for use in rice transformation.Plant Cell2163–171. 10.1105/tpc.2.2.163
18
MelcherK.NgL. M.ZhouX. E.SoonF. F.XuY.Suino-PowellK. M.et al (2009). A gate-latch-lock mechanism for hormone signaling by abscisic acid receptors.Nature462602–608. 10.1038/nature08613
19
MizoguchiM.UmezawaT.NakashimaK.KidokoroS.TakasakiH.FujitaY.et al (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression.Plant Cell Physiol.51842–847. 10.1093/pcp/pcq041
20
MustilliA.MerlotS.VavasseurA.FenziF.GiraudatJ. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production.Plant Cell143089–3099. 10.1105/tpc.007906
21
NishimuraN.HitomiK.ArvaiA. S.RamboR. P.HitomiC.CutlerS. R.et al (2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.Science3261373–1379. 10.1126/science.1181829
22
OdellJ. T.NagyF.ChuaN. H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter.Nature313810–812. 10.1038/313810a0
23
RaiM.HeC.WuR. (2009). Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice.Transgenic Res.18787–799. 10.1007/s11248-009-9263-2
24
SheardL. B.ZhengN. (2009). Plant biology: signal advance for abscisic acid.Nature462575–576. 10.1038/462575a
25
SheltonA. M.ZhaoJ. Z.RoushR. T. (2002). Economic, ecological, food safety, and social consequences of the deployment of bt transgenic plants.Annu. Rev. Entomol.47845–881. 10.1146/annurev.ento.47.091201.145309
26
TianS. J.MaoX. G.ZhangH. Y.ChenS. S.ZhaiC. C.YangS. M.et al (2013). Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat.J. Exp. Bot.642063–2080. 10.1093/jxb/ert072
27
UmezawaT.YoshidaR.MaruyamaK.Yamaguchi-ShinozakiK.ShinozakiK. (2004). SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana.Proc. Natl. Acad. Sci. U.S.A.10117306–17311. 10.1073/pnas.0407758101
28
Yamaguchi-ShinozakiK.MinoM.MundyJ.ChuaN. H. (1990). Analysis of an ABA-responsive rice gene promoter in transgenic tobacco.Plant Mol. Biol.15905–912. 10.1007/BF00039429
29
ZavalloD.BilbaoM. L.HoppH. E.HeinzR. (2010). Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.).Plant Cell Rep.29239–248. 10.1007/s00299-010-0816-x
30
ZhangH. Y.LiW. Y.MaoX. G.JingR. L.JiaH. F. (2016). Differential activation of the wheat SnRK2 family by abiotic stresses.Front. Plant Sci.7:420. 10.3389/fpls.2016.00420
31
ZhangH. Y.MaoX. G.JingR. L.XieH. M. (2011). Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses.J. Exp. Bot.62975–988. 10.1093/jxb/erq328
32
ZhangH. Y.MaoX. G.WangC. S.JingR. L. (2010). Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis.PLoS ONE5:e16041. 10.1371/journal.pone.0016041
33
ZhangH. Y.MaoX. G.ZhangJ. N.ChangX. P.JingR. L. (2013). Single-nucleotide polymorphisms and association analysis of drought-resistance gene TaSnRK2.8 in common wheat.Plant Physiol. Biochem.70174–181. 10.1016/j.plaphy.2013.04.010
Summary
Keywords
TaSnRK2.8, inducible promoter, abiotic stress, abscisic acid, deletion analysis
Citation
Zhang H, Jing R and Mao X (2017) Functional Characterization of TaSnRK2.8 Promoter in Response to Abiotic Stresses by Deletion Analysis in Transgenic Arabidopsis. Front. Plant Sci. 8:1198. doi: 10.3389/fpls.2017.01198
Received
30 April 2017
Accepted
26 June 2017
Published
13 July 2017
Volume
8 - 2017
Edited by
Avinash Mishra, Central Salt & Marine Chemicals Research Institute (CSIR), India
Reviewed by
Charanpreet Kaur, Jawaharlal Nehru University, India; An-Ping Chen, Mayo Clinic, Mayo Medical School, United States; Vivekanand Tiwari, Weizmann Institute of Science, Israel
Updates
Copyright
© 2017 Zhang, Jing and Mao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongying Zhang, zhangying198215@163.com
†These authors have contributed equally to this work.
This article was submitted to Plant Abiotic Stress, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.